Abstract
Early embryonic arrest is one of the major causes of female infertility. However, because of difficulties in phenotypic evaluation, genetic determinants of human early embryonic arrest are largely unknown. With the development of assisted reproductive technology, the phenotype of early human embryonic arrest can now be carefully evaluated. Here, we describe a consanguineous family with a recessive inheritance pattern of female infertility characterized by recurrent early embryonic arrest in cycles of in vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI). We have identified a homozygous PADI6 nonsense mutation (c.1141C>T [p.Gln381∗]) that is responsible for the phenotype. Mutational analysis of PADI6 in a cohort of 36 individuals whose embryos displayed developmental arrest identified two affected individuals with compound-heterozygous mutations (c.2009_2010del [p.Glu670Glyfs∗48] and c.633T>A [p.His211Gln]; c.1618G>A [p.Gly540Arg] and c.970C>T [p.Gln324∗]). Immunostaining indicated a lack of PADI6 in affected individuals’ oocytes. In addition, the amount of phosphorylated RNA polymerase II and expression levels of seven genes involved in zygotic genome activation were reduced in the affected individuals’ embryos. This phenotype is consistent with Padi6 knockout mice. These findings deepen our understanding of the genetic basis of human early embryonic arrest, which has been a largely ignored Mendelian phenotype. Our findings lay the foundation for uncovering other genetic causes of infertility resulting from early embryonic arrest.
Main Text
It has been estimated that more than 48.5 million couples worldwide are infertile, which has a widespread global impact.1 Assisted reproductive technology (ART) is now routine in the treatment of infertile individuals. With the development of ART, the number of in vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI) cycles has increased every year, and now more than five million babies have been delivered by ART.2
In an IVF or ICSI cycle, the cumulus-oocyte complex is first isolated from the individual’s follicular fluid, and if ICSI is pursued, the cumulus cells surrounding the oocytes are further removed. The oocytes are then cultured in vitro. Fertilization occurs within 14–16 hr after the IVF or ICSI treatment. Normal fertilized oocytes continue to be cultured, and embryo quality is assessed 3 days after fertilization on the basis of cell number and morphology. Normal embryonic development is the key to establishing a successful pregnancy. It is estimated that about 40%–70% of the human embryos produced in IVF are viable embryos, whereas others arrest at different early stages of development.3 Developmentally arrested embryos are discarded, whereas viable embryos are cryopreserved, cultivated to blastocysts, or implanted directly into the individual’s uterus. IVF and ICSI cycles fail if all of an individual’s embryos are arrested at early stages of development (preimplantation embryonic lethality [MIM: 616814]). Some infertile individuals have this kind of phenotype after multiple failed IVF and ICSI attempts. It is commonly thought that much of human embryonic developmental potential is determined before fertilization by the content of the oocyte and is encoded by so-called maternal-effect genes.4, 5 Recently, a mutation in TLE6 (MIM: 612399), which encodes a protein participating in the subcortical maternal complex, was reported to cause the earliest known human embryonic lethality.6 However, until now, the genetic determinants that cause early human embryonic arrest have remained largely unknown.
In this study, we identified homozygous premature nonsense and compound-heterozygous mutations in PADI6 (MIM: 610363; GenBank: NM_207421.4) in infertile individuals with early embryonic arrest. We also investigated the expression level of PADI6 in individuals’ oocytes and the genes involved in zygotic genome activation (ZGA) in their embryos.
During IVF and ICSI, the morphology of embryos at different developmental stages was determined by light microscopy and time-lapse microscopy. Embryo grading and the definition of viable embryos followed the system developed by the Society for Assisted Reproductive Technology and two other references.7, 8, 9 In our study, metaphase I (MI) oocytes from donors were cultivated in vitro for maturation into control metaphase II (MII) oocytes. Control oocytes of good quality were then fixed and used for immunostaining. Control embryos were supernumerary embryos from donors undergoing successful IVF. All oocytes and embryos from control and affected individuals were obtained with written informed consent signed by the donor couples. This study was approved by the reproductive study ethics committee of Shanghai Ninth Hospital, the Shanghai Ji Ai Genetics and IVF Institute affiliated with the Obstetrics and Gynecology Hospital of Fudan University, and the ethics committee of the Medical College of Fudan University.
The proband (individual IV-5) in the consanguineous family (family 1) was 34 years old at examination, and she had received a diagnosis of primary infertility at the age of 32 years. Her parents are cousins and have six children (Figure 1A). She was married at the age of 27 years and has not conceived even though she engages in unprotected sexual intercourse twice per week on average. She has a normal menstrual cycle and has no endocrine disorders. Infertility-related examination did not reveal any physical abnormalities. She has two older sisters (44 and 46 years old, respectively), who were also diagnosed with primary infertility for unexplained reasons and who have never conceived despite several years of trying. Her two brothers and another sister with normal fertility have their own children. Thus, three of four sisters in this family suffer from primary infertility.
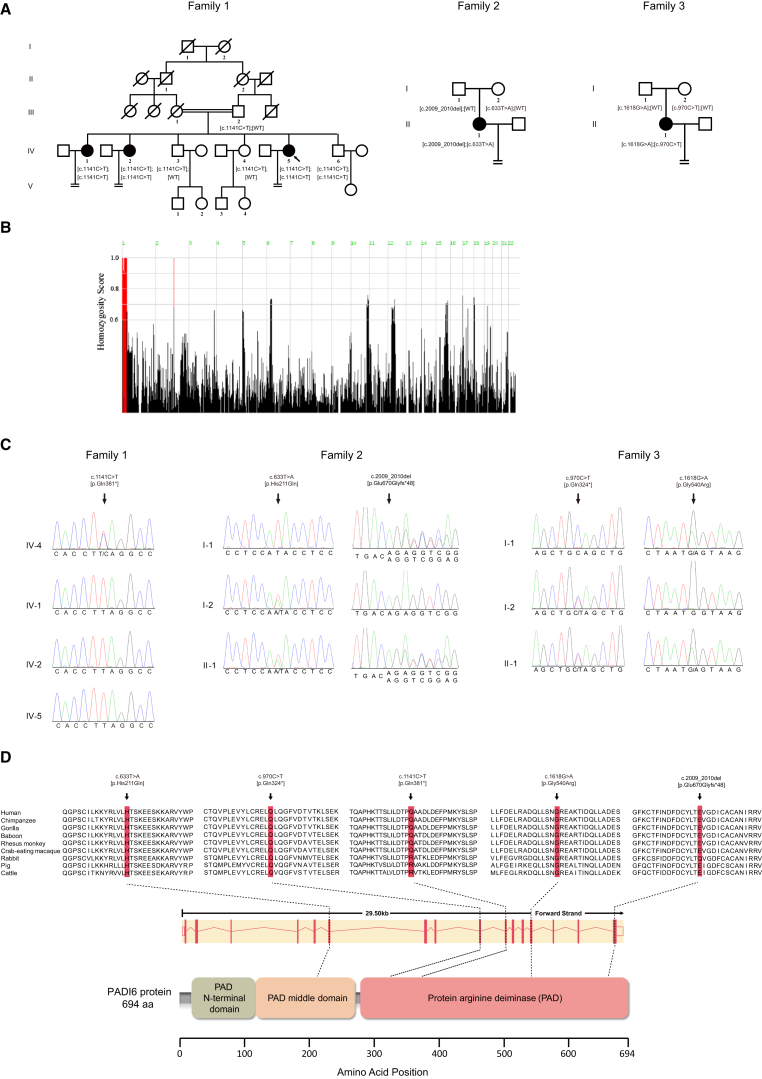
Figure 1.
Identification of Mutations in PADI6
(A) Pedigrees of the three families affected by infertility. Squares denote male family members, circles denote female members, solid symbols represent affected members, and slashes indicate deceased members. Equal signs indicate infertility, and the double line represents consanguinity. The arrow points to the proband.
(B) Homozygosity mapping of family 1; homozygous regions with the strongest signal are indicated in red.
(C) Sanger sequencing confirmation of PADI6 mutations in three families.
(D) The conservation of amino acid residues affected by mutations in different species, the structure of PADI6, and the known domains of the gene product. Exons are depicted as red vertical bars, introns are depicted as dashed lines, and the open rectangles at each end indicate noncoding exons.
Individual IV-5 underwent six failed IVF and ICSI attempts between June 2014 and January 2016. In the first attempt, which used a long-protocol treatment, nine oocytes (including seven MII oocytes) were retrieved. Six of these were successfully fertilized and underwent normal cleavage. After cultivation, only two viable embryos with many fragments (a grade IV 5-cell embryo and a grade III 6-cell embryo) remained on day 3. The other embryos were arrested at the 2- to 4-cell stage. The two viable embryos were implanted, but she did not become pregnant. In the second attempt, which used a short protocol, six MII oocytes out of eight oocytes were obtained, and five MII oocytes were successfully fertilized. However, no embryos were viable on day 3, and all of them were arrested at the 2- to 4-cell stage. In the third attempt, which used a prolonged protocol, four MII oocytes out of six oocytes were obtained. Three oocytes were successfully fertilized, and two embryos had normal cleavage. One embryo was arrested at the 2-cell stage, whereas the other was a viable 6-cell embryo on day 3. After extended cultivation, the viable embryo failed to reach the blastocyst stage and was discarded. In the fourth attempt, which used a mild stimulation protocol, seven MII oocytes were retrieved. Five were fertilized, but none were viable on day 3. All embryos were arrested at the 2- to 3-cell stage. In the fifth attempt, which used a mild stimulation protocol, four MII oocytes were retrieved, but only one oocyte was successfully fertilized and proceeded to divide to form an embryo. After cultivation, the embryo arrested at the 3-cell stage on day 3. In her last attempt, which used a mild stimulation protocol, five MII oocytes were retrieved, and two were fertilized. Unfortunately, however, none of them were viable on day 3 (Figure S1A).
The affected individual in family 2 was 34 years old at examination. She was diagnosed with primary infertility for unexplained reasons at 30 years of age and underwent three IVF and ICSI attempts. She had a total of 18 successfully fertilized oocytes, but most of them were arrested at the 1- to 2-cell stage. Although two embryos had five and six cells on day 3, they were arrested when they were cultivated in vitro until day 5.
The affected individual in family 3 was 35 years old at examination and was diagnosed with primary infertility for unexplained reasons at 30 years of age. She underwent a total of five IVF and ICSI attempts. Some embryos were uncleaved, but most were arrested between the 2- and 5-cell stages on day 3. Among the 22 embryos with normal cleavage, four embryos between the 5- and 7-cell stages on day 3 were implanted, but she failed to establish a pregnancy. Table 1 summarizes the specific IVF and ICSI clinical information of the affected individuals.
Table 1.
Oocyte and Embryo Characteristics of IVF and ICSI Attempts of Three Affected Individuals
| Individual | IVF and ICSI Attempts | Protocol | Total Oocytes | MII Oocytes | Fertilized Oocytes | Normal Cleavage Embryos | Viable Embryos on Day 3 | Outcomes |
|---|---|---|---|---|---|---|---|---|
| IV-5 in family 1 | first IVF | long | 9 | 7 | 6 | 6 | 2 | the two viable embryos (a grade IV 5-cell embryo and a grade III 6-cell embryo) were transferred into the uterus but failed to establish pregnancy; the remaining four embryos were arrested at the 2- to 4-cell stage on day 3 |
| second ICSI | short | 8 | 6 | 5 | 4 | 0 | all embryos were arrested at the 2- to 4-cell stage on day 3 | |
| third ICSI | prolonged | 6 | 4 | 3 | 2 | 1 | the only viable embryo proceeded to be cultivated but failed to form a blastocyst on day 5; the other embryo was arrested at the 2-cell stage on day 3 | |
| fourth ICSI | mild stimulation | 8 | 7 | 5 | 4 | 0 | all embryos were arrested at the 2- to 3-cell stage on day 3 | |
| fifth ICSI | mild stimulation | 6 | 4 | 1 | 1 | 0 | the embryo was arrested at the 3-cell stage on day 3 | |
| sixth ICSI | mild stimulation | 5 | 5 | 2 | 2 | 0 | all embryos were arrested at the 2- to 4-cell stage on day 3 | |
| II-1 in family 2 | first IVF | short | 6 | 6 | 3 | 3 | 1 | the only viable (6-cell) embryo on day 3 was further cultured but failed to form a blastocyst on day 5; the other embryos were arrested at the 2- and 5-cell stages |
| second ICSI | mild stimulation | 7 | 7 | 7 | 1 | 0 | the embryo was arrested at the 2-cell stage on day 3 | |
| third ICSI | mild stimulation | 12 | 8 | 8 | 6 | 0 | all embryos were arrested at the 3- to 4-cell stage on day 3 | |
| II-1 in family 3 | first ICSI | mild stimulation | 5 | 4 | 4 | 3 | 2 | one embryo was arrested at the 2-cell stage; two grade III 6-cell embryos were transferred into the uterus but failed to establish pregnancy |
| second ICSI | short | 7 | 5 | 5 | 3 | 0 | three embryos were arrested at the 4-cell stage on day 3 | |
| third ICSI | mild stimulation | 12 | 10 | 9 | 5 | 1 | four embryos were arrested at the 2- to 4-cell stage on day 3; one grade II 5-cell embryo was transferred into the uterus but failed to establish pregnancy | |
| fourth ICSI | mild stimulation | 13 | 11 | 11 | 9 | 1 | eight embryos were arrested at the 2- to 5-cell stage, whereas one viable embryo at the 7-cell stage was transferred into the uterus but failed to establish pregnancy | |
| fifth ICSI | mild stimulation | 7 | 2 | 2 | 2 | 0 | one embryo was arrested at the 2-cell stage on day 3, and another embryo was arrested at the 5-cell stage on day 3 |
Genomic DNA from all affected individuals and their family members was extracted from peripheral blood via standard methods. Exome capture and sequencing were performed in individuals III-2, IV-1, IV-2, IV-4, and IV-5 in family 1 (Figure 1A) by Agilent SureSelect whole-exome capture and Illumina sequencing technology. Because family 1 is consanguineous and has a recessive inheritance pattern, homozygous or compound-heterozygous variants were selected for further analysis. A homozygous or compound-heterozygous variant was considered to be a candidate mutation if it was (1) shared by all three affected individuals, (2) absent in other family members, and (3) not previously reported or reported to have a frequency below 0.1% (for homozygous variants) or 1% (for compound-heterozygous variants) in three public databases: 1000 Genomes, the NHLBI Exome Sequencing Project Exome Variant Server (EVS), and the Exome Aggregation Consortium (ExAC) Browser. Meanwhile, homozygosity mapping was carried out with the HomozygosityMapper.10 Homozygous regions greater than 2.0 Mb were considered to be candidate regions. Homozygous variants located in these regions were considered with priority. The variants were then validated by Sanger sequencing in the affected individuals and other family members. In addition to the consanguineous family, 36 additional infertile individuals with early embryonic arrest and 100 women with normal fertility were recruited from Shanghai Ninth Hospital and the Shanghai Ji Ai Genetics and IVF Institute.
Homozygosity mapping identified the strongest signal in regions on chromosome 1 in this family, demonstrating that homozygous mutations in a disease-associated gene might be located in these regions (Figure 1B). By performing whole-exome sequencing in the three affected and two healthy control individuals in this family and using the filtering criteria, we identified two homozygous variants. The first is a homozygous nonsense mutation (c.1141C>T [p.Gln381∗] [GenBank: NM_207421.4]) in exon 10 in PADI6, and it is predicted to result in a truncated protein. Alternatively, this mutation could also trigger nonsense-mediated decay. The second is a homozygous missense mutation (c.1043G>A [p.Arg348His] [GenBank: NM_004442.7]) in exon 5 in EPHB2 (MIM: 600997). Because EPHB2 is not expressed in all stages of human oocyte and early embryonic development (according to our in-house database), it was ruled out as a candidate gene in this pedigree. Thus, PADI6 p.Gln381∗ is the only variant responsible for the phenotype in this pedigree. This homozygous mutation was confirmed by Sanger sequencing (Figure 1C) and found to co-segregate with female infertility in this family. All three infertile individuals and their youngest brother (IV-6) carry the homozygous mutation, whereas the other family members with normal fertility have a heterozygous mutation. This mutation is not present in the three public databases (1000 Genomes, the EVS, or the ExAC Browser), and we did not find it in a sample of 100 healthy Chinese women with normal fertility.
In addition, we performed targeted resequencing of PADI6 in an additional 36 infertile individuals whose embryos displayed developmental arrest in multiple IVF and ICSI attempts. Compound-heterozygous mutations in PADI6 (GenBank: NM_207421.4) were identified in two affected individuals. A compound-heterozygous mutation in PADI6 in the affected individual in family 2 consisted of a 2 bp exon 16 deletion (c.2009_2010del) resulting in a prolonged protein (p.Glu670Glyfs∗48) and an exon 6 point mutation (c.633T>A) leading to a missense substitution (p.His211Gln). Meanwhile, another compound-heterozygous mutation in PADI6 in the affected individual in family 3 consisted of an exon 13 point mutation (c.1618G>A) leading to a missense substitution (p.Gly540Arg) and an exon 9 nonsense mutation (c.970C>T [p.Gln324∗]) predicted to result in a truncated protein. The nonsense mutation in PADI6 might also induce nonsense-mediated decay. In both cases, we found that the individuals had inherited one of the compound-heterozygous mutations from each of their parents (Figure 1C). We performed an in silico analysis of the c.633T>A (p.His211Gln) and c.G1618A (p.Gly540Arg) mutations by using PolyPhen-2, SIFT, and MutationTaster. However, the effects of both missense mutations could not be predicted by SIFT or MutationTaster because no corresponding transcripts were recorded. Although PolyPhen-2 shows that both missense mutations are benign, the fact that they change the amino acids from alkaline to polar or from polar to alkaline lead us to suggest that the mutations might have some impact on protein structural stability and that the effects might be even more severe in the background of another existing variant that eliminates the function of the protein (p.Glu670Glyfs∗48 in family 2 and p.Gln324∗ in family 3). PADI6 contains 16 exons encoding a 694 aa protein (Figure 1D). The nearby sequence of all PADI6 mutant residues is highly conserved in many vertebrate species (Figure 1D). Interestingly, it is not conserved in other species. This indicates that PADI6 might play a unique evolutionary and essential role in early embryonic development in advanced vertebrate species. The peptidylarginine deiminase (PADI) family includes calcium-dependent enzymes involved in the process of citrullination.11 The PADI family consists of five members (PADI1–PADI4 and PADI6), among which PADI1, PADI2, PADI4, and PADI6 are most highly localized in female reproductive tissues, indicating their roles in female fertility.11 Compared with other family members, PADI6 is uniquely localized in oocytes and early embryos.12
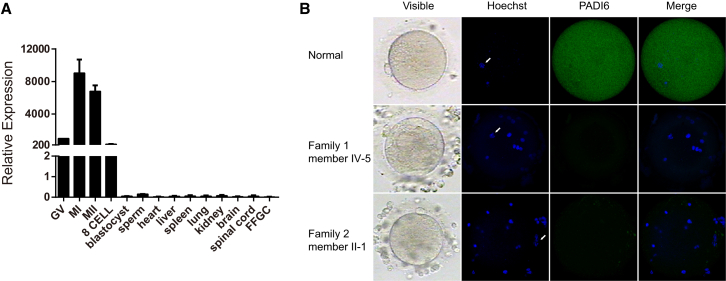
We examined PADI6 expression in human germinal vesicle (GV), MI, and MII oocytes, day 3 embryos, blastocysts, granulosa cells, mature sperm, and 3- to 4-month-old fetal tissues, including heart, liver, spleen, lung, kidney, brain, and spinal cord. Tissues were obtained from aborted fetuses provided by the Obstetrics and Gynecology Hospital of Fudan University after informed-consent forms were signed by the donor’s parents. Total RNA was extracted with an RNeasy Mini Kit (QIAGEN). Genomic DNA was depleted, and reverse transcription was performed with the PrimeScript RT Reagent Kit with the gDNA Eraser (Takara). The expression level of PADI6 was determined with the use of specific primers (Table S1) and was normalized to the expression level of an internal GAPDH (MIM: 138400) control. Real-time qPCRs were performed in triplicate on a 7900HT Fast Real-Time PCR System (Applied Biosystems). The relative expression level is k2 − ΔCt, where ΔCt = Ct (PADI6) − Ct (GAPDH), and k is a constant. As indicated in Figure 2A, PADI6 was highly expressed in oocytes and was weakly expressed in other somatic tissues and in sperm.
Figure 2.
Expression Pattern of PADI6 and Immunostaining of PADI6 in Oocytes
(A) The relative expression of PADI6 mRNA in developing human oocytes, early embryos, and several somatic tissues was measured by qRT-PCR and normalized to that of GAPDH mRNA (control). PADI6 was abundantly present in human germinal vesicle (GV), metaphase I (MI), metaphase II (MII), and 8-cell oocyte embryos. The bars show the mean of three separate measurements, and error bars denote SDs.
(B) The morphology and immunostaining results of a normal oocyte from a volunteer and the affected individuals’ oocytes. Oocytes from control and affected individuals (IV-5 in family 1 and II-1 in family 2) were fixed, immunolabeled, and examined by confocal microscopy with the use of antibodies against PADI6 (shown in green) and were counterstained with Hoechst 33342 (shown in blue) for DNA visualization. White arrows point to the nuclei of the oocytes. Hoeschst-stained foci and the oocyte nucleus are from the nuclei of cumulus cells around the oocytes.
We next performed an immunofluorescence analysis to detect PADI6 localization in the affected individuals’ oocytes. Control oocytes and the affected individuals’ oocytes were fixed and incubated with an anti-PADI6 antibody for the determination of PADI6 localization. The anti-PADI6 antibody was purchased from Abcam (ab16480, 1:9,000 dilution). Hoechst 33342 (1:700 dilution, BD Biosciences) was used to label DNA. Oocytes were mounted on glass slides and examined with a confocal laser-scanning microscope (Leica). In the control oocytes, the immunofluorescence signal was observed in the cytoplasm, whereas the signal was completely absent in the oocytes of affected individuals with either the homozygous nonsense mutation or compound-heterozygous mutations (Figure 2B). This indicates that these mutations completely eliminate PADI6 activity. It has been demonstrated that Padi6 knockout mice are infertile when developmental arrest occurs at the 2-cell stage.13 For the affected individual in family 2, in her last ICSI attempt, fertilized oocytes were monitored by a time-lapse microscope. Two embryos were uncleaved (Figure S1B), whereas three embryos had a high percentage of fragmentation and showed developmental arrest (Figure 3A). In all three affected individuals, most embryos arrested at the 2- to 5-cell stage (Table 1 and Figure S1B), indicating a slight difference in the phenotype of embryonic development between Padi6 knockout mice and affected individuals with PADI6 mutations.
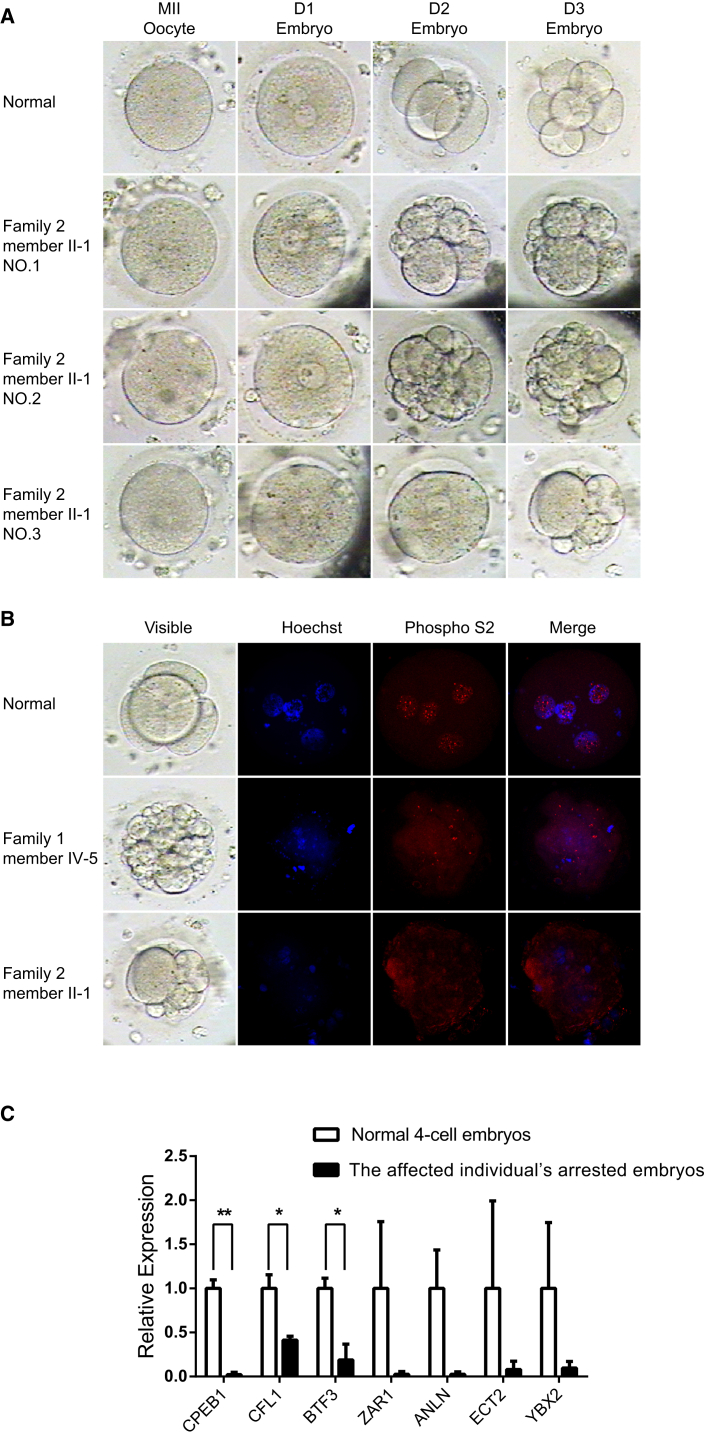
Figure 3.
Morphology, Immunostaining, and Expression Analysis of Genes Involved in Transcriptional Activation in Normal and Affected Individuals’ Embryos
(A) The morphology of a control oocyte embryo and three embryos from affected individual II-1 (family 2) was examined by light microscopy on days 1 (D1), 2 (D2), and 3 (D3) after fertilization.
(B) The immunostaining result of a normal 4-cell embryo and the arrested embryos from affected individuals IV-5 (family 1) and II-1 (family 2). Embryos from the control and affected individuals were fixed, immunolabeled, and examined by confocal microscopy with the use of antibodies against phosphorylated RNA polymerase II (Phospho S2, shown in red) and were counterstained with Hoechst 33342 (shown in blue) for DNA visualization.
(C) The relative expression of genes involved in transcriptional activation in normal 4-cell embryos and the arrested embryos from affected individual II-1 (family 2). Arrested embryos displayed significantly decreased expression of CPEB1, CFL1, and BTF3. Although expression of the rest of the genes was not significant as a result of the large variation in control embryos, expression was still obviously lower in arrested embryos than in normal embryos. The error bars denote SDs. ∗p < 0.05 and ∗∗p < 0.01.
It has been demonstrated that early embryonic arrest in Padi6 knockout mice is caused by disruption of ZGA.13 The levels of phosphorylated RNA polymerase II, a well-characterized marker for ZGA, are dramatically reduced in 2-cell embryos of Padi6 knockout mice, suggesting that ZGA is affected in these mice.13 We next detected the localization of phosphorylated RNA polymerase II in the affected individuals’ embryos. Embryos were fixed and incubated with mouse anti-RNA polymerase II CTD phosphoserine 2 antibody (Abcam ab24758, 1:500 dilution). Hoechst 33342 (1:700 dilution, BD Biosciences) was used to label DNA. As in the embryos of Padi6 knockout mice, the immunofluorescence signal of phosphorylated RNA polymerase II was significantly reduced in the affected individuals’ developmentally arrested embryos (Figure 3B), indicating defective ZGA.
It has been documented that several specific genes involved in transcriptional activation have significantly reduced expression levels in arrested embryos.14 Therefore, we further analyzed the expression of genes involved in cytokinesis (CFL1 [MIM: 601442], ECT2 [MIM: 600586], and ANLN [MIM: 616027]), transcription (BTF3 [MIM: 602542] and ZAR1 [MIM: 607520]), and RNA processing (YBX2 [MIM: 611447] and CPEB1 [MIM: 607342]) in both normal and arrested embryos. We pooled two arrested embryos from the affected individual (II-1 in family 2) and four normal control 4-cell embryos. RNA was extracted with an RNeasy Mini Kit (QIAGEN). Reverse transcription and amplification was performed with the SMARTer Universal Low Input RNA Kit (Clontech). The expression levels of genes involved in transcriptional activation were determined with the use of specific primers (Table S1) and were normalized to the expression level of an internal ACTB (MIM: 102630) control. Compared with normal embryos, arrested embryos displayed significantly decreased expression of CPEB1, CFL1, and BTF3, whereas an obvious trend of reduced expression in ZAR1, ANLN, ECT2, and YBX2 was observed in arrested embryos (Figure 3C). Thus, we conclude that ZGA is defective in affected individuals’ embryos with mutations in PADI6.
The fusion of two gametes generates a totipotent embryo that will undergo cleavage, differentiation, and development. During IVF and ICSI treatment, the embryos with good quality on day 3 or 5 will be transferred into the individual’s uterus to establish pregnancy. Several factors in the procedure can result in failed pregnancy; for example, aneuploidy is commonly known to result in failure of blastocyst formation, and implantation and is a primary factor in recurrent miscarriages.15, 16 In clinical IVF and ICSI cycles, some infertile individuals have embryos that are all arrested at earlier developmental stages. They therefore have no viable embryos on day 3, despite several IVF and ICSI attempts. The genetic determinant of this phenotype is largely unknown. Beginning with the transition from maternal to embryonic control, several genes regulate embryonic transcription after fertilization.17 This process involves transcriptional activation of the newly formed embryonic genome, which is referred to as ZGA. ZGA is a universal event during embryogenesis in all animal species and is critical for establishing totipotency and maintaining normal embryonic development.18 In mice, ZGA is observed in the early 1-cell stage and undergoes a major burst during the 2-cell stage,19 whereas in humans, ZGA occurs between the 4- and 8-cell stages.14, 20 The inhibition of ZGA in animals results in morphological defects in the embryo, arrest in early development, and ultimately sterility or decreased fecundity.18 Mutations or deficiencies in a short list of murine maternal genes have been shown to result in arrested development phenotypes with disrupted ZGA.5
It has been demonstrated that Padi6-deficient mice are infertile as a result of arrested embryonic development at the 2-cell stage.21 A lack of Padi6 in mice results in the disappearance of the oocyte cytoplasmic lattices that store the maternal contribution of ribosomes in the early embryo and causes defective ZGA in the 2-cell embryo, and this explains the infertility observed in these mice.13 In the present study, most of the embryos from the individuals with mutations in PADI6 led to arrest at the 2- to 5-cell stage. Moreover, the amount of phosphorylated RNA polymerase II and expression levels of a few genes involved in transcription activation were significantly reduced, suggesting impaired ZGA in their embryos. The minor differences in the arrested stage of embryos between humans and mice could be explained by the different time of ZGA in mice and humans.22, 23
PADI6 is thought to encode a protein participating in the subcortical maternal complex (SCMC), which is essential for embryonic progression past the 2-cell stage in mice.5 The SCMC also includes other protein members encoded by Mater,24 Floped,25 Tle6,25 and Filia.26 Recently, a TLE6 (MIM: 612399) mutation was shown to cause the earliest known human embryonic lethality by affecting progression of oocyte meiosis II.6 In this study, we found that PADI6 mutations caused early embryonic arrest by impairing ZGA. Thus, we provide further evidence that the SCMC is essential in human early embryonic development and that mutations in genes encoding SCMC members cause female infertility by early embryonic arrest through different molecular mechanisms.
Although a great number of Mendelian phenotypes have been extensively described and investigated, many more Mendelian conditions have yet to be recognized,27 and the application of IVF and ICSI has provided an unprecedented opportunity for discovering and investigating the genetic basis of previously unknown Mendelian phenotypes during spermatogenesis, oogenesis, and early embryonic development. Such discoveries not only contribute to the recognition of novel gene function, pathways, and genetic mechanisms in human reproduction but also help in the development of genetic diagnoses and suggest possible therapeutic targets for treating infertility.
In conclusion, we have identified PADI6 mutations as the cause of female infertility characterized by early embryonic arrest in multiple IVF and ICSI cycles in a family affected by a homozygous premature nonsense mutation and in two individuals with compound-heterozygous mutations in PADI6. It is worthwhile to conduct further screening of this gene and other potential genes in individuals with infertility caused by early embryonic arrest in IVF and ICSI. These findings not only deepen our understanding of the genetic basis of human early embryonic arrest but also provide the basis for genetic diagnoses of clinically infertile individuals with this phenotype and lay the foundation for uncovering other genetic causes of female infertility caused by early embryonic arrest.
Acknowledgments
This work was supported by the National Key R&D Program of China (2016YFC1000600), the National Basic Research Program of China (2015CB943300), and the National Natural Science Foundation of China (81270747, 81571501, and 81571506). We want to thank Dr. Scott Coonrod from Cornell University and Dr. Xueseng Zhang from Nan Jing Medical University for their help.
Published: August 18, 2016
Footnotes
Supplemental Data include one figure and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.06.024.
Contributor Information
Xiaoxi Sun, Email: xiaoxi_sun@aliyun.com.
Lei Wang, Email: wangleiwanglei@fudan.edu.cn.
Web Resources
1000 Genomes, http://browser.1000genomes.org
ExAC Browser, http://exac.broadinstitute.org/
MutationTaster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Sever, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2, http://www.genetics.bwh.harvard.edu/pph2/
Supplemental Data
References
- 1.Mascarenhas M.N., Flaxman S.R., Boerma T., Vanderpoel S., Stevens G.A. National, regional, and global trends in infertility prevalence since 1990: a systematic analysis of 277 health surveys. PLoS Med. 2012;9:e1001356. doi: 10.1371/journal.pmed.1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson G., Tabangin M., Macaluso M., De Mouzon J. The number of babies born globally after treatment with the assisted reproductive technologies (ART) Fertil. Steril. 2013;3:S42. [Google Scholar]
- 3.Gardner D.K., Lane M. Culture and selection of viable blastocysts: a feasible proposition for human IVF? Hum. Reprod. Update. 1997;3:367–382. doi: 10.1093/humupd/3.4.367. [DOI] [PubMed] [Google Scholar]
- 4.Yanez L.Z., Han J., Behr B.B., Reijo Pera R.A., Camarillo D.B. Human oocyte developmental potential is predicted by mechanical properties within hours after fertilization. Nat. Commun. 2016;7:10809. doi: 10.1038/ncomms10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L., Zheng P., Dean J. Maternal control of early mouse development. Development. 2010;137:859–870. doi: 10.1242/dev.039487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alazami A.M., Awad S.M., Coskun S., Al-Hassan S., Hijazi H., Abdulwahab F.M., Poizat C., Alkuraya F.S. TLE6 mutation causes the earliest known human embryonic lethality. Genome Biol. 2015;16:240. doi: 10.1186/s13059-015-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum. Reprod. 2011;26:1270–1283. doi: 10.1093/humrep/der037. [DOI] [PubMed] [Google Scholar]
- 8.Veek L., Rosenwaks Z. Parthenon Publishing Group; Carnforth, UK: 1999. An atlas of human gametes and conceptuses: an illustrated reference for assisted reproductive technology; pp. 46–51. [Google Scholar]
- 9.Hardarson T., Hanson C., Sjögren A., Lundin K. Human embryos with unevenly sized blastomeres have lower pregnancy and implantation rates: indications for aneuploidy and multinucleation. Hum. Reprod. 2001;16:313–318. doi: 10.1093/humrep/16.2.313. [DOI] [PubMed] [Google Scholar]
- 10.Seelow D., Schuelke M., Hildebrandt F., Nürnberg P. HomozygosityMapper--an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:W593–W599. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horibata S., Coonrod S.A., Cherrington B.D. Role for peptidylarginine deiminase enzymes in disease and female reproduction. J. Reprod. Dev. 2012;58:274–282. doi: 10.1262/jrd.2011-040. [DOI] [PubMed] [Google Scholar]
- 12.Wright P.W., Bolling L.C., Calvert M.E., Sarmento O.F., Berkeley E.V., Shea M.C., Hao Z., Jayes F.C., Bush L.A., Shetty J. ePAD, an oocyte and early embryo-abundant peptidylarginine deiminase-like protein that localizes to egg cytoplasmic sheets. Dev. Biol. 2003;256:73–88. doi: 10.1016/s0012-1606(02)00126-4. [DOI] [PubMed] [Google Scholar]
- 13.Yurttas P., Vitale A.M., Fitzhenry R.J., Cohen-Gould L., Wu W., Gossen J.A., Coonrod S.A. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development. 2008;135:2627–2636. doi: 10.1242/dev.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong C.C., Loewke K.E., Bossert N.L., Behr B., De Jonge C.J., Baer T.M., Reijo Pera R.A. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat. Biotechnol. 2010;28:1115–1121. doi: 10.1038/nbt.1686. [DOI] [PubMed] [Google Scholar]
- 15.Vanneste E., Voet T., Le Caignec C., Ampe M., Konings P., Melotte C., Debrock S., Amyere M., Vikkula M., Schuit F. Chromosome instability is common in human cleavage-stage embryos. Nat. Med. 2009;15:577–583. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- 16.Harper J., Sermon K., Geraedts J., Vesela K., Harton G., Thornhill A., Pehlivan T., Fiorentino F., SenGupta S., de Die-Smulders C. What next for preimplantation genetic screening? Hum. Reprod. 2008;23:478–480. doi: 10.1093/humrep/dem424. [DOI] [PubMed] [Google Scholar]
- 17.Li L., Lu X., Dean J. The maternal to zygotic transition in mammals. Mol. Aspects Med. 2013;34:919–938. doi: 10.1016/j.mam.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee M.T., Bonneau A.R., Giraldez A.J. Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 2014;30:581–613. doi: 10.1146/annurev-cellbio-100913-013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schultz R.M. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum. Reprod. Update. 2002;8:323–331. doi: 10.1093/humupd/8.4.323. [DOI] [PubMed] [Google Scholar]
- 20.Niakan K.K., Han J., Pedersen R.A., Simon C., Pera R.A. Human pre-implantation embryo development. Development. 2012;139:829–841. doi: 10.1242/dev.060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esposito G., Vitale A.M., Leijten F.P., Strik A.M., Koonen-Reemst A.M., Yurttas P., Robben T.J., Coonrod S., Gossen J.A. Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Mol. Cell. Endocrinol. 2007;273:25–31. doi: 10.1016/j.mce.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Warner C.M., Versteegh L.R. In vivo and in vitro effect of α-amanitin on preimplantation mouse embryo RNA polymerase. Nature. 1974;248:678–680. doi: 10.1038/248678a0. [DOI] [PubMed] [Google Scholar]
- 23.Braude P., Bolton V., Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature. 1988;332:459–461. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]
- 24.Tong Z.-B., Gold L., Pfeifer K.E., Dorward H., Lee E., Bondy C.A., Dean J., Nelson L.M. Mater, a maternal effect gene required for early embryonic development in mice. Nat. Genet. 2000;26:267–268. doi: 10.1038/81547. [DOI] [PubMed] [Google Scholar]
- 25.Li L., Baibakov B., Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev. Cell. 2008;15:416–425. doi: 10.1016/j.devcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng P., Dean J. Role of Filia, a maternal effect gene, in maintaining euploidy during cleavage-stage mouse embryogenesis. Proc. Natl. Acad. Sci. USA. 2009;106:7473–7478. doi: 10.1073/pnas.0900519106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T., Centers for Mendelian Genomics The genetic basis of Mendelian phenotypes: discoveries, challenges, and opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.