Abstract
The diffusion of molecules such as nutrients and oxygen through densely packed cells is impeded by blockage and consumption by cells, resulting in a limited depth of penetration. This has been a major hurdle to a bulk (3-D) culture. Great efforts have been made to develop methods for generating branched microchannels inside hydrogels to support mass exchange inside a bulk culture. These previous attempts faced a common obstacle: researchers tried to fabricate microchannels with gels already loaded with cells, but the fabrication procedures are often harmful to the embedded cells. Herein, we present a universal strategy to create microchannels in different types of hydrogels, which effectively avoids cell damage. This strategy is based on a freestanding alginate 3-D microvascular network prepared by in-situ generation of copper ions from a sacrificial copper template. This alginate network could be used as implants to create microchannels inside different types of hydrogels. This approach effectively addresses the issue of cell damage during microfabrication and made it possible to create microchannels inside different types of gels. The microvascular network produced with this method is (1) strong enough to allow handling, (2) biocompatible to allow cell culturing, and (3) appropriately permeable to allow diffusion of small molecules, while sufficiently dense to prevent blocking of channels when embedded in different types of gels. In addition, composite microtubules could be prepared by simply pre-loading other materials, e.g., particles and large biomolecules, in the hydrogel. Compared with other potential strategies to fabricate freestanding gel channel networks, our method is more rapid, low-cost and scalable due to parallel processing using an industrially mass-producible template. We demonstrated the use of such vascular networks in creating microchannels in different hydrogels and composite gels, as well as with a cell culture in a nutrition gradient based on microfluidic diffusion. In this way, the freestanding hydrogel vascular network we produced is a universal functional unit that can be embedded in different types of hydrogel; users will be able to adopt this strategy to achieve vascular mass exchange in the bulk culture without changing their current protocol. The method is readily implementable to applications in vascular tissue regeneration, drug discovery, 3-D culture, etc.
I. INTRODUCTION
While showing their great potential in further benefiting human health, several major research fields of medicine and biology, such as regenerative therapy and drug discovery, are sharing a common demand of practical strategies for bulk or 3-D cell culture.1,2 For example, artificial tissues based on bulk culture could be used not only for tissue/organ level biology research and drug discovery3–5 but also for regenerative medicine;6,7 on the other hand, even for “cell-level” researches, which were traditionally based on 2-D cultures, an increasing number of studies revealed that the tested cells may behave differently in a 3-D culture environment, which is more similar to real situations,8–11 suggesting that 3-D culture is the better approach as it mimics real situations more than the 2-D culture approach.
Culturing cells in a 3-D environment, on the other hand, is not easy to achieve. In general, there are two major requirements for establishing a 3-D culture. The first is to use a scaffold that mimics the extracellular matrix (ECM) to immobilize cells and to construct the 3-D cell stacks.12 Many porous scaffolds have been developed for this purpose such as those made of poly(d,l-lactide-co-glycolide) (PLGA),13 hyaluronic acid,14 collagen,15,16 polyacrylic acid (PAA),17 poly(ethylene glycol),10 chitosan,18 alginate,19,20 and others.21–23 These scaffolds can immobilize cells and support cell growth.
The second requirement of a 3-D cell culture, however, is still unmet. The challenge is how to effectively deliver nutrients to and remove metabolic wastes from the cells, which are densely packed in the culture. Without fulfilling this requirement, cells, hundreds of microns deep inside the bulk culture, will starve to death.24 Although there have been attempts using alternative strategies,25–29 it seems that the most effective way to address this challenge is to construct branched semi-permeable capillary networks inside the volume of a cell culture, just like that of vascularized blood vessel systems. Aiming at this demand, some previous studies proposed methods to create microchannels inside the bulk cell-laden hydrogel matrix, which can be grouped into: (1) casting-peeling-bonding scheme commonly used in fabricating microfluidic devices, (2) sacrificial template scheme, and (3) 3-D printing technologies.26,30–34
However, these methods are still unsuitable for constructing the vascular structures needed for supporting a 3-D culture for the following reasons: (1) for those strategies based on the casting–and-bonding scheme, multiple layers of channels and complicated connections have to be fabricated and stacked carefully to realize a certain thickness of the culture; (2) the bioactive hydrogels that are suitable for embedding cells are usually very soft, making it difficult to 3-D print such gels with sufficient resolution to match the scale of capillary blood vessels; and (3) the low mechanical strength of bioactive hydrogels and limited methods for sealing the channels make channels apt to leak and difficult to connect to tubes.31–33 More importantly, some of the microfabrication processes involved in these aforementioned techniques, such as heating, photopolymerization, metal coordination, and dissolving of sacrificial template, are often harmful to the embedded cells; also, the fabrication methods are not universal, meaning that each method only supports very limited types of hydrogels.
Herein, we propose a universal strategy to meet the aforementioned demand. In our strategy, a freestanding hydrogel tubule network is prepared in advance and used as an implant in cell-laden hydrogels to create microchannels in a bulk culture. In this way, the cells in the bulk culture would not be affected by microchannel fabrication, and the gel to support cell culture can be chosen with complete freedom. The key part in this strategy is a branched tubular network strong enough to allow handling and is biocompatible to allow cell culture, appropriately permeable to allow diffusion of small molecules, while sufficiently dense to prevent blocking of channels inside when embedded in different types of gels. In this work, we created a method to fabricate such freestanding 3-D tubular structures and demonstrated the implementation of these functional units for creating microchannels in different types of hydrogels. It is worth noting that prior to our work, there were reported methods for generating fibers or tubes in hydrogel, but those methods are incapable of producing branched freestanding tubular networks.35–38 To create branched 3-D tubular structures made of hydrogel, we employed a copper template that could be prepared by reported mass-production strategies39–41 or recent 3-D printing strategies.42–44 We immersed the copper template in alginate, a natural polysaccharide derived from brown sea algae, which has been widely used to fabricate bio-scaffolds in biomedical and biological applications;19,45–49 then we conducted electrolysis, during which the released copper ions from the template quickly cross-linked the alginate in the solution and formed gel. The reaction was so quick that all the generated Cu2+ ions were consumed before diffusing away, resulting in a uniform thin layer of hydrogel coating covering the copper template. After completely dissolving the template with Fe3+, and subsequently replacing the Fe3+ ions inside the alginate gel by Ca2+, we obtained a 3-D tubular network made of calcium alginate hydrogel. The entire process took only a few hours. Moreover, it is scalable at low cost through parallel processing. In contrast, even if 3-D printing of hydrogel has the potential to reach similar resolution in the future to create such freestanding tubular networks, it will still be restricted by the speed and cost owing to its serial-processing nature. It is worth noting that there were two types of non-printing methods to create microtubes: one was to cast from capillary tubes, and the other was to form tubes through microfluidic laminar flow.36,50 Neither of the methods were suitable for generating branched tube networks like in our work. Besides, other materials and biomolecules could be pre-loaded in our hydrogel tubular networks by mixing them with the alginate solution beforehand, and the thickness of the tubule wall could be easily controlled by tuning the time span of electrolysis. To evaluate the properties of the produced vascular networks, we demonstrated the selective permeability of the vessel structures to molecules of different sizes and illustrated the function of nutrition delivery in the 3-D cell culture. What is more important is that the tubular structures formed using our method are strong enough to support themselves and could be used as a microfluidic system alone, allowing greater flexibility in different applications. By de-convolving the “microchannel” fabrication step from the bulk gel generation in which cells are embedded, our strategy leads to at least two advantages: (1) completely avoiding the potential damages caused during the microchannel fabrication step to the embedded cells in the bulk gel and (2) allowing generation of microchannels in arbitrary hydrogels regardless of the inherent properties of the hydrogel.
II. EXPERIMENTAL SECTIONS
A. Materials and equipment
Alginate, agar, gelatin, fluorescein powder, fluorescein isothiocyanate-labeled bovine serum albumin, and other chemicals were purchased from Sigma-Aldrich (Hong Kong, China). All reagents were of analytical grade and used without further purification. Copper wire was from Monster, US. DC power supply used in the experiment was from Shenzhen HongSheng Electronic Co., Ltd. (DPS-305CF, China). Heating plate was from Xinruiqi Electronic, Inc. (LKTC-B1-T, China). The syringe pump was purchased from Cole-Parmer Instrument Company (74900 Cole Parmer, USA). The fluorescence microscope used in the experiment was from Micro-shot Technology Limited (ML-30, China) and was equipped with Infinity 2 digital camera (Lumenera Corporation, Canada) to capture images.
B. Fabrication of alginate hydrogel tubular networks
First, copper scaffolds were handmade by folding and twisting copper wires with a diameter of 100 μm. Then, each scaffold was electrolyzed at 3.5 V in 3% (w/v) alginate solution. Upon electrolysis, copper ions were released from the surface of the copper scaffold, which cross-linked the nearby alginate molecules and formed a thin layer of alginate hydrogel coating on the copper wires. After that, the gel-coated copper scaffold was immersed in 1 M CaCl2 solution for further electrolysis to dissolve most of the copper. The remaining copper was dissolved completely in 0.1 M FeCl3 solution at 50 °C for 1 h. Finally, the Fe3+ ions in the alginate tubules were replaced by Ca2+ ions (1 M CaCl2 and 0.02 M EDTA, pH 5) at 50 °C for 1 h.
C. Characterization of alginate hydrogel tubular networks
Scanning electron microscope (LEO 1530 VP, Zeiss) and confocal fluorescence microscope (C1, Nikon) were used to characterize the microstructure and morphology of hydrogel tubules.
D. Diffusion tests
Fluorescein and fluorescein isothiocyanate-labeled bovine serum albumin (FITC-BSA) were used for the corresponding diffusion tests in hydrogel tubular networks. 1.25 mg/ml fluorescein solution and 2.5 mg/ml FITC-BSA were injected into the hydrogel tubules, which were embedded in agar gel, and the diffusion of the dye molecules was observed under fluorescence microscope equipped with a digital camera (Infinity 2, Lumenera).
E. Perfusion experiment of HepG2
The cell suspension (1 × 107 cells/ml HepG2) was mixed with 3% (w/v) warm agar solution prepared by Dulbecco's modified Eagle's medium (DMEM). After embedding the microvascular tubule in the cell-mixed agar gel, the DMEM medium was introduced into the tubule by a syringe pump at a flow rate of 2 μl/min for 3 days in a 5.0% CO2 cell incubator. For cell viability assessments, the cell-embedded agar gel was sliced into 1-mm thick pieces, which were submerged in a live/dead staining solution (10 μg/ml Fluorescein diacetate and 10 μg/ml propidium iodide in DMEM), followed by 20 min of incubation at 37 °C and 5.0% CO2. Finally, the slices were washed with the DMEM medium three times for further observation. Images were taken with a fluorescence microscope (Eclipse Ti with D-Eclipse C1, Nikon Instruments). ImageJ (V1.50) was used for merging green and red fluorescence images and counting the live and dead cells.
III. RESULTS AND DISCUSSION
In this work, we proposed a method to prepare a freestanding microvascular network and employ it into cell culture experiments as an implanted unit. However, to be used as the implant, the branched freestanding vascular structures need to fulfill several requirements: (1) the network should be made of semipermeable, hydrophilic material with suitable porosity to allow diffusion of small molecules, while being sufficiently dense to prevent blockage of channels when embedded in different types of gels; (2) the network should be strong enough to allow handling during the fabrication process; (3) the network should be biocompatible to allow cell culture; and (4) it should also be affordable, mass-producible, and stable for long-term storage. Accordingly, we chose alginate as the basic material for the fabrication mainly because it is biocompatible, inexpensive, moderately porous, and has sufficient mechanical strength once crosslinked. To generate the branched tubular networks with controllable thickness of the tube wall, we employed a new strategy to produce the hydrogel microstructures, which is different from the previously reported strategies on microfabrication of hydrogels.30–34 A key component of our strategy is the copper scaffold, which was chosen with the following considerations. First, the copper can be electrolyzed to generate copper ions, which can crosslink alginate molecules to form the hydrogel structure, while the remaining copper can be fully dissolved afterwards to create hollow structure inside. Second, the copper structure serves as a template to precisely define the shape and size of the channel inside the hydrogel tubular networks. Third, the available techniques39–44 to mass-produce 3-D microstructures in copper make our method scalable (however, in this work, we simply used hand-made copper templates for demonstration). Finally, we successfully developed a convenient and cost-efficient method to replace the copper ions with calcium ions to eliminate the potential toxicity that the copper ions may have on cells.
It is worth noting that although we employed a sacrificial template (i.e., the copper scaffold), our method is clearly different from the previous methods based on sacrificial templates. One key difference in our method is that our sacrificial template is used not for the creation of channels directly in the bulk gel with cells loaded but for generating the freestanding tubule structures with no cell involved. Another key difference is that previous methods used the sacrificial template mainly to define the internal volume of the channel structure (passive), but our sacrificial template is used for in-situ generation of ions for crosslinking the hydrogel (active); because of this difference, our method generates a thin coating layer with controllable thickness owing to the diffusion limit of generated ions before they are consumed by the crosslinking reaction, and as such only our method can generate hydrogel tubular structures. The other methods generate a bulk gel.
Moreover, the dissolution of sacrificial template may affect the cells in that the dissolving agent as well as the substances from the dissolved template may harm the cells. In previous reports, it has been frequently discussed that the dissolving of sacrificial templates inside the cell-laden gel may hurt the cells.29,31,33,50 However, continuous efforts until now have not yet fully addressed this common problem, probably because of the inherent drawback of the strategy that involves microfabrication process in the presence of the live cells. For instance, in a recent report, Huang and his co-workers tried to reduce the damage to cells caused by the fabrication process by using a biocompatible sacrificial template made of sodium alginate; however, the reagent used for dissolving the template, ethylenediaminetetraacetic acid (EDTA), is not biocompatible.50,51 In another work, Chen et al. reported a smart design using a 3-D-printed carbohydrate glass sacrificial template that dissolves in water; however, although this template can be removed by a biocompatible buffer, the dissolution of carbohydrate will cause osmotic shock that damages the cells.31 Thus, the authors needed to coat the carbohydrate glass lattice with a thin layer of poly(D-lactide-co-glycolide) (PDLGA); and because the template was highly hygroscopic, it had to be reinforced by dextrans, and stored at 45 °C or in a vacuum chamber before use. Our method is free of these problems because the cells are not present during the fabrication of the microchannels.
Fig. 1 provides an illustration of the fabrication process, which was completed within 3 h (Fig. 2). The thickness of the tubule wall is controlled by adjusting the duration of electrolysis, while branched tubular networks of various shapes can be made by designing different features of the copper scaffold (Fig. 3). Also, other materials and biomolecules were loaded in the alginate hydrogel tubular networks by mixing them with alginate solution beforehand (Fig. 4).
FIG. 1.
Schematic diagram of the fabrication process of 3-D tubular networks in alginate hydrogel. (a) A thin layer of alginate hydrogel was formed on the surface of a copper wire by Cu2+ released through electrolysis; (b) further electrolysis in CaCl2 solution was conducted to dissolve most of the copper scaffold; (c) the remaining copper was dissolved by Fe3+ ions; (d) the Fe3+ ions were then replaced by Ca2+ ions; (e) a capillary tube was inserted into the hydrogel tubule; and (f) the connected hydrogel tubule was imbedded in agar gel for further experiment.
FIG. 2.
Results of each step in the fabrication of freestanding 3-D tubular networks in alginate hydrogel. The four images represent the four steps of the fabrication process. A thin, transparent layer of alginate gel coated the copper scaffold after electrolysis in alginate solution (a), the tubular networks turned light blue (b) after further electrolysis in CaCl2. Then the tubular networks became yellow (owing to iron (III) ions) after the remaining Cu pieces dissolved in FeCl3 (c). In the last step, the color of the tubules returned to light white after the Fe3+ ions were replaced with Ca2+ ions in a mixture solution of CaCl2 and EDTA acidic solution (d). Inset shows freestanding 3D microvascular networks. The scale bars are 10 mm long.
FIG. 3.
Plot of the thickness of the hydrogel coating versus the period of applied potential for electrolysis. The error bars represent standard deviations for N = 3 (inset: corresponding microscopic images, scale bar is 200 μm).
FIG. 4.
Microscopic images of hydrogel tubular networks. (a) A tubular network made of fluorescent particle-doped alginate gel; inset is confocal fluorescence image of cross section of the tubule. (b) and (c) are SEM images of partially dried tubular networks. (d) Cross section of a tubule with thinner wall fabricated by shorter period of electrolysis in alginate solution; inset is the tubule imbedded in agar gel.
A key point of our fabrication strategy is the in-situ generation and exchange of metal ions. Fig. 2 shows photos of the hydrogel tubular networks after each step in the fabrication process. First, we conducted electrolysis for 10 s at 3.5 V, which gave a thin, transparent layer of alginate hydrogel coated on the copper scaffold (Fig. 2(a)). We carried out further electrolysis in 1 M CaCl2 solution to dissolve the copper scaffold as completely as possible, after which the color of the hydrogel tubules turned light blue (Fig. 2(b)) due to Cu2+ ions. Then, we completely dissolved the remaining copper by Fe3+ ions (in 0.1 M FeCl3 solution) and the color changed to yellow brown, owing to Fe3+ ions (Fig. 2(c)). Compared with Cu2+ ions, Fe3+ ions are trivalent and thus better at cross-linking the alginate monomers, resulting in largely increased rigidity of the hydrogel structure. When we replaced the Fe3+ ions in the alginate hydrogel with Ca2+ ions in 0.02 M ethylenediaminetetraacetic acidic (EDTA) solution (pH 5), the hydrogel tubules reverted to a soft and semi-transparent white material (Fig. 2(d)). Since copper and iron ions were involved in the fabrication process, it is important to make sure that these ions are completely removed to avoid any heavy metal-induced toxicity in further in vivo studies. We used a classical colorimetric approach using ammonia52 to test residual copper and thiocyanate ions53 to test residual iron in the hydrogel tubules. No signal was detected by the spectrophotometer for either copper or iron ions, which indicated that the as-prepared hydrogel tubules do not contain significant amounts of copper or iron. Alternatively, for the final step of replacing Fe3+ ions with Ca2+ in an EDTA acidic solution, there are two biocompatible alternatives (FDA approved drug additives) of EDTA (see supplementary material, Fig. S1), which would be even safer for in vivo applications. As illustrated in Figs. 1(e) and 1(f), after all the steps, we inserted a capillary tube into the prepared hydrogel tubular network for injection of a solution, and then imbedded the setup in 3% (w/v) agar gel for further use (see supplementary material, Fig. S2).
Parameters including gel concentration, as well as the voltage and duration of electrolysis, were also found to affect fabrication. We observed that the mechanical strength of the hydrogel structures produced was enhanced by increased concentration of alginate. Therefore, we used a high concentration of 3% (w/v), close to the maximum concentration possible to prepare. As illustrated in Fig. 1, the wall of hydrogel tubules is formed by the electrolysis of copper that generates Cu2+ ions. To understand the dynamics of this process, we investigated the relationship between the thicknesses of the hydrogel tubule wall and the time of electrolysis. We chose 3.5 V as the voltage for electrolysis, since voltages are higher than these produced uneven thicknesses of the wall. The plot in Fig. 3 shows the trend of increasing thickness of hydrogel tubule wall along with the increasing time of electrolysis, and the inset shows the corresponding bright-field microscope images. The hydrogel tubule wall quickly reached 200-μm thick within 10 s and exceeded 400-μm in 60 s, indicating that our method is controllable and very rapid in generating the hydrogel tubular networks. Figs. 4(b) and 4(c) show the environmental-SEM images of partially dehydrated hydrogel tubular networks. The images show that the thickness of the tubule wall could be effectively controlled by changing the time of electrolysis, and the tubule wall was uniform without any pinholes even when the wall was thin (Fig. 4(d)).
Fig. 4(a) shows fluorescent microparticles uniformly entrapped inside the wall of the tubule, demonstrating that composite (doped) hydrogel tubules could be conveniently prepared by preloading particles in the starting material. According to our diffusion-cutoff test (Fig. 6), biomacromolecules (e.g., growth factors) can also be loaded in the hydrogel tubule in the same way, which can be useful for applications involving mammalian cell cultures.
FIG. 6.
Diffusion tests in hydrogel tubular networks imbedded in agar using fluorescein (a) and (c) and FITC-BSA (b) and (d) as samples. (a1) and (b1) are bright-field images before introducing samples. Fluorescence images (a2) and (a3) were taken at 0 min and 5 min, respectively, after injection of 1.25 mg/ml fluorescein. Fluorescence images (b2) and (b3) were taken at 0 min and 5 min, respectively, after injection of 2.5 mg/ml FITC-BSA. (c) and (d) Fluorescence intensity profiles across the channel, for fluorescein (c) and FITC-BSA (d). Scale bars are 200 μm long.
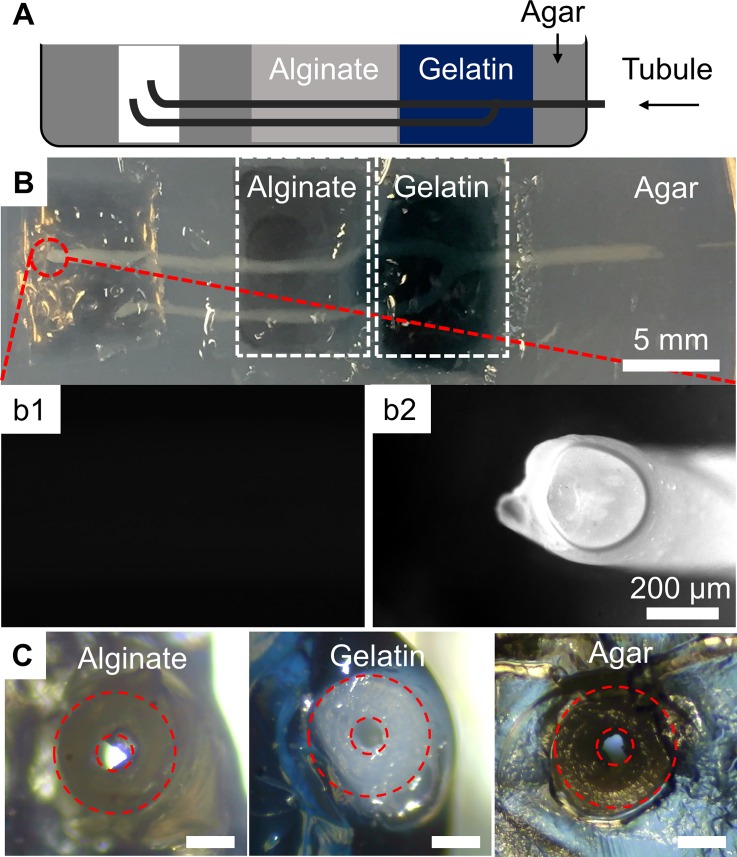
A nice feature of our method is that the produced 3-D tubular networks are mechanically strong enough to support themselves, even when suspended in air. Shaking at 80 rpm and 50 °C in a water bath shaker for 2 h did not damage the integrity of the 3-D tubular networks. This is mainly because of the relatively high rigidity of calcium alginate compared to other hydrogels, which allows great convenience in manipulation.54 After 1 year of storage in water, there was no observable change in the network even under microscopic inspection, indicating that it possesses adequate stability for commercial use. Owing to this good mechanical strength, our vascular structures can be used as microfluidic channels alone or as universal functional units to create microchannels inside other gels. When the gel network is used alone as microfluidic channels immersed in a buffer solution, it allows seeding of different cell types from both the inside and the outside of the tubule structures to form coaxial layered structures similar to blood capillaries, which is impossible to achieve using previously reported methods that generate channels in a bulk hydrogel.30–34 This could be useful in the long run, for generating man-made blood vessel structures. We then imbedded the fabricated alginate tubular networks in calcium alginate, genipin-crosslinked gelatin, and agar gel (see Fig. 5) and confirmed in all cases that the channels inside the alginate tubular networks were not blocked. These hydrogels were employed as models of three different gelation mechanisms—phase transition, chemical cross-linking, and physical cross-linking. The results demonstrated that different gels can be used to fill the space outside the tubule, allowing great convenience and flexibility in choosing the most suitable matrix for constructing a 3-D cell culture. This would be difficult to achieve without our method.
FIG. 5.
(a) Scheme (sideview) and (b) photo (topview) of embedding hydrogel tubular networks in different types of hydrogel including agar, gelatin, and alginate gel in (b). (b1) and (b2) are fluorescence images of the end of a hydrogel tubule before and after injecting 1 mg/ml fluorescein, respectively. (c) Images showing the cross-sections of the alginate tube embedded inside different types of hydrogels. The scale bars in (c) are 200 μm long.
To validate the feasibility and functionality of the prepared channels in hydrogel, we conducted several tests. A specific aim of our design is to support effective transportation of nutrients to cells, which means while supporting the fluidic flow through the channel inside the tubules, the hydrogel network should also allow diffusion of the nutrient molecules out of the tubule wall. As alginate is a copolymer composed of blocks of mannuronic and guluronic acids, the as-prepared calcium alginate hydrogel is porous and allows molecules smaller than its pores to pass through.55 To investigate the permeability of the hydrogel channels, we used fluorescein and fluorescein isothiocyanate-labeled bovine serum albumin (FITC-BSA) as models for small and large molecules, respectively, in a diffusion test of the hydrogel tubule, and the results are presented in Figure 6. We observed that FITC-BSA was restricted within the tubule while fluorescein can penetrate the tubule wall, which is consistent with the principle of the sieving effect of hydrogels. These results indicate that our alginate hydrogel tubules can mimic the function of blood vessels to a certain degree for mass exchange in a 3-D cell culture. On the other hand, the permeability of alginate is reported to be tunable. The pore size of alginate gel is mainly dependent on the concentration of alginate, as well as the type and concentration of the cross-linking ions.54,55 It is reported that the average pore size of 1.5% alginate is around 5 nm.56 And the pore size of 0.5% (w/v) sodium-alginate is as large as 20 nm, large enough for most proteins to diffuse through. Generally, molecules smaller than the pore size of a gel can diffuse through the gel; however, the penetration of macromolecules is often more complicated. For instance, some molecules such as vascular endothelial growth factor can bind to alginate and thus exhibit delayed release, which may in turn be beneficial for the immobilization of growth factors.57 Moreover, external stimuli such as temperature, pH, and enzymes can also influence the transport of large molecules.58,59 Finally, there is biodegradable alginate available to use, and as previously discussed the gel for making tubules can be doped, if further tunability of the permeability is needed.55
To further demonstrate the new functions realized by creating channels inside commonly used hydrogels, we conducted an experiment using cell-growth medium with a gradient of nutrient concentrations to support the cell culture. We embedded the tubule networks in HepG2 cell-seeded agar gel to deliver nutrient-rich medium to support cell culture. As our hydrogel tubules are permeable to small molecules, the culture medium will diffuse out when flowing through the tubules. Cells near a tubule will grow under sufficient medium supply, while cells far away from the tubule will lack these nutrients. A control experiment was also performed by loading the same density of cells in agar gel without culture medium supplied, although the moisture was still maintained. After culturing for 3 days, the agar gel was sliced into sections to determine cell viability. Based on the results of the live/dead assay shown in Figure 7, the viability of cells seeded near the tubule was higher than that of the cells far away from the tubule, while most of the cells died in the control group. The results indicated that our hydrogel microvascular networks are biocompatible and can be used for effective delivery of nutrient-rich medium to support a 3-D cell culture. The alginate tubule we prepared was sufficiently dense, and therefore, we did not observe any penetration of the tubule walls by cells, avoiding blockage of the channel by chemotaxis. On the other hand, the hydrodynamic character of our vessel-assisted diffusion culture system is different from those of the microfluidic cell culture systems that culture cells inside microchannels. Whilst the use of microfluidic channels can allow for the flexible control/change of concentrations of solutes and thus overcome the limitations of traditional static culture systems, the fluidic flow in the channels may harm or even flush away the cells.60,61 In contrast, our vessel system allows well-controlled delivery of fluid within the tubules, and thus eliminates the disturbance of the flow on the cultured cells. As a result, we have a flow-free environment to conveniently track the change of each specific cell during the culturing process. Moreover, after completion of the culture, our system allows us to collect the cells by cutting the gel.
FIG. 7.
Perfusion experiment of HepG2. (a) Schematic illustration of the setup for the test. Dashed line boxes indicate the positions for taking image B (near the tubule), C (about 300-μm from the tubule), and D (at the edge of the slice, about 3-mm away from the tubule). (b)–(d) live/dead fluorescence images of the sliced agar (green: live; red: dead). (e) Live/dead fluorescence image of a control experiment taken with a cell-laden agar incubated without supplying culture medium (kept wet in PBS). (f) The statistics of cell viability versus distance away from the tubule. Scale bars are 200 μm long (error bar = ± SD; n = 3).
IV. CONCLUSION
In this work, we developed a method to produce freestanding 3-D microtubule networks made of calcium alginate that can be used as implants to create microchannels inside different types of hydrogels, including those with living cells embedded. A unique idea of this strategy is to de-convolve the “microchannel” fabrication step from the bulk gel generation where cells are embedded. By doing so, not only are we able to avoid cell injury that commonly happens during microchannel fabrication in cell-laden hydrogels but we also gain the freedom to create channels in any gel, including those that are not easy to microfabricate. The formation of such hydrogel tubule structures was based on in-situ generated copper ions from the surface of copper sacrificial templates. We can freely load the walls of the hydrogel tubules with particles and biomacromolecules during preparation, and control the thickness of the hydrogel tubule walls. The generated 3-D tubular microstructures are biocompatible, self-supportive, and can serve as flow delivery channels alone or be implanted in different types of gels to support diffusive mass transfer. Perfusion culture of HepG2 cells in agar gel was demonstrated with this system. With this method, the users will gain the benefit of vascular mass exchange in the bulk culture without changing their current gel-based culture protocol. The self-supporting character of the tubular network allows broader functionality and applications, including the ability to seed cells in coaxial layers both inside and outside the tubule walls to form structures that can better mimic blood vessels in both structure and functionality. The tubule networks are cost efficient and mass producible, and show great stability for storage; they have the ability to become commercial products for convenient use in different bio-applications.
SUPPLEMENTARY MATERIAL
See supplementary material for the structure of two alternatives (FDA-approved drug additives) of EDTA for extracting Fe3+ in replacing process, microscopic images of imbedding hydrogel tubular networks in agar gel and alginate gel, and the comparison between our method and previous conventional methods for fabrication of microchannels in cell-laden hydrogels.
ACKNOWLEDGMENTS
This work was supported by the Hong Kong RGC (No. #22200515), NSFC (No. 21505110), and Hong Kong Baptist University (FRG2/14-15/072 and SDF 03-17-096). The authors would like to thank Dr. Maria T. Dulay at Stanford University for her kind help in editing the language.
References
- 1. Simons M. and Ware J. A., Nat. Rev. Drug Discovery , 863 (2003). 10.1038/nrd1226 [DOI] [PubMed] [Google Scholar]
- 2. Justice B. A., Badr N. A., and Felder R. A., Drug Discovery Today , 102 (2009). 10.1016/j.drudis.2008.11.006 [DOI] [PubMed] [Google Scholar]
- 3. Liao S. W., Rawson J., Omori K., Ishiyama K., Mozhdehi D., Oancea A. R., Ito T., Guan Z., and Mullen Y., Biomaterials , 3984 (2013). 10.1016/j.biomaterials.2013.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Discher D. E., Janmey P., and Wang Y., Science , 1139 (2005). 10.1126/science.1116995 [DOI] [PubMed] [Google Scholar]
- 5. Murphy S. V. and Atala A., Nat. Biotechnol. , 773 (2014). 10.1038/nbt.2958 [DOI] [PubMed] [Google Scholar]
- 6. Lei Y. and Schaffer D. V., Proc. Natl. Acad. Sci. U.S.A. , 5039 (2013). 10.1073/pnas.1309408110 [DOI] [Google Scholar]
- 7. Koehler K. R., Mikosz A. M., Molosh A. I., Patel D., and Hashino E., Nature , 217 (2013). 10.1038/nature12298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Derda R., Laromaine A., Mammoto A., Tang S. K., Mammoto T., Ingber D. E., and Whitesides G. M., Proc. Natl. Acad. Sci. U.S.A. , 18457 (2009). 10.1073/pnas.0910666106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pampaloni F., Reynaud E. G., and Stelzer E. H., Nat. Rev. Mol. Cell Biol. , 839 (2007). 10.1038/nrm2236 [DOI] [PubMed] [Google Scholar]
- 10. Cushing M. C. and Anseth K. S., Science , 1133 (2007). 10.1126/science.1140171 [DOI] [PubMed] [Google Scholar]
- 11. Fennema E., Rivron N., Rouwkema J., van Blitterswijk C., and de Boer J., Trends Biotechnol. , 108 (2013). 10.1016/j.tibtech.2012.12.003 [DOI] [PubMed] [Google Scholar]
- 12. Kharka P. M., Kiick K. L., and Kloxin A. M., Chem. Soc. Rev. , 7335 (2013). 10.1039/C3CS60040H [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee J., Bhang S. H., Park H., Kim B.-S., and Lee K. Y., Pharm. Res. , 767 (2010). 10.1007/s11095-010-0067-0 [DOI] [PubMed] [Google Scholar]
- 14. Giavaresi G., Torricelli P., Fornasari P. M., Giardino R., Barbucci R., and Leone G., Biomaterials , 3001 (2005). 10.1016/j.biomaterials.2004.08.027 [DOI] [PubMed] [Google Scholar]
- 15. Zhao Y., Xu Y., Zhang B., Wu X., Xu F., Liang W., Du X., and Li R., Tissue Eng., Part C , 653 (2010). 10.1089/ten.tec.2009.0053 [DOI] [PubMed] [Google Scholar]
- 16. Wang X.-Y., Pei Y., Xie M., Jin Z.-H., Xiao Y.-S., Wang Y., Zhang L.-N., Li Y., and Huang W.-H., Lab Chip , 1178 (2015). 10.1039/C4LC00973H [DOI] [PubMed] [Google Scholar]
- 17. Dimitrov M., Lambov N., Shenkov S., Dosseva V., and Baranovski V. Y., Acta. Pharm. , 25 (2003). [PubMed] [Google Scholar]
- 18. Kumar P. T., Lakshmanan V. K., Anilkumar T. V., Ramya C., Reshmi P., Unnikrishnan A. G., Nair S. V., and Jayakumar R., ACS Appl. Mater. Interfaces , 2618 (2012). 10.1021/am300292v [DOI] [PubMed] [Google Scholar]
- 19. Betz J. F., Cheng Y., Tsao C.-Y., Zargar A., Wu H.-C., Luo X., Payne G. F., Bentley W. E., and Rubloff G. W., Lab Chip , 1854 (2013). 10.1039/c3lc50079a [DOI] [PubMed] [Google Scholar]
- 20. Martinez C. J., Kim J. W., Ye C., Ortiz I., Rowat A. C., Marquez M., and Weitz D., Macromol. Biosci. , 946 (2012). 10.1002/mabi.201100351 [DOI] [PubMed] [Google Scholar]
- 21. Xiong B., Ren K. N., Shu Y. W., Chen Y., Shen B., and Wu H. K., Adv. Mater. , 5525 (2014). 10.1002/adma.201305348 [DOI] [PubMed] [Google Scholar]
- 22. Ren K. N., Chen Y., and Wu H. K., Curr. Opin. Biotechnol. , 78 (2014). 10.1016/j.copbio.2013.09.004 [DOI] [PubMed] [Google Scholar]
- 23. Ren K. N., Zhou J. H., and Wu H. K., Acc. Chem. Res. , 2396 (2013). 10.1021/ar300314s [DOI] [PubMed] [Google Scholar]
- 24. Gillette B. M., Jensen J. A., Tang B., Yang G. J., Bazargan-Lari A., Zhong M., and Sia S. K., Nat. Mater. , 636 (2008). 10.1038/nmat2203 [DOI] [PubMed] [Google Scholar]
- 25. Bhatia S. N. and Ingber D. E., Nat. Biotechnol. , 760 (2014). 10.1038/nbt.2989 [DOI] [PubMed] [Google Scholar]
- 26. Choi N. W., Cabodi M., Held B., Gleghorn J. P., Bonassar L. J., and Stroock A. D., Nat. Mater. , 908 (2007). 10.1038/nmat2022 [DOI] [PubMed] [Google Scholar]
- 27. Zheng Y., Chen J., Craven M., Choi N. W., Totorica S., Diaz-Santana A., Kermani P., Hempstead B., Fischbach-Teschl C., López J. A., and Stroock A. D., Proc. Natl. Acad. Sci. U.S.A. , 9342 (2012). 10.1073/pnas.1201240109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meyerbröker N., Kriesche T., and Zharnikov M., ACS Appl. Mater. Interfaces , 2641 (2013). 10.1021/am400065f [DOI] [PubMed] [Google Scholar]
- 29. He J., Mao M., Liu Y., Shao J., Jin Z., and Li D., Adv. Healthcare Mater. , 1108 (2013). 10.1002/adhm.201200404 [DOI] [PubMed] [Google Scholar]
- 30. Golden A. P. and Tien J., Lab Chip , 720 (2007). 10.1039/b618409j [DOI] [PubMed] [Google Scholar]
- 31. Miller J. S., Stevens K. R., Yang M. T., Baker B. M., Nguyen D. H., Cohen D. M., Toro E., Chen A. A., Galie P. A., Yu X., Chaturvedi R., Bhatia S. N., and Chen C. S., Nat. Mater. , 768 (2012). 10.1038/nmat3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kolesky D. B., Truby R. L., Gladman A., Busbee T. A., Homan K. A., and Lewis J. A., Adv. Mater. , 3124 (2014). 10.1002/adma.201305506 [DOI] [PubMed] [Google Scholar]
- 33. Lee J. B., Wang X., Faley S., Baer B., Balikov D. A., Sung H. J., and Bellan L. M., Adv. Healthcare Mater. , 781 (2016). 10.1002/adhm.201500792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chan H. N., Shu Y., Tian Q., Chen Y., Chen Y., and Wu H., Mater. Horiz. , 309 (2016). 10.1039/C6MH00058D [DOI] [Google Scholar]
- 35. Hwang C. M., Sant S., Masaeli M., Kachouie N. N., Zamanian B., Lee S.-H., and Khademhosseini A., Biofabrication , 035003 (2010). 10.1088/1758-5082/2/3/035003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee K. H., Shin S. J., Park Y., and Lee S.-H., Small , 1264 (2009). 10.1002/smll.200801667 [DOI] [PubMed] [Google Scholar]
- 37. Su J., Zheng Y., and Wu H., Lab Chip , 996 (2009). 10.1039/B813518E [DOI] [PubMed] [Google Scholar]
- 38. Cheng Y., Yu Y., Fu F., Wang J., Shang L., Gu Z., and Zhao Y., ACS Appl. Mater. Interfaces , 1080 (2016). 10.1021/acsami.5b11445 [DOI] [PubMed] [Google Scholar]
- 39. Hidber P. C., Nealey P. F., Helbig W., and Whitesides G. M., Langmuir , 5209 (1996). 10.1021/la960238u [DOI] [Google Scholar]
- 40. Yang H., Love J. C., Arias F., and Whitesides G. M., Chem. Mater. , 1385 (2002). 10.1021/cm011513n [DOI] [Google Scholar]
- 41. Siegel A. C., Bruzewicz D. A., Weibel D. B., and Whitesides G. M., Adv. Mater. , 727 (2007). 10.1002/adma.200601787 [DOI] [Google Scholar]
- 42. Visser C. W., Pohl R., Sun C., Römer G.-W., Huisin't Veld B., and Lohse D., Adv. Mater. , 4087 (2015). 10.1002/adma.201501058 [DOI] [PubMed] [Google Scholar]
- 43. Zenou M., Sa'ar A., and Kotler Z., Small , 4082 (2015). 10.1002/smll.201500612 [DOI] [PubMed] [Google Scholar]
- 44. Zenou M., Sa'ar A., and Kotler Z., Sci. Rep. , 17265 (2015). 10.1038/srep17265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Choi Y. S., Hong S. R., Lee Y. M., Song K. W., Park M. H., and Nam Y. S., Biomaterials , 409 (1999). 10.1016/S0142-9612(98)00180-X [DOI] [PubMed] [Google Scholar]
- 46. Luckanagul J., Lee L. A., Nguyen Q. L., Sitasuwan P., Yang X., Shazly T., and Wang Q., Biomacromolecules , 3949 (2012). 10.1021/bm301180c [DOI] [PubMed] [Google Scholar]
- 47. Luo Y., Lode A., Wu C., Chang J., and Gelinsky M., ACS Appl. Mater. Interfaces , 6541 (2015). 10.1021/am508469h [DOI] [PubMed] [Google Scholar]
- 48. Du X. W., Zhou J., Shi J. F., and Xu B., Chem. Rev. , 13165 (2015). 10.1021/acs.chemrev.5b00299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Verhulsel M., Vignes M., Descroix S., Malaquin L., Vignjevic D. M., and Viovy J.-L., Biomaterials , 1816 (2014). 10.1016/j.biomaterials.2013.11.021 [DOI] [PubMed] [Google Scholar]
- 50. Wang X.-Y., Jin Z.-H., Gan B.-W., Lv S.-W., Xie M., and Huang W.-H., Lab Chip , 2709 (2014). 10.1039/c4lc00069b [DOI] [PubMed] [Google Scholar]
- 51. Oviedo C. and Rodríguez J., Quim. Nova , 901 (2003). 10.1590/S0100-40422003000600020 [DOI] [Google Scholar]
- 52. Mehlig J., Ind. Eng. Chem., Anal. Ed. , 533 (1941). 10.1021/i560096a006 [DOI] [Google Scholar]
- 53. J. F. Below, Jr. , Connick R. E., and Coppel C. P., J. Am. Chem. Soc. , 2961 (1958). 10.1021/ja01545a015 [DOI] [Google Scholar]
- 54. West E. R., Xu M., Woodruff T. K., and Shea L. D., Biomaterials , 4439 (2007). 10.1016/j.biomaterials.2007.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee K. Y. and Mooney D. J., Prog. Polym. Sci. , 106 (2012). 10.1016/j.progpolymsci.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Simpliciano C., Clark L., Asi B., Chu N., Mercado M., Diaz S., Goedert M., and Mobed-Miremadi M., J. Surf. Eng. Mater. Adv. Technol. , 1 (2013). 10.4236/jsemat.2013.34A1001 [DOI] [Google Scholar]
- 57. Lee K. Y., Peters M. C., and Mooney D. J., J. Controlled Release , 49 (2003). 10.1016/S0168-3659(02)00349-8 [DOI] [PubMed] [Google Scholar]
- 58. Klein J., Stock J., and Vorlop K., Eur. J. Appl. Microbiol. Biotechnol. , 86 (1983). 10.1007/BF00500829 [DOI] [Google Scholar]
- 59. Martinsen A., Storrø I., and Skjårk-Braek G., Biotechnol. Bioeng. , 186 (1992). 10.1002/bit.260390210 [DOI] [PubMed] [Google Scholar]
- 60. Zhou J. H., Ren K. N., Dai W., Zhao Y. H., Ryan D., and Wu H. K., Lab Chip , 2288 (2011). 10.1039/c0lc00466a [DOI] [PubMed] [Google Scholar]
- 61. Ren K. N., Banaei N., and Zare R. N., ACS Nano , 6031 (2013). 10.1021/nn401768s [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for the structure of two alternatives (FDA-approved drug additives) of EDTA for extracting Fe3+ in replacing process, microscopic images of imbedding hydrogel tubular networks in agar gel and alginate gel, and the comparison between our method and previous conventional methods for fabrication of microchannels in cell-laden hydrogels.