Abstract
In principle, the inhibition of candidate gain-of-function genes defined through genomic analyses of large patient cohorts offers an attractive therapeutic strategy. In this study, we focused on changes in expression of CD24, a well validated clinical biomarker of poor prognosis and a driver of tumor growth and metastasis, as a benchmark to assess functional relevance. Through this approach, we identified GON4L as a regulator of CD24 from screening a pooled shRNA library of 176 candidate gain-of-function genes. GON4L depletion reduced CD24 expression in human bladder cancer cells, blocked cell proliferation in vitro and tumor xenograft growth in vivo. Mechanistically, GON4L interacted with transcription factor YY1, promoting its association with the androgen receptor to drive CD24 expression and cell growth. In clinical bladder cancer specimens, expression of GON4L, YY1 and CD24 was elevated compared to normal bladder urothelium. This pathway is biologically relevant in other cancer types as well, where CD24 and the androgen receptor are clinically prognostic, given that silencing of GON4L and YY1 suppressed CD24 expression and growth of human lung, prostate and breast cancer cells. Overall, our results define GON4L as a novel driver of cancer growth, offering new biomarker and therapeutic opportunities.
Keywords: androgen receptor, CD24, functional genomics, GON4L, YY1
Introduction
Hundreds of genes have been identified as candidate biomarkers of disease (1), patient prognosis (2) or response to therapy (3). Those with predicted gain-of-function through copy number alterations, gene expression or mutations make attractive therapeutic targets if they regulate cancer development, growth or progression. Here we used a pooled shRNA library and DNA sequencing to identify genes that are both novel and clinically relevant drivers of growth in human bladder cancer. For this library, we selected 176 genes with DNA or copy number alterations in bladder cancer patients, and whose gene expression was positively associated with malignancy or progression in human disease. To further ensure the identified genes have robust clinical relevance (4) and a high likelihood of driving tumor growth, the endpoint of the screen was reduction in expression of cluster of differentiation 24 (CD24), a known driver of cancer progression and metastasis (5,6).
CD24, a glycosylated mucin-like protein, is not expressed in most normal tissues (7) but gains expression in many cancers including those of breast, ovarian, colorectal, pancreatic, prostate, and bladder origin (6). Furthermore, CD24 expression levels positively correlate with tumor aggressiveness and poor prognosis (6). In a BBN-induced carcinogenesis mouse model that mimics muscle-invasive human bladder cancer, CD24 knockout mice developed fewer bladder tumors and in mice with cancer, fewer had metastases compared to wild type mice (5). Homozygous deletion of CD24 in APC (Min/+) mice causes complete abolishment of tumor formation in all sections of small intestine (8). Knockdown of CD24 also retards the growth, progression and metastasis of prostate cancer (9), gastric cancer (10), and breast cancer (11). Targeting CD24 by anti-CD24 antibody inhibits growth and/or metastasis in models of osteosarcoma (12), lung (13), pancreatic, colorectal (14) and bladder (15) cancer. Thus, identifying genes whose depletion reduces CD24 expression are likely to be clinically relevant and provide additional and targetable regulators of tumor growth.
Our screen discovered GON4L as a driver of both CD24 expression and cancer growth. It also defined GON4L as a novel therapeutic target across multiple tumor types. By showing that androgen receptor (AR) and YY1 associate with GON4L to drive growth, we established anti-androgen receptor therapy as a potential therapeutic angle for patients with expression of GON4L and AR. The screening method detailed here can be adapted to most cancer types to determine which patient tumor genomic alterations regulate tumor growth, thus providing novel therapeutic opportunities.
Materials and Methods
Human cancer cell lines
Bladder cancer cell lines UMUC3, TCCSUP, RT4, CRL2742, SW780, MGHU4, and UMUC9 were previously described (4). Lung and prostate cancer cell lines, A549 (16) and 22Rv1 (17) were obtained from the cell line repository at University of Colorado Cancer Center PPSR. Breast cancer cell lines HCC1806 and BT549 (18) were a gift from Dr. Richer at the University of Colorado. HEK293T cells were purchased from ATCC. All reagents and media were purchased from Invitrogen. All lines were authenticated by the University of Colorado Cancer Center PPSR core using an Applied Biosystems Profiler Plus Kit that analyzed 9 loci (Life Technologies 4303326). Cells were used in experiments within 6 weeks of thawing and were tested to be free of mycoplasma.
Cell proliferation and soft agar colony formation assays
For proliferation assay, cells were plated onto 96-well plates and allowed to grow for 5 days with freezing one plate each day. All plates were then processed with CyQuant assay kit (Life Technologies). For soft agar assays, 20,000 cells were suspendedin 0.4% agar and plated on 1% agar hard base in each well of 6-well plate. Colonies formed after 9–18 days in culture were stained with Nitro-BT (Sigma-Aldrich) at 37°C and counted using software ImageJ.
Subcutaneous Xenograft Tumor Growth
All animals used were treated according to University of Colorado and Institutional Animal Care and Use Committee (IACUC) guidelines (IACUC protocol number B-93413(12)1E).. 4–5 week-old NCr nu/nu male mice (NCI-Frederick) were injected with UMUC3 cells stably expressing non-target, GON4L, or YY1 shRNAs in the right and left flanks of each mouse. Mice were injected with 500,000 and 50,000 cells/site at 3 or 4 sites/mice and 5 mice/group. Each mouse was injected with cells harboring distinct shRNAs. Tumors were measured and volumes calculated as described (19).
qRT-PCR, Western Blot and Immunoprecipitation
For determination of changes in mRNA expression as measured by qRT-PCR, the ΔΔCT method was used.. Expression was normalized to internal control β-actin and gene knockdown was determined by comparing to control-treated cells.
Whole cell lysates were subjected to SDS-PAGE using Novex 4–20% Tris-glycine gels (Life technologies) and western blotting was performed as described (19). Since CD24 protein is heavily glycosylated, it runs differently on SDS-PAGE for individual cell lines as shown in Supplementary Fig. S1A.
For immunoprecipitations (IPs), cells were suspended in a lysis buffer containing 50mM Tris-HCl, pH 8.0, 120mM NaCl, 0.5% Nonidet P-40, phosphatase and protease inhibitor mixture (Roche Applied Sciences). Lysates were incubated with either Flag-conjugated agarose beads (F2426, Sigma-aldrich), or antibodies conjugated to protein A/G agarose beads (sc-2003, Santa Cruz Biotechnology) at 4°C overnight. Antibodies used for IP were anti-HA (MMS-101P, Covance), anti-AR (sc-7305), and anti-YY1 (sc-1703) from Santa Cruz Biotechnology. A matched isotype antibody was used as a negative control.
CD24 promoter reporter assay
We used the promoter from UMUC3 cells called “CD24-1896” and “CD24-1896 AREmut” as described (5). UMUC3 cells were transfected with siRNA using RNAiMax and 24h later with DNA using Lipofectamine-2000. Renilla luciferase plasmid was transfected at 1/50 conc. of CD24 promoter plasmid containing firefly luciferase, as an internal control. After 48h of DNA transfection, cells were lysed and assayed for dual-luciferase activity according to the manufacturer’s instructions (Promega). Firefly luciferase values were normalized to Renilla luciferase values. Transcription factor binding site analysis was done using TRANSFAC database (courtesy of BIOBASE) (20).
Gene expression, patient cohorts and statistics
Putative "gain-of-function" genes were identified, in part, by analysis of 13 bladder tumor patient cohorts (N = 1451), using the Bladder Cancer Biomarker Evaluation Tool (21). Here, p-values for differential expression of genes (22–25) were calculated using the non-parametric Wilcoxon rank-sum test, while log-rank p-values for survival were calculated comparing patients with high expression (top 50%) to patients with low expression (bottom 50%) for each gene. Later in the work, we further analyzed an additional set of 4 bladder cancer patient cohorts with both normal bladder tissue and primary tumor profiles for specific genes. These cohorts included 50 patients profiled at Aarhus University Hospital, Skejby (Dyrskjot et al. cohort (26)); downloaded from the Gene Expression Omnibus GEO accession no. GSE3167 (27,28); 175 patients profiled at Chungbuk National University Hospital (Kim et al. cohort (29)), and 129 patients profiled at Memorial Sloan Kettering Cancer Center (Sanchez-Carbayo et al. cohort (30), downloaded from supplementary material to publication). There were also a total of 19 patients in the TCGA bladder cancer cohort with matched tumor and normal RNA sequencing (23). For gene expression data measured by microarray, GON4L, YY1, and CD24 probes were identified based on current Affymetrix (Dyrskjot et al. and Sanchez-Carbayo et al. cohorts) and Illumina (Kim et al. cohort) annotation. Specifically, the hgu133a.db package (version 3.1.3) from Bioconductor (www.bioconductor.org) was used for Affymetrix annotation and the GPL6102 platform data from GEO (last updated Feb 2013) was used for Illumina annotation. For all cohorts, when multiple probe sets corresponded to a gene, we used the probe set with highest mean expression (31). For TCGA dataset, gene expression was processed according to the TCGA protocols (23). The normalized expression of CD24, GON4L, and YY1 were compared using a Wilcoxon rank sum test. The CNA and mutation results taken from cBioPortal (http://cbioportal.org) (32) are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov (32,33).
Data were analyzed using a two-tailed Student’s t-test with unequal variances. All experiments were done at least three independent times.
Results
GON4L is a positive regulator of CD24 expression
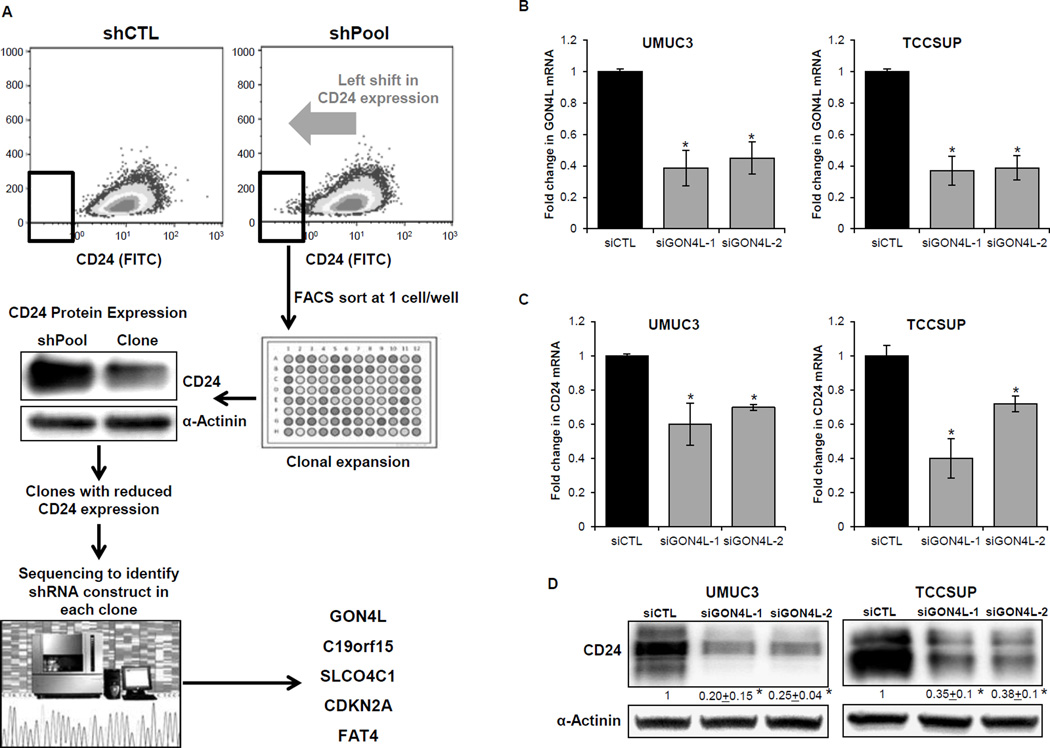
Human bladder cancer cell lines MGHU4 and UMUC3 that stably expressed either non-target shRNA (shCTL) or a pooled library of shRNAs (shPool) were FACS sorted based on surface CD24 expression. The shPool cells showed a slight shift towards undetectable CD24 staining compared to shCTL cells (Fig. 1A, top). MGHU4 shPool cells that showed a shift in CD24 staining were then sorted into a 96-well plate at 1 cell/well for clonal expansion. The clones were then examined for reduction in CD24 expression compared to the shPool cells (Fig. 1A). Five clones showing reduction in CD24 protein levels were sequenced to identify the specific shRNA molecule being expressed. The identified constructs targeted the genes C19orf15, SLCO4C1, CDKN2A, FAT4 and GON4L. Identification of CDKN2A, whose loss has been reported to negatively regulate CD24 in melanoma cells (34) suggested our approach had the ability to find regulators of CD24 expression.
Figure 1.
Identification of GON4L as a regulator of CD24 expression. A, schematic diagram showing the screening strategy. MGHU4 shPool cells shifted towards low FITC-CD24 intensity as indicated by the arrow and cells in box were FACS sorted, clonally expanded, screened for low CD24 expression and shRNAs present were identified by sanger sequencing. Cells were transfected with non-target control (siCTL) or GON4L siRNAs and after 96h, B, mRNA levels of GON4L were measured; C, mRNA levels of CD24 and D, representative western blot images showing CD24 protein (densitometric analysis was done using ImageJ software, values were normalized to α-actinin) in cells from (B). Results are shown as mean ± S.D., *p<0.05 by Student’s t-test.
Next, we depleted these 5 target genes in 5 different bladder cancer cell lines (MGHU4, UMUC9, CRL2742, TCCSUP, and UMUC3) using shRNA constructs different from those used in the library (Supplementary Table S1). Depletion of GON4L showed the most consistent reduction in CD24 expression across the cell lines ranging from 25% to 77% decrease compared to shCTL treated cells (Supplementary Fig. S1B). To confirm our findings, we transiently depleted GON4L in two bladder cancer cell lines, UMUC3 and TCCSUP, using pools of 4 siRNAs. Both lines showed 76–80% reduction in CD24 levels with knockdown of GON4L (Supplementary Fig. S1C). Next, we used two individual GON4L siRNAs (GON4L-1 and GON4L-2) to evaluate the effect of GON4L knockdown on CD24 mRNA and protein level in both UMUC3 and TCCSUP cells. Both GON4L siRNAs showed almost 60% reduction in GON4L mRNA levels in both cells lines used (Fig. 1B). We observed the significant reduction in CD24 expression at both mRNA (Fig. 1C) and protein levels (Fig. 1D) with both siRNAs. These results positioned GON4L as a putative novel transcriptional regulator of CD24.
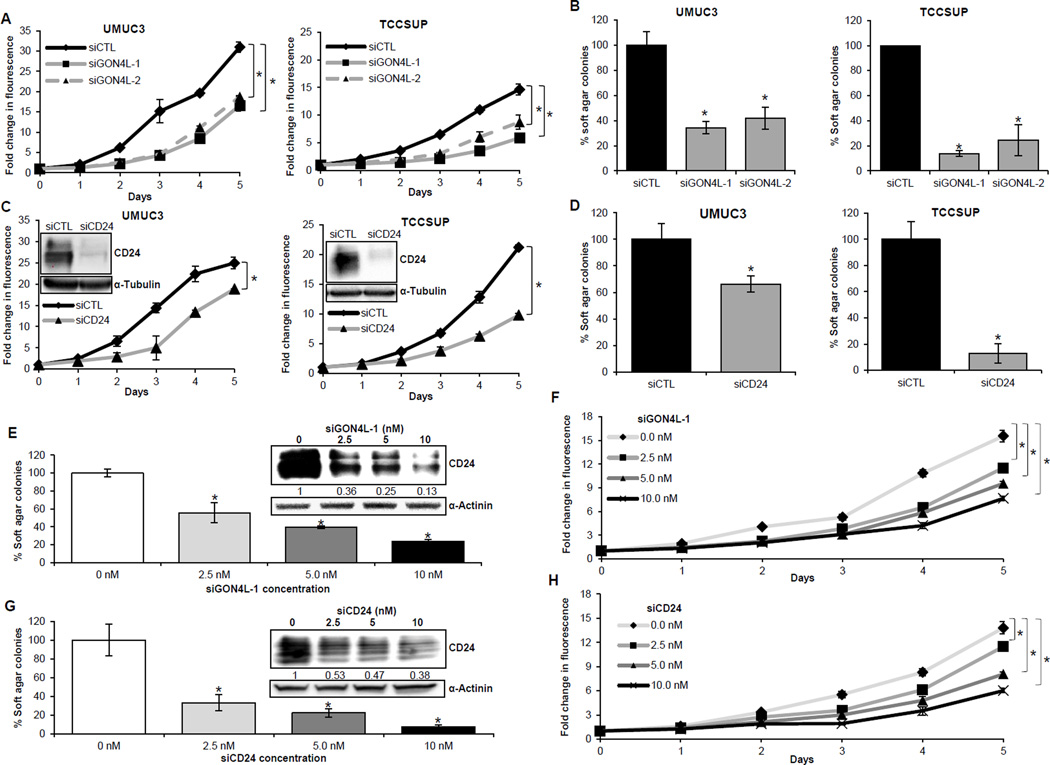
We next explored the role of GON4L on bladder cancer cell proliferation. Knockdown with two different siRNAs showed a significant reduction in monolayer (Fig. 2A) and anchorage-independent (Fig. 2B) cell proliferation of UMUC3 and TCCSUP lines. To build on the evidence suggesting GON4L is a regulator of CD24 expression, we investigated whether CD24 depletion elicited phenotypes similar to GON4L depletion. Indeed, CD24 knockdown suppresses both monolayer proliferation and anchorage-independent growth (Fig. 2C and D). To gain additional support for this model, GON4L-1 siRNA was titrated into TCCSUP cells at 2.5, 5.0 and 10.0nM and anchorage dependent and independent proliferation measured along with CD24 expression. We observed a siRNA dose-dependent reduction in GON4L mRNA (Supplementary Fig. S2A), CD24 protein expression (Fig. 2E inset), and anchorage independent (Fig. 2E) and dependent (Fig. 2F) cell proliferation. This mirrored the dose-dependent effect of CD24 siRNA on growth of TCCSUP cells (Fig. 2G and H). Noticeably, even 2.5nM of GON4L-1 siRNA showed significant reduction in cell growth and CD24 levels, suggesting that the effect of GON4L is specific and suggests few off-target effects.
Figure 2.
GON4L and CD24 depletion inhibit growth of bladder cancer cells. After 48h of transfection with control and GON4L siRNAs, cells were processed for (A) cell proliferation. Graphs show fold change in CyQuant dye fluorescence intensity compared to day 0 of siCTL and siGON4L, respectively. B, cells from (A) split onto soft agar plates. Graphs show % change in number of soft agar colonies. C and D, cells depleted of CD24 by siRNA knockdown were processed for (C) cell proliferation, CD24 western blot (inset) and (D) soft agar assays. E and F, TCCSUP cells treated with increasing concentrations of GON4L siRNA have dose-dependent reduction in (E) soft agar colonies, CD24 protein levels (inset) and (F) cell proliferation rate. G and H, TCCSUP cells treated with increasing concentrations of CD24 siRNA have dose-dependent reduction in (G) soft agar colonies, CD24 protein levels (inset) and (H) cell proliferation rate. Results are shown as mean ± S.D., *p<0.05 by Student’s t-test.
GON4L and YY1 interact to regulate CD24 expression and cell growth
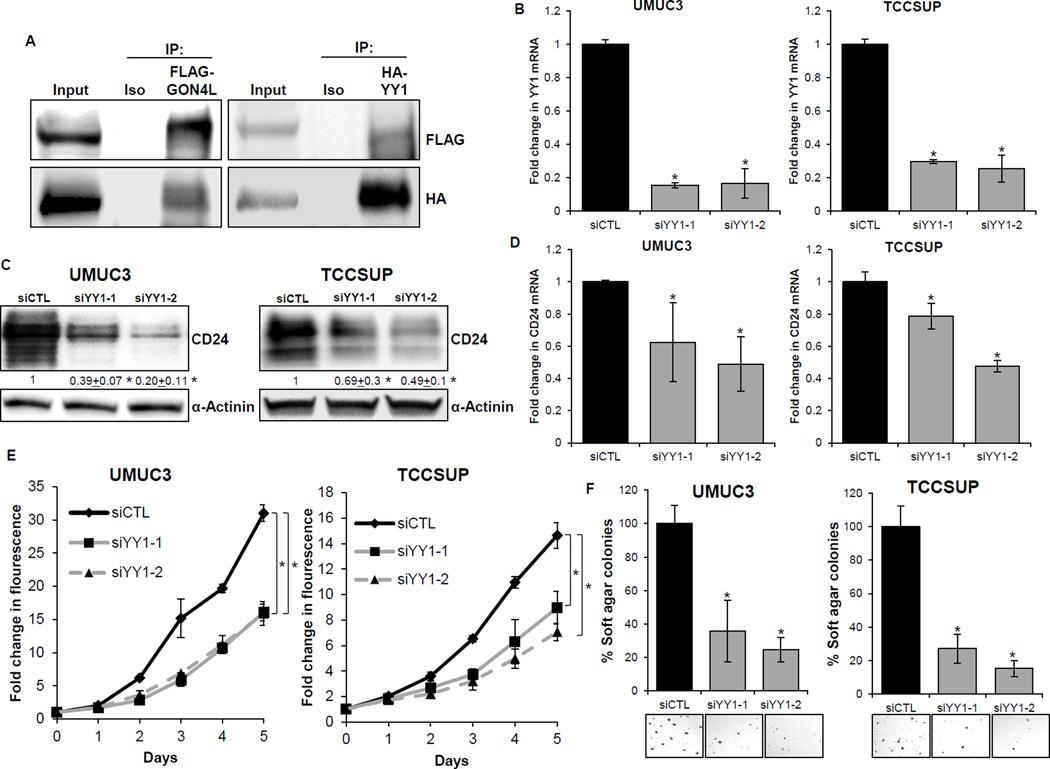
To understand how GON4L regulates CD24 transcription, we used a two stage computational approach to identify putative interacting partners of GON4L that could mediate its effects on CD24 transcription. First we used the STRING database (35) and found 10 proteins either known or predicted to bind GON4L (Supplementary Fig. S2B). Next, using the TRANSFAC database (20), we examined if any of these proteins bound the CD24 promoter in a known or predicted fashion. This analysis revealed that the transcription factor YY1 (Yin Yang 1) is known to interact with GON4L in murine cells at both endogenous and exogenous levels (36) and has predicted binding sites on the CD24 promoter. Hence, we hypothesized that GON4L binds YY1 and this leads to activation of CD24 transcription.
To test this hypothesis, we first examined whether GON4L is found in complex with YY1 in human bladder cancer cells. Since, a well-characterized GON4L antibody is not available commercially, we decided to examine their exogenous interactions using tagged-proteins. We generated a UMUC3 cell line that stably expresses Flag-tagged GON4L and then transiently transfected these cells with HA-tagged YY1. A co-immunoprecipitation (Co-IP) experiment with either Flag or HA-conjugated beads showed that GON4L and YY1 form a complex (Fig. 3A). Next we examined the effect of YY1 depletion on CD24 levels and in vitro cellular phenotypes. Depletion of YY1 with two different siRNAs (YY1-1 and YY1-2) showed 70–85% knockdown of YY1 (Fig. 3B) and significantly reduced CD24 expression at both mRNA and protein levels (Fig. 3C and D). Both YY1 siRNAs showed almost 50% reduction in cell proliferation rate and 3–6 fold reduction in soft agar colony formation in both UMUC3 and TCCSUP cell lines (Fig. 3E and F). We also confirmed that knockdown of YY1 had no effect on GON4L mRNA expression (Supplementary Fig. S3). Taken together, these data suggest that GON4L interacts with YY1 in human bladder cancer cells and this association drives expression of CD24 and cell growth.
Figure 3.
YY1 interacts with GON4L and regulates CD24 expression and growth of bladder cancer cells. A, Co-IP for the GON4L-YY1 interaction in UMUC3 cells stably expressing Flag-GON4L and transiently transfected for HA-YY1. Cells were lysed with NP-40 lysis buffer after 96h of transfection. Cells were transfected with siCTL and two different siRNAs for YY1 (YY1-1 and YY1-2), after 96h were processed for (B) YY1 mRNA expression, (C) Protein levels of CD24 and (D) CD24 mRNA expression. After 48h of transfection, cells were processed for cell proliferation and soft agar colony formation. E, fold changes in CyQuant dye fluorescence intensity as compared to Day 0 of siCTL and siYY1 cells. F, percent change in soft agar colonies. Results are shown as mean ± S.D., *p<0.05 by Student’s t-test.
Expression of GON4L and YY1 affects tumor formation in vivo and is associated with human bladder cancer
To study the effect of GON4L and YY1 knockdown on subcutaneous tumor growth, we used lentivirus to deliver constructs expressing non-target control, GON4L or YY1 shRNAs. After selection with puromycin, stably transduced cells were analyzed for gene knockdowns by qRT-PCR and for effect on CD24 expression by western blot (Fig. 4A). These stable cell lines were then injected subcutaneously at various doses into mice. Compared to shCTL, both shGON4L and shYY1 cells had a 40–80% decrease in tumor incidence (Fig. 4B and Supplementary Fig. S4A) and delayed tumor outgrowth (Fig. 4C and Supplementary Fig. S4B). Reported in TCGA (23) and by Iyer et al. (37), 45% and 34% of bladder cancer patients showed gain or amplification in GON4L (Fig. 4D). Hence we analyzed RNA expression of GON4L, YY1 and CD24 together in additional patient cohorts and found that all three genes were elevated in tumor samples compared to normal bladder urothelium in three different cohorts (Fig. 4E and Supplementary Fig. S4C and S4D). Notably, all three genes have elevated expression in matched tumor-normal urothelium of patient samples from TCGA (23) (Fig. 4F). These human bladder cancer patient data support our experimental cell line data showing that GON4L, YY1, and CD24 are bladder cancer drivers.
Figure 4.
GON4L and YY1 affect tumor formation in vivo and their expression is associated with bladder cancer. UMUC3 cells transduced with non-target, GON4L and YY1 shRNAs and selected with puromycin for 4 days before use. A, knockdown of GON4L and YY1 confirmed by qRT-PCR, inset showing CD24 protein levels after GON4L and YY1 knockdown. Results are shown as mean ± S.D. B, differences in tumor incidence with the bars representing percentage of tumors formed in nude mice injected with 500,000 cells/site (n = 15). C, subcutaneous tumor growth over time. Results are shown as mean ± S.E., *p<0.05 by Student’s t-test. D, genomic alterations in GON4L in 4 human bladder cancer patient datasets (indicated), as reported in cBioPortal (http://www.cbioportal.org). CNA-copy number alterations. E and F, GON4L, YY1 and CD24 mRNA expression in human urothelial tumors compared to normal human bladder tissue. F, normal tissues were “matched” to tumors in the same patient in TCGA set (gray). Results in (E) and (F) were analyzed using different gene expression platforms as mentioned in the Materials and Methods section, statistical comparisons were done using the Wilcoxon rank sum test. Horizontal lines in boxes represent median of gene expression. Box and whisker represents the first and third quartiles..
YY1 regulates CD24 through the androgen receptor
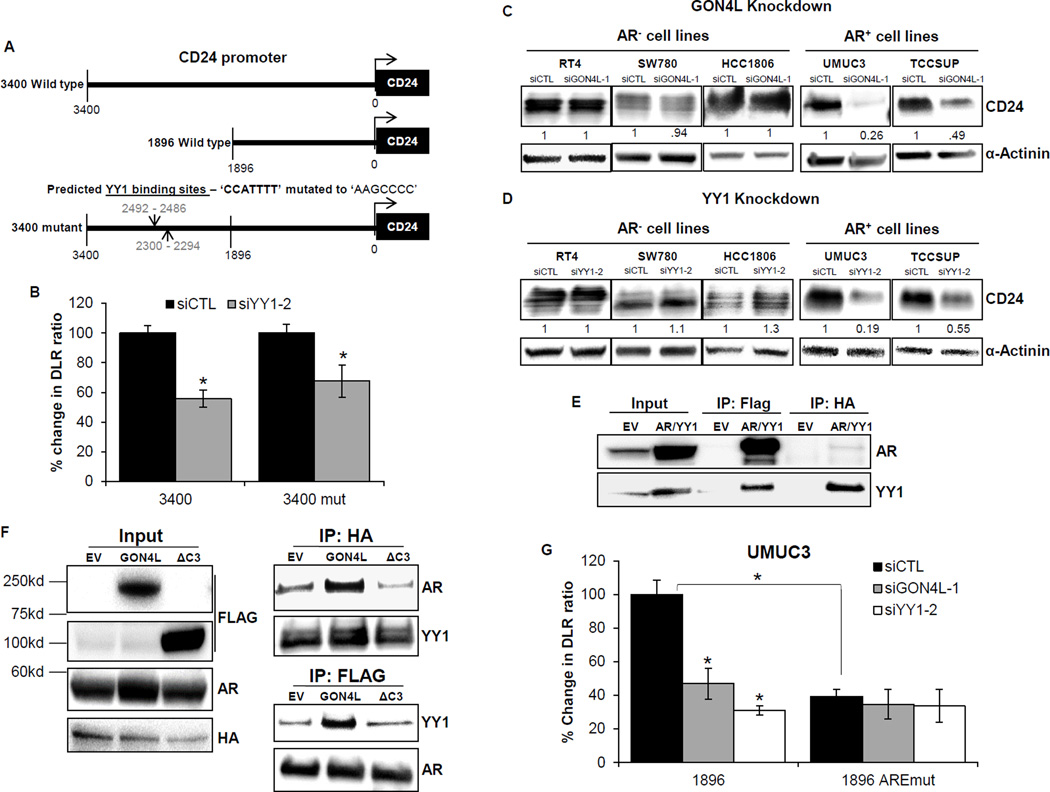
As with GON4L knockdown (Fig. 1C), depletion of YY1 also reduced CD24 mRNA levels in both UMUC3 and TCCSUP cells (Fig. 3D). Therefore, we sought to examine if YY1 regulates CD24 transcriptionally by binding to its predicted binding sites at 2492-2486 bp and 2300-2294 bp upstream of the CD24 transcription start site. We used a reporter construct containing 3400bp of the CD24 promoter and mutated the two predicted YY1 binding sites (Fig. 5A). GON4L and YY1 knockdown reduced luciferase activity of both the wild-type and mutant promoter reporters (Fig. 5B and G). Control reporter constructs consisting of a promoterless basic reporter or a CMV promoter reporter were unaffected (Supplementary Fig. S5A). These results suggest that GON4L-YY1 complex is not binding the CD24 promoter at the predicted sites.
Figure 5.
GON4L and YY1 regulate CD24 transcription via AR. A, diagram of 3400bp, 1896bp wild type and 3400bp CD24 promoter with both YY1 binding sites mutated. B, percent change in DLR ratio (firefly/renilla luciferase activity) representing CD24 promoter activity in control and YY1 knockdown cells. After 96h of transfection with (C) GON4L and (D) YY1 siRNA, AR− and AR+ cells were blotted for CD24 and α-actinin as loading control. Non-target siRNA was used as control. Densitometric analysis was done using ImageJ software, values were normalized with α-actinin. E, Co-IP in UMUC3 cells overexpressing empty vector (EV) or Flag-AR and HA-YY1 (AR/YY1). F, Co-IP in 293T cells transfected with EV, full length GON4L or deletion mutant GON4L (ΔC3) along with Flag-AR and HA-YY1. G, cells were transfected with control, GON4L and YY1 siRNA, 24h later transfected with wild type or ARE mutant 1896bp CD24 promoter and activity was measured after 48h. Results are shown as mean ± S.D., *p<0.05 by Student’s t-test.
YY1 has been shown to directly interact with AR and co-activate its transcriptional activity (38). AR has previously been shown to up-regulate CD24 mRNA and protein in bladder cancer (5). Thus, we hypothesized that GON4L regulates CD24 by affecting the binding of YY1 to AR. To examine the dependency of CD24 expression on the ability of GON4L-YY1 to bind AR, we transiently depleted GON4L and YY1 in AR− (RT4, SW780 and HCC1806) and AR+ (UMUC3 and TCCSUP) cancer cell lines. Only the AR+ lines showed reduced CD24 levels while AR− lines had no significant change in CD24 expression (Fig. 5C and D, and Supplementary Fig. S6A). We also confirmed that knockdown of GON4L and YY1 is not affecting the AR protein expression in both AR+ lines (Supplementary Fig. S5C and S5D). Thus, GON4L and YY1 depletion appear to decrease CD24 expression only when AR is present. Next we examined if YY1 interacts with AR in bladder cancer by Co-IP of exogenous (Flag-AR and HA-YY1) and endogenous proteins in bladder cancer cells and found this to occur in both settings (Fig. 5E and Supplementary Fig. S5B). To determine if GON4L has the ability to influence YY1-AR association, we transiently transfected GON4L, either full length or a deletion mutant (ΔC3) lacking the YY1-binding domain. GON4L ΔC3 mutant has been shown previously not to interact with YY1 (36). We found that the GON4L mutant failed to enhance YY1-AR interaction in stark contrast to full length GON4L (Fig. 5F). Further, to test whether GON4L and YY1 knockdown are reducing CD24 promoter activity as a result of less AR activity, we used two different 1896bp CD24 promoter reporters. One reporter was wild-type while the other had an AR responsive element (ARE) mutation (5). In siCTL treated cells, and consistent with previously published data (5), the ARE mutant reporter itself has decreased activity relative to wild-type (Fig. 5G). Further analysis revealed that depletion of GON4L or YY1 had no additional decrease in activity (Fig. 5G), strongly suggesting that GON4L-YY1 regulates CD24 at the transcriptional level by controlling binding of AR to CD24 promoter at the ARE.
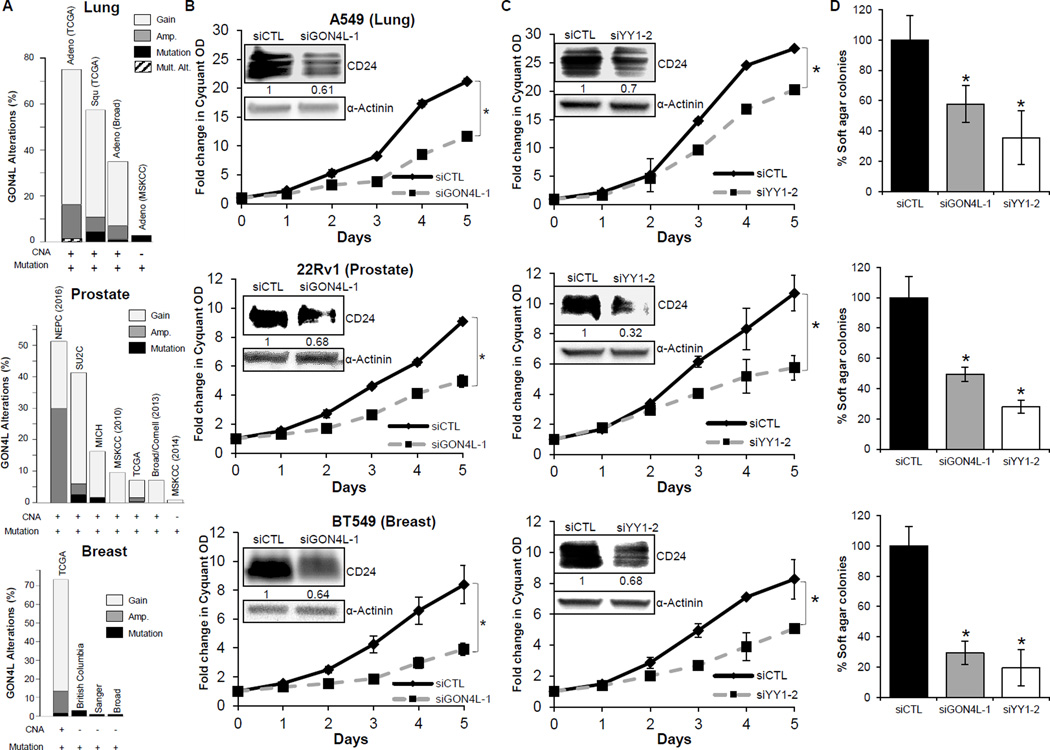
GON4L is altered and drives growth of multiple androgen sensitive tumor types
The biological and human pathological relevance of GON4L, YY1 and CD24 in bladder cancer encouraged us to examine GON4L in other cancer types, particularly types where CD24 and AR are known to be biologically and clinically relevant, such as prostate (9,39), breast (6,39) and lung (13,39). GON4L is also mostly either gained or amplified in these cancer types in multiple cohorts (Fig. 6A). We selected cell lines A549, 22Rv1 and BT549 to represent lung, prostate and breast cancer, respectively, based on the expression of both AR (Supplementary Fig. S6B) and CD24. Knockdown of GON4L or YY1 (Supplementary Fig. S6C and S6D) in these cells led to a significant reduction in CD24 levels and both anchorage dependent and independent growth (Fig. 6B–D). These results indicate a role for GON4L and the GON4L-YY1-AR-CD24 interaction in not just bladder cancer but also in numerous other tissue types.
Figure 6.
GON4L and YY1 knockdown affect CD24 expression and growth of human lung, prostate and breast cancer cell lines. A, genomic alterations in GON4L in lung, prostate and breast cancer patient datasets, as taken from cBioPortal (http://www.cbioportal.org). CNA-copy number alterations. B–D, cells were transfected with non-target, GON4L and YY1 siRNAs. 48h later, cells were split to measure (B and C) proliferation and (D) soft agar colonies. Cells were also processed for CD24 western blot after 96h of transfection shown in inset. Densitometric analysis was done using ImageJ software, values were normalized with α-actinin. Results are shown as mean ± S.D., *p<0.05 by Student’s t-test.
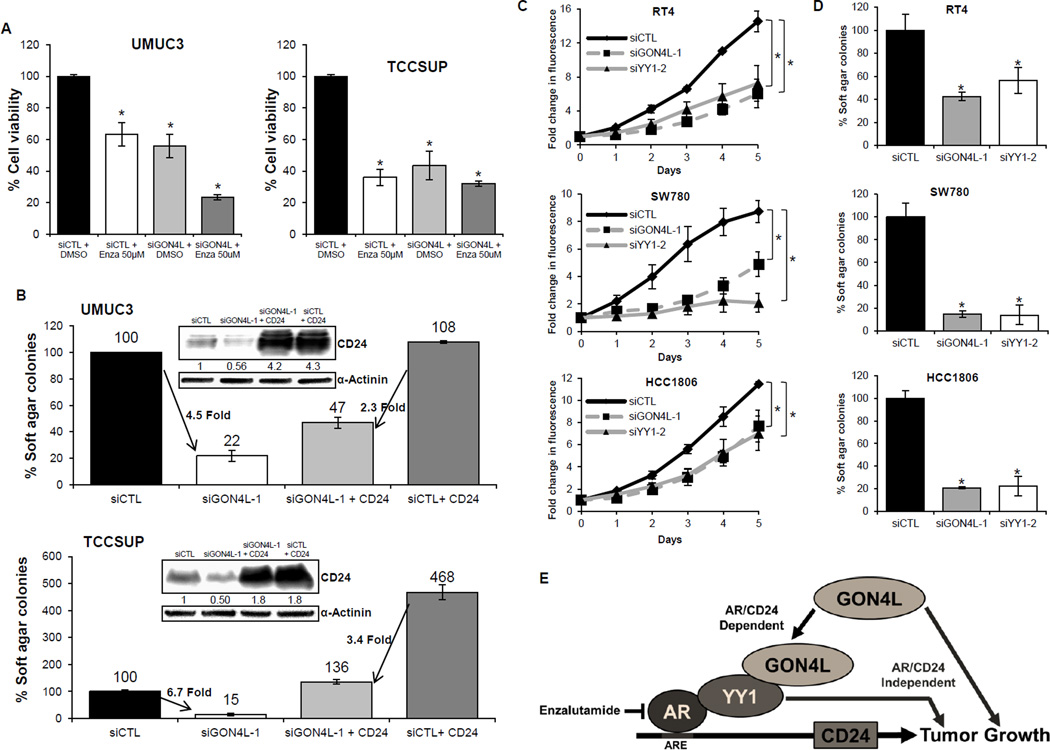
GON4L also has an AR/CD24 independent effect on cancer cell growth
To determine if the ability of GON4L to control cell growth is completely dependent on AR, we compared the treatment of bladder cancer cells with GON4L siRNA to treatment with enzalutamide, an AR inhibitor (40). GON4L siRNA alone reduced cell proliferation to the same extent as enzalutamide treatment alone, in both UMUC3 and TCCSUP cells. However, in UMUC3 cells we observed a further reduction in cell growth when siGON4L and enzalutamide were added simultaneously (Fig. 7A). This suggests that in addition to using AR, GON4L affects cell growth in AR/CD24 independent ways. We took two approaches to test this hypothesis. First, to determine if the growth effects of GON4L are exclusively mediated via its effect on CD24 expression, we overexpressed CD24 in cells that were depleted of GON4L. We found that overexpression of CD24 can partially rescue the anchorage-independent growth suppression induced by GON4L depletion in both UMUC3 and TCCSUP cells (Fig. 7B). These results suggest CD24 is a major but not unique effector of GON4L regulated growth. Second, we knocked down GON4L and YY1 in AR− cell lines (Supplementary Fig. S7) those were positive for CD24 and measured cell proliferation. Interestingly, knockdown of either GON4L or YY1 significantly reduced the anchorage dependent and independent growth of all three cancer cell lines tested (Fig. 7C and D) while having no significant change in CD24 expression (Fig. 5C and D). Based on the compilation of all of our results, we propose a model where GON4L and YY1 are strong mediators of cancer cell growth and do so via both AR/CD24 dependent and independent pathways (Fig. 7E).
Figure 7.
AR and CD24 independent effect of GON4L and YY1 on cell growth. A, cells transfected with siGON4L or siCTL treated with either DMSO as solvent control or 50µM Enzalutamide (Enza) were processed for cell viability by MTT assay after 72h. Graphs show the percent change in optical density at 530nm. B, UMUC3 and TCCSUP cells stably overexpressing CD24 or empty vector were transfected with non-target or GON4L siRNA and processed for soft agar colony formation. Empty vector + siCTL transfected cells were used as control. Representative CD24 western blots are shown in inset. Densitometric analysis was done using ImageJ software, values were normalized with loading control. Values on the arrows indicate fold decrease in number of colonies formed. C and D, graphs showing changes in (C) cell proliferation rate and (D) percent soft agar colonies formed by siCTL, siGON4L and siYY1 transfected AR− cells. Results are shown as mean ± S.D., *p<0.05 by Student’s t-test. E, proposed mechanism of action of GON4L regulated tumor growth.
Discussion
GON4L is a nuclear protein and, as is the case with CD24 (41), plays a role in B-lymphopoiesis (42). GON4L also regulates gene expression in developmental pathways of C. elegans, D. melanogaster, and D. rerio (zebrafish). Absence of GON4L causes cell cycle arrest and increased cellular apoptosis in developing zebrafish embryos (43–45). Despite these roles, no evidence of its association with human cancer growth or its mechanism of action in this setting had been shown until our work. We showed that depletion of GON4L suppresses the growth of human cancer cells both in vitro and in vivo. Analysis of GON4L expression in bladder cancer patients revealed that GON4L is elevated in tumor samples compared to normal bladder urothelium supporting and establishing our findings in human cancer cells.
GON4L has been shown to co-immunoprecipitate with the transcription factor YY1 endogenously in mouse M12 B cells and exogenously in 293T cells (36). Our co-immunoprecipitation studies also confirmed that GON4L interacts with YY1 in human bladder cancer cells. Knockdown of YY1 have similar effects on cell growth and CD24 levels as with GON4L knockdown. YY1 itself was not targeted in our 176 gene shRNA library since we had not detected it in patient mutations (24,46). YY1 can act as an activator or a repressor through directly binding to promoters, or as a cofactor by interacting with other proteins such as p53, MDM2, AR, EZH2, R6, caspases and HDACs (38),(47),(48). This is particularly interesting in bladder cancer since functional loss of p53 has been implicated in the most aggressive forms of the disease (49). YY1 itself does not appear to bind the CD24 promoter as mutation of the predicted YY1-binding sites had no effect in our CD24 promoter reporter assays. YY1 has been shown to physically interact with transcription factor AR both in a cell-free system and in cultured cells. YY1 is required for the optimal transcriptional activity of AR in promoting the transcription of the prostate-specific antigen (PSA) promoter. It has also been reported that YY1–AR interaction and not YY1-DNA interaction is essential to YY1-mediated transcription activity of AR (38). In addition, we have shown previously that AR binds to the ARE on the CD24 promoter and activates its transcription in human bladder cancer cells (5). Here we show that YY1 interacts with AR in bladder cancer cells and acts as a co-activator for AR in enhancing the transcription of CD24 promoter. Furthermore, we discovered that GON4L facilitates the binding of YY1 to AR, thereby enhancing the AR-mediated transactivation of CD24 promoter. YY1 is also known to interact with other cofactors such as EP300 and CREB-binding protein (CREBBP) (48). Both of these proteins have histone acetyltransferase activity (50) and are frequently mutated in bladder cancer (46).
Our experiments with the AR inhibitor drug Enzalutamide and partial rescue of GON4L knockdown effect with CD24 overexpression suggests that GON4L also regulates cell growth via AR and CD24 independent pathways. Further investigation revealed that GON4L and YY1 can both promote cancer cell growth in an AR and CD24 independent manner. The exact mechanisms underlying this are currently unknown but worth future investigation given the dramatic effects of GON4L or YY1 depletion on tumor growth in vivo. The biological function of GON4L is essentially unknown, however, the bioinformatics analysis of mouse Gon4l gene structure suggests its multifunctional role. It has the putative nuclear localization sequence (NLS) at the N-terminus and adjacent to NLS has a region with weak homology to nucleoplasmins. Nucleoplasmins are known to be involved in chromatin remodeling and genomic stability (51). It is reasonable to postulate that GON4L might affect chromatin remodeling allowing access to transcriptional regulatory proteins to control gene expression, cell cycle progression and differentiation. The central domain of mouse GON4L has homology to human YY1AP1 (YY1 associated protein 1) through which it interacts with YY1 and influence YY1-mediated transcriptional regulation. Since, we have found that GON4L enhances the binding of YY1 to AR, it may be possible that GON4L-YY1 interaction can also enhance the activity of other transcription factors as well. The C-terminal region of mouse GON4L has paired amphiphathic helix (PAH) repeat sequence and SANT domain (SW13, ADA2, N-CoR, and TFIIIB). Both PAH and SANT domains mediate protein-protein interactions important for transcriptional regulation (36). In fact, it has been shown that GON4L complexes with SIN3a and HDAC1 in mouse M12 B cells (36). SIN3a acts as both transcriptional activator and co-repressor, and also affects protein stability through protein-protein interactions (52). HDAC1 acts as a transcriptional regulator via histone deacetylation and is also reported to interact with YY1 (48). By interacting with these proteins, GON4L may regulate their activity and thus the expression of growth-promoting genes. It is intriguing to postulate, for example, that tumors with AR and CD24 expression may be treated more effectively when AR inhibitor therapy is combined with therapy aimed at disrupting GON4L-YY1 or other GON4L complexes.
Supplementary Material
Acknowledgments
We thank University of Colorado Denver Animal and flow cytometry facilities staff for their technical assistance and John D. Colgan (University of Iowa) for providing GON4L plasmids and useful information.
Grant Support: This work was supported in part by US National Institutes of Health (NIH) grants CA075115 and CA104106 (DT), CA046934 supporting the Functional Genomics Shared Resource at University of Colorado Comprehensive Cancer Center and Cancer League of Colorado research grant (to NA).
Footnotes
Conflicts of Interest: The authors declared no conflict of interest.
References
- 1.Borah S, Xi L, Zaug AJ, Powell NM, Dancik GM, Cohen SB, et al. Cancer. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science. 2015;347(6225):1006–1010. doi: 10.1126/science.1260200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith SC, Baras AS, Dancik G, Ru Y, Ding KF, Moskaluk CA, et al. A 20-gene model for molecular nodal staging of bladder cancer: development and prospective assessment. Lancet Oncol. 2011;12(2):137–143. doi: 10.1016/S1470-2045(10)70296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JK, Havaleshko DM, Cho H, Weinstein JN, Kaldjian EP, Karpovich J, et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci U S A. 2007;104(32):13086–13091. doi: 10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dancik GM, Ru Y, Owens CR, Theodorescu D. A framework to select clinically relevant cancer cell lines for investigation by establishing their molecular similarity with primary human cancers. Cancer Res. 2011;71(24):7398–7409. doi: 10.1158/0008-5472.CAN-11-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Overdevest JB, Knubel KH, Duex JE, Thomas S, Nitz MD, Harding MA, et al. CD24 expression is important in male urothelial tumorigenesis and metastasis in mice and is androgen regulated. Proc Natl Acad Sci U S A. 2012;109(51):E3588–E3596. doi: 10.1073/pnas.1113960109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JH, Kim SH, Lee ES, Kim YS. CD24 overexpression in cancer development and progression: a meta-analysis. Oncol Rep. 2009;22(5):1149–1156. doi: 10.3892/or_00000548. [DOI] [PubMed] [Google Scholar]
- 7.Kristiansen G, Denkert C, Schluns K, Dahl E, Pilarsky C, Hauptmann S. CD24 is expressed in ovarian cancer and is a new independent prognostic marker of patient survival. Am J Pathol. 2002;161(4):1215–1221. doi: 10.1016/S0002-9440(10)64398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naumov I, Zilberberg A, Shapira S, Avivi D, Kazanov D, Rosin-Arbesfeld R, et al. CD24 knockout prevents colorectal cancer in chemically induced colon carcinogenesis and in APC(Min)/CD24 double knockout transgenic mice. Int J Cancer. 2014;135(5):1048–1059. doi: 10.1002/ijc.28762. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Liu R, Ye P, Wong C, Chen GY, Zhou P, et al. Intracellular CD24 disrupts the ARF-NPM interaction and enables mutational and viral oncogene-mediated p53 inactivation. Nat Commun. 2015;6:5909. doi: 10.1038/ncomms6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujikuni N, Yamamoto H, Tanabe K, Naito Y, Sakamoto N, Tanaka Y, et al. Hypoxia-mediated CD24 expression is correlated with gastric cancer aggressiveness by promoting cell migration and invasion. Cancer Sci. 2014;105(11):1411–1420. doi: 10.1111/cas.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee KM, Ju JH, Jang K, Yang W, Yi JY, Noh DY, et al. CD24 regulates cell proliferation and transforming growth factor beta-induced epithelial to mesenchymal transition through modulation of integrin beta1 stability. Cell Signal. 2012;24(11):2132–2142. doi: 10.1016/j.cellsig.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Zhang YH, Wang ZY, Hao FY, Zhang L. Cluster of differentiation 24 monoclonal antibody induces apoptosis in the osteosarcoma cells. Eur Rev Med Pharmacol Sci. 2014;18(14):2038–2041. [PubMed] [Google Scholar]
- 13.Salnikov AV, Bretz NP, Perne C, Hazin J, Keller S, Fogel M, et al. Antibody targeting of CD24 efficiently retards growth and influences cytokine milieu in experimental carcinomas. Br J Cancer. 2013;108(7):1449–1459. doi: 10.1038/bjc.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sagiv E, Starr A, Rozovski U, Khosravi R, Altevogt P, Wang T, et al. Targeting CD24 for treatment of colorectal and pancreatic cancer by monoclonal antibodies or small interfering RNA. Cancer Res. 2008;68(8):2803–2812. doi: 10.1158/0008-5472.CAN-07-6463. [DOI] [PubMed] [Google Scholar]
- 15.Overdevest JB, Thomas S, Kristiansen G, Hansel DE, Smith SC, Theodorescu D. CD24 offers a therapeutic target for control of bladder cancer metastasis based on a requirement for lung colonization. Cancer Res. 2011;71(11):3802–3811. doi: 10.1158/0008-5472.CAN-11-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren J, Agata N, Chen D, Li Y, Yu WH, Huang L, et al. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anticancer agents. Cancer Cell. 2004;5(2):163–175. doi: 10.1016/s1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomlins SA, Rhodes DR, Yu J, Varambally S, Mehra R, Perner S, et al. The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell. 2008;13(6):519–528. doi: 10.1016/j.ccr.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balasubramani M, Nakao C, Uechi GT, Cardamone J, Kamath K, Leslie KL, et al. Characterization and detection of cellular and proteomic alterations in stable stathmin-overexpressing, taxol-resistant BT549 breast cancer cells using offgel IEF/PAGE difference gel electrophoresis. Mutat Res. 2011;722(2):154–164. doi: 10.1016/j.mrgentox.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guin S, Pollard C, Ru Y, Ritterson Lew C, Duex JE, Dancik G, et al. Role in tumor growth of a glycogen debranching enzyme lost in glycogen storage disease. J Natl Cancer Inst. 2014;106(5) doi: 10.1093/jnci/dju062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34(Database issue):D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dancik GM. An online tool for evaluating diagnostic and prognostic gene expression biomarkers in bladder cancer. BMC Urol. 2015;15(1):59. doi: 10.1186/s12894-015-0056-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balbas-Martinez C, Sagrera A, Carrillo-de-Santa-Pau E, Earl J, Marquez M, Vazquez M, et al. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat Genet. 2013;45(12):1464–1469. doi: 10.1038/ng.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo G, Sun X, Chen C, Wu S, Huang P, Li Z, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45(12):1459–1463. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solomon DA, Kim JS, Bondaruk J, Shariat SF, Wang ZF, Elkahloun AG, et al. Frequent truncating mutations of STAG2 in bladder cancer. Nat Genet. 2013;45(12):1428–1430. doi: 10.1038/ng.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dyrskjot L, Kruhoffer M, Thykjaer T, Marcussen N, Jensen JL, Moller K, et al. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64(11):4040–4048. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- 27.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang YD, Shan W, Zeng L, Wu Y. Screening of differentially expressed genes related to bladder cancer and functional analysis with DNA microarray. Asian Pac J Cancer Prev. 2013;14(8):4553–4557. doi: 10.7314/apjcp.2013.14.8.4553. [DOI] [PubMed] [Google Scholar]
- 29.Kim WJ, Kim EJ, Kim SK, Kim YJ, Ha YS, Jeong P, et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Mol Cancer. 2010;9:3. doi: 10.1186/1476-4598-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24(5):778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 31.Miller JA, Cai C, Langfelder P, Geschwind DH, Kurian SM, Salomon DR, et al. Strategies for aggregating gene expression data: the collapse Rows R function. BMC Bioinformatics. 2011;12:322. doi: 10.1186/1471-2105-12-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bloethner S, Hemminki K, Thirumaran RK, Chen B, Mueller-Berghaus J, Ugurel S, et al. Differences in global gene expression in melanoma cell lines with and without homozygous deletion of the CDKN2A locus genes. Melanoma Res. 2006;16(4):297–307. doi: 10.1097/01.cmr.0000222597.50309.05. [DOI] [PubMed] [Google Scholar]
- 35.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu P, Hankel IL, Hostager BS, Swartzendruber JA, Friedman AD, Brenton JL, et al. The developmental regulator protein Gon4l associates with protein YY1, co-repressor Sin3a, and histone deacetylase 1 and mediates transcriptional repression. J Biol Chem. 2011;286(20):18311–18319. doi: 10.1074/jbc.M110.133603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iyer G, Al-Ahmadie H, Schultz N, Hanrahan AJ, Ostrovnaya I, Balar AV, et al. Prevalence and co-occurrence of actionable genomic alterations in high-grade bladder cancer. J Clin Oncol. 2013;31(25):3133–3140. doi: 10.1200/JCO.2012.46.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng Z, Wan M, Cao P, Rao A, Cramer SD, Sui G. Yin Yang 1 regulates the transcriptional activity of androgen receptor. Oncogene. 2009;28(42):3746–3757. doi: 10.1038/onc.2009.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang C, Lee SO, Yeh S, Chang TM. Androgen receptor (AR) differential roles in hormone-related tumors including prostate, bladder, kidney, lung, breast and liver. Oncogene. 2014;33(25):3225–3234. doi: 10.1038/onc.2013.274. [DOI] [PubMed] [Google Scholar]
- 40.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324(5928):787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu L, Chappel MS, Humphries RK, Osmond DG. Regulation of cell survival during B lymphopoiesis: increased pre-B cell apoptosis in CD24-transgenic mouse bone marrow. Eur J Immunol. 2000;30(9):2686–2691. doi: 10.1002/1521-4141(200009)30:9<2686::AID-IMMU2686>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 42.Lu P, Hankel IL, Knisz J, Marquardt A, Chiang MY, Grosse J, et al. The Justy mutation identifies Gon4-like as a gene that is essential for B lymphopoiesis. J Exp Med. 2010;207(7):1359–1367. doi: 10.1084/jem.20100147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedman L, Santa Anna-Arriola S, Hodgkin J, Kimble J. gon-4, a cell lineage regulator required for gonadogenesis in Caenorhabditis elegans. Dev Biol. 2000;228(2):350–362. doi: 10.1006/dbio.2000.9944. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Du L, Osato M, Teo EH, Qian F, Jin H, et al. The zebrafish udu gene encodes a novel nuclear factor and is essential for primitive erythroid cell development. Blood. 2007;110(1):99–106. doi: 10.1182/blood-2006-11-059204. [DOI] [PubMed] [Google Scholar]
- 45.Lim CH, Chong SW, Jiang YJ. Udu deficiency activates DNA damage checkpoint. Mol Biol Cell. 2009;20(19):4183–4193. doi: 10.1091/mbc.E09-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43(9):875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng Z, Cao P, Wan MM, Sui G. Yin Yang 1: a multifaceted protein beyond a transcription factor. Transcription. 2010;1(2):81–84. doi: 10.4161/trns.1.2.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25(8):1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 49.Dancik GM, Theodorescu D. Pharmacogenomics in bladder cancer. Urol Oncol. 2014;32(1):16–22. doi: 10.1016/j.urolonc.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87(5):953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 51.Gillespie PJ, Blow JJ. Nucleoplasmin-mediated chromatin remodelling is required for Xenopus sperm nuclei to become licensed for DNA replication. Nucleic Acids Res. 2000;28(2):472–480. doi: 10.1093/nar/28.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kadamb R, Mittal S, Bansal N, Batra H, Saluja D. Sin3: insight into its transcription regulatory functions. Eur J Cell Biol. 2013;92(8–9):237–246. doi: 10.1016/j.ejcb.2013.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.