Abstract
Background
Plasmodium knowlesi and Plasmodium cynomolgi are two malaria parasites naturally transmissible between humans and wild macaque through mosquito vectors, while Plasmodium inui can be experimentally transmitted from macaques to humans. One of their major natural hosts, the long-tailed macaque (Macaca fascicularis), is host to two other species of Plasmodium (Plasmodium fieldi and Plasmodium coatneyi) and is widely distributed in Southeast Asia. This study aims to determine the distribution of wild macaques infected with malarial parasites by examining samples derived from seven populations in five countries across Southeast Asia.
Methods
Plasmodium knowlesi, P. cynomolgi, P. coatneyi, P. inui and P. fieldi, were detected using nested PCR assays in DNA samples from 276 wild-caught long-tailed macaques. These samples had been derived from macaques captured at seven locations, two each in the Philippines (n = 68) and Indonesia (n = 70), and one each in Cambodia (n = 54), Singapore (n = 40) and Laos (n = 44). The results were compared with previous studies of malaria parasites in long-tailed macaques from other locations in Southeast Asia. Fisher exact test and Chi square test were used to examine the geographic bias of the distribution of Plasmodium species in the macaque populations.
Results
Out of 276 samples tested, 177 were Plasmodium-positive, with P. cynomolgi being the most common and widely distributed among all long-tailed macaque populations (53.3 %) and occurring in all populations examined, followed by P. coatneyi (20.4 %), P. inui (12.3 %), P. fieldi (3.4 %) and P. knowlesi (0.4 %). One P. knowlesi infection was detected in a macaque from Laos, representing the first documented case of P. knowlesi in wildlife in Laos. Chi square test showed three of the five parasites (P. knowlesi, P. coatneyi, P. cynomolgi) with significant bias in prevalence towards macaques from Malaysian Borneo, Cambodia, and Southern Sumatra, respectively.
Conclusions
The prevalence of malaria parasites, including those that are transmissible to humans, varied among all sampled regional populations of long-tailed macaques in Southeast Asia. The new discovery of P. knowlesi infection in Laos, and the high prevalence of P. cynomolgi infections in wild macaques in general, indicate the strong need of public advocacy in related countries.
Keywords: Plasmodium knowlesi, Plasmodium cynomolgi, Macaca fascicularis, Geographic distribution, Biased infection rate
Background
Most malaria parasites are considered to be host-specific, in that most of them infect only one host species, though a single host can be infected by multiple Plasmodium species [1]. Humans are the natural hosts for four Plasmodium species: Plasmodium falciparum, Plasmodium malariae, Plasmodium vivax, and Plasmodium ovale [2]. Since 1930, three other species of Plasmodium that typically infect wild macaques in Southeast Asia had been known to be transmissible to humans. This was first demonstrated by passage with blood infected with Plasmodium knowlesi [3], and later with Plasmodium cynomolgi [4] and Plasmodium inui [5]. Furthermore, accidental infections of humans in research laboratories with P. cynomolgi through mosquito bites were reported in 1960 [4, 6] and 1980 [7], and also human infections through mosquito-transmission experiments in the 1960s with P. cynomolgi [4, 6, 8] and P. inui [9].
In 1965, the first case of a human naturally infected with knowlesi malaria was reported [10] and human infections in nature were considered extremely rare until in 2004, when 120 of 208 malaria patients at Kapit Hospital in Sarawak, Malaysian Borneo were found to be infected with P. knowlesi using nested PCR assays [11]. Additional human knowlesi malaria cases were subsequently reported throughout Southeast Asian countries, including Thailand, Singapore, Peninsular Malaysia, Cambodia, Vietnam, Indonesia and the Philippines [12], and in the Nicabar and Andaman Islands of India [13]. In 2014, another malaria parasite of long-tailed macaques, P. cynomolgi, was reported to have naturally infected a woman in Peninsular Malaysia [14]. These recent discoveries of humans infected with what were previously thought to be non-human malarias implied the potential larger distribution of human cases that may have not yet been confirmed and highlighted the potentially high risks of more zoonotic malaria infections.
The natural hosts of P. knowlesi and P. cynomolgi are long-tailed macaques (Macaca fascicularis) and pig-tailed macaques (Macaca nemestrina) [2]. Long-tailed macaques are widely distributed in Southeast Asia and exhibit the third most widespread geographical distribution among all primates after humans and rhesus macaques (Macaca mulatta) [15]. Their natural range extends southward and eastward from Southeastern Bangladesh and Burma, through the Southern part of the Indochinese Peninsula (Thailand, Cambodia, Laos, and Vietnam), and into the Malaysian Peninsula including Singapore, and the islands of Sumatra, Borneo, Java, and the Philippines [16].
Other than P. knowlesi and P. cynomolgi, long-tailed macaques are also natural hosts of three other Plasmodium species, namely P. coatneyi, P. fieldi and P. inui [11, 17]. The prevalence of these three parasites and their distribution in regional long-tailed macaque populations, however, had only been characterized for macaques in Malaysia and Singapore [18–20]. Other studies have examined blood samples from macaques by either molecular detection assays solely for P. knowlesi in Thailand, Peninsular Malaysia and Indonesia [21–25], or by sequencing of the Plasmodium mitochondrial genome in Malaysian Borneo [26]. A thorough investigation of Plasmodium infections in long-tailed macaques is necessary to determine their potential as a public health threat. The present study utilizes sensitive molecular detection methods, and characterizes the prevalence of five Plasmodium species in seven long-tailed macaque populations across the geographic range of these non-human primates in Southeast Asia.
Methods
DNA samples
DNA samples of 276 wild-caught long-tailed macaques from seven populations in Southeast Asian countries were employed for this study. These samples had been collected for genetic studies that were reported elsewhere [27–29]. These included 50 samples from Southern Sumatra, 54 samples from Cambodia, 40 samples from Zamboanga in Southern Philippines, 28 samples from Batangas in Northern Philippines, 40 samples from Singapore, 20 samples from Bintan island in Indonesia (near Singapore), and 44 samples from Laos. These DNA samples were obtained from single time point blood collection and were stored at −20 °C in the Molecular Anthropology Laboratory at the University of California, Davis. DNA samples were obtained from long-tailed macaques that were all originally wild-caught. A map illustrating the geographic distribution and the origins of these samples is shown as Fig. 1.
Fig. 1.

Locations of origin of capture of long-tailed macaques
Nested PCR assays
Malaria parasites have distinct small subunit ribosomal RNA (SSU rRNA) genes that are developmentally regulated [30]. Nested PCR assays targeting the distinct SSU rRNA genes of Plasmodium species were used to amplify DNA from macaque blood samples [31]. Samples were initially examined using Plasmodium-specific primers and those that were positive were subsequently examined with species-specific primers. The PCR parameters and primers used to detect Plasmodium, P. knowlesi, P. cynomolgi, P. inui, P. fieldi and P. coatneyi have been described previously [21, 32]. Plasmids containing the SSU rRNA genes of P. knowlesi, P. cynomolgi, P. inui, P. fieldi and P. coatneyi were used as positive controls and were provided by the Malaria Research Centre at Universiti Malaysia Sarawak [19]. The amplification products were analyzed by gel electrophoresis and data of Plasmodium infections of the samples were compared with published data in Plasmodium infections from wild long-tailed macaques in Sarawak, Malaysian Borneo, and multiple sites from Peninsular Malaysia, Thailand, Singapore and Indonesia [19–26].
Sequencing of PCR amplified fragment and sequence analysis
One of the PCR amplified fragments, generated from a macaque from Laos with P. knowlesi-specific primers, was sequenced using the Sanger dideoxy sequencing method [33] and analyzed on the ABI 3730 Capillary Electrophoresis Genetic Analyzer. The DNA sequences were aligned by Clustal W and analysed using DNASTAR software (Lasergene, USA) and DnaSP version 4.10.8. The phylogenetic analysis was inferred by the Neighbor-joining method [34] with Kimura 2-parameter, including transitions and transversions, using MEGA version 3.1. The reliability of the internal nodes of the tree was assessed by the bootstrap method after 1000 replicates.
Analysis of prevalence of malaria parasites
To determine whether the prevalence of each Plasmodium species is geographically biased, Fisher exact test was performed on software R (version 3.2.4) on the distribution table. The null hypothesis under the Fisher exact test is, “prevalence and region are independent, i.e., prevalence of malaria parasite does not change as geographic location changes”. For the test, 107 random tables with the same column and row totals as the observed data, assuming row and column independence, were generated. The P value cited is the sum of the probabilities of all generated tables that have individual probabilities smaller than those in the observed table. To compare each observed value with its expected value under null hypothesis, Chi square tests were performed in R on the observed table to generate each expected value under the null hypothesis.
Results
Out of 276 long-tailed macaques screened for Plasmodium DNA using nested PCR assays, 177 were found to be positive (Table 1). Multiple infections with different species were observed in each of 32 samples. Among the positive samples, the frequency of single, double and triple infections was 81.9, 17.0, and 1.1 %, respectively. Plasmodium cynomolgi was detected in at least one macaque from all seven sampled locations, showing the highest prevalence (53.3 %) and most widespread distribution among macaques across Southeast Asia, followed by P. coatneyi (20.4 %), P. inui (12.3 %), P. fieldi (3.4 %), and P. knowlesi (0.4 %). The prevalence of each parasite species among the macaque populations varied by location (Table 2; Fig. 1), and these were compared with previous published studies conducted in Malaysian Borneo [21, 28], Thailand [24, 25], Peninsular Malaysia [20, 27], Singapore [22], and Indonesia [26].
Table 1.
Malaria parasite infections in long-tailed macaques by location
| Infection | Plasmodium species | Number of long-tailed macaques infected | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Laos | Singapore | Vanny, Cambodia | Batangas, Philippines | Zamboanga, Philippines | Southern Sumatra | Bintan Island | Total | ||
| Single | Pk | 1 | 1 | ||||||
| Pcy | 28 | 23 | 24 | 2 | 1 | 39 | 9 | 126 | |
| Pct | 7 | 1 | 1 | 9 | |||||
| Pin | 2 | 1 | 2 | 1 | 2 | 8 | |||
| Pfi | 1 | 1 | |||||||
| Double | Pcy, Pin | 1 | 2 | 9 | 2 | 15 | |||
| Pin, Pfi | 1 | 1 | |||||||
| Pct, Pfi | 1 | 1 | 2 | ||||||
| Pcy, Pfi | 1 | 1 | 2 | ||||||
| Pct, Pcy | 1 | 1 | 1 | ||||||
| Pin, Pct | 9 | 9 | |||||||
| Triple | Pcy, Pin, Pfi | 1 | 1 | 2 | |||||
| Total Plasmodium-positive | 30 | 31 | 44 | 3 | 4 | 49 | 16 | 177 (64.1 %) | |
| Total Plasmodium-negative | 14 | 9 | 10 | 25 | 36 | 1 | 4 | 99 (35.9 %) | |
| Total number of macaques | 44 | 40 | 54 | 28 | 40 | 50 | 20 | 276 (100 %) | |
Pk P. knowlesi, Pcy P. cynomolgi, Pin P. inui, Pct P. coatneyi, Pfi P. fieldi
Table 2.
Comparison of prevalence of species of Plasmodium in long-tailed macaques
| Sample origin | Total number | No. Pk (χ2 Exp.) | % Pk | No. Pcy (χ2 Exp.) | % Pcy | No. Pin (χ2 Exp.) | % Pin | No. Pct (χ2 Exp.) | % Pct | No. Pfi (χ2 Exp.) | % Pfi | Year |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Batangas, Philippines | 28 | 0 (1) | 0 | 3 (2) | 10.7 | 0 (1) | 0 | 1 (1) | 3.6 | 0 (0) | 0 | 2012 |
| Zamboanga, Philippines | 40 | 0 (1) | 0 | 1 (2) | 2.5 | 2 (1) | 5 | 1 (1) | 2.5 | 0 (0) | 0 | 2012 |
| Vanny, Cambodia | 54 | 0 (12) | 0 | 27 (22) | 50 | 12 (12) | 22.2 | 16 (8) | 29.6 | 1 (2) | 1.9 | 2011 |
| Southern Sumatra | 50 | 0 (13) | 0 | 48 (23) | 96 | 10 (12) | 20 | 0 (8) | 0 | 0 (2) | 0 | 2010 |
| Guidong/Laos | 44 | 1 (7) | 2.3 | 29 (12) | 65.9 | 0 (6) | 0 | 0 (4) | 0 | 1 (1) | 2.3 | 2013 |
| Bintan Island, Indonesia | 20 | 0 (5) | 0 | 13 (8) | 65 | 5 (4) | 25 | 1 (3) | 5 | 2 (1) | 10 | 2007 |
| Singapore | 40 | 0 (8) | 0 | 26 (15) | 65 | 5 (8) | 12.5 | 2 (5) | 5 | 4 (1) | 10 | 2007 |
| Total | 276 | 1 | 0.4 | 147 | 53.3 | 34 | 12.3 | 20.4 | 12 | 3.4 | ||
| Sarawak, Malaysian Borneo [21] | 82 | 71 (54) | 86.6 | 52 (99) | 63.4 | 69 (51) | 84.1 | 52 (35) | 63.4 | 4 (9) | 4.9 | 2011 |
| Sabah, Malaysian Borneo [26] | 26 | 4 (4) | 15.4 | 3 (7) | 11.5 | 8 (4) | 30.8 | 1 (2) | 3.8 | 1 (1) | 3.8 | 2014 |
| Singapore [22] | 157 | 45 (21) | 28.7 | 40 (39) | 25.5 | 1 (20) | 0.6 | 2 (14) | 1.3 | 11 (4) | 7 | 2011 |
| Hulu Selangor, Malaysia [20] | 70 | 21 (17) | 30.0 | 18 (31) | 25.7 | 23 (16) | 32.9 | 16 (11) | 22.9 | 1 (3) | 1.4 | 2015 |
| Kuala Lumpur and Selangor, Malaysia [27] | 41 | 0 | 0 | − | − | − | − | − | − | − | − | 2008 |
| Kuala Lipis, Pahang, Malaysia [27] | 75 | 10 | 13.3 | − | − | − | − | − | − | − | − | 2008 |
| Thailand [25] | 195 | 1 | 0.5 | − | − | 7 | 3.6 | 3 | 1.5 % | 0 | 0 | 2010 |
| Thailand [24] | 105 | 0 | 0 | − | − | − | − | − | − | − | − | 2008 |
| Indonesia [26] | N/A | N/A | 3.2 % | − | − | − | − | − | − | − | − | 2007 |
The figures in brackets are expected number of infections estimated by Chi square test under the assumption of “Plasmodium parasite prevalence independent from the geographic location”
Pk P. knowlesi, Pcy P. cynomolgi, Pin P. inui, Pct P. coatneyi, Pfi P. fieldi, No number, χ 2 Exp. Chi square test expected values, − not done, N/A not available
The P value for the Fisher exact test is 10−7, indicating that the observed geographical distribution of Plasmodium infection is extremely unlikely under the null hypothesis; i.e., the prevalence of each species of Plasmodium exhibits bias among different geographic locations.
According to the comparison between observed number of infected macaques with what is expected by the Chi square test under the null hypothesis (Table 2), P. inui and P. fieldi both exhibited relatively uniform distributions over geographical locations. The prevalence of P. knowlesi and P. cynomolgi in macaques from Sarawak is higher and lower, respectively than all other sites. The prevalence of P. knowlesi in macaques from all other locations is lower than expected. A much higher prevalence of P. cynomolgi in the macaque samples from Southern Sumatra and Singapore was also observed, along with relatively higher prevalence of P. coatneyi in macaques from Sarawak and Cambodia, and lower prevalence of P. coatneyi in macaques from Sumatra and Laos. Macaques from Bintan Island and Singapore, two locations in close geographical proximity, showed close similarities in the infection profiles. In addition, the prevalence for all Plasmodium species were unusually low in the macaques from the Philippines compared to those from other geographic regions.
Only one sample from Laos was positive for P. knowlesi. The nested PCR assay with P. knowlesi-specific primers gave a similar result for this sample when the assay was repeated, using new nest 1 and nest 2 PCR amplifications. The sequence of the SSU rRNA gene fragment amplified in the nested PCR assay was aligned and compared with Plasmodium SSU rRNA sequences in GenBank [21]. It was identical with the sequence of the S-type SSU rRNA gene of P. knowlesi isolate KH33, and 99.6 % similar with sequences from P. knowlesi isolates KH50, KH96, KH107, KH43 and KH 131. Phylogenetic analysis confirmed that the macaque from Laos was infected with P. knowlesi (Fig. 2).
Fig. 2.
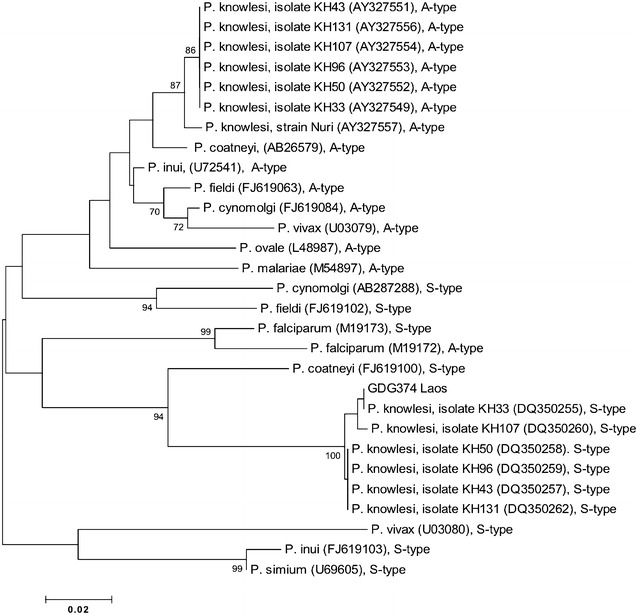
Phylogenetic tree based on the SSU rRNA genes of Plasmodium species produced by the Neighbor-joining method. Accession numbers for each species of Plasmodium are stated within brackets; Only bootstrap values above 70 % are shown; ‘S-type’ = sexually transcribed form and ‘A-type’ = asexually transcribed form
Discussion
Long-tailed macaques are widely distributed in both suburban areas and dense forests in Southeast Asia [15]. Historically and culturally, long-tailed macaques particularly have an extensive interface with humans, especially locals who share spatial overlap with them and also tourists who come in contact with them. The close contact and geographic range overlap between humans and long-tailed macaques introduce to both species a situation of mutual disease transmission. Humans in Southeast Asian countries who are naturally exposed to long-tailed macaques are at potential risks of being infected with a number of simian pathogens, including simian T cell lymphotropic viruses (STLV), simian retrovirus (SRV), Macacine herpesvirus 1 (B virus), and the zoonotic malarias [35–38]. Understanding the distribution patterns of macaques that carry malaria parasites especially the ones transmissible to humans facilitates an effective assessment of this human health threat.
The present study demonstrates that two macaque malaria parasites naturally transmissible to humans, P. cynomolgi, and P. inui, exist among all the 7 sampled regional populations of wild long-tailed macaques. Specifically, P. cynomolgi showed distinctively and uniformly high prevalence in the macaques, except for the two populations from the Philippines. The low prevalence of Plasmodium in macaques from Zamboanga in the southern Philippines and Batangas in the northern Philippines does not suggest that host genetic differences are responsible, because long-tailed macaques in the latter geographic region are assigned to a uniquely different subspecies (M. f. philippensis) than those (M. f. fascicularis) inhabiting all other regions studied [29]. It is only recently confirmed that P. cynomolgi can also be transmitted to humans naturally and thus represents a potential new zoonotic malaria [14]. The observation of the high prevalence of P. cynomolgi in macaques from this study indicates the strong need of public advocacy in the affected areas.
A complex nature of Plasmodium infections in natural hosts was noted in this study. Among the 177 malaria-positive macaques, 18.1 % were each found to have multiple (double and triple) Plasmodium species infections. More multiple infections, including co-infections with four or five species of Plasmodium in each long-tailed macaque were reported in other studies [19, 20]. This complex nature of infections represents challenges for accurate identification by microscopy, as well as a potential underestimation of prevalence and distribution of Plasmodium parasites in macaques and other primates.
In previous studies, it was shown that in peri-domestic and urban areas of Singapore, Peninsular Malaysia, and Thailand, the parasitaemia and the presence of Plasmodium infections in free-ranging macaques were low or zero, hypothetically due to the absence or limited presence of known competent vectors of malaria parasites [17, 25, 39]. A comparison with multiple studies using molecular methods to detect Plasmodium infections in macaques from other locations indicates that the prevalence of malaria and of the different species of Plasmodium vary according to the sampling site, even on a small island like Singapore [18–20, 22–26]. Comparing the Singaporean samples screened from this study, which were collected from 2007, with the Singaporean samples reported from 2011 [20], there was a decrease in the prevalence of P. knowlesi infection. The exact location from where the macaques were trapped in Singapore is not known for the current study and the differences in the prevalence of P. knowlesi between the two studies could be due to different sampling locations in Singapore. In addition, in a study conducted in Sabah, Malaysian Borneo, Fornace et al. [40] have shown spatial heterogeneity in P. knowlesi incidence depending on the land type. They found that the incidence of P. knowlesi varied between village, forests, and historical forest loss areas, which further indicates that environmental changes influence the transmission of P. knowlesi. More recently, Moyes and co-workers used a newly developed incidence–environment assessment model to investigate the distributions of reservoir hosts and Anopheline vectors of P. knowlesi in Southeast Asia and assessed their relationships with the environment [41]. Their analyses showed that the predicted distribution of both the long-tailed macaques and vectors of knowlesi malaria encompassed a wide range of habitats, most significantly in the northern part of their study area which included Myanmar, Thailand, Laos, Cambodia and Vietnam. These recent studies highlight that environmental variables possibly are the key driving force in determining the distribution of Plasmodium parasites.
It has been noted that the parasitaemia of malaria parasites in a human or non-human primate host could change on a daily basis [2]. Because the DNA samples used in this study were from single time points, further studies are warranted to investigate and follow up the change of infections over time in macaques.
The detection of P. knowlesi infection from a long-tailed macaque from Laos is the first report of P. knowlesi infection in either humans or macaques in that country. This finding should encourage further investigations of P. knowlesi using molecular detection methods among macaques and humans in Laos.
Conclusions
The prevalence of malaria parasites that naturally infect long-tailed macaques, including those that are transmissible to humans, varies among regional populations of wild long-tailed macaques in Southeast Asia. Plasmodium cynomolgi, which can be naturally acquired by humans, exists in all wild long-tailed macaque populations studied with varying prevalence. This is the first report of a natural infection of P. knowlesi in a macaque from Laos. The presence of malaria parasites underscores the risk of potential zoonotic infections in the local human populations.
Authors’ contributions
XZ, BS and DGS conceived the study, interpreted the data, and wrote the paper. XZ conducted laboratory experiments, and performed data analysis. LFQ assisted XZ and conducted laboratory experiments. KAK provided plasmid control samples, detailed laboratory protocols and analyzed the data. JV provided samples from Singapore and Bintan Island. HD provided samples from Laos/Guidong. PH provided all remaining samples. All authors read and approved the final manuscript.
Acknowledgements
The authors thank Dr. Mark Grote at Department of Anthropology at University of California Davis for excellent statistical analysis support and Joshua Noyer for map design. We also thank support from the DNA Sequencing Facility at UC Davis.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The DNA sequence data obtained for this research are available in GenBank with Accession number: KU693334.
Consent for publication
All authors have seen and approved the manuscript and its contents, and agreed to submission for publication.
Ethics approval and consent to participate
Approval for this study was provided by the Institutional Animal Care and Use Committee (IACUC: Protocol #18684) of University of California Davis.
Fundings
This research was financially supported by NIH Grants RR005090 and RR025781, and a grant from University Malaysia Sarawak (01[TD03]/1003/2013[01]).
Abbreviations
- PCR
Polymerase chain reaction
- Nest1
Nested PCR amplification round 1
- Nest2
Nested PCR amplification round 2
- SSU rRNA
Small subunit ribosomal RNA
- STLV
Simian T cell lymphotropic viruses
- SRV
Simian retrovirus
Contributor Information
Xinjun Zhang, Email: zxjzhang@ucdavis.edu.
Khamisah Abdul Kadir, Email: akkhamisah@unimas.my.
Leslie Fabiola Quintanilla-Zariñan, Email: lfquintanilla@ucdavis.edu.
Jason Villano, Email: jvillano@med.umich.edu.
Paul Houghton, Email: paulhoughton@primateproducts.com.
Hongli Du, Email: hldu@scut.edu.cn.
Balbir Singh, Email: bsingh@unimas.my.
David Glenn Smith, Email: dgsmith@ucdavis.edu.
References
- 1.Garnham PCC. Malaria parasites and other haemosporidia. Oxford: Blackwell Scientific Publications Ltd.; 1966. [Google Scholar]
- 2.Coatney GR, Collins WE, Warren M, Contacos PG. The primate malarias. U.S. Government Printing Office. 1971.
- 3.Knowles R, Gupta B. A study of monkey-malaria, and its experimental transmission to man. Indian Med Gaz. 1932;67:301–320. [PMC free article] [PubMed] [Google Scholar]
- 4.Eyles DE, Coatney GR, Getz ME. Vivax-type parasite of macaques transmissible to man. Science. 1960;132:1812–1813. doi: 10.1126/science.131.3416.1812. [DOI] [PubMed] [Google Scholar]
- 5.Das Gupta BM. Transmission of P. inui to man. Proc Natl Inst Sci India. 1938;4:241–244. [Google Scholar]
- 6.Schmidt LH, Greenland R, Genther CS. The transmission of Plasmodium cynomolgi to man. Am J Trop Med Hyg. 1960;10:679–688. doi: 10.4269/ajtmh.1961.10.679. [DOI] [PubMed] [Google Scholar]
- 7.Druilhe P, Trape JF, Leroy JP, Godard C, Gentilini M. Deux cas d’infection humaine accidentelle par Plasmodium cynomolgi bastianellii. Etude clinique sérologique. Ann Soc Belge Med Trop. 1980;60:349–354. [PubMed] [Google Scholar]
- 8.Coatney GR, Elder HA, Contacos PG, Getz ME, Greenland R, Rossan RN, et al. Transmission of the M strain of Plasmodium cynomolgi to man. Am J Trop Med Hyg. 1961;10:673–678. doi: 10.4269/ajtmh.1961.10.673. [DOI] [PubMed] [Google Scholar]
- 9.Coatney GR, Chin W, Contacos PG, King HK. Plasmodium inui, a quartan-type malaria parasite of Old World monkeys transmissible to man. J Parasitol. 1966;52:660–663. doi: 10.2307/3276423. [DOI] [PubMed] [Google Scholar]
- 10.Chin W, Contacos PG, Coatney GR, Kimball HR. A naturally acquired quotidian-type malaria in man transferable to monkeys. Science. 1965;149:865. doi: 10.1126/science.149.3686.865. [DOI] [PubMed] [Google Scholar]
- 11.Singh B, Sung LK, Matusop A, Radhakrishnan A, Shamsul SSG, Cox-Singh J, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 12.Singh B, Daneshvar C. Human infections and detection of Plasmodium knowlesi. Clin Microbiol Rev. 2013;26:165–184. doi: 10.1128/CMR.00079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyagi RK, Das MK, Singh SS, Sharma YD. Discordance in drug resistance-associated mutation patterns in marker genes of Plasmodium falciparum and Plasmodium knowlesi during coinfections. J Antimicrob Chemother. 2013;68:1081–1088. doi: 10.1093/jac/dks508. [DOI] [PubMed] [Google Scholar]
- 14.Ta TH, Hisam S, Lanza M, Jiram AI, Ismail N, Rubio JM. First case of a naturally acquired human infection with Plasmodium cynomolgi. Malar J. 2014;13:68. doi: 10.1186/1475-2875-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fooden J. Systematic review of Southeast Asian longtail macaques, Macaca fascicularis (Raffles, 1821) Fieldiana Zool. 1995;81:1–206. [Google Scholar]
- 16.Eudey AA. The crab-eating macaque (Macaca fascicularis): widespread and rapidly declining. Primate Conserv. 2008;23:129–132. doi: 10.1896/052.023.0115. [DOI] [Google Scholar]
- 17.Jeslyn WPS, Huat TC, Vernon L, Irene LMZ, Sung LK, Jarrod LP, et al. Molecular epidemiological investigation of Plasmodium knowlesi in humans and macaques in Singapore. Vector Borne Zoonotic Dis. 2011;11:131–135. doi: 10.1089/vbz.2010.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akter R, Vythilingam I, Khaw LT, Qvist R, Lim YAL, Sitam FT, et al. Simian malaria in wild macaques: first report from Hulu Selangor district, Selangor, Malaysia. Malar J. 2015;14:386. doi: 10.1186/s12936-015-0856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee KS, Divis PC, Zakaria SK, Matusop A, Julin RA, Conway DJ, et al. Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog. 2011;7:e1002015. doi: 10.1371/journal.ppat.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Meizhi I. Identification and molecular characterisation of simian malaria parasites in wild monkeys of Singapore. National University of Singapore, MSc thesis. 2011:184.
- 21.Jongwutiwes S, Buppan P, Kosuvin R, Seethamchai S, Pattanawong U, Sirichaisinthop J, et al. Plasmodium knowlesi malaria in humans and macaques, Thailand. Emerg Infect Dis. 2011;17:1799–1806. doi: 10.3201/eid1710.110349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seethamchai S, Putaporntip C, Malaivijitnond S, Cui L, Jongwutiwes S. Malaria and Hepatocystis species in wild macaques, southern Thailand. Am J Trop Med Hyg. 2008;78:646–653. [PubMed] [Google Scholar]
- 23.Putaporntip C, Jongwutiwes S, Thongaree S, Seethamchai S, Grynberg P, Hughes AL. Ecology of malaria parasites infecting Southeast Asian macaques: evidence from cytochrome b sequences. Mol Ecol. 2010;19:3466–3476. doi: 10.1111/j.1365-294X.2010.04756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones-Engel L, Engel G, Schillaci M, Pacheco M, Escalante A. Malarial monkeys: reservoir for zoonotic infection? Am J Primatol. 2007;69:40–41. [Google Scholar]
- 25.Vythilingam I, Noorazian YM, Huat TC, Jiram AI, Yusri YM, Azahari AH, et al. Plasmodium knowlesi in humans, macaques and mosquitoes in peninsular Malaysia. Parasit Vectors. 2008;1:26. doi: 10.1186/1756-3305-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muehlenbein MP, Pacheco MA, Taylor JE, Prall SP, Ambu L, Nathan S, et al. Accelerated diversification of nonhuman primate malarias in Southeast Asia: adaptive radiation or geographic speciation? Mol Biol Evol. 2015;32:422–439. doi: 10.1093/molbev/msu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng J, Trask JS, Houghton P, Smith DG, Kanthaswamy S. Use of genome-wide heterospecific single-nucleotide polymorphisms to estimate linkage disequilibrium in Rhesus and Cynomolgus macaques. Comp Med. 2015;65:62–69. [PMC free article] [PubMed] [Google Scholar]
- 28.Kanthaswamy S, Ng J, Ross CT, Trask JS, Smith DG, Buffalo VS, et al. Identifying human-rhesus macaque gene orthologs using heterospecific SNP probes. Genomics. 2013;101:30–37. doi: 10.1016/j.ygeno.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith DG, Ng J, George D, Trask JS, Houghton P, Singh B, et al. A genetic comparison of two alleged subspecies of Philippine cynomolgus macaques. Am J Phys Anthropol. 2014;155:136–148. doi: 10.1002/ajpa.22564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers MJ, Li J, McCutchan F. The Plasmodium rRNA genes: developmental regulation and drug target. In: Sherman IW, editor. Malaria: parasite biology, pathogenesis and protection. Washington, DC: American Society for Microbiology; 1998. pp. 203–217. [Google Scholar]
- 31.Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- 32.Daneshvar C, Davis TME, Cox-Singh J, Rafa’ee MZ, Zakaria SK, Divis PCS, et al. Clinical and laboratory features of human Plasmodium knowlesi infection. Clin Infect Dis. 2009;49:852–860. doi: 10.1086/605439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitou NM. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 35.Engel GA, Jones-engel L, Schillaci MA, Suaryana KG, Putra A, Fuentes A, et al. Human exposure to Herpesvirus B-seropositive macaques, Bali, Indonesia. Emerg Infect Dis. 2002;8:789–795. doi: 10.3201/eid0808.010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuentes A. Human culture and monkey behavior: assessing the contexts of potential pathogen transmission between macaques and humans. Am J Primatol. 2006;68:880–896. doi: 10.1002/ajp.20295. [DOI] [PubMed] [Google Scholar]
- 37.Jones-Engel L, Engel GA, Schillaci MA, Rompis A, Putra A, Suaryana KG, et al. Primate-to-human retroviral transmission in Asia. Emerg Infect Dis. 2005;11:1028–1035. doi: 10.3201/eid1107.040957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallis J, Lee DR. Prevention of disease transmission in primate conservation. Int J Primatol. 1999;20:803–826. doi: 10.1023/A:1020879700286. [DOI] [Google Scholar]
- 39.Putaporntip C, Hongsrimuang T, Seethamchai S, Kobasa T, Limkittikul K, Cui L, et al. Differential prevalence of Plasmodium infections and cryptic Plasmodium knowlesi malaria in humans in Thailand. J Infect Dis. 2009;199:1143–1150. doi: 10.1086/597414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fornace KM, Abidin TR, Alexander N, Brock P, Grigg MJ, Murphy A, et al. Association between landscape factors and spatial patterns of Plasmodium knowlesi infections in Sabah, Malaysia. Emerg Infect Dis. 2016;22:201–208. doi: 10.3201/eid2202.150656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moyes CL, Shearer FM, Huang Z, Wiebe A, Gibson HS, Nijman V, et al. Predicting the geographical distributions of the macaque hosts and mosquito vectors of Plasmodium knowlesi malaria in forested and non-forested areas. Parasites Vectors. 2016;9:242. doi: 10.1186/s13071-016-1527-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The DNA sequence data obtained for this research are available in GenBank with Accession number: KU693334.


