SUMMARY
The epidemiological investigation of a foodborne outbreak, including identification of related cases, source attribution, and development of intervention strategies, relies heavily on the ability to subtype the etiological agent at a high enough resolution to differentiate related from nonrelated cases. Historically, several different molecular subtyping methods have been used for this purpose; however, emerging techniques, such as single nucleotide polymorphism (SNP)-based techniques, that use whole-genome sequencing (WGS) offer a resolution that was previously not possible. With WGS, unlike traditional subtyping methods that lack complete information, data can be used to elucidate phylogenetic relationships and disease-causing lineages can be tracked and monitored over time. The subtyping resolution and evolutionary context provided by WGS data allow investigators to connect related illnesses that would be missed by traditional techniques. The added advantage of data generated by WGS is that these data can also be used for secondary analyses, such as virulence gene detection, antibiotic resistance gene profiling, synteny comparisons, mobile genetic element identification, and geographic attribution. In addition, several software packages are now available to generate in silico results for traditional molecular subtyping methods from the whole-genome sequence, allowing for efficient comparison with historical databases. Metagenomic approaches using next-generation sequencing have also been successful in the detection of nonculturable foodborne pathogens. This review addresses state-of-the-art techniques in microbial WGS and analysis and then discusses how this technology can be used to help support food safety investigations. Retrospective outbreak investigations using WGS are presented to provide organism-specific examples of the benefits, and challenges, associated with WGS in comparison to traditional molecular subtyping techniques.
INTRODUCTION
Foodborne pathogens are major causes of morbidity and mortality throughout the world, and the ability to conduct epidemiological investigations and intervene in foodborne illnesses is a critical part of the existing public health infrastructure. Molecular subtyping of the etiological microorganisms is an important tool for such investigations. At any time, several strains of a given foodborne pathogen may be cocirculating through a population and this redundancy can confound epidemiological investigations. Subtyping strategies are used to identify organisms to a higher phylogenetic resolution than the species level so that cooccurring outbreaks can be differentiated, sources of contamination can be identified, and intervention strategies can be enacted. To be useful, subtyping schemes must be able to clearly and accurately resolve the relatedness of several microbial isolates, so that linked cases are identified and included in the investigation. It is equally important that concurrent, nonrelated, and sporadic cases can be differentiated from outbreak cases so as not to confound the investigation. The latter is becoming increasingly important since the complexity of the modern food supply chain is vast and food products and ingredients can be sourced from locations distributed over entire continents and unrelated outbreaks may geographically and temporally overlap. High-resolution subtyping should result in more cases linked to defined outbreaks and more confidence in the identification of outbreak sources. Second- and third-generation sequencing platforms have advanced microbial whole-genome sequencing (WGS) to the point where it has become available for routine use in research and reference laboratories and etiological microorganisms can be sequenced in real time during an outbreak. The data, once analyzed and interpreted, can then be readily used in investigations. WGS surveillance of foodborne pathogens is already being applied routinely by several national authorities, including the Food and Drug Administration (FDA) in the United States, Public Health England, and the Statens Serum Institut in Denmark (1). WGS is fast and cheap and produces subtyping and phylogenetic resolution on a scale that was never before achievable, even when combining the results of several other molecular subtyping schemes. In this review, we examine numerous ways in which WGS can contribute to the management of foodborne microbial hazards.
SUBTYPING TECHNIQUES
In outbreak investigations of bacterial foodborne illnesses, culture-dependent methods are generally used to obtain an isolate from the implicated food products, production facilities, and affected individuals—isolation is still critical because of the legal implications associated with any regulatory actions or public health interventions taken. After this initial step for culturable organisms, and as a starting point for nonculturable organisms (e.g., norovirus [NoV]) phage typing, serotyping, or molecular methods can be used to subtype the strain. Molecular subtyping techniques can be divided up into techniques that use amplification, restriction digestion, or DNA sequencing, as deemed appropriate to the pathogen under investigation.
Serotyping
Serotyping is a method of grouping microorganisms on the basis of the reaction between a given antiserum and cell surface antigens that allows classification to the subspecies level. The serotyping of most bacterial species is based on the detection of flagellar and somatic antigens (2), though capsular antigens may also be used (3). Serotyping of Clostridium botulinum, however, relies on the detection of different types of neurotoxin but also uses serology (4).
Phage Typing
Phage typing has been used as an epidemiologic tool to differentiate isolates of Salmonella and Escherichia coli O157:H7 beyond the serotype level for several decades (5–7). Phage typing relies on the detection of bacterial lysis by a specific standardized phage (6). Though of limited discrimination, phage typing has been used in combination with verotoxin typing to identify linked cases of E. coli O157:H7 infection (8). Highly clonal Salmonella serovars require additional subtyping during large-scale surveillance efforts, and phage typing was useful for this (9). However, the use of phage typing was ultimately limited because it has poor resolving ability, is expensive, and requires technical expertise (10).
Amplification-Based Techniques
Amplification-based techniques include variable-number tandem repeat (VNTR), amplified fragment length polymorphism (AFLP), and PCR amplification methods. In addition to subtyping, PCR amplification can also be used to identify specific interesting genes, such as virulence factors or antimicrobial resistance (AMR) genes. VNTR utilizes highly repetitive DNA regions in bacterial genomes (11). Repetitive regions are highly variable in terms of the number of copies, even in related strains, and differences in the number of repeats can be used to delineate clonal and nonclonal isolates; although to gain increased discrimination, multiple loci must be used for what is referred to as a multiple-locus VNTR analysis (MLVA) (12). In this technique, nonvarying PCR primers bind to sequences that flank the repeat array to allow for efficient amplification of the repeat motif, the PCR products are separated, and strains are differentiated on the basis of amplicon size. VNTR/MLVA was commonly used during the early to mid-2000s, and there were several method comparison studies that indicated that VNTR/MLVA produced more discriminatory results than pulsed-field gel electrophoresis (PFGE), which is still widely used (13–15). AFLP combines PCR amplification with restriction enzyme (RE) digestion. The bacterial genome is first digested with a RE, and then primers that contain a sequence complementary to the restriction site are used to amplify the restriction fragments. To reduce the number of restriction fragments, primers typically contain one to three additional random nucleotides on the 3′ end to interact with nucleotides inside the adapter fragment (16). In addition, a feature of amplification profiling is that important genes such as virulence factors or AMR genes can be incorporated into a typing scheme (17, 18).
Restriction Digestion-Based Techniques
Restriction digestion-based techniques include two commonly used techniques based on restriction fragment length polymorphism (RFLP): ribotyping and PFGE. In RFLP subtyping, bacterial genomic DNA is digested with a restriction endonuclease and the DNA fragments are separated by gel electrophoresis. However, depending on the enzyme, >100 fragments can be produced, making the comparison of strains problematic. There are two ways to deal with this issue: using a rare-cutting enzyme and specialized electrophoresis to separate large fragments (e.g., PFGE) or transferring the DNA fragments to membranes and then hybridizing them with a labeled probe for specific fragments such as ribosomal RNA genes (e.g., ribotyping). PFGE was introduced 2 decades ago and remains the “gold standard” for molecular typing methods for foodborne pathogens by PulseNet International, which is discussed in detail in the next section. Using PFGE, bacterial isolates that differ by a single genetic event (two or three band differences) are defined as closely related, while multiple band differences are indicative of unrelated strains (19). In addition, the background history of the isolates under investigation is taken into consideration, for example, organism variability, the prevalence of PFGE patterns, and the length of an ongoing outbreak (20). However, since PFGE relies on gel electrophoresis and the resolution of large bands, it is not sensitive enough to detect single nucleotide polymorphisms (SNPs).
Ribotyping relies on differences in the genomic location and number of rRNA genes for genotyping. Unlike PFGE, ribotyping uses a frequently cutting enzyme, but probe DNA is used to label certain bands, making them distinguishable (21). Variability in the number of rRNA genes, as well as size variability in the detected bands, leads to discrimination between bacterial strains. It is generally well accepted that the resolution available through ribotyping is not as high as that of PFGE (22).
PulseNet
There are two basic requirements of source attribution; the first is subtyping, and the second is a system that allows for effective communication of typing data so that authorities have access to real-time information on national (or international) outbreak events. This led to the development of PulseNet. BioNumerics (Applied Maths) is a software package that is used to consistently analyze PFGE patterns, and these patterns are then uploaded to PulseNet. The power of the PulseNet system is that all participating laboratories perform analysis with the same algorithm(s), molecular standards, and protocols so that comparisons of genetic profiles across jurisdictions and countries can be easily performed (23). Under this model, international databases of the molecular profiles of a variety of foodborne pathogens can be generated and maintained. It is also important to note that the attribution of a particular PFGE molecular profile to an outbreak cluster is weighed against additional evidence such as epidemiological data.
In 1993, >700 people fell ill in the United States after consuming burger patties contaminated with E. coli O157:H7. PFGE was used to determine that the bacteria sourced from hamburger matched the strain causing illness in humans, and scientists at the U.S. Centers for Disease Control (CDC) realized that the outbreak could have been identified earlier if public health laboratories had been able conduct PFGE and share results in real time (24, 25). In 1995, the CDC and the Association of Public Health Laboratories (APHL) selected state laboratories to participate in a network pilot aimed at disease surveillance; this pilot ultimately lead to the beginnings of PulseNet (26). Currently, PulseNet USA is headquartered at the CDC in Atlanta, GA, and consists of >83 state public health laboratories in seven regions. PulseNet is able to quickly group individuals that probably consumed the same contaminated food or that were exposed in some way to a foodborne pathogen. Later, it was recognized that food supply globalization could play a major role in foodborne illness and PulseNet International was created to include other countries. Currently, PulseNet International consists of members from Africa, the Asia Pacific region, Europe, Latin America and the Caribbean, the Middle East, the United States, and Canada.
PulseNet generally tracks PFGE patterns but also supplements PFGE with additional MLVA subtyping; it is also currently considering how WGS will be integrated into its platform. E. coli, Campylobacter, Listeria monocytogenes, Salmonella, Shigella, Vibrio parahaemolyticus, and Cronobacter isolates are currently characterized through PulseNet.
Sequencing-Based Techniques
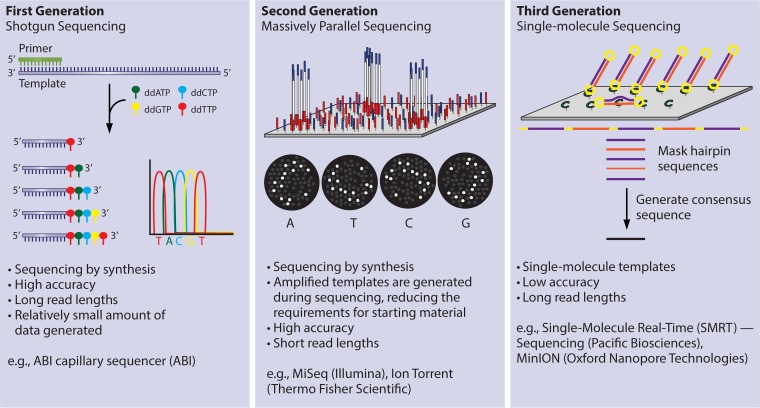
Microbial genome sequences contain variability between isolates because of mutation accumulation and recombination; and the sequence variability that exists can be exploited when developing molecular typing schemes. Each subtyping scheme based on DNA sequencing relies on the detection and cataloguing of SNPs—variations that occur at single base positions when comparing multiple genomes. When a SNP within a gene is found in at least two isolates, the version of the gene with the SNP is described as an allele. Multilocus sequence typing (MLST) is based on allele variance in a small set of housekeeping genes. Alleles typically correspond to phylogeny (27). MLST is typically carried out by a low-throughput technique such as Sanger sequencing, where each of the genes in the typing scheme will be sequenced from end to end. The gene sequences are compared against an established database, such as pubMLST/BIGSdb, to determine the individual alleles and thereafter the multilocus sequence type (ST) of the isolate. It is now cheaper to sequence an entire genome by massively parallel sequencing than to sequence the MLST genes individually (Fig. 1); therefore, there are several user-friendly platforms that are able to calculate STs from a whole genome sequence, such as the Center for Genomic Epidemiology (28). Isolates having matching STs are defined as clonal—having a common ancestor (29).
FIG 1.
First-, second-, and third-generation sequencing platforms are currently available, and each has distinct advantages and disadvantages. First-generation sequencers, including the ABI capillary sequencer, are characterized by high accuracy and a relatively long read length; however, these systems are not amenable to high-throughput sequencing. Second-generation platforms include MiSeq, HiSeq, and NextSeq from Illumina, as well as Ion Torrent from Thermo Fisher Scientific. These platforms use massively parallel sequencing to achieve high throughput and have high base-calling accuracy; however, sequencing reads are short and this can result in split contigs in repetitive regions during sequence assembly. Third-generation sequencers, including PacBio from Pacific Biosciences and MinION, PromethION, and SmidgION from Oxford Nanopore Technologies, are able to sequence single-molecule templates, which results in very long read length at a high throughput. Third-generation sequencers have a very high error rate relative to the other technologies. Currently, the optimal sequencing platform is highly dependent on the desired applications.
Single Nucleotide Polymorphism-Based Analysis
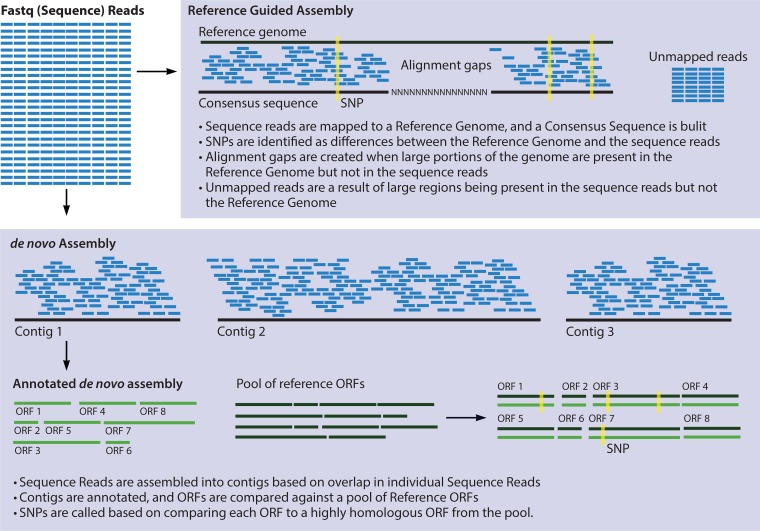
SNPs can occur in genes, in noncoding regions, or in the mobile genetic elements of a genome. SNPs that occur within genes can be synonymous if they do not change the coding sequence or nonsynonymous if a SNP changes the amino acid sequence. If there are more nonsynonymous than synonymous SNPs, the population is said to be under diversifying selection. The term informative SNP, also called parsimony informative SNP, is used to describe a SNP that is shared by two or more strains in an alignment and therefore supports the phylogenetic grouping of two or more isolates (30). To detect SNPs, a comparison must be made via a pairwise alignment of two or more genomes or sections of multiple genomes (i.e., all open reading frames [ORFs], MLST genes, or random 1,000-bp fragments) (Fig. 2), and several programs are available for this purpose (i.e., BLAST, Mauve, Muscle, ClustalW, FSA, MAFFT). Aligning whole genomes is possible, although it is computationally intensive and minimally informative if the genomes being aligned are not nearly identical. One of the least computationally intensive ways to detect SNPs is reference-guided assembly. After the reads have been mapped to a reference genome, a SNP caller can be used to identify the SNPs between the genomes; however, this introduces the issue of reference bias, and the choice of reference genome is extremely important (31). It is also possible to do a SNP analysis of de novo-assembled genomes. This approach requires that the assembled contigs are annotated, and then ORFs are compared to related ORFs and screened for SNPs (30) (Fig. 2).
FIG 2.
Analysis of SNP variation within a genome sequence can be used to compare isolates on a phylogenetic basis and draw conclusions about the relatedness of strains. However, there are various methods of detecting SNPs, and different methodologies can result in different conclusions. After sequencing, genomes can be assembled by either a reference-guided or a de novo approach. A common method of SNP detection based on a reference-guided assembly is to assemble sequencing reads to the reference genome and then detect SNPs based on nucleotide differences between the reference and the assembly. After de novo assembly, a commonly used approach is to use a concatenated sequence of each of the core genes in a genome and call SNPs based on this pseudoreference genome. The optimal SNP detection approach will depend on the desired application of the data. Reference-guided assembly is a much less computationally intensive method. However, in a reference-guided assembly, reads from regions in the genome being assembled that are not present in the reference genome will be discarded. In addition, regions present in the reference genome that are not present in the genome being assembled will result in alignment gaps.
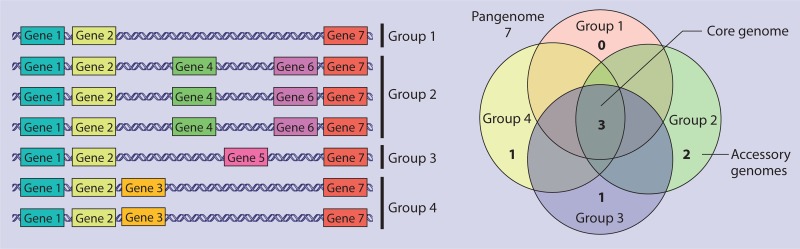
SNP-based analysis is an accurate way to determine the phylogenetic relationships of two or more genomes. Traditional MLST relies heavily on the presence SNPs among a few conserved genes to assign a ST to a given genome. However, new sequencing technologies now support the development of extended MLST frameworks. Core genome MLST (cgMLST) compares all of the genes present in every genome of microbes of a specific phylotype, known as the core genome, against each other and identifies the presence of SNPs, and then isolates are subtyped on the basis of this larger data set (32, 33) (Fig. 3). SNPs within the accessory genome can also be compared; however, this type of analysis requires that the isolates being compared both contain the accessory genes in question and could only be used to delineate closely related isolates (34) (Fig. 3).
FIG 3.
Pangenome, core genome, and accessory genome are commonly used terms in genomics. Pangenome refers to all of the genes that occur in a given phylotype. For example, each gene identified in any L. monocytogenes genome is part of the L. monocytogenes pangenome. In this visualization, genes 1 to 7 are part of the pangenome. The core genome is defined as all of the genes that are present in all of the members of a given phylotype. In this example, genes 1, 2, and 7 are all part of the core genome. The core genome will always be smaller than the pangenome. The term accessory genome is used to refer to genes present in an organism, or group of organisms, that are unique to that organism or group. Accessory genes are part of the pangenome but not the core genome. In the image, genes 4 and 6 are part of the accessory genome of group 2, gene 5 is the accessory genome of group 1, gene 3 is the accessory genome of group 4, and group 1 does not have any accessory genes.
Extended MLST can provide insight into the phylogeny of a group of isolates. However, it raises an important question: how many SNPs are required to conclude that two strains are related? Although several studies have been done surrounding this topic, the answers are complex and no consensus has been established. For example, epidemiologically linked L. monocytogenes isolates examined in one retrospective study differed by <10 core genome SNPs (35). In the same study, two isolates recovered a day apart from the same patient differed by 21 SNPs (35). Conversely, a different study reported that two L. monocytogenes isolates that originated in the same processing plant but were isolated 12 years apart varied by as little as one core genome SNP, though a phage sequence included in both genomes differed by 1,274 SNPs (34). A retrospective study of 183 sequenced E. coli O157:H7 isolates with known epidemiological linkages defined linked isolates as having fewer than five SNP differences, and on average, only one SNP difference was found between isolates from patients belonging to the same family (36). A precise criterion for ruling isolates in or out of an outbreak on the basis of SNP analysis has yet to be defined and universally accepted. In considering the examples listed above, it is important to note that SNP calling varies on the basis of several factors, including definition of the core genome, species, choice of reference genome, choice of SNP caller and SNP calling parameters, sequencing metrics, how many isolates are included in the analysis, and the nature of the outbreak, so care must be taken when comparing the numbers of SNPs being found through retrospective work across studies because of the variability of the ways in which they are found. Standardization is required. Several questions have also been asked about the reliability and reproducibility of SNP-based analysis, what level of variability is expected from instrumentation error, and how much variation arises during subculturing. Allard et al. tried to address these questions by passaging a single Salmonella isolate four times on solid medium, sequencing four colonies from each passage, and finally sequencing the same colony four times. No SNP variation was observed in this set of experiments after homopolymeric sequences and SNPs adjacent to breakpoints were removed (30).
METHODS OF MICROBIAL WGS
First Generation—the Sanger Shotgun Approach
The shotgun approach of WGS and assembly was carried out by Sanger sequencing, which relies on the selective incorporation of chain-terminating dideoxynucleotides by DNA polymerase during DNA replication (37) (Fig. 1). During the early days of this technology, dideoxynucleotides were radiolabeled to allow for detection and manual determination of the consensus sequence (37); however, Applied Biosystems (ABI) later introduced fluorescent base labeling, which allowed automation of the process and subsequently higher throughput (38). Automated Sanger sequencing was the sequencing platform of choice for almost 20 years and was used to sequence the first finished human genome. In 1995, the first bacterial genome sequence, for Haemophilus influenzae, was published, and it was sequenced by the Sanger method (39). Specifically, in this approach, genomic DNA from H. influenzae was isolated, randomly sheared, and cloned into a plasmid vector. The resulting vectors were transformed into host E. coli cells and propagated, plasmid DNA was extracted, and insert DNA from H. influenzae was sequenced from both ends (called “random shotgun” sequencing), producing sequence lengths of about 460 bp per clone. This was a monumental task and required the production and sequencing of 9,500 E. coli clones to obtain a draft genome with 5× consensus coverage. This work also knowingly left 37% of the genome unsequenced (39). Despite this method's being labor intensive, hundreds more bacterial genomes, including those of Mycobacterium tuberculosis, Yersinia pestis, E. coli K-12, Bacillus subtilis, and Treponema pallidum, were sequenced this way over the next few years (40–44).
The relatively high throughput of automated the Sanger sequencing led to the birth of bioinformatics as a mechanism to assemble the shotgun sequences into genomes and later to help make sense of rapidly accumulating sequence data. During the first generation of microbial WGS, there was a strong emphasis on closing or finishing genomes and the first software packages were designed to assist in this endeavor (45). First-generation assembly programs (Allora, Celera Assembler, and TIGR Assembler) used an overlap-layout-consensus algorithm and were optimized to work with the high-accuracy long reads produced during Sanger sequencing (46). The first H. influenzae bacterial genome (1.83 Mbp) was closed after initial assembly with the TIGR Assembler into 140 contigs with 98 sequence gaps, and various algorithms were then used to map and fill physical gaps, while sequence gaps were closed by primer walking (39).
Second Generation—Massively Parallel Sequencing
Massively parallel or next-generation sequencing (NGS) was first introduced by Roche in the form of the 454 GS20 in 2005 (47) and was followed by Illumina's genetic analyzer (GA) II in 2007 (Fig. 1). The early versions of NGS platforms came with several hurdles to routine WGS of microorganisms. For example, short sequencing reads, 110 bp for the 454 GS20 system and 35 bp for the GA II, required the development of novel assemblers optimized for the assembly of short-read sequences. In addition, each of these platforms had a massive footprint and price tag. However, by 2010, benchtop versions of these and other sequencing platforms (454 GS-FLX Titanium and 454 GS-FLX Junior [Roche], MiSeq and HiSeq [Illumina], SOLiD 3 [Life Technologies], and Ion Torrent [Thermo Fisher Scientific]) were available to most microbiology laboratories, leading to an explosion in the number of microbial sequences available for comparative analysis, epidemiological investigations, and ecological studies. Each NGS platform had a slightly different approach to DNA preparation, sequencing, imaging, and analysis (reviewed extensively in reference 48). Variation in approaches resulted in each platform having different strengths and weaknesses. The 454 platforms produced longer reads, which improved mapping in repetitive regions; however, they suffered because of high reagent cost and high error rates in homopolymer regions and produced less data per run than their competitors. By the end of 2013, Roche had announced that 454 Life Science was shutting down and that 454 GS FLX Titanium system support would stop in mid-2016. As of early 2016, the Ion Torrent S5 platform is able to produce 400-bp reads but produces about half as much data per run (6 to 8 Gb) as the Illumina MiSeq platform, which produces only slightly shorter reads (300 bp) and is now very widely used in the field (48).
The major shortcoming of all second-generation sequencing platforms is a relatively short read length, and genome assembly algorithms built for the assembly of Sanger data do not perform well with the short-read data produced by second-generation platforms (49). In addition, the closed genomes produced during the first wave of microbial sequencing can now be used as a scaffold on which to guide the assembly of short-read data, for the first time giving scientists the choice between de novo and reference-guided genome assembly methods (Fig. 2). De novo genome assembly can be done by a variety of assemblers that are based on de Bruijn graph assembly (50), such as SPAdes (51) and Velvet (52). Reference-guided assembly maps short sequence reads by assessing the placement of each read against the reference genome and calculating the probability of its match with the reference. These alignments are then used to construct a novel consensus sequence for the sequence data. There are several different reference-guided assemblers, including Burrows-Wheeler Aligner (53), Novoalign, MOSAIK, and SMALT, and each uses a different algorithm: Burrows-Wheeler Transform, global Needleman-Wunsch (54), banded Smith-Waterman (55), and short-word hashing/Smith-Waterman, respectively (56). Reference-guided assembly is much less memory intensive and requires less computing power; in addition, more data are provided than when using de novo assemblies, particularly when sequence coverage is low (57). However, reference-guided assembly introduces large biases toward the reference genome and several types of data are missed, including some SNPs (31) (Fig. 2), structural variations (rearrangements) (58), and repetitive regions, making downstream synteny analysis inaccurate. In response, newer tools (i.e., reference-assisted chromosome assembly [59] and Ragout [58]) have been designed to reduce bias by simultaneous alignment with multiple reference genomes. Illumina has recently introduced a new library preparation technique combined with a downstream software package that has had success making synthetic long reads up to about 18 kbp in length (60), and this may help to counter problems with assembling complex, highly repetitive genomes and help Illumina compete with the newer third-generation sequencing techniques.
Second-generation sequencing gave rise to the proliferation of available draft genomes, where genomes that are sequenced to a high coverage depth but not closed, are submitted to databases, and are used for downstream analysis. This has generated an ongoing debate in the field of microbial genetics about the allocation of resources: is it better to close a lower number of genomes (45, 61), or should a higher number of genomes be sequenced to draft status (62)? Draft genomes are sufficient for the majority of analyses, including virulence factor identification, phylogenetics, and MLST; however, closed genomes are required for genomic island identification (63), characterization of repetitive elements, and sometimes structural rearrangements (64).
Third Generation—Single-Molecule Sequencing
Third-generation sequencing platforms can address the problems inherent to short sequence reads by sequencing long single molecules in real time (Fig. 1). Single-molecule sequencing was introduced in 2008 by Helicos BioSciences Corporation and was initially used to sequence a viral genome (65) but quickly progressed into parallel sequencing and the sequencing of a human genome (66). However, the short reads produced by the Helicos platform (averaging 32 bp) (66) did not differentiate this technology from its second-generation counterparts, and it was no match for the single-molecule real-time (SMRT) sequencing platform (PacBio) introduced by Pacific Biosciences in 2009, which was able to regularly produce reads exceeding 1 kb (67), and Helicos filed for bankruptcy in late 2012. In 2014, Oxford Nanopore launched the MinION access program. Oxford Nanopore's mission is to allow routine WGS anywhere with minimal reagents and sequencing equipment. The advantages of having real-time sequencing available anywhere, even given limited resources, were demonstrated during the West African 2014-2015 Ebola outbreak, where the Nanopore platform was used to support monitoring and surveillance of the transmission chains and the evolution of the virus at the height of the outbreak (68, 69).
However, what second-generation sequencing lacked in sequence read length, third-generation sequencing appears to lack in accuracy. In comparison, the second-generation Illumina MiSeq platform has an error rate of 0.58% and the PacBio platform's error rate is 14%. This has led to the development of bioinformatic approaches to appropriately call insertion, deletion, and SNP variations despite the high error rate. Third-generation sequencers rely on high-coverage consensus to correct for sequencing error and correctly call variants. As an example, during the Ebola outbreak, variants were called only on the basis of a log likelihood ratio of >200 and a coverage depth of >50, and SNPs were called only in regions covered by >20 nanopore reads where the SNP was seen in at least 20% of the reads (68).
Despite the rapid emergence of third-generation technologies for routine studies and surveillance, in 2016, these second- and third-generation technologies should be viewed as complementary rather than successive for several reasons. Short read length limits the ability of second-generation sequencers to successfully assemble highly repetitive regions and closed bacterial genomes. SMRT sequencing is currently more expensive than second-generation sequencing, has high sequencing error, and generates significantly less coverage. However, SMRT sequencing is able to produce long enough reads that even challenging bacterial genomes, with several repeat regions or low GC content, can readily be sequenced to a closed status (48, 70). Therefore, a potential strategy would be to generate a closed-genome scaffold using a third-generation technology and then generate deep coverage by using a second-generation technique for accurate SNP analysis. This is referred to as hybrid sequencing. Several assemblers have been built to work with hybrid data (i.e., PBcR), and bacterial genomes have been successfully closed by using this approach (71). However, using hybrid sequencing requires the added work of preparing two libraries, and when the appropriate amount of data is gathered by SMRT sequencing, these data independently yield better closed assemblies than hybrid data do (72). The optimal ways of combining sequencing data may shift in the future if the cost associated with PacBio sequencing declines, if the Nanopore becomes widely adopted by microbiologists, or if a separate technology that can combine high accuracy with long reads emerges.
Standards for Quality and Quantity
The field of microbial genetics currently lacks universal standards regarding WGS quality, including acceptable sequencing results, coverage depth, and assembly quality. The National Center for Biotechnology Information (NCBI) has minimum requirements for submission, including that vector and linker DNA be removed from the final assembly and that contigs be at least 200 bp in length, but data that conform to these standards can differ drastically in quality—and the quality of assembly can be hard to judge. The Genomics Standards Consortium is an open-membership group that is helping to drive standardization activities (73). The N50, which is similar to the mean of lengths but assigns more weight to larger contigs, is sometimes used to assess assembly quality (74, 75). However, the practical implications of this value, such as what N50 value is required to yield an assembly with a complete gene for every gene in a core genome, have yet to be determined. Attempts have been made to make recommendations such as the best assembly software and what parameters to use. However, the answers to these questions depend on the species, the sequencing platform, and the assembler; changing any of these parameters can cause the quality of the assembly to vary widely (74, 75).
EXAMPLES OF WGS IN EPIDEMIOLOGICAL INVESTIGATIONS
L. monocytogenes
L. monocytogenes causes serious infections that can result in a range of clinical illnesses, including invasive bacteremia, meningoencephalitis, spontaneous abortion in pregnant females, and potentially death (76). Epidemiological investigations of L. monocytogenes present several specific challenges. The incubation period of listeriosis is long; while it is approximately 21 days on average, it can be up to 70 days after exposure. This aspect is significant, as it can affect the accuracy of food histories patients are able to provide, as well as the availability of food samples (77). L. monocytogenes also has the ability to persist in food processing for years, and persistent contamination has been liked to intermittent illnesses that can span years (78, 79). Outbreaks that include few cases and occur years apart are difficult to link via epidemiological investigation, and PFGE offers little help, since mobile genetic elements, present in the L. monocytogenes accessory genome, change frequently and confound analysis through overdiscrimination (80). L. monocytogenes also has a highly stable core genome, and this means that finding high similarity in a core genome SNP-based analysis does not necessarily establish a link between strains (34, 81). However, retrospective studies have shown that WGS has clear advantages over all other molecular tools in epidemiological investigations and specific analyses are being developed to overcome existing issues.
Since case fatality rates can be high during L. monocytogenes outbreaks, infections are generally monitored by public health facilities (82). The resources available to these monitoring systems have, in several instances, been leveraged to conduct several retrospective and prospective surveillance studies that compare the accuracy of SNP-based subtyping to that of traditional methods. The Australian Listeria reference laboratory compared the typing results for 97 isolates obtained by WGS to those of PFGE, MLST, MLVA, and PCR serotyping and found that SNP analysis could easily differentiate between epidemiologically linked and unlinked cases with identical PFGE patterns (35). In addition, this study verified that in silico tools could be used to generate data comparable to PFGE, MLST, and PCR serotyping results from a sequence, but because of the short length of the Illumina MiSeq sequence reads and the highly repetitive nature of MLVA regions, in silico MLVA was not feasible (35). In Austria and Germany, seven cases of listeriosis occurred between April 2011 and July 2013. Isolates from these cases all shared a serotype (1/2b), a PFGE pattern, and an AFLP pattern that were indistinguishable from those of isolates obtained from five food producers, making it impossible to differentiate linked and sporadic cases or elucidate the source of the outbreak on the basis of traditional techniques (83). WGS of each of the seven human isolates, as well as 10 food isolates, was performed. On the basis of cgMLST, four cases were linked to each other, as well as a soft cheese product and a ready-to-eat (RTE) meat product, both of which were found on the grocery bills of each of the outbreak patients. Three cases were clearly distinguishable as a separate outbreak (83).
Overdiscrimination by PFGE can occur when L. monocytogenes isolates with a close ancestor in common differ by three or fewer bands. This shift in PFGE bands can result from a single genetic event, like the movement of a mobile genetic element. WGS has been shown to readily overcome problems associated with PFGE overdiscrimination in L. monocytogenes. As an example, in 2008, Canada experienced a large outbreak of listeriosis associated with RTE cold cut meat products. During the outbreak investigation, two distinct AscI PFGE patterns emerged; however, WGS analysis revealed that a 33-kbp prophage and a 50-kbp putative mobile genetic element accounted for the different AscI patterns and led to the conclusion that three distinct but clonal strains were involved in this outbreak and all originated from the same source (80). PFGE overdiscrimination also makes differentiating persistent contamination by L. monocytogenes from reintroduction in food-associated environments difficult. Different PFGE patterns suggest reintroduction; however, if WGS reveals that the difference in PFGE pattern is caused by a single mobile element, this suggests persistent contamination. A retrospective study examined persistent L. monocytogenes contamination in a deli setting and showed several PFGE patterns, suggesting reintroduction. However, cgMLST of the same samples showed that the strains were clonal and secondary analysis of the sequence data revealed that differing PFGE patterns were due to loss or gain of prophage regions—allowing the authors to conclude that persistent colonization was the likely issue (81). Resolving these differences would help food-processing facilities identify problems in their biosecurity and lead to a better understanding of underlying problems to take the necessary steps to prevent future contamination events.
Lack of SNP diversity within the L. monocytogenes core genome means that high cgMLST similarity between human and food isolates is not necessarily confirmation of a causal link (34). At the same time, horizontal gene transfer (HGT) and mobile genetic elements can make significant contributions to genomic diversity within the accessory genome (84, 85) (Fig. 3). Therefore, WGS results must be carefully analyzed and interpreted. For example, in the United States, a sporadic case of listeriosis in 1988 was linked to an outbreak in 2000, and both originated from the same processing plant. Analysis by cgMLST showed that a single synonymous SNP was the only difference between a 1988 human isolate and a 2000 food isolate, and only 11 SNPs differed among the four genomes sequenced in the study (34). However, 1,274 SNPs were observed between the 1988 and 2000 comK accessory prophage sequences—which allows the strains to be clearly differentiated (34). A separate retrospective study of persistent L. monocytogenes contamination in a deli setting also revealed that L. monocytogenes isolates from geographically and temporally different delis had identical or nearly identical (0 to 1 SNP difference) cgMLST results, further cautioning against establishing linkages based solely on low SNP variance within the core genome (81). Here it is important to note that other analyses that incorporate the whole genome—such as synteny, clustered regularly interspaced palindromic repeat (CRISPR)-associated locus subtype sequences, and the sequences of mobile genetic elements—can be used to supplement cgMLST analysis to interpret WGS. This interpretation, along with epidemiological data, can delineate L. monocytogenes strains during outbreak investigations (86).
Salmonella enterica
The genus Salmonella is divided into two species, S. enterica and S. bongori. S. enterica, is one of the most prevalent foodborne pathogens in the world and causes 11% of all food-related deaths globally (87). There are six subspecies of S. enterica (I, II, IIIa, IIIb, IV, and VI); however, most nontyphoidal salmonellosis is caused by subspecies I, which is additionally divided into >1,500 serovars defined by the detection of flagellar and somatic antigens (2). The antigen used in serovar typing seems to be reflective of evolutionary relatedness in some serovars but not others. For example, S. Newport has at least three distinct lineages and is polyphyletic (88), while S. Enteritidis (89), S. Typhimurium (90, 91), and S. Montevideo (30) are highly clonal. The genomic homogeneity implicit in highly clonal serovars makes traditional molecular subtyping methods inadequate for differentiation in outbreak investigations and makes WGS particularly attractive for subtyping this pathogen (92). As an example, S. Enteritidis is the most common cause of salmonellosis (93), and 85% of isolates can be classified into just five PFGE patterns. Anecdotally, this means that, for example, the New York State Department of Health receives 350 to 500 S. Enteritidis isolates a year, ∼50% of which are of a single PFGE type (JEGX01.0004); additionally, ∼30% are of a single MLVA type (89). Subtyping methods that use WGS have been shown to successfully delineate clonal S. enterica serovars where traditional techniques cannot; several examples are discussed in this section. WGS has also been used to link historical cases of salmonellosis to current outbreaks, which was not previously possible through the use of PFGE (94).
During an S. Enteritidis outbreak in Connecticut and New York in 2010, several isolates were obtained during the defined outbreak period and all had the same PulseNet (JEGX01.0004) PFGE pattern; WGS, followed by SNP-based phylogenetic analysis, was able to produce a well-defined clade for epidemiologically defined outbreak isolates and readily differentiate between outbreak and concurrent sporadic cases (92). In Belgium in 2014, there were several cases of infection with S. Enteritidis; the isolates were of phage type 4a and had the same MLVA profile, though the epidemiological investigation indicated that there may have been two independent sources. cgMLST analysis placed the isolates into two separate clades and confirmed that two overlapping outbreaks were taking place. A maximum of two SNP difference based on pairwise alignment of human and food isolates was observed in the first clade, no SNP differences were observed in the second, and 53 SNPs between the two were detected (95). In a retrospective study, 55 isolates from seven S. Enteritidis outbreaks could be accurately grouped into clades by cgMLST; each outbreak had four or fewer SNP differences between isolates, but there was an average of 42.5 SNPs differentiating outbreak clades (96). In a different study, 52 S. Enteritidis isolates from 16 outbreaks were analyzed by cgMLST, MLVA, CRISPR-MVLST, and PFGE in tandem to compare the effectiveness of cgMLST with that of established techniques. Phylogenetic inference based on cgMLST accurately predicted which strains belonged to each of the 16 outbreaks; in comparison, the next most accurate technique, MLVA, correctly grouped only 6 of the 16 outbreaks (97). From the 10 outbreaks that were not correctly grouped by MLVA, 8 grouped with isolates from other outbreaks and in 2 instances isolates from the same outbreak did not form a group with each other (97). In 2014, a hospital network in the United Kingdom observed a spike in S. Enteritidis infections in several hospitals and within communities serviced by the hospitals. Prospective WGS using the Illumina MiSeq platform was able to determine if cases were part of the outbreak and establish phylogenetic information after only 18 h (98). Retrospective analysis of the same samples was done with the Oxford Nanopore to evaluate this emerging technology. Nanopore data were analyzed in real time during sequencing, and identification to the species level was available after 20 min, the serotype was available after 40 min, and within 2 h, it could be determined if the strain was part of the outbreak (98).
In a Danish study, 18 isolates from six S. Typhimurium outbreaks were compared to 16 unrelated strains. Phylogenetic analysis based on cgMLST retrospectively identified outbreak cases with 100% accuracy, and outbreak clades differed by 5 to 12 SNPs, while unrelated isolates differed by 15 to 344 SNPs (99). In a larger study of 57 isolates of S. Typhimurium from five outbreaks, cgMLST demonstrated high-resolution subtyping, as all of the isolates from four outbreaks differed by only one or two SNPs, although isolates from the fifth outbreak differed by only 12 SNPs (100). cgMLST has also been evaluated for the ability to group outbreak cases from nonrelated isolates within the same phage type—in this case, DT 8. DT 8 isolates were highly clonal, with only 342 SNPs differentiating all DT 8 strains; however, outbreak clades were clearly defined and differed by a maximum pairwise distance of 3 SNPs (101). WGS has also been shown to have value for the identification and source attribution of laboratory-acquired salmonellosis, which is complex, given the number of strains to which a technician can be exposed (102).
The S. Heidelberg serovar is also clonal, and most isolates have the same PFGE profiles. A retrospective study conducted in Quebec, Canada, compared 46 isolates from three outbreaks and found that SNP-based analysis was able to correctly place the outbreak isolates into three clades. Within the outbreak clades, isolates differed by 1 to 4 SNPs, while >59 SNPs were observed among the three previously indistinguishable outbreaks, which had the same PFGE and phage type (103).
S. Montevideo was the causative agent of a large outbreak in the United States in 2009-2010 that reportedly affected nearly 300 people and confounded conventional epidemiological traceback. On the basis of patient histories, spiced meat appeared to be the causative agent of this outbreak; however, each isolate had a single PFGE pattern (JIXX01.0011) that was also associated with a 2008 outbreak caused by contaminated pistachios (104, 105). SNP-based analysis was able to readily differentiate clinical, environmental, and foodborne isolates from the spiced meat outbreak from other S. Montevideo isolates with the same PFGE patterns, including isolates from the pistachio outbreak (30).
S. Newport is polyclonal and has a high degree of genomic diversity, and phylogenetically distinct lineages of S. Newport can be more closely related to other serovars than to each other (106). However, cgMLST had been demonstrated to provide more accurate delineation of S. Newport than serovar identification, PFGE, or MLST (89). For example, in Europe, the whole genomes of 24 clinical S. Newport isolates involved in an outbreak caused by contaminated melon were sequenced. Nineteen were identical after SNP-based analysis, and the remaining five differed by only a single SNP, while nonoutbreak S. Newport strains differed by several thousand SNPs (107).
Since serotyping remains a gold standard in food safety management of Salmonella, several software packages that are capable of accurately predicting Salmonella serotypes on the basis of WGS data have been developed. SeqSero is a web application that is able to determine the serotype of Salmonella isolates from both raw sequencing reads and whole-genome assemblies with an accuracy of 92 to 99% (108). The Salmonella In Silico Typing Resource (SISTR) is an online platform that allows users to upload assembled genomic data and then predicts the serotype (with 94.6% accuracy), performs MLST and cgMLST, and allows users to view their sequence in a broad phylogenetic context (109). While this software currently provides typing information, it is also being updated to provide AMR profiling and detect virulence genes (109). The Metric-Orientated Sequence Typer can also estimate a serovar on the basis of a short-read sequence, but users must download the program from github and run it from a command line interface, making it less accessible (110).
E. coli
E. coli can be an innocuous part of the intestinal microbiome, but certain isolates have the pathogenic potential to cause significant illness. Diarrheal E. coli can be divided into six major pathotypes: enteropathogenic E. coli (EPEC), Shiga toxin-producing E. coli (STEC), Shigella/enteroinvasive E. coli, enteroaggregative E. coli, diffusely adherent E. coli, and enterotoxigenic E. coli (ETEC) (111). Pathotypes do not form phylogenetic clades (112), but rather, E. coli genomes are particularly plastic and rapid gains and losses of genetic material are associated with equally rapid changes in virulence potential (113). The dynamic nature of the E. coli genome leads to an exceedingly small core genome but a large pan-genome, relative to other foodborne pathogens (114). Most of the definable virulence factors in E. coli are on mobile genetic elements and readily travel between E. coli strains by HGT. For example, while most toxin genes and colonization factors required for ETEC pathogenesis are carried by plasmids (115), the locus of enterocyte effacement, which is required for EPEC pathogenesis and is also carried by some STEC isolates, is located on a pathogenicity island (116) and the Shiga toxin involved in STEC pathogenesis is encoded by a bacteriophage (117). While WGS and SNP-based analysis are able to delineate E. coli outbreaks and rapidly determine if isolates belong to a particularly virulent lineage (118), currently, the most valuable role for WGS in E. coli management is the rapid and reliable detection of virulence genes.
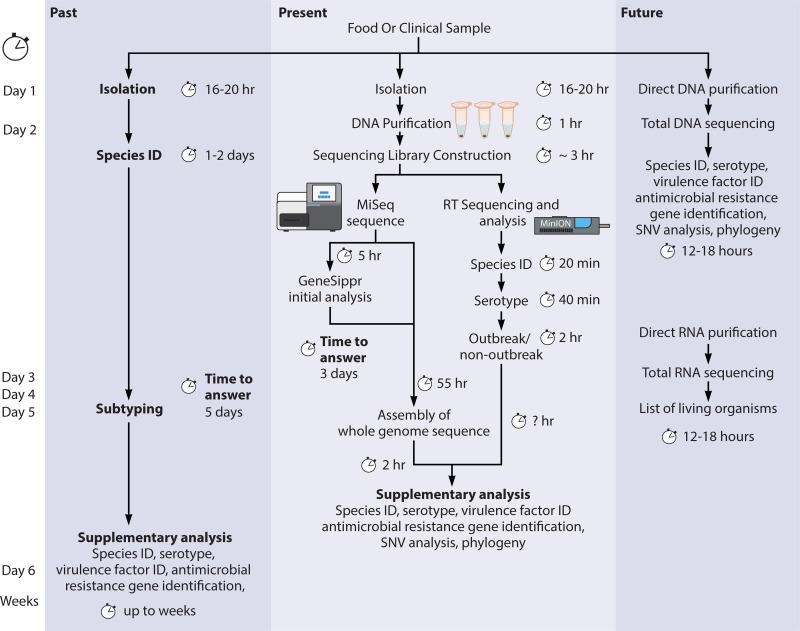
The detection of virulence genes in E. coli typically has, until recently, relied on phenotypic tests such as hemolysis, the cell culture assay for toxins, or PCR-based amplification of virulence genes (119). Completion of these typing tests (including PFGE, MLST, PCR, and serotyping) would generally take >5 days, while WGS can provide analogous data in 3 days (120). In the latter scenario, a novel approach is used to interpret data during an ongoing Illumina MiSeq run and can correctly identify virulence genes in STEC only 4.5 h after the sequencing run is started (121). Studies that compare routine virulence gene typing with WGS and in silico typing have found that the two methods produce almost identical results (>90% concordance) (120, 122). The discordant results that do occur are attributed to limitations in the assembly of short-read sequences (122), and this issue will probably be resolved by third-generation sequencing or the improvement of short-read assembly. User-friendly websites are available for automatic detection of virulence gene presence from WGS data; a popular one is VirulenceFinder (120). SuperPhy is an online platform that also detects virulence determinants, including Shiga toxin subtypes, in E. coli genomes but also goes several steps further and identifies AMR markers and known statistical correlations with geographic source, genotype, host, source, and phylogenetic clade (123). Web tools are also available for in silico serotyping of E. coli, and these tools report very accurate results compared to traditional serotyping (98 to 99% agreement) (124). WGS data can also be used in comparative genomic approaches to identify gene clusters that are responsible for changes in virulence in a particular lineage. As an example, a comparison of STEC associated with an outbreak with a high proportion of cases developing hemolytic-uremic syndrome (HUS) and STEC not associated with HUS identified several novel plasmids and fimbrial genes that were later investigated for roles in the observed increase in virulence (125).
Beyond the rapid detection of virulence genes, WGS, followed by SNP-based analysis, can also be used to build high-resolution phylogenies that provide insight into the relatedness of strains during outbreak investigations. MLVA has the highest discriminatory power of traditional typing methods for E. coli; however, SNP-based analysis had been consistently shown to provide better resolution (122). For example, in 2009, the United Kingdom had a large outbreak of STEC serotype O157:H7, which infected 93 people (126). Retrospective WGS was able to help determine that contamination by a single successful strain had spread through an entire farm to several animals before the first human case of EHEC infection (127). Each isolate involved in this outbreak was of serotype O157:H7 and ST11; therefore, the resolution necessary to make this conclusion by either of these techniques would not have been possible (127). Furthermore, only 9 to 25% of the STEC isolates in the United Kingdom are linked to an identified outbreak, with the rest of the cases assumed to be sporadic, and sporadic cases are rarely attributed to a source (36). Each clinical E. coli isolate is phage typed in the United Kingdom; however, the majority of isolates are PT8 or PT21/28, and therefore, this technique also provides insufficient resolution to connect sporadic cases (128). However, when 242 isolates responsible for apparently sporadic cases were retrospectively subjected to WGS, 136 were shown to be linked to an outbreak and differed by fewer than five SNPs (36). These results indicate that WGS can identify E. coli outbreaks that occur below the limit of detection by other typing tools. In a different retrospective analysis, SNPs were detected in a sample set of 11 groups of two or more epidemiologically linked isolates. Epidemiologically linked isolates formed phylogenetic clades with 100% bootstrap support that differed by fewer than four core genome SNPs (122).
The whole genomes of isolates from the largest E. coli O157:H7 outbreak in Alberta, Canada, were retrospectively sequenced, and the results of a SNP-based phylogenetic analysis were compared to those of MLVA, PFGE, and gene profiling of 49 STEC virulence genes. Isolates from the outbreak contained multiple yet closely related PFGE and MLVA patterns, while gene profiling was unable to differentiate outbreak isolates from unrelated sporadic cases (129). SNP-based phylogeny was able to place all epidemiologically linked outbreak isolates into one well-defined clade where isolates differed by 0 to 5 SNPs and differed from clades of concurrent but epidemiologically unlinked isolates by 231 to 257 SNPs (129).
Campylobacter
Campylobacteriosis, caused by Campylobacter jejuni (90%) and Campylobacter coli (10%), is the most common cause of self-limiting bacterial gastroenteritis globally and is also probably highly underreported (130). Campylobacter isolates are highly genetically diverse and regularly undergo horizontal genetic exchange; this diversity confounds the development of reproducible typing schemes, which are essential in both epidemiology and disease control (131). However, despite high levels of genetic exchange, Campylobacter populations are highly structured into clonal complexes of related bacteria that have an ancestor in common and have other properties, like host association range and virulence potential, in common (132, 133). It is likely that finding ways to intervene in the food chain to reduce the prevalence of Campylobacter in food-producing animals is the only way to reduce infections. To assess the effectiveness of interventions, WGS and subsequent SNP analysis are invaluable (134). Traditional MLST is useful in assigning isolates to clonal complexes, particularly because the same MLST scheme is used for both C. jejuni and C. coli—which allows comparison of inter- and intraspecies diversity (135); however, it is unable to establish relationships within and between clonal complexes (134). cgMLST is able to provide resolution within clonal complexes and information on the relatedness of clonal complexes and therefore provides better genetic attribution with respect to the source than MLST does, and this leads to a better understanding of the effectiveness of interventions (134).
WGS typing has been repeatedly shown to be more discriminatory than the traditional Campylobacter MLST, PFGE, and flaA typing methods (136). A retrospective study from Finland showed that 80% of C. jejuni clinical isolates are associated with only three clonal complexes (ST45, ST283, and ST677), demonstrating that MLST has a limited ability to determine if cases are sporadic or associated with other cases (137). However, cgMLST can provide enough resolution to reveal distinct clades within clonal complexes, genetically link apparent sporadic infections, and identify a common infection source (137). For example, comparative genomics was used to identify the proportion of human cases originating from chicken consumption versus swimming water. Since only four STs (ST45, ST230, ST267, and ST677) covered most of the isolates, MLST was unable to help identify the source of the human infections. However, WGS was able to link 24% of human cases directly to contaminated chicken slaughter batches and no cases were linked to swimming water (138), indicating that other sources of C. jejuni infection probably exist. While it was previously thought that epidemiologically linked isolates with the same or similar PFGE profiles were closely related and were expected to be the expansion of a single clone, it is now known that PFGE tends to overestimate the clonal relationships between Campylobacter isolates (139–141). For example, in a large waterborne outbreak of C. jejuni in 1998, several isolates were obtained from patients and the environment that were all of Penner serotype 12 and ST45 and had nearly identical KpnI and ScaII profiles (142). However, WGS of these isolates revealed that two patient strains, which were both epidemiologically related to the contaminated water, were genetically distinct enough to be considered different strains, indicating that the water probably harbored multiple contaminants (141).
Vibrio cholerae
On 12 January 2010, a magnitude 7.0 earthquake, centered near Port-au-Prince, struck Haiti and resulted in infrastructure damage including crippled sanitation systems that contributed to a devastating cholera outbreak starting in October. According to the Pan American Health Organization, this outbreak, which is still ongoing, has sickened >750,000 Haitians and killed 9,068 to date (143). In the first few weeks of the outbreak, the U.S. CDC analyzed the strain by PFGE and evidence suggested that the etiological strain originated in South Asia and was probably brought to the region by United Nations (UN) workers; however, several argued that this was not conclusive, as PFGE does not yield a detailed enough fingerprint for such a conclusion (144). Independent researchers proposed that, instead of human introduction, the cause was climate change that led to increases in temperature and salinity in the river estuaries around the Bay of Saint Marc in Haiti, leading to the competing climate hypothesis (145, 146). The main challenge in Vibrio outbreak source tracing is that the most common PFGE patterns tend to drift over the course of an outbreak, indicating that multiple concurrent outbreaks may be occurring, a possibility that, in this case, also challenged the single-source introduction hypothesis (147). Given the political and legal ramifications of aid workers being the source of this outbreak, a more detailed analysis of this outbreak by WGS was required.
V. cholerae can be classified into serogroups on the basis of the somatic (O) antigen, and at least 155 serogroups have been reported (148). Serogroup O1 isolates were responsible for all epidemic and endemic cholera cases until 1993, when serogroup O139 emerged and was linked to a cholera outbreak centering in Bangladesh and India (149). These serotypes can be further defined by biotyping into El Tor (hemolytic) and classical (nonhemolytic) (150). With only two circulating serogroups of V. cholerae responsible for the majority of illness, serotyping is not specific enough to trace outbreaks caused by this species. PFGE has been studied and validated extensively for use in the epidemiological surveillance of V. cholerae (151). The SfiI PFGE pattern of the Haitian strains was KZGS12.0088, and the NotI pattern was KZGN11.0092; however, from 2005 to 2010, strains with the same PFGE pattern had been isolated in Sri Lanka, India, Cameroon, Nepal, and Pakistan (152); therefore, PFGE results could not independently lead to a definitive conclusion about the origin of the cholera outbreak. The use of whole-genome MLST demonstrated that Haitian outbreak strains clearly formed a clade with strains isolated from Bangladesh (153). However, the strains used for comparison were isolated in Latin America in 1991 and South Asia in 2002 and 2008 and there was no guarantee that the strains circulating at those times were the same as the strains circulating in 2010. WGS of 24 V. cholerae genomes that were circulating in Nepal in the months leading up to the outbreak was performed (154). On the basis of a phylogenetic analysis of 184 parsimony-informative SNPs, three Nepalese isolates were differentiated from the Haitian isolates—which were identical to each other—by a single SNP, providing strong evidence that this clonal group was the source of the 2010 Haitian outbreak (154). As the outbreak continued, by 2012, various PFGE pattern combinations, serotypes, and antibiotic susceptibility patterns had been identified in V. cholerae isolates from Haiti; which led some investigators to question if multiple simultaneous outbreaks were taking place (146). However, the results of a WGS analysis of 23 V. cholerae isolates from 2010 to 2012 in Haiti supported the conclusion that the outbreak was clonal and that Nepalese isolates were the closest relatives, despite the presence of multiple PFGE patterns (147). This study was also important because WGS was performed in conjugation with bioinformatic analysis and the results were used to infer a molecular clock, which determined that the most recent common ancestor of the Haitian and Nepalese strains was estimated to be between 23 July and 17 October 2010. This aligned the cholera outbreak in Nepal and the arrival of Nepalese soldiers in Haiti with the start of the Haitian outbreak (145). WGS provided particularly strong evidence that Nepalese UN peacekeeping troops brought cholera to Haiti (154–156). The intricacies of this outbreak could not have been solved and the source of the contamination would not have been conclusively identified without the use of a WGS typing approach.
Foodborne Viruses
Viruses are the greatest reservoirs of genetic diversity on the planet (157), and they are constantly evolving under strong selection pressure (158). Viral WGS is now being used to provide information about the total viral population within infected organisms or environmental samples, and because of deep coverage at each position, second- and third-generation sequencing can be employed to identify the source and direction of transmission in an outbreak situation, which can aid in forensic investigation and intervention (159). This became apparent during the 2014-2015 Ebola outbreak (68, 69).
NoV is a highly infectious and rapidly evolving RNA virus that is responsible for the majority of acute gastroenteritis cases around the world (160–162). There is no vaccine or therapeutic intervention available to prevent or control NoV infections. Therefore, the development of a control strategy requires understanding of the sources of contamination and the mechanisms of new transmissions. To investigate the NoV transmission events and to examine interhost dynamics of NoVs, WGS was used to analyze genomic variations among three linked patients (163). Each recipient's major nonsynonymous variant was found to be identical to a minor variant isolated from the donor. In other words, a donor's minor variant may become the major variant in the recipient. This finding indicates that a strong bottleneck effect occurs in person-to-person transmission of NoVs. Viral WGS also has been used in source attribution during nosocomial transmission of NoV to immunocompromised patients in a United Kingdom hospital (164). Phylogenetic patterns and SNP-based analysis demonstrated that two out of three patients on the same ward were infected with closely related viruses, which indicated either nosocomial transmission or a single source of contamination of these two patients (164).
Hepatitis A virus (HAV)-associated foodborne outbreaks are frequently reported worldwide (165, 166). HAV is a nonenveloped, single-stranded, positive-sense RNA virus that belongs to the Picornaviridae family. In 2013, a large outbreak of HAV was reported in Italy that involved 1,202 cases (167, 168). Further study led to the hypothesis that the outbreak could be linked to frozen berries from two independent sources. To understand the relationship between these two potential sources, viral WGS was used. HAV was propagated on fetal rhesus monkey kidney cells inoculated with each of two samples and then sequenced via amplicon sequencing (169). SNP differences were not detected in the variable region (VP1-2A) when the two berry samples were compared to the clinical sequence, strongly indicating that the berries were the source of the outbreak (169).
Recently, there has been a suggestion of a correlation of beef consumption with an increased risk of colorectal cancer (170, 171). A potential explanation for this link is the presence in beef products of oncogenic viruses, such as polyomaviruses, that survive cooking (171). The presence of polyomaviruses was investigated by sequencing the metagenome of retail meat products after virion enrichment (172). Three polyomaviruses were identified by this study: bovine polyomavirus 1 (BoPyV1), BoPyV2, and BoPyV3 (172). BoPyV2 is phylogenetically related to Merkel cell polyomavirus and raccoon polyomavirus, both of which are shown to cause cancer in their native hosts (172). BoPyV2 has also been identified in retail meat products in San Francisco; however, it should be noted that neither of these studies has proven a conclusive link between virus infection and cancer (173).
Human adenoviruses (HAdVs) are among the most abundant enteric viruses in water. HAdV is a nonenveloped DNA virus with a genome composed of one double-stranded DNA chain containing 35 to 37 kbp, depending on the virus type (174). The clinical manifestations of HAdV infections vary from an absence of symptoms in healthy carriers to death in immunocompromised individuals. HAdVs can also be associated with enteric diseases (174). In the past, PCR and Sanger sequencing approaches were employed to detect HAdV contaminations in waste and river water matrices. However, these methods identify only a limited number predominant species, while water samples may contain multiple viral strains (175). Amplicon sequencing of the nonconserved hexon gene was used to improve the detection and identification of HAdV diversity in wastewater and river water matrices, and this provided high enough resolution to discriminate all 54 subtypes of HAdV (176).
Bridging the Gap with Historical Techniques
As sequencing-based technologies progress and bioinformatic analyses become commonplace, WGS is set to become the dominant typing method for investigating foodborne outbreaks and microbial contamination. Thus, it will become increasingly important to link WGS data to data in traditional typing databases such as PulseNet and PubMLST. This will allow for new WGS data to be placed into the proper historical context and enhance the capability to respond to outbreak events. One method of accomplishing this is to retrospectively sequence culture collections, as the FDA is doing as part of their GenomeTrakr network (177). Another method is to generate in silico results for traditional typing methods from WGS data, and several software packages are now available to do this (35). As mentioned earlier, MLST data can be readily extracted from assembled WGS data (28). The Microbial In Silico Typer (MIST) is a highly customizable tool that can be used to yield in silico results for one or more typing assays that are user defined. The MIST has been used successfully to generate VNTR and MLVA data for L. monocytogenes and to determine the pathotypes of E. coli isolates (178). Other software packages previously discussed in earlier sections, such as SISTR and SuperPhy, also have functions that can generate serovar/serotype predictions and help to compare current isolates in a historical context (109, 123). Beyond generating comparable results for historical techniques, several software platforms that attempt to predict if an isolate is pathogenic on the basis of its whole genome sequence are now becoming available and include PathogenFinder 1.1 (179) and the NCBI Pathogen Detection pipeline (http://ncbi.nlm.nih.gov/pathogens/).
USES OF WGS BEYOND HIGH-RESOLUTION SUBTYPING
Microbial Risk Assessment
Microbial risk assessments support the process of deciding whether to withdraw a food from the market during an outbreak event or assessing the effectiveness of intervention strategies along the farm-to-fork continuum (1). Several phenotypic markers, including virulence factors, host adaptation, stress resistance, and AMR, are important to microbial risk assessment, since they focus attention on microbial subpopulations that pose the most risk to human health (180). However, the potential uses of WGS in microbial risk assessment, beyond strain identity and clustering, are still unclear. The number of hazards identified by traditional phenotypic methods is dwarfed by those extracted from WGS data (181), and determining which information is most useful is difficult. However, for genotypic data to be useful to food safety authorities and policy makers, there must be an established way to use genotypic data to predict a phenotype that can accurately produce a true measure of risk, which is an active area of research (1). In addition, appropriate statistical methodology for the application of complex WGS data in risk assessment must be developed (181).
Geographic Source Attribution
Globalization of the food supply has created a situation where food- and waterborne outbreaks can take place on an international scale. In addition, the ingredients in a single food product may be sourced from different international locations, making source attribution particularly complicated in these situations. There is, however, substantial evidence that WGS data can be used to help predict the geographic origins of pathogenic isolates—and that this information can aid substantially in outbreak delineation. The idea that geographic source attribution could be successful came in 2012 during retrospective WGS. In that study, a Salmonella Bareilly outbreak in the United States was successfully linked by WGS to a scraped tuna product imported from India on the basis of sequence similarity to a 5-year-old historical isolate from the FDA archives that was collected from a processing facility <8 km away from the source of the 2012 outbreak (182). In addition to this initial observation, later studies have shown that geographic source attribution can be useful as a tool for delineating geographically dispersed outbreaks.
C. botulinum represents a group of spore-forming bacteria that are capable of causing fatal foodborne infection/intoxication. A recent study demonstrated that SNP analysis could resolve isolates by the geographic location of the origin of the outbreak (183).
On the basis of the evidence that WGS data can be used in geographic source attribution, the FDA has introduced the Genome Trakr network, which is predicated on the idea that the FDA's historical isolate collection could be sequenced, archived, and ultimately used to provide investigators with geographic clues surrounding the sources of large outbreaks (177). This would provide faster responses and intervention during large outbreaks. The SISTR platform has also been expanded to provide geographic predictions based on WGS of Salmonella isolates (109). However, it should be noted that these tools are only as good as the databases, and continual expansion of genome sequences from new and historical isolates is critical to future accuracy.
In Silico AMR Prediction
In addition to providing high-resolution typing schemes, WGS data are immediately available for secondary analyses. While traditional AMR testing is time-consuming and expensive, foodborne bacterial isolates can now be examined in silico for the presence of AMR genes immediately after sequencing. Several online software packages have been created to identify AMR genes in whole or partially assembled genomes, including the Resistance Gene Identifier (RGI) in the Comprehensive Antimicrobial Resistance Database (CARD) (184), the Antibiotic Resistance Database (ARDB) (185), and the Resistance Gene Finder (ResFinder) database (186). These databases vary in size and specificity. The ARDB contains 23,137 known resistance genes from 1,737 bacterial species, the CARD contains 4,221 gene that are responsible for resistance to many antibiotic classes, and the ResFinder database contains 1,800 resistance genes from 12 different antimicrobial classes (187). These tools have been reported in additional studies to have high concordance with phenotypic resistance. For example, a Danish study in which 200 foodborne isolates were phenotypically tested for susceptibility to 14 to 17 antibiotics showed that The ResFinder database was 99.7% accurate when predicting AMR and only struggled when predicting spectinomycin resistance in E. coli (188). AMR gene databases can also be downloaded, and genomes can be searched for resistance markers with custom scripts; this method appears to have good accuracy. For example, a study of Campylobacter AMR reported a 95.4 to 100% correlation between phenotypic and genotypic testing results, dependent on which antibiotic was evaluated (189). Different work on E. coli demonstrated a 97.8% correlation between phenotypic and genotypic testing results, where again the only discordant results were attributed to streptomycin (190). In addition to the fact that resistance to some antibiotics is harder to detect via genotypic methods, there are other limitations to WGS for antibiotic resistance prediction that must be addressed. For example, in members of the family Enterobacteriaceae, carbapenem resistance can result because of a combination of porin loss and extended-spectrum beta-lactamases enzymes but not because of either separately, and in silico detection algorithms struggle with situations like this (191). The genes underlying novel resistance mechanisms must also be identified in the laboratory before being added to the databases (192), and this represents a substantial amount of work.
METAGENOMICS AND CULTURE INDEPENDENCE
Metagenomic analysis, culture-independent analysis of the genetic material of all of the microbial DNA in a given environment, is often used to refer both to 16S rRNA gene amplification and subsequent sequencing and to “true” metagenomics, where all of the DNA extracted from a sample is sequenced. For food safety testing, current 16S rRNA sequencing protocols are not useful, since the short sequence reads provided by current next-generation sequencers are too short to correctly resolve closely related pathogenic and nonpathogenic species (e.g., L. monocytogenes and L. innocua [193]). This may change as third-generation sequencers are optimized for 16S rRNA sequencing or other more phylogenetically informative markers are further developed for amplicon sequencing. However, true metagenomic sequencing is already offering a culture-independent alternative for direct detection of pathogens in food in the case of nonculturable viruses and parasites (194). This technique may eventually be extended to culturable bacteria to decrease detection time and cost; however, there are numerous valid concerns about receiving positive signals because of the presence of DNA rather than a viable organism (195).
Culture-independent diagnostic testing will introduce major challenges to platforms that allow real-time communication of outbreak information, such as PulseNet. The concern is that as clinical settings switch to the use of rapid, nonculture tests, there will be fewer (or no) isolates from patients with foodborne illnesses. The CDC is currently working with the APHL, regulatory agencies, diagnostic kit manufacturers, and clinicians to ensure that positive test results will be followed by the collection of samples to allow for recovery of the bacterial pathogen.
CONCLUSIONS
The molecular landscape in food safety investigation is rapidly changing from the use of traditional molecular subtyping methods to WGS-based typing methods (Fig. 4). This change represents a shift from techniques that use discrete categorical indexing (i.e., a PFGE pattern, multilocus ST, or serotype) to a more continuous, custom-designed, and arguably infinitely flexible index (number of SNP differences) to define the relatedness of two isolates. This has brought about a transition period. This transition period is giving researchers the opportunity to address important issues such as standardization, quality control, methodology, and regulatory use. Currently, interpretation of WGS data requires the judgement of experts in the field such as epidemiologists, bioinformaticians, and microbiologists. It is also noteworthy that WGS has not and will not replace a good epidemiological investigation, but instead, the two must be thought of as complementary data sets for rapid delineation of outbreak events. The increased resolution provided by WGS provides epidemiologists with information that will help to link cases that would have been overlooked as sporadic in the past. WGS data can also be used for secondary analysis such as evaluating the effectiveness of contamination intervention strategies in the farm-to-fork-to-flush continuum, detection of AMR genes, and geographic source attribution. Several software packages are now available to aid in this analysis, and novel ones are continually being developed. In addition, from a strictly technical perspective, we are years away from being able to conduct culture-independent investigations and it is important that sequencing not completely displace culturing, as obtaining an isolate is still an important part of microbiology and secondary analysis. Because of the declining costs, increased resolution, and value-added secondary analysis NGS approaches offer to food safety, these modern approaches will continue to replace traditional molecular subtyping methods and drive improvements in global food safety.
FIG 4.
Past, present, and potential future workflows of pathogen detection by WGS and traditional subtyping techniques. Although times are approximate and vary according to the pathogen being analyzed, in all instances, WGS offers substantial time savings in comparison to traditional techniques. Future techniques using culture-independent metagenomic and high-throughput RNA sequencing technologies have the potential to offer faster detection times. SNV, SNP variation.
ACKNOWLEDGMENTS
We are very grateful to Sabah Bidawid for suggesting the topic for this review. We also thank Alex Gill, Erling Rud, Enrico Buenaventura, Kyle Ganz, and Amalia Martinez of the Bureau of Microbial Hazards, as well as Morag Graham of the Public Health Agency of Canada, for critically reviewing the manuscript and offering very helpful suggestions and comments. We also thank Angela Catford, who generously donated her time and photography skills to take and edit the images in the Author Bios section.
J.R. and N.N. acknowledge support from the Visiting Fellow in a Government Laboratory Program.
Biographies

Jennifer Ronholm is a visiting fellow in Health Canada's Bureau of Microbial Hazards. She completed her B.Sc. (Honors) in microbiology at the University of Waterloo and her Ph.D. in microbiology and immunology at the University of Ottawa under the supervision Dr. Min Lin and Dr. Xudong Cao. She received a postdoctoral fellowship from the Canadian Astrobiology Training Program and carried out her research at McGill University under the supervision of Dr. Lyle Whyte and Dr. Edward Cloutis. Her current research focuses on using NGS strategies for the detection and delineation of foodborne pathogens in outbreak situations. Additionally, she is interested in using culture-independent metagenomic strategies for pathogen detection and for understanding how indigenous bacterial communities affect food quality.

Neda Nasheri is currently a visiting fellow in Health Canada's Bureau of Microbial Hazards. She received her Ph.D. in Microbiology and Immunology from the University of Ottawa in 2015 studying host-virus interaction of the hepatitis C virus and earned her M.Sc. in Microbiology and Immunology for her work on the polymerase activity of paramyxoviruses. She received her B.Sc. Honors in Human Biology and Medical Sciences from the Baha'i Institute for Higher Education (Iran). Her current research interests are detection, characterization, and inactivation of foodborne viruses such as NoV, hepatitis E virus, and HAV.

Nicholas Petronella is a bioinformatician who first completed an honors double major undergraduate degree in computer science and biology at Queen's University in Kingston, Ontario, Canada. In 2011, he obtained a master's degree in bioinformatics at the University of Ottawa. While employed as a student at Health Canada under the Federal Student Work Experience Program, he built, managed, and utilized a computer lab to assist with heavy computational tasks. After completion of his master's degree, he was hired by the Food Directorate at Health Canada as a bioinformatician. Currently, his focus is on WGS, specializing in writing custom software and tools, performing robust genomic analyses, and training Health Canada's premarket evaluators and health risk assessors, in addition to being a driving force behind the continuous scientific innovations at Health Canada.

Franco Pagotto is a research scientist at Health Canada in the Bureau of Microbial Hazards. Dr. Pagotto graduated from the University of Ottawa's Medical School in the Department of Biochemistry, Microbiology, and Immunology. He oversees the research activities of the Listeria and Cronobacter laboratory, and some of the projects under way in his laboratory include the microbiological safety and control of fresh-cut produce, hazard identification and risk assessment of L. monocytogenes and Cronobacter species in foods, molecular diagnostics, and genomic characterization of foodborne bacterial pathogens. Dr. Pagotto is the codirector of the Listeriosis Reference Centre for Canada.
REFERENCES
- 1.Franz E, Gras LM, Dallman T. 2016. Significance of whole genome sequencing for surveillance, source attribution and microbial risk assessment of foodborne pathogens. Curr Opin Food Sci 8:74–79. doi: 10.1016/j.cofs.2016.04.004. [DOI] [Google Scholar]
- 2.Grimont PAD, Weill F-X. 2007. Antigenic formulae of the Salmonella serovars, 9th ed WHO Collaborating Centre for Reference and Research on Salmonella, Paris, France: http://www.scacm.org/free/Antigenic%20Formulae%20of%20the%20Salmonella%20Serovars%202007%209th%20edition.pdf. [Google Scholar]
- 3.McCallum KL, Whitfield C. 1991. The rcsA gene of Klebsiella pneumoniae O1:K20 is involved in expression of the serotype-specific K (capsular) antigen. Infect Immun 59:494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoki KR. 2001. Pharmacology and immunology of botulinum toxin serotypes. J Neurol 248:I3–I10. doi: 10.1007/PL00007816. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed R, Bopp C, Borczyk A, Kasatiya S. 1987. Phage-typing scheme for Escherichia coli O157:H7. J Infect Dis 155:806–809. doi: 10.1093/infdis/155.4.806. [DOI] [PubMed] [Google Scholar]
- 6.Hickman-Brenner FW, Stubbs AD, Farmer J. 1991. Phage typing of Salmonella enteritidis in the United States. J Clin Microbiol 29:2817–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ward LR, de Sa JDH, Rowe B. 1987. A phage-typing scheme for Salmonella enteritidis. Epidemiol Infect 99:291–294. doi: 10.1017/S0950268800067765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willshaw G. 2001. Use of strain typing to provide evidence for specific interventions in the transmission of VTEC O157 infections. Int J Food Microbiol 66:39–46. doi: 10.1016/S0168-1605(00)00511-0. [DOI] [PubMed] [Google Scholar]
- 9.Laconcha I, López-Molina N, Rementeria A, Audicana A, Perales I, Garaizar J. 1998. Phage typing combined with pulsed-field gel electrophoresis and random amplified polymorphic DNA increases discrimination in the epidemiological analysis of Salmonella enteritidis strains. Int J Food Microbiol 40:27–34. doi: 10.1016/S0168-1605(98)00007-5. [DOI] [PubMed] [Google Scholar]
- 10.Demczuk W, Soule G, Clark C, Ackermann HW, Easy R, Khakhria R, Rodgers F, Ahmed R. 2003. Phage-based typing scheme for Salmonella enterica serovar Heidelberg, a causative agent of food poisonings in Canada. J Clin Microbiol 41:4279–4284. doi: 10.1128/JCM.41.9.4279-4284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindstedt B-A. 2005. Multiple-locus variable number tandem repeats analysis for genetic fingerprinting of pathogenic bacteria. Electrophoresis 26:2567–2582. doi: 10.1002/elps.200500096. [DOI] [PubMed] [Google Scholar]
- 13.Keys C, Kemper S, Keim P. 2005. Highly diverse variable number tandem repeat loci in the E. coli O157:H7 and O55:H7 genomes for high-resolution molecular typing. J Appl Microbiol 98:928–940. doi: 10.1111/j.1365-2672.2004.02532.x. [DOI] [PubMed] [Google Scholar]
- 14.Torpdahl M, Sørensen G, Lindstedt B-A, Nielsen EM. 2007. Tandem repeat analysis for surveillance of human Salmonella Typhimurium infections. Emerg Infect Dis 13:388–395. doi: 10.3201/eid1303.060460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang SY, Watanabe H, Terajima J, Li CC, Liao JC, Tung SK, Chiou CS. 2007. Multilocus variable-number tandem-repeat analysis for molecular typing of Shigella sonnei. J Clin Microbiol 45:3574–3580. doi: 10.1128/JCM.00675-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savelkoul PH, Aarts HJ, de Haas J, Dijkshoorn L, Duim B, Otsen M, Rademaker JL, Schouls L, Lenstra JA. 1999. Amplified-fragment length polymorphism analysis: the state of an art. J Clin Microbiol 37:3083–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynne AM, Rhodes-Clark BS, Bliven K, Zhao S, Foley SL. 2008. Antimicrobial resistance genes associated with Salmonella enterica serovar Newport isolates from food animals. Antimicrob Agents Chemother 52:353–356. doi: 10.1128/AAC.00842-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng H, Sun Y, Mao Z, Jiang B. 2008. Investigation of virulence genes in clinical isolates of Yersinia enterocolitica. FEMS Immunol Med Microbiol 53:368–374. doi: 10.1111/j.1574-695X.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- 19.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett TJ, Gerner-Smidt P, Swaminathan B. 2006. Interpretation of pulsed-field gel electrophoresis patterns in foodborne disease investigations and surveillance. Foodborne Pathog Dis 3:20–31. doi: 10.1089/fpd.2006.3.20. [DOI] [PubMed] [Google Scholar]
- 21.Chisholm SA, Crichton PB, Knight HI, Old DC. 1999. Molecular typing of Salmonella serotype Thompson strains isolated from human and animal sources. Epidemiol Infect 122:33–39. doi: 10.1017/S0950268898001836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foley SL, Lynne AM, Nayak R. 2009. Molecular typing methodologies for microbial source tracking and epidemiological investigations of Gram-negative bacterial foodborne pathogens. Infect Genet Evol 9:430–440. doi: 10.1016/j.meegid.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 23.Centers for Disease Control and Prevention. 2004. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis training. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/pulsenet/resources/training-and-outreach.html. [Google Scholar]
- 24.Barrett TJ, Lior H, Green JH, Khakhria R, Wells JG, Bell BP, Greene KD, Lewis J, Griffin PM. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J Clin Microbiol 32:3013–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV, CDC PulseNet Task Force . 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg Infect Dis 7:382–389. doi: 10.3201/eid0703.017303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephenson J. 1997. New approaches for detecting and curtailing foodborne microbial infections. JAMA 277:1337–1340. [DOI] [PubMed] [Google Scholar]
- 27.Kotetishvili M, Stine OC, Kreger A, Morris JG, Sulakvelidze A. 2002. Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J Clin Microbiol 40:1626–1635. doi: 10.1128/JCM.40.5.1626-1635.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spratt BG. 1999. Multilocus sequence typing: molecular typing of bacterial pathogens in an era of rapid DNA sequencing and the Internet. Curr Opin Microbiol 2:312–316. doi: 10.1016/S1369-5274(99)80054-X. [DOI] [PubMed] [Google Scholar]
- 30.Allard MW, Luo Y, Strain E, Li C, Keys CE, Son I, Stones R, Musser SM, Brown EW. 2012. High resolution clustering of Salmonella enterica serovar Montevideo strains using a next-generation sequencing approach. BMC Genomics 13:32. doi: 10.1186/1471-2164-13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pightling AW, Petronella N, Pagotto F. 2014. Choice of reference sequence and assembler for alignment of Listeria monocytogenes short-read sequence data greatly influences rates of error in SNP analyses. PLoS One 9:e104579-11. doi: 10.1371/journal.pone.0104579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pightling AW, Petronella N, Pagotto F. 2015. The Listeria monocytogenes Core-Genome Sequence Typer (LmCGST): a bioinformatic pipeline for molecular characterization with next-generation sequence data. BMC Microbiol 15:224. doi: 10.1186/s12866-015-0526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruppitsch W, Pietzka A, Prior K, Bletz S, Fernandez HL, Allerberger F, Harmsen D, Mellmann A. 2015. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J Clin Microbiol 53:2869–2876. doi: 10.1128/JCM.01193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orsi RH, Borowsky ML, Lauer P, Young SK, Nusbaum C, Galagan JE, Birren BW, Ivy RA, Sun Q, Graves LM, Swaminathan B, Wiedmann M. 2008. Short-term genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genomics 9:539. doi: 10.1186/1471-2164-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwong JC, Mercoulia K, Tomita T, Easton M, Li HY, Bulach DM, Stinear TP, Seemann T, Howden BP. 2016. Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J Clin Microbiol 54:333–342. doi: 10.1128/JCM.02344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dallman TJ, Byrne L, Ashton PM, Cowley LA, Perry NT, Adak G, Petrovska L, Ellis RJ, Elson R, Underwood A, Green J, Hanage WP, Jenkins C, Grant K, Wain J. 2015. Whole-genome sequencing for national surveillance of Shiga toxin-producing Escherichia coli O157. Clin Infect Dis 61:305–312. doi: 10.1093/cid/civ318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanger F, Sicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith LM, Sanders JZ, Kaiser RJ, Hughes P, Dodd C, Connell CR, Heiner C, Kent SB, Hood LE. 1986. Fluorescence detection in automated DNA sequence analysis. Nature 321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- 39.Fleischmann R, Adams M, White O, Clayton R, Kirkness E, Kerlavage A, Bult C, Tomb J, Dougherty B, Merrick J, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae. Science 269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 40.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 41.Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MT, Prentice MB, Sebaihia M, James KD, Churcher C, Mungall KL, Baker S, Basham D, Bentley SD, Brooks K, Cerdeño-Tárraga AM, Chillingworth T, Cronin A, Davies RM, Davis P, Dougan G, Feltwell T, Hamlin N, Holroyd S, Jagels K, Karlyshev AV, Leather S, Moule S, Oyston PC, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- 42.Blattner FR, Guy Plunkett I, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 43.Kunst F, Ogasawara N, Moszer I, Albertini AM. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 44.Fraser CM. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 45.Parkhill J. 2000. In defense of complete genomes. Nat Biotechnol 18:493–494. doi: 10.1038/75346. [DOI] [PubMed] [Google Scholar]
- 46.Koren S, Phillippy AM. 2015. One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Curr Opin Microbiol 23:110–120. doi: 10.1016/j.mib.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 47.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman K, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Metzker ML. 2010. Sequencing technologies—the next generation. Nat Rev Genet 11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 49.Schatz MC, Delcher AL, Salzberg SL. 2010. Assembly of large genomes using second-generation sequencing. Genome Res 20:1165–1173. doi: 10.1101/gr.101360.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Compeau PEC, Pevzner PA, Tesler G. 2011. How to apply de Bruijn graphs to genome assembly. Nat Biotechnol 29:987–991. doi: 10.1038/nbt.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Needleman SB, Wunsch CD. 1970. A general method applicable to the search for similarities in the amino acid sequence [sic] of two proteins. J Mol Biol 48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 55.Smith TF, Waterman MS, Fitch WM. 1981. Comparative biosequence metrics. J Mol Evol 18:38–46. doi: 10.1007/BF01733210. [DOI] [PubMed] [Google Scholar]
- 56.Smith TF, Waterman MS. 1981. Identification of common molecular subsequences. J Mol Biol 147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 57.Alkan C, Sajjadian S, Eichler EE. 2011. Limitations of next-generation genome sequence assembly. Nat Methods 8:61–65. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kolmogorov M, Raney B, Paten B, Pham S. 2014. Ragout—a reference-assisted assembly tool for bacterial genomes. Bioinformatics 30:i302–i309. doi: 10.1093/bioinformatics/btu280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim J, Larkin DM, Cai Q, Asan Zhang Y, Ge RL, Auvil L, Capitanu B, Zhang G, Lewin HA, Ma J. 2013. Reference-assisted chromosome assembly. Proc Natl Acad Sci U S A 110:1785–1790. doi: 10.1073/pnas.1220349110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCoy RC, Taylor RW, Blauwkamp TA, Kelley JL, Kertesz M, Pushkarev D, Petrov DA, Fiston-Lavier AS. 2014. Illumina TruSeq synthetic long-reads empower de novo assembly and resolve complex, highly repetitive transposable elements. PLoS One 9:e106689. doi: 10.1371/journal.pone.0106689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fraser CM, Eisen JA, Nelson KE, Paulsen IT, Salzberg SL. 2002. The value of complete microbial genome sequencing (you get what you pay for). J Bacteriol 184:6403–6405. doi: 10.1128/JB.184.23.6403-6405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Selkov E, Overbeek R, Kogan Y, Chu L, Vonstein V, Holmes D, Silver S, Haselkorn R, Fonstein M. 2000. Functional analysis of gapped microbial genomes: amino acid metabolism of Thiobacillus ferrooxidans. Proc Natl Acad Sci U S A 97:3509–3514. doi: 10.1073/pnas.97.7.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dhillon BK, Laird MR, Shay JA, Winsor GL, Lo R, Nizam F, Pereira SK, Waglechner N, McArthur AG, Langille MGI, Brinkman FSL. 2015. IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res 43(W1):W104–W108. doi: 10.1093/nar/gkv401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomma BPHJ, Seidl MF, Shi-Kunne X, Cook DE, Bolton MD, van Kan JAL, Faino L. 2016. Mind the gap; seven reasons to close fragmented genome assemblies. Fungal Genet Biol 90:24–30. doi: 10.1016/j.fgb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 65.Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, Causey M, Colonell J, Dimeo J, Efcavitch JW, Giladi E, Gill J, Healy J, Jarosz M, Lapen D, Moulton K, Quake SR, Steinmann K, Thayer E, Tyurina A, Ward R, Weiss H, Xie Z. 2008. Single-molecule DNA sequencing of a viral genome. Science 320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 66.Pushkarev D, Neff NF, Quake SR. 2009. Single-molecule sequencing of an individual human genome. Nat Biotechnol 27:847–850. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. 2009. Real-time DNA sequencing from single polymerase molecules. Science 323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 68.Quick J, Loman NJ, Duraffour S, Simpson JT, Severi E. 2016. Real-time, portable genome sequencing for Ebola surveillance. Nature 530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Check Hayden E. 2015. Pint-sized DNA sequencer impresses first users. Nature 521:15–16. doi: 10.1038/521015a. [DOI] [PubMed] [Google Scholar]
- 70.Koren S, Harhay GP, Smith TP, Bono JL, Harhay DM, Mcvey SD, Radune D, Bergman NH, Phillippy AM. 2013. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol 14:R101. doi: 10.1186/gb-2013-14-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G, Wang Z, Rasko DA, McCombie WR, Jarvis ED, Phillippy AM. 2012. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol 30:693–700. doi: 10.1038/nbt.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liao Y-C, Lin S-H, Lin H-H. 2015. Completing bacterial genome assemblies: strategy and performance comparisons. Sci Rep 5:8747. doi: 10.1038/srep08747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Field D, Field D, Amaral-Zettler L, Cochrane G, Cole JR, Dawyndt P, Garrity GM, Gilbert J, Glöckner FO, Hirschman L, Karsch-Mizrachi I, Klenk H-P, Knight R, Kottmann R, Kyrpides N, Meyer F, San Gil I, Sansone SA, Schriml LM, Stark P, Tatusova T, Ussery DW, White O, Wooley J. 2011. The Genomic Standards Consortium. PLoS Biol 9:e1001088. doi: 10.1371/journal.pbio.1001088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Salzberg SL, Phillippy AM, Zimin A, Puiu D, Magoc T, Koren S, Treangen TJ, Schatz MC, Delcher AL, Roberts M, Marcais G, Pop M, Yorke JA. 2012. GAGE: a critical evaluation of genome assemblies and assembly algorithms. Genome Res 22:557–567. doi: 10.1101/gr.131383.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Magoc T, Pabinger S, Canzar S, Liu X, Su Q, Puiu D, Tallon LJ, Salzberg SL. 2013. GAGE-B: an evaluation of genome assemblers for bacterial organisms. Bioinformatics 29:1718–1725. doi: 10.1093/bioinformatics/btt273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Farber JM, Peterkin PI. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev 55:476–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goulet V, King LA, Vaillant V, de Valk H. 2013. What is the incubation period for listeriosis? BMC Infect Dis 13:11. doi: 10.1186/1471-2334-13-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carpentier B, Cerf O. 2011. Review—persistence of Listeria monocytogenes in food industry equipment and premises. Int J Food Microbiol 145:1–8. doi: 10.1016/j.ijfoodmicro.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 79.Ferreira V, Wiedmann M, Teixeira P, Stasiewicz MJ. 2014. Listeria monocytogenes persistence in food-associated environments: epidemiology, strain characteristics, and implications for public health. J Food Prot 77:150–170. doi: 10.4315/0362-028X.JFP-13-150. [DOI] [PubMed] [Google Scholar]
- 80.Gilmour MW, Graham M, Van Domselaar G, Tyler S, Kent H, Trout-Yakel KM, Larios O, Allen V, Lee B, Nadon C. 2010. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 11:120. doi: 10.1186/1471-2164-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stasiewicz MJ, Oliver HF, Wiedmann M, den Bakker HC. 2015. Whole-genome sequencing allows for improved identification of persistent Listeria monocytogenes in food-associated environments. Appl Environ Microbiol 81:6024–6037. doi: 10.1128/AEM.01049-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hamon M, Bierne H, Cossart P. 2006. Listeria monocytogenes: a multifaceted model. Nat Rev Microbiol 4:423–434. doi: 10.1038/nrmicro1413. [DOI] [PubMed] [Google Scholar]
- 83.Schmid D, Allerberger F, Huhulescu S, Pietzka A, Amar C, Kleta S, Prager R, Preußel K, Aichinger E, Mellmann A. 2014. Whole genome sequencing as a tool to investigate a cluster of seven cases of listeriosis in Austria and Germany, 2011–2013. Clin Microbiol Infect 20:431–436. doi: 10.1111/1469-0691.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bergholz TM, den Bakker HC, Katz LS, Silk BJ, Jackson KA, Kucerova Z, Joseph LA, Turnsek M, Gladney LM, Halpin JL, Xavier K, Gossack J, Ward TJ, Frace M, Tarr CL. 2016. Determination of evolutionary relationships of outbreak-associated Listeria monocytogenes strains of serotypes 1/2a and 1/2b by whole-genome sequencing. Appl Environ Microbiol 82:928–938. doi: 10.1128/AEM.02440-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kuenne C, Billion A, Mraheil MA, Strittmatter A, Daniel R, Goesmann A, Barbuddhe S, Hain T, Chakraborty T. 2013. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics 14:47. doi: 10.1186/1471-2164-14-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Q, Holmes N, Martinez E, Howard P, Hill-Cawthorne G, Sintchenko V. 2015. It is not all about single nucleotide polymorphisms: comparison of mobile genetic elements and deletions in Listeria monocytogenes genomes links cases of hospital-acquired listeriosis to the environmental source. J Clin Microbiol 53:3492–3500. doi: 10.1128/JCM.00202-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Timme RE, Pettengill JB, Allard MW, Strain E, Barrangou R, Wehnes C, Van Kessel JS, Karns JS, Musser SM, Brown EW. 2013. Phylogenetic diversity of the enteric pathogen Salmonella enterica subsp. enterica inferred from genome-wide reference-free SNP characters. Genome Biol Evol 5:2109–2123. doi: 10.1093/gbe/evt159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Allard MW, Luo Y, Strain E, Pettengill J, Timme R, Wang C, Li C, Keys CE, Zheng J, Stones R, Wilson MR, Musser SM, Brown EW. 2013. On the evolutionary history, population genetics and diversity among isolates of Salmonella enteritidis PFGE pattern JEGX010004. PLoS One 8:e55254. doi: 10.1371/journal.pone.0055254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sangal V, Harbottle H, Mazzoni CJ, Helmuth R, Guerra B, Didelot X, Paglietti B, Rabsch W, Brisse S, Weill FX, Roumagnac P, Achtman M. 2010. Evolution and population structure of Salmonella enterica serovar Newport. J Bacteriol 192:6465–6476. doi: 10.1128/JB.00969-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Trujillo S, Keys CE, Brown EW. 2011. Evaluation of the taxonomic utility of six-enzyme pulsed-field gel electrophoresis in reconstructing Salmonella subspecies phylogeny. Infect Genet Evol 11:92–102. doi: 10.1016/j.meegid.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 92.den Bakker HC, Allard MW, Bopp D, Brown EW, Fontana J, Iqbal Z, Kinney A, Limberger R, Musser KA, Shudt M, Strain E, Wiedmann M, Wolfgang WJ. 2014. Rapid whole-genome sequencing for surveillance of Salmonella enterica serovar Enteritidis. Emerg Infect Dis 20:1306–1314. doi: 10.3201/eid2008.131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chai SJ, White PL, Lathrop SL, Solghan SM, Medus C, McGlinchey BM, Tobin-D'Angelo M, Marcus R, Mahon BE. 2012. Salmonella enterica serotype Enteritidis: increasing incidence of domestically acquired infections. Clin Infect Dis 54:S488–S497. doi: 10.1093/cid/cis231. [DOI] [PubMed] [Google Scholar]
- 94.Mohammed M, Delappe N, O'Connor J, McKeown P, Garvey P, Cormican M. 2016. Whole genome sequencing provides an unambiguous link between Salmonella Dublin outbreak strain and a historical isolate. Epidemiol Infect 144:576–581. doi: 10.1017/S0950268815001636. [DOI] [PubMed] [Google Scholar]
- 95.Wuyts V, Denayer S, Roosens NHC, Mattheus W, Bertrand S, Marchal K, Dierick K, De Keersmaecker SCJ. 11 September 2015. Whole genome sequence analysis of Salmonella Enteritidis PT4 outbreaks from a national reference laboratory's viewpoint. PLoS Curr doi: 10.1371/currents.outbreaks.aa5372d90826e6cb0136ff66bb7a62fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taylor AJ, Lappi V, Wolfgang WJ, Lapierre P, Palumbo MJ, Medus C, Boxrud D. 2015. Characterization of foodborne outbreaks of Salmonella enterica serovar Enteritidis with whole-genome sequencing single nucleotide polymorphism-based analysis for surveillance and outbreak detection. J Clin Microbiol 53:3334–3340. doi: 10.1128/JCM.01280-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deng X, Shariat N, Driebe EM, Roe CC, Tolar B, Trees E, Keim P, Zhang W, Dudley EG, Fields PI, Engelthaler DM. 2015. Comparative analysis of subtyping methods against a whole-genome-sequencing standard for Salmonella enterica serotype Enteritidis. J Clin Microbiol 53:212–218. doi: 10.1128/JCM.02332-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fernandez-Cuesta L, Sun R, Menon R, George J, Lorenz S, Meza-Zepeda LA, Peifer M, Plenker D, Heuckmann JM, Leenders F, Zander T, Dahmen I, Koker M, Schöttle J, Ullrich RT, Altmüller J, Becker C, Nürnberg P, Seidel H, Böhm D, Göke F, Ansén S, Russell PA, Wright GM, Wainer Z, Solomon B, Petersen I, Clement JH, Sänger J, Brustugun OT, Helland Å, Solberg S, Lund-Iversen M, Buettner R, Wolf J, Brambilla E, Vingron M, Perner S, Haas SA, Thomas RK. 2015. Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella. Genome Biol 16:7. doi: 10.1186/s13059-014-0558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leekitcharoenphon P, Nielsen EM, Kaas RS, Lund O, Aarestrup FM. 2014. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS One 9:e87991. doi: 10.1371/journal.pone.0087991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Octavia S, Wang Q, Tanaka MM, Kaur S, Sintchenko V, Lan R. 2015. Delineating community outbreaks of Salmonella enterica serovar Typhimurium by use of whole-genome sequencing: insights into genomic variability within an outbreak. J Clin Microbiol 53:1063–1071. doi: 10.1128/JCM.03235-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ashton PM, Peters T, Ameh L, McAleer R, Petrie S, Nair S, Muscat I, de Pinna E, Dallmand T. 10 February 2015. Whole genome sequencing for the retrospective investigation of an outbreak of Salmonella Typhimurium DT 8. PLoS Curr doi: 10.1371/currents.outbreaks.2c05a47d292f376afc5a6fcdd8a7a3b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alexander DC, Fitzgerald SF, DePaulo R, Kitzul R, Daku D, Levett PN, Cameron ADS. 2016. Laboratory-acquired infection with Salmonella enterica serovar Typhimurium exposed by whole-genome sequencing. J Clin Microbiol 54:190–193. doi: 10.1128/JCM.02720-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bekal S, Berry C, Reimer AR, Van Domselaar G, Beaudry G, Fournier E, Doualla-Bell F, Levac E, Gaulin C, Ramsay D, Huot C, Walker M, Sieffert C, Tremblay C. 2016. Usefulness of high-quality core genome single-nucleotide variant analysis for subtyping the highly clonal and the most prevalent Salmonella enterica serovar Heidelberg clone in the context of outbreak investigations. J Clin Microbiol 54:289–295. doi: 10.1128/JCM.02200-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lienau EK, Strain E, Wang C, Zheng J, Ottesen AR, Keys CE, Hammack TS, Musser SM, Brown EW, Allard MW, Cao G, Meng J, Stones R. 2011. Identification of a salmonellosis outbreak by means of molecular sequencing. N Engl J Med 364:981–982. doi: 10.1056/NEJMc1100443. [DOI] [PubMed] [Google Scholar]
- 105.Bakker HC, Moreno Switt AI, Cummings CA, Hoelzer K, Degoricija L, Rodriguez-Rivera LD, Wright EM, Fang R, Davis M, Root T, Schoonmaker-Bopp D, Musser KA, Villamil E, Waechter H, Kornstein L, Furtado MR, Wiedmann M. 2011. A whole-genome single nucleotide polymorphism-based approach to trace and identify outbreaks linked to a common Salmonella enterica subsp. enterica serovar Montevideo pulsed-field gel electrophoresis type. Appl Environ Microbiol 77:8648–8655. doi: 10.1128/AEM.06538-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Didelot X, Bowden R, Street T, Golubchik T, Spencer C, McVean G, Sangal V, Anjum MF, Achtman M, Falush D, Donnelly P. 2011. Recombination and population structure in Salmonella enterica. PLoS Genet 7:e1002191. doi: 10.1371/journal.pgen.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Byrne L, Fisher I, Peters T, Mather A, Thomson N, Rosner B, Bernard H, McKeown P, Cormican M, Cowden J, Aiyedun V, Lane C, International Outbreak Control Team . 2014. A multi-country outbreak of Salmonella Newport gastroenteritis in Europe associated with watermelon from Brazil, confirmed by whole genome sequencing: October 2011 to January 2012. Euro Surveill 19:6–13. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang S, Yin Y, Jones MB, Zhang Z, Deatherage Kaiser BL, Dinsmore BA, Fitzgerald C, Fields PI, Deng X. 2015. Salmonella serotype determination utilizing high-throughput genome sequencing data. J Clin Microbiol 53:1685–1692. doi: 10.1128/JCM.00323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VPJ, Nash JHE, Taboada EN. 2016. The Salmonella In Silico Typing Resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ashton PM, Nair S, Peters TM, Bale JA, Powell DG, Painset A, Tewolde R, Schaefer U, Jenkins C, Dallman TJ, de Pinna EM, Grant KA, Salmonella Whole Genome Sequencing Implementation Group . 2016. Identification of Salmonella for public health surveillance using whole genome sequencing. PeerJ 4:e1752 https://doi.org/10.7717/peerj.1752. doi: 10.7717/peerj.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. 2013. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev 26:822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lan R, Alles MC, Donohoe K, Martinez MB, Reeves PR. 2004. Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp. Infect Immun 72:5080–5088. doi: 10.1128/IAI.72.9.5080-5088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui El M, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguénec C, Lescat M, Mangenot S, Martinez-Jéhanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Saint Ruf C, Schneider D, Tourret J, Vacherie B, Vallenet D, Médigue C, Eduardo Rocha PC, Denamur E. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kaas RS, Friis C, Ussery DW, Aarestrup FM. 2012. Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes. BMC Genomics 13:577. doi: 10.1186/1471-2164-13-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Qadri F, Svennerholm AM, Faruque ASG, Sack RB. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin Microbiol Rev 18:465–483. doi: 10.1128/CMR.18.3.465-483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A 92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Asadulghani M, Ogura Y, Ooka T, Itoh T, Sawaguchi A, Iguchi A, Nakayama K, Hayashi T. 2009. The defective prophage pool of Escherichia coli O157: prophage-prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog 5:e1000408. doi: 10.1371/journal.ppat.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Manning SD, Motiwala AS, Springman AC, Qi W, Lacher DW, Ouellette LM, Mladonicky JM, Somsel P, Rudrik JT, Dietrich SE, Zhang W, Swaminathan B, Alland D, Whittam TS. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc Natl Acad Sci U S A 105:4868–4873. doi: 10.1073/pnas.0710834105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fratamico PM, DebRoy C, Liu Y, Needleman DS, Baranzoni GM, Feng P. 2016. Advances in molecular serotyping and subtyping of Escherichia coli. Front Microbiol 7:644. doi: 10.3389/fmicb.2016.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lambert D, Carrillo CD, Koziol AG, Manninger P, Blais BW. 2015. GeneSippr: a rapid whole-genome approach for the identification and characterization of foodborne pathogens such as priority Shiga toxigenic Escherichia coli. PLoS One 10:e0122928. doi: 10.1371/journal.pone.0122928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Holmes A, Allison L, Ward M, Dallman TJ, Clark R, Fawkes A, Murphy L, Hanson M. 2015. Utility of whole-genome sequencing of Escherichia coli O157 for outbreak detection and epidemiological surveillance. J Clin Microbiol 53:3565–3573. doi: 10.1128/JCM.01066-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Whiteside MD, Laing CR, Manji A, Kruczkiewicz P, Taboada EN, Gannon VPJ. 2016. SuperPhy: predictive genomics for the bacterial pathogen Escherichia coli. BMC Microbiol 16:65. doi: 10.1186/s12866-016-0680-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Joensen KG, Tetzschner AMM, Iguchi A, Aarestrup FM, Scheutz F. 2015. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol 53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ihekweazu C, Carroll K, Adak B, Smith G, Pritchard GC, Gillespie IA, Verlander NQ, Harvey-Vince L, Reacher M, Edeghere O, Sultan B, Cooper R, Morgan G, Kinross PTN, Boxall NS, Iversen A, Bickler G. 2012. Large outbreak of verocytotoxin-producing Escherichia coli O157 infection in visitors to a petting farm in South East England, 2009. Epidemiol Infect 140:1400–1413. doi: 10.1017/S0950268811002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Underwood AP, Dallman T, Thomson NR, Williams M, Harker K, Perry N, Adak B, Willshaw G, Cheasty T, Green J, Dougan G, Parkhill J, Wain J. 2013. Public health value of next-generation DNA sequencing of enterohemorrhagic Escherichia coli isolates from an outbreak. J Clin Microbiol 51:232–237. doi: 10.1128/JCM.01696-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Khakhria R, Duck D, Lior H. 1990. Extended phage-typing scheme for Escherichia coli O157:H7. Epidemiol Infect 105:511–520. doi: 10.1017/S0950268800048135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Berenger BM, Berry C, Peterson T, Fach P, Delannoy S, Li V, Tschetter L, Nadon C, Honish L, Louie M, Chui L. 2015. The utility of multiple molecular methods including whole genome sequencing as tools to differentiate Escherichia coli O157:H7 outbreaks. Euro Surveill 20:30073. doi: 10.2807/1560-7917.ES.2015.20.47.30073. [DOI] [PubMed] [Google Scholar]
- 130.Strachan NJ, Forbes KJ. 2010. The growing UK epidemic of human campylobacteriosis. Lancet 376:665–667. doi: 10.1016/S0140-6736(10)60708-8. [DOI] [PubMed] [Google Scholar]
- 131.Sheppard SK, Jolley KA, Maiden MCJ. 2012. A gene-by-gene approach to bacterial population genomics: whole genome MLST of Campylobacter. Genes 3:261–277. doi: 10.3390/genes3020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sheppard SK, Colles F, Richardson J, Cody AJ, Elson R, Lawson A, Brick G, Meldrum R, Little CL, Owen RJ, Maiden MCJ, McCarthy ND. 2010. Host association of Campylobacter genotypes transcends geographic variation. Appl Environ Microbiol 76:5269–5277. doi: 10.1128/AEM.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sheppard SK, Dallas JF, Wilson DJ, Strachan NJC, McCarthy ND, Jolley KA, Colles FM, Rotariu O, Ogden ID, Forbes KJ, Maiden MCJ. 2010. Evolution of an agriculture-associated disease causing Campylobacter coli clade: evidence from national surveillance data in Scotland. PLoS One 5:e15708. doi: 10.1371/journal.pone.0015708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cody AJ, McCarthy ND, Jansen van Rensburg M, Isinkaye T, Bentley SD, Parkhill J, Dingle KE, Bowler ICJW, Jolley KA, Maiden MCJ. 2013. Real-time genomic epidemiological evaluation of human Campylobacter isolates by use of whole-genome multilocus sequence typing. J Clin Microbiol 51:2526–2534. doi: 10.1128/JCM.00066-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Miller WG, On SLW, Wang G, Fontanoz S, Lastovica AJ, Mandrell RE. 2005. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C upsaliensis, and C helveticus. J Clin Microbiol 43:2315–2329. doi: 10.1128/JCM.43.5.2315-2329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pendleton S, Hanning I, Biswas D, Ricke SC. 2013. Evaluation of whole-genome sequencing as a genotyping tool for Campylobacter jejuni in comparison with pulsed-field gel electrophoresis and flaA typing. Poultry Sci 92:573–580. doi: 10.3382/ps.2012-02695. [DOI] [PubMed] [Google Scholar]
- 137.Kovanen SM, Kivisto RI, Rossi M, Schott T, Karkkainen UM, Tuuminen T, Uksila J, Rautelin H, Hanninen ML. 2014. Multilocus sequence typing (MLST) and whole-genome MLST of Campylobacter jejuni isolates from human infections in three districts during a seasonal peak in Finland. J Clin Microbiol 52:4147–4154. doi: 10.1128/JCM.01959-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kovanen S, Kivistö R, Llarena A-K, Zhang J, Kärkkäinen U-M, Tuuminen T, Uksila J, Hakkinen M, Rossi M, Hänninen M-L. 2016. Tracing isolates from domestic human Campylobacter jejuni infections to chicken slaughter batches and swimming water using whole-genome multilocus sequence typing. Int J Food Microbiol 226:53–60. doi: 10.1016/j.ijfoodmicro.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 139.Kennedy AD, Otto M, Braughton KR. 2008. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci U S A 105:1327–1332. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Harrison EM, Paterson GK, Holden MTG, Larsen J, Stegger M, Larsen AR, Petersen A, Skov RL, Christensen JM, Bak Zeuthen A, Heltberg O, Harris SR, Zadoks RN, Parkhill J, Peacock SJ, Holmes MA. 2013. Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. EMBO Mol Med 5:509–515. doi: 10.1002/emmm.201202413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Revez J, Llarena A-K, Schott T, Kuusi M, Hakkinen M, Kivistö R, Hänninen M-L, Rossi M. 2014. Genome analysis of Campylobacter jejuni strains isolated from a waterborne outbreak. BMC Genomics 15:768. doi: 10.1186/1471-2164-15-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kuusi M, Nuorti JP, Hanninen ML, Koskela M, Jussila V, Kela E, Miettinen I, Ruutu P. 2005. A large outbreak of campylobacteriosis associated with a municipal water supply in Finland. Epidemiol Infect 133:593–601. doi: 10.1017/S0950268805003808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pan American Health Organization. 2015. Epidemiological update: cholera, 12 August 2015. Pan American Health Organization, Washington, DC: http://www.paho.org/hq/index.php?option=com_docman&task=doc_view&Itemid=270&gid=31105&lang=en. [Google Scholar]
- 144.Enserink M. 2010. Haiti's outbreak is latest in cholera's new global assault. Science 330:738–739. doi: 10.1126/science.330.6005.738. [DOI] [PubMed] [Google Scholar]
- 145.Orata FD, Keim PS, Boucher Y. 2014. The 2010 cholera outbreak in Haiti: how science solved a controversy. PLoS Pathog 10:e1003967. doi: 10.1371/journal.ppat.1003967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hasan NA, Choi SY, Eppinger M, Clark PW, Chen A, Alam M, Haley BJ, Taviani E, Hine E, Su Q, Tallon LJ, Prosper JB, Furth K, Hoq MM, Li H, Fraser-Liggett CM, Cravioto A, Huq A, Ravel J, Cebula TA, Colwell RR. 2012. Genomic diversity of 2010 Haitian cholera outbreak strains. Proc Natl Acad Sci U S A 109:E2010–E2017. doi: 10.1073/pnas.1207359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Katz LS, Petkau A, Beaulaurier J, Tyler S, Antonova ES, Turnsek MA, Guo Y, Wang S, Paxinos EE, Orata F, Gladney LM, Stroika S, Folster JP, Rowe L, Freeman MM, Knox N, Frace M, Boncy J, Graham M, Hammer BK, Boucher Y, Bashir A, Hanage WP, Van Domselaar G, Tarr CL. 2013. Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. mBio 4:e00398-13. doi: 10.1128/mBio.00398-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shimada DT, Arakawa E, Itoh K, Okitsu T, Matsushima A, Asai Y, Yamai S, Nakazato T, Nair GB, Albert MJ, Takeda Y. 1994. Extended serotyping scheme for Vibrio cholerae. Curr Microbiol 28:175–178. doi: 10.1007/BF01571061. [DOI] [Google Scholar]
- 149.Bhattacharya MK, Bhattacharya SK, Garg S, Saha PK, Dutta D, Nair GB, Deb BC, Das KP. 1993. Outbreak of Vibrio cholerae non-O1 in India and Bangladesh. Lancet 341:1346–1347. [DOI] [PubMed] [Google Scholar]
- 150.Barrett TJ, Blake PA. 1981. Epidemiological usefulness of changes in hemolytic activity of Vibrio cholerae biotype El Tor during the seventh pandemic. J Clin Microbiol 13:126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cameron DN, Khambaty FM, Wachsmuth IK, Tauxe RV, Barrett TJ. 1994. Molecular characterization of Vibrio cholerae O1 strains by pulsed-field gel electrophoresis. J Clin Microbiol 32:1685–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Reimer AR, Van Domselaar G, Stroika S, Walker M, Kent H, Tarr C, Talkington D, Rowe L, Olsen-Rasmussen M, Frace M, Sammons S, Dahourou GA, Boncy J, Smith AM, Mabon P, Petkau A, Graham M, Gilmour MW, Gerner-Smidt P, V cholerae Outbreak Genomics Task Force . 2011. Comparative genomics of Vibrio cholerae from Haiti, Asia, and Africa. Emerg Infect Dis 17:2113–2121. doi: 10.3201/eid1711.110794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chin C-S, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, Bullard J, Webster DR, Kasarskis A, Peluso P, Paxinos EE, Yamaichi Y, Calderwood SB, Mekalanos JJ, Schadt EE, Waldor MK. 2011. The origin of the Haitian cholera outbreak strain. N Engl J Med 364:33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hendriksen RS, Price LB, Schupp JM, Gillece JD, Kaas RS, Engelthaler DM, Bortolaia V, Pearson T, Waters AE, Upadhyay BP, Shrestha SD, Adhikari S, Shakya G, Keim PS, Aarestrup FM. 2011. Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak. mBio 2:e00157-11. doi: 10.1128/mBio.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Robins WP, Mekalanos JJ. 2014. Genomic science in understanding cholera outbreaks and evolution of Vibrio cholerae as a human pathogen. Curr Top Microbiol Immunol 379:211–229. doi: 10.1007/82_2014_366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Frerichs RR, Keim PS, Barrais R, Piarroux R. 2012. Nepalese origin of cholera epidemic in Haiti. Clin Microbiol Infect 18:E158–E163. doi: 10.1111/j.1469-0691.2012.03841.x. [DOI] [PubMed] [Google Scholar]
- 157.Suttle CA. 2005. Viruses in the sea. Nature 437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 158.Sironi M, Cagliani R, Forni D, Clerici M. 2015. Evolutionary insights into host-pathogen interactions from mammalian sequence data. Nat Rev Genet 16:224–236. doi: 10.1038/nrg3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ladner JT, Beitzel B, Chain PSG, Davenport MG, Donaldson E, Frieman M, Kugelman J, Kuhn JH, O'Rear J, Sabeti PC, Wentworth DE, Wiley MR, Yu GY, Threat Characterization Consortium, Sozhamannan S, Bradburne C, Palacios G. 2014. Standards for sequencing viral genomes in the era of high-throughput sequencing. mBio 5:e01360-14. doi: 10.1128/mBio.01360-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lindsay L, Wolter J, De Coster I, Van Damme P, Verstraeten T. 2015. A decade of norovirus disease risk among older adults in upper-middle and high income countries: a systematic review. BMC Infect Dis 15:425. doi: 10.1186/s12879-015-1168-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Moore MD, Goulter RM, Jaykus L-A. 2015. Human norovirus as a foodborne pathogen: challenges and developments. Annu Rev Food Sci Technol 6:411–433. doi: 10.1146/annurev-food-022814-015643. [DOI] [PubMed] [Google Scholar]
- 162.Verhoef L, Hewitt J, Barclay L, Ahmed SM, Lake R, Hall AJ, Lopman B, Kroneman A, Vennema H, Vinjé J, Koopmans M. 2015. Norovirus genotype profiles associated with foodborne transmission, 1999–2012. Emerg Infect Dis 21:592–599. doi: 10.3201/eid2104.141073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bull RA, Eden JS, Luciani F, McElroy K, Rawlinson WD, White PA. 2012. Contribution of intra- and interhost dynamics to norovirus evolution. J Virol 86:3219–3229. doi: 10.1128/JVI.06712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kundu S, Lockwood J, Depledge DP, Chaudhry Y, Aston A, Rao K, Hartley JC, Goodfellow I, Breuer J. 2013. Next-generation whole genome sequencing identifies the direction of norovirus transmission in linked patients. Clin Infect Dis 57:407–414. doi: 10.1093/cid/cit287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Aggarwal R, Goel A. 2015. Hepatitis A: epidemiology in resource-poor countries. Curr Opin Infect Dis 28:488–496. doi: 10.1097/QCO.0000000000000188. [DOI] [PubMed] [Google Scholar]
- 166.Chi H, Haagsma EB, Riezebos-Brilman A, van den Berg AP, Metselaar HJ, de Knegt RJ. 2014. Hepatitis A related acute liver failure by consumption of contaminated food. J Clin Virol 61:456–458. doi: 10.1016/j.jcv.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 167.Rizzo C, Alfonsi V, Bruni R, Busani L, Ciccaglione AR, De Medici D, Di Pasquale S, Equestre M, Escher M, Montano-Remacha MC, Scavia G, Taffon S, Carraro V, Franchini S, Natter B, Augschiller M, Tosti ME, Central Task Force on Hepatitis A . 2013. Ongoing outbreak of hepatitis A in Italy: preliminary report as of 31 May 2013. Euro Surveill 18:20518 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20518. [PubMed] [Google Scholar]
- 168.Tavoschi L, Severi E, Niskanen T, Boelaert F, Rizzi V, Liebana E, Gomes Dias J, Nichols G, Takkinen J, Coulombier D. 2015. Food-borne diseases associated with frozen berries consumption: a historical perspective, European Union, 1983 to 2013. Euro Surveill 20:21193 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=21193. doi: 10.2807/1560-7917.ES2015.20.29.21193. [DOI] [PubMed] [Google Scholar]
- 169.Chiapponi C, Pavoni E, Bertasi B, Baioni L, Scaltriti E, Chiesa E, Cianti L, Losio MN, Pongolini S. 2014. Isolation and genomic sequence of hepatitis A virus from mixed frozen berries in Italy. Food Environ Virol 6:202–206. doi: 10.1007/s12560-014-9149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hang J, Cai B, Xue P, Wang L, Hu H, Zhou Y, Ren S, Wu J, Zhu M, Chen D, Yang H, Wang L. 2015. The joint effects of lifestyle factors and comorbidities on the risk of colorectal cancer: a large Chinese retrospective case-control study. PLoS One 10:e0143696. doi: 10.1371/journal.pone.0143696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.zur Hausen H. 2012. Red meat consumption and cancer: reasons to suspect involvement of bovine infectious factors in colorectal cancer. Int J Cancer 130:2475–2483. doi: 10.1002/ijc.27413. [DOI] [PubMed] [Google Scholar]
- 172.Peretti A, FitzGerald PC, Bliskovsky V, Buck CB, Pastrana DV. 2015. Hamburger polyomaviruses. J Gen Virol 96:833–839. doi: 10.1099/vir.0.000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhang W, Li L, Deng X, Kapusinszky B, Delwart E. 2014. What is for dinner? Viral metagenomics of US store bought beef, pork, and chicken. Virology 468-470: 303–310. doi: 10.1016/j.virol.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Arnberg N. 2012. Adenovirus receptors: implications for targeting of viral vectors. Trends Pharmacol Sci 33:442–448. doi: 10.1016/j.tips.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 175.Mena KD, Gerba CP. 2009. Waterborne adenovirus. Rev Environ Contam Toxicol 198:133–167. doi: 10.1007/978-0-387-09647-6_4. [DOI] [PubMed] [Google Scholar]
- 176.Ogorzaly L, Walczak C, Galloux M, Etienne S, Gassilloud B, Cauchie HM. 28 April 2015. Human adenovirus diversity in water samples using a next-generation amplicon sequencing approach. Food Environ Virol doi: 10.1007/s12560-015-9194-4. [DOI] [PubMed] [Google Scholar]
- 177.Allard MW, Strain E, Melka D, Bunning K, Musser SM, Brown EW, Timme R. 2016. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J Clin Microbiol 54:1975–1983. doi: 10.1128/JCM.00081-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Krukczkiewicz P, Mutschall S, Barker D, Thomas J, Domselaar GV, Gannon VPJ, Carrillo CD, Taboada EN. 2013. MIST: a tool for rapid in silico generation of molecular data from bacterial genome sequences, p 316–323. In Pastor O, Sinoquet C, Plantier G, Schultz T, Fred A, Gamboa H (ed), Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOSTEC 2013), 3–6 March 2014, Angers, France. [Google Scholar]
- 179.Cosentino S, Voldby Larsen M, Møller Aarestrup F, Lund O. 2013. PathogenFinder—distinguishing friend from foe using bacterial whole genome sequence data. PLoS One 8:e77302. doi: 10.1371/journal.pone.0077302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Franz E, van Hoek AHAM, Wuite M, van der Wal FJ, de Boer AG, Bouw EI, Aarts HJM. 2015. Molecular hazard identification of non-O157 Shiga toxin-producing Escherichia coli (STEC). PLoS One 10:e0120353. doi: 10.1371/journal.pone.0120353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pielaat A, Boer MP, Wijnands LM, van Hoek AHAM, Bouw E, Barker GC, Teunis PFM, Aarts HJM, Franz E. 2015. First step in using molecular data for microbial food safety risk assessment; hazard identification of Escherichia coli O157:H7 by coupling genomic data with in vitro adherence to human epithelial cells. Int J Food Microbiol 213:130–138. doi: 10.1016/j.ijfoodmicro.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hoffmann M, Luo Y, Monday SR, González-Escalona N, Ottesen AR, Muruvanda T, Wang C, Kastanis G, Keys C, Janies D, Senturk IF, Catalyurek UV, Wang H, Hammack TS, Wolfgang WJ, Schoonmaker-Bopp D, Chu A, Myers R, Haendiges J, Evans PS, Meng J, Strain EA, Allard MW, Brown EW. 2016. Tracing origins of the Salmonella bareilly strain causing a food-borne outbreak in the United States. J Infect Dis 213:502–508. doi: 10.1093/infdis/jiv297. [DOI] [PubMed] [Google Scholar]
- 183.Weedmark KA, Mabon P, Hayden KL, Lambert D, Van Domselaar G, Austin JW, Corbett CR. 2015. Clostridium botulinum group II isolate phylogenomic profiling using whole-genome sequence data. Appl Environ Microbiol 81:5938–5948. doi: 10.1128/AEM.01155-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O'Brien JS, Pawlowski AC, Piddock LJV, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Liu B, Pop M. 2009. ARDB—antibiotic resistance genes [sic] database. Nucleic Acids Res 37:D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bakour S, Sankar SA, Rathored J, Biagini P, Raoult D, Fournier P-E. 2016. Identification of virulence factors and antibiotic resistance markers using bacterial genomics. Future Microbiol 11:455–466. doi: 10.2217/fmb.15.149. [DOI] [PubMed] [Google Scholar]
- 188.Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agersø Y, Lund O, Larsen MV, Aarestrup FM. 2013. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother 68:771–777. doi: 10.1093/jac/dks496. [DOI] [PubMed] [Google Scholar]
- 189.Zhao S, Tyson GH, Chen Y, Li C, Mukherjee S, Young S, Lam C, Folster JP, Whichard JM, McDermott PF. 2016. Whole-genome sequencing analysis accurately predicts antimicrobial resistance phenotypes in Campylobacter spp. Appl Environ Microbiol 82:459–466. doi: 10.1128/AEM.02873-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tyson GH, McDermott PF, Li C, Chen Y, Tadesse DA, Mukherjee S, Bodeis-Jones S, Kabera C, Gaines SA, Loneragan GH, Edrington TS, Torrence M, Harhay DM, Zhao S. 2015. WGS accurately predicts antimicrobial resistance in Escherichia coli. J Antimicrob Chemother 70:2763–2769. doi: 10.1093/jac/dkv186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Martínez-Martínez L, Pascual A, Hernández-Allés S, Alvarez-Díaz D, Suárez AI, Tran J, Benedí VJ, Jacoby GA. 1999. Roles of beta-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob Agents Chemother 43:1669–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Liu Y-Y, Wang Y, Walsh TR, Yi L- X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 193.Collins M. 1991. Phylogenetic analysis of the genus Lactobacillus and related lactic acid bacteria as determined by reverse transcriptase sequencing of 16S rRNA. FEMS Microbiol Lett 77:5–12. doi: 10.1111/j.1574-6968.1991.tb04313.x. [DOI] [Google Scholar]
- 194.Kawai T, Sekizuka T, Yahata Y, Kuroda M, Kumeda Y, Iijima Y, Kamata Y, Sugita-Konishi Y, Ohnishi T. 2012. Identification of Kudoa septempunctata as the causative agent of novel food poisoning outbreaks in Japan by consumption of Paralichthys olivaceus in raw fish. Clin Infect Dis 54:1046–1052. doi: 10.1093/cid/cir1040. [DOI] [PubMed] [Google Scholar]
- 195.Bergholz TM, Moreno Switt AI, Wiedmann M. 2014. Omics approaches in food safety: fulfilling the promise? Trends Microbiol 22:275–281. doi: 10.1016/j.tim.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]