Abstract
Purpose
CLL-associated gene mutations that influence CLL cell fitness and chemotherapy resistance should increase in clonal representation when measured before therapy and at relapse.
Experimental Design
To uncover mutations associated with CLL relapse, we have performed whole exome sequencing (WES) in a discovery cohort of sixty-one relapsed CLL patients identifying eighty-six recurrently mutated genes. The variant allele fractions (VAFs) of nineteen genes with mutations in ≥ 3/61 cases were measured in fifty-three paired pre- and post-treatment CLL samples sorted to purity using panel-based deep re-sequencing or by droplet digital PCR (ddPCR).
Results
We identify mutations in TP53 as the dominant subclonal gene driver of relapsed CLL often demonstrating substantial increases in VAFs. Subclonal mutations in SAMHD1 also recurrently demonstrated increased VAFs at relapse. Mutations in ATP10A, FAT3, FAM50A and MGA, although infrequent, demonstrated enrichment in ≥2 cases each. In contrast, mutations in NOTCH1, SF3B1, POT1, FBXW7, MYD88, NXF1, XPO1, ZMYM3 or CHD2 were predominantly already clonal prior to therapy indicative of a pre-treatment pathogenetic driver role in CLL. Quantitative analyses of clonal dynamics uncovers rising, stable and falling clones and subclones without clear evidence that gene mutations other than in TP53 and possibly SAMHD1 are frequently selected for at CLL relapse.
Conclusion
Data in aggregate support a provisional categorization of CLL-associated recurrently mutated genes into three classes i) often subclonal pre-therapy and strongly enriched after therapy, or, ii) mostly clonal pre-therapy or without further enrichments at relapse, or, iii) subclonal before and after therapy and enriching only in sporadic cases.
Keywords: relapsed CLL, whole exome sequencing, subclonal gene mutations
INTRODUCTION
Chronic lymphocytic leukemia is a heterogeneous disease caused by marked differences in biology and manifested in varied clinical presentations (1-4). Substantial efforts have been directed at defining the molecular and cellular underpinnings that cause variations in CLL. These efforts have identified molecular traits that associate with initial disease progression, the response durations to up-front therapies and ultimately, differences in overall survival. Most of these efforts have been directed at the characterization of CLL at diagnosis(5-7).
In contrast, comparatively less information is available characterizing CLL that has relapsed from frontline chemo-immunotherapies. What are the determinants or drivers of CLL cell accumulation after therapy? What is the frequency of CLL clonal evolution under therapy and what are the common drivers of therapy resistance? What role do gene mutations as opposed to other CLL traits serve in relapsed CLL? A refined understanding will facilitate development of novel research directions, better diagnostics, risk-adapted counseling and therapy approaches.
With the goal of identifying genomic drivers of CLL progression, investigators have compared the frequencies of selected molecular characteristics in relapsed or refractory CLL patient cohorts with frequencies in unrelated CLL cohorts analyzed at diagnosis. Such efforts, although prone to biases, have implicated TP53 mutations/del17p in acquired therapy resistance and have generated hypothesis-generating findings about the involvement of specific factors in CLL disease aggressiveness(8-10).
The analysis of paired longitudinal samples procured before and after therapy reduces biases in the discovery of factors driving CLL relapse and clonal evolution. Such paired longitudinal analysis has uncovered the acquisition of selected genomic aberrations as detected through clinical FISH testing in CLL patients over time as well as acquisition of novel acquired copy number aberrations (aCNA) and loss-of-heterozygosity (LOH) in CLL when assayed longitudinally by karyotyping or on high-resolution SNP array platforms(11-18).
With the goals of identifying gene mutations that drive relapsed CLL and to further clarify the biological roles of gene mutations in CLL in general(19-22), we have performed a longitudinal genomic analysis of paired CLL samples procured before and after chemo immunotherapies. Using an experimental approach that combines WES in a discovery cohort of 61 relapsed CLL, followed by deep panel-based re-sequencing in paired pre- and post-treatment samples complemented with droplet digital PCR-measurements of mutated genes, we substantially qualify the role of gene mutations in the pathogenesis of CLL and relapsed CLL.
Data in aggregate support a provisional categorization of CLL-associated recurrently mutated genes into three classes i) often subclonal pre-therapy and strongly enriched after therapy, or, ii) mostly clonal or major subclonal pre-therapy and without further enrichments at relapse, or, iii) subclonal before and after therapy and enriched only in sporadic cases. Combined, our results suggest mutations in TP53 and possibly SAMHD1 as relapse gene drivers in a minority subset (20%) of CLL relapsing after chemo-immunotherapy. The data based on complementary genomic analyses support the refined concept that the majority of CLL undergo clonal evolution after therapy. Importantly, we identify an early pre-treatment pathogenetic role for many known recurrently mutated genes in CLL and provide limits to the hypothesis that sub clonal gene mutations other than in TP53 frequently drive CLL relapse.
METHODS
Patients
Between January 2005 and June 2011, 300 patients evaluated at the University of Michigan Comprehensive Cancer Center were enrolled onto this study and pre-treatment samples analyzed here were procured at enrollment. As specified in the protocol, patients were resampled, where applicable, at multiple time points following initial enrollment. The trial was approved by the University of Michigan Institutional Review Board (IRBMED #2004-0962) and written informed consent was obtained from all patients prior to enrollment. DNA from 61 relapsed CLL patients that was subjected to WES constituted the discovery cohort. Of these 61 patients, 53 patients had available paired samples procured before therapy and at subsequent relapse from prior chemotherapy (chemo immunotherapy was administered to 93% of these patients) and 8 patients had longitudinal samples analyzed without receiving intercurrent therapy. Of the 53 patients with available paired samples, 41 were untreated at first sampling, while 12 had relapsed at trial enrollment and had undergone additional round(s) of chemotherapy followed by a subsequent relapse (see Tables 1 and 2; Supplementary Table 1).
Table 1.
Characteristics of fifty-three relapsed CLL patients analyzed by WES, deep custom panel-based resequencing and ddPCR.
| Patient Characteristics | Intercurrent therapy no. (%) |
|---|---|
| Sample size N= 53 patients | 53 (100%) |
| Age at enrollment, years | |
| Median | 64 |
| Range | 39 - 84 |
| Gender | |
| Female | 18 (34%) |
| Male | 35 (66%) |
| Rai stage at enrollment | |
| Low, 0 | 12 (23%) |
| Intermediate, I-II | 36 (67%) |
| High, III-IV | 5 (10%) |
| NOTCH1 exon 34 mutations at enrollment (Sanger) | |
| Wild-type | 43 (81%) |
| Mutated | 10 (19%) |
| SF3B1 exons 13-17 mutations at enrollment (Sanger) | |
| Wild-type | 45 (85%) |
| Mutated | 8 (15%) |
| P53 exons 2-10 mutations at enrollment (Sanger) | |
| Wild-type | 51 (96%) |
| Mutated | 2 (4%) |
| Prioritized interphase FISH-25* | |
| 17p deletion | 2 (4%) |
| 11q deletion | 6 (11%) |
| Trisomy 12 | 11 (21%) |
| Normal FISH | 13 (25%) |
| 13q deletion (sole abnormality) | 21 (40%) |
| IgVH mutational status | |
| Unmutated (≥ 98% homology to germline) | 31 (58%) |
| Mutated (< 98% homology to germline) | 18 (34%) |
| Not evaluable | 4 (8%) |
| ZAP-70 expression | |
| Positive (> 20%) | 33 (62%) |
| Negative (≤ 20%) | 16 (30%) |
| Not available | 4 (8%) |
| Treatment status at enrollment | |
| Not treated | 41 (77%) |
| Treated | 12 (23%) |
| Number of prior therapies | |
| 0 | 41 (77%) |
| 1 | 7 (13%) |
| 2 | 3 (6%) |
| >2 | 2 (4%) |
FISH findings in ≥25% of nuclei. Order of prioritization: 17p > 11q > trisomy 12 > 13q > Ig translocations > normal FISH. Please see Supplementary Table 1 for details.
Table 2.
Listing of therapies received by CLL patients between sample procurements, response types and clonal evolution events.
| CLL ID | Treatment status at trial enrollment | Intercurrent therapy | Response type to intercurrent therapy | Clonal evolution (CE) or stability? |
|---|---|---|---|---|
| CLL002 | T | R → FCR | PR | CE (+aCNA; +M) |
| CLL004 | T | FR | PR | stable |
| CLL007 | UT | FR | CR | CE (+aCNA, −aCNA; +M) |
| CLL008 | UT | FR | CR | CE (+aCNA; +M) |
| CLL010 | UT | FR | PR | CE (+aCNA; +M) |
| CLL011 | UT | FR | CR | CE (+aCNA; +M) |
| CLL013 | T | R-CVP | PR | stable |
| CLL017 | T | FR → FCR | SD/PR | CE (+M) |
| CLL021 | UT | R-CVP → BEXXAR | CR/CR | CE (+M) |
| CLL024 | UT | FR | CR | CE (+aCNA) |
| CLL027 | UT | FR | PR | CE (+M) |
| CLL029 | UT | FR | CR | CE (+M) |
| CLL035 | UT | FR | PR | stable |
| CLL036 | UT | FR | PR | CE (+cnLOH) |
| CLL058 | UT | FR | CR | CE (+M) |
| CLL060 | T | PCR | PR | stable |
| CLL064 | T | PCR | PR | stable |
| CLL066 | UT | R → PCR | PR | stable |
| CLL072 | UT | FR | PD | CE (+M) |
| CLL074 | UT | FR | CR | CE (+aCNA; +M) |
| CLL078 | UT | FR | CR | stable |
| CLL085 | T | FCR | CR | CE (+M) |
| CLL090 | T | BR | CR | CE (+aCNA; +M) |
| CLL094 | T | A | PR | stable |
| CLL104 | UT | F → A | PR/CR | CE (+aCNA, −aCNA; +M) |
| CLL107 | UT | PCR | CR | CE (+M) |
| CLL109 | UT | FR | PR | stable |
| CLL112 | UT | FR | CR | stable |
| CLL116 | UT | FR | CR | CE (+cnLOH; +M) |
| CLL117 | UT | FCR → BR | CR/CR | CE (+aCNA, +M, +cnLOH, −M) |
| CLL120 | UT | FR | PR | stable |
| CLL121 | UT | FCR | PR | CE (+aCNA; +M) |
| CLL125 | UT | F → B | SD/SD | CE (+M) |
| CLL135 | UT | FR | PR | stable |
| CLL155 | UT | FCR | CR | stable |
| CLL170 | T | PCR → flavopiridol → RICE | PD/PD/PD | stable |
| CLL176 | UT | FCR | CR | stable |
| CLL185 | UT | FR | CR | CE (+aCNA, −aCNA; +M) |
| CLL186 | UT | FR, BR | PR,PR | CE (+M) |
| CLL189 | UT | FR | PR | CE (+M) |
| CLL195 | UT | FCR | PR | CE (+M) |
| CLL205 | UT | FCR | PR | CE (+M) |
| CLL209 | T | PCR | PR | CE (+aCNA; +M) |
| CLL211 | UT | FCR | CR | CE (−M) |
| CLL212 | T | BR | PR | stable |
| CLL218 | UT | FR | PR | CE (+M) |
| CLL219 | UT | R-CVP → B | CR/CR | CE (+M) |
| CLL230 | UT | BR → BR | PR/PR | stable |
| CLL242 | UT | F → FC → BR | PD/PR | stable |
| CLL270 | UT | R-CHOP | PD | CE (+M) |
| CLL271 | UT | B | PR | stable |
| CLL279 | UT | FR | CR | stable |
| CLL296 | UT | BR | CR | CE (+M) |
| CLL018 | T | NONE | stable | |
| CLL045 | T | NONE | CE (+M) | |
| CLL077 | T | NONE | stable | |
| CLL123 | T | NONE | stable | |
| CLL164 | T | NONE | stable | |
| CLL165 | T | NONE | stable | |
| CLL188 | T | NONE | stable | |
| CLL220 | T | NONE | stable |
Please see Supplementary Table 1 for details.
F: fludarabine; C: cyclophosphamide; R: rituximab; A: alemtuzumab; O: ofatumumab; V: vincristine; P: prednisone; P: pentostatin; M: methylprednisolone; I: ifosphamide; E: etoposide. CR: complete remission; PR: partial remission; SD: stable disease; PD: progressive disease. CE: clonal evolution; aCNA: acquired genomic copy number aberration; (+M): acquisition of a gene mutations; cnLOH: copy-neutral LOH. UT: untreated at sample procurement; T: previously treated prior to sample procurement.
CLL treatment was defined as cytotoxic chemotherapy (usually fludarabine, pentostatin, bendamustine or cyclophosphamide) with or without monoclonal antibody therapy for CLL. Clinical information, including Rai stage and all treatments given, was collected on all patients. Patient samples were characterized for selected CLL-associated chromosomal aberrations on the day of trial enrollment as a routine clinical test at the Mayo Clinic (Rochester, MN) using FISH (CLL-FISH). Measurements of CLL-associated molecular characteristics were as described(6).
Cell Isolation
Flow cytometry sorting of CLL specimens
Cryopreserved PBMCs (frozen after Ficoll-gradient purification) from CLL blood specimens were prepared for FACS sorting into CD19+ and CD3+ cells as previously described(6).
Preparation of Sample DNA
DNA used for SNP 6.0 profiling was extracted from FACS-sorted CD19+ and CD3+ cells as described(6).
Solution-based exome capture and HiSeq2000-based massively parallel sequencing
Solution-based exome capture and HiSeq2000-based massively parallel sequencing was performed as described(23).
Bioinformatic pipeline analysis of WES data
The exome sequencing data was analyzed by the variant calling pipeline developed by the University of Michigan Bioinformatics Core. For each of the samples, paired-end reads were aligned to the hg19 reference genome using BWA v0.7.8(24), followed by removal of sequence duplicates using PicardTools v1.79 (http://picard.sourceforge.net), local realignment around INDELs and base quality score recalibration using GATK v3.2-2(25). Read coverage on exome capture target regions was calculated using BEDTools v2.20.1(26). Normal-Tumor paired alignment files were submitted to MuTect v1.1.4(27), Strelka v1.0.14(28) and Varscan v2.3.7 (with its false-positive filter)(29) for the detection of somatic and germline SNPs and INDELs.
Candidate variant calls across all samples and patients were merged using Jacquard(30) into a single VCF file that included all variant loci whose filter field passed in MuTect or Strelka or VarScan (VarScan calls were limited to somatic variants confirmed in false-positive filter). Variants were annotated using SnpEff v4.0/hg19(31), dbNSFP v2.4(32), dbSNP v138 (Database of Single Nucleotide Polymorphisms, National Center for Biotechnology Information, National Library of Medicine), and 1000 Genomes v3(33). For variants associated with multiple effects or multiple transcripts, a single “top effect” annotation was nominated based on annotation confidence, predicted impact, gene region, and transcript length. Common variants (at or above 5% overall population allele frequency as reported by 1000 Genomes) were excluded.
Exon resequencing of nominated variants in CLL samples
Primers to amplify and sequence all variants of interest were designed using the primer 3 program (http://primer3.ut.ee/) and sequence information generated using direct sequencing as described(23). Sequence variants were detected using Mutation Surveyor and visible inspection of sequence traces. Mutations were confirmed to be somatically acquired using unamplified CLL CD19+ cell-derived DNA and paired CD3+ cell-derived DNA from sorted cells as templates.
Deep coverage massively parallel re-sequencing of 17 genes and bioinformatics analysis
A customized multiplexed primer panel (Qiagen Gene Read Panel # CNGHS-00586X-849) was used to amplify all coding exons of the genes: TP53, NOTCH1, SF3B1, XPO1, BIRC3, MYD88, NXF1, POT1, CACNA1E, CHD2, EGR2, FAM50A, FAT3, FBXW7, MGA, SAMHD1 and ZMYM. PCR products were pooled and sequencing libraries prepared using barcoded adapters. Sequencing was done on a HiSeq2000 sequencer. Bioinformatics nomination of sequence variants was performed using Broad GATK HaplotypeCaller v3.3.0 and Varscan. Discordant variant caller results were qualified through ddPCR measurements.
For details of the bioinformatics analyses please see Supplementary Methods.
Measurements of VAFs using droplet digital PCR (ddPCR)
Allele-specific fluorescent oligonucleotide probes using minor groove-binding (Life Technologies, Grand Island, NY) or locked nucleic acid (Integrated DNA Technologies [IDT], Coralville, IA) chemistries were developed for each mutation using Primer Express (Life Technologies) and Oligoanalyzer (IDT), respectively. Wildtype and mutant allele-specific probes (5 uM) were combined with allele-independent forward and reverse PCR primers (20 uM) to generate a probe mix. Each probe mix was optimized for allele discrimination using quantitative PCR to derive an optimal annealing temperature (TA) as follows: 2X Taqman Genotyping Master Mix (Life Technologies) was combined with 40X probe mix and either 30 ng of mutant gDNA derived from a patient sample harboring the mutation of interest or homozygous wildtype genomic DNA (gDNA) derived from the Ly18 cell line and then amplified with a Bio-Rad CFX96 Real-Time System using a TA gradient for 45 cycles and analyzed with Bio-Rad CFX Manager. The temperature at which both wildtype and mutant probes were amplified in the presence of DNA with a heterozygous mutation but at which the mutant probe was not amplified in the presence of wildtype-only DNA was selected as the optimal TA to carry forward for ddPCR.
ddPCR was then performed in duplicate for the paired diagnosis and relapsed samples of each case with a mutation of interest as well as a wildtype-only control derived from the Ly18 cell line as follows: a master mix containing 2x Taqman Genotyping Master Mix, 40X probe mix, 25X droplet stabilizer (RainDance Techonologies) and 50X restriction endonuclease (HaeIII or MseI, New England Biolabs) was added to separate wells containing 50 ng of gDNA for a total reaction volume of 25 uL and then incubated at room temperature for 15 mins. Samples were then pipetted into a RainDrop Source chip (RainDance Technologies) and 3.5-4.5 million droplets were generated with the RainDrop Source machine. The resulting emulsion was amplified for 45 cycles at the previously derived optimal TA with the following settings: 95o for 10 min, then 45 cycles of 95° for 15 s and TA for 60 s, then 98° for 10 min. End-point fluorescence was then measured with a RainDrop Sense machine and analyzed with RainDrop Analyst software. The wildtype and mutant signals of each relapsed sample were used to draw positive gates to then analyze VAF of the diagnosis samples and wildtype-only controls. To call the presence of mutant signal, the mutant gate must have contained at least 5 droplets as well as a ten-fold greater number of mutant droplets than the wildtype control.
RESULTS
Patient Characteristics
To identify gene mutations that may have contributed to CLL that has relapsed from prior chemo immunotherapy and to further qualify the roles served by recurrent gene mutations in CLL pathogenesis and disease evolution, we subjected DNA isolated from FACS-sorted CD19+ cells and paired CD3+ cells from 61 relapsed CLL patients (the discovery cohort) to whole exome sequencing (WES). Characteristics of these 61 CLL patients are summarized in Supplementary Table 1. Of these 61 patients, 53 patients had available paired samples procured before therapy and at subsequent relapse from prior chemotherapy (chemoimmunotherapy was administered to 93% of these patients; characteristics of these 53 patients and treatment given are summarized in Tables 1 and 2; Supplementary Table 1) and 8 patients had longitudinal samples analyzed without receiving intercurrent therapy.
Massively parallel sequencing of the coding genome of relapsed CLL samples
To further our understanding of the genetic basis of relapsed CLL, we used solution exon capture of sheared and processed genomic DNA isolated from highly purified flow-sorted CD19+ B-cells and paired CD3+ T-cells isolated from 61 cases of relapsed CLL followed by paired-end massively parallel sequencing. The very high purity of CLL B-cells and paired CD3+ T-cells afforded by cell sorting improved sensitivity of mutation detection and largely eliminated the distorting effect of cell impurity on VAF estimates. The WES data were characterized by a range of de-duplicated mapped reads per DNA sample of 47,521,667 to 98,812,508 (mean of 71,967,091) and a mean depth of coverage of 72 (range of 52 to 102).
The landscape of recurrently mutated genes in relapsed CLL
We identified and subsequently confirmed through Sanger sequencing in paired CD19+ and CD3+ cell-derived DNA 86 genes that were recurrently mutated in the discovery cohort of 61 relapsed CLL. The confirmed mutations were also analyzed through Sanger sequencing in paired pre-treatment DNA. Details of these results are summarized in Supplementary Table 2. Of the 86 genes, 19 were mutated in ≥ 3/61 (~≥5%) of cases in rCLL. In descending order of frequency of CLL cases involved these are: NOTCH1 (13/61), xTP53 (8/61), SF3B1 (8/61), POT1 (8/61), XPO1 (7/61), SAMHD1 (6/61), CHD2 (6/61), FAM50A (4/61), MYD88 (4/61), NXF1 (4/61), ZMYM3 (4/61), APT10A (3/61), ATRX (3/61), CACNA1E (3/61), EGR2 (3/61), FAT3 (3/61), IRF4 (3/61), MGA (3/61) and FBXW7 (3/61). Some of these genes have not previously been recognized as recurrently mutated in CLL.
To complement the WES data, we re-sequenced RPS15 and NFKBIE exon 1 spanning the previously identified c.759_762delTTAC in 294 consecutively enrolled, unselected CLL patients: We detected 7/294 (2.4%) RPS15 mutations and 3/294 (1%) NFKBIE mutations(34-36). We had previously reported on sporadic (<2%) BRAF mutations in CLL(37).
In the 61 relapsed CLL subjected to WES we detected 2/61 (3%) RPS15 mutations, both of which were present before and after therapy. We detected 1/61 BRAF mutation (we sequenced the hotspot exons 11 - 14), which was acquired after therapy. We detected no (0/61) NFKBIE mutations. Finally, review of ATM mutations as detected by WES and confirmed by Sanger sequencing uncovered 2/61 mutations, one acquired at relapse and one present before and after therapy.
Measurement of variant allele fractions in paired pre- and post-treatment CLL samples of 19 recurrently mutated genes
Aided by cell sorting to achieve maximal CLL cell and DNA purity, we proceeded with custom primer panel-based deep re-sequencing of all coding exons of NOTCH1, TP53, SF3B1, POT1, XPO1, SAMHD1, CHD2, FAM50A, MYD88, NXF1, ZMYM3, BIRC3, CACNA1E, EGR2, FAT3, MGA and FBXW7 in 53 paired DNA samples isolated from CLL samples procured before and at relapse from chemo immunotherapy. For two genes (APT10A and ATRX), we resorted to ddPCR-based VAF estimations. For the analysis of deep panel re-sequencing data and VAF estimations we employed two separate bioinformatics approaches in parallel that overall demonstrated excellent concordance. For selected gene mutations and for instances of discordant bioinformatics results, these estimates were further qualified using ddPCR. Finally, a few selected mutations were manually identified using review of BAM files and the fraction of mutated reads was used as a VAF estimate. A summary of VAFs in paired pre- and post-treatment samples for the gene mutations analyzed is outlined in Supplementary Table 3.
We proceeded with SNP 6.0 array profiling of the CLL cases that were subjected to WES analysis, extending previously published measurements(38). We find that almost all gene mutations in CLL (other than in TP53 or ATM) are not associated with either copy loss or gain of the wild type alleles, providing additional evidence that these lesion types independently influence CLL pathogenesis. VAF estimates for gene mutations as presented here were therefore not in need of correction for aCNAs and can be used to directly estimate clone sizes.
The recurrent emergence of clonal TP53 and SAMHD1 mutations at relapse due to enrichment of pre-existing subclones
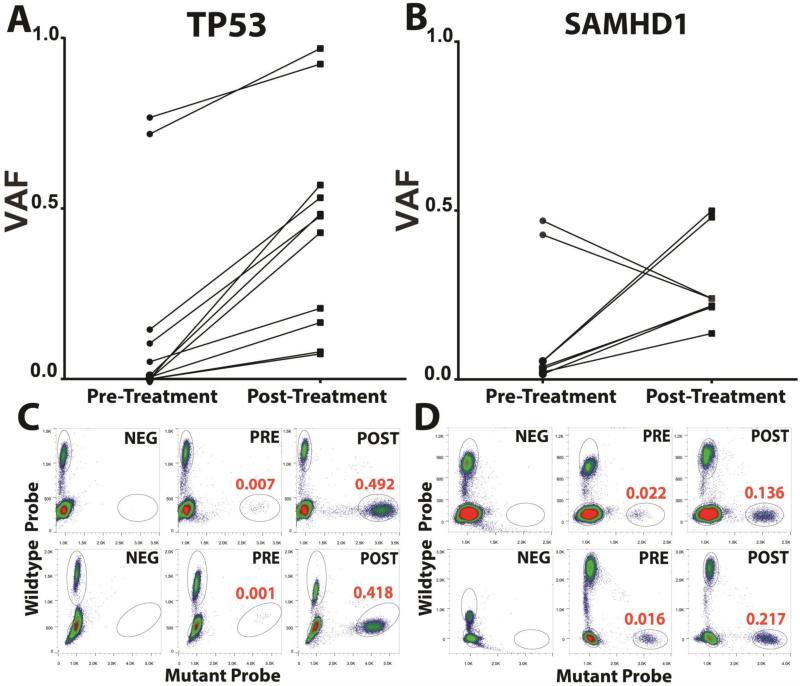
A comparison of VAFs pre- and post-therapy and the identification of VAF increases post-therapy can identify candidate genes involved in chemotherapy resistance resulting in preferential clonal outgrowth at relapse. In this large longitudinal CLL genomics study, we identified only two genes that frequently enriched at relapse: TP53 and SAMHD1 (Figure 1)(39, 40). Overall, 13% (7/53) of rCLL demonstrated enrichment of TP53 mutations from preexisting subclones at relapse and no case demonstrated a decline (please note that 2 CLL cases carried more than one TP53 mutation: CLL90 acquired a del17p at relapse and carried two TP53 mutations in two distinct clones, while CLL117 acquired a del17p at relapse and carried three TP53 mutations in 2 or possibly 3 distinct clones; see Supplementary Table 3). Mutations in TP53 detected at relapse were all present at diagnosis allowing for the novel conclusion that CLL therapy did not induce such mutations directly. Most such pre-treatment TP53 mutations existed as subclones at times characterized by very minor clonal representation (VAF range 0.0002 – 0.1448) as measured through ddPCR. In two CLL cases, TP53 mutations that were already clonal did enrich further at relapse, indicating conversion to full homozygosity. One CLL case that was assayed without receiving intercurrent therapy maintained a subclonal TP53 mutation status and showed no enrichment (Supplementary Figure 1).
Figure 1. Frequent enrichment of mutations in TP53 and SAMHD1 after therapy in CLL.
Displayed are variant allele fractions (VAFs) for A: TP53 (N=7 CLL carrying a combined total of 11 mutations, CLL90 carried 2 mutations and CLL117 carried 3 mutations) and, B: SAMHD1 (N=4 CLL carrying a combined total of 8 mutations; CLL 17 carried 4 mutations and CLL 218 carried 2 mutations; the two declining SAMHD1 VAFs are both from CLL 17) in paired CLL samples procured before and after therapy. VAFs for TP53 were not corrected for presence of del17p. C: Representative droplet digital PCR results for NEG: unmutated control DNA, PRE: CLL DNA procured pre-therapy and POST: CLL DNA procured post-therapy. Red numbers indicate the mean VAFs based on duplicate measurements. Results for two representative CLL cases are shown each for TP53 and SAMHD1.
The second mutated gene that frequently enriched after therapy was SAMHD1(40). Four CLL cases demonstrated substantial enrichment from pre-existing subclones of SAMHD1 mutations at relapse. Two additional cases that were analyzed without having received intercurrent therapy but were already in relapse from chemo immunotherapy at first sampling were already clonal and did not enrich further (Supplementary Figure 1). Finally, one case (CLL17) that contained 4 distinct SAMHD1 mutations demonstrated enrichment for 2 such mutations and depletion of two other ones, indicative of two co-existing subclones carrying such mutations.
To provide additional context to these findings and to identify potential interactions with other genomic events, we reviewed the six CLL cases with SAMHD1 mutations in greater detail: CLL17 carried 4 SAMHD1 mutations likely in two subclones and harbored no aCNAs in either disease phase. The rising subclone was also identified by mutations in ATP10A and CACNA1E (see below); CLL 77 (no intercurrent therapy) carried 1 SAMHD1 mutation, displaying a VAF of ~ 0.5 in both phases and harbored no aCNA in either phase; CLL 188 (no intercurrent therapy) carried 1 SAMHD1 mutation, displaying a VAF of ~0.4 in both phases and harbored no aCNA in the diagnosis sample; CLL 205 had 1 SAMHD1 mutation at increasing VAFs of 0.02 and 0.14 and carried a short del13q-I. Interestingly, this case also demonstrated enrichment in an ATP10A mutation. CLL 218 carried 2 SAMHD1 mutations each displaying strong VAF gains (both ~0.05 to 0.5) indicative of one clone and lacked aCNAs in either phase. CLL218 at relapse also lost a clone containing a SF3B1 mutation. Finally, CLL 296 had 1 SAMHD1 mutation with increasing VAFs (0.02 – 0.22) and carried a del11q pre-treatment but no post treatment SNP array data were available. CLL296 also carried a separate rising dominant clone marked by a NOTCH1 mutation.
These data in aggregate identify SAMHD1 mutations as a candidate gene conferring some degree of in vivo resistance to standard CLL therapies.
Of additional interest is the fact that both TP53 and SAMHD1 mutations were still often present in subclones (VAFs <0.5) post therapy, suggesting that a true bottleneck does not exists in the evolution of CLL but instead multiple CLL clones survived and emerged in parallel post-therapy together constituting the clinically apparent relapsed disease.
Gene drivers of early CLL pathogenesis identified through high VAFs pre-therapy
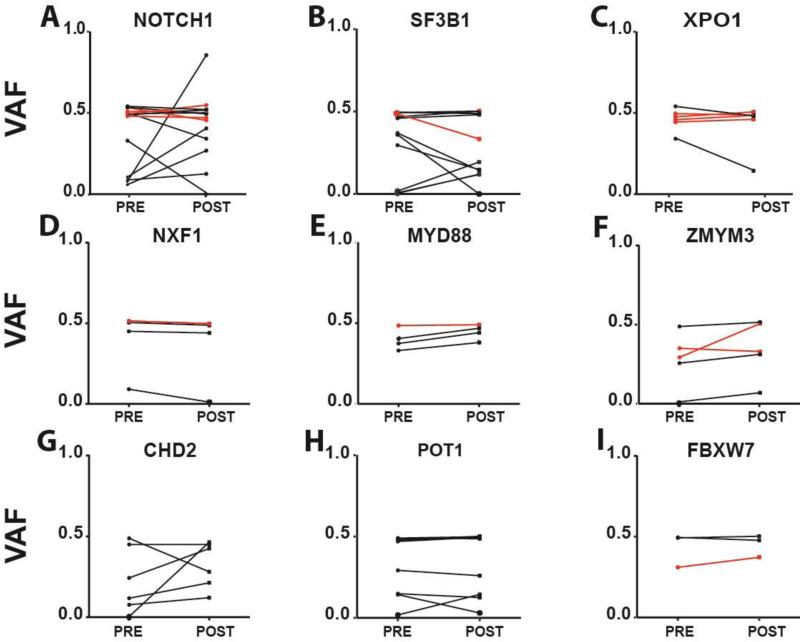
One of the important findings resulting from our VAF measurements pre- and post-therapy in a large CLL cohort relates to the common pre-therapy clonal representation of the following gene mutations: NOTCH1, SF3B1, XPO1, NXF1, MYD88, ZMYM3, CHD2, POT1 and FBXW7 (Figure 2). The quantitatively most common genotype was a clonal (as opposed to subclonal) mutation status pre-therapy, providing support to a revised model in which most of these gene mutations serve an early therapy-independent role in CLL pathogenesis. Although occasional CLL cases for some genes showed enrichment at relapse other cases revealed depletion of pre-exiting mutations thus demonstrating no consistent trends (for instance see data for NOTCH1 or SF3B1).
Figure 2. Evidence for an early pathogenetic role of various recurrent gene mutations in CLL.
Displayed are variant allele fractions (VAFs) for A: NOTCH1; B: SF3B1; C: XPO1; D: NXF1; E: MYD88; F: ZMYM3; G: CHD2; H: POT1 and, I: FBXW7 in paired samples procured before and after therapy. Black: CLL samples untreated at first measurements; Red: CLL samples that had relapsed at first measurement and that received additional therapy followed by relapse.
To provide additional context to these later findings we reviewed the five CLL cases that demonstrated substantial changes in NOTCH1 mutation VAFs before and after therapy. CLL117 lost the NOTCH1 mutant clone and acquired a dominant TP53 mutant clone; CLL270 demonstrated a drop in NOTCH1 VAFs from 0.48 to 0.32 and expanded a minor clone identified through a mutation in MGA that must have been NOTCH1 wt; CLL185 expanded a NOTCH1 mutant clone (VAFs: 0.048-0.255) while acquiring a large del13q-II lesion, a lesion type previously implicated in CLL relapse(38). CLL195 demonstrated a substantial expansion of the NOTCH1 mutated clone (VAFs: 0.095-0.389) while also carrying a del11q at both disease phases. By FISH del11q was 43% pre-therapy indicating it was acquired before the NOTCH1 mutation. Finally, CLL296 demonstrated a very strong NOTCH1 mutant clone expansion (VAFs: 0.07-0.83).
For the six CLL cases that demonstrated substantial changes in SF3B1 mutation VAFs we find the following genomic context: CLL11 (declining mutant SF3B1 VAFs: 0.296-0.147) maintained a clonal POT1 mutation in both phase and gained a separate clone identified through a mutation in the gene TENM2 (0.02-0.238); CLL36 (declining mutant SF3B1 VAFs: 0.396-0.143) had no additional markers for the rising SF3B1 wt clone identified; CLL209 (declining mutant SF3B1 VAFs: 0.476-0.329) maintained clonal NOTCH1 and XPO1 mutations and gained an 11q deletion; CLL218 largely lost the SF3B1 mutant clone (declining mutant SF3B1 VAFs: 0.387-0.002) and acquired a clone marked by mutations in SAMHD1, MGA and FAT3. Finally, two CLL cases (CLL72 and CLL78) demonstrated minor enrichments in SF3B1 mutant VAFs post therapy without attaining clonal representation (see Supplementary Table 3).
Further, we find that the VAFs for occasional CLL cases demonstrated subclonal status pre-therapy (for instance, see data for CHD2 or POT1) but very little or no enrichment was seen for most such cases post-therapy. Finally CLL #219 demonstrated a strong CHD2 mutation enrichment that occurred together with a TP53 mutation enrichment.
Identification of candidate subclonal gene drivers of relapsed CLL
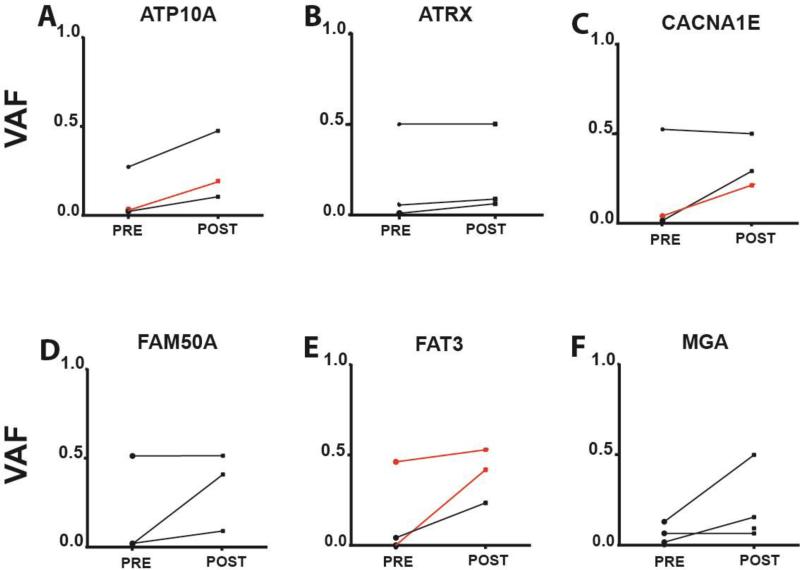
Within the group of infrequently mutated genes for which we determined VAFs at diagnosis and relapse we identified a subset that demonstrated enrichment at relapse in ≥2 cases for each gene. These are: ATP10A, ATRX, CACNA1E, FAT3, FAM50A and MGA (Figure 3) and together they constitute candidate genes that may contain true, albeit infrequent drivers of CLL relapse. Once we excluded gene mutations demonstrating VAF increases in CLL cases that also demonstrated the simultaneous emergence of a TP53 mutation, only the genes ATP10A, ATRX, FAM50A, FAT3 and MGA remained, and as outlined above, ATP10A mutations occurred twice in the setting of acquired SAMHD1 mutations. Notwithstanding these caveats, some of these candidate genes are deserving of further study in much larger relapsed CLL cohorts.
Figure 3. Candidate gene drivers of CLL relapse.
Displayed are variant allele fractions (VAFs) for A: ATP10A; B: ATRX; C: CACNA1E; D: FAM50A; E: FAT3 and, F: MGA in paired samples procured before and after therapy. Black: CLL samples untreated at first measurements; Red: CLL samples that had relapsed at first measurement and that received additional therapy followed by relapse.
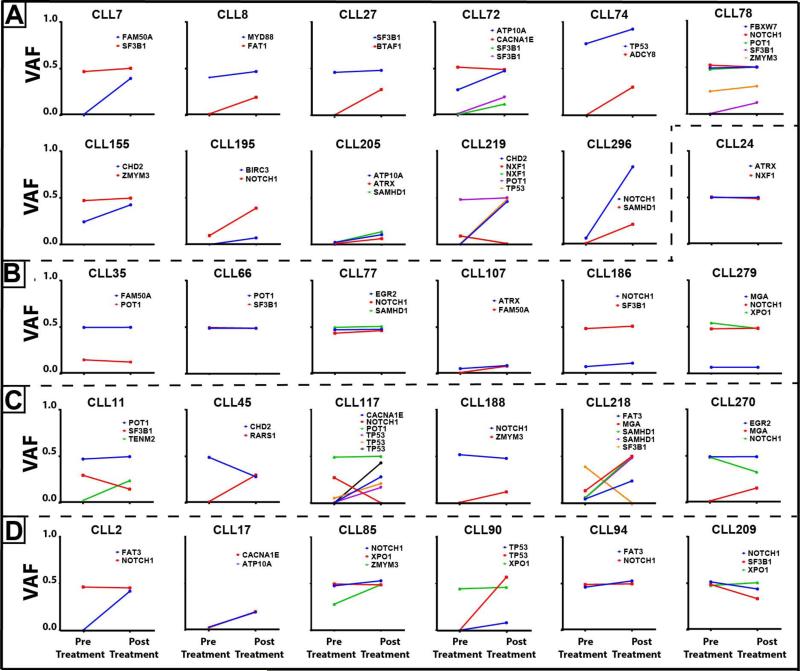
Analyses of clonal dynamics uncovers rising, stable and falling clones without clear evidence that recurrent subclonal gene mutations other than in TP53 frequently drive CLL relapse
The ultra-high purity of our isolated CLL cell DNA paired with the fact that the commonly mutated genes other than TP53 were almost never affected by genomic gains or losses as measured through SNP 6.0 profiling (Supplementary Table 3) allowed us to track clonal dynamics directly through the VAF measurements. For heterozygous gene mutations, which constitute the vast majority of genes in this study, the cell fraction carrying the mutations is twice the VAF measurement (e.g. VAF = 0.5; cell fraction with mutations 1; the VAF for X-linked genes in males were corrected by a factor of 0.5). Representative results for CLL cases are displayed in Figure 4 (A: rising subclone scenario; B: stable clone scenario and C: complex and falling clones scenario). From this data and the summary data displayed in Figure 2 it is clear that genes commonly mutated in CLL, including NOTCH1, SF3B1, XPO1, NXF1, MYD88, ZMYM3, CHD2, POT1 and FBXW7 serve early pre-treatment roles in CLL pathogenesis and often are already present in almost all tumor cells pre-therapy.
Figure 4. CLL clone dynamics in individual patients identifies rising, stable and falling clones and subclones post therapy.
A-C: All CLL cases were untreated at first measurements. A: CLL cases with rising subclones; B: CLL cases with stable clones; C: CLL cases with complex clone dynamics including rising and falling clones. D: CLL samples that had relapsed at first measurement and that received additional therapy.
Review of the various clone tracking scenarios displayed in Figure 4A-D also clearly demonstrates limits to the hypothesis that recurrent subclonal gene mutations frequently are responsible for CLL relapse because i) rising, stable and falling clones were identified in multiple cases and ii) subclonal CLL relapse drivers other than TP53 and possibly SAMHD1 were not commonly detected, and, iii) rising CLL subclones were marked by a variety of uncommon or non-recurrent gene mutations likely indicative of passenger status(22, 41).
The majority of relapsed CLL demonstrates clonal evolution from pre-treatment states
We proceeded with SNP 6.0 array profiling of the CLL cases that were subjected to sequence analysis, extending previously published measurements(38). Subsequently, we combined the CLL sequencing data with the SNP array profiling data and categorized the emergence or loss of any genomic lesion at relapse as a clonal evolution event. Of the 53 CLL patients subjected to longitudinal analysis that received intercurrent therapy, 62% (33/53) developed genomic changes (almost always in the form of acquisitions) in aCNAs or copy neutral LOHs or gene mutations, while 38% (20/53) did not demonstrate such changes (Table 2). Therefore, CLL relapse in the majority of CLL cases is due to the emergence of novel clones that differ from the quantitatively dominant pre-treatment clones usually through acquisition of additional genomic changes. In a minority of cases the CLL relapse is apparently due to proportional regrowth of the pre-existing dominant clone or to changes not captured through WES and SNP array profiling.
DISCUSSION
In this study, we have performed WES followed by Sanger sequence validation to interrogate the exome of 61 relapsed CLL patients for somatically acquired gene mutations. Next, we employed custom panel-based deep re-sequencing of all exons of the genes most frequently mutated in this discovery cohort on paired CLL samples procured before therapy and at relapse to measure variant allele fractions (VAFs). For selected genes we validated and/or resorted to droplet digital PCR for VAF estimations allowing for confirmation of rare subclones at a depth not previously reported. Quantitative measurements further allowed for analyses of clone and subclone dynamics in 30 informative cases.
Summarizing the major findings from this study, we i) identify recurrent strong enrichment of TP53 mutations from pre-existing rare subclones in CLL at relapse(42, 43); suggesting TP53 mutations were the dominant gene driver of rCLL; ii) recurrent enrichment of subclonal SAMDH1 mutations at relapse nominating this gene as an in vivo therapy resistance factor in CLL; iii) identify a few candidate CLL relapse-associated mutated genes that enriched at relapse albeit present individually in only a few cases each. Some of these genes are deserving of expansion studies once much larger cohorts of rCLL become available; iv) importantly, we identify many of the known recurrently mutated genes in CLL as often already clonal prior to therapy (NOTCH1, SF3B1, POT1, FBXW7, MYD88, NXF1, XPO1, ZMYM3 or CHD2), or without consistent enrichment/depletion trends at relapse providing clear evidence that these genes serve an early therapy-independent role in CLL pathogenesis(35), v) through analyses of clone and subclone dynamics we identify patterns of clonal enrichment, or stability or complex patterns including clonal decline that together do not support recurrent subclonal gene drivers (other than in TP53) as frequent causes of CLL relapse, and, vi) confirm through a more complete genomic analysis the frequent occurrence of genomically altered CLL clones that dominate the disease at relapse.
This study is characterized by various methodological strengths that support the overall conclusions drawn, including: i) the use of high-purity, flow-sorted CD19+ and CD3+ cells as a source of DNA therefore effectively eliminating confounding effects of impure CLL tumor cell populations on estimations of VAFs and clone sizes; the very high tumor cell purity is evidenced by VAFs clustering around 0.5 for many of the studied genes (equal to 100% of cells carrying heterozygous mutations given the lack of genomic alterations affecting mutated gene loci), ii) a large patient cohort that was relatively uniformly treated and sampled longitudinally before and after therapy reducing the effect of biases; iii) genomic aCNA/LOH and gene resequencing analyses that were based on paired DNA samples (tumor and paired normal) for all CLL samples; and, iv); use of various complementary highly sensitive techniques to quantify VAFs in paired CLL diagnosis and relapse samples.
The understanding of the role of gene mutations in the pathogenesis of CLL is undergoing steady refinements. Here we demonstrate that many previously identified recurrently mutated genes are already clonal pre- therapy in CLL, which implies a role in disease initiation possibly from pre-leukemic cells or outgrowth of dominant CLL clones from MBL(35, 44, 45). Findings may partly inform conflicting reports of prognostic implications of gene mutations given that prognosis is influenced by factors governing cell accumulation and therapy efficaciousness and less likely so by early biological events in CLL(46-50). The overarching finding that only a minority of relapsed CLL acquired recurrent clonal gene mutations after potent chemotherapy supports the conclusion that gene mutations as a group, with the exceptions of TP53 and possibly SAMHD1, do not markedly affect chemotherapy efficaciousness in patients and that molecular contributors to relapse reside within acquired copy number aberrations and hitherto unidentified molecular changes worthy of additional studies.
It is however worth mentioning that future studies could aim at unbiased exome-wide ultra-deep analysis of genomic lesions and subclones in CLL at diagnosis and again at relapse. While such studies are highly likely to identify additional complexity, the critical question would be how much of that complexity translates into disease progression and subclonal outgrowth or, alternatively, just represents subclonal diversity present in all cancers.
In summary, based on the largest reported longitudinal multi-dimensional genomic analysis in relapsed CLL to date, we demonstrate that the emergence of novel dominant clones is frequent in relapsed CLL. The origin of most genomic changes, including mutations in genes like TP53 or SAMHD1, is likely therapy-independent and due to random mutagenesis. Our data support a revised appreciation of an early pathogenetic role for many of the more commonly mutated genes in the pathogenesis of CLL and qualify the roles of subclones as marked by gene mutations in CLL pathogenesis.
Supplementary Material
TRANSLATIONAL RELEVANCE.
The molecular determinants that influence outgrowth of CLL from monoclonal B cell lymphocytosis (MBL) pre-therapy and the determinants of relapse following therapy are under active investigations. In particular, the discovery of multiple recurrently mutated genes in CLL motivates research into their contribution to CLL disease biology and progression after therapy. In this study, we have used complementary experimental approaches to study recurrent gene mutations in paired CLL samples procured before and after therapy. Using DNA isolated from flow-sorted CLL cells for all analyses, we find that many gene mutations in CLL are already clonal prior to therapy or show no consistent enrichment post-therapy indicative of an early pathogenetic and therapy-unrelated role in CLL. In contrast, through use of highly sensitive droplet digital PCR able to detect mutations at 0.01% allele frequency, we find that clones carrying TP53 or SAMHD1 mutation enrich post-therapy often from deep subclones and are not therapy-induced.
Acknowledgement
Supported by the National Institutes of Health through CA136537 (SM), a Clinical Scholars Award of the Leukemia and Lymphoma Society of America (SM), and a research grant by Janssen Pharmaceuticals (SM). This research is supported (in part) by the National Institutes of Health through the University of Michigan's Cancer Center Support Grant (5 P30 CA46592). We are grateful for services provided by the microarray core of the University of Michigan Comprehensive Cancer Center.
Footnotes
Individual contributions:
Nisar Amin, Kamlai Saiya-Cork, Brian Parkin and Sami Malek performed the laboratory research.
Kerby Shedden assisted with statistical analysis and software development for data analysis.
Erlene Seymour assisted with database management.
Nisar Amin and Sami Malek wrote the paper.
Sami Malek conceived the study and supervised the work.
Conflict of Interest:
None to declare
REFERENCES
- 1.Malek SN. The biology and clinical significance of acquired genomic copy number aberrations and recurrent gene mutations in chronic lymphocytic leukemia. Oncogene. 2013;32:2805–17. doi: 10.1038/onc.2012.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010;10:37–50. doi: 10.1038/nrc2764. [DOI] [PubMed] [Google Scholar]
- 3.Strefford JC. The genomic landscape of chronic lymphocytic leukaemia: biological and clinical implications. Br J Haematol. 2014 doi: 10.1111/bjh.13254. [DOI] [PubMed] [Google Scholar]
- 4.Gaidano G, Foa R, Dalla-Favera R. Molecular pathogenesis of chronic lymphocytic leukemia. J Clin Invest. 2012;122:3432–8. doi: 10.1172/JCI64101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–6. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 6.Ouillette P, Collins R, Shakhan S, Li J, Peres E, Kujawski L, et al. Acquired genomic copy number aberrations and survival in chronic lymphocytic leukemia. Blood. 2011;118:3051–61. doi: 10.1182/blood-2010-12-327858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oscier DG, Gardiner AC, Mould SJ, Glide S, Davis ZA, Ibbotson RE, et al. Multivariate analysis of prognostic factors in CLL: clinical stage, IGVH gene mutational status, and loss or mutation of the p53 gene are independent prognostic factors. Blood. 2002;100:1177–84. [PubMed] [Google Scholar]
- 8.Messina M, Del Giudice I, Khiabanian H, Rossi D, Chiaretti S, Rasi S, et al. Genetic lesions associated with chronic lymphocytic leukemia chemo-refractoriness. Blood. 2014;123:2378–88. doi: 10.1182/blood-2013-10-534271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zenz T, Habe S, Denzel T, Mohr J, Winkler D, Buhler A, et al. Detailed analysis of p53 pathway defects in fludarabine-refractory chronic lymphocytic leukemia (CLL): dissecting the contribution of 17p deletion, TP53 mutation, p53-p21 dysfunction, and miR34a in a prospective clinical trial. Blood. 2009;114:2589–97. doi: 10.1182/blood-2009-05-224071. [DOI] [PubMed] [Google Scholar]
- 10.Guieze R, Robbe P, Clifford R, de Guibert S, Pereira B, Timbs A, et al. Presence of multiple recurrent mutations confers poor trial outcome of relapsed/refractory CLL. Blood. 2015;126:2110–7. doi: 10.1182/blood-2015-05-647578. [DOI] [PubMed] [Google Scholar]
- 11.Shanafelt TD, Hanson C, Dewald GW, Witzig TE, LaPlant B, Abrahamzon J, et al. Karyotype evolution on fluorescent in situ hybridization analysis is associated with short survival in patients with chronic lymphocytic leukemia and is related to CD49d expression. J Clin Oncol. 2008;26:e5–6. doi: 10.1200/JCO.2008.16.7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knight SJ, Yau C, Clifford R, Timbs AT, Sadighi Akha E, Dreau HM, et al. Quantification of subclonal distributions of recurrent genomic aberrations in paired pre-treatment and relapse samples from patients with B-cell chronic lymphocytic leukemia. Leukemia. 2012;26:1564–75. doi: 10.1038/leu.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stilgenbauer S, Sander S, Bullinger L, Benner A, Leupolt E, Winkler D, et al. Clonal evolution in chronic lymphocytic leukemia: acquisition of high-risk genomic aberrations associated with unmutated VH, resistance to therapy, and short survival. Haematologica. 2007;92:1242–5. doi: 10.3324/haematol.10720. [DOI] [PubMed] [Google Scholar]
- 14.Braggio E, Kay NE, VanWier S, Tschumper RC, Smoley S, Eckel-Passow JE, et al. Longitudinal genome-wide analysis of patients with chronic lymphocytic leukemia reveals complex evolution of clonal architecture at disease progression and at the time of relapse. Leukemia. 2012;26:1698–701. doi: 10.1038/leu.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunnarsson R, Mansouri L, Isaksson A, Goransson H, Cahill N, Jansson M, et al. Array-based genomic screening at diagnosis and during follow-up in chronic lymphocytic leukemia. Haematologica. 2011;96:1161–9. doi: 10.3324/haematol.2010.039768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grubor V, Krasnitz A, Troge JE, Meth JL, Lakshmi B, Kendall JT, et al. Novel genomic alterations and clonal evolution in chronic lymphocytic leukemia revealed by representational oligonucleotide microarray analysis (ROMA). Blood. 2009;113:1294–303. doi: 10.1182/blood-2008-05-158865. [DOI] [PubMed] [Google Scholar]
- 17.Ojha J, Ayres J, Secreto C, Tschumper R, Rabe K, Van Dyke D, et al. Deep sequencing identifies genetic heterogeneity and recurrent convergent evolution in chronic lymphocytic leukemia. Blood. 2015;125:492–8. doi: 10.1182/blood-2014-06-580563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shanafelt TD, Witzig TE, Fink SR, Jenkins RB, Paternoster SF, Smoley SA, et al. Prospective evaluation of clonal evolution during long-term follow-up of patients with untreated early-stage chronic lymphocytic leukemia. J Clin Oncol. 2006;24:4634–41. doi: 10.1200/JCO.2006.06.9492. [DOI] [PubMed] [Google Scholar]
- 19.Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puente XS, Bea S, Valdes-Mas R, Villamor N, Gutierrez-Abril J, Martin-Subero JI, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–24. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 22.Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526:525–30. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Kaminski MS, Li Y, Yildiz M, Ouillette P, Jones S, et al. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood. 2014;123:1487–98. doi: 10.1182/blood-2013-05-500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–7. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 29.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gates C, Bene J, Bhasi A, Ulintz P, Meng K, Cavalcoli J. Jacquard: A practical approach to integrating complex somatic variant data sets.. ISMB: Intelligent Systems in Molecular Biology Conference Proceedings; Dublin, Ireland. 2015. [Google Scholar]
- 31.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34:E2393–402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mansouri L, Sutton LA, Ljungstrom V, Bondza S, Arngarden L, Bhoi S, et al. Functional loss of IkappaBepsilon leads to NF-kappaB deregulation in aggressive chronic lymphocytic leukemia. J Exp Med. 2015;212:833–43. doi: 10.1084/jem.20142009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Damm F, Mylonas E, Cosson A, Yoshida K, Della Valle V, Mouly E, et al. Acquired initiating mutations in early hematopoietic cells of CLL patients. Cancer Discov. 2014;4:1088–101. doi: 10.1158/2159-8290.CD-14-0104. [DOI] [PubMed] [Google Scholar]
- 36.Domenech E, Gomez-Lopez G, Gzlez-Pena D, Lopez M, Herreros B, Menezes J, et al. New mutations in chronic lymphocytic leukemia identified by target enrichment and deep sequencing. PLoS One. 2012;7:e38158. doi: 10.1371/journal.pone.0038158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Reis M, Khoriaty R, Li Y, Ouillette P, Samayoa J, et al. Sequence analysis of 515 kinase genes in chronic lymphocytic leukemia. Leukemia. 2011;25:1908–10. doi: 10.1038/leu.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ouillette P, Saiya-Cork K, Seymour E, Li C, Shedden K, Malek SN. Clonal evolution, genomic drivers, and effects of therapy in chronic lymphocytic leukemia. Clin Cancer Res. 2013;19:2893–904. doi: 10.1158/1078-0432.CCR-13-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trbusek M, Malcikova J. TP53 aberrations in chronic lymphocytic leukemia. Adv Exp Med Biol. 2013;792:109–31. doi: 10.1007/978-1-4614-8051-8_5. [DOI] [PubMed] [Google Scholar]
- 40.Clifford R, Louis T, Robbe P, Ackroyd S, Burns A, Timbs AT, et al. SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood. 2014;123:1021–31. doi: 10.1182/blood-2013-04-490847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–26. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossi D, Khiabanian H, Spina V, Ciardullo C, Bruscaggin A, Fama R, et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123:2139–47. doi: 10.1182/blood-2013-11-539726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malcikova J, Stano-Kozubik K, Tichy B, Kantorova B, Pavlova S, Tom N, et al. Detailed analysis of therapy-driven clonal evolution of TP53 mutations in chronic lymphocytic leukemia. Leukemia. 2015;29:877–85. doi: 10.1038/leu.2014.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rawstron AC, Bennett FL, O'Connor SJ, Kwok M, Fenton JA, Plummer M, et al. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med. 2008;359:575–83. doi: 10.1056/NEJMoa075290. [DOI] [PubMed] [Google Scholar]
- 45.Kikushige Y, Ishikawa F, Miyamoto T, Shima T, Urata S, Yoshimoto G, et al. Self- renewing hematopoietic stem cell is the primary target in pathogenesis of human chronic lymphocytic leukemia. Cancer Cell. 2011;20:246–59. doi: 10.1016/j.ccr.2011.06.029. [DOI] [PubMed] [Google Scholar]
- 46.Stilgenbauer S, Schnaiter A, Paschka P, Zenz T, Rossi M, Dohner K, et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood. 2014;123:3247–54. doi: 10.1182/blood-2014-01-546150. [DOI] [PubMed] [Google Scholar]
- 47.Jeromin S, Weissmann S, Haferlach C, Dicker F, Bayer K, Grossmann V, et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia. 2014;28:108–17. doi: 10.1038/leu.2013.263. [DOI] [PubMed] [Google Scholar]
- 48.Baliakas P, Hadzidimitriou A, Sutton LA, Rossi D, Minga E, Villamor N, et al. Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia. 2015;29:329–36. doi: 10.1038/leu.2014.196. [DOI] [PubMed] [Google Scholar]
- 49.Rossi D, Rasi S, Spina V, Bruscaggin A, Monti S, Ciardullo C, et al. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood. 2013;121:1403–12. doi: 10.1182/blood-2012-09-458265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oscier DG, Rose-Zerilli MJ, Winkelmann N, Gonzalez de Castro D, Gomez B, Forster J, et al. The clinical significance of NOTCH1 and SF3B1 mutations in the UK LRF CLL4 trial. Blood. 2013;121:468–75. doi: 10.1182/blood-2012-05-429282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.