SUMMARY
Bacterial pathogens have coevolved with humans in order to efficiently infect, replicate within, and be transmitted to new hosts to ensure survival and a continual infection cycle. For enteric pathogens, the ability to adapt to numerous host factors under the harsh conditions of the gastrointestinal tract is critical for establishing infection. One such host factor readily encountered by enteric bacteria is bile, an innately antimicrobial detergent-like compound essential for digestion and nutrient absorption. Not only have enteric pathogens evolved to resist the bactericidal conditions of bile, but these bacteria also utilize bile as a signal to enhance virulence regulation for efficient infection. This review provides a comprehensive and up-to-date analysis of bile-related research with enteric pathogens. From common responses to the unique expression of specific virulence factors, each pathogen has overcome significant challenges to establish infection in the gastrointestinal tract. Utilization of bile as a signal to modulate virulence factor expression has led to important insights for our understanding of virulence mechanisms for many pathogens. Further research on enteric pathogens exposed to this in vivo signal will benefit therapeutic and vaccine development and ultimately enhance our success at combating such elite pathogens.
INTRODUCTION
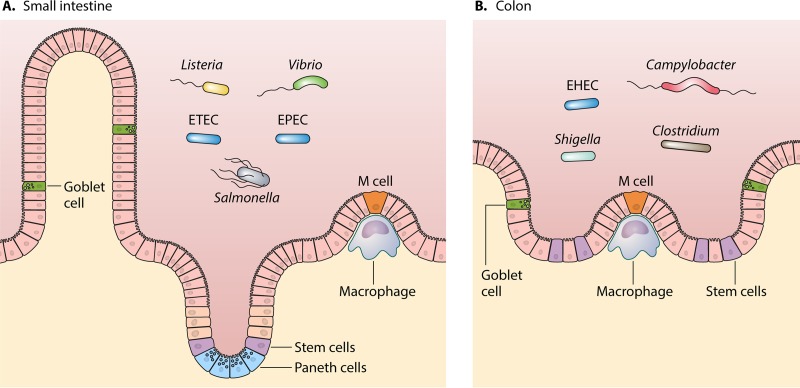
Enteric pathogenic bacteria represent a unique set of pathogens, as these bacteria must overcome numerous challenges in order to successfully establish infection in the small intestine or colon (Fig. 1). These challenges include pathogen-specific immune responses (1) and several bactericidal conditions of the gastrointestinal tract, including the low pH of the stomach (2), the presence of bile in the small intestine (3, 4), low iron availability (5, 6), and an established commensal microbiome consisting of billions of bacteria from hundreds of different species (7). The human gastrointestinal tract is a challenging environment that prevents many bacteria from surviving under these harsh conditions; however, enteric bacterial pathogens have evolved not only to survive these severe exposures but also to successfully colonize and establish infection in the host.
FIG 1.
Infection sites of enteric pathogens in the gastrointestinal tract. Enteric bacterial pathogens have been shown to infect either the small intestine (represented with microvillus protrusions) (A) or the colon (B). In both settings, goblet cells produce mucus, stem cells initiate maturation of the enterocytes, and specialized microfold (M) cells serve as the point of antigen sampling that can be exploited by invasive pathogens to access epithelial cells. Paneth cells in the small intestine are important for initiating innate mucosal defenses. Infection mechanisms include adherence to or invasion of the epithelial cells lining the gastrointestinal tract, with Listeria, Vibrio, Salmonella, and some pathogenic Escherichia coli strains (EPEC and ETEC) infecting the small intestine and Campylobacter, EHEC, Shigella, and Clostridium infecting the colon.
Given the length of the small intestine, bile is one of the more long-term exposures encountered by bacteria, both pathogenic and commensal, in the early part of the gastrointestinal tract. For the host, bile is essential for digestion and nutrient absorption and is composed of proteins (globulins and albumins), lipids (phospholipids, cholesterol, and fatty acids), carbohydrates, vitamins, mineral salts, and other trace elements (8). In the liver, the primary bile acids cholic acid and chenodeoxycholic acid are synthesized by hepatocytes and are further metabolized in the liver by conjugation to glycine or taurine through N-acyl amidation. Conjugated bile acids have reduced bile acid toxicity and increased solubility, are almost fully ionized at physiological pH, and are termed bile salts. Bile salts are maintained at high concentrations in the duodenum, jejunum, and proximal ileum to solubilize, digest, and absorb lipids and lipid-soluble vitamins (8, 9). The facultative and anaerobic commensal bacteria in the small intestine metabolize bile salts through deconjugation and hydroxy group oxidation, resulting in the production of secondary bile salt compounds. In the distal ileum, bile salts are absorbed into the bloodstream, complexed to plasma proteins, and returned to the liver (9). The majority of bile (95%) is recycled back into the small intestine and does not enter the colon (10). Concentrations of bile salts range from 0.2% to 2% (wt/vol), depending on the time of day, diet, and individual (11). As reviewed by Ridlon et al., bacterial colonization in the small intestine increases from ∼103 bacteria/ml in the duodenum to 104 bacteria/ml in the jejunum and 106 to 108 bacteria/ml in the ileum. Lactobacillus and Streptococcus are the predominant genera in the duodenum and jejunum, while Enterococcus, Bacteroides, Clostridium, and other commensals, in addition to Lactobacillus, are able to colonize the ileum (9). Human and rodent studies have demonstrated that the bactericidal properties of bile components likely prevent overgrowth of the commensal bacteria in the small intestine. The absence of this mechanism is problematic for individuals who have reduced bile output, such as patients with hereditary defects like intrahepatic cholestasis (12–15).
Depending on the organism and the site of infection, pathogenic bacteria have developed resistance mechanisms to counteract the bactericidal conditions of bile. In fact, a number of enteric pathogens use bile as a signal to regulate virulence gene expression to either colonize or maintain infection in the human gastrointestinal tract (Tables 1 and 2). For pathogens that colonize the small intestine, resistance to bile is critical to ensure long-term survival in the host. Here we review the bile-associated regulation of virulence mechanisms in enteric bacterial pathogens and provide an analysis of the bile-related research performed to date.
TABLE 1.
Summary of bile resistance mechanisms in enteric pathogens
| Pathogen | Function of outer membrane proteins (reference[s]) | Mechanism of induction of stress response genes (reference[s]) | Efflux pump(s) (reference[s]) | Additional resistance mechanism(s) (reference[s]) |
|---|---|---|---|---|
| Escherichia coli | Repression of ompF (23) | sulA to correct DNA damage (35) | AcrAB (23, 38) | MqsR/MqsA toxin/antitoxin system (34); basRS two-component system (23) |
| Shigella | To be determined | |||
| Vibrio | Regulated expression of ompU and ompT (76) | RpoS in V. vulnificus (102) | AcrAB (77, 78), BreAB (79, 80), VexAB (81, 101), VprAB (82), Vme pumps (99, 100) | Induction of transcriptional regulator LeuO (75) and biofilm formation (84) |
| Salmonella | Repression of OmpF and OmpC (3); utilization of TolA and TolC (3, 125) | SoxRS (132) and OxyR (133) | AcrAB (119 – 121); AcrEF, MdtABC, MdsCBA, EmrAB, MdtK, and MacAB (118) | Alterations of LPS, including remodeling of lipid A (3), elongation of O-chains through fepE (129), and Vi antigen production (130) |
| Campylobacter | CmeABC and CmeDEF (154, 155) | Utilization of the two-component regulator CbrR (159) | ||
| Clostridium | To be determined; spores are naturally resistant (169) | |||
| Listeria | Induced stress response genes encoding DNA repair proteins, protein folding chaperones, and oxidative stress response proteins (176) | MdrT (177) | BrtA transcriptional regulator (177); bile salt hydrolase enzyme (178); biofilm formation genes induced (35) |
TABLE 2.
Summary of altered virulence gene expression in the presence of bile
| Pathogen | Adhesin/flagellum (reference[s]) | Invasin/secretion system (reference[s]) | Toxin(s) (reference[s]) | Additional factor(s) or description (reference[s]) |
|---|---|---|---|---|
| Escherichia coli | ||||
| ETEC | Induced colonization factors such as CS19 (45) and CS5 (44) | Altered estA and eltA enterotoxin expression (24) | ||
| EPEC | Induction of the bundle-forming pilus, increased adherence, and decreased motility (25) | Reduced bile reabsorption by the apical sodium-dependent bile acid transporter in enterocytes (50) | ||
| EHEC | Repressed flagellar motor genes (39) | Repressed Shiga toxin expression (23) | Repressed LEE pathogenicity island (39) | |
| Shigella | Induced adherence mediated by OspE1/OspE2 (65) | Induced invasion through IpaD interaction (61) | Induced protein secretion (58, 65), and 51 induced genes in the presence of bile salts (65) | |
| Vibrio | ||||
| V. cholerae | Repressed toxin-coregulated pilus and increased motility (88, 89) | Induced expression of T3SS genes (94); induction of VopX, a T3SS effector (95); diminished T6SS activity (97) | Repression of cholera toxin (88, 89), induction of the related TRH (96) | Effects on transcriptional regulators ToxR (90), TcpP (91), H-NS and FlrA (92), and ToxT (93) |
| V. parahaemolyticus | Induced expression of T3SS2 (105) | Induced expression of TDH (105) | Induced transcriptional regulators VtrA and VtrB (105); 69 genes induced in the presence of bile (104) | |
| V. vulnificus | To be determined | |||
| Salmonella | Downregulation of motility; regulation of fimbrial adherence factors (128, 137, 142) | Repression of T3SS through SPI-1 inhibition (126, 136); SipD interacts with bile (140, 141) | Repression of SirC and InvF (3, 117); gallbladder colonization | |
| Campylobacter | Increased flagellin A expression (166), biofilm formation mediated through adhesion with the flagellar filament (165) | Induced expression of Cia (164) | Increased chemotaxis through induced CheV protein and ATP synthase expression (166) | |
| Clostridium | Enhanced germination of spores with greater abundance of primary bile acids following antimicrobial killing of the microbiota (170, 172) | |||
| Listeria | Possible enhanced fecal shedding following growth in the gallbladder (174) |
ESCHERICHIA COLI
Escherichia coli is a Gram-negative, facultative, anaerobic bacterium that naturally resides in the gastrointestinal tract of mammals and composes 0.1% of the microbiota found in the cecum and colon of humans (16). E. coli strains can include both nonpathogenic commensals or pathovars associated with gastrointestinal, urogenital, and extraintestinal infections (17). As E. coli bacteria traverse the gastrointestinal tract, they encounter signals provided by the host that trigger the expression of numerous colonization mechanisms, a process known as interkingdom signaling (18). These signals include factors that facilitate gastrointestinal adherence of the bacteria to the small intestinal or colonic epithelial cells (17, 19). Studies investigating bile responses in E. coli include both commensals and diarrheagenic pathovars, including enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), and enterohemorrhagic E. coli (EHEC) (20–28).
Commensal E. coli and Common Bile Resistance Mechanisms Shared with Pathovars
Since bile is a major host-produced component encountered by E. coli in the small intestine (29), resistance to bile acids in both commensal and pathogenic E. coli strains has long been known to be an important property for overcoming the environmental stress of the small intestine, where bile is most abundant (30, 31). A study comparing 72 E. coli reference collection (ECOR) strains and 10 EHEC O157:H7 strains demonstrated that commensal and pathogenic strains had similar growth capabilities in sodium dodecyl sulfate (SDS), a detergent often used to mimic bile salts exposure (32). Additionally, a study in mice demonstrated that the presence of bile salts in the gastrointestinal tract is a selective pressure for E. coli that confers a fitness advantage to strains that can survive and colonize in the presence of bile (33). In order to counteract the detergent-like properties of bile, both commensal and pathogenic E. coli strains activate stress responses, efflux pumps, and toxin/antitoxin systems to remove bile compounds (21, 30, 34). Stress response genes induced by bile salts, such as sulA, are used for the cessation of cell division in order to correct damage to DNA and the membrane following oxidative stress (21, 35–37), while efflux pump mechanisms, such as AcrAB-TolC, overcome bile acid toxicity and confer resistance to bile (38). A recent study demonstrated that the MqsR/MqsA toxin/antitoxin system degrades ygiS mRNA, which encodes a periplasmic protein that promotes the uptake of the bile salt deoxycholate by the bacteria and increases stress on the bacterial membrane (34). ygiS deletion mutants demonstrated improved growth and survival and reduced membrane damage in the presence of the bile salt deoxycholate (34). Both commensal and pathogenic E. coli strains utilize these mechanisms to resist bile; however, the pathogens also carry virulence genes that are tightly regulated in response to bile and other environmental signals present in the gastrointestinal tract in order to outcompete commensal organisms.
Pathogenic E. coli
Genome sequencing has demonstrated that the majority of pathogenic strains associated with gastrointestinal infection contain virulence genes often carried on mobile genetic elements that are lacking in commensal strains. Pathogenic E. coli bacteria that infect and colonize the gastrointestinal tract encode virulence factors, typically fimbriae and enterotoxins, that aid in bacterial colonization and allow the pathogen to outcompete commensals (17, 19, 25, 39). Diarrheagenic E. coli strains can be grouped into six well-defined pathovars characterized by the presence of canonical virulence factors that have been demonstrated to be essential for infection. Isolates of the different pathovars preferentially colonize regions of the gastrointestinal tract, including the small intestine and/or large intestine, to cause disease. Bile responses have been characterized for three of these pathovars: ETEC, EPEC, and EHEC (17, 19, 20, 22–24).
Enterotoxigenic E. coli.
ETEC, a significant cause of diarrheal illness around the world, is characterized by the production of heat-stable (ST) and/or heat-labile (LT) enterotoxins (40). ETEC isolates can encode >25 described colonization factors for colonization of the small intestine (41–43). Studies of bile responses in ETEC have established that bile salts are an important signal for inducing colonization factor expression in some isolates (24, 44–46). Sommerfelt et al. demonstrated that bile salts exposure significantly increased the fimbriated cell percentage (FCP) in numerous wild-type ETEC strains that were lacking the virulence regulator CfaD (46). Other in vitro studies demonstrated bile-dependent fimbrial expression of CS19 (coli surface antigen 19) in ETEC strain F595C via electron microscopy (45), while a transcriptional study by Nicklasson et al. further identified mRNA expression of CS5 fimbriae in response to a bile salt component, sodium glycocholate hydrate, in four different ETEC strains (44). With regard to toxin production, preincubation with crude bile has been demonstrated to have direct inhibitory effects on the ability of the heat-labile toxin protein to bind to the GM1 receptor (47). Overall, these results highlight the significance of the upregulation of colonization mechanisms by ETEC in response to bile before the initiation of toxin and virulence gene expression in the gastrointestinal tract. Interestingly, a more recent study by Sahl and Rasko used global transcriptomics techniques to further verify bile salts as a trigger for virulence genes by using quantitative reverse transcription-PCR (qRT-PCR) and transcriptome sequencing (RNA-Seq) in a strain-dependent manner (24). Variations in enterotoxin gene expression between two severe ETEC prototype isolates in response to bile were observed. Prototype ETEC strain H10407 had a significantly greater increase in the expression level of the heat-labile enterotoxin gene eltA than did strain E24377A, while the heat-stable enterotoxin gene estA was induced in E24377A. Moreover, the colonization factors CS1 and CS3 were downregulated in E24377A in the presence of bile. Analysis of growth at different time points hinted that transcriptional responses were potentially intertwined with a quorum-sensing mechanism (24). This study suggests that some ETEC isolates encode diverse transcriptional networks that can regulate virulence genes differently in response to bile. In all, the responses of ETEC to bile will require further functional studies to describe the complex interactions in detail, and understanding the bile response pathway in ETEC should prove useful considering the propensity of ETEC to infect the small intestine.
Enteropathogenic E. coli.
EPEC, an E. coli pathovar that is associated with infant diarrhea and the production of attaching and effacing lesions on enterocytes, also colonizes the small intestine (17, 19). EPEC has been demonstrated in numerous studies to respond to bile salts by increasing adhesion to epithelial cells (25, 35, 48). de Jesus et al. demonstrated that bile exposure enhanced the adhesion of typical EPEC strain E2348/69 to Hep2 epithelial cells between 1.3- and 2.6-fold. There was also an increase in bundle-forming pilus protein expression as determined via Western blot analysis (25). Other studies using the major bile component deoxycholate observed decreased flagellar protein levels in EPEC strain E2348/69 and unaltered intimin expression (25, 31). The data suggest that colonization factors other than fimbriae were utilized to enhance binding events in typical EPEC strains in response to bile. Conversely, enhanced adhesion was not observed with atypical EPEC strain 1551-2, which lacks the bundle-forming pilus, when grown on HeLa cells (49). A second interesting observation is that EPEC responses to bile have been demonstrated to impact host cell physiology, as described by Annaba et al. in a study showing that EPEC inhibits enterocyte bile absorption in the ileum and leads to increased bile concentrations in the colon. Caco-2 or Hek293 cells infected with EPEC prototype strain E2348/69 exhibited reduced uptake of taurocholic acid by the apical sodium-dependent bile acid transporter (ASBT), a transporter important for bile absorption (50). Bile malabsorption by enterocytes can result in high bile concentrations in the colon, which can result in mucosal damage and may be associated with chronic inflammation in the gastrointestinal tract (51). However, bile malabsorption was ablated with mutations in the EPEC type III secretion system (T3SS) effectors espNABD (known inhibitors of macrophage phagocytosis) or in the presence of protein tyrosine phosphatase inhibitors, suggesting that these effectors function as phosphatases to inhibit the ASBT (50, 52). The interaction of EPEC with host enterocytes appears to be quite dynamic in response to bile and may also impact subsequent disease manifestations. The clinical implication of bile malabsorption during and after EPEC infection requires more investigation in order to understand important implications for therapeutic and vaccine development.
Enterohemorrhagic E. coli.
EHEC is a diarrheagenic pathovar that has an infectious dose as low as 100 cells and causes disease in the large intestine. EHEC appears to employ responses that are adapted to the colon, a region that contains only 5% of the bile concentrations present in the small intestine (17, 53, 54). Unlike EPEC and ETEC, EHEC isolates do not appear to upregulate colonization factors required for adherence in response to bile but instead have a stress response that is similar to those observed for commensal E. coli strains (32). A study evaluating bile salt responses of EHEC O157:H7 using microarrays identified increased expression levels of virulence genes involved in the acrAB efflux pump and basRS two-component signal transduction system but downregulation of Shiga toxin genes and the outer membrane porin gene ompF. Additionally, lipid A modification genes of the arn operon were upregulated in bile salts, which, together with the basRS system, appear to allow EHEC to resist both bile and the antimicrobial peptide polymyxin B during colonization of the gastrointestinal tract (23). A recent study by Hamner et al. reported similar results, demonstrating Shiga toxin gene downregulation when interrogating the bile response transcriptome of O157:H7 by using qRT-PCR. This study observed a significant downregulation of genes in the locus of enterocyte effacement (LEE) pathogenicity island, including the eae intimin gene (39). However, this finding was not observed by Kus et al., and the lack of concordance of the results was attributed to variations in the EHEC strains and growth time points used. Furthermore, Hamner et al. identified an increased regulation of genes encoding the iron acquisition and flagellar body segments; however, a 2-fold decrease was detected for genes associated with the flagellum filament, stator motor, and chemotaxis mechanisms. Moreover, no significant transcriptional alteration was observed for the adhesin gene ompA, which encodes a protein important for Gram-negative membrane integrity and adherence (39, 54). The downregulation of the LEE and flagellar motor genes coupled with the lack of a change in outer membrane protein genes indicate that O157:H7 may have additional, as-yet-unidentified virulence factors that respond to bile during early-stage pathogenesis or in preparation for colonization of the large intestine. Alternatively, the response to bile may indicate a mechanism used by EHEC as an environmental signal to delay the transcription of genes associated with colonization until the bacteria reach the more permissive environment of the colon. Future transcriptomic studies will enable us to understand the full extent of the response of EHEC to bile and the role that the response plays during EHEC infection.
Future Studies of Escherichia coli
There are several areas of research that will improve our understanding of bile responses in both pathogenic and commensal E. coli strains. First, future studies should seek to understand whether other understudied pathovars, such as diffuse-adhering E. coli (DAEC) and enteroinvasive E. coli (EIEC), alter the expression of colonization factors following bile exposure. These studies should also identify the potential link between bile exposure and colonic inflammation that may be associated with the newly defined adherent-invasive E. coli (AIEC) pathovar, especially since recent work has demonstrated that bile salts induce the expression of long polar fimbriae to facilitate host cell adhesion (55). Additional genomic and transcriptomic comparisons will provide insight into which components of the bacterium are responsible for bile resistance and the regulation of bile-mediated virulence factors. Furthermore, investigations of how diseases are associated with opportunistic or enteric pathogens could provide clues toward understanding the clinical relevance of E. coli in Crohn's disease and other inflammatory bowel diseases. Comparative studies of various diarrheagenic pathogens should seek to address colonization efficiency to identify if these differences account for the establishment of the different niches along the gastrointestinal tract exploited by enteric pathogens. Finally, it remains to be seen if the ability of E. coli to resist bile is significantly different between commensal and pathogenic strains, as very few commensal isolates have been studied in detail. Future studies should account for differences in nutrient acquisition and resistance to detergents as well as differences in colonization or infection locations (small intestine or colon) when considering whether there are fitness advantages among strains following bile exposure.
SHIGELLA
Shigella species are Gram-negative, facultative, intracellular pathogens that cause a significant global burden each year by causing millions of cases of watery diarrhea or bacillary dysentery, typically in children <5 years of age in developing countries. Infection develops from the ability of the bacteria to invade epithelial cells lining the colon (56, 57). A 220-kb virulence plasmid is required for infection and encodes a T3SS, the Ipa invasion proteins, and additional T3SS effector proteins important for enhancing infection and intracellular survival. As few as 100 cells can cause infection after ingestion. Progression of shigellosis requires the bacteria to transit through the small intestine, where bile exposure occurs, before reaching the site of infection in the colon (56).
Most research involving increased virulence due to bile exposure has focused on S. flexneri. The first observations that bile increases the virulence of S. flexneri came from an initial study by Pope et al., who identified that adherence, protein secretion, and invasion are induced after the bacteria are exposed to the bile salt deoxycholate (58). Increased adherence was demonstrated to be independent of invasion, since an ΔipaB mutant, which is unable to invade, also had increased adherence to HeLa cells after exposure to bile salts. The virulence factor required for induced adherence was never identified in this study; however, the authors stated that the factor most likely required the T3SS. Increased invasion and adherence were also reported for Shigella dysenteriae; however, interestingly, EIEC did not have induced phenotypes following deoxycholate exposure. As stated by the authors, this finding is surprising and warrants further investigation given that the virulence plasmids of Shigella and EIEC are highly homologous (58, 59). Indeed, comparisons of the effects of bile and bile salts between Shigella and EIEC not only will enhance our understanding of bile-induced virulence for each pathogen but also will allow us to understand key differences between the two pathogens. Further investigation into the effect of bile salts on invasion led to the discovery that deoxycholate interacts with the IpaD invasion protein at the tip of the type III (T3) needle complex. This interaction induces a conformational change to allow another invasion protein, IpaB, to bind, leading to the increased invasion of epithelial cells (60–63). This observation is significant and indicates that IpaD acts as an environmental sensor to prepare the bacteria for induced T3 secretion and subsequent invasion once contact with colonic epithelial cells is made (60). Interestingly, deoxycholate has been utilized to fully investigate the interaction of IpaD and IpaB to elucidate the assembly of the T3 needle complex required for the invasion process (64).
Recently, Faherty et al. identified the Shigella flexneri effectors required for bile-induced bacterial adherence (65). By using polarized epithelial cells and a 1:1 mixture of the bile salts cholate and deoxycholate at 0.4% (wt/vol), this study determined that the 99% identical T3SS effector proteins OspE1 and OspE2 were necessary for bile salt-induced adherence, since a double mutant lost the phenotype. The study further demonstrated that ospE1 and ospE2 were among the 51 genes induced in response to bile salts, and investigation of the outer membrane identified the localization of the OspE1 and OspE2 proteins following bile salts exposure. Future work will determine the nature of this outer membrane protein localization and to which eukaryotic structures binding occurs. In addition, this study detected increases in T3 and non-T3 protein secretion, as was observed by Pope et al. (58). The mechanism behind the induced protein secretion requires further investigation. It is interesting that in this study, S. flexneri was exposed to bile salts during the subculture step, prior to the addition of bacteria to eukaryotic cells in the adherence assay. No differences in adherence were detected when bile salts were added to the cell culture medium during infection (65). This preexposure mimics in vivo conditions in that 95% of bile salts are recycled back into the small intestine and do not enter the colon (10). Therefore, these conditions highlight the changes in virulence factor expression that occur during transit in the small intestine prior to invasion at the site of infection in the colon.
In all, studies evaluating the effects of bile salt exposure on S. flexneri virulence provide significant evidence that these bacteria respond to environmental changes in the small intestine to enhance virulence prior to reaching the site of infection in the colon. Future work utilizing bile extracts to account for additional factors, analyzing the effects of exposure on other Shigella isolates, and determining the mechanisms of bile resistance and bile-regulated virulence will enhance our understanding of the importance of bile in the pathogenesis of Shigella.
VIBRIO
Vibrio species are Gram-negative, curved, motile bacteria that are inhabitants of coastal and aquatic environments. Approximately 12 species are pathogenic, with Vibrio cholerae, V. parahaemolyticus, and V. vulnificus causing the majority of infections, outbreaks, and epidemics worldwide (66, 67). V. cholerae is thought to survive in brackish water between epidemics (68), while V. parahaemolyticus and V. vulnificus are found in raw seafood (69). Infections typically result from the consumption of contaminated food or water and range from mild to severe gastroenteritis characterized by watery diarrhea, severe abdominal cramps, and dehydration (68). Wound infections and septicemia have also been associated with V. vulnificus and V. parahaemolyticus (70). The bacteria penetrate the mucus layer of the gut and adhere to epithelial cells in the small intestine. Virulence factors required to cause infection include cholera toxin (CT), other enterotoxins, adherence factors such as the toxin-coregulated pilus (TCP), motility systems, and hemolysins (66). The CT and TCP virulence factors are regulated by the transmembrane DNA-binding protein ToxR regulatory cascade (71). ToxR activates the expression of toxT in a coordinated effort with TcpP, another transmembrane transcriptional activator. ToxT subsequently activates the transcription of virulence genes, including CT and TCP genes, to facilitate colonization of and disease in the gastrointestinal tract (71, 72). As outlined below, the regulation and coordinated expression of these virulence factors in the presence of bile have enabled Vibrio to adapt to and survive within the human host following a lifestyle in aquatic environments.
Vibrio cholerae
V. cholerae represents a well-studied pathogen with regard to bile exposure. There are several mechanisms by which V. cholerae resists the bactericidal conditions of bile. First, the general diffusion porins OmpU and OmpT, located in the bacterial outer membrane, have been demonstrated to be critical for bile resistance (72). The expression of ompU and ompT is mediated by ToxR, which results in induced ompU expression but repressed ompT expression in the presence of bile salts. It is hypothesized that OmpU is expressed in the presence of deoxycholate since it is an anion-selective porin that restricts the passage of negatively charged compounds such as those found in bile (73–76). Increased ompU expression appears to be unique to V. cholerae, as other pathogens such as EHEC and Salmonella (see below) repress genes encoding the Omp proteins in the presence of bile. Second, several efflux pumps are required for bile resistance, including the AcrAB-TolC system (77, 78), BreAB (79, 80), VexAB (81), and VprAB (82). Indeed, given the constant exposure of Vibrio to bile, the utilization of several efflux pumps is necessary to ensure survival and establish infection in the small intestine. Finally, recent work identified the transcriptional regulator LeuO, also regulated by ToxR, as being important for bile resistance. While the authors this study demonstrated that leuO expression was a specific response to bile and required the ToxR periplasmic signaling domain, a clear role for LeuO in bile resistance remains to be identified (75).
Formation of biofilms is a well-characterized phenotype in Vibrio species (69). Biofilms are homogenous or heterogeneous communities of bacteria embedded on a matrix of extracellular polymeric substances (EPSs) composed mostly of polysaccharides, with additional molecules such as proteins, lipids, and nucleic acids. Biofilm formation is a common approach for several bacterial pathogens to resist harsh environmental conditions such as exposure to antibiotics, UV radiation, and pH stress (83). Hung et al. investigated biofilm formation in the presence of crude bile and the bile acid sodium cholate. Biofilm formation following sodium cholate treatment was dependent upon the induction of the Vibrio polysaccharide synthesis (vps) genes and posttranscriptional activation of the regulator VpsR. Moreover, biofilm formation led to an increased ability of bacteria to resist the cytotoxic effects of sodium cholate (84). Recent work demonstrated that the regulation of the second messenger molecule cyclic di-GMP (c-di-GMP) is important for biofilm formation in the presence of bile, as deletion of genes encoding enzymes important for c-di-GMP turnover abolishes biofilm formation (85, 86). Finally, it is worth mentioning that the bile salt taurocholate was recently demonstrated to disperse ingested mature V. cholerae biofilms. Since V. cholerae biofilms are known to form in aquatic environments, exposure to taurocholate in the proximal small intestine may degrade the environmental biofilm prior to V. cholerae virulence gene activation in the new host environment (87). How this observation relates to the above-mentioned observations of biofilm formation in the presence of crude bile and sodium cholate requires further investigation. Nevertheless, V. cholerae has clearly utilized biofilm formation to resist the bactericidal conditions of the small intestine and establish infection.
V. cholerae Virulence Factor Expression
The differential expression of several key V. cholerae virulence factors following bile exposure has been well documented. One of the more interesting observations is that the V. cholerae ctxAB toxin genes and tcpA, a component of the TCP, are repressed at high bile concentrations; however, expression is increased once the bacterium reaches the site of infection at the epithelial surface, where the concentration of bile is lower (88, 89). In conjunction with this expression pattern, motility genes are induced significantly in bile and repressed as the bacteria make contact with the epithelium. The authors of this study hypothesized that this coordinated virulence factor expression pattern enables V. cholerae to move to the more permissible colonization environment at the apical surface of epithelial cells, which then triggers CT and TCP expression (88, 89).
The role of ToxR and ToxT in the repression of CT and TCP in the presence of bile is quite complex. Early work demonstrated that ToxT was involved in CT and TCP inhibition (89). However, in light of observations that bile induced OmpU expression through ToxR (72), Hung and Mekalanos performed further analyses to demonstrate that bile acids actually induced in vitro CT expression in a ToxR-dependent, ToxT-independent manner (90). These authors stated that the analysis was initiated by the hypothesis that the heterogenous composition of bile had pleiotropic effects on Vibrio. The alkaline pH and temperature of 37°C used in this study better represented in vivo conditions encountered by these bacteria. Furthermore, a separate study utilizing the V. cholerae El Tor biotype, an ex vivo intestinal model, and the bile salt taurocholate found that bile salts induced disulfide bond formation in the transcriptional activator TcpP, which in turn led to toxT expression and induced virulence (91). While the use of the ex vivo model mimics a more physiologically relevant environment for bacteria, the use of the El Tor biotype strain as opposed to the classical biotype strain of V. cholerae may explain the different findings regarding ToxT and virulence factor induction. Finally, in light of data from the study by Hung and Mekalanos, further work by Chatterjee et al. with fractionated crude bile demonstrated that the presence of unsaturated fatty acid components of bile, namely, arachidonic acid, linoleic acid, and oleic acid, repressed both the ctxAB and tcpA genes (92). This repression was mediated through the histone-like nucleoid-structuring transcriptional regulator (H-NS), and recently, it was shown that linoleic acid decreases the binding affinity of ToxT at the promoters of ToxT-controlled genes (93). Motility was also significantly induced in the presence of linoleic acid, which was regulated through the transcriptional activator flrA (92). While the individual components of bile have various effects on CT and TCP, bile clearly serves as a signal for V. cholerae to optimize virulence factor expression for efficient infection. Indeed, the coordinated virulence factor expression profile in response to bile exemplifies the complex regulatory network required for V. cholerae in different environments.
Additional V. cholerae virulence factors have been shown to be affected by bile or bile salts. In a TCP/CT-negative clinical isolate of V. cholerae belonging to the O39 serogroup, both bile and deoxycholate were found to induce the expression of T3SS genes, including the ToxR-like regulatory genes vttRA and vttRB (94). Some of the secreted effectors of the T3SS have been defined, with VopX being shown to be induced by bile and regulated by VttRA and VttRB (95). Furthermore, the trh gene encoding the related thermostable direct hemolysin (TRH) was induced in the presence of bile, which was regulated by VttRA, VttRB, and ToxR (96). A clear role for TRH in V. cholerae virulence requires further investigation. Finally, bile acids were recently shown to affect the type VI secretion system (T6SS) of V. cholerae (97), which has been demonstrated to be important for environmental competition among bacterial species, particularly in Gram-negative bacteria (98). By using a pandemic strain of V. cholerae lacking CT, Bachmann et al. demonstrated that mucin activated the T6SS, but the activity was repressed by deoxycholic acid. Interestingly, cholic acid had no effect on the repression of the T6SS. This study also demonstrated that modification of bile acids by the microbiota diminished the bacterial killing activity of the T6SS (97). This study not only highlights an important mechanism by which V. cholerae competes with commensal bacteria in the gut to establish infection but also demonstrates the importance of intestinal microbiota that modifies bile acids in an attempt to disrupt T6SS-mediated killing. In all, the virulence factors induced during bile exposure clearly demonstrate the ability of V. cholerae to alter gene expression in response to host signals to ensure survival and cause disease.
Vibrio parahaemolyticus and Vibrio vulnificus
V. parahaemolyticus and V. vulnificus are also able to resist the bactericidal conditions of bile. Several efflux pumps required for bile acid resistance have been identified in V. parahaemolyticus, including VmeAB, VmeCD, and VmeTUV (99, 100). An analysis of V. vulnificus efflux pumps identified VexAB as being important for bile acid resistance despite the lack of significant gene expression induction in the presence of bile acids (101). Additionally, the stress response of the RpoS protein is required for V. vulnificus bile resistance (102), and bile-adapted strains have been shown to respond to and survive low-salinity stress (103). These studies highlight mechanisms of bile resistance and tolerance similar to those of V. cholerae. As mentioned above, efflux pumps are an important mechanism by which several Vibrio species resist bile and adapt to infection in the small intestine, where bile is abundant.
Like V. cholerae, virulence factors encoded by V. parahaemolyticus include two T3SSs and the accompanying transcriptional regulators, thermostable direct hemolysin (TDH), two flagellar systems, and a T6SS (104, 105). Gotoh et al. demonstrated that the T3SS encoded on chromosome II (T3SS2), which is essential for intestinal colonization and enterotoxicity, was induced during crude bile exposure along with TDH. Furthermore, microarray analysis identified 77 genes induced by bile (105). The induced TDH expression confirms previously reported findings that TDH is secreted more in the presence of bile acids (106). To further highlight the importance of the induced gene profile in V. parahaemolyticus during bile exposure, Gotoh et al. demonstrated that absorptive removal of endogenous bile salts in the rabbit ileal loop assay, a measure of enterotoxicity and subsequent intestinal fluid accumulation, significantly reduced fluid accumulation, confirming induced virulence gene expression in the presence of bile. Additionally, these authors determined that the transcriptional regulators VtrA and VtrB were induced by bile and required for induced T3SS2 and TDH expression (105). Recently, an RNA sequencing analysis of V. parahaemolyticus exposed to bile and compared to bacteria isolated from an infant rabbit model of infection was performed. Of the 69 genes induced in the presence of bile, 53 were also induced in the rabbit model, and induction of T3SS2 was confirmed (104). These studies not only identify important gene expression profiles of V. parahaemolyticus under different conditions but also exemplify how bile exposure can mimic the in vivo conditions encountered by bacterial pathogens.
Future Studies of Vibrio
Extensive studies of Vibrio species related to bile exposure, resistance, and virulence gene activation have allowed a deeper understanding of virulence mechanisms and infections in pathogens that cause a significant global disease burden each year. These studies have also aided researchers who study other bacterial pathogens with similar virulence factors in understanding host-pathogen interactions. Future studies should be directed at further elucidating virulence gene expression of V. cholerae following bile exposure, especially given the complex regulatory pathways and variability in the related response to the various components of bile. Indeed, a consensus viewpoint and understanding of virulence gene expression will facilitate future antibiotic and vaccine development. With regard to V. parahaemolyticus and V. vulnificus, efforts should be focused on improving the comprehension of bile responses and related virulence gene activation. While these pathogens may be similar to V. cholerae, identifying differences in the responses to bile for each species may lead to significant advances in the scientific community that will help control infection. Finally, studies that include other in vivo signals combined with bile will truly allow researchers to appreciate the complexity of survival in the gastrointestinal tract.
SALMONELLA
Salmonella is a Gram-negative facultative anaerobe with motility conferred by peritrichous flagella. Closely related to E. coli but possessing distinct genomic content, there are two Salmonella species: S. bongori and S. enterica. S. bongori strains predominantly colonize cold-blooded reptiles, whereas S. enterica strains are capable of infecting both humans and mammals (107). S. enterica strains fall into six subspecies and are divided into two serovars based on human disease phenotype: nontyphoidal strains cause self-limiting, localized infection through invasion of macrophages and epithelial cells lining the small intestine, while typhoidal strains cause systemic illness due to dissemination to the lymph nodes, liver, and spleen through the lymphatic system (108). Typhoidal strains encompass S. enterica serovars Typhi and Paratyphi. Typhoid illness has a clinical presentation consisting of headache, fever, and transient diarrhea or constipation that can lead to systemic pathologies and persistent infection in the respiratory, digestive, or nephrology systems if left untreated. The nontyphoidal S. enterica serovars Typhimurium and Enteritidis are represented by numerous subspecies differentiated by O- and H-antigen typing of the lipopolysaccharide (LPS) structure (107, 109). All serovars possess at least five distinct Salmonella pathogenicity islands encoding several virulence factors. These genetic elements are denoted Salmonella pathogenicity island 1 (SPI-1) to SPI-5, with additional serovar-specific islands, such as SPI-7, encoding the Vi antigen, a capsular polysaccharide that covers the O-antigen of LPS, specific to typhoidal serovars (110–112). The gene clusters encompassed in the pathogenicity islands encode virulence factors such as the immune evasion proteins SptP and SseL; two separate coding regions for T3SSs; and host cell membrane manipulation machinery proteins, including SipA and SopE (113). Despite the fact that typhoidal serovars have smaller genomes than those of nontyphoidal serovars, S. enterica serovar Typhi contains additional potent virulence factors such as typhoid toxin and the Vi antigen (112, 114, 115). As highlighted below, bile has been demonstrated to serve as an extremely important physiological stimulus that regulates changes in gene expression for both survival and virulence gene activation.
Salmonella Bile Resistance Mechanisms
The ability of Salmonella to survive in bile is a longstanding observation and is utilized in selective media such as MacConkey agar, which contains bile salts, to promote the isolation of Salmonella and other Gram-negative bacteria (116, 117). Salmonella serovars have several approaches for survival in the presence of bile salts, which include defense against membrane destruction and DNA damage facilitated by the detergent activity of bile compounds (3). To prevent bile-mediated destruction, removal of intracellular bile compounds is achieved through the coordinated use of efflux pumps and additional outer membrane proteins. The S. enterica serovar Typhimurium genome encodes at least nine efflux pumps for resistance to several antimicrobial compounds, including bile (118). The other Salmonella serovars have homologous efflux pump genes; however, there is significant variation between serovars (Table 3). One well-defined efflux pump is the acridine orange removal pump encoded by acrAB, an operon that is also carried by EHEC (119, 120). Knockdown of S. enterica serovar Typhimurium acrAB resulted in hypersensitivity to bile salts, other detergents, and antibiotics (121). Expression of acrAB is enhanced after S. enterica serovar Typhimurium is exposed to cholic acid, deoxycholic acid, or a mixture of bile salts, a phenotype which was also shown for the EHEC bile response. Regulation of acrAB is achieved by activation with RamA or through repression by RamR (119). Interestingly, bile salts stimuli increase the expression of both Ram proteins. Cholate has been shown to directly bind RamA in Salmonella Typhimurium, and despite enhanced ramR expression under the same conditions, the interaction of RamR with the promoter-binding region is blocked in the presence of bile (119, 122, 123). Additional efflux pump systems identified in S. enterica serovar Typhimurium include acrEF, mdtABC, mdsCBA, emrAB, mdtABC, mdtK, and macAB, many of which have homologs in E. coli or Pseudomonas aeruginosa (118). There may be additional efflux pumps that are yet to be identified since no other serovar possesses all nine of the efflux pumps identified in S. enterica serovar Typhimurium (Table 2). As seen for Vibrio, the constant exposure of Salmonella to bile results in the utilization of several efflux pumps to survive and establish infection in the small intestine.
TABLE 3.
Comparison of Salmonella efflux pump genese
| Efflux pump gene(s) | Presence in S. enterica serovar: |
|||
|---|---|---|---|---|
| Enteritidisa | Typhimuriumb | Typhic | Paratyphid | |
| acrAB | No | Yes | Yes | Yes |
| acrEF | Partial | Yes | No | Partial |
| mdtABC | Yes | Yes | Yes | Partial |
| mdsCBA | Yes | Yes | Yes | Partial |
| emrAB | Yes | Yes | Partial | Yes |
| mdtABC | Yes | Yes | Yes | Yes |
| mdtK | Yes | Yes | Yes | Yes |
| macAB | Yes | Yes | Yes | Partial |
| tolC | Yes | Yes | Yes | Yes |
Strain P125109 (GenBank accession number NC_011294.1).
Strain LT2 (GenBank accession number NC_003197.1).
Strain CT18 (GenBank accession number NC_003198.1).
Strain ATCC 9150 (GenBank accession number NC_006511.1).
S. enterica serovars Enteritidis, Typhi, and Paratyphi carry 78%, 78%, and 56% of the genes carried by S. enterica serovar Typhimurium, respectively.
Most efflux pump genes require an associated outer membrane protein in the tol family (TolA and TolC) to protect against membrane destruction following bile salts exposure (3). There is significant genetic sequence diversity of the TolA peptide sequences between S. enterica serovars Typhi and Typhimurium. S. enterica serovar Typhi TolA is 27 amino acids shorter due to the lack of 3 additional tandem repeats in the C-terminal membrane-binding region that exist in S. enterica serovar Typhimurium. This amino acid difference suggests a potential explanation for the differences in bile resistance observed between the two serovars. Analysis of the MIC of bile for each serovar demonstrated that the MIC for S. enterica serovar Typhimurium is higher (18%) than that for S. enterica serovar Typhi (12%) (117, 124). However, dose curve studies indicate that S. enterica serovar Typhi may have a survival advantage at higher concentrations of bile (8%) (125). This difference may be due to the ability of S. enterica serovar Typhi to survive in biofilms that are frequently observed in the gallbladder of chronic carriers (see below). Nevertheless, deletion of tolA in both S. enterica serovars Typhi and Typhimurium renders the bacteria susceptible to bile (125). Conversely, NCBI BLAST analysis reveals that the TolC proteins maintain between 99% and 100% amino acid and DNA sequence identities across the sequenced Salmonella species, confirming the conserved nature of the structure and function of TolC. Exposure to bile salts stimulates immediate modifications of the outer membrane to prevent additional damage. To enhance the bile-removing processes of the efflux pumps, outer membrane pore-forming complexes are downregulated to prevent additional bile from entering the bacterial cell. For example, reducing the expression of genes encoding the outer membrane proteins OmpF and OmpC, which normally permit bile transport into the bacterial cell, is one strategy by which Salmonella prevents bile uptake (3). This strategy is similar to the repression of ompT in Vibrio in response to bile exposure; however, it remains unknown if Salmonella harbors a V. cholerae ompU homolog for which expression would be increased following bile exposure. Additionally, it has been shown that exposure to the bile salt deoxycholate resulted in a dose-dependent increase in the expression of the regulatory locus marAB in S. enterica serovar Typhimurium (126), conferring survival in the presence of bile salts and antibiotics. The role of marAB is independent of the acrAB efflux pump, suggesting that marAB regulates a different mechanism for survival during bile-induced stress (4, 126). Taken together, the data establish the importance of efflux pumps and outer membrane pore-forming complexes in maintaining Salmonella survival in the presence of bile.
Other modifications to the outer membrane are achieved by targeting LPS. First, the activation of the two-component regulatory system PhoPQ results in the remodeling of the lipid A structure of LPS (3, 117, 127). Although PhoPQ does not sense bile directly, it regulates several genes upon stimulation with bile, including central metabolism and respiration genes (124, 128). Deletion of phoP decreases the viability of both S. enterica serovars Typhi and Typhimurium in bile, and furthermore, constitutively expressed PhoP increases bacterial fitness in media containing bile (124). However, a recent microarray analysis using physiological murine bile obtained from mice found that phoP in S. enterica serovar Typhimurium was repressed in the presence of bile following 24 h of exposure, and this repression was independent of PhoPQ signaling. While physiological bovine bile also repressed phoP expression, commercial bovine bile and bile acids did not have the same effect. Therefore, different components of bile may be responsible for different responses in vitro and in vivo (128). Data from this recent analysis parallel the various findings of Vibrio research upon the utilization of different sources or components of bile, all of which highlight the importance of the use of physiological conditions to understand the complex interactions of each pathogen with bile. Certainly, future research analyzing PhoPQ regulation in physiological bile in both S. enterica serovars Typhi and Typhimurium will enhance our understanding of this regulation. Second, the addition of aminoarabinose to the lipid A component or the addition of phosphoethanolamine to either the core or lipid A region of LPS alters the outer membrane structure to minimize bile salt destruction (3, 117). Finally, the fepE gene of S. enterica serovar Typhimurium regulates the production of long O-chains, specifically responsible for the production of O-antigen repeats of >100 units. Mice infected with bacteria lacking the fepE gene did not develop colitis as observed with a wild-type strain. The authors of this study hypothesized that modification of the LPS structure through fepE may be an important bile survival strategy contributing to S. enterica serovar Typhimurium-associated mouse colitis (129). For typhoidal serovars, expression of the Vi antigen capsule is a strategy to mask the LPS structure and prevent immune detection (130). Although expression has not been demonstrated to be regulated by bile exposure, the Vi antigen has been observed in bacteria interacting with the ileal epithelium (131). Future studies should be directed toward determining if bile regulates the expression of the Vi antigen to influence immune evasion prior to contact with host cells. In all, modifications of the bacterial outer membrane occur rapidly after bile exposure to mitigate and prevent damage incurred from bile exposure.
Despite efforts to prevent bile uptake, some bile is still internalized in the bacterial cell, resulting in DNA damage (3, 117). In Salmonella, bile exposure increases the frequency of G-C-to-A-T transitions, which has been demonstrated to increase the expression of genes regulated by the stress-induced sigma factors SoxRS and OxyR (132, 133). Specifically, increased expression of DNA adenine methylase (dam) after bile salts exposure may serve to repair damage caused by exposure to bile salts, since knockout of dam abolished Salmonella viability after exposure to bile salts (132). It has also been hypothesized that S. enterica serovar Typhi bacteria colonizing the gallbladder in chronic carriers may be more prone to mutagenesis due to the high concentrations of bile salts (133). The constant exposure to high concentrations of bile increases the likelihood of mutagenesis through bile-induced DNA damage. A recent study demonstrated that S. enterica serovar Typhi infection of the gallbladder was sufficient to cause gallbladder carcinoma (GBC) in genetically predisposed cells. This association was investigated after the identification of an overlap in populations at risk for both typhoidal infection and GBC, with a high occurrence of both diseases in India (134). Scanu et al. demonstrated that bacterially mediated AKT and mitogen-activated protein kinase (MAPK) activation is required for the initiation of cellular transformation and tumor growth but is not required for sustained tumorigenesis, a conclusion concordant with previously reported hypotheses linking bacterially mediated downregulation of apoptosis and cancer initiation (135). Overall, membrane reorganization to prevent bile salt internalization and DNA damage serves as an important defense mechanism permitting Salmonella to survive under bile salt conditions.
Virulence Factor Regulation in Salmonella
Exposure of Salmonella to bile has been demonstrated to repress the expression of the T3SS through the inhibition of SPI-1, which is essential for bacterial invasion of epithelial cells. The sirC and invF genes encoding the transcriptional regulators for SPI-1 and SPI-1 effector proteins are downregulated following bile exposure (3, 117, 136, 137). Since murine infection models reveal Salmonella localization to microfold cells (M cells) within Peyer's patches in the ileum, it is believed that areas of high concentrations of bile, such as in the duodenum, do not serve as optimal invasion locations for Salmonella (3, 107, 138, 139). Therefore, by repressing the expression of the bacterial invasion machinery at high bile concentrations, Salmonella is able to invade cells in a more permissible environment along the gastrointestinal tract, where bile concentrations are decreased (136, 137). Despite data demonstrating the repression of the T3SS in response to bile, the invasion protein SipD, which is the Salmonella homolog of the Shigella invasion protein IpaD, has been shown to bind bile directly (140, 141). The importance of this binding is not yet known, but it represents a curious observation for how Salmonella interacts with the environment during infection. Additional virulence factors repressed by bile in S. enterica serovar Typhimurium include the flagellar flhC, flgC, and fliC genes; the fimbrial fimI and fimZ genes; and other virulence genes (128, 137). Unlike for Vibrio, motility appears to be repressed in the presence of bile, which could be linked to gallbladder colonization (see below). Future in-depth analyses linking virulence gene regulation to the duration and composition of bile exposure along the various segments of the gastrointestinal tract will certainly enhance our understanding of Salmonella pathogenesis.
Chronic Salmonella Infection May Be Attributed to Bile Activation of Survival Genes
Colonization of the gallbladder may serve as a long-term survival strategy for Salmonella, as indicated by the downregulation of motility genes and upregulation of stress response genes in response to bile (137). Salmonella serovars Typhimurium, Enteritidis, and Typhi have all been isolated from gallbladder biofilms. PCR-based identification of Salmonella species in the gallbladder of patients with cholecystitis, a condition characterized by inflammation of the gallbladder, suggests an association between inflammation and gallstone formation (142). Despite the identification of several Salmonella serovars in the gallbladder, only Salmonella serovar Typhi appears to be capable of long-term survival in the gallbladder, leading to an asymptomatic carrier state that facilitates ongoing bacterial transmission, as exemplified by the famous case of Typhoid Mary (117, 142, 143). Bacteria isolated from gallbladder biofilms show increased expression levels of flagellar filament and fimbrial proteins for colonization of gallstones (117, 137, 142). However, the expression of invasion and motility proteins, such as the flagellar motor, is not necessary for gallstone biofilm formation by Salmonella serovars, suggesting that colonization and invasion components are differentially regulated depending on the bile salt concentration of the environment (142). Interestingly, there have been conflicting reports regarding the Vi antigen of Salmonella serovar Typhi in gallbladder biofilms. One report demonstrated that EPS matrix production was independent of the Vi antigen, given that a mutant of the regulator for the Vi antigen, a ΔtviB mutant, was able to form a biofilm over the course of 18 days (144). However, a recent study demonstrated that active bile-induced biofilms stain positively for Vi antigen expression (145). As stated above, future studies of the regulation of the Vi antigen in the presence of bile will enhance our understanding of this important virulence factor. Certainly, the ability to resist bile and establish a biofilm during gallbladder colonization in the chronic infection state is essential to the life cycle of Salmonella to maintain a survival niche and eventually facilitate transmission to new hosts.
Future Research on Salmonella
In summary, while there are several common traits that explain how Salmonella serovars respond to and interact with bile, there also appears to be a diverse set of bile responses that distinguishes each serovar. These unique bile responses may serve to provide critical insight into the pathogenicity, infection strategies, and host specificity of each of these diverse serovars. Further research into bile resistance proteins such as TolA, which differ in sequence and function between Salmonella serovars Typhimurium and Typhi, will provide insights into bile regulation of Salmonella pathogenesis. Conversely, further research into proteins that are homologous between serovars, such as the transcriptional regulator MarA, could reveal critical similarities in gene activation in response to bile. Future research should incorporate in vivo models of Salmonella colonization and survival under conditions of high and low bile concentrations to enhance our understanding of Salmonella infection. Additionally, since bile composition is altered across mammals (146), comparisons of bile compositions among susceptible host species may reveal a host-specific signal responsible for regulating Salmonella infection.
CAMPYLOBACTER JEJUNI
Campylobacter jejuni is a Gram-negative, spiral-shaped, microaerophilic bacterium that is a common cause of diarrheal disease around the world. C. jejuni colonizes and invades the distal ileum and colonic epithelium, causing cramping, bloody stools, and diarrhea (147). Outbreaks of C. jejuni occur in both industrialized and developing nations, typically following the consumption of contaminated food and water; however, developing countries also tend to be areas where the disease is endemic, and high rates of coinfection or asymptomatic carriage are also common (147–149). Although most patients recover after self-limiting infection, 1 in every 2,000 infected individuals develops Guillain-Barré syndrome, an acute disease that leads to muscle weakness and ascending paralysis of the peripheral nervous system and is believed to be an autoimmune response associated with bacterial or viral infections (150). Research with C. jejuni has been challenging due to the high degree of genomic plasticity and high rates of genetic transformation found within this naturally competent microorganism (149). However, strains that carry or overexpress flagellum genes, adherence structures, and invasion mechanisms are associated with increased virulence (148–151). As summarized below, pathogenic studies reveal that bile exposure acts as a stimulus for the upregulation of many virulence mechanisms in C. jejuni.
Bile Resistance in C. jejuni
Similar to other enteric pathogens, C. jejuni has developed bile resistance to survive within the human gastrointestinal tract (152, 153). The CmeABC multidrug efflux pump is one of the most studied bile response systems in C. jejuni (154). This system is encoded by the cmeABC operon, encoding a periplasmic protein (CmeA), an inner membrane transporter (CmeB), and an outer membrane protein (CmeC), respectively. The operon has been demonstrated to be constitutively expressed but is also controlled by a TetR family repressor (CmeR) that recognizes the cmeABC promoter region, resulting in decreased transcription (155). Bile salts appear to inhibit the CmeR-cmeABC promoter interaction, which results in increased cmeABC operon transcription (153). Once expression is triggered in bile salts, the CmeABC efflux pump increases the ability of the bacteria to withstand the bactericidal conditions of bile, antibiotics, and other antimicrobial substances such as SDS (156). Mutants defective in cmeABC expression have a decreased ability to survive in bile salts and colonize chicken intestinal tracts (154). An additional efflux pump, CmeDEF, works in conjunction with CmeABC to confer an additional level of resistance to bile and other antimicrobials (155). However, CmeDEF appears to be a secondary resistance mechanism since bile resistance is dominated by the activity of CmeABC, and CmeDEF may preserve bile resistance only in the absence of CmeABC (157).
Additional mechanisms implicated in the C. jejuni bile response are two-component regulatory systems (TCRSs) that sense and respond to bile salts. TCRS are often utilized by bacteria after exposure to environmental stimuli as a means of altering gene expression to cope with changing conditions (158). Raphael et al. identified the Campylobacter bile resistance regulator (CbrR) through in silico analyses and subsequent genetic mutagenesis experiments (159). Response regulators, such as CbrR, are vital TCRS components that either directly or indirectly alter gene expression after phosphorylation via a histidine/sensor kinase receptor (158). A CbrR mutant could not survive in the presence of medium supplemented with bile salts and had a diminished ability to colonize chickens (159). Although the CbrR-binding partners and the exact mechanism of bile resistance are unknown, studies have also demonstrated that the response regulator has a critical role in C. jejuni survival in bile. Recently, Okoli et al. utilized a bioinformatic approach with protein and domain architecture analyses to assess genomes of members of the order Campylobacterales for the presence of homologous proteins important for bile responses in other bacteria (152). This study identified 151 proteins conserved across species, as well as species-specific genes, implicated in bile tolerance. This cross-species analysis also revealed putative proteins that may be important for the C. jejuni bile response (152). Future studies will determine if these genes encode proteins that regulate virulence mechanisms in C. jejuni in the presence of bile.
C. jejuni Virulence Factor Expression in the Presence of Bile
In addition to survival and resistance genes, bile exposure increases the expression of key virulence factors in C. jejuni. The Campylobacter invasive antigens (Cia) are proteins secreted directly into the epithelial cell cytoplasm through a flagellar apparatus, resulting in host membrane ruffling, alterations in signaling and intracellular trafficking, and increased bacterial uptake (160–162). Mutagenesis studies that blocked Cia secretion revealed significantly reduced C. jejuni internalization by epithelial cells (161). The Cia proteins are thought to be synthesized and secreted in response to stimulatory substances provided by the host. Moreover, the Cia proteins have been induced after exposure to bile acids such as deoxycholate, cholate, and chenodeoxycholate (163, 164). Exposure to other molecules, such as heat-stable proteins in fetal bovine serum, epithelial cell lysates, and extracellular matrix components, has also been found to increase the synthesis of Cia. However, secretion of the Cia proteins through the flagellar apparatus appears to be dependent on bacterial contact with cellular components of host epithelia (164).
Two additional virulence mechanisms in C. jejuni that may be regulated by bile exposure are biofilm formation and motility genes. One study identified that flagella are necessary for enhanced C. jejuni biofilm formation following bile salts exposure, with the flagellar filament mediating attachment for the biofilm (165). Another study used proteomic two-dimensional (2D) gel electrophoresis to demonstrate that ox bile may increase the expression of flagellin A, the chemotaxis protein CheV, and the ATP synthase, suggesting that bile components may induce motility and act as chemotactic attractants for C. jejuni (166). Li et al. further confirmed that bile components are chemotactic attractants after observing that methyl-accepting chemotaxis proteins are needed for chemotaxis and that the absence of these proteins, or the absence of bile, severely impairs colonization in mice (167). In contrast, a study examining motility via zone-of-migration assays found that deoxycholate exposure did not alter the motility of C. jejuni in vitro, and deoxycholate did not affect adherence to epithelial cells (168). These divergent findings for both bile salt-dependent motility and adherence could be due to the genomic plasticity and phase variation found in C. jejuni isolates. Future research is needed to elucidate the relationships that bile exposure has to the colonization, internalization, and motility of C. jejuni. Furthermore, transcriptomic assessment of bile salts exposure over various time points is needed to describe the impact of bile on C. jejuni pathogenesis during biofilm formation and planktonic stages.
GRAM-POSITIVE BACTERIA
While the majority of enteric pathogens studied in detail are Gram-negative bacteria, a few Gram-positive bacteria can cause a significant burden of infection in the gastrointestinal tract. This section highlights recent findings of studies investigating bile responses in Clostridium difficile and Listeria monocytogenes. As outlined below, and like Gram-negative pathogens, these Gram-positive bacteria are able to survive and adapt in the presence of bile while also enhancing infection and survival in the gastrointestinal environment.
Clostridium difficile
C. difficile is a spore-forming bacterium responsible for recurring gastrointestinal colitis associated with recent antibiotic treatment (169). Several C. difficile studies have demonstrated that bile has a significant impact on C. difficile germination, which can be either enhanced or inhibited by different bile acids. Primary bile acids have been shown to promote the germination of C. difficile spores, while secondary bile acids inhibit the germination of spores into vegetative bacteria (170–172). Although only 5% of the bile acids along the gastrointestinal tract typically enter the colon in a healthy individual, during C. difficile infection, bile composition and microbiota homeostasis are significantly altered to increase the levels of primary bile acids in the colon and potentially promote the germination of C. difficile spores (29, 173). Antibiotic treatment perturbs the commensal microbiota that converts the primary bile acids into secondary bile acids, which normally inhibit C. difficile proliferation (170, 172, 173). The loss of the microbiota therefore results in a greater abundance of primary bile acids that can germinate C. difficile spores (170, 172). However, the increased concentration of primary bile acids in the colon during recurring C. difficile infections can be reversed following fecal microbiota transplantation (173). Much like other enteric bacteria, C. difficile appears to have found a mechanism for overcoming the inhibitory effects of bile by exploiting the dynamics between the compositions of microbiota and bile in the gastrointestinal tract. It will be interesting to see if additional C. difficile virulence factors are induced by bile.
Listeria monocytogenes
L. monocytogenes is a foodborne pathogen that causes large outbreaks due to colonization of prepared or prepackaged food sources such as deli meat and produce and exists in the environment as a saprophyte that can gain access to food sources either directly or through carriage in farm animals (174). This pathogen can efficiently adapt to the different host environments, including the gastrointestinal tract (174, 175). As reviewed by Gahan and Hill, bile induces stress responses (176), efflux pumps and related transcriptional regulators (177), as well as a bile salt hydrolase enzyme (178), with each factor contributing to L. monocytogenes survival during bile exposure (174). The bile stress response includes the induced expression of DNA repair enzymes, chaperones, and oxidative stress proteins (176, 179). Furthermore, a recent study compared the abilities of different isolates to survive bile exposure under aerobic and anaerobic conditions at acidic pH. White et al. demonstrated that acidic pH reduced bile resistance but that anaerobic conditions improved bile resistance at pH 5.5, particularly in virulent isolates from humans. The authors hypothesized that oxygen availability may influence stress response genes (180), which requires further investigation. Finally, numerous studies have identified gallbladder infection and colonization in both animal models and, in rare cases, human gallbladder infections. It is hypothesized that growth in the bile-rich environment of the gallbladder enhances fecal shedding during infection (174). Taken together, the survival of and potentially enhanced infection by L. monocytogenes as a result of bile exposure highlight the ability of this pathogen to maintain a successful infectious life cycle. Future work identifying additional virulence factors induced by bile will enhance our understanding of the virulence mechanisms of this foodborne opportunistic pathogen.
CONCLUDING REMARKS
In closing, enteric pathogens, and even some commensal bacteria, have adapted to the environment of the gastrointestinal tract to efficiently colonize the host and cause infection. As a whole, enteric bacteria have modified outer membrane proteins, utilized porins, and relied on efflux mechanisms to resist the bactericidal conditions of bile. It is fascinating how unrelated species have utilized similar mechanisms, particularly with regard to efflux pumps, as a prime example of convergent evolution. Furthermore, as these bacteria are able to survive in the presence of bile, enteric pathogens have evolved to exploit this host signal to regulate virulence factor gene expression. Whether each species activates virulence genes to colonize the small intestine or delays the expression of virulence factors until a more permissible environment in the ileum, colon, or epithelial surface of the gastrointestinal tract is reached, the coordination of virulence gene expression in the presence of bile is quite remarkable and provides fertile ground for further studies using the global transcriptional analysis tools of RNA-Seq. The studies outlined here have highlighted the ability of these pathogens to adapt to and successfully colonize the host and cause infection. As researchers move forward with investigative studies (Table 4) and vaccine development for enteric pathogens, we hope that future experiments will utilize conditions that better represent the in vivo environment encountered by these bacteria. Even the simple addition of bile or bile salts to culture media has significantly enhanced our understanding of enteric bacterial pathogenesis. We hope that the gains in the awareness of bile-regulated virulence in enteric bacteria will finally result in successful vaccine development for many of these formidable pathogens.
TABLE 4.
Future bile-related research goals for each pathogen
| Pathogen | Future research goal(s) |
|---|---|
| Escherichia coli | Analysis of additional pathovars such as EIEC, AIEC, and DAEC |
| Further analysis of bile resistance and regulation of bile-mediated virulence | |
| Further analysis of commensal organisms | |
| ETEC | Functional studies to characterize the bile response and coordination of virulence factor expression |
| EPEC | Characterization of the mechanism and clinical implications of bile malabsorption during infection |
| EHEC | Future transcriptomic studies to understand the complete bile response |
| Shigella | Identification of resistance mechanisms and regulation of virulence factors |
| Utilization of bile extract in future analyses | |
| Vibrio | |
| V. cholerae | Further investigation into the complex regulatory network of the bile response |
| Use of consistent sources of bile | |
| V. parahaemolyticus | Improved understanding of the bile response |
| Further analysis of bile-mediated virulence | |
| V. vulnificus | Identification of virulence factors affected by bile exposure |
| Salmonella | Comparison of serovars to understand various bile responses and related infection strategies |
| Utilization of bile in analyses of infection models | |
| Campylobacter | Clarification of the relationship between bile exposure and induction of adherence and motility |
| Additional transcriptomic analyses of bile exposure | |
| Clostridium | Identification of resistance mechanisms and additional virulence factors |
| Listeria | Identification of virulence factors affected by bile exposure |
ACKNOWLEDGMENTS
This project was funded in part by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, from grants T32 5T32AI095190-04 (J.R.S.), U19AI110820 (D.A.R.), and 1K22AI104755-01 (C.S.F.).
Biographies

Jeticia R. Sistrunk received her bachelor's degree in Microbiology from the University of Oklahoma in 2008. She is currently working on comparative genomics and transcriptomics of enterotoxigenic Escherichia coli isolates in Dr. David A. Rasko's laboratory for her Ph.D. in Molecular Microbiology and Immunology from the University of Maryland School of Medicine.

Kourtney P. Nickerson received her Ph.D. in Molecular Medicine from the Cleveland Clinic Lerner College of Medicine at Case Western Reserve University in 2014. She is currently a postdoctoral scientist at Massachusetts General Hospital, conducting research at the Mucosal Immunology and Biology Research Center in the laboratories of Dr. Christina S. Faherty and Dr. Alessio Fasano. Her research focuses on host-pathogen interactions in Shigella flexneri and Salmonella Typhi.

Rachael B. Chanin received her bachelor's degree in psychology from Brandeis University in 2010. She became a research technician at the Mucosal Immunology and Biology Research Center at Massachusetts General Hospital in 2014 and is currently researching the early infection process of Shigella flexneri. In the fall of 2016, she will begin a Ph.D. program at the UT Southwestern Medical Center in the Division of Basic Sciences.

David A. Rasko is an Associate Professor at the Institute for Genome Sciences, University of Maryland School of Medicine. His research focuses on utilizing comparative genomics of closely related isolates to examine pathogen evolution, with a particular interest in Acinetobacter baumannii, Escherichia coli, and Shigella. Dr. Rasko earned a Ph.D. in Medical Microbiology and Immunology at the University of Alberta, Canada, in 2000. He did a postdoctoral fellowship at the University of Maryland School of Medicine and has served as both a GenBank Biologist at the National Center for Biotechnology Information (NCBI) and a staff scientist at the Institute for Genomic Research (TIGR). Following a Research Assistant Professor position at the University of Texas Southwestern Medical Center at Dallas, Dr. Rasko became an Assistant Professor at the University of Maryland School of Medicine in 2008 and was promoted to Associate Professor in 2013.

Christina S. Faherty is an Assistant Professor at the Mucosal Immunology and Biology Research Center at Massachusetts General Hospital with an academic appointment at Harvard Medical School. Her research focuses on host-pathogen interactions in enteric bacterial pathogens, and she is specifically interested in how Shigella utilizes host signals to alter virulence gene expression prior to invasion of colonic epithelial cells. Dr. Faherty received her Ph.D. in Infectious Diseases at the Uniformed Services University of the Health Sciences in Bethesda, MD, in 2009. She did her postdoctoral work at the Center for Vaccine Development, University of Maryland School of Medicine, and became an Assistant Professor at Massachusetts General Hospital/Harvard Medical School in 2013.
REFERENCES
- 1.Elliott DE, Siddique SS, Weinstock JV. 2014. Innate immunity in disease. Clin Gastroenterol Hepatol 12:749–755. doi: 10.1016/j.cgh.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hong W, Wu YE, Fu X, Chang Z. 2012. Chaperone-dependent mechanisms for acid resistance in enteric bacteria. Trends Microbiol 20:328–335. doi: 10.1016/j.tim.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Begley M, Gahan CG, Hill C. 2005. The interaction between bacteria and bile. FEMS Microbiol Rev 29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Merritt ME, Donaldson JR. 2009. Effect of bile salts on the DNA and membrane integrity of enteric bacteria. J Med Microbiol 58:1533–1541. doi: 10.1099/jmm.0.014092-0. [DOI] [PubMed] [Google Scholar]
- 5.Kortman GA, Boleij A, Swinkels DW, Tjalsma H. 2012. Iron availability increases the pathogenic potential of Salmonella Typhimurium and other enteric pathogens at the intestinal epithelial interface. PLoS One 7:e29968. doi: 10.1371/journal.pone.0029968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandrini S, Masania R, Zia F, Haigh R, Freestone P. 2013. Role of porin proteins in acquisition of transferrin iron by enteropathogens. Microbiology 159:2639–2650. doi: 10.1099/mic.0.071928-0. [DOI] [PubMed] [Google Scholar]
- 7.Yu LC, Wang JT, Wei SC, Ni YH. 2012. Host-microbial interactions and regulation of intestinal epithelial barrier function: from physiology to pathology. World J Gastrointest Pathophysiol 3:27–43. doi: 10.4291/wjgp.v3.i1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reshetnyak VI. 2013. Physiological and molecular biochemical mechanisms of bile formation. World J Gastroenterol 19:7341–7360. doi: 10.3748/wjg.v19.i42.7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ridlon JM, Kang DJ, Hylemon PB. 2006. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47:241–259. [DOI] [PubMed] [Google Scholar]
- 10.Hamer HM, De Preter V, Windey K, Verbeke K. 2012. Functional analysis of colonic bacterial metabolism: relevant to health? Am J Physiol Gastrointest Liver Physiol 302:G1–G9. doi: 10.1152/ajpgi.00048.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kristoffersen SM, Ravnum S, Tourasse NJ, Okstad OA, Kolsto AB, Davies W. 2007. Low concentrations of bile salts induce stress responses and reduce motility in Bacillus cereus ATCC 14579 [corrected]. J Bacteriol 189:5302–5313. doi: 10.1128/JB.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding JW, Andersson R, Soltesz V, Willen R, Bengmark S. 1993. The role of bile and bile acids in bacterial translocation in obstructive jaundice in rats. Eur Surg Res 25:11–19. doi: 10.1159/000129252. [DOI] [PubMed] [Google Scholar]
- 13.Slocum MM, Sittig KM, Specian RD, Deitch EA. 1992. Absence of intestinal bile promotes bacterial translocation. Am Surg 58:305–310. [PubMed] [Google Scholar]
- 14.Lisowska A, Kobelska-Dubiel N, Jankowska I, Pawlowska J, Moczko J, Walkowiak J. 2014. Small intestinal bacterial overgrowth in patients with progressive familial intrahepatic cholestasis. Acta Biochim Pol 61:103–107. [PubMed] [Google Scholar]
- 15.Mayo SA, Song YK, Cruz MR, Phan TM, Singh KV, Garsin DA, Murray BE, Dial EJ, Lichtenberger LM. 2016. Indomethacin injury to the rat small intestine is dependent upon biliary secretion and is associated with overgrowth of enterococci. Physiol Rep 4:e12725. doi: 10.14814/phy2.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 18.Hughes DT, Sperandio V. 2008. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol 6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. 2013. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev 26:822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torres AG, Tutt CB, Duval L, Popov V, Nasr AB, Michalski J, Scaletsky IC. 2007. Bile salts induce expression of the afimbrial LDA adhesin of atypical enteropathogenic Escherichia coli. Cell Microbiol 9:1039–1049. doi: 10.1111/j.1462-5822.2006.00850.x. [DOI] [PubMed] [Google Scholar]
- 21.Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol Microbiol 48:1609–1619. doi: 10.1046/j.1365-2958.2003.03531.x. [DOI] [PubMed] [Google Scholar]
- 22.Thanassi DG, Cheng LW, Nikaido H. 1997. Active efflux of bile salts by Escherichia coli. J Bacteriol 179:2512–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kus JV, Gebremedhin A, Dang V, Tran SL, Serbanescu A, Barnett Foster D. 2011. Bile salts induce resistance to polymyxin in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol 193:4509–4515. doi: 10.1128/JB.00200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahl JW, Rasko DA. 2012. Analysis of global transcriptional profiles of enterotoxigenic Escherichia coli isolate E24377A. Infect Immun 80:1232–1242. doi: 10.1128/IAI.06138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Jesus MC, Urban AA, Marasigan ME, Barnett Foster DE. 2005. Acid and bile-salt stress of enteropathogenic Escherichia coli enhances adhesion to epithelial cells and alters glycolipid receptor binding specificity. J Infect Dis 192:1430–1440. doi: 10.1086/462422. [DOI] [PubMed] [Google Scholar]
- 26.Barnett Foster D. 2013. Modulation of the enterohemorrhagic E. coli virulence program through the human gastrointestinal tract. Virulence 4:315–323. doi: 10.4161/viru.24318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arenas-Hernandez MM, Rojas-Lopez M, Medrano-Lopez A, Nunez-Reza KJ, Puente JL, Martinez-Laguna Y, Torres AG. 2014. Environmental regulation of the long polar fimbriae 2 of enterohemorrhagic Escherichia coli O157:H7. FEMS Microbiol Lett 357:105–114. doi: 10.1111/1574-6968.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Froelich JM, Tran K, Wall D. 2006. A pmrA constitutive mutant sensitizes Escherichia coli to deoxycholic acid. J Bacteriol 188:1180–1183. doi: 10.1128/JB.188.3.1180-1183.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hofmann AF, Hagey LR. 2008. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell Mol Life Sci 65:2461–2483. doi: 10.1007/s00018-008-7568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kramer VC, Nickerson KW, Hamlett NV, O'Hara C. 1984. Prevalence of extreme detergent resistance among the Enterobacteriaceae. Can J Microbiol 30:711–713. doi: 10.1139/m84-106. [DOI] [PubMed] [Google Scholar]
- 31.D'Mello A, Yotis WW. 1987. The action of sodium deoxycholate on Escherichia coli. Appl Environ Microbiol 53:1944–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobsen L, Durso L, Conway T, Nickerson KW. 2009. Escherichia coli O157:H7 and other E. coli strains share physiological properties associated with intestinal colonization. Appl Environ Microbiol 75:4633–4635. doi: 10.1128/AEM.00003-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Paepe M, Gaboriau-Routhiau V, Rainteau D, Rakotobe S, Taddei F, Cerf-Bensussan N. 2011. Trade-off between bile resistance and nutritional competence drives Escherichia coli diversification in the mouse gut. PLoS Genet 7:e1002107. doi: 10.1371/journal.pgen.1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwan BW, Lord DM, Peti W, Page R, Benedik MJ, Wood TK. 14 February 2015. The MqsR/MqsA toxin/antitoxin system protects Escherichia coli during bile acid stress. Environ Microbiol doi: 10.1111/1462-2920.12749. [DOI] [PubMed] [Google Scholar]
- 35.Bernstein C, Bernstein H, Payne CM, Beard SE, Schneider J. 1999. Bile salt activation of stress response promoters in Escherichia coli. Curr Microbiol 39:68–72. doi: 10.1007/s002849900420. [DOI] [PubMed] [Google Scholar]
- 36.Foster PL. 2007. Stress-induced mutagenesis in bacteria. Crit Rev Biochem Mol Biol 42:373–397. doi: 10.1080/10409230701648494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kandell RL, Bernstein C. 1991. Bile salt/acid induction of DNA damage in bacterial and mammalian cells: implications for colon cancer. Nutr Cancer 16:227–238. doi: 10.1080/01635589109514161. [DOI] [PubMed] [Google Scholar]
- 38.Sengupta C, Ray S, Chowdhury R. 2014. Fine tuning of virulence regulatory pathways in enteric bacteria in response to varying bile and oxygen concentrations in the gastrointestinal tract. Gut Pathog 6:38. doi: 10.1186/s13099-014-0038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamner S, McInnerney K, Williamson K, Franklin MJ, Ford TE. 2013. Bile salts affect expression of Escherichia coli O157:H7 genes for virulence and iron acquisition, and promote growth under iron limiting conditions. PLoS One 8:e74647. doi: 10.1371/journal.pone.0074647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qadri F, Svennerholm AM, Faruque AS, Sack RB. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin Microbiol Rev 18:465–483. doi: 10.1128/CMR.18.3.465-483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Binsztein N, Jouve MJ, Viboud GI, Lopez Moral L, Rivas M, Orskov I, Ahren C, Svennerholm AM. 1991. Colonization factors of enterotoxigenic Escherichia coli isolated from children with diarrhea in Argentina. J Clin Microbiol 29:1893–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaastra W, Svennerholm AM. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol 4:444–452. doi: 10.1016/0966-842X(96)10068-8. [DOI] [PubMed] [Google Scholar]
- 43.von Mentzer A, Connor TR, Wieler LH, Semmler T, Iguchi A, Thomson NR, Rasko DA, Joffre E, Corander J, Pickard D, Wiklund G, Svennerholm AM, Sjoling A, Dougan G. 2014. Identification of enterotoxigenic Escherichia coli (ETEC) clades with long-term global distribution. Nat Genet 46:1321–1326. doi: 10.1038/ng.3145. [DOI] [PubMed] [Google Scholar]
- 44.Nicklasson M, Sjoling A, von Mentzer A, Qadri F, Svennerholm AM. 2012. Expression of colonization factor CS5 of enterotoxigenic Escherichia coli (ETEC) is enhanced in vivo and by the bile component Na glycocholate hydrate. PLoS One 7:e35827. doi: 10.1371/journal.pone.0035827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grewal HM, Valvatne H, Bhan MK, van Dijk L, Gaastra W, Sommerfelt H. 1997. A new putative fimbrial colonization factor, CS19, of human enterotoxigenic Escherichia coli. Infect Immun 65:507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sommerfelt H, Grewal HM, Svennerholm AM, Gaastra W, Flood PR, Viboud G, Bhan MK. 1992. Genetic relationship of putative colonization factor O166 to colonization factor antigen I and coli surface antigen 4 of enterotoxigenic Escherichia coli. Infect Immun 60:3799–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chatterjee A, Chowdhury R. 2008. Bile and unsaturated fatty acids inhibit the binding of cholera toxin and Escherichia coli heat-labile enterotoxin to GM1 receptor. Antimicrob Agents Chemother 52:220–224. doi: 10.1128/AAC.01009-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gabig M, Herman-Antosiewicz A, Kwiatkowska M, Los M, Thomas MS, Wegrzyn G. 2002. The cell surface protein Ag43 facilitates phage infection of Escherichia coli in the presence of bile salts and carbohydrates. Microbiology 148:1533–1542. doi: 10.1099/00221287-148-5-1533. [DOI] [PubMed] [Google Scholar]
- 49.Romao FT, Hernandes RT, Yamamoto D, Osugui L, Popi A, Gomes TA. 2014. Influence of environmental factors in the adherence of an atypical enteropathogenic Escherichia coli strain to epithelial cells. BMC Microbiol 14:299. doi: 10.1186/s12866-014-0299-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Annaba F, Sarwar Z, Gill RK, Ghosh A, Saksena S, Borthakur A, Hecht GA, Dudeja PK, Alrefai WA. 2012. Enteropathogenic Escherichia coli inhibits ileal sodium-dependent bile acid transporter ASBT. Am J Physiol Gastrointest Liver Physiol 302:G1216–G1222. doi: 10.1152/ajpgi.00017.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alrefai WA, Gill RK. 2007. Bile acid transporters: structure, function, regulation and pathophysiological implications. Pharm Res 24:1803–1823. doi: 10.1007/s11095-007-9289-1. [DOI] [PubMed] [Google Scholar]
- 52.Goosney DL, Celli J, Kenny B, Finlay BB. 1999. Enteropathogenic Escherichia coli inhibits phagocytosis. Infect Immun 67:490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bauer ME, Welch RA. 1996. Characterization of an RTX toxin from enterohemorrhagic Escherichia coli O157:H7. Infect Immun 64:167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Torres AG, Kaper JB. 2003. Multiple elements controlling adherence of enterohemorrhagic Escherichia coli O157:H7 to HeLa cells. Infect Immun 71:4985–4995. doi: 10.1128/IAI.71.9.4985-4995.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chassaing B, Etienne-Mesmin L, Bonnet R, Darfeuille-Michaud A. 2013. Bile salts induce long polar fimbriae expression favouring Crohn's disease-associated adherent-invasive Escherichia coli interaction with Peyer's patches. Environ Microbiol 15:355–371. doi: 10.1111/j.1462-2920.2012.02824.x. [DOI] [PubMed] [Google Scholar]
- 56.Schroeder GN, Hilbi H. 2008. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev 21:134–156. doi: 10.1128/CMR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 58.Pope LM, Reed KE, Payne SM. 1995. Increased protein secretion and adherence to HeLa cells by Shigella spp. following growth in the presence of bile salts. Infect Immun 63:3642–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hale TL, Sansonetti PJ, Schad PA, Austin S, Formal SB. 1983. Characterization of virulence plasmids and plasmid-associated outer membrane proteins in Shigella flexneri, Shigella sonnei, and Escherichia coli. Infect Immun 40:340–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olive AJ, Kenjale R, Espina M, Moore DS, Picking WL, Picking WD. 2007. Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect Immun 75:2626–2629. doi: 10.1128/IAI.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stensrud KF, Adam PR, La Mar CD, Olive AJ, Lushington GH, Sudharsan R, Shelton NL, Givens RS, Picking WL, Picking WD. 2008. Deoxycholate interacts with IpaD of Shigella flexneri in inducing the recruitment of IpaB to the type III secretion apparatus needle tip. J Biol Chem 283:18646–18654. doi: 10.1074/jbc.M802799200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barta ML, Guragain M, Adam P, Dickenson NE, Patil M, Geisbrecht BV, Picking WL, Picking WD. 2012. Identification of the bile salt binding site on IpaD from Shigella flexneri and the influence of ligand binding on IpaD structure. Proteins 80:935–945. doi: 10.1002/prot.23251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dickenson NE, Zhang L, Epler CR, Adam PR, Picking WL, Picking WD. 2011. Conformational changes in IpaD from Shigella flexneri upon binding bile salts provide insight into the second step of type III secretion. Biochemistry 50:172–180. doi: 10.1021/bi101365f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dickenson NE, Arizmendi O, Patil MK, Toth RT IV, Middaugh CR, Picking WD, Picking WL. 2013. N-terminus of IpaB provides a potential anchor to the Shigella type III secretion system tip complex protein IpaD. Biochemistry 52:8790–8799. doi: 10.1021/bi400755f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Faherty CS, Redman JC, Rasko DA, Barry EM, Nataro JP. 2012. Shigella flexneri effectors OspE1 and OspE2 mediate induced adherence to the colonic epithelium following bile salts exposure. Mol Microbiol 85:107–121. doi: 10.1111/j.1365-2958.2012.08092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cary JW, Linz JE, Bhatnagar D. 1999. Microbial foodborne diseases: mechanisms of pathogenesis and toxin synthesis. CRC Press, Boca Raton, FL. [Google Scholar]
- 67.Austin B. 2010. Vibrios as causal agents of zoonoses. Vet Microbiol 140:310–317. doi: 10.1016/j.vetmic.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 68.Faruque SM, Albert MJ, Mekalanos JJ. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev 62:1301–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yildiz FH, Visick KL. 2009. Vibrio biofilms: so much the same yet so different. Trends Microbiol 17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Butt AA, Aldridge KE, Sanders CV. 2004. Infections related to the ingestion of seafood part I: viral and bacterial infections. Lancet Infect Dis 4:201–212. doi: 10.1016/S1473-3099(04)00969-7. [DOI] [PubMed] [Google Scholar]
- 71.Peterson KM. 2002. Expression of Vibrio cholerae virulence genes in response to environmental signals. Curr Issues Intest Microbiol 3:29–38. [PubMed] [Google Scholar]
- 72.Provenzano D, Schuhmacher DA, Barker JL, Klose KE. 2000. The virulence regulatory protein ToxR mediates enhanced bile resistance in Vibrio cholerae and other pathogenic Vibrio species. Infect Immun 68:1491–1497. doi: 10.1128/IAI.68.3.1491-1497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Duret G, Delcour AH. 2006. Deoxycholic acid blocks Vibrio cholerae OmpT but not OmpU porin. J Biol Chem 281:19899–19905. doi: 10.1074/jbc.M602426200. [DOI] [PubMed] [Google Scholar]
- 74.Simonet VC, Basle A, Klose KE, Delcour AH. 2003. The Vibrio cholerae porins OmpU and OmpT have distinct channel properties. J Biol Chem 278:17539–17545. doi: 10.1074/jbc.M301202200. [DOI] [PubMed] [Google Scholar]
- 75.Ante VM, Bina XR, Howard MF, Sayeed S, Taylor DL, Bina JE. 2015. Vibrio cholerae leuO transcription is positively regulated by ToxR and contributes to bile resistance. J Bacteriol 197:3499–3510. doi: 10.1128/JB.00419-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Provenzano D, Lauriano CM, Klose KE. 2001. Characterization of the role of the ToxR-modulated outer membrane porins OmpU and OmpT in Vibrio cholerae virulence. J Bacteriol 183:3652–3662. doi: 10.1128/JB.183.12.3652-3662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bina JE, Mekalanos JJ. 2001. Vibrio cholerae tolC is required for bile resistance and colonization. Infect Immun 69:4681–4685. doi: 10.1128/IAI.69.7.4681-4685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chatterjee A, Chaudhuri S, Saha G, Gupta S, Chowdhury R. 2004. Effect of bile on the cell surface permeability barrier and efflux system of Vibrio cholerae. J Bacteriol 186:6809–6814. doi: 10.1128/JB.186.20.6809-6814.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cerda-Maira FA, Kovacikova G, Jude BA, Skorupski K, Taylor RK. 2013. Characterization of BreR interaction with the bile response promoters breAB and breR in Vibrio cholerae. J Bacteriol 195:307–317. doi: 10.1128/JB.02008-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cerda-Maira FA, Ringelberg CS, Taylor RK. 2008. The bile response repressor BreR regulates expression of the Vibrio cholerae breAB efflux system operon. J Bacteriol 190:7441–7452. doi: 10.1128/JB.00584-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bina JE, Provenzano D, Wang C, Bina XR, Mekalanos JJ. 2006. Characterization of the Vibrio cholerae vexAB and vexCD efflux systems. Arch Microbiol 186:171–181. doi: 10.1007/s00203-006-0133-5. [DOI] [PubMed] [Google Scholar]
- 82.Herrera CM, Crofts AA, Henderson JC, Pingali SC, Davies BW, Trent MS. 2014. The Vibrio cholerae VprA-VprB two-component system controls virulence through endotoxin modification. mBio 5:e02283-14. doi: 10.1128/mBio.02283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gupta P, Sarkar S, Das B, Bhattacharjee S, Tribedi P. 16 September 2015. Biofilm, pathogenesis and prevention—a journey to break the wall: a review. Arch Microbiol doi: 10.1007/s00203-015-1148-6. [DOI] [PubMed] [Google Scholar]
- 84.Hung DT, Zhu J, Sturtevant D, Mekalanos JJ. 2006. Bile acids stimulate biofilm formation in Vibrio cholerae. Mol Microbiol 59:193–201. doi: 10.1111/j.1365-2958.2005.04846.x. [DOI] [PubMed] [Google Scholar]
- 85.Koestler BJ, Waters CM. 2014. Bile acids and bicarbonate inversely regulate intracellular cyclic di-GMP in Vibrio cholerae. Infect Immun 82:3002–3014. doi: 10.1128/IAI.01664-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koestler BJ, Waters CM. 2014. Intestinal GPS: bile and bicarbonate control cyclic di-GMP to provide Vibrio cholerae spatial cues within the small intestine. Gut Microbes 5:775–780. doi: 10.4161/19490976.2014.985989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hay AJ, Zhu J. 2015. Host intestinal signal-promoted biofilm dispersal induces Vibrio cholerae colonization. Infect Immun 83:317–323. doi: 10.1128/IAI.02617-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gupta S, Chowdhury R. 1997. Bile affects production of virulence factors and motility of Vibrio cholerae. Infect Immun 65:1131–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schuhmacher DA, Klose KE. 1999. Environmental signals modulate ToxT-dependent virulence factor expression in Vibrio cholerae. J Bacteriol 181:1508–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hung DT, Mekalanos JJ. 2005. Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc Natl Acad Sci U S A 102:3028–3033. doi: 10.1073/pnas.0409559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang M, Liu Z, Hughes C, Stern AM, Wang H, Zhong Z, Kan B, Fenical W, Zhu J. 2013. Bile salt-induced intermolecular disulfide bond formation activates Vibrio cholerae virulence. Proc Natl Acad Sci U S A 110:2348–2353. doi: 10.1073/pnas.1218039110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chatterjee A, Dutta PK, Chowdhury R. 2007. Effect of fatty acids and cholesterol present in bile on expression of virulence factors and motility of Vibrio cholerae. Infect Immun 75:1946–1953. doi: 10.1128/IAI.01435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Plecha SC, Withey JH. 2015. Mechanism for inhibition of Vibrio cholerae ToxT activity by the unsaturated fatty acid components of bile. J Bacteriol 197:1716–1725. doi: 10.1128/JB.02409-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alam A, Tam V, Hamilton E, Dziejman M. 2010. vttRA and vttRB encode ToxR family proteins that mediate bile-induced expression of type three secretion system genes in a non-O1/non-O139 Vibrio cholerae strain. Infect Immun 78:2554–2570. doi: 10.1128/IAI.01073-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alam A, Miller KA, Chaand M, Butler JS, Dziejman M. 2011. Identification of Vibrio cholerae type III secretion system effector proteins. Infect Immun 79:1728–1740. doi: 10.1128/IAI.01194-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miller KA, Hamilton E, Dziejman M. 2012. The Vibrio cholerae trh gene is coordinately regulated in vitro with type III secretion system genes by VttR(A)/VttR(B) but does not contribute to Caco2-BBE cell cytotoxicity. Infect Immun 80:4444–4455. doi: 10.1128/IAI.00832-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bachmann V, Kostiuk B, Unterweger D, Diaz-Satizabal L, Ogg S, Pukatzki S. 2015. Bile salts modulate the mucin-activated type VI secretion system of pandemic Vibrio cholerae. PLoS Negl Trop Dis 9:e0004031. doi: 10.1371/journal.pntd.0004031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Russell AB, Peterson SB, Mougous JD. 2014. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol 12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Matsuo T, Nakamura K, Kodama T, Mikami T, Hiyoshi H, Tsuchiya T, Ogawa W, Kuroda T. 2013. Characterization of all RND-type multidrug efflux transporters in Vibrio parahaemolyticus. Microbiologyopen 2:725–742. doi: 10.1002/mbo3.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Matsuo T, Ogawa W, Tsuchiya T, Kuroda T. 2014. Overexpression of vmeTUV encoding a multidrug efflux transporter of Vibrio parahaemolyticus causes bile acid resistance. Gene 541:19–25. doi: 10.1016/j.gene.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 101.Lee S, Yeom JH, Seo S, Lee M, Kim S, Bae J, Lee K, Hwang J. 2015. Functional analysis of Vibrio vulnificus RND efflux pumps homologous to Vibrio cholerae VexAB and VexCD, and to Escherichia coli AcrAB. J Microbiol 53:256–261. doi: 10.1007/s12275-015-5037-0. [DOI] [PubMed] [Google Scholar]
- 102.Chen WL, Oliver JD, Wong HC. 2010. Adaptation of Vibrio vulnificus and an rpoS mutant to bile salts. Int J Food Microbiol 140:232–238. doi: 10.1016/j.ijfoodmicro.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 103.Wong HC, Liu SH. 2006. Susceptibility of the heat-, acid-, and bile-adapted Vibrio vulnificus to lethal low-salinity stress. J Food Prot 69:2924–2928. [DOI] [PubMed] [Google Scholar]
- 104.Livny J, Zhou X, Mandlik A, Hubbard T, Davis BM, Waldor MK. 2014. Comparative RNA-Seq based dissection of the regulatory networks and environmental stimuli underlying Vibrio parahaemolyticus gene expression during infection. Nucleic Acids Res 42:12212–12223. doi: 10.1093/nar/gku891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gotoh K, Kodama T, Hiyoshi H, Izutsu K, Park KS, Dryselius R, Akeda Y, Honda T, Iida T. 2010. Bile acid-induced virulence gene expression of Vibrio parahaemolyticus reveals a novel therapeutic potential for bile acid sequestrants. PLoS One 5:e13365. doi: 10.1371/journal.pone.0013365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Osawa R, Yamai S. 1996. Production of thermostable direct hemolysin by Vibrio parahaemolyticus enhanced by conjugated bile acids. Appl Environ Microbiol 62:3023–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fabrega A, Vila J. 2013. Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin Microbiol Rev 26:308–341. doi: 10.1128/CMR.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.de Jong HK, Parry CM, van der Poll T, Wiersinga WJ. 2012. Host-pathogen interaction in invasive salmonellosis. PLoS Pathog 8:e1002933. doi: 10.1371/journal.ppat.1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Beltran P, Musser JM, Helmuth R, Farmer JJ III, Frerichs WM, Wachsmuth IK, Ferris K, McWhorter AC, Wells JG, Cravioto A. 1988. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc Natl Acad Sci U S A 85:7753–7757. doi: 10.1073/pnas.85.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fuentes JA, Jofre MR, Villagra NA, Mora GC. 2009. RpoS- and Crp-dependent transcriptional control of Salmonella Typhi taiA and hlyE genes: role of environmental conditions. Res Microbiol 160:800–808. doi: 10.1016/j.resmic.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 111.Vernikos GS, Parkhill J. 2006. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 22:2196–2203. doi: 10.1093/bioinformatics/btl369. [DOI] [PubMed] [Google Scholar]
- 112.Zhang H, Zhou Y, Bao H, Liu HW. 2006. Vi antigen biosynthesis in Salmonella Typhi: characterization of UDP-N-acetylglucosamine C-6 dehydrogenase (TviB) and UDP-N-acetylglucosaminuronic acid C-4 epimerase (TviC). Biochemistry 45:8163–8173. doi: 10.1021/bi060446d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Srikanth CV, Mercado-Lubo R, Hallstrom K, McCormick BA. 2011. Salmonella effector proteins and host-cell responses. Cell Mol Life Sci 68:3687–3697. doi: 10.1007/s00018-011-0841-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Deng L, Song J, Gao X, Wang J, Yu H, Chen X, Varki N, Naito-Matsui Y, Galan JE, Varki A. 2014. Host adaptation of a bacterial toxin from the human pathogen Salmonella Typhi. Cell 159:1290–1299. doi: 10.1016/j.cell.2014.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Song J, Gao X, Galan JE. 2013. Structure and function of the Salmonella Typhi chimaeric A(2)B(5) typhoid toxin. Nature 499:350–354. doi: 10.1038/nature12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Atlas RM. 1996. Handbook of microbiological media, 2nd ed CRC Press, Boca Raton, FL. [Google Scholar]
- 117.Alvarez-Ordonez A, Begley M, Prieto M, Messens W, Lopez M, Bernardo A, Hill C. 2011. Salmonella spp. survival strategies within the host gastrointestinal tract. Microbiology 157:3268–3281. doi: 10.1099/mic.0.050351-0. [DOI] [PubMed] [Google Scholar]
- 118.Nishino K, Latifi T, Groisman EA. 2006. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol Microbiol 59:126–141. doi: 10.1111/j.1365-2958.2005.04940.x. [DOI] [PubMed] [Google Scholar]
- 119.Nikaido E, Yamaguchi A, Nishino K. 2008. AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhimurium by RamA in response to environmental signals. J Biol Chem 283:24245–24253. doi: 10.1074/jbc.M804544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Martins A, Amaral L. 2012. Screening for efflux pump systems of bacteria by the new acridine orange agar method. In Vivo 26:203–206. [PubMed] [Google Scholar]
- 121.Buckley AM, Webber MA, Cooles S, Randall LP, La Ragione RM, Woodward MJ, Piddock LJ. 2006. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol 8:847–856. doi: 10.1111/j.1462-5822.2005.00671.x. [DOI] [PubMed] [Google Scholar]
- 122.Bailey AM, Ivens A, Kingsley R, Cottell JL, Wain J, Piddock LJ. 2010. RamA, a member of the AraC/XylS family, influences both virulence and efflux in Salmonella enterica serovar Typhimurium. J Bacteriol 192:1607–1616. doi: 10.1128/JB.01517-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Baucheron S, Nishino K, Monchaux I, Canepa S, Maurel MC, Coste F, Roussel A, Cloeckaert A, Giraud E. 2014. Bile-mediated activation of the acrAB and tolC multidrug efflux genes occurs mainly through transcriptional derepression of ramA in Salmonella enterica serovar Typhimurium. J Antimicrob Chemother 69:2400–2406. doi: 10.1093/jac/dku140. [DOI] [PubMed] [Google Scholar]
- 124.van Velkinburgh JC, Gunn JS. 1999. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect Immun 67:1614–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lahiri A, Ananthalakshmi TK, Nagarajan AG, Ray S, Chakravortty D. 2011. TolA mediates the differential detergent resistance pattern between the Salmonella enterica subsp. enterica serovars Typhi and Typhimurium. Microbiology 157:1402–1415. doi: 10.1099/mic.0.046565-0. [DOI] [PubMed] [Google Scholar]
- 126.Prouty AM, Brodsky IE, Falkow S, Gunn JS. 2004. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium. Microbiology 150:775–783. doi: 10.1099/mic.0.26769-0. [DOI] [PubMed] [Google Scholar]
- 127.Dalebroux ZD, Miller SI. 2014. Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Curr Opin Microbiol 17:106–113. doi: 10.1016/j.mib.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Antunes LC, Wang M, Andersen SK, Ferreira RB, Kappelhoff R, Han J, Borchers CH, Finlay BB. 2012. Repression of Salmonella enterica phoP expression by small molecules from physiological bile. J Bacteriol 194:2286–2296. doi: 10.1128/JB.00104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Crawford RW, Keestra AM, Winter SE, Xavier MN, Tsolis RM, Tolstikov V, Baumler AJ. 2012. Very long O-antigen chains enhance fitness during Salmonella-induced colitis by increasing bile resistance. PLoS Pathog 8:e1002918. doi: 10.1371/journal.ppat.1002918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wilson RP, Raffatellu M, Chessa D, Winter SE, Tukel C, Baumler AJ. 2008. The Vi-capsule prevents Toll-like receptor 4 recognition of Salmonella. Cell Microbiol 10:876–890. doi: 10.1111/j.1462-5822.2007.01090.x. [DOI] [PubMed] [Google Scholar]
- 131.Tran QT, Gomez G, Khare S, Lawhon SD, Raffatellu M, Baumler AJ, Ajithdoss D, Dhavala S, Adams LG. 2010. The Salmonella enterica serotype Typhi Vi capsular antigen is expressed after the bacterium enters the ileal mucosa. Infect Immun 78:527–535. doi: 10.1128/IAI.00972-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Prieto AI, Ramos-Morales F, Casadesus J. 2004. Bile-induced DNA damage in Salmonella enterica. Genetics 168:1787–1794. doi: 10.1534/genetics.104.031062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Prieto AI, Ramos-Morales F, Casadesus J. 2006. Repair of DNA damage induced by bile salts in Salmonella enterica. Genetics 174:575–584. doi: 10.1534/genetics.106.060889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Scanu T, Spaapen RM, Bakker JM, Pratap CB, Wu LE, Hofland I, Broeks A, Shukla VK, Kumar M, Janssen H, Song JY, Neefjes-Borst EA, te Riele H, Holden DW, Nath G, Neefjes J. 2015. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell Host Microbe 17:763–774. doi: 10.1016/j.chom.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 135.Faherty CS, Maurelli AT. 2008. Staying alive: bacterial inhibition of apoptosis during infection. Trends Microbiol 16:173–180. doi: 10.1016/j.tim.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Prouty AM, Gunn JS. 2000. Salmonella enterica serovar Typhimurium invasion is repressed in the presence of bile. Infect Immun 68:6763–6769. doi: 10.1128/IAI.68.12.6763-6769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Prouty AM, Brodsky IE, Manos J, Belas R, Falkow S, Gunn JS. 2004. Transcriptional regulation of Salmonella enterica serovar Typhimurium genes by bile. FEMS Immunol Med Microbiol 41:177–185. doi: 10.1016/j.femsim.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 138.Takeuchi A. 1967. Electron microscope studies of experimental Salmonella infection. I. Penetration into the intestinal epithelium by Salmonella typhimurium. Am J Pathol 50:109–136. [PMC free article] [PubMed] [Google Scholar]
- 139.Jones BD, Ghori N, Falkow S. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J Exp Med 180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kaniga K, Trollinger D, Galan JE. 1995. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J Bacteriol 177:7078–7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang Y, Nordhues BA, Zhong D, De Guzman RN. 2010. NMR characterization of the interaction of the Salmonella type III secretion system protein SipD and bile salts. Biochemistry 49:4220–4226. doi: 10.1021/bi100335u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gonzalez-Escobedo G, Marshall JM, Gunn JS. 2011. Chronic and acute infection of the gall bladder by Salmonella Typhi: understanding the carrier state. Nat Rev Microbiol 9:9–14. doi: 10.1038/nrmicro2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Soper GA. 1939. The curious career of Typhoid Mary. Bull N Y Acad Med 15:698–712. [PMC free article] [PubMed] [Google Scholar]
- 144.Prouty AM, Schwesinger WH, Gunn JS. 2002. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect Immun 70:2640–2649. doi: 10.1128/IAI.70.5.2640-2649.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Marshall JM, Flechtner AD, La Perle KM, Gunn JS. 2014. Visualization of extracellular matrix components within sectioned Salmonella biofilms on the surface of human gallstones. PLoS One 9:e89243. doi: 10.1371/journal.pone.0089243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kararli TT. 1995. Comparison of the gastrointestinal anatomy, physiology, and biochemistry of humans and commonly used laboratory animals. Biopharm Drug Dispos 16:351–380. doi: 10.1002/bdd.2510160502. [DOI] [PubMed] [Google Scholar]
- 147.Silva J, Leite D, Fernandes M, Mena C, Gibbs PA, Teixeira P. 2011. Campylobacter spp. as a foodborne pathogen: a review. Front Microbiol 2:200. doi: 10.3389/fmicb.2011.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Crushell E, Harty S, Sharif F, Bourke B. 2004. Enteric Campylobacter: purging its secrets? Pediatr Res 55:3–12. doi: 10.1203/01.PDR.0000099794.06260.71. [DOI] [PubMed] [Google Scholar]
- 149.Young KT, Davis LM, Dirita VJ. 2007. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol 5:665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- 150.Wassenaar TM, Blaser MJ. 1999. Pathophysiology of Campylobacter jejuni infections of humans. Microbes Infect 1:1023–1033. doi: 10.1016/S1286-4579(99)80520-6. [DOI] [PubMed] [Google Scholar]
- 151.Platts-Mills JA, Kosek M. 2014. Update on the burden of Campylobacter in developing countries. Curr Opin Infect Dis 27:444–450. doi: 10.1097/QCO.0000000000000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Okoli AS, Wadstrom T, Mendz GL. 2007. MiniReview: bioinformatic study of bile responses in Campylobacterales. FEMS Immunol Med Microbiol 49:101–123. doi: 10.1111/j.1574-695X.2006.00194.x. [DOI] [PubMed] [Google Scholar]
- 153.Lin J, Martinez A. 2006. Effect of efflux pump inhibitors on bile resistance and in vivo colonization of Campylobacter jejuni. J Antimicrob Chemother 58:966–972. doi: 10.1093/jac/dkl374. [DOI] [PubMed] [Google Scholar]
- 154.Lin J, Sahin O, Michel LO, Zhang Q. 2003. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect Immun 71:4250–4259. doi: 10.1128/IAI.71.8.4250-4259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lin J, Akiba M, Sahin O, Zhang Q. 2005. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob Agents Chemother 49:1067–1075. doi: 10.1128/AAC.49.3.1067-1075.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mavri A, Smole Mozina S. 2013. Resistance to bile salts and sodium deoxycholate in macrolide- and fluoroquinolone-susceptible and resistant Campylobacter jejuni and Campylobacter coli strains. Microb Drug Resist 19:168–174. doi: 10.1089/mdr.2012.0217. [DOI] [PubMed] [Google Scholar]
- 157.Akiba M, Lin J, Barton YW, Zhang Q. 2006. Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J Antimicrob Chemother 57:52–60. [DOI] [PubMed] [Google Scholar]
- 158.Gunn JS. 2000. Mechanisms of bacterial resistance and response to bile. Microbes Infect 2:907–913. doi: 10.1016/S1286-4579(00)00392-0. [DOI] [PubMed] [Google Scholar]
- 159.Raphael BH, Pereira S, Flom GA, Zhang Q, Ketley JM, Konkel ME. 2005. The Campylobacter jejuni response regulator, CbrR, modulates sodium deoxycholate resistance and chicken colonization. J Bacteriol 187:3662–3670. doi: 10.1128/JB.187.11.3662-3670.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Konkel ME, Klena JD, Rivera-Amill V, Monteville MR, Biswas D, Raphael B, Mickelson J. 2004. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J Bacteriol 186:3296–3303. doi: 10.1128/JB.186.11.3296-3303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Christensen JE, Pacheco SA, Konkel ME. 2009. Identification of a Campylobacter jejuni-secreted protein required for maximal invasion of host cells. Mol Microbiol 73:650–662. doi: 10.1111/j.1365-2958.2009.06797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Eucker TP, Konkel ME. 2012. The cooperative action of bacterial fibronectin-binding proteins and secreted proteins promote maximal Campylobacter jejuni invasion of host cells by stimulating membrane ruffling. Cell Microbiol 14:226–238. doi: 10.1111/j.1462-5822.2011.01714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Konkel ME, Kim BJ, Rivera-Amill V, Garvis SG. 1999. Bacterial secreted proteins are required for the internalization of Campylobacter jejuni into cultured mammalian cells. Mol Microbiol 32:691–701. doi: 10.1046/j.1365-2958.1999.01376.x. [DOI] [PubMed] [Google Scholar]
- 164.Rivera-Amill V, Kim BJ, Seshu J, Konkel ME. 2001. Secretion of the virulence-associated Campylobacter invasion antigens from Campylobacter jejuni requires a stimulatory signal. J Infect Dis 183:1607–1616. doi: 10.1086/320704. [DOI] [PubMed] [Google Scholar]
- 165.Svensson SL, Pryjma M, Gaynor EC. 2014. Flagella-mediated adhesion and extracellular DNA release contribute to biofilm formation and stress tolerance of Campylobacter jejuni. PLoS One 9:e106063. doi: 10.1371/journal.pone.0106063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Fox EM, Raftery M, Goodchild A, Mendz GL. 2007. Campylobacter jejuni response to ox-bile stress. FEMS Immunol Med Microbiol 49:165–172. doi: 10.1111/j.1574-695X.2006.00190.x. [DOI] [PubMed] [Google Scholar]
- 167.Li Z, Lou H, Ojcius DM, Sun A, Sun D, Zhao J, Lin X, Yan J. 2014. Methyl-accepting chemotaxis proteins 3 and 4 are responsible for Campylobacter jejuni chemotaxis and jejuna colonization in mice in response to sodium deoxycholate. J Med Microbiol 63:343–354. doi: 10.1099/jmm.0.068023-0. [DOI] [PubMed] [Google Scholar]
- 168.Malik-Kale P, Parker CT, Konkel ME. 2008. Culture of Campylobacter jejuni with sodium deoxycholate induces virulence gene expression. J Bacteriol 190:2286–2297. doi: 10.1128/JB.01736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, Kuijper EJ, Wilcox MH. 2010. The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev 23:529–549. doi: 10.1128/CMR.00082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sorg JA, Sonenshein AL. 2010. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol 192:4983–4990. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Giel JL, Sorg JA, Sonenshein AL, Zhu J. 2010. Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLoS One 5:e8740. doi: 10.1371/journal.pone.0008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wilson KH. 1983. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J Clin Microbiol 18:1017–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, Nelson VM, Sadowsky MJ, Khoruts A. 2014. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol 306:G310–G319. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gahan CG, Hill C. 2014. Listeria monocytogenes: survival and adaptation in the gastrointestinal tract. Front Cell Infect Microbiol 4:9. doi: 10.3389/fcimb.2014.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cossart P. 2011. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc Natl Acad Sci U S A 108:19484–19491. doi: 10.1073/pnas.1112371108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Payne A, Schmidt TB, Nanduri B, Pendarvis K, Pittman JR, Thornton JA, Grissett J, Donaldson JR. 2013. Proteomic analysis of the response of Listeria monocytogenes to bile salts under anaerobic conditions. J Med Microbiol 62:25–35. doi: 10.1099/jmm.0.049742-0. [DOI] [PubMed] [Google Scholar]
- 177.Quillin SJ, Schwartz KT, Leber JH. 2011. The novel Listeria monocytogenes bile sensor BrtA controls expression of the cholic acid efflux pump MdrT. Mol Microbiol 81:129–142. doi: 10.1111/j.1365-2958.2011.07683.x. [DOI] [PubMed] [Google Scholar]
- 178.Dussurget O, Cabanes D, Dehoux P, Lecuit M, Buchrieser C, Glaser P, Cossart P, European Listeria Genome Consortium. 2002. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol Microbiol 45:1095–1106. doi: 10.1046/j.1365-2958.2002.03080.x. [DOI] [PubMed] [Google Scholar]
- 179.van der Veen S, Abee T. 2011. Contribution of Listeria monocytogenes RecA to acid and bile survival and invasion of human intestinal Caco-2 cells. Int J Med Microbiol 301:334–340. doi: 10.1016/j.ijmm.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 180.White SJ, McClung DM, Wilson JG, Roberts BN, Donaldson JR. 2015. Influence of pH on bile sensitivity amongst various strains of Listeria monocytogenes under aerobic and anaerobic conditions. J Med Microbiol 64:1287–1296. doi: 10.1099/jmm.0.000160. [DOI] [PMC free article] [PubMed] [Google Scholar]