Summary
Protein maturation in the endoplasmic reticulum is controlled by multiple chaperones, but how they recognize and determine the fate of their clients remains unclear. We developed an in vivo peptide library covering substrates of the ER Hsp70 system: BiP, Grp170, and three of BiP’s DnaJ-family co-factors (ERdj3, ERdj4, and ERdj5). In vivo binding studies revealed that sites for pro-folding chaperones BiP and ERdj3 were frequent and dispersed throughout the clients, whereas Grp170, ERdj4, and ERdj5 specifically recognized a distinct type of rarer sequence with a high predicted aggregation potential. Mutational analyses provided insights into sequence recognition characteristics for these pro-degradation chaperones, which could be readily introduced or disrupted, allowing the consequences for client fates to be determined. Our data reveal unanticipated diversity in recognition sequences for chaperones, establish a sequence-encoded interplay between protein folding, aggregation, and degradation, and highlight the ability of clients to co-evolve with chaperones ensuring quality control.
Introduction
To become biologically active, most proteins must adopt a defined three-dimensional structure. In the cell this process is guided and monitored by molecular chaperones (Hartl et al., 2011), which is of particular importance when large quantities of proteins must be folded and failures in protein folding are not easily remedied. This applies to the endoplasmic reticulum (ER), which processes one third of the proteins encoded in the human genome, and is the major site of quality control for them (Braakman and Hebert, 2013). The fidelity of chaperone-aided protein folding in the ER is indispensable and failures in this process are associated with numerous human diseases (Guerriero and Brodsky, 2012).
Several major chaperone families have been identified, members of which exist in organisms from bacteria to humans, and most are able to interact transiently with a vast variety of seemingly sequence- and structure-unrelated immature proteins. Insights into the apparent promiscuity of chaperones initially came from studies on the mammalian ER Hsp70 family member, BiP, which binds 7-9 amino acid sequences primarily composed of alternating aliphatic residues that are often buried in proteins in their native state (Blond-Elguindi et al., 1993; Flynn et al., 1991). Similar patterns were found for the E. coli Hsp70 protein, DnaK (Gragerov and Gottesman, 1994; Rudiger et al., 1997). This sequence preference explains how chaperones like BiP can bind so many diverse clients, and even multiple sequences on a given protein, that are in most cases restricted to unfolded/unassembled proteins (Kaloff and Haas, 1995; Ng et al., 1992; Schmitz et al., 1995).
All Hsp70 proteins act as central chaperones in a hub of multiple (co-)chaperones and folding enzymes (Hartl et al., 2011). Most prominent among these are DnaJ co-factors; some of which also bind directly to proteins and can deliver them to their Hsp70 partner. Peptide interaction studies on DnaJ proteins from E. coli (Rudiger et al., 2001) and S. cerevisiae (Kota et al., 2009) identified a similar preference for sequences rich in aliphatic residues, and in fact many peptides that interact with Hsp70s also interact with DnaJ proteins. This rather generic recognition pattern has shaped our perception of chaperone binding preferences. However, in eukaryotes a large number of different DnaJ co-factors exist to presumptively confer specifity for Hsp70 chaperone functions (Cyr and Ramos, 2015; Kampinga and Craig, 2010), but their substrate binding preferences are not well understood . In the mammalian ER, 4 of the 8 known DnaJ family members (ERdj3-6) bind to unfolded protein clients and can either facilitate the pro-folding or pro-degradation functions of BiP (Dong et al., 2008; Ushioda et al., 2008; Wen and Damania, 2010). How they help execute this key checkpoint in protein folding remains poorly understood. While ERdj3 is structurally related to DnaK and Ydj1 (Jin et al., 2009) and is likely to have a similar client recognition pattern, nothing is known about the client interaction surface on other ERdj proteins or their sequence preferences/substrate specificity. To better understand how various ER (co-)chaperones recognize clients and contribute to distinct outcomes for their substrates, we designed an in vivo system to determine the sequence-specific binding preferences of the ER Hsp70 system members in their native cellular environment. For these studies, we identified two secretory pathway proteins; immunoglobulin γ1 heavy chain and NS-1 κ light chain that are poised between folding and degradation. Both proteins are natural clients of BiP (Bole et al., 1986; Ma et al., 1990) and ERdj3 (Shen and Hendershot, 2005) but also interact directly with ERdj4 (Shen et al., 2002) and ERdj5, as well as with the resident ER Hsp110 cognate, Grp170 (Behnke and Hendershot, 2014). Our data show that unlike BiP and ERdj3, which recognized multiple sequences in both clients, ERdj4, ERdj5 and Grp170 bound specifically to aggregation-prone sequences that occurred less frequently in these clients. This reveals unexpected diversity in substrate binding specificity of the various ER chaperones. Our approach further allowed us to introduce or delete defined (co-)chaperone binding sites, providing insights into how different ER (co-)chaperones act to affect the fate of clients. Lastly, our results revealed a provocative ability of clients to co-evolve with the ER chaperone machinery to coordinate maximum folding opportunities while maintaining rigorous quality control of the process.
Results
Establishing an in vivo peptide expression system
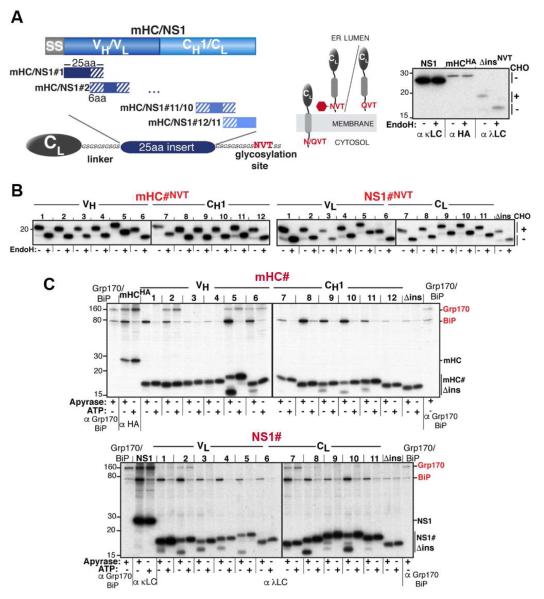
To analyze (co-)chaperone binding sequences in a cellular context, we chose two natural clients of the ER Hsp70 chaperone system: a non-secreted mouse κ antibody light chain (NS1 LC) and a truncated version of a mouse γ1 antibody heavy chain (mini-HC: mHC), which contains the critical regions of ER quality control for this protein. These substrates have been well-characterized in terms of their folding status in vitro (Feige et al., 2009) and in vivo (Knittler et al., 1995; Lee et al., 1999), as well as their ER quality control checkpoints and the chaperones involved (Hendershot et al., 1987; Meunier et al., 2002; Skowronek et al., 1998), providing an excellent starting point for our analysis. To engineer an in vivo peptide expression library, a well-folded, ER-targeted λ LC constant domain (λ CL), which does not significantly bind to any of the chaperones being queried (Hellman et al., 1999); this study), was tethered to a GS linker and culminated in a single N-linked glycosylation site (NVT). Since neither NS1 nor mHC are glycosylated, this C-terminal NVT site could be used to monitor full transport of the peptide sequence into the ER (Figure 1A). A series of 25 amino acid, overlapping peptides covering the two model proteins were inserted into the GS linker; yielding an initial set of 12 constructs for the mHC (mHC#1-12) and 11 for NS1 (NS1#1-11) (Figure S1A). A construct without a peptide insert (Δins) served as a negative control for (co-)chaperone binding. The single C-terminal engineered glycosylation site was modified on all constructs when they were expressed in COS-1 cells, indicating complete transport into the ER lumen (Figure 1B). Having established ER import, the two libraries were recreated with the NVT sequence changed to QVT to avoid glycan-mediated interactions with the ER lectin chaperones. The QVT library constructs were used for all subsequent experiments.
Figure 1. An in vivo peptide library reveals distinct substrate binding patterns for BiP and Grp170.
(A) A series of 25 amino acid overlapping peptides corresponding to the mHC and NS1 were inserted into a flexible GS-linker downstream of an ER-targeted λ LC constant domain (CL) ending with a C-terminal NVT glycosylation site or QVT. Only upon ER entry is the NVT site modified (red hexagon). EndoH resistance confirmed that NS1 and mHC are glycoproteins (right). (B) After transfection into COS-1 cells, metabolically labeled peptide constructs were immunoprecipitated with anti-λ antiserum, treated with (+) or without (−) EndoH and analyzed by SDS-PAGE. (C) COS-1 cells co-expressing Grp170, BiP, and the indicated peptide constructs were metabolically labeled, lysed with ATP or apyrase added. Peptide constructs were isolated with anti-λ and separated on SDS gels. Cells transfected with only Grp170 and BiP serve as markers and the full-length client for each set of peptides is included as a reference for chaperone binding.
BiP and Grp170 have distinct substrate binding patterns
We co-expressed BiP and Grp170 with the peptide library and examined their ability to be co-immunoprecipitated with our peptides (Figure 1C). BiP bound to multiple sequences above the Δins background signal in apyrase supplemented lysates, which was readily disrupted by ATP addition as expected for bona fide substrate binding to an Hsp70 chaperone. The frequent number of binding sites observed is consistant with in silico predictions (Knarr et al., 1995; Schneider et al., 2016) and in vivo experiments (Kaloff and Haas, 1995) revealing that BiP can bind multiple antibody domains. Unfortunately the apyrase treatment used to stabilize BiP binding resulted in partial cleavage of some of the peptides due to a contaminating protease activity. In spite of this limitation, our screen not only detected both BiP binding sites previously identified on an isolated recombinant CH1 domain (corresponding to mHC#7 and mHC#9) (Marcinowski et al., 2013), but also revealed two novel BiP-interacting peptides (mHC#8 and mHC#10) in this domain. The in vivo binding of BiP to mHC#7 could be disrupted by mutating the same residue that inhibited BiP binding in vitro (Marcinowski et al., 2013), whereas this approach was not sufficient to completely abolish BiP binding to the mHC#9 peptide in vivo (Figure S1B). Further attempts to disrupt BiP binding to peptides in the CH1 domain, which represents the stable site of BiP interaction in unassembled γHC (Hendershot et al., 1987), were largely unsuccessful (Figure S1C-E and data not shown). Our inability to readily disrupt BiP binding via mutations is in keeping with the pronounced promiscuity of BiP revealed by in vitro studies (Blond-Elguindi et al., 1993).
Unlike BiP, Grp170 interacted with peptides in the VH but not the CH1 domain of the mHC. This binding was resistant to ATP addition (Figure 1C and S2), like its binding to full-length clients (Behnke and Hendershot, 2014). While the 25-mer peptides that bound to Grp170 also interacted with BiP, they did not necessarily correlate with the strongest BiP binding peptides. The detection of a Grp170 binding site in both the variable (VL) and constant (CL) domains of the NS1 LC is consistent with data showing that in vivo Grp170 interacts with the partially oxidized form of the NS1 LC in which only VL is unfolded, and to a greater extent with the fully reduced form where the CL domain is also unfolded (Behnke and Hendershot, 2014). Together these data suggested the interactions of BiP and Grp170 with our peptide library were faithfully reporting on their associations with full-length clients in the ER and revealed intriguing differences in the binding patterns for these two ER Hsp70 chaperones.
Evidence for substrate specificity within the ERdj proteins
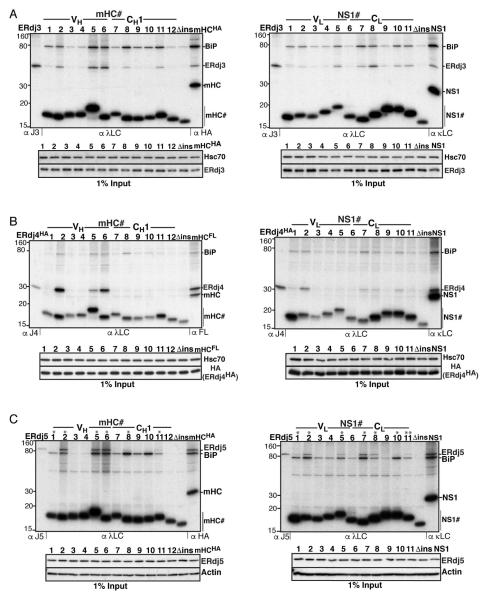
Unlike other BiP (co-)chaperones, ERdj3’s binding to clients is sensitive to detergent lysis and requires cross-linking to maintain this association (Meunier et al., 2002). Thus, peptide chimeras were DSP-crosslinked in cells prior to lysis and examined for co-immunoprecipation of ERdj3 (Figure 2A). This revealed that ERdj3 bound to multiple peptides throughout both domains of the clients at levels above background (Δins). In most cases these peptides coincided with ones that bound BiP, but not all BiP binding peptides interacted with ERdj3.
Figure 2. ERdj3 binds frequently throughout mHC and NS1, while ERdj4 and ERdj5 possess fewer, mostly shared binding motifs.
(A) COS-1 cells were co-transfected with ERdj3 and BiP together with the indicated peptide constructs and metabolically labeled, DSP-crosslinked and lysed in RIPA buffer. Peptide constructs were immunoprecipitated with anti-λ antiserum and analyzed by reducing SDS-PAGE. Cells transfected with ERdj3 and BiP alone were immunoprecipitated with antiserum specific for ERdj3 as molecular weight marker, and full-length clients were included as a reference for chaperone binding. Levels of ERdj3 expression in each sample by immunoblotting are shown under the panel. (B) Binding of mHC and NS1 peptides to ERdj4 was performed as in (A), except that HA-tagged ERdj4 was co-transfected and cells were lysed directly in NP40-lysis buffer. (C) Binding of ERdj5 to mHC and NS1 peptides was performed as in (B), except that ERdj5 was co-transfected and the lysis/washing buffers contained 20 mM NEM.
Neither the client binding surfaces nor possible sequence recognition patterns were known for ERdj4 or ERdj5. Since the interaction of these (co-)chaperones with clients is not affected by detergent or ATP, we could monitor their binding to peptides independent of their association with BiP by including ATP in the lysis buffer. When ERdj4 was co-expressed with the peptide library, we readily detected its binding to three peptides in the VH domain of the mHC but none in the CH1 domain (Figure 2B). In the case of the NS1 library, signals for ERdj4 were detected in both domains, with peptides #2 and #7 showing strongest binding to ERdj4 when susbstrate expression levels were taken into account. ERdj5 interacted with the same peptides in both clients but also bound weakly to several others (Figure 2C). It is noteworthy that all of the ERdj5-binding peptides had a cysteine (denoted by asterisks) in keeping with its role as an oxidoreductase, but not all peptides containing a cysteine interacted significantly with ERdj5 (e.g., mHC#1 and NS1#10). Surprisingly, while Grp170, ERdj4 and ERdj5 have no known structural homology that might contribute to client recognition, we observed a nearly complete overlap of peptides they bound.
Elucidation of sequence recognition patterns for BiP (co-)chaperones
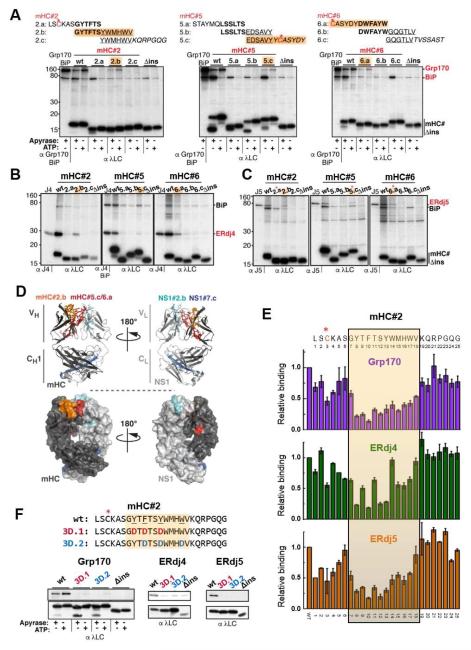
To further define the binding sites for Grp170, ERdj4, and ERdj5, we subdivided the peptides that interacted with these (co-)chaperones into three overlapping segments. While we were able to detect interacting peptides upon subdivision for all three (co-)chaperones, subdivision generally weakened binding (Figures 3A-C and S3A-D). BiP and Grp170 sites could be separated in the subdivided mHC#2 and mHC#6 peptides, indicating some distinction in their recognition properties (Figure 3A). ERdj5 interacted with the exact same subdivided fragments as Grp170, arguing at least in these cases neither one was likely to interact via BiP. Most of the subdivided fragments ERdj5 bound contained the cysteine, except for mHC#2 where ERdj5 bound the fragment immediately following the cysteine (Figures 3A and C), and in the case of NS1#2 ERdj5 bound two overlapping peptides after subdivision; only one of which contained the cysteine (Figure S3D). Together these data suggest ERdj5 may be targeted to a subset of cysteine containing peptides through other sequence-specific interactions. Where a signal could be detected, ERdj4 bound the same subdivided peptides as Grp170 and ERdj5 (Figures 3B and S3C). Mapping the smaller peptides that these (co-)chaperones interacted with on a modelled mHC/NS1 antibody Fab fragment revealed that all of the core recognition sequences were largely buried in the folded and assembled Fab fragment (Figure 3D).
Figure 3. Grp170/ERdj4/ERdj5 recognize shared motifs that are buried after protein folding or subunit assembly.
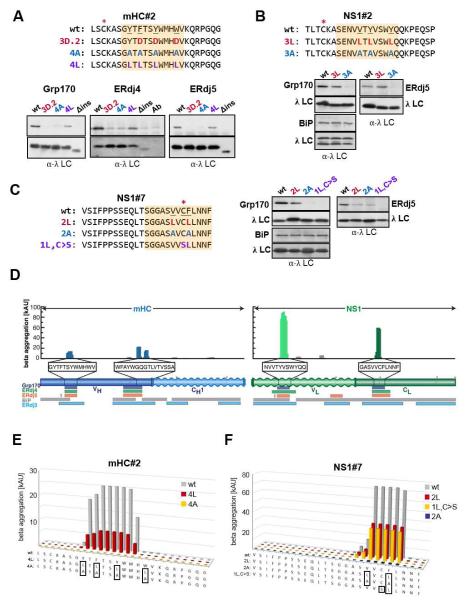
(A) The three 25 amino acid peptides in the mHC recognized by Grp170 were further subdivided into 12 amino acid overlapping peptides as shown above the gel images. COS-1 cells were co-transfected with Grp170 and BiP along with the indicated reporter constructs and analyzed as in Figure 1C. Peptides recognized by Grp170 are highlighted in orange, and cysteines are marked by red asterisks. (B-C) The same subdivided peptides were examined for ERdj4 (B) or ERdj5 (C) binding as described in Figure 2. Interacting peptides are indicated in orange, and in (C) cysteine containing peptides are marked with a red asterisk. (D) Binding sites shared by Grp170, ERdj4 and ERdj5 were mapped on a model of a mHC-NS1 heterodimer (top: ribbon representation, bottom: surface representation). The mHC is shown in dark grey with mHC#2.b in orange and #5.c/#6.a in red. NS1 is shown in light grey with NS1#2.b in turquois and #7.c in blue. (E) Each amino acid in the mHC#2 peptide was individually mutated to Asp. The subdivided peptide that bound all three (co-)chaperones is indicated with an orange box. COS-1 cells transfected with Grp170, ERdj4 or ERdj5 together with BiP and each of the constructs were metabolically labeled and analyzed as before. The levels of the co-precipitated (co-)chaperones were quantified as described in Extended Experimental Procedures, and are shown ±SEM (n≥3). The value obtained for the parent mHC#2 (wt) was set to 1. (F) (Co-)chaperone binding to the mHC#2 wild-type and mutant constructs was determined by immunoprecipitation with α-λ and blotted with the appropriate (co-)chaperone (top) or λ (bottom) antiserum.
For a more in-depth analysis of the Grp170/ERdj4/ERdj5 recognition preferences we chose mHC#2, which was a particularly strong binder for all three (Figures 1C, 2B and 2C). Each of the 25 amino acids in this peptide was individually replaced with an aspartate, and the effect on (co-)chaperone binding was measured. While mutation of any residue within the 12 amino acid core identified by subdivision (shaded in orange) significantly disrupted binding of Grp170 and ERdj5 to mHC#2, in the case of ERdj4 it appeared that every other residue had a larger impact on its association (Figure 3E). In keeping with the reduced binding of the (co-)chaperones to the subdivided sequences, we found some mutations outside the 12aa core sequence also impaired their binding. Among those, mutation of the cysteine residue most strongly impacted Grp170, ERdj4 and ERdj5 binding. In an attempt to completely disrupt (co-)chaperone binding to this peptide, we made two mHC#2 mutant constructs in which sets of three aromatic residues were altered to aspartates, because two peptides rich in aromatic residue had been reported to bind yeast Sse1, a cytosolic orthologue of Grp170, (Goeckeler et al., 2008; Xu et al., 2012). Both mHC#2 mutants disrupted binding of all three (co-)chaperones to nearly undetectable levels (Figure 3F).
Grp170, ERdj4, and ERdj5 recognize aggregation-prone sequences
As all the peptides that bound Grp170, ERdj4 and ERdj5 were rich in aromatic residues, we continued to test their importance. Towards this end, all four aromats in the mHC#2 peptide, whose mutation to aspartates abolished binding (Figure 3F), were substituted with either alanine (4A) or leucine (4L). The 4A mutant was nearly as deficient in binding the three (co-)chaperones as an aspartate mutant, whereas their binding to the 4L mutant was only slightly diminished (Figure 4A). Thus, even though our initial binding sequences were rich in aromatic residues, these are not necessarily required for binding, which is similar to in vitro data obtained for Sse1 (Xu et al., 2012). To generalize these findings, we similarly mutated the (co-)chaperone binding sites in the NS1#2 and NS1#7 peptides. Substitution of aromatic and aliphatic residues with alanine in these peptides also reduced binding of Grp170 and ERdj5 to barely detectable levels, whereas leucines were more readily tolerated (Figure 4B and 4C). The interaction of ERdj4 with the mutated peptides was more difficult to call due to the background bands caused by the precipitating antibody (data not shown). For both peptides, aspartate substitutions prevented (co-)chaperone binding (Figure S4A and S4C). Since we found the cysteine residue in mHC#2 was important for binding of all three (co-)chaperones, we made an additional mutant in NS1#7 in which the cysteine was substituted with a serine and a single aromat was altered to leucine. We found this combination completely abrogated binding to both Grp170 and ERdj5, while binding of BiP to all of the mutants was maintained (Figure 4B and 4C).
Figure 4. Grp170, ERdj4 and ERdj5 binding sites correspond to aggregation-prone regions in their clients.
(A-C) Cells expressing the indicated (co-)chaperones together with mHC#2 (A), NS1#2 (B) or NS1#7 (C) peptide constructs were analyzed by immunoprecipitation-coupled western blotting as described in Figure 3F. The 12-aa fragments identified in Figure 3 and S3 are indicated with orange boxes. The top panel in each set shows anti-(co-)chaperone western blots and the bottom anti-λ blots. For ERdj4, a separate lane with the anti-λ antibody was included and samples were run under-non-reducing conditions to reduce background signal from the precipitating antibody. (D) Sequences for the mini-HC (blue) and NS1 LC (green) were subjected to analysis by the TANGO algorithm, which identifies regions that are predicted to be prone to β-aggregate formation. Wavy segments indicate unfolded domains in each client (when expressed in isolation). The core binding peptides for Grp170 (purple), ERdj4 (green), and ERdj5 (orange) are indicated below, as are binding sites for BiP (grey) and ERdj3 (blue). (E-F) The TANGO algorithm was used to calculate the β-aggregation potential for the indicated mHC#2 (E) and NS1#7 (F) constructs. Each mutant is color-coded with sequences indicated below and mutated residues boxed.
Based on the contribution of leucine and aromatic residues to protein aggregation (Ahmed and Kajava, 2013), we considered that Grp170, ERdj4 and ERdj5 might directly recognize aggregation-prone regions in their clients. To test this idea, we subjected mHC and NS1 sequences to analysis by the TANGO algorithm, which identifies regions in proteins that are predicted to be prone to β-aggregate formation (Fernandez-Escamilla et al., 2004), and to the Zipper DB algorithm, which predicts sequences prone to form β-amyloids (Thompson et al., 2006). The TANGO algorithm identified five regions in our two clients that are predicted to be particularly aggregation-prone. Strikingly, these corresponded to the same five peptides that bound Grp170, ERdj4 and ERdj5 (Figure 4D). In contrast, the amyloidogenic sequences identified by Zipper DB did not show any systematic overlap with these five peptides (data not shown).
To further explore the possibility that the three (co-)chaperones recognized aggregation-prone peptides, we examined our peptide mutants with the TANGO algorithm. The mHC#2, NS1#2, and NS1#7 mutants that no longer bound Grp170, ERdj4 or ERdj5 were not predicted to aggregate, and correspondingly the mutants with the leucine substitutions, which retained some binding, were still predicted to have some aggregation potential (Figures 4E, 4F and S4B-D). The single outlier was the NS1#7 1L,C>S mutant, which no longer supported (co-)chaperone binding but was still predicted to be aggregation-prone, indicating some further constraints on sequence recognition requirements for all three (co-)chaperones. Based on the strong concordance between aggregation and binding, we attempted to disrupt binding of (co-)chaperones with mutations predicted to abolish aggregation. A single amino acid change in mHC#2 (TSY → TPY) resulted in a peptide that was no longer predicted to aggregate (Figure S4D), and correspondingly, the binding of all three (co-)chaperones was significantly reduced (Figure S4D).
(Co-)chaperone binding can be introduced in peptides and proteins by minimal mutations that increase predicted aggregation potential
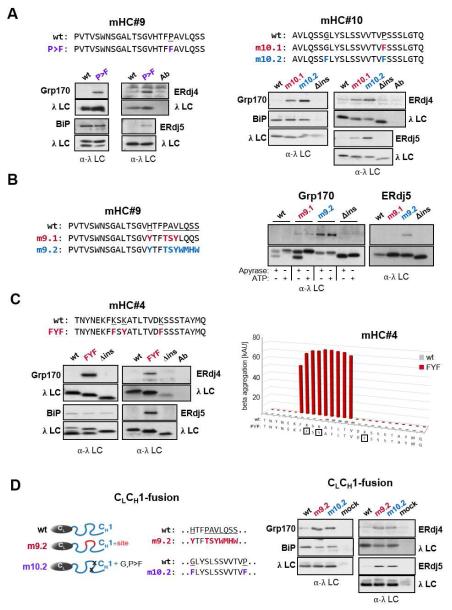
To further explore the significance of these findings, we attempted to use TANGO predictions to insert a (co-)chaperone binding site into CH1 domain-derived peptides of the mHC. A single change from FPA → FFA in mHC#9 and VPS → VFS within the mHC#10 sequence were each predicted to change the peptides from ones that were not aggregation-prone to ones that were, and the combination of the VPS → VFS and SGL → SFL alterations in mHC#10 resulted in an even higher aggregation potential (Figure S5). We found both the mHC#9 and #10 mutants now bound all three (co-)chaperones including ERdj5, even though no cysteines are present in either the mHC#9 or #10 peptides (Figure 5A). We made two additional mutants in the mHC#9 sequence; one in which HTFPAV was changed to YTFTSY, which provided a 6 amino acid sequence identical to that found in the first half of peptide mHC#2.2, and in a second mutant we included a total of 10 amino acids from the core of mHC#2.2 (Figure 5B). Both mutants were predicted to be aggregation-prone, although the second mutant was particularly so (Figure S5), and both bound to Grp170. The larger set of mutations, which also led to stronger Grp170 binding, was required to observe binding to ERdj5 (Figure 5B). We were unable to conclusively detect ERdj4 interaction with either mutant (data not shown).
Figure 5. Mutations predicted to increase aggregation lead to (co-)chaperone binding and can result in aggregation of full-length clients.
(A) Putative (co-)chaperone binding sites were introduced into mHC#9 and mHC#10 with the indicated substitutions and analyzed by immunoprecipitation-coupled western blotting for (co-)chaperone binding as before. (B) The indicated substitutions were made in mHC#9 to create a partial (m9.1) or the complete (m9.2) binding site identified in mHC#2. Constructs were analyzed as in (A). In the case of Grp170 binding samples were lysed with apyrase or ATP added. (C) The TANGO algorithm was used to predict amino acid changes that would lead to an aggregation-prone region in mHC#4, which was negative for both BiP and (co-)chaperone binding. The wild-type and mutant constructs were examained for BiP and (co-)chaperone binding as above. (D) Mutations that led to (co-)chaperone binding in either mHC#9 or #10 were engineered into the corresponding region of the CH1 domain in a chimeric protein composed of the ER targeted λ CL domain and the γ1 CH1 domain. (Co-)chaperone interactions and BiP binding were detected by immunoprecipitation-coupled western blotting.
As all of our (co-)chaperone binding sites at least partially overlapped with BiP binding sites, we sought to insert a ERdj4/ERdj5/Grp170 site in the absence of BiP binding. For this experiment we chose the mHC#4 peptide, which did not bind BiP or any of the (co-)chaperones, and was not predicted to be aggregation-prone (Figure 4D). We found that substitution of 3 lysines in this peptide to phenylalanine or tyrosine dramatically increased the predicted aggregation potential (Figure 5C). This mutant readily interacted with all three (co-)chaperones, but its binding to BiP remained at the low background observed for the parent (mHC#4) and the Δins constructs, further arguing that the binding of the (co-)chaperones is not dependent on BiP binding. Having successfully introduced a novel binding site for the (co-)chaperones, we attempted to introduce (co-)chaperone binding into the CH1 domain of the γHC. To reduce any confounding effects due to the binding of these (co-)chaperones to other portions of the HC, we first tethered the complete CH1 domain to the λ CL domain (CL-CH1-fusion) and inserted the indicated mutations into the corresponding region of the CH1 domain. While the CL-CH1-fusion did not significantly bind any of the three (co-)chaperones as expected, the mutants did (Figure 5D). Our data revealed that a relatively small sequence predicted to be aggregation-prone can provide a core binding site for Grp170, ERdj4 and ERdj5, which can be readily disrupted or introduced with minimal mutations.
Disruption or introduction of (co-)chaperone sites reveals a (co-)chaperone-modulated balance between protein foldability and aggregation
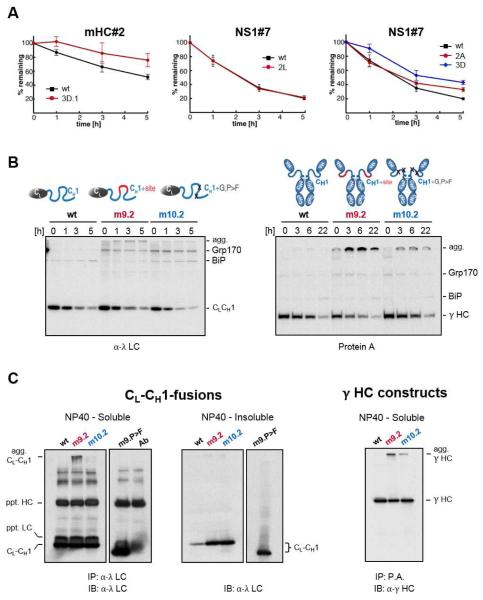
Since Grp170, ERdj4 and ERdj5 have all been linked to ERAD, we examined the effect of disrupting their interaction on the turnover of the wild-type and mutant peptides. Indeed loss of (co-)chaperone binding extended the half-life of the mutant mHC#2 and NS1#7 peptides, whereas the half-life of a mutant that retained (co-)chaperone binding was unaltered (Figure 6A). This suggests binding of these (co-)chaperones could target proteins with exposed aggregation-prone regions for degradation if folding or assembly did not bury them. We next examined the effects of introducing a (co-)chaperone binding site into the unfolded CH1 domain, which is devoid of such sites. When turnover of these CL-CH1 chimera constructs was examined, we found the introduction of a (co-)chaperone binding site into the CH1 domain did not enhance the turnover of the mutant clients as might have been expected. Instead these altered clients began to accumulate as high molecular weight species (Figure 6B), which was particularly prominent in the m9.2 mutant. This same (co-)chaperone binding site is naturally present and well-tolerated in the folded VH domain of the HC. When similar sites were introduced into the CH1 domain of an actual γHC, pulse-chase experiments revealed an even more dramatic accumulation of slow mobility species (Figure 6B). Western blotting confirmed that in both cases these species represented client aggregates, which in the case of the CL-CH1-fusion proteins were also present in the NP40-insoluble fraction (Figure 6C). These data demonstrate that the sequences identified by TANGO and recognized by Grp170, ERdj4 and ERdj5 are indeed aggregation-prone in cells.
Figure 6. Introduction or disruption of (co-)chaperone binding sites alters the fate of client proteins in vivo.
(A) COS-1 cells expressing the indicated mHC#2 and NS1#7 constructs were pulse-labeled and chased for the indicated times. Immunoprecipitated peptide constructs were analyzed by SDS-PAGE, signals were quantified by phosphorimaging and the amount of remaining constructs over time was calculated ±SEM (n≥3). (B) Grp170 and BiP were co-expressed with the wild-type CL-CH1-fusion protein (left) and with the corresponding mutants in the full-length γ HC (right). Cells were pulse-labeled and chased for the indicated times. Immunoprecipitated material was analyzed by reducing SDS-PAGE and migration of clients and chaperones are marked. Slower migrating aggregated clients are indicated (agg). (C) COS-1 cells were co-transfected with Grp170, wild-type (D), and the indicated CL-CH1-fusion proteins. Cells were lysed in NP40 lysis buffer and soluble material from the indicated transfectants was immunoprecipitated with anti-λ (CL-CH1-fusion) and blotted with anti-λ antisera. The antibody control reveals bands present in the antiserum used for immunoprecipation, which are recognized when the same anti-λ serum is used for blotting. Equal amounts of NP40-insoluble material was directly analyzed by western blotting and the images of soluble and insoluble for each mutant are from the same exposure.
Discussion
We developed a tool that allowed binding sequences for five members of the ER Hsp70 chaperone system to be determined in their native setting; four of which had not been previously investigated. Our system confirmed and extended BiP binding sequences identified in antibody molecules using in vitro and in silico methods, and revealed them to be particularly tolerant to mutations. This screen also identified the first sites for ERdj3 in any client. The presence of multiple potential BiP and ERdj3 sites throughout the entire sequence of a client likely allows for binding of these (co-)chaperones during Sec61-mediated polypeptide entry into the ER lumen, where both, BiP and ERdj3, have been localized (Hamman et al., 1998; Guo and Snapp, 2013). It is notable that the BiP and ERdj3 substrate binding preferences observed within the complex environment of the cell are quite similar to those determined in vitro, where E. coli DnaJ was shown to bind sequences that are similar to and overlap with those recognized by the Hsp70 DnaK, both of which also occur frequently throughout the model proteins investigated (Rudiger et al., 2001).
Our identification and characterization of binding sites for Grp170, ERd4, and ERdj5 revealed a number of unanticipated and particularly intriguing findings. First, Grp170, ERd4, and ERdj5 all recognized nearly identical sequences with some minor distinctions, even though they are structurally unrelated. Grp170 is the mammalian ER orthologue of the yeast cytosolic Hsp110 protein, SseI, which possesses an N-terminal ATPase domain similar to other Hsp70 proteins and a C-terminal domain that likely encodes a client binding site (Liu and Hendrickson, 2007). ERdj5 is comprised of a J domain and six thioredoxin domains, which are similar to those found in PDI (Hagiwara et al., 2011), and as such could encode a peptide binding region as has been shown for PDI (Ellgaard and Ruddock, 2005). ERdj4 is structurally uncharacterized but it is significantly smaller than the other two (co-)chaperones and is not expected to resemble either Grp170 or ERdj5. Second, all of the sequences these three (co-)chaperones recognized are uniquely predicted to be prone to β-aggregate formation, as first indicated by their near complete correspondence to sequences identified by the TANGO algorithm (Fernandez-Escamilla et al., 2004). We also analyzed our sequences using the Zipper DB algorithm, which predicts sequences with a propensity to form amyloid fibrils (Thompson et al., 2006), but several of the sequences these (co-)chaperones bound were not recognized in Zipper DB and others that were predicted to form amyloid structures were not recognized by the three (co-)chaperones. Together this suggests these three (co-)chaperones evolved convergently to recognize sequences prone to forming more amorphous aggregates. This possibility was further supported when we inserted novel (co-)chaperone binding sites into the CH1 domain of an Ig HC. The propensity to aggregate was particularly apparent when the CH1 domain sequence was modified to contain the mHC#2 (co-)chaperone binding site that is naturally present in the VH domain of this HC. While this sequence is well-tolerated in the VH domain that folds independently, its presence in the CH1 domain, which remains unstructured prior to assembly with LC, results in aggregation. This confirms that sequences recognized by the three (co-)chaperones are indeed aggregation-prone but also suggests why the mouse γ1 CH1 domain, is devoid of such sites. In fact, the CH1 domain of human IgM and IgG subclasses are also devoid of sequences predicted by TANGO to be aggregation-prone, whereas most other domains in Ig HCs contain such regions (data not shown). The CH1 domains may have specifically evolved to exclude aggregation-prone sequences, as HCs are expressed first developmentally and at high levels in plasma cells, but CH1 domains cannot fold autonomously (Feige et al., 2009; Lee et al., 1999).
Although the less frequent (co-)chaperone binding sites overlapped with a subset of those that bound BiP, their binding did not occur through interactions with BiP. Instead their association required distinct residues within these sequences, as sites for the (co-)chaperones could be disrupted or introduced in peptides without significantly altering BiP binding. Our mutational analyses further provided insights into sequence preferences and constraints in the recognition sites on clients for the (co-)chaperones (Figure S6). The presence of aromatic residues was important for binding but leucines could be substituted in three different peptides without significant consequences for binding. On the other hand alanines were not well-tolerated, which is comparable to preferences observed for peptide binding to Sse1, the yeast cytosolic orthologue of Grp170. Furthermore, we found the presence of cysteines in the peptides enhanced association with all three (co-)chaperones. In fact, one mutant peptide (NS1#7 1L,C>S) was still predicted to be aggregation prone but no longer bound any of the (co-)chaperones. While this might be expected for the oxidoreductase ERdj5, there was no data to suggest this would be true for Grp170 or ERdj4. Cysteines generally increased the affinity of several sequences for the (co-)chaperones, but they were not always required, as we were able to insert sites using TANGO as a guide that did not include cysteines (i.e, mutant constructs mHC#4, mHC#9 and mHC#10). Our finding that ERdj5 also interacted with all of these mutant peptides supports the idea that it binds directly to certain amino acid sequences that may target it to specific disulfide bonds. We found that prolines and lysines were not present in any of the subdivided fragments that bound to the (co-)chaperones, and introduction of either amino acid both reduced the aggregation potential and their binding. This is consistent with the fact that these residues are often considered “gatekeepers” that reduce aggregation propensity. These two amino acids however can be a component of BiP binding sites (Marcinowski et al., 2013), which further distinguishes their binding preferences from those of BiP. While there was a great deal of similarity in the recognition sequences for the three (co-)chaperones and these were in general longer than for BiP, we found that ERdj4 binding was most sensitive to mutational disruption and showed an alternating binding pattern consistent with β-sheet structure. ERdj5 was the most promiscuous in its binding, as it interacted weakly with several cysteine containing peptides that did not appear to support either Grp170 or ERdj4 binding.
It is noteworthy that the three (co-)chaperones that recognize the same aggregation-prone sequences have all been linked to the degradation of various ERAD clients (Dong et al., 2008; Ushioda et al., 2008). In keeping with this, mutations that disrupted (co-)chaperone binding to peptide constructs measurably reduced their turnover, arguing that (co-)chaperone recognition of specific sequences contributes to targeting the clients for ERAD. These sequences are largely buried in the assembled and folded LC-HC dimer (Figure 3D), and thus would not be recognized by these three (co-)chaperones once the protein adopted a folded conformation (model in Figure 7). However, the NS1 VL domain, which possesses a (co-)chaperone binding site, does not fold stably, and recognition of this site thus may assist in rapid turnover of the NS1 LC (Skowronek et al., 1998). The VH domain of our model HC contains the only identified (co-)chaperone binding sites within mHC, but in this case it is oxidized and stably folded, which may contribute to the longevity of the unassembled HC (Vanhove et al., 2001). Based on the TANGO algorithm, we were also able to introduce (co-)chaperone binding sites into the CH1 domain of a CL-CH1 chimeric protein and full-length HCs. However, instead of accelerating turnover of these clients, a significant portion of the altered proteins now aggregated. The reason why (co-)chaperone binding was insufficient to target these proteins for degradation is not clear at this point, but it is possible that sequences for recognition by other components of the ERAD machinery are also lacking in the CH1 domain. Alternatively, the aggregation we observed could in part be due to a concomitant release of BiP and Grp170 as a consequence of simultaneous binding to these clients (model in Figure 7), which will be a less likely scenario for our peptides due to their much shorter length. In keeping with this possibility, a recent study with cytosolic orthologues of BiP and Grp170 found that Hsp110 (Apg2) promoted a coordinated dissociation of Hsp70 and a DnaJ protein from α-synuclein amyloid fibrils in vitro (Gao et al., 2015). Further studies are needed to resolve this point.
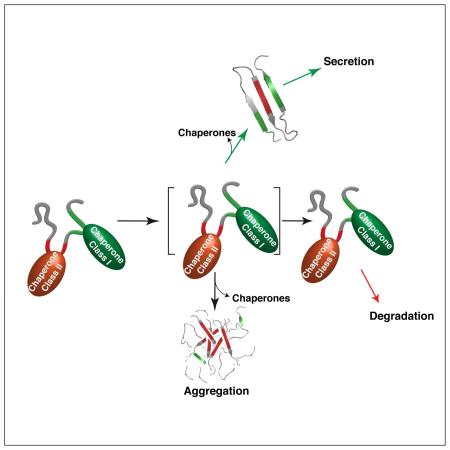
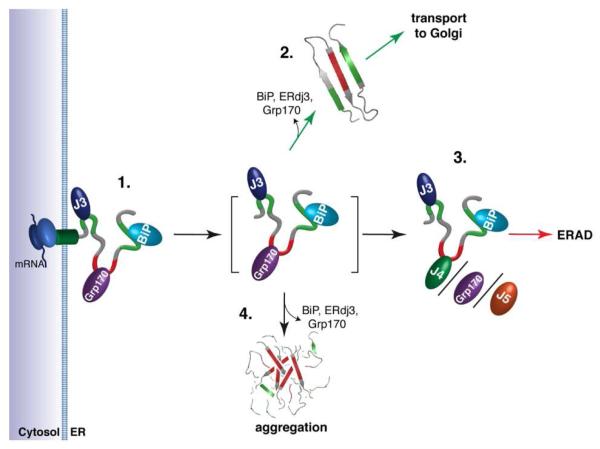
Figure 7. A model predicting how members of the ER Hsp70 chaperone system might interact to control the fate of client proteins.
(1.) Proteins entering the ER can encounter the (co-)chaperones ERdj3, BiP, and Grp170, which reside in proximity to the translocon. Grp170 recognizes sequences (indicated in red) that are distinct from BiP and ERdj3 binding sequences (indicated in green). (2.) As folding procedes, these (co-)chaperones are released allowing the properly matured client to be transported to the Golgi. Grp170/ERdj4/ERdj5 binding sites (indicated in red) become buried during folding, protecting them from aggregation. (3.) If folding fails, the sites recognized by Grp170/ERdj4/ERdj5 remain exposed, leading the client to be targeted for ERAD. (4.) Release of BiP, ERdj3, and Grp170 in the absence of folding can lead to aggregation. Thus, either rapid burial of the Grp170/ERdj4/ERdj5 recognition sites upon folding or their detection for the purpose of ERAD is critical, as these sites possess high aggregation propensity.
In summary, our data provided important insights into molecular chaperone recognition in the cellular environment and revealed an unanticipated specialization among Hsp70 system members. Chaperones like BiP and ERdj3 were capable of binding many diverse, generic sequences on their clients, which is typical of patterns previously identified for other chaperones, that were resistant to mutational disruption (Class I chaperones). In contrast, Grp170, ERdj4 and ERdj5 specifically recognized aggregation-prone regions, which if continuously exposed would signal imminent danger to the cell and necessitate disposal of the protein. Once folded, these sequences are buried in the clients we investigated. These Class II chaperone binding sequences were readily introduced or disrupted by point mutations. Together our work identified fine-tuned sequence recognition patterns for multiple ER chaperones, some of which might easily be disrupted in human disease.
Experimental Procedures
Library and chaperone constructs
To produce an in vivo peptide library, an ER-targeted λ LC constant domain followed by a GS-linker and ending in either a NVT or QVT sequence was used as a template (Δins) (Feige and Hendershot, 2013). Eleven 25 amino acid overlapping sequences from NS1 κ LC (Skowronek et al., 1998) and twelve from a truncated γHC (mHC) (Lee et al., 1999) were inserted into the GS-linker by restriction-free cloning (van den Ent F. and Lowe, 2006) or genes were completely synthesized (Thermo Fisher Scientific) and cloned into the pSVL vector (Amersham). Dissection of the peptides and introduction of point mutations were performed via restriction-free cloning. All sequences were verified. Previously used molecular chaperone expression vectors are described in the Supplemental Information.
Detection of ER chaperone:peptide library interactions
COS-1 cells were maintained and transfected using GeneCellin (BioCellChallenge) as described (Behnke and Hendershot, 2014). Interaction of peptides with individual chaperones was determined by co-expressing the peptide constructs with the indicated chaperone(s) and monitored by metabolic labeling or immunoprecipitation-coupled western blotting. Experimental details and sources of antibodies used are described the Supplemental Information.
Structural modeling
A structural model of an mHC-NS1 Fab fragment was built with Yasara structure. First, a homology model of the mHC was built using the pdb file 4WUK as a template. Subsequently, the modeled structure was aligned with the Fd fragment of the pdb entry 4KVC, which contains a light chain with the NS1 sequence, and the 4KVC Fd fragment was then replaced with the modelled mHC structure – giving rise to a mHC-NS1 dimer. The obtained heterodimer was energy-minimized in explicit solvent using Yasara structure. Figures were prepared with PyMOL.
Bioinformatic analyses
Sequences were analyzed using the TANGO and Zipper DB algorithms, which predict amino acid sequences with a propensity to form β-aggregates (Fernandez-Escamilla et al., 2004) or amyloid fibrils (Thompson et al., 2006), respectively.
Supplementary Material
Acknowledgments
JB gratefully acknowledges support by a Boehringer Ingelheim Fonds PhD fellowship. MJF gratefully acknowledges funding through the Marie Curie COFUND program and the Technische Universität München Institute for Advanced Study, funded by the German Excellence Initiative and the European Union Seventh Framework Program under Grant Agreement 291763 and by the Center for Integrated Protein Science Munich. This work was supported by National Institutes of Health Grant R01 GM54068 to LMH and by the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital. We are very grateful for discussions and experimental assistance from Walid Awad, Viraj Ichhaporia, Miranda Jarrett, Christina Oikonomou, and Rachael Wood.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, JB, MJF and LMH; Investigation, JB, MJM, MJF; Writing Original Draft, LMH; Writing, Review and Editing, MJF, JB, and MJM; Funding Acquistion, JB, MJF and LMH; Resources, JB and F-LS
References
- Ahmed AB, Kajava AV. Breaking the amyloidogenicity code: methods to predict amyloids from amino acid sequence. FEBS Lett. 2013;587:1089–1095. doi: 10.1016/j.febslet.2012.12.006. [DOI] [PubMed] [Google Scholar]
- Behnke J, Hendershot LM. The large hsp70 grp170 binds to unfolded protein substrates in vivo with a regulation distinct from conventional hsp70s. J Biol. Chem. 2014;289:2899–2907. doi: 10.1074/jbc.M113.507491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, Gething MJ. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- Bole DG, Hendershot LM, Kearney JF. Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J. Cell. Biol. 1986;102:1558–1566. doi: 10.1083/jcb.102.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Hebert DN. Protein folding in the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 2013;5:a013201. doi: 10.1101/cshperspect.a013201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr DM, Ramos CH. Specification of Hsp70 function by Type I and Type II Hsp40. Subcell. Biochem. 2015;78:91–102. doi: 10.1007/978-3-319-11731-7_4. [DOI] [PubMed] [Google Scholar]
- Dong M, Bridges JP, Apsley K, Xu Y, Weaver TE. ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol. Biol. Cell. 2008;19:2620–2630. doi: 10.1091/mbc.E07-07-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige MJ, Groscurth S, Marcinowski M, Shimizu Y, Kessler H, Hendershot LM, Buchner J. An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol. Cell. 2009;34:569–579. doi: 10.1016/j.molcel.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige MJ, Hendershot LM. Quality control of integral membrane proteins by assembly-dependent membrane integration. Mol. Cell. 2013;51:297–309. doi: 10.1016/j.molcel.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- Flynn GC, Pohl J, Flocco MT, Rothman JE. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- Gao X, Carroni M, Nussbaum-Krammer C, Mogk A, Nillegoda NB, Szlachcic A, Guilbride DL, Saibil HR, Mayer MP, Bukau B. Human Hsp70 Disaggregase Reverses Parkinson's-Linked alpha-Synuclein Amyloid Fibrils. Mol. Cell. 2015;59:781–793. doi: 10.1016/j.molcel.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeckeler JL, Petruso AP, Aguirre J, Clement CC, Chiosis G, Brodsky JL. The yeast Hsp110, Sse1p, exhibits high-affinity peptide binding. FEBS Lett. 2008;582:2393–2396. doi: 10.1016/j.febslet.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gragerov A, Gottesman ME. Different peptide binding specificities of hsp70 family members. J. Mol. Biol. 1994;241:133–135. doi: 10.1006/jmbi.1994.1482. [DOI] [PubMed] [Google Scholar]
- Guerriero CJ, Brodsky JL. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol Rev. 2012;92:537–576. doi: 10.1152/physrev.00027.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Snapp EL. ERdj3 regulates BiP occupancy in living cells. J Cell Sci. 2013;126:1429–1439. doi: 10.1242/jcs.118182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara M, Maegawa K, Suzuki M, Ushioda R, Araki K, Matsumoto Y, Hoseki J, Nagata K, Inaba K. Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5. Mol. Cell. 2011;41:432–444. doi: 10.1016/j.molcel.2011.01.021. [DOI] [PubMed] [Google Scholar]
- Hamman BD, Hendershot LM, Johnson AE. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell. 1998;92:747–758. doi: 10.1016/s0092-8674(00)81403-8. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hellman R, Vanhove M, Lejeune A, Stevens FJ, Hendershot LM. The in vivo association of BiP with newly synthesized proteins is dependent on their rate and stability of folding and simply on the presence of sequences that can bind to BiP. J. Cell. Biol. 1999;144:21–30. doi: 10.1083/jcb.144.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot L, Bole D, Kohler G, Kearney JF. Assembly and secretion of heavy chains that do not associate posttranslationally with immunoglobulin heavy chain-binding protein. J. Cell. Biol. 1987;104:761–767. doi: 10.1083/jcb.104.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Zhuang M, Hendershot LM. ERdj3, a luminal ER DnaJ homologue, binds directly to unfolded proteins in the mammalian ER: identification of critical residues. Biochem. 2009;48:41–49. doi: 10.1021/bi8015923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaloff CR, Haas IG. Coordination of immunoglobulin chain folding and immunoglobulin chain assembly is essential for the formation of functional IgG. Immunity. 1995;2:629–637. doi: 10.1016/1074-7613(95)90007-1. [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knarr G, Gething MJ, Modrow S, Buchner J. BiP binding sequences in antibodies. J. Biol. Chem. 1995;270:27589–27594. doi: 10.1074/jbc.270.46.27589. [DOI] [PubMed] [Google Scholar]
- Knittler MR, Dirks S, Haas IG. Molecular chaperones involved in protein degradation in the endoplasmic reticulum: Quantitative interaction of the heat shock cognate protein BiP with partially folded immunoglobulin light chains that are degraded in the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 1995;92:1764–1768. doi: 10.1073/pnas.92.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota P, Summers DW, Ren HY, Cyr DM, Dokholyan NV. Identification of a consensus motif in substrates bound by a Type I Hsp40. Proc. Natl. Acad. Sci. U. S. A. 2009;106:11073–11078. doi: 10.1073/pnas.0900746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-K, Brewer JW, Hellman R, Hendershot LM. BiP and Ig light chain cooperate to control the folding of heavy chain and ensure the fidelity of immunoglobulin assembly. Mol. Biol. Cell. 1999;10:2209–2219. doi: 10.1091/mbc.10.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Hendrickson WA. Insights into hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell. 2007;131:106–120. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Kearney JF, Hendershot LM. Association of transport-defective light chains with immunoglobulin heavy chain binding protein. Mol. Immunol. 1990;27:623–630. doi: 10.1016/0161-5890(90)90004-j. [DOI] [PubMed] [Google Scholar]
- Marcinowski M, Rosam M, Seitz C, Elferich J, Behnke J, Bello C, Feige MJ, Becker CF, Antes I, Buchner J. Conformational selection in substrate recognition by Hsp70 chaperones. J Mol. Biol. 2013;425:466–474. doi: 10.1016/j.jmb.2012.11.030. [DOI] [PubMed] [Google Scholar]
- Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DT, Watowich SS, Lamb RA. Analysis in vivo of GRP78-BiP/substrate interactions and their role in induction of the GRP78-BiP gene. Mol. Biol. Cell. 1992;3:143–155. doi: 10.1091/mbc.3.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO. J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger S, Schneider-Mergener J, Bukau B. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 2001;20:1042–1050. doi: 10.1093/emboj/20.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A, Maintz M, Kehle T, Herzog V. In vivo iodination of a misfolded proinsulin reveals co- localized signals for Bip binding and for degradation in the ER. EMBO. J. 1995;14:1091–1098. doi: 10.1002/j.1460-2075.1995.tb07092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Rosam M, Glaser M, Patronov A, Shah H, Back KC, Daake MA, Buchner J, Antes I. BiPPred: Combined sequence- and structure-based prediction of peptide binding to the Hsp70 chaperone BiP. Proteins. 2016 doi: 10.1002/prot.25084. [DOI] [PubMed] [Google Scholar]
- Shen Y, Hendershot LM. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP's interactions with unfolded substrates. Mol. Biol. Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Meunier L, Hendershot LM. Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J. Biol. Chem. 2002;277:15947–15956. doi: 10.1074/jbc.M112214200. [DOI] [PubMed] [Google Scholar]
- Skowronek MH, Hendershot LM, Haas IG. The variable domain of non-assembled Ig light chains determines both their half-life and binding to BiP. Proc. Natl. Acad. Sci. U. S. A. 1998;95:1574–1578. doi: 10.1073/pnas.95.4.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MJ, Sievers SA, Karanicolas J, Ivanova MI, Baker D, Eisenberg D. The 3D profile method for identifying fibril-forming segments of proteins. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4074–4078. doi: 10.1073/pnas.0511295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Lowe J. RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem. Biophys. Methods. 2006;67:67–74. doi: 10.1016/j.jbbm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Vanhove M, Usherwood Y-K, Hendershot LM. Unassembled Ig heavy chains do not cycle from BiP in vivo, but require light chains to trigger their release. Immunity. 2001;15:105–114. doi: 10.1016/s1074-7613(01)00163-7. [DOI] [PubMed] [Google Scholar]
- Wen KW, Damania B. Hsp90 and Hsp40/Erdj3 are required for the expression and anti-apoptotic function of KSHV K1. Oncogene. 2010;29:3532–3544. doi: 10.1038/onc.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Sarbeng EB, Vorvis C, Kumar DP, Zhou L, Liu Q. Unique peptide substrate binding properties of 110-kDa heat-shock protein (Hsp110) determine its distinct chaperone activity. J Biol. Chem. 2012;287:5661–5672. doi: 10.1074/jbc.M111.275057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.