SUMMARY
The number of large-scale genomics projects is increasing due to the availability of affordable high-throughput sequencing (HTS) technologies. The use of HTS for bacterial infectious disease research is attractive because one whole-genome sequencing (WGS) run can replace multiple assays for bacterial typing, molecular epidemiology investigations, and more in-depth pathogenomic studies. The computational resources and bioinformatics expertise required to accommodate and analyze the large amounts of data pose new challenges for researchers embarking on genomics projects for the first time. Here, we present a comprehensive overview of a bacterial genomics projects from beginning to end, with a particular focus on the planning and computational requirements for HTS data, and provide a general understanding of the analytical concepts to develop a workflow that will meet the objectives and goals of HTS projects.
INTRODUCTION
High-throughput sequencing (HTS) has transformed biomedical research. Declining costs and development of accessible computing options have resulted in the widespread adoption of these technologies in the scientific community. PCR and Sanger sequencing (often referred to as “traditional sequencing” methods) required proportionally more time generating the data than was needed for downstream analysis; in contrast, HTS platforms can produce massive amounts of data relatively quickly compared to the time needed for analysis and interpretation. The bottleneck between data generation and meaningful interpretation has generated a need for new, efficient, and innovative data management and analysis methods.
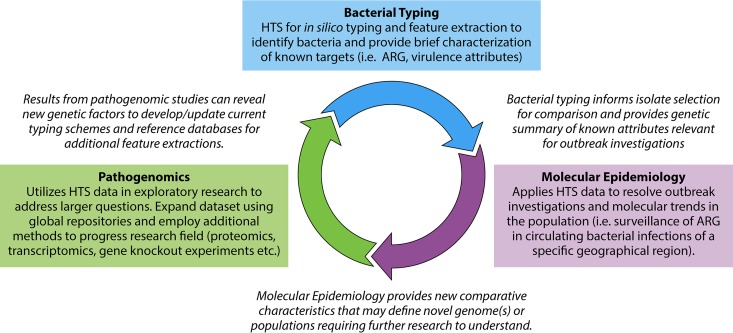
Here, we provide a comprehensive review on how to conduct an HTS project in bacterial genomics with particular emphasis on infectious disease microbiology. Although basic scientific processes and experimental design have not changed, the additional steps and scale of data generation have caused a paradigm shift in the time and resource allocations required to successfully complete HTS projects. We present the process in the context of three applications with various scopes, with the goal that this review will be relevant and scalable to many areas of infectious disease genomics research. The three applications include (i) bacterial typing, (ii) molecular epidemiology, and (iii) pathogenomics. Figure 1 illustrates how the use of HTS for whole-genome sequencing (WGS) can apply to scalable projects in a feedback loop. The whole-genome data can be mined for comparison with current typing schemes or used to create expanded “fingerprints” of the bacteria (bacterial typing), which in turn can contribute to investigating a larger defined bacterial population (molecular epidemiology). The comparative information regarding population trends, identification of novel strains, or genomic features can be studied in more depth by employing complementary research methods to understand pathogenic mechanisms (pathogenomics).
FIG 1.
Example of three HTS applications for infectious disease bacterial genomics. These applications use HTS data to answer common questions regarding bacterial pathogenesis in a public health/clinical microbiological research setting from bacterial typing to molecular epidemiology and in-depth pathogenomic investigations. These applications are shown in a feedback loop to demonstrate that HTS provides data that can be analyzed to various degrees (both depth and breadth) based on the hypotheses under test and the number of isolates included for comparative genomics.
To overcome the bottleneck associated with big data analysis, a shift in resource allocation is needed to ensure that adequate computational resources and expertise are available to efficiently produce high-quality data and results. Therefore, proper planning and a multidisciplinary team are essential to successfully execute large-scale HTS projects. This review provides a resource for conducting HTS projects from beginning to end, based on expertise from successful infectious disease genomics projects in the literature and personal experiences.
PREPARATION
Reallocation of resources to efficiently handle the increasing sample sizes and large amounts of HTS data produced presents new challenges to researchers. The amount of data generated often exceeds the computational storage and computing capacity of local systems, requiring researchers to find additional resources to organize and manage it all through their analysis workflows. Therefore, an end-to-end understanding of microbial HTS projects and available options will better equip researchers to anticipate bottlenecks and prepare sufficient resources to mitigate them.
HTS technologies enhance our ability to characterize and differentiate clinically relevant bacterial populations, understand and predict epidemiological trends, and create new analytical tools or improve existing non-HTS molecular tests (Fig. 1). The timeline for project completion depends on many variables such as the scope of the project (i.e., number of samples, size of the research team, and depth of research questions), biological characteristics of bacteria under study, sequencing platform(s) used, and outcome goals. Figure 2 illustrates a generalized timeline of the major stages in a large-scale HTS project. We have placed a large emphasis on the planning stage prior to data generation and the need for ongoing project management to maintain continual forward progression of tasks through each stage. The analysis has been separated into three stages: primary, secondary, and tertiary. Primary analysis is the first analytical pass: quality assurance (QA) and control of the HTS data. Secondary analysis employs common (likely automated) workflows typically performed on newly generated genomes, which can include reference mapping and de novo assembly. Tertiary analysis is the “sense-making” stage of the project, where interpretations and conclusions are drawn from comparative analyses, and it includes more specialized or focused processes.
FIG 2.
General HTS project stages and timeline. The importance and time requirements for the project planning stage and ongoing project management are sometimes underestimated but are invaluable for large-scale HTS projects for which hundreds of samples and terabytes of data are produced. The stages are divided by natural timeline progression and also increasing depth of investigation and specialized analysis requirements.
Project Management
The project manager role is often filled by the lead principal investigator or may be divided among senior members of the project. For some large-scale projects, a dedicated project manager may be assigned. In general terms, the project manager is responsible for organizing and controlling performance as the project progresses (1). HTS project-specific considerations are summarized in Table 1. Project management tasks can be categorized into communication, logistical facilitation (i.e., transfer of materials/data), and data management. For more detailed information on data management, we refer readers to recent publications that summarize the need for data management throughout the data life cycle in HTS projects (i.e., raw, intermediate, and result data) and propose some best-practice guidance to develop policies for the management, analysis, and sharing of data within HTS projects (2, 3).
TABLE 1.
Project management principles and how they relate to HTS research projects
| Principle | General components | HTS project-specific considerations |
|---|---|---|
| Planning | Identify questions to be addressed; identify stakeholders; identify tasks; identify outcomes, identify risks and plan for them; establish roles and responsibilities | Funding and ethics timelines; equipment costs/availability for all stages; choose sequencing platform and configurations; computational resources and analysis plan; appropriate strain selection; metadata organization and coding of samples; anticipate risks or bottlenecks in the workflow |
| Organizing | Organize resources; organize data | Staff training if required; data management; procurement; data collection |
| Controlling performance | Organize, focus, and motivate personnel; track work and results; communicate; update stakeholders; monitor and mitigate risks | Curate data on an ongoing basis; identify bottlenecks/issues and facilitate progression to next steps; confirm and assess work against milestones; progress reports as required |
Experimental Design
The experimental design should be established during the planning stage and encompass the entire project from the initial question/hypothesis through the sampling strategy, data generation methodology, and analysis plans to defining the outcome goals and deliverable endpoints. There are many legitimate sampling strategies in scientific research; the application will dictate which strategy is appropriate. Table 2 describes some general examples of sampling strategies for the three applications targeted in this review.
TABLE 2.
Sampling strategy examples for 3 applications of HTS projects
| Example of sampling strategy | Application(s)a |
|---|---|
| Unbiased prospective or retrospective (or combined) sampling—sampling of all strains meeting a specific definition (i.e., over a time period or region) for unbiased discovery (typically population-based studies) | |
| ∙ Characterization of genome population: what is circulating? | BT, ME |
| ∙ Sample and reveal trends for strains based on geography or time | ME |
| Differential or niche sampling—categorically biased sampling to assess anticipated population differences (typical case-vs-control or cohort studies) | |
| ∙ Compare genomic differences between epidemiologically defined groups (i.e., community vs hospital acquired; presence vs absence of a pathogenic phenotype or clinical outcome) | BT, ME, P |
| ∙ Characterize and compare genomes of closely related strains from different environmental niches | BT, ME, P |
| ∙ Pathogenic vs commensal isolates within the same species | ME, P |
| ∙ Characterization of new pathogen genotype(s) and/or novel strains | P |
BT, bacterial typing; ME, molecular epidemiology; P, pathogenomics.
The data describing each sample are known as the “metadata” and are crucial to the extraction of meaningful biological interpretation from the analysis results. Minimal information such as the source, location, and collection date should be supplied for each sample to ensure that results can be correctly inferred for the surveyed population. A metadata assessment metric, the Metadata Coverage Index (MCI), has been suggested as a standardized metric for quantifying database annotation richness. In the future, the MCI might be used to ascertain the richness of metadata coverage for genomics standard compliance, quality filtering, and reporting (4), and yet it remains unlikely that manual curation of metadata can be eliminated. Researchers persevere globally to establish performance specifications and to fit HTS within existing communities of practice (regulatory or professional standards) (5, 6). Hence, in this changing context, HTS processing and quality guidelines will remain a “space to watch” for the foreseeable future.
One of the first and most important steps in any scientific investigation is the generation of a hypothesis. Although the large data sets generated by genomics technologies do permit data-driven research, these studies are typically designed to help sharpen a broad hypothesis, not to resolve it (7). Once the project has a defined question or hypothesis, the outcome goals and deliverables can be established. These desired goals will guide the course that the analysis workflow should follow. There are likely multiple paths that analysis can pursue; thus, establishing a clear objective and defined endpoint early will help ensure that the project is successfully completed in a timely fashion and that resources will be applied most efficiently. Depending on the purpose and nature of the study, deliverables may include publications, presentations, regulatory/response action, or policy changes. Knowledge translation in the form of sharing data publicly should be recognized with high priority to enhance global data repository resources and analysis tool development.
Computational infrastructure resources.
The large amount of data generated by HTS and the processing required to perform comparative genomics require a substantial computing infrastructure and sophisticated software. Before undertaking an HTS project, careful consideration should be given to the computing requirements and qualified experts (i.e., computational biologists and bioinformaticians) necessary to complete the data analysis. For example, a single Illumina MiSeq run can produce up to 15 gigabases and many contemporary, large-scale projects require multiple MiSeq runs or the use of larger-capacity platforms. Consequently, analysis of such output data sets can take a significant amount of time and resources. Although some of the most rudimentary analyses for a single genome can be achieved on modern desktop computers with the proper software and configuration, generating accurate and timely results for hundreds of simultaneously analyzed genomes requires considerably more computational “muscle.” A standard desktop computer may have only 8 gigabytes (GB) of memory, 4 processing cores, and 1 terabyte (TB) of storage space, whereas high-end machines found in large data centers likely have hundreds of gigabytes of memory, as many as 64 processing cores per machine, and access to hundreds or thousands of terabytes of storage. These high-end machines can be linked together to construct high-performance computing clusters capable of simultaneously analyzing hundreds or thousands of genomes. Computing on this scale typically has its own administrator and requires housing within a data center with redundant, uninterruptable power supplies and industrial-scale cooling systems. Although such large-scale computing clusters may not be required for small or even some medium-size HTS projects, the HTS project planning stage should include advance estimates for the computational resources required. If the requisite computing infrastructure is not available locally or as a shared resource within an institution, a popular alternative is commercial cloud computing services, in which large-scale computational resources are provided on demand for a fee.
(i) Estimating computational resources.
Estimating computational resources should include attention to items such as physical memory of the machine (random access memory [RAM]) and processing power (central processing unit [CPU] cores and speed), as well as network bandwidth for large data transfers (e.g., transfer of data from the HTS instrument to its interim data storage location or final archive).
Computational resource comparisons are often made with respect to secondary processes such as reference mapping (the alignment/mapping of HTS sequence reads to a reference genome) or de novo assembly (the process of combining sequence reads to reconstruct the original genome without the guidance of a reference). Both processes are fully described in the Secondary Analysis section. While resource requirements can vary between tasks and software chosen, they are often on the order of several gigabytes of memory and several hours per genome (8). For small numbers of genomes, secondary analyses can be performed sequentially on a single workstation or even a high-end laptop; however, large projects with high sample numbers multiply these resource requirements. Thus, adjustments may require different software and/or upgraded computers depending on the software's computational time requirements, the number of genomes to be analyzed, and the project deadlines.
Network bandwidth is another important consideration, particularly if cloud computing or offsite computing resources are used, or if additional data are required from external resources such as NCBI's Sequence Read Archive (SRA) (9). Hence, the time to transfer these data should be taken into account before initiating an HTS project. Gigabit networking cards are affordable, and comparable Internet speeds are increasingly becoming available from most service providers, which are adequate for timely transfers of HTS data.
(ii) Data storage requirements.
Storage requirements for an HTS project include both storage of the initial sequence reads and the necessary space for performing data analysis. Although storage is relatively inexpensive, with most standard hard drives capable of storing 1 TB or more, the inherent large file sizes of raw sequence data, as well as the incorporation of publicly available sequence read data for many analysis pipelines, can take up significant storage space. Common file formats used to store sequence reads include FASTQ (10), BAM (11), and the SRA (12) format. These formats store both the individual bases for each sequence read (ATCG) and a Phred quality score encoding the probability of an error in the base (13), often in a compressed form. An estimate of the sequence read storage requirements for a single Escherichia coli genome stored in FASTQ format may be on the order of several hundred megabytes. Hence, permanent raw data storage requirements must be scaled accordingly for larger numbers of bacterial genomes.
Estimation of the storage requirements for data analysis is even more challenging owing to the numerous analytical possibilities and the large temporary interim files generated. These analysis steps often produce multiple redundant copies of the compressed reads along with large internal temporary files, expanding the initial storage requirements by severalfold. Although many of these large temporary files can eventually be deleted, maintaining these copies over the course of an investigation may be desirable for troubleshooting and validation of the results. When considering the tens, hundreds, or thousands of genomes to be processed in parallel, for example, when generating large-scale whole-genome phylogenies, one quickly realizes the impact of HTS data volume on data storage and on the computing and qualified personnel required to manage it.
(iii) Cloud-based computing.
Cloud-based analysis environments, where computational resources are provided by large-scale commercial data centers, have become increasingly commonplace and can provide high-performance computing on demand. Cloud computing can be divided into three different service models: Infrastructure as a Service (IaaS), which provides physical computing resources (e.g., 40 CPU cores and 160 GB of memory) and complete control over the operating system and software installed; Platform as a Service (PaaS), which provides a preinstalled operating system and suite of standard software; and Software as a Service (SaaS), which provides access to specific software applications through a common interface such as a Web browser. Amazon Web Services (Amazon.com Inc., Seattle, WA, USA), Google Cloud Platform (Google, Mountain View, CA, USA), and Microsoft Azure (Microsoft, Redmond, WA, USA), shown in Table 3, provide a mixture of IaaS and PaaS cloud services and have been used for large-scale bioinformatics analysis (14–16). However, the setup and configuration of an HTS cloud-based analysis environment may still require considerable time and expertise. SaaS providers, such as Illumina's BaseSpace (San Diego, CA, USA), impart value by removing the required setup and maintenance of HTS computing environments. For those lacking resources or time for a local HTS data analysis environment, SaaS may be the preferred option so long as requisite analysis software is available to achieve project goals.
TABLE 3.
List of bioinformatics analysis resources
| Type | Name | Cost | Comments |
|---|---|---|---|
| Cloud services (IaaS/PaaS) | Amazon Web Services (Amazon.com Inc., Seattle, WA, USA) | Commercial | Commercial cloud environments providing resources to construct customized high-performance computing environments; acts as a base from which additional software (e.g., Galaxy) can be utilized |
| Microsoft Azure (Microsoft, Redmond, WA, USA) | Commercial | ||
| Google Cloud Platform (Google, Mountain View, CA, USA) | Commercial | ||
| Cloud services (SaaS) | Illumina BaseSpace (San Diego, CA, USA) | Commercial | Commercial cloud-based bioinformatics analysis environments associated with different sequencing instruments; provides analysis tools and data management fine-tuned for each sequencing instrument; often integrates free and open-source bioinformatics tools described in this review (e.g., FastQC for quality control of sequence reads) |
| Thermo Fisher Cloud (South San Francisco, CA, USA) | Commercial | ||
| Metrichor (Oxford, UK) | Commercial | ||
| DNAnexus (Mountain View, CA, USA) | Commercial | Cloud-based bioinformatics environment not specifically tied to any sequencing platform | |
| Web services | Galaxy (24) | Free | Free and open-source bioinformatics analysis environment available at https://galaxyproject.org/; private instances can be installed on local hardware or within a cloud-based environment; |
| RAST (29) | Free | Web service focused on genome annotation; available at http://rast.nmpdr.org/ | |
| Center for Genomic Epidemiology (30–32) | Free | Available at http://www.genomicepidemiology.org/; provides access to free tools related to genomic epidemiology (e.g., genome sequence typing or construction of phylogenetic trees) | |
| Desktop based | CLC Genomics Workbench (CLC Bio, Aarhus, Denmark) | Commercial | Commercial desktop-based bioinformatics environments; may also provide support for integration with high-performance computing environments; often integrates existing free and open-source bioinformatics tools (e.g., Velvet for de novo assembly) |
| BioNumerics (Applied Maths, Sint-Martens-Latem, Belgium) | Commercial | ||
| Ridom SeqSphere+ (Ridom GmbH, Münster, Germany) | Commercial |
For any cloud-based solution, data privacy and security become a consideration. In the United States, the Health Insurance Portability and Accountability Act (HIPAA) defines a set of standards for the protection and security of electronic health information (http://www.hhs.gov/hipaa/). In particular, the HIPAA Privacy Rule establishes standards for the use of “protected health information” (i.e., individually identifiable information) managed by “covered entities” (i.e., health care providers) (17). The promising use of cloud computing services within health services has generated an interest in developing HIPAA-compliant or other privacy-compliant systems in a cloud environment, often requiring the use of technical solutions such as well-defined access controls, data encryption, and auditing (18). Most cloud providers will advertise their privacy and security policies, and interested readers are encouraged to review these policies for additional information.
While HIPAA is concerned with the protection of personally identifiable information, such as clinical records, there are fewer restrictions on the use or disclosure of deidentified health information (17). DNA has previously been excluded from being regarded as personally identifiable (17), although this is increasingly being called into question for human-derived data wherein there may be a risk of deducing identifiable information in certain circumstances (19). As reported by “Pathogen Genomics into Practice” from the PHG Foundation (20), such risk (of being personally identifiable) is much lower for microbial HTS data generated from isolated microbial cultures unless there is (unlikely) contaminating human DNA carryover. HTS sequences from uncultured samples sourced from humans (i.e., metagenomics) are perceived as a higher risk owing to the presence of human genomic information. Such human-derived data can and should be removed in the primary data processing stage. Metadata associated with clinical samples (e.g., description of isolate source) have the highest risk, as they often include personally identifiable information.
Thus, for the use of cloud computing services, or more broadly for sharing data into the public archives, a clear definition of what constitutes personally identifiable information should be preestablished. For privacy compliance purposes, only deidentified information (e.g., HTS data) should be shared with and stored within cloud services where possible, with more sensitive information kept separate. The GenomeTrakr network in the United States mitigates this challenge by segregating the HTS data away from the sensitive metadata. In that system, only HTS data from microbial cultures and a minimal set of metadata are deposited in public archives to facilitate efficient monitoring of foodborne pathogens nationally and globally, while the more sensitive information is kept confidential (21). However, even in the absence of storing identifiable information within cloud services, plans should be made in advance related to data control, security, and accountability in the event of a cloud service failure (22).
Software and workflow management.
Analysis of HTS data requires the execution of a large collection of software through a series of stages, called workflows or pipelines, before the final result is produced and interpretation can begin. The individual software components at each analysis stage are made available through a variety of sources such as free and open-source downloadable packages, Web services, or commercial software. Organizing these software components into a data analysis workflow can be challenging. For example, software outputs are often needed as input to the next step (but may not conform); thus, these workflows need appropriate transformation and management. Software to assist in this process has been developed, spanning a spectrum from generic scientific workflow managers to extremely customized data analysis pipelines. As part of the HTS planning stage, examination of these software solutions should be performed, keeping in mind the desired results of the project and the cost, including expertise for the setup and maintenance of any software selected. A number of available software options are described below and also shown in Table 3. Readers are encouraged to refer to additional reviews (23) or the software-specific citations for further details.
Galaxy (24) is a popular Web-based bioinformatics data and workflow management platform. Galaxy provides a large collection of data analysis and statistical software as well as data manipulation tools that can be executed through a standard Web browser. Software and tools required for the analysis can be linked together within Galaxy to create automated workflows. The customized workflow can subsequently be configured to run multiple data sets in parallel using high-performance computing environments. Many workflows are publicly available, making it easier for novice Galaxy users to run standard analysis pipelines without having to create complex workflows themselves. Tools, software, and workflows are continually being added by a large community of bioinformaticians and software developers through the Galaxy ToolShed (25). Galaxy is publicly available online (https://usegalaxy.org/) but may not provide the required storage and rapid processing time for large-scale data analysis or offer requisite data privacy. Galaxy is free, open-source software allowing anyone to download and install it on a local computing environment, be it a desktop/laptop or high-performance computing cluster. Unfortunately, the setup and maintenance of such an environment require considerable expertise well beyond the skill set and interest of most nonbioinformaticians. CloudMan (26) provides a method to alleviate some of the setup and maintenance difficulties by simplifying the process of deploying Galaxy within a cloud environment and has been used successfully, for example, by the University of Melbourne researchers to develop the Genomics Virtual Laboratory (27). However, the varying quality of documentation and support for individual tools may leave Galaxy less suited for clinical applications. Commercially supported Galaxy environments, such as Globus Genomics (28) and the BioTeam Galaxy Appliance (BioTeam Inc., Middleton, MA, USA), are available and attempt to address some of these shortcomings for a cost.
Alternatively, many cloud-based SaaS platforms have been developed with fine-tuned pipelines for HTS data analysis. This software requires no installation or local computational infrastructure and is commonly used through a standard Web browser. Illumina provides BaseSpace, while Thermo Fisher (South San Francisco, CA, USA) provides Thermo Fisher Cloud. Pacific Biosciences (PacBio; Menlo Park, CA, USA) provides its single molecular real-time (SMRT) analysis software in the form of a downloadable virtual machine image that can be executed locally or in cloud-based environments and provides additional analysis support through partner companies. Oxford Nanopore (Oxford, United Kingdom) provides cloud-based analysis for its nanopore sequencers such as the portable MinION and high-throughput PromethION via the company Metrichor. In addition to sequencer-specific cloud-based analysis platforms, companies such as DNAnexus (Mountain View, CA, USA) can provide alternative options.
For many SaaS platforms, HTS data can be directly uploaded to the cloud either via a Web interface or directly from compatible sequencing platforms. Once uploaded, a variety of software applications can be executed on these data for tasks such as de novo assembly or variant identification. These software applications may be linked together to form complex scientific workflows. Unfortunately, not all data analysis types (such as constructing whole-genome phylogenies) or pipeline operating procedures are supported. Alternative, user-supplied solutions may be required. Additionally, many of these solutions are provided only commercially and associated costs may be prohibitive.
As an alternative to Web-based cloud software, a variety of commercial desktop applications have been developed. Unlike cloud-based software, desktop applications are installed on a specific local machine and interaction is via a (point-and-click) graphical user interface (GUI). Data analysis can be performed locally, or data can be submitted to a preconfigured high-performance computational cluster for more complex analysis procedures. The list of desktop-based bioinformatics software for analysis of HTS data is large and growing; however, some popular options include CLC Genomics Workbench (CLC Bio, Aarhus, Denmark), BioNumerics (Applied Maths, Sint-Martens-Latem, Belgium), and Ridom SeqSphere+ (Ridom GmbH, Münster, Germany). Built-in analysis modules are provided by each application for standard analysis types, such as de novo assembly; however, more advanced analysis modules may be available. In particular, BioNumerics and Ridom SeqSphere+ have both been developing whole-genome and core genome multilocus sequence typing (MLST) modules (as wgMLST and cgMLST, respectively), thereby enabling rapid phylogenomic comparisons of many genomes. However, the associated cost of some of these applications may be prohibitive for smaller-scale HTS projects or some investigators.
Another set of software includes the variety of free bioinformatics Web services. These are operated using a standard Web browser with data analysis performed on remote computing infrastructure. However, unlike generic SaaS providers, these services are often focused on a particular analysis type, such as the RAST server (29) for genome annotation, and provide minimal data management capabilities. The Center for Genomic Epidemiology provides a large collection of free Web services for analysis types such as in silico MLST typing (30), identification of antimicrobial resistance genes (31), and construction of whole-genome phylogenies (32). These services can provide a rapid method for data analysis; however, minimal control is provided over the operating procedures of each pipeline, caps may be implemented on the amount of data that can be uploaded, and no guarantee is provided as to when results will be completed. Data are generally processed on demand, but there is limited retention of the analysis or the results other than for a short duration.
(i) Data analysis reproducibility.
Reproducibility of analysis results is an important aspect of scientific research (33); however, reproducibility in the data sciences can be challenging owing to the use of complex analysis workflows and incomplete recording of details and software necessary to replicate a study (34, 35). The use of HTS data for infectious disease analysis is a growing field, with a large collection of data analysis software and pipelines actively under development. Use of the previously mentioned workflow managers and analysis software is useful, but there exists no single software package that can handle all data types and all analyses of interest to the typical research laboratory (although there is effort being made in this area; see, for example, http://irida.ca). Thus, it is common to analyze HTS data using a variety of different software from multiple sources, either desktop based or Web based, commercial or open source, before a final result can be generated (23). Data transformations between software are also common, often requiring custom-written scripts. Reference databases used in many types of analysis (e.g., genome annotation) are often changing over time, and software is continually being revised. This complexity leads to difficulties in repeating analyses as well as potential for introducing and propagating errors through to the final result. Differences in the choices of bioinformatics software, databases, and analysis strategies for the same data sets have been shown to lead to differences in the final results and potential misinterpretations (36, 37). At minimum, a thorough record of all software (versions), databases, data transformations, and software operating parameters used to generate the final results is necessary for identification of errors and to assess analysis reproducibility.
Laboratory resources: choosing an HTS platform.
The term “laboratory” in this review refers to the wet laboratory component of HTS projects, which is the preanalytical steps, including the sample processing and data generation. Sample processing includes the thawing of archived strains and/or isolation of the bacteria through culturing and DNA extraction, of which the majority of molecular biology or microbiology laboratories are well equipped to execute. Some laboratories may already have sequencers or ready access to HTS platforms, whereas other projects may need to incur the cost of purchasing such equipment or sending the samples to a third-party sequencing service center.
The HTS field is fluid with regular updates and technology developments; thus, we present general terminology and considerations for those embarking on HTS projects and refer readers to several excellent reviews on the currently available HTS platforms (38–41). Additionally, the “NGS Field Guide” (first published in 2011 [42]) is now updated online, providing a comprehensive comparison of HTS platforms (http://www.molecularecologist.com/next-gen-fieldguide-2016/). Beyond the restraints of cost and accessibility, selecting the optimal HTS platform(s) to meet the project outcome goals should take additional key features into consideration: (i) read length, (ii) read type, (iii) error types and rates, and (iv) coverage and run output. It should be noted that these features are not necessarily exclusive or fixed; modifications can be made to improve affordability and to meet the project needs within the constraints of one platform or by combining technologies.
(i) Read length.
Read length is a general but distinguishing feature of the currently available platforms, with short-read sequencers producing reads between 75 and 1,000 bp and long-read sequencers producing reads from 1,000 to >30,000 bp; however, by the time that this review is published, these numbers may have changed. The most common short-read HTS platforms include the HiSeq, NextSeq, and MiSeq platforms from Illumina and the Ion PGM and S5 platforms from Thermo Fisher (South San Francisco, CA, USA). The longer-read, single-molecule sequencing technologies are the Sequel and RSII systems from Pacific Biosciences and the MinION, PromethION, and SmidgION by Oxford Nanopore Technologies. The outcome goals and biology of the microbes being sequenced will dictate the read lengths required to provide accurate data to traverse repetitive DNA elements and unambiguously resolve the order and orientation of genomic sequences flanked by such repetitive elements. If only short-read sequencers are available, modifying the read type (see below) may be one avenue to traverse low-complexity regions.
(ii) Read type.
Once the sequencing platform is chosen, there are additional options for how the template libraries are prepared and/or how the instrument is run to optimize the data toward the project goals. With respect to the ubiquitous Illumina technology (as a short-read sequencer example), libraries can be prepared and indexed as single-end (SE) reads, paired-end (PE) reads, or mate-pairs (MP). The choice of library will impact how one elects to fragment or shear the DNA. The sequencing kits have a “cycle” number, which is the number of times that the instrument will add a nucleotide to the DNA fragment copy. For example, a “600-cycle kit” could theoretically synthesize up to 600-bp-length sequences in massively paralleled clusters. If a single-read library is chosen, the user would set the instrument parameters to sequence a 600-bp fragment in one direction only. A PE library would reduce the individual read lengths achievable with the same sequencing kit but would read the same template library fragment from both directions (similar in concept to the forward- and reverse-strand sequencing reads on the Sanger platform). Therefore, if the DNA fragments are 1,000 bp in length (insert size), a PE run could generate 300-bp reads from either end of the 1,000-bp library fragment, leaving an intervening gap (inner distance) of 400 bp that remains unsequenced. The known inner distance between the PE reads can be applied algorithmically to traverse repeat regions larger than the single-read length alone. MP libraries are also known as “long-insert paired-end,” as procedural differences in the library preparation utilize much longer DNA fragments and leave a greater inner distance between the two PE sequences, enabling one to effectively traverse larger repetitive regions in the genome.
Knowing some genome biology for the microbes being sequenced can aid greatly in the selection and design of the HTS library. For example, monomorphic organisms such as Bacillus anthracis or Mycobacterium tuberculosis containing small-scale variations may be suitably sequenced using short single reads. Organisms with highly promiscuous genomes and those with multiple internally repeated sequences (e.g., ribosomal operons and insertion sequence [IS] elements) and foreign acquired DNA (e.g., prophage and genomic islands) may require multiple data types—PE, MP, and/or long-read sequence data—from a complementary platform in order to suitably assemble the genome.
The long-read sequencing technologies are evolving quickly as fast, accurate data with the ability to traverse repetitive or low-complexity genomic regions are in high demand (43, 44). Several library approaches may be applied for Pacific Biosciences single molecular real-time (SMRT) technology and be used to produce continuous long reads (CLR; 1,000 to 25,000 bp) and shorter, more accurate circular consensus reads (CCS; 500 to 1,000 bp). An optimal approach is to combine the longer but more error-prone reads with the shorter but more accurate, higher-coverage data from the same platform (45) or another platform. In another useful development, the Oxford Nanopore system offers longer reads with new real-time flexibility options such as resequencing regions for higher coverage or stopping the sequencer in midrun to focus on specific microorganisms in a metagenomic sample (46). Illumina, meanwhile, also offers a library preparation kit to produce synthetic long reads (Moleculo) that have been shown to improve resolution of low-complexity genomic regions (47).
(iii) Error types and rates.
Error types and rates vary between platform technologies, with the short-read technologies such as Illumina having lower error rates, more comparable to those of traditional Sanger sequencing at ∼2%. Despite this low overall error rate, Illumina sequences are prone to single nucleotide substitutions (48–50). Substitution errors can usually be overcome with sufficient coverage depth (essentially sequencing redundancy at each base) (51) and an adequate number of replicates to identify true variants between genomes (52). In contrast, ion-measuring sequencers remain prone to insertions/deletions (indels) owing to base calling errors in homonucleotide regions. The ion-based sequencers also have lower error rates (∼4%) relative to the long reads produced by platforms such as Pacific Biosciences and Oxford Nanopore, which are more prone to deletions and can have a higher frequency of deletion errors (∼18%). However, as mentioned above, options within the long-read platforms have been developed to improve their consensus base call accuracy. In all cases, the key to overcoming most platform error types remains related to ensuring that one acquires sufficient depth of read coverage.
(iv) Coverage.
Based on the experimental design and outcome goals, the depth of coverage and quality of the resultant assembled genome(s) should be a major focus when choosing a sequencing platform. The term “coverage” is often used interchangeably with “depth” or “sequence redundancy” and refers to the number of times that a base is represented in the raw sequencing data (51). The sequences produced by the instrument are not equally distributed across the genome, and thus, the term coverage is often reported as the average coverage (e.g., 10× coverage) and is used to plan in advance the number of samples placed simultaneously on a sequencing run. The theoretical average coverage (C) can be calculated with the Lander-Waterman equation as C = LN/G, where L is the length of the read, N is the number of reads, and G is the length of the genome in base pairs (53). Knowing that the reads will not be evenly distributed across the entire genome, it may be wise to overestimate the coverage required for each sample so that lower-coverage regions are sufficient for downstream analyses such as variant calling (i.e., if a minimum 50× coverage is deemed required for confident variant calling, calculate the expected coverage for each sample to be 75 to 90× to ensure that all regions meet the minimum coverage requirement). All HTS platform vendors have resources to provide the theoretical run output information needed to calculate the number of genomes that can be combined on a run (i.e., multiplexed) once the desired coverage has been stipulated. Note that owing to wet lab inefficiencies and operational complexity, it is not always be possible to achieve theoretical run outputs per vendor specification, and so conservative estimates are recommended at the stage of configuring runs.
SAMPLE PROCESSING AND DATA GENERATION
DNA Extraction and Template Assessment
Steps to avoid contamination, ongoing programs of staff competency training, and proactive method improvement procedures are considered good standards of practice. Similar to Sanger sequencing, the input template is often the cause of HTS failure. Poorly prepared samples rarely make good libraries for HTS. Quality monitoring and control in the wet lab workflow begin with quantification and assessment of extracted nucleic acid template quality (yield, purity, and integrity [size]).
Accurate quantitation is critical to successful HTS. Most library preparation protocols are very sensitive to DNA input concentration (libraries may generate poor yields or smaller fragment sizes); therefore, it is important to achieve accurate template quantification. One should measure template concentrations via two methods of quantitation, such as absorbance (e.g., spectrophotometer or NanoDrop) and fluorescence (e.g., Qubit) systems. Fluorescence approaches (e.g., Picogreen) are more precise than UV absorbance-based methods; hence, templates quantified with fluorescence will yield more accurate measures of template concentration. However, if concentration measurements from the two approaches are grossly different, the sample is likely contaminated and will need to be cleaned up.
HTS is exceptionally more sensitive than Sanger sequencing to contaminants carried over in the templates. Impurities are problematic as they negatively impact many enzymatic stages during HTS library preparation; hence, all templates should be assessed for the presence of excess proteins, organics, and/or other enzyme inhibitors such as bile salts or carbohydrates (e.g., bacterial capsular slime), a problem which demonstrates the benefit of employing absorbance measurements. Template purity is assessed by calculating absorbance ratios, namely, A260/A280 (the ratio of the absorbance at 260 nm divided by the reading at 280 nm) and A260/A230; lower ratios indicate that more contaminants are present. Low A260/A280 ratios (below 1.8) suggest the presence of contaminating protein, phenol, or surfactant micelles; nucleic acids that are not fully resuspended can scatter light, also resulting in low A260/A280 ratios. Elevated absorption at 230 nm is caused by contamination with particulates (e.g., silica particles), precipitates such as carryover of chaotropic salt crystals (i.e., guanidine thiocyanate, LiCl, or NaI), phenolate ions, solvents, and other organic compounds, which also may cause abnormal A260/A280 ratios. Although A260/A280 ratios lack sensitivity for protein contamination in nucleic acids, a DNA sample is considered sufficiently pure when an A230/A260/A280 ratio of at least 1:1.8:1 is achieved (54). Elevated A260/A280 ratios (higher than 2.1) usually indicate the presence of RNA; this can be tested by running the sample (∼1 μg) on an agarose gel. Protein or phenol contamination is indicated by A230/A260 ratios greater than 0.5. Additional RNase treatment after nucleic acid template extraction and postextraction cleanup of templates may be required. Lastly, although isolation of virtually intact high-molecular-weight genomic DNA (gDNA) is not essential for short-read HTS technologies (such as Illumina), it is crucial for longer-read platforms (e.g., PacBio and Oxford Nanopore). Hence, template integrity (size of extracted gDNA) should be qualitatively assessed by performing electrophoresis in an agarose gel or similar device (e.g., Agilent Tapestation device or equivalent) before proceeding to HTS library generation. Although templates will appear as smears, the predominant DNA species should be located very high in the gel or digital image (appearing close to the loading well), which is indicative of high-molecular-weight (intact) template.
HTS Library Preparation and Sequencing
As discussed in the Preparation section, the outcome goals will determine the sequencing data needed (i.e., read length, read type [SE, MP, PE], and average coverage) and the chosen HTS platform(s) will dictate the options available for library preparation to generate said data. Consequently, all HTS platforms as well as commercial library preparation kit vendors provide protocols, with appropriate procedural stopping points as opportunities for library quality monitoring and control. There are diverse library preparation methods, each of which comes with its own set of nuances. Detailed commentary will not be made here as such decisions are based not only on the project goals but also on the laboratory equipment available; instead, motivated readers are referred to the appropriate proprietary protocols for their chosen library kit(s) and HTS platform(s). Users are urged to think carefully about these protocols, weighing them against their own experience and training, and consider appropriate stopping points to apply controls and quality checks, even beyond what may be minimally recommended by the manufacturers.
PRIMARY ANALYSIS
For meaningful, confident biological inference and interpretation, all HTS users should implement robust quality assurance (QA) and quality control (QC) procedures, formalized in a quality management system (QMS) for reproducibility. QA specifies the laboratory operational measures taken to produce data of documented accuracy, whereas QC procedures are applied to demonstrate that the process is robust. For example, QC processes are designed to immediately detect errors caused, for example, by HTS (the test system) failure, adverse environmental conditions, or operator error. In HTS, QA procedures are implemented for determining the quality of laboratory data (measured against internal and external quality control measures), as in proficiency panel comparisons or training, and for monitoring the accuracy and precision of the method's performance over time. Although quality best practices for microbial genomics/forensics deploying HTS have lagged behind the clinical genetics field (5, 55), significant global efforts such as the Global Coalition for Regulatory Science Research (GCRSR) (56), the OIE Ad Hoc Working Group on High Throughput Sequencing and Bioinformatics and Computational Biology (HTS-BCG; Massimo Palmorini, personal communication), and the Global Microbial Identifier (GMI) (57) are under way, aiming to formalize such standards and quality metrics for infectious disease surveillance, food regulatory activities, and clinical diagnostics (58).
This section describes general quality practices for wet lab workflows and data generation for HTS. Quality practices for the analyses of resultant data are described in subsequent sections of the review.
The computational analysis of HTS sequence data can be conceptualized in primary, secondary, and tertiary stages. Primary HTS data analysis may be performed on-instrument (i.e., the HTS sequencer) or directly after the data have been generated. On-instrument primary analysis output includes reports and visualizations of HTS run metrics that are proprietary to each HTS platform. These primary data analysis outputs summarize run characteristics for monitoring platform performance and assessing HTS data quality; some are provided even before all data are collected (i.e., cluster density for Illumina). At minimum, metrics for a completed run should meet performance specifications established by the HTS platform manufacturer. Ideally, any HTS run should yield close to the instrument's expected specification for the numbers of raw (unprocessed) output reads and for on-instrument quality-filtered reads (i.e., percent Q score of >30). Additional run performance metrics may include density or number of read-generating templates, G+C content template bias, or first base read success.
HTS runs should be assessed not only to ascertain whether the sequencer performed and collected sound data but also to assess whether project requirements/expectations of the data will be met by the data generated (59). Assessment of the HTS read quality with respect to base call quality scores is but one important consideration. So, too, is the read signal intensity plotted over the read length: an expected decline over cumulative bases is observed for most HTS platforms, affecting the accuracy of individual base calls. Thus, base calling error rates are typically dependent on the length of read and where (within the read) the base error rate is measured. Regardless of the HTS application or platform, representative additional metrics that should be evaluated include depth of coverage, uniformity of coverage, and whether multiplexed libraries were well balanced or if particular genomic regions or sequences are under- or overrepresented.
All HTS platforms will provide said metrics as described above; however, third-party software also has been developed to assess raw HTS data before beginning downstream analysis. These tools are important when the sequencing run assessment metrics are unavailable or for applying standardized quality checks across a large and varied set of data. This is of particular importance when incorporating publicly available sequence read data. NCBI's SRA (12) provides a few common quality assessments, such as base-quality charts of the reads, but sequencer-specific quality metrics are missing, and quality standards for data may have been inconsistently applied before the data were deposited.
FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) is a popular open-source software package that can be used for a general overview of the sequence read data quality. FastQC produces a summary report consisting of a series of charts for aspects such as base quality and G+C content of the sequenced reads. The report is evaluated by FastQC and given a grade of “pass,” “warning,” or “failure” based on built-in criteria. Guidance for interpreting such reports is available on the FastQC website.
In addition to quality reports, cleaning of the reads may be performed to generate higher-quality read sets for more stringent downstream analyses. Cleaning of reads is accomplished by removing low-quality reads, masking (replacing low-quality bases with an “N” to represent an “undetermined” base), trimming low-quality ends of reads, and removing adaptors and other sequencing artifacts. Software for cleaning reads includes the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/), Trimmomatic (60), and PEAT (61). The effectiveness of read cleaning methods has been studied, and the methods have been shown by some to have a positive impact on de novo assembly, reference mapping, and variant calling (62, 63). However, not all read cleaning methods are equally effective. While the method of read trimming has been shown by some to aid in variant calling (62), other studies suggest that read trimming can also increase the number of misaligned reads, leading to an increase in the number of false-positive variants called (64, 65). This is supported by others (63), who recommend masking as a more effective read cleaning method due to the removal of low-quality base calls while still maximizing the information retained within each read.
Another quality control step for sequencing reads is the detection and possible removal of contaminated DNA sequences. Adaptor sequences, ligated onto the ends of DNA fragments during library preparation, can sometimes be included in the read data set if the DNA fragments are smaller than the sequencing read length capabilities. These can be detected and removed by software such as FastQC or Cutadapt (66). Other sources of contamination include the control plasmid (e.g., phiX used in Illumina) (67) or a potentially contaminated/mixed sample. Programs designed to identify and, for some, filter contaminants include Kraken, Deconseq, MGA, QC-Chain, and Genome Peek (68–72). Ensuring that the input data are high quality and cleaned of potential contamination will increase the quality of downstream analysis results.
SECONDARY ANALYSIS
Reference Mapping and Variant Calling
One of the most common types of analysis performed on newly generated HTS data is to compare them with and identify the variations observed between other, similar genomes. This type of analysis is carried out by reference mapping followed by variant calling. Reference mapping is the process of determining the optimal placement of reads along a previously assembled, closely related reference genome, and variant calling is the process of detecting variation from the reference genome in the form of single nucleotide variants (SNVs), insertions/deletions (indels), or other types of structural variation. The output of the reference mapping process is a file called a “pileup,” containing the optimal placement of the sequence reads along the chosen reference genome, often stored using sequence alignment/map (SAM) or the binary version (BAM) formatted files (11). The aligned reads are further processed in a subsequent variant calling stage, which examines the pileup and produces a list of identified variants often stored in a variant call format (VCF) or binary version (BCF) file (73, 74). Table 4 provides a sample of popular software used for reference mapping and variant calling, and readers are encouraged to refer to additional reviews (75, 76) for more details.
TABLE 4.
List of reference mapping and variant calling software
| Type | Name (reference[s]) | Comments |
|---|---|---|
| Reference mapping | Bowtie 2 (83) | Available at http://bowtie-bio.sourceforge.net/bowtie2/ |
| BWA-SW (84) | Available at http://bio-bwa.sourceforge.net/ | |
| SMALT | Available at http://www.sanger.ac.uk/science/tools/smalt-0 | |
| Variant calling | GATK (Unified Genotyper) (90) | Part of the GATK toolkit for variant calling of nondiploid organisms |
| FreeBayes | Uses Bayesian inference to detect variants; available at https://github.com/ekg/freebayes | |
| BreakDancer (86) | Detects structural variation from anomalous read pairs produced by mate-pair sequencing; available at http://breakdancer.sourceforge.net/ | |
| MetaSV (245) | Executes and combines results from many different structural variation detection software; available at http://bioinform.github.io/metasv/ | |
| Variant annotation | SnpEff (88) | Available at http://snpeff.sourceforge.net/ |
| TRAMS (89) | Available at https://sourceforge.net/projects/strathtrams/files/Latest/ or as integrated within Galaxy; free for academic use, requires written consent from authors for commercial use | |
| Toolkits | GATK (77, 90) | All-in-one toolkit for reference mapping and variant analysis; free for academic use, nonfree for commercial use; available at https://www.broadinstitute.org/gatk/ |
| SAMtools (11, 73) | Toolkit for working with sequence alignments in SAM/BAM formats; can also be used for variant calling but assumes a diploid model; available at http://www.htslib.org/ | |
| BCFtools (11, 73) | Toolkit for working with variants in VCF/BCF formats; available at http://www.htslib.org/ | |
| VCFtools (74) | Toolkit for working with variants in VCF format; available at https://vcftools.github.io/ | |
| Picard | Toolkit for working with sequence alignments and variants in SAM/BAM or VCF formats; available at http://broadinstitute.github.io/picard/ | |
| Visualization | IGV (85) | Available at https://www.broadinstitute.org/igv/; visualization of multiple tracks of information and multiple genomes |
| Tablet (246) | Available at https://ics.hutton.ac.uk/tablet/; visualization of sequence alignments, variants, or genes | |
| iobio (247) | Web service for upload and visualization of sequence alignments (BAM format), or variants (VCF format); available at http://iobio.io/ |
Guidelines for variant calling, such as the Genome Analysis Toolkit (GATK) best practices (77), have previously been published. However, these guidelines often default to giving instructions for variant calling in human and other eukaryotic data sets and so contain subtle differences that are not suitable for variant calling in microbial genomes. In particular, the assumption of variant calling with diploid organisms is often made, such as is the case with the SAMtools package (73). Variant calling software that assumes a diploid model may produce heterozygous variant calls, which are unexpected for haploid organisms and can be indicative of false positives introduced owing to read misalignment or copy number variation of repetitive regions (78). For the GATK best practices, it is recommended that the Unified Genotyper be applied, as opposed to the Haplotype Caller, when dealing with nondiploid organisms (77).
Reference mapping issues.
There are a number of common issues that can impact the results of reference mapping. One such issue is the presence of repetitive regions in the sequenced genome, the reference genome, or both. A combination of short read lengths for existing HTS technologies (on the order of hundreds of base pairs) and repetitive regions on the reference genome will result in ambiguity in selecting the best location to align matching reads (79). Approaches aimed at mitigating such ambiguity include completely ignoring reads aligning to multiple locations, picking a random location for reads with equal-scoring mapping locations, or reporting all nonunique read alignment locations. Potential caveats of these approaches range from excluding potential variation in the final results to misidentifying variants. Repetitive regions within the sequenced genome that are not present in the reference genome will lead to an unusually large pileup of reads in the repeat region (e.g., a genome sequenced to 50× coverage will show 100× coverage or more in the repetitive region depending on the number of extra copies harbored by the sequenced genome). Treangen and Salzberg describe in more detail the effect of reference mapping in repetitive regions (79).
In addition to repeat regions, structural variation (e.g., deletions or translocations) and additional mobile elements (e.g., plasmids, transposons, and prophage) can be problematic for reference mapping. Structural variation can cause reads to be mapped in an incorrect manner, while mobile elements can be excluded from reference mapping analysis altogether if absent from the reference genome. One approach to capture mobile elements not present in the reference genome is to perform de novo assembly, gene prediction, and annotation of a newly sequenced genome's unmapped reads. The presence of many mobile genes after this analysis will be an indication that a putative mobile element exists in the sequenced genome but is absent in the chosen reference genome. Approaches for structural variation often require alternative analysis strategies and potentially alternative sequencing methods (i.e., long-read sequencing or mate-pair sequencing) (80).
Selecting a reference genome.
Selecting an appropriate reference genome is an important first step to reference mapping and yet can be a nuanced decision. Ideally, the reference genome chosen should have no gaps or errors in the sequence data and should be genetically a very close match to the sequenced genome. NCBI (http://www.ncbi.nlm.nih.gov/) provides access to a large collection of previously published reference genomes that can be used, and yet caution should be exercised since publicly available genomes may be too genetically dissimilar for use as a reference in one's own investigation. Generating a high-quality reference genome in an ad hoc manner is possible, especially with longer-read technologies such as PacBio's SMRT sequencing, which can often produce completely or nearly ungapped genomes. Closely related draft genomes can be used as a reference; however, it should be noted that contiguous consensus sequence (contig) breaks and collapsed repeats in such draft genomes are problematic for mapping HTS reads, and extra consideration, such as manual inspection of the pileup in these regions and possibly masking of these regions, should be conducted before variant selection.
Quality control of input data.
As mentioned in the Primary Analysis section above, inspection of the sequence reads should be done to verify that they pass standard quality checks before proceeding with any secondary analysis. Additionally, assessment of whether or not an appropriate depth of coverage has been achieved for sequencing should be conducted. Low coverage can lead to false-negative variant calls, while excessive coverage is wasteful and can lead to performance issues such as longer running time or higher memory usage (76). A read coverage of at least 50× has been recommended for the best results (37, 81). Sequenced genomes that do not pass these quality checks can be excluded from further analysis or resequenced to generate a better-quality data set. Once a raw data set has been selected, cleaning of the sequence reads (as described in the Primary Analysis section) can be performed to verify that the data are of sufficient quality for downstream use.
Generating a read pileup.
After quality control of input sequence reads, an alignment of the quality-filtered reads is generated to produce a collection of mapped reads (along with their optimal placement on a reference genome), resulting in a read pileup against the reference. A large collection of software has been developed for efficiently aligning HTS reads to a reference genome (76, 82); popular options include Bowtie2 (83) and BWA-SW (84). Standard input files include sequence reads (in FASTQ format) and a reference genome. The output is a read pileup often stored in the SAM (text-based, uncompressed) or BAM (binary, compressed) file format (11).
Quality control of a read alignment pileup.
Following the generation of a sequence read alignment pileup, validation should occur to verify that the pileup is correct. A large collection of bioinformatics toolkits, such as SAMtools (11), have been developed for inspection of read alignment files and generating summary statistics. Additional visual inspection and quality analysis of the pileup can be performed with software such as the Integrative Genomics Viewer (IGV) (85).
One important issue to evaluate is whether a high percentage of unmapped reads exists, which can indicate quality issues with the read data or contamination or could indicate a large number of unique regions in the sequenced genome (i.e., a mobile element). SAMtools along with other tools have the capability to check for the percentage of unmapped reads. High numbers of unmapped reads may also indicate that the reference genome selection was inappropriate; in this case, selection of a new reference genome is advised.
Variant calling, filtering, and annotation.
Variant calling is the process of scanning the SAM/BAM file and searching for areas of significant variation from the reference genome. This is often limited to SNVs, insertions/deletions, and other small regions of variation due to the shorter read length of the sequenced reads. Larger-scale variant detection is possible when using appropriate sequencing techniques, such as mate-pair sequencing with longer insert sizes. Here, each pair of reads is mapped, and anomalies in the distances between pairs of reads or the orientation of pairs of reads are used to detect larger structural variations such as insertions, deletions, or inversions (86). Variant callers typically produce a report of potential variants using the VCF (text-based variant call format) or BCF (smaller, more-efficient binary) file formats (73, 74). Examples of variant calling software are given in Table 4, with additional reviews (75) providing more details.
Variant filtering involves removing variants that do not match defined thresholds to remove false positives from further analysis. Many metrics can be used for filtering variants, such as the depth of coverage or the QUAL field of a VCF file, which provides a Phred-scaled quality score for the listed variant (74). The GATK best practices describe a process known as variant quality score recalibration, which requires a known set of true variant calls used to calibrate the variant quality scores followed by removal of variants with low scores (77). For novel variant discovery in microbial genomes, these known variant calls may not be available, limiting the use of variant quality scores due to unknown thresholds. Instead, the use of other hard-filtering thresholds to remove poor-quality variants can be used, such as a minimum depth of coverage or a minimum proportion of reads supporting a variant call (e.g., minimum of 10 reads and 75% of all reads supporting a variant call) (77, 87).
Once there is adequate evidence that the variants are true, they can be annotated with relation to an annotated reference genome. Variant annotation is the process of placing the variation in the context of the genomic features that contain those variants and their effects on those features such as amino acid changes and frameshifts. Software for variant annotation includes snpEff (88), TRAMS (89), and GATK (90). Each program requires an annotated reference genome as input along with a list of variant calls, in VCF/BCF format, and will produce a list of the effects of these variants. Although the variant calling process can be automated, it is important to note that variants should be manually inspected to ensure that the gene annotations are accurate, and ideally, those inferred to alter metabolic processes or virulence mechanisms would be further confirmed with laboratory experimentation, as described under “Bacterial Pathogenomics.”
De Novo Assembly
De novo assembly is defined as the reconstruction of a genome from sequence reads without the aid of a reference. More technically, de novo assembly is the computational process of reconstructing longer contiguous consensus sequences (contigs) by determining the longest overlap and optimal placement of shorter reads. The result of this initial automated approach is considered a draft genome. If additional information such as optical mapping data, mate-pair, or long-read sequences is available, these contigs can be ordered into larger scaffolds; the resulting assembly is classified as a “high-quality draft genome.” A designation of “closed genome” requires that the gaps between these scaffolds be resolved. A “finished genome” requires the resolution of any misassemblies or other sequencing anomalies and uncertainties. The level of closure or finishing (sequence polishing) pursued for the genomes in a project will depend on requirements of the sequencing project as defined in the project planning phase. Additionally, the sequencing data for each isolate should be of sufficient quantity, quality, and type (e.g., paired-end or single short reads, long reads, or a combination of data types) to generate a de novo assembly that satisfies the project objectives determined in the planning phase.
Choosing de novo assembly software.
As HTS technology evolves, so too does the development of new and/or improved de novo assembly programs. There are detailed reviews of assembly software found elsewhere (48, 91–94); however, we have included a comparative list of some popular assemblers within each of the major assembly algorithms, greedy, overlap-layout-consensus (OLC), and de Bruijn graphs, in Table 5. Assemblers have evolved in roughly this order with early assemblers such as TIGR using the greedy approach during the Human Genome Project (95). The OLC assemblers organize reads into graph structures with each read being a node in the graph connected by an edge to other overlapping reads (48). This paradigm was more commonly used with Sanger data and HTS longer reads as the process is computationally intensive and, in the past, has not performed as well with high volumes of short, high-coverage HTS reads, although advances have been made to improve the performance of OLC-based assemblers (93). For example, the AMOS suite of assembly tools remains a popular choice for OLC-based assembly of HTS data (96). De Bruijn graph assemblers partition the reads into overlapping subsequences of length k, called k-mers, to create the nodes for efficient graph structures, allowing programs to computationally handle larger data sets. Early de Bruijn-based assemblers such as Euler (97) and Velvet (98) popularized the use of these methods for bacterial genomes. Algorithms have since evolved and expanded upon these original paradigms to improve assemblies of long-read data such as HGAP, Edena, and SGA (99–101) and short, high-coverage data such as SOAPdenovo and SPAdes (102–104).
TABLE 5.
List of de novo assembly software
| Type | Name | Read type | Comments |
|---|---|---|---|
| OLC | String Graph Assembler (SGA) | Illumina (>200-bp reads) | Performs best on larger genomes with high coverage; has a built-in error correction module; https://github.com/jts/sga |
| MIRA | Sanger, Ion Torrent, Illumina, PacBio (CCS reads or error-corrected long CLR reads) | Can combine multiple libraries/sequencing technologies into a single, hybrid assembly; slower run times than other assemblers; capable of producing high-quality assemblies; requires higher level of expertise to set run parameters; https://sourceforge.net/p/mira-assembler/wiki/Home/ | |
| Hierarchical Genome Assembly Process (HGAP) | PacBio | Long-read de novo assembler for PacBio SMRT sequencing data; only one long-insert shotgun DNA library required; uses short reads to correct long reads within the same library; de novo assembly using Celera assembler; includes assembly polishing with Quiver; https://github.com/PacificBiosciences/Bioinformatics-Training/wiki/HGAP | |
| De Bruijn graph | Velvet | Illumina, 454, Ion Torrent, Sanger | High memory requirement; easy to run; can map reads onto a reference sequence(s) to help guide the assembly (Columbus module); user must select a single k-mer length to use (VelvetOptimiser can be used to select the optimal k-mer length); https://www.ebi.ac.uk/∼zerbino/velvet/ |
| Velvet-SC | Illumina | Adaptation of Velvet assembler for single-cell sequencing data; no error-correction module; http://bix.ucsd.edu/projects/singlecell/ | |
| SOAPdenovo | Illumina | Has an error correction, scaffolder, and gap-filler module; relatively fast compared to other assemblers; http://soap.genomics.org.cn/soapdenovo.html | |
| Ray | Illumina, 454 | Can combine different technologies to create a hybrid assembly; well documented; http://denovoassembler.sourceforge.net/ | |
| A5-MiSeq | Illumina | Uses the IDBA-UD algorithm; easy to use-little bioinformatics experience required; relatively fast with low memory requirements; https://sourceforge.net/projects/ngopt/ | |
| ALLPATHS | Illumina, PacBio | Requires at least 2 specialized libraries (e.g., Illumina fragment and PacBio long-read or Illumina jump library); has an error correction module; http://www.broadinstitute.org/software/allpaths-lg/blog/ | |
| Assembly by Short Sequences (ABySS) | Illumina, 454, Sanger | http://www.bcgsc.ca/platform/bioinfo/software/abyss | |
| SPAdes | Illumina, Ion Torrent, PacBio, Nanopore | Can support single-cell sequencing input data; can handle nonuniform coverage; has an error correction module (BayesHammer/IonHammer) and scaffolder; uses multiple k-mer lengths; capable of producing high-quality assemblies; relatively fast assembler; most widely used assembler for bacterial genome assembly; http://bioinf.spbau.ru/spades | |
| Maryland Super-Read Celera Assembler (MaSuRCA) | Illumina only or mixture of short and long reads (Sanger, 454) | Attempts to create superreads using the paired-end reads; http://www.genome.umd.edu/masurca.html | |
| OLC/de Bruijn hybrid | CLC Assembly Cell (CLC Bio, Aarhus, Denmark) | Illumina, 454, Ion Torrent | Commercial software with licensing fee; easy to use with point-and-click graphical user interface; contains a scaffolder module; fast |
| Proprietary algorithm | SeqMan NGen (DNAStar Inc., Madison, WI, USA) | Illumina, PacBio, 454, Ion Torrent | Commercial software with licensing fee; easy to use with point-and-click graphical user interface; fully integrated with Lasergene's SeqMan Pro; patented algorithm (black box) |
As this review is aimed at researchers working with bioinformaticians on HTS projects (not bioinformaticians themselves), we want to stress that it is not essential to understand the mathematical theory behind all de novo assemblers. It is, however, important to understand that all assemblers have their strengths and limitations. The performance of an assembler is influenced by the biology of the genome (e.g., repetitive elements, overall size, multiple extrachromosomal plasmids, etc.), the nature of the data (e.g., sequence length, orientation, coverage depth, and uniformity), and the computational resources available (105).
Evaluating de novo assemblies.
Without knowing the true genome structure of an organism, a de novo assembly is a hypothesis formed by short DNA segments compiled into contigs through computed mathematical models. Contiguity and correctness are two attributes of the resultant assembly that can be assessed. There have recently been reports in the literature focused on comparing the performances of assembly workflows (8, 106–109). Common summary statistics for genome assemblies include the total number and lengths of the contigs. An additional popular measure is the N50 statistic. The N50 refers to the median contig length of which 50% of the assembled nucleotides are found to be; this definition extends to the N50 scaffold and the NG50, which incorporates the expected genome size (107). These summary statistics, however, assess only the contiguity of the assembled sequences, not their correctness.
The correctness or accuracy of an assembly can be evaluated by mapping the original reads back onto the assembly to identify regions with unusually high coverage (possibly a repeat collapse) or low coverage (possibly indicating an incorrect join) (94). There are a growing number of programs compiled to aid with assembly evaluations such as Amosvalidate, Quast, and REAPR software, described in more detail elsewhere (110–113). There have also been genome assembly competitions, such as Assemblathon 1 and 2 (106, 107) or GAGE-B (109), where several researchers were tasked with constructing de novo assemblies with the same data. These studies concluded that there was no one assembler that performed best for all organisms and metrics used to evaluate the assembly quality. Therefore, it may be useful to test a few de novo assemblers and evaluate the workflow that meets project goals for assembly quality and can perform efficiently within the available computational resources.
TERTIARY ANALYSIS
Tertiary analysis includes the processes required to “make sense” of the data or interpret the results to gain a broader understanding of genome content (e.g., annotation and mobile genetic element identification) and of how the genomes compare to each other and larger populations (e.g., molecular epidemiology and phylogenomics) and for further characterization of the bacteria, host-pathogen interactions, and bacterial behavior (i.e., pathogenomics). The level of tertiary analysis required is dependent on the project objectives and is not limited to the analytical methods described below, nor are the methods necessarily performed in the order that we have elected to present them.
Bacterial Genome Annotation
Genome annotation is the process of identifying the biologically important features contained in a genome and attaching descriptive information to those features. Genome annotation is typically one of the first steps applied after sequence assembly and can be performed on draft or closed sequences, although the latter is preferred when conducting a detailed comparative analysis of a group of genomes, since features that exist in the actual genome may not be present in the draft assembly (owing to gaps) or may be misassembled, which can result in spurious relationships and invalid conclusions regarding genomic structure and organismal function.
The features typically annotated in bacterial genomes are the protein coding genes, referred to as coding sequences (CDS), and the noncoding genes, such as the rRNA and tRNA. Other biologically important features, such as pseudogenes, operons, clustered regularly interspaced short palindromic repeats (CRISPRs), transposons, integrons, and other genomic features, also fit into this feature annotation category; we do not cover these types of annotations here (mobile element detection is covered below). The focus of this section is on the annotation of entire prokaryotic (bacterial and archaeal) genomes, which tend to range in size (∼700 to ∼10,000 genes), have variable gene content, and have a predictable gene structure and organization that lend themselves well to automated approaches. We provide only a brief overview of the process of annotating bacterial genomes; for an in-depth discussion of microbial genome annotation, we refer the reader to the many existing excellent reviews (114–116).
Genome annotation can be divided into two main tasks: structural annotation and functional annotation. Structural annotation, commonly referred to as gene finding or gene prediction, involves the identification of the location of the protein coding genes (CDS) and the noncoding genes (tRNA and rRNA). The functions of the noncoding genes are self-evident; however, the functions of the protein coding sequences are diverse and not straightforward to determine. These genes must undergo functional annotation to infer their probable biological function. A flowchart outlining the bacterial genome annotation process is provided in Fig. 3.
FIG 3.
Overview of bacterial genome annotation. Structural annotation identifies the location of genes on the contigs of an assembled bacterial genome. Protein-encoding locations are identified, followed by automated assignment of gene function by comparison to existing databases. Non-protein-encoding genes are annotated by identifying key signatures for each type of gene. The resulting annotations are combined with an optional manual curation that can be performed before the final annotated genome is produced.
Bacterial gene prediction programs come in two flavors: ab initio, or intrinsic gene finders, which attempt to identify coding sequences based solely on the information contained within the newly sequenced genome or contig, and extrinsic gene finders, which use a database of previously identified and verified bacterial protein coding sequences to aid in the identification of genes in a newly sequenced genome. Extrinsic gene finders, such as ORPHEUS (117) and CRITICA (118), enjoyed early popularity; however, they have largely been supplanted by ab initio gene finders, which rival extrinsic approaches in their accuracy since they learn from known gene content information already within the target genome and thus can be better tuned to find the remaining genes. In contrast, extrinsic gene finders rely on gene content typically harvested from model organisms such as Escherichia coli, which may be evolutionarily distant to the newly sequenced genome, resulting in reduced prediction accuracy.
The central task for ab initio gene finders is to distinguish coding from noncoding sequences within a set of all open reading frames (ORFs; contiguous stretches of genomic sequence flanked by in-frame stop codons), which may or may not contain coding sequences, within the newly sequenced genome. These gene finders use extensive heuristics combined with sophisticated computational approaches, such as state machines, dynamic programming algorithms, and hidden Markov models (HMMs), to carry out this task. HMMs have become especially popular for this task since they have the ability to “learn” the attributes for genome features, such as coding sequence composition, and distinguish it from sequences that lack the attributes, such as noncoding sequence composition, and can even predict the start and endpoints of a feature within a larger genomic region. GeneMark.hmm (119) and GLIMMER (120) are two well-known, highly accurate ab initio gene predictors. Prodigal is another highly accurate gene finder, although it forgoes HMMs in favor of a combination of dynamic programming and heuristics to distinguish coding sequences from noncoding sequences (121). These programs have predictive accuracies in the range of 95 to 98%, although they can have trouble identifying genes acquired from foreign DNA and in genomes of high G+C content. Each of these programs varies in its approach; for a detailed review, the reader is referred to reference 122.
The noncoding rRNA and tRNA genes can also be structurally annotated using automated approaches. The standard method for predicting tRNA uses tRNAscan-SE (123), which calculates the likelihood of the presence of a tRNA by scoring the presence of the well-defined substructures inherent in each tRNA molecule and boasts an impressively low false-positive rate of just one falsely called tRNA molecule in 3,000 average-size bacterial genomes. Several rRNA gene prediction software programs exist and can identify the 16S, 23S, and 5S genes normally collated in a single operon and existing typically in one to 15 copies in the average bacterial genome. Of these, the most popular is probably Infernal (124), which uses stochastic covariance models to score primary and secondary structure. Also popular is RNAmmer (125), which uses HMMs to identify rRNA genes. For RNAmmer, the ability to identify the presence of these genes is respectable (above 95%), but its accuracy suffers in predicting the exact starts and stops of the genes since, unlike coding sequences and tRNA sequences, there exist no well-defined signals demarking them; hence, due caution should be exercised when analyzing ribosomal genes obtained from automated prediction software.
The set of coding sequences contained in the bacterial genome defines its biology, and so it is of great interest and importance to characterize the functions of these coding sequences. This is especially true in the prediction/assessment of pathogen virulence and risk. Modern genome annotation approaches use a combination of sequence similarity searches, HMM-based searches, and a variety of biochemical property searches to infer the function of genes in newly sequenced genomes.
Similarity searches rely on the assumption that similar gene sequences possess identical functions; it should be recognized that this is generally a good assumption but does not always hold, since even a single base pair change may alter or even abolish the function of the resulting protein product. In this approach, the BLAST family of similarity searching programs is used to determine the degree of similarity between a newly sequenced gene and a database of reference genomes. Annotations from reference genes with sufficient similarity and length are then transitively applied to the newly sequenced gene. The choice of reference databases used to compare sequences is key to the success of the functional annotation process based on similarity search. Highly curated reference sequences with lab-validated functions, such as HAMAP (126), are preferred for maximal accuracy. However, these curated databases lack breadth, and many genes will be missed. Databases containing large coverage, such as the NCBI nr (nonredundant) database and the EMBL Nucleotide Sequence Database (127), are effective at annotating as many genes as possible but are the least reliable, since their annotation and curation are the responsibility of the submitter and lack any validation. Analysis of the nr database has shown that it likely contains a substantial number of noncoding ORFs misannotated as actual coding sequences (128). Transitive annotation from these databases without additional verification can result in the misannotation of genes and propagation of error. Semicurated, high-quality reference sequences, such as those contained in the NCBI microbial RefSeq database (129), which contains a combination of high-quality, manually curated genomes and other uncurated but consistently annotated genomes, provide a good compromise between accuracy and scope. If possible, a stratified annotation process should be used wherein reference sequences are searched, and annotations applied, in series from highest to lowest acceptable accuracy.
An alternate approach to similarity searching using BLAST is to employ profile HMMs that contain information describing the content, length, and variation in groups of related sequences with defined function, called a sequence family. The TIGRFAM collection (130) represents a manually curated, experimentally validated set of profile HMMs that contain validated functional characterization of protein function. Hits to TIGRFAMs can be used to reliably infer the same or highly similar function in target coding sequence. The TIGRFAM collection can typically annotate around 30% of the genes in a typical bacterial genome. FIGfams (131) are similar to TIGRFAMs but are automatically generated and describe sequences from the same protein family. The functional annotations in FIGfams are of high quality and, like TIGRFAMs, can be used to reliably infer functional annotation of a target coding sequence. The Pfam database (132) is a third database of manually curated and automatically generated profile HMMs that can be used to infer function, although this database includes functional information for protein domain families in addition to overall protein families, so care should be taken to ensure that hits to the HMMs in this database are valid for the entire target protein under study and not to a smaller functional domain.
Despite the existence and growth of these reference databases, many newly sequenced genes may have no matching counterpart contained in these databases, and the databases themselves can contain a substantial number of genes without a known function (i.e., hypothetical and conserved hypothetical genes). For these sequences, it may be possible to identify the partial function(s) of the protein products by an examination of their inferred biochemical properties. As mentioned, the Pfam database can be searched to look for matching functional and structural domains. Motif sequences are short, conserved sequences that impart a significant biological function to that protein, such as DNA binding, metal binding, or phosphorylation motifs. Sequence motifs can be searched against the PROSITE database (133) with the ScanProsite tool (134). Transmembrane domains are sections of a protein that span cell membranes. Proteins harboring transmembrane domains can be involved in cell signaling, cell adhesion, catalysis, or transport of substances across the cell membrane. A popular HMM-based transmembrane domain predictor is TMHMM (135). Protein subcellular location (e.g., cytoplasm, cytoplasmic membrane, periplasm, and extracellular space) can be used to assist in a protein's function and can be predicted with PSORTb (136).
Organizing and running this menagerie of genome annotation tools and methods to generate high-quality annotations are beyond the typical researcher's capability; instead, automated genome annotation systems have been developed that compile these tools into coordinated pipelines that remove much of the complexity of performing bacterial genome annotation. Early pipelines such as MAGPIE (137), GenDB (138), BASys (116), MaGe (139), RAST (29, 140), and IMG-ER (141), are available as Web applications and provide a wide variety of annotation services, including manual review and correction, and submission to public archives such as NCBI. More recently, downloadable annotation systems such as DIYA (142) and Prokka (143) have been made available that take advantage of the availability of high-performance workstations. These systems allow additional customization of the pipeline workflows and databases and are rapidly gaining popularity due to their high customizability. The choice of annotation pipeline to apply for an annotation job depends on a number of factors and requires an understanding of their strategies and relative advantages and disadvantages; a detailed review of these and other annotation systems is available in reference 122.
Recombination and Mobile Elements
DNA sometimes harbors features that allow it to rearrange, resulting in a change in the gene content for the organism that contains that DNA. The mechanisms that impart these rearrangements are naturally occurring and found across the spectrum of life. In this review, we restrict the scope of our discussion to the types of rearrangements occurring in prokaryotes, which come in two forms: bacterial genetic recombination and mobile elements.
Recombination.
Genetic recombination refers to the exchange of two segments of DNA contained on the same chromosome or on different chromosomes, resulting in new combinations of genes and other genomic structures. Recombination is a method for bacteria to acquire diversity that may aid in survival, and bacterial organisms often take advantage of recombination for immune system evasion (144) or to acquire antimicrobial resistance (145). Because bacterial recombination is acquired asexually, it does not follow hereditary evolution and, hence, must be taken into account when generating phylogenies and conducting population structure analysis.
Early programs for identifying recombination in bacteria such as ClonalFrame (146) use MLST data but are not scalable to WGS. More recently developed programs such as ClonalFrameML (147), Gubbins (148), and BRAT NextGen (149) are designed to more efficiently analyze HTS data and yet still take considerable time to analyze large data sets and are difficult to install and use. Scalable, easy-to-use systems for recombination detection in bacterial genomes are yet to be realized, but given their desirability, we anticipate the introduction of such tools in the near future.
Mobile genetic elements.
Mobile genetic elements, often simply referred to as “mobile elements,” are segments of DNA with the ability to move around within a genome and between genomes. Among bacteria, mobile elements play a critical role in shuttling genes that confer survivability in a particular ecological niche. This includes virulence factors and antimicrobial resistance genes and, as such, plays a critical role in human health and disease. Here, we describe the main types of mobile elements found in prokaryotes.
(i) Transposons.
Transposons are genetic elements that rearrange their position within a bacterial genome or between two separate genomes. Transposons carry genetic elements that control their own movement. Transposition requires at minimum a transposase enzyme and a pair of flanking sequence elements called terminal inverted repeats. These minimal transposons are referred to as insertion sequence (IS) elements. The ISMapper program (150) can be used to identify IS elements from genome sequence data. Insertion sequences can be classified by their similarity, and currently, over 1,500 insertion sequences grouping into 20 families have been identified. The ISsaga software suite (151) can be used to identify and classify IS elements. If two IS elements exist with an intervening set of genes between them, they can form a composite transposon. Composite transposons can transpose these “cargo” genes, which frequently contain genes encoding virulence or antibiotic resistance, and their flanking IS elements as a combined unit. Conjugative transposons, also referred to as integrative and conjugative elements (ICEs), possess additional genes that facilitate the transfer and integration of the transposon into the genome of a different bacterial cell. Mobile element detection programs such as IslandViewer (152) are available for detecting conjugative transposons along with other genomic islands of probable horizontal origin.
(ii) Plasmids.
Plasmids are smaller, mostly circular, double-stranded extrachromosomal DNA molecules that exist and replicate independently of the main bacterial chromosome. Importantly, they often carry genes that confer a selective advantage in a given environment, such as antibiotic resistance or sanitizer or metal resistance, and virulence genes. Bacteria can harbor multiple distinct plasmids, and each plasmid can exist in multiple copies. Some plasmids known as episomes can reversibly integrate into the chromosome. Many plasmids can transfer themselves from one cell to another via asexual conjugation. As such, plasmids serve as a major mechanism of horizontal gene transfer between bacteria. Given their importance and ubiquity, it is perhaps surprising that not many software tools exist to identify and comprehensively characterize bacterial plasmids. This paucity arises from the difficulty of distinguishing plasmids from chromosomal sequences (and distinguishing plasmids containing similar content) using draft whole-genome data generated with short-read sequencing technologies. Although most plasmids do have identifiable signals present in their sequences, such as the replicon region, there is sufficient variation in this region so as to make their in silico detection difficult; as well, it is possible that plasmid segments without identifiable signals may exist on different contigs in draft genome sequence data. Plasmid sequences can be compared to databases of known plasmids, but the high inherent plasticity found in plasmids gives rise to extensive mosaicism that makes this approach unreliable. The PlasmidFinder website (153) can be used to detect and type plasmids from the Enterobacteriaceae family and can detect multiple plasmids within a bacterial genome, although it cannot fully extract and assemble full plasmid sequences from draft contig data. The use of mate-pair technologies can help to scaffold contigs belonging to plasmids but cannot guarantee a full plasmid sequence. Currently, the most popular approach for completely assembling and characterizing plasmids relies on long-read sequencing technologies such as those generated by the Pacific Biosciences and Oxford Nanopore platforms. Long-read technologies often can yield fully closed plasmid sequences as single contigs, even from genomes containing multiple plasmids (154).
(iii) Prophage.
Bacteriophage, commonly referred to simply as phage, represent viruses that specifically infect bacterial cells. Phage can have a lytic life cycle stage, wherein their genomic material exists and replicates within the bacterial cell but separately from the host bacterial genome, or they can exist in a dormant lysogenic life cycle stage, wherein they become integrated within the chromosomal DNA of the host bacterial cell (or sometimes may be inserted in a plasmid); this lysogenic phage state is referred to as (an integrated) prophage. Prophage often contain genetic features of interest to microbiologists, such as those conferring pathogenicity factors, antimicrobial resistance, or means for altering bacterial cell surface structures that allow invasive bacteria to evade the host immune system. As noted for plasmids, prophage also are a major vector of horizontal gene transfer in prokaryotes. Prophage can be identified by similarity to known phage or by analyzing the G+C content and nucleotide composition differences from the surrounding bacterial chromosomal DNA. Prophage are also known to preferentially insert and thus disrupt tRNA genes. Popular tools for the identification of prophage include ProphageFinder (155) and PHAST (156).
Phylogenetics to Phylogenomics
Comparative analysis of sequences or genomes often includes phylogenetic methods. Phylogenetics is the study of the evolutionary relationships among organisms. Phylogenetics is a very active field of study and plays an important role in many life sciences. The literature on phylogenetics is extensive, and many excellent books and review articles have already been written (157–159). Here, we aim strictly to introduce the main concepts and technologies used in phylogenetics that can be applied for comparison of single genes, a subset of concatenated DNA segments, or even full genomes; the latter is referred to as phylogenomics and will be discussed with respect to infectious disease bacterial genomics applications.
General phylogenetic concepts.
The central concept in phylogenetics is the phylogenetic tree, also known as an evolutionary tree or a phylogeny (Fig. 4A). The phylogenetic tree depicts the inferred evolutionary relationship among the organisms under study. The tree consists of nodes and branches. The “leaves” of the tree (i.e., nodes without descendants) are referred to as terminal nodes. Nodes with descendants are referred to as internal nodes; each internal node represents the most recent common ancestor (MRCA) of those descendants. Branches connect the descendants to their ancestors. The ancestor of all the descendants in the tree is called the root, although not all trees have a root. Subgroups within a tree consisting of a common ancestor and its descendants are referred to as clades.
FIG 4.
General phylogenetic concepts. (A) The general structure of a phylogenetic tree consisting of nodes and branches. A terminal node represents a particular organism (or sequence) under study. Terminal nodes are connected to internal nodes representing hypothetical ancestors. A common ancestor and all of its descendants are referred to as a clade. The ancestor to all descendants in the tree is called the root. (B) A cladogram showing the relative recency of common ancestry. Branch lengths are not informative in a cladogram. (C) A phylogram showing both the relative recency of common ancestry and evolutionary distance. Branch lengths are scaled to reflect the evolutionary distance between an organism and its ancestors.
Phylogenetic trees are depicted either as cladograms (Fig. 4B) or as phylograms (Fig. 4C). Cladograms show only the relative recency of common ancestry. The evolutionary distance between ancestors and descendants is not depicted; therefore, only the tree topology is informative in cladograms, not the branch lengths. Cladograms are often used to represent a hypothetical relationship between species. In contrast, phylograms show the relative recency of common ancestry along with the evolutionary distances (such as time or genetic mutations), which are depicted by scaling the branches to reflect the distance between ancestors and their descendants. Branch length values are often presented along the branch or by adding a scale bar. In contrast to cladograms, phylograms often are used to represent, to the degree possible, the “real” evolutionary relationship among species. Note that only the tree topology and the relative branch lengths (for phylograms) are meaningful in a standard phylogenetic tree. Both phylograms and cladograms can have a vertical or horizontal orientation, and the relative order of the terminal nodes can be adjusted by “rotating” the internal branches in the tree without changing the relationships depicted by the tree; that is, the relationships between terminal nodes are defined by the internal nodes linking them, not by the left-to-right (or top-to-bottom) order in which they appear in the tree.
Inferring phylogenetic trees.
Phylogenetic trees can be inferred by analyzing the physical variation or the genetic variation present in the organisms under study. These genetic data are normally available only for living or recently living organisms; thus, the available data pertain only to the leaf nodes in the tree. However, owing to the lack of data for the ancestors of these terminal nodes, their placement in the tree must be inferred by tree-building techniques. Standard tree-building methods typically assume a bifurcating tree structure where each internal node is connected to two descendants. If the ancestral node is not known, as is often the case, the result is an unrooted tree. Unrooted trees depict only the relative relatedness of the leaf nodes but do not illustrate the ancestry. Trees can be rooted by including an outgroup (i.e., a leaf node with data from an organism that is known to be ancestral to the other organism in the tree); this allows the tree-building programs to impute the MRCA of all the leaves in the tree and results in a rooted tree where the direction of evolution and therefore the absolute ancestry are inferred.
Many different tree-building methods exist; however, the most popular for building phylogenies from genetic data can be grouped into two classes: distance-based methods and character-based methods. For both classes, the input normally comes in the form of an aligned set of genetic data, also called a multiple-sequence alignment (MSA). Distance-based methods, such as the neighbor-joining (NJ) method and the unweighted pair group method with arithmetic mean (UPGMA), calculate the number of genetic differences (polymorphisms) between every sequence and every other sequence in the comparator group and then use these distances to infer the tree. Distance methods are simple to implement and efficient to run but do not incorporate evolutionary models. Character-based methods, such as the maximum parsimony (MP) method and the maximum likelihood (ML) method, examine the actual mutations present in the sequence data and use this information to infer the best tree (160). They can incorporate sophisticated evolutionary models that generate more accurate branch lengths than the simpler distance-based methods; however, they can require substantial processing power to run, especially for large trees.
The robustness of a phylogenetic tree (i.e., the likelihood that it is “correct”) is often estimated by bootstrapping. In this method, the columns of the input multiple alignment are randomly sampled with replacement to generate a new multiple alignment that will have some columns repeated and others absent relative to the original, and a tree is inferred from this new alignment. This process is repeated a certain number of times (e.g., 100 or 1,000). The resulting trees are compared for their concordance by calculating the number of times that each ancestor grouped the descendants repeatedly in the same clade in each run. The result is a semistatistical measure of how robustly each internal node in the tree supports the inferred evolutionary relationship of the organisms under study. The nodes are often labeled with their bootstrap values in the resulting tree.
Phylogenomics.
Phylogenomics is the application of WGS data to the study of evolution. Traditional phylogenetic methods often make use of one or a few genes for tracing the evolutionary history of an organism. HTS provides the capability to extend these methods with information from the entire genome, enabling reconstruction of extremely high-resolution phylogenies. This has applications in infectious disease surveillance and outbreak response, where reconstructed phylogenies from genomes have the potential to complement or replace traditional typing methods such as MLST, pulsed-field gel electrophoresis (PFGE), or serotyping.
Although constructing whole-genome phylogenies has shown many successes, there are still many challenges. Constructing whole-genome phylogenies on highly recombinant organisms is problematic, as recombination can confound phylogenetic methods by obscuring the signal of vertically inherited variation (161). For homologous recombination, where similar sequences of DNA are exchanged, if the source of recombination is external to the study population, the exchanged regions can contain a higher number of SNVs than observed elsewhere in the population. This has the consequence of increasing genetic distance and also branch lengths with most phylogenetic methods and could lead to falsely excluding direct transmission within an epidemiological study (147). If recombination occurs with a source internal to the population under study, then inconsistencies in the phylogenetic signal can occur, leading to an incorrect tree topology (147, 161). Managing homologous recombination often involves identifying and excluding phylogenetic signals from regions having undergone recombination, leaving only those regions arising from vertical descent—that is, the clonal frame (161). For nonhomologous recombination, introduction of paralogous sequence can lead to inflation of genetic distance, for example, by introducing false-positive SNVs due to incorrect read mapping (161).
Additional concerns include data management and scalability. Handling these issues has led to the development and adoption of many phylogenetic analysis methods, which can be broadly categorized as alignment-based, alignment-free, and gene-by-gene methods. A categorized list of software can be found in Table 6, while additional in-depth reviews can be found in references 159 and 162.
TABLE 6.
List of phylogenomics softwarea
| Type | Subtype | Name (reference[s]) | Input | Dist. | Comments |
|---|---|---|---|---|---|
| Alignment based | Whole-/core genome alignment | Gubbins (148) | WGA | L | Recombination detection and removal; requires independent generation of a whole-genome alignment (e.g., with de novo assemblies or reference mapping) |
| ClonalFrameML (147) | WGA | L | Recombination detection and removal; requires independent generation of a whole-genome alignment | ||
| Harvest suite (166) | AG | L | All-in-one package for genome alignment, variant detection, recombination removal, and visualization; alignments restricted to the core genome | ||
| Concatenated gene alignments | Osiris (248) | SR/AG | L/W | Galaxy-based pipeline; for sequence read input, assemblies can be performed as part of the pipeline; demonstration server at http://galaxy-dev.cnsi.ucsb.edu/osiris/ | |
| Reference based | CFSAN SNP Pipeline (174) | SR | L | Available at http://snp-pipeline.readthedocs.io/en/latest/ | |
| SNVPhyl | SR | L | Galaxy-based pipeline; available at http://snvphyl.readthedocs.io/ | ||
| Lyve-SET | SR | L | Available at https://github.com/lskatz/lyve-SET | ||
| Snippy | SR | L | Available at https://github.com/tseemann/snippy | ||
| CSIPhylogeny (32) | SR | W | Available at https://cge.cbs.dtu.dk/services/CSIPhylogeny/ | ||
| REALPHY (176) | SR/AG | L/W | Makes use of multiple reference genomes and includes invariant sites in alignment (similar to whole-genome alignments); available at http://realphy.unibas.ch/fcgi/realphy | ||
| Reference free | SISRS (180) | SR | L | Composite reference genome assembled from sequence reads and used to identify variation | |
| kSNP (178, 179) | SR/AG | L | Identification of SNVs through k-mer comparisons; reference genomes can optionally be included to annotate SNV functions | ||
| Alignment free | Word based | FFP (182) | AG | L | Suite of small, command-line-based tools for constructing phylogenies |
| CVTree3 (249) | AG | W | Available at http://tlife.fudan.edu.cn/archaea/cvtree/cvtree3/ | ||
| Match lengths | Andi (184) | AG | L | Available at https://github.com/evolbioinf/andi/ | |
| Gene by gene | rMLST | PubMLST rMLST (188) | AG | W | Available at http://pubmlst.org/rmlst/; only for academic and noncommercial use, requires emailing software maintainers for an account |
| rMLST/cgMLST | Ridom SeqSphere+ | AG | L | Commercial software, used in studies such as reference 195; includes modules for genome assembly among others | |
| cgMLST/wgMLST | BioNumerics | AG | L | Commercial software, used for example by reference 175; includes modules for genome assembly and reference-based SNV phylogenies among others | |
| SISTR (200) | AG | W | Allows for rapid global comparison with Salmonella genomes from NCBI; available at https://lfz.corefacility.ca/sistr-app/ |
Abbreviations: WGA, whole-genome alignment; AG, assembled genome; SR, sequence reads; Dist., distribution of software; L, locally installed software; W, web service.
(i) Alignment-based phylogenies.
Alignment-based whole-genome phylogenies typically rely on the generation and analysis of a multiple-sequence alignment (MSA), that is, an alignment of nucleotide or amino acid characters where each row represents an isolate and each column in the alignment represents a hypothetical homology. As described under “Inferring phylogenetic trees,” the generated MSA is analyzed using distance-based or character-based methods to produce a phylogenetic tree. Although originally designed for the use of multiple alignments of single genes, these methods have been adapted to make use of whole genomes. Some of the more popular methods involve the generation of a whole-genome multiple-sequence alignment, the generation of a concatenated alignment of genes, or the construction of a whole-genome SNV-based alignment.
Whole-genome alignments, where homologous regions of entire genomes are aligned, are a straightforward method for constructing an MSA to be used for phylogenomic analysis. Once constructed, a whole-genome alignment can be used for phylogenetic inference either at the small scale by using nucleotide variation or at the larger scale by examining rearrangement or duplication events (163). Additionally, many modern recombination-detection and phylogenetic inference software programs make use of whole-genome alignments. Software such as Gubbins (148) makes use of whole-genome alignments to scan for regions of elevated SNV density occurring on branches of an initially constructed phylogenetic tree of all nucleotide substitutions. Significantly highly SNV-dense regions are reported as potential recombination events and removed to construct a phylogenetic tree with nucleotide variation from only the clonal frame. Other software such as ClonalFrameML (147) makes use of whole-genome alignments along with a model of recombination and maximum likelihood methods to identify and mask recombinant regions and then reconstruct a phylogenetic tree with variation from the clonal frame.
Constructing a whole-genome multiple alignment often starts with an initial set of de novo-assembled genomes. A common approach, as taken by software such as progressiveMauve (164), first identifies smaller homologous and colinear (i.e., no genomic structural variation) regions of each genome and then combines these regions into a whole-genome alignment. An in-depth description of whole-genome alignment methods can be found in Colin Dewey's book chapter on whole-genome alignments (163).
While the use of de novo-assembled genomes is common for constructing whole-genome alignments, scalability and running time can become a concern with large data sets of hundreds of genomes. As an example, the authors of progressiveMauve report a running time of 24 h for 20 genomes and 70 h for 40 genomes (164), not including the time spent for the assembly of each genome. For generation of larger-scale whole-genome alignments, some studies (165) have instead used a reference mapping approach. A reference genome is selected, and SNVs, indels, and other variations are identified and used to generate a consensus sequence for each genome. These consensus sequences are combined into a whole-genome alignment. Alternatively, more recent software, such as the Harvest suite (166), takes a different approach. Instead of whole-genome alignments being constructed, only the core genome among a set of isolates is aligned, taking many orders of magnitude less time than whole-genome alignments (minutes compared to hours with progressiveMauve on the same data set) (166). Additionally, the Harvest suite includes software for removal of recombinant regions, generation of a phylogenetic tree, and visualization of the phylogenetic tree alongside identified variants, providing an all-in-one package for phylogenomic analysis. However, the Harvest suite requires high-quality assemblies to achieve these results, and variation in noncore regions of the genome is excluded compared to whole-genome alignment methods (166).
Concatenated gene alignments, sometimes called a “supermatrix” or “supergene,” offer an alternative to whole-genome alignment methods for phylogenetic analysis. While similar to whole-genome alignments, concatenated gene alignments are constructed from separate multiple alignments of homologous genes, specifically orthologous genes, to generate trees reflecting vertical descent, which are concatenated to produce an overall alignment (159, 167, 168). Alignments with paralogous genes should be removed, while missing genes within some isolates can be either coded as missing data or excluded altogether (159). Identification of orthologous genes often involves the identification of highly similar pairs of genes, such as through all-versus-all BLAST comparisons used by OrthoMCL (169), followed by graph-based analysis to cluster these pairs of genes into larger orthologous gene groups (168). Following ortholog identification, multiple alignment of each set of genes can be performed with software such as Clustal Omega (170). As an advantage, concatenated gene alignments provide the additional capability of detecting gene duplication, gene loss, or recombination events by comparing the species tree (i.e., the underlying phylogeny of a species) with the gene trees (i.e., the phylogeny of individual genes) (168). However, these methods often require a priori knowledge of the underlying species tree (168), which for closely related bacteria is often unknown (albeit it could be estimated with other phylogenetic methods described here). Also, for any concatenated gene alignment method, there is the requirement to first assemble and annotate each genome followed by orthologous gene identification, which can be computationally costly.
SNV-based phylogenies are generated by identifying SNVs from a set of genomes and producing a multiple-sequence alignment of variant-only sites. The reduction in size of the alignment to only variant-containing sites provides for a shorter computation time for generating a phylogeny. However, removing invariant sites can cause overestimation of branch lengths, and proper correction when generating the phylogeny should be applied (171, 172).
Identifying SNVs is accomplished through either a reference-based approach or a reference-free approach. In a reference-based approach, an assembled reference genome is used as a basis for identifying SNVs. This is often accomplished through reference mapping and variant calling; however, assembled genomes also can be used with software such as MUMMer (173) for identifying SNVs. The CFSAN SNP pipeline (174) is an example of a locally installable pipeline that uses a reference mapping and variant calling approach that is currently in use by the U.S. Centers for Disease Control and Prevention (CDC) and U.S. Food and Drug Administration (FDA) for outbreak detection and investigation (175).
Reference-based approaches provide an advantage of being able to identify the exact location and corresponding gene for each variant with respect to the reference, validating any variants identified using the sequence read alignments, and applying recombination detection and masking techniques. However, the requirement for choosing a proper reference can be problematic as distantly related reference genomes can bias the generated phylogeny (176). The software REALPHY (176) provides both a Web service and downloadable software that attempt to address this reference genome issue through the use of multiple reference genomes.
In contrast to a reference-based approach, a reference-free approach does not require a reference genome but instead identifies SNVs directly from the sequence data. This eliminates any biases potentially introduced due to the selection of a reference and allows for the detection of SNVs not present in the reference genome. However, as noted by Pettengill et al., a reference-free approach may lead to a higher SNV false discovery rate without appropriate thresholds (177). The software package kSNP (178, 179) takes a reference-free approach to identifying SNVs by breaking up each genomic data set into k-mers and comparing these k-mers. Another software package, SISRS (180), assembles a composite genome from the sequencing data and uses this assembled composite as a reference for variant calling.
(ii) Alignment-free phylogenies.
Alignment-free methods for constructing whole-genome phylogenies do not require the use of a multiple-sequence alignment. Instead, they are constructed by defining and measuring a quickly computable pairwise distance between each genome. Once distances are computed, they are run through previously mentioned clustering algorithms such as neighbor joining or UPGMA. This enables the rapid generation of phylogenies with many hundreds or thousands of genomes without the costly computation time of generating a multiple-sequence alignment.
The Genome BLAST Distance Phylogeny (181) method calculates a distance based on alignments between each genome, in this case using pairwise BLAST alignments. Word-based methods, such as Feature Frequency Profiles (182, 183), break each genome up into k-mers and compare the frequencies of these k-mers to define a genomic distance. Other software, such as Andi, computes rapid local alignments between each genome and defines the distance based on mismatches within each alignment (184). Haubold (185), as well as Bonham-Carter et al. (186), has written excellent reviews of alignment-free methods that can be referred to for additional details.
The advantages of alignment-free phylogenies are speed and scalability; phylogenetic trees can be computed quickly for a large number of genomes. However, care must be taken to properly deal with many genomic events, such as large insertions or horizontal gene transfer, which can confound the phylogeny. Methods include restriction of sequence data for analysis to only those within the core genome, removal of repetitive sequence data, and application of appropriate phylogenetic distance models (183).
(iii) Gene-by-gene phylogenies.
An alternative approach to alignment-based and alignment-free methods is the gene-by-gene approach. This method is an extension of traditional MLST from a small set of housekeeping genes to larger collections of genes, enabling much higher resolution than traditional MLST methods (187). Ribosomal MLST (rMLST) extends the limited 6 or 7 housekeeping genes used by traditional MLST to a set of 53 genes encoding the bacterial ribosomal protein subunits and enables resolution across the entire bacterial domain down to the individual sequence type level (188). An example of an rMLST database and Web service for classification of genomes available for academic and noncommercial use is PubMLST (http://pubmlst.org/rmlst/). Whole-genome MLST (wgMLST) extends this concept to encompass all genes within a given genome, on the order of thousands, while core genome MLST (cgMLST) restricts this gene set to the core genome loci common among a group of isolates. Relatedness is often based on a distance between each genome defined by the number of shared alleles for each gene in the extended MLST set (189). These distances can be organized into a distance matrix and analyzed using standard clustering methods, such as neighbor joining, or methods to account for conflicting phylogenetic signals (i.e., recombination and horizontal gene transfer), such as Neighbor-net (190), SplitsTree (191), and PHYLOViZ (192).
Recent publications (189, 193–195) have demonstrated the usefulness of this gene-by-gene approach for rapid identification and classification of closely related isolates, comparable to classification generated from alignment-based phylogenies. The software BIGSdb (196) has been developed for defining gene loci and grouping into arbitrary schemata and is used to power many databases hosted at PubMLST. Commercial software also making use of the gene-by-gene approach is listed in Table 6.
(iv) Choosing a method for phylogeny generation.
The selection of which phylogenomic analysis method to apply depends primarily on the intended use of the generated phylogeny as well as considerations on the available computational resources to complete the analysis. For organisms thought to be highly recombinant, or where recombination detection is a focus, a whole-genome alignment method would be most useful. This was used in a study of 240 Streptococcus pneumoniae strains (165) where a reference mapping approach was used to construct a whole-genome alignment followed by phylogenomic analysis with methods later packaged into the software Gubbins (148).
Alternatively, where there is less focus on recombination analysis, an SNV-only alignment may be most useful. This was applied in studies on the outbreak of Vibrio cholerae in Haiti in 2010 (197, 198) and for a study on real-time surveillance and outbreak detection of verocytotoxin-producing E. coli in Denmark in 2012 (199) and is currently in use by the CDC, along with wgMLST methods, for real-time surveillance and outbreak detection (175). However, with highly divergent genomes, overestimation of branch lengths can occur (171, 176), leaving this method most applicable for a rapid in-depth analysis where the population under study is closely related.
Gene-by-gene methods, primarily wgMLST and cgMLST, have resolution comparable to that of alignment-based methods and have the benefit of a standard gene schema (and associated classification nomenclature that encourages data sharing compatibility) (193, 195). Thus, gene-by-gene approaches are particularly useful for integration of newly sequenced data into a global context. To date, use of wgMLST/cgMLST methods has been limited to custom schemata developed and curated for individual sequencing projects (193, 195), free Web services specific to a particular organism (200), or large institutions (175). However, with commitment by some, such as the CDC (175), to expand availability of wgMLST tools, these methods will become more and more relevant in the near future.
Alignment-free methods have been successfully applied in studies such as a phylogenetic analysis of Escherichia coli/Shigella (183) and are currently being used by NCBI's Pathogen Detection project to construct large-scale phylogenetic trees based on k-mer analysis (http://www.ncbi.nlm.nih.gov/projects/pathogens/about/). However, in particular for genomic epidemiology, a large focus has been on the use of alignment-based or gene-by-gene methods (175). The NCBI's Pathogen Detection project itself plans to implement SNV-based methods for further comparisons of isolates in the future (http://www.ncbi.nlm.nih.gov/projects/pathogens/about/). Thus, while able to rapidly produce phylogenies, alignment-free methods have become most useful for a first, qualitative look at how sequenced bacteria are related to one another.
Recently, there has been a large focus on comparing these phylogenomic methods as well as comparing the performances of different software for the purpose of assessing their accuracy and consistency characteristics. Proficiency testing for constructing whole-genome phylogenies has been ongoing through the GMI (http://www.globalmicrobialidentifier.org/Workgroups/About-the-GMI-Proficiency-Tests), and a newly initiated ASM conference on NGS and Bioinformatics Pipelines included a Pathogen Surveillance Software Demonstration method exercise for reconstructing whole-genome phylogenies (https://github.com/katholt/NGSchallenge). The results of these software comparisons should provide useful information on the compatibility and accuracy of different methods for reconstructing whole-genome phylogenies.
HTS IN THE CONTEXT OF SPECIFIC APPLICATIONS
At the beginning of this review, we introduced three common applications of HTS within the context of infectious disease bacterial genomics: bacterial typing, molecular epidemiology, and pathogenomics (Fig. 1). These applications are not mutually exclusive but are often progressions within a project to provide nuanced information or additional in-depth knowledge; however, consideration of the primary application(s) is important during the planning stage to ensure that the sampling strategy (Table 2) and analysis plans can effectively meet project objectives.
Generally, once the planning stage is complete, the early workflow steps (data generation and primary and secondary analysis) are typically completed within an automated pipeline. Commonly used tertiary analyses may also be automated; however, this stage of analysis often requires more specialized processes tailored to the application. For the purpose of this review, this issue will be discussed in the context of the three abovementioned applications.
Bacterial Typing
In silico typing or feature extraction can be considered a secondary analysis as many bacteria have validated molecular typing schemes with existing databases for PCR-generated amplicons that can be extracted from HTS data for comparison. These databases may contain Sanger-sequenced products of a single comparator gene (e.g., 16S rRNA gene, rpoB, and cpn60) or a panel of housekeeping genes as in MLST. Multiple efforts are under way globally for each organism to expand such typing schemes to take into account more genomic loci and are certainly going to be leveraged extensively in the future. Additionally, categorical features such as virulence factors, antimicrobial resistance genes (ARGs), or transposable elements also can be extracted to achieve more detailed molecular subtyping or to compare bacterial genomes based on the project objectives, moving feature extraction and in-depth characterization into tertiary analysis efforts.
The Basic Local Alignment Search Tool (BLAST) is a generic tool that can be used to extract features from large HTS data sets (201). The query sequence is compared to a database, searching for regions of local sequence similarity. Therefore, a reference sequence or extracted feature (query) could be compared to the HTS-derived assemblies (database) to extract similar sequences, or vice versa, an unknown sequence/contig could be queried against a database of annotated features. Parameters can be set to adjust the stringency of the BLAST, and results are presented as presence/absence or the aligned database matches or “hits” for further analysis. BLAST is publicly available and can be used online (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to search a variety of public databases, or the software can be downloaded to search a personalized, local database. Extracting certain feature classes can be achieved using specialized Web-accessible databases such as the multiple “Finder” applications on the Center for Genomic Epidemiology website (ResFinder [31], VirulenceFinder [199], PathogenFinder [202]), PHAST (156) to search for integrated prophage, and CARD to identifiy antimicrobial resistance genes (203). The latter programs require assembled contigs or annotations as input; however, recently developed tools such as GeneSippr and SRST2 identify gene markers using a mapping approach from quality-filtered sequence reads as input instead of assembled contigs (204, 205).
Historically, bacterial molecular typing offered rapidly communicated designations for public health and clinical microbiology applications such as spa typing of S. aureus (206) and NG-MAST for Neisseria gonorrhoeae (207). Moreover, typing schemes are often used as categorical variables to influence strain selection in research applications. Although these targeted typing approaches are informative, they are not always consistently used, making epidemiological comparisons difficult (208).
As bacterial typing is enhanced by the finer resolution of genome-wide data, revealing novel alleles and gene functions, it provides opportunities for new protocol development (209, 210). Furthermore, the ability to mine WGS data and extract multiple typing schemes makes global comparisons achievable, which is essential for global surveillance and epidemiological investigations. For example, the SISTR platform (200) uses WGS draft assemblies to rapidly extract multiple typing schemes, including both molecular (MLST) and phenotypic (serotyping) schemes.
Molecular Epidemiology
Molecular epideomiology (ME) uses molecular biological methods to investigate the source, transmission, and pathogenesis of disease. High-throughput sequencing has had an enormous impact on ME studies of bacterial pathogens. Prior to HTS, ME investigations relied on classical subtyping techniques to discover and track microbial pathogens with a common molecular subtype or “fingerprint” suspected of being implicated in a disease outbreak. Many of these technologies, such as pulsed-field gel electrophoresis (PFGE) and MLST, are established, validated, and considered “gold-standard” technologies for subtyping bacterial pathogens; however, they use only a minuscule fraction of the information available in the typical bacterial genome. For some highly clonal, slowly evolving organisms such as Mycobacterium tuberculosis, Bacillus anthracis, or Salmonella enterica serovar Enteritidis, the diversity captured by these techniques is often insufficient to discriminate between an outbreak-implicated isolate and a sporadic, unrelated isolate. In contrast, HTS can (in theory) discriminate between isolates differing by a single nucleotide out of the several million contained in most bacterial genomes. Thus, genome-based molecular epidemiology, termed genomic epidemiology, represents a powerful new method with vastly improved resolution over current gold-standard techniques.
Bacterial pathogen genomic epidemiology established itself as a bona fide approach for public health investigations during the high-profile Haiti cholera outbreak which began in 2010. It was hypothesized to have been imported to Haiti with arriving United Nations (UN) peacekeepers from Nepal. The current, standard typing technology for V. cholerae, namely, PFGE, had inadequate discriminatory power to distinguish environmental isolates from outbreak-related strains in Southeast Asia. Through multiple, independent genomic epidemiological investigations (197, 198, 211), the source of the outbreak was conclusively determined to be imported to Haiti by the Nepalese UN peacekeepers, thereby solving an important epidemiological controversy that prior methods could not. The application of genomic epidemiology to other high-profile events, such as the 2011 German E. coli O104:H4 outbreak (212), has cemented the reputation of genomic epidemiology as a powerful new method for outbreak investigation, and it is currently positioned to replace the existing gold-standard methods as the main tool for both surveillance and outbreak response by public health laboratories around the world. Indeed, some early adopters, such as the CDC and the FDA, are already using HTS to assist in their real-time foodborne disease detection, surveillance, and outbreak response activities via the GenomeTrakr network (21). Bacterial pathogen genomics employs the methods described under “Phylogenomics.” k-mer trees can be used to quickly assess the evolutionary relationship of a group of genomes within the context of a larger population of genomes, which can be useful, for example, in selecting a reference genome for subsequent SNV-based phylogenomics or for identifying and removing outliers that may be derived from contamination or isolate misclassification. For routine surveillance, both the SNV-based approach and gene-by-gene-based approaches have found application. The selection of approach can depend on several factors. For example, for low-diversity organisms, an SNV-based approach, such as the CFSAN pipeline adopted by the FDA's GenomeTrakr project, may be desired. However, the current SNV-based approaches do not allow the set of discovered SNVs for an organism to be collapsed into a simple categorical subtype. In contrast to SNV-based phylogenies, a set of reported alleles can be easily assigned to a simple subtype category; therefore, for organisms with more inherent variation, such as Listeria monocytogenes, the gene-by-gene approach has been adopted by PulseNet International (C. Nadon, personal communication). The main shortcoming with gene-by-gene-based approaches is the requirement to generate and curate large schemata consisting of the loci and alleles for each organism, many of which remain works in progress.
Routine surveillance with genomic epidemiology may require the sequencing and analysis of voluminous bacterial genomes; the large-scale GenomeTrakr project, for example, sequences over 1,000 isolates each month. Surveillance at such a scale requires thousands of CPU cores and petabytes of storage. Sequencing and computing requirements for single outbreak investigations are not as computationally onerous as routine surveillance; however, the generation and analysis of the sequence data may need to be performed under extreme time pressures, and thus, the available resources will need to provide this “surge capacity.” Acute outbreaks can vary in scale from international foodborne disease investigations (212) to nosocomial outbreaks where the threat of frequent transmission events and antimicrobial resistance is a major concern for immunocompromised hospital patients (213–215). For both routine surveillance and outbreak investigation, draft genome data with short-read technologies appear to be sufficient for genomic epidemiological investigation of most pathogens (144, 216).
Bacterial Pathogenomics
Pathogenomics is a field of study that uses genomic sequence data to understand how genomic variation influences microbial diversity and how this diversity influences host-microbial interactions and other bacterial behaviors that result in the development or inheritance of virulence factors involved in disease. The introduction of HTS and related comparative genomics approaches has vastly improved our ability to conduct bacterial pathogenomics studies and has revealed new insights into bacterial genome structure and dynamics. Perhaps most surprising is the observation that the gene contents of some species such as E. coli can differ from each other by as much as 30% (217). Such “open pan-genomes” in turn have important implications for the variety and complexity of virulence factors that can influence disease, requiring investigators to sample and sequence large populations of bacteria in order to understand pathogenicity even within a single species. In this section, we outline the main methods for bacterial pathogenomics research using HTS. Obviously, a sequence-plus-bioinformatics analysis can only give rise to hypotheses about the mechanisms of pathogenesis; follow-up studies involving forward and reverse genetic screens that satisfy molecular Koch's postulates are necessary to unambiguously assign causality for a genomic feature's contribution to bacterial pathogenicity.
Three main forces govern bacterial genome evolution: gene loss, gene gain, and genome rearrangement. The interaction of these forces results in a variety of bacterial genome dynamics, including SNV, gene duplication, gene shedding/loss (gene content is lost in entirety), gene decay (in which gene sequence or function is changed through partial loss), recombination, and horizontal gene transfer (leading to gene acquisition and/or allelic diversification). Different bacterial pathogens have adopted various evolutionary strategies that are manifest in their genome dynamics, and knowledge of a specific organism's genome dynamics is important for its proper contextual analysis.
The smallest-scale variation is the SNV. Organisms that employ SNV changes as their primary method of evolution include intracellular obligate parasites such as Chlamydia trachomatis and Mycobacterium tuberculosis (218, 219). SNV discovery and SNV annotation methods, such as those described under “Reference Mapping and Variant Calling,” are appropriate for the analysis of these genomically monomorphic organisms, and examples of such studies abound. For example, SNV-based methods were used to identify a critical virulence gene, CT135 in C. trachomatis (218). Certain SNV mutations in this gene led to considerably long clearance times and increased virulence, with further studies showing that the CT135 virulence gene is stable in vivo but quickly mutates in vitro (220). SNV methods also can be used for the analysis of the pathogenicity of clonally related organisms, one notable example being the study of the accumulation over time of pathogenicity in Burkholderia dolosa in chronically infected cystic fibrosis patients (221).
Other organisms, such as Streptococcus pneumoniae and Neisseria spp., can take up and recombine homologous chromosomal DNA, resulting in the generation of allelic diversity. Analysis of recombination can be important for the pathogenomics study of highly recombinant organisms; illustrative examples include vaccine escape analysis of S. pneumoniae (149, 165, 222) and the adaptive evolution of outbreak-associated Legionella pneumophila (223).
Horizontal gene transfer is by far the most effective means of acquisition of genomic variation that can influence microbial virulence. Indeed, many organisms employ horizontal gene transfer to acquire and share virulence factors that enable colonization, immune suppression or aberration, immune evasion, host cell invasion, and other genomic features involved in persistence and infection. Pathogenomics investigations focusing on horizontal gene transfer involve the tools and methods already described under “Mobile genetic elements.” In addition, it can be valuable to partition the genes from pathogenic bacteria into core and accessory genes using ortholog analysis programs such as OrthoMCL (169). Genes in the accessory genome and contained within mobile elements can be mined for virulence factors by searching against virulence factor databases such as VFDB (224), MvirDB (225), VirulenceFinder (199), and the PATRIC virulence factor library (226). It is important to use caution in this type of investigation, however, since the factors that impart virulence can be highly organism specific; thus, it is important to have a strong understanding of the biology as well as the molecular genetics associated with a given organism in order to properly conduct a pathogenomic investigation.
Horizontal gene transfer allows organisms to gain potentially large amounts of genes, especially those with open pan-genomes, such as E. coli (217) and Campylobacter spp. (227). Despite this, the average genome size remains approximately the same, implying that genes are lost at about the same rate at which they are gained. The evolutionary mechanisms that drive gene loss include large segmental deletions of genomic regions that no longer provide a selective advantage (228) and the creation and subsequent deletion of pseudogenes. The latter mechanism is often observed in recently emerged pathogens that have evolved to live in a new host. Salmonella enterica serovar Typhi, for example, has been observed to contain many hundreds of pseudogenes (229) that once generated are rapidly shed from the genome, suggesting that these pseudogenes are under selection (230). This gene shedding serves to modulate pathogenicity, making these pseudogenes interesting targets for pathogenomic studies. Automated systems for bacterial pseudogene detection exist but are typically tuned to the detection of pseudogenes of a given species (231); most pseudogene detection involves the manual alignment and inspection of orthologous genes and their pseudogene counterparts.
Methods for genome-wide association studies (GWAS) of bacteria also have been developed (232). Thorough GWAS require the tools and techniques developed for nearly all the main methods of analyzing genomic variation and correlating these variations with biological traits. Pipelines such as PhenoLink (233) can assist in this effort, although they can be cumbersome and still require large amounts of manual analysis. Newer programs like Neptune (234), although not a replacement for true GWAS, are automated and have been demonstrated to quickly find genomic loci that are associated with biological traits.
In addition to GWAS, pathogenomic results can fuel extension projects to further characterize the novel strains or genomic features identified within earlier tertiary analysis. As illustrated in Fig. 1, extension projects may include complementary methods such as proteomics, transcriptomics, metabolomics, gene knockout experiments, or animal models, etc.; however, finding expertise in such a broad spectrum of scientific disciplines may not be possible within the scope of a single project. However, sharing of the raw data in addition to the publication of significant findings can benefit the larger scientific community and make such extension studies possible.
GLOBAL ACCESSIBILITY OF GENOMICS DATA
Data sharing is increasingly being recognized as a major benefit to the scientific and medical communities at large as it allows data to be fully vetted by other researchers and collated for reuse and further evaluations, thereby achieving even greater global impact. As modern science produces data at ever increasing rates, open data offer the best opportunity to ensure that the data remain transparent (available) and fully supported (credible) into the foreseeable future. Open data also provide researchers with data to develop, enhance, and benchmark analytical methods. Consequently, there is increasing pressure to provide open data through initiatives such as the STROME-ID (Strengthening the Reporting of Molecular Epidemiology for Infectious Diseases) statement (235). Many journals have adopted mandatory open data policies, meaning that all supporting data must be submitted to a relevant publicly accessible depository. Many science funding bodies have similarly followed suit, requiring that all data generated as a result of funding must be made publicly available within a reasonable time frame.
Data sharing within the context of infectious disease genomics from cultured bacteria may never reach the same heightened level of privacy concerns as human sequence data; however, the impact of timely data sharing on public health and the scientific community is comparable. Infectious disease outbreaks have already led to adoption of HTS and warrant timely data generation, analysis, and sharing to intervene in order to stop the spread of infection and save lives (236, 237). There are an increasing number of global outbreaks where accessible genomics data were vital to the investigation, due not only to the speed of data production but to the global-scale collaboration that was sparked by the data release. Recent examples include the 2011 German E. coli O104:H4 outbreak, in which the open-access data and crowd-sourced analysis resulted in valuable epidemiological results in less than 1 week (238). Other recent examples of globally collaborative outbreak investigations attributable to data sharing include the H1N1 influenza A (swine flu) virus outbreak of 2009 (239), the Haitian V. cholerae outbreak in 2010 (240, 241), and the West African Ebola outbreak in 2014 (242).
Although these listed scenarios exemplify the benefits of sharing data, there are risks and barriers that can potentially have detrimental consequences on many levels. These risks are generally tied to the associated metadata describing the characteristics of the isolate source. The inferred/suspected infectious source and transmission routes may prematurely prompt trade embargos and travel bans, stigmatizing specific geographic areas, countries, or individuals before appropriate source attribution vetting has occurred. In addition to the ethical challenges, there are also significant logistical challenges that require, for example, infrastructure decisions to be made and mutually agreed upon between researchers and multiple levels of government in order to effectively share and retain data while protecting the privacy rights of individual parties (be they persons, corporations, or countries) (236, 243, 244). The ideal of freely sharing data for scientific advancement and public health (i.e., monitoring and control of infectious diseases) is admirable and in many cases a reality, and yet there remain many challenges to which there are likely no quick solutions. Therefore, details regarding the release of project data should be addressed in the planning stage, including timing and the selection of data repository and necessary agreement among project partners.
CONCLUSIONS
The adoption of HTS methods for applications such as bacterial typing, molecular epidemiology, and pathogenomics is growing in frequency and magnitude. These new technologies pose new challenges for researchers as the growing scale of HTS projects require a paradigm shift in experimental design and resource planning due to the quickly produced, large amounts of data generated and the requirement for enhanced computational infrastructure and bioinformatic support for meaningful interpretation. This review was intended to highlight these new challenges and provide a foundational understanding of the terminology and concepts for nonbioinformatician investigators to explore before venturing into the use of HTS technologies for infectious disease research and potential mainstream usage in the areas of public health and clinical microbiology.
APPENDIX
Glossary
- antimicrobial resistance gene (ARG)
An acquired gene or gene variant encoding an antimicrobial resistance phenotype.
- BAM
Binary version of a SAM file that contains sequence alignment data.
- CCS
Circular consensus reads produced by PacBio sequencing, shorter and more accurate than CLR.
- core genome MLST (cgMLST)
An extension of traditional MLST to include genes from the core genome of a group of bacteria.
- cloud computing
A computing model where scalable computational resources are provided on demand from large data centers.
- CLR
Continuous long reads produced by PacBio sequencing, longer but more error prone than CCS reads.
- contig
Contiguous consensus sequence.
- de novo assembly
The process of combining sequence reads to reconstruct a sequenced genome without the aid of a reference genome.
- FASTQ
FASTQ files are text files containing sequence data with a quality (Phred) score for each base represented as an ASCII character.
- gigabyte (GB)
1,024 megabytes.
- Infrastructure as a Service (IaaS)
A cloud computing model that provides only the low-level physical computing resources.
- indel
Insertion/deletion.
- k-mer
A short fragment of sequence data of length “k” produced and used by many bioinformatics algorithms.
- mate-pair (MP) sequencing
Also called “long-insert paired-end.” A sequencing process where a DNA fragment is sequenced from both ends but has been constructed such that each end is further apart.
- megabyte (MB)
1,024 kilobytes.
- overlap-layout-consensus (OLC)
A method of sequence assembly.
- Platform as a Service (PaaS)
A cloud computing model that provides a computing environment with a suite of standard software.
- paired-end (PE) sequencing
A sequencing process where a DNA fragment is read from both ends.
- Phred
Phred or Q score is an integer representing the estimated probability of an error (probability that the base is incorrect).
- reference mapping
The process of aligning sequence reads to a reference genome.
- ribosomal MLST (rMLST)
A variation on MLST making use of the genes encoding the ribosomal protein subunits.
- Software as a Service (SaaS)
A cloud computing model that provides access to specific software applications.
- Sequence Alignment/Map (SAM)
A text-based format used to store sequence reads aligned to a reference genome.
- scaffold
The result of ordering and possibly merging contigs into larger sequences with additional data such as mate-pair or long-read sequencing.
- single-end (SE) sequencing
A sequencing process where a DNA fragment is read from only one end.
- single nucleotide variant (SNV)
Any single nucleotide variation within a population.
- Sequence Read Archive (SRA)
An archive of publicly available biological sequence read data. Can also refer to the file format for storing such data.
- terabyte (TB)
1,024 gigabytes.
- variant call format (VCF)
Compact text file to store variations in sequence data with respect to a reference.
- whole-genome MLST (wgMLST)
An extension of traditional MLST to include genes from an entire bacterial genome for typing.
Biographies

Tarah Lynch, Ph.D., is a computational biologist at Calgary Laboratory Services working on the application of infectious disease genomics in clinical microbiology and holds a Clinical Assistant Professor appointment in the Department of Pathology and Laboratory Medicine at the University of Calgary. She received her Ph.D. from the University of Calgary in Microbiology and Infectious Diseases before completing postdoctoral training through an ASM/CCID fellowship at the Centers for Disease Control and Prevention (Fort Collins, Colorado, USA) and an NSERC Visiting Fellowship in Canadian Government Laboratories program at the Canadian Science Centre for Human and Animal Health (CSCHAH, Winnipeg, Manitoba, Canada). Her current research interests include HIV evolution, bacterial genomics, and antimicrobial resistance.
Aaron Petkau is a bioinformatician at the CSCHAH in Winnipeg, Manitoba, Canada. Initially starting work in bioinformatics as a student at the CSCHAH in 2008 and continuing after receiving his Bachelor of Computer Science in 2010 at the University of Manitoba, Aaron has developed and applied software for the investigation of disease outbreaks using genomics data. He currently leads the integration of existing and novel software into a system developed at the CSCHAH for the routine use of whole-genome sequencing data for epidemiology.
Natalie Knox, Ph.D., is a computational biologist at the CSCHAH in Winnipeg, Manitoba, Canada. She is head of Bacterial Genomics in the Bioinformatics Core at the CSCHAH. In her graduate studies, she developed an interest in metagenomics and large-scale bioinformatics analyses that led her to pursue a career in computational biology and bioinformatics. Her current research activities include applying novel comparative bacterial genomic methods for cluster detection of foodborne diseases and leading bioinformatics analysis of large-scale bacterial whole-genome sequencing projects. She received her Bachelor of Science degree and Ph.D. in Animal Sciences from the University of Manitoba, Canada.

Morag Graham, Ph.D., is Chief of Genomics at the CSCHAH—the national infectious disease hub for the Public Health Agency of Canada in Winnipeg, Manitoba. She is also a University of Manitoba Medical Microbiology Department Adjunct Professor and a Research Affiliate with the Canadian Centre for Agri-Food Research in Health and Medicine (CCARM). Dr. Graham has a longstanding interest in microbial pathogenesis; her research applies pathogenomics and genome sequencing to better understand emerging pathogens. She currently leads several large-scale bacterial whole-genome sequencing projects and actively works to translate microbial genomic data into genomic epidemiology for the benefit of Canada's public health surveillance and outbreak response front lines. She also applies metagenomics to investigate human health and disease. She received her Bachelor of Science (Biochemistry) from the University of Waterloo and Ph.D. (Microbiology) from the University of Guelph in Canada.

Gary Van Domselaar, Ph.D. (University of Alberta), is the Chief of the Bioinformatics Laboratory at the CSCHAH and Adjunct Professor in Medical Microbiology at the University of Manitoba. Dr. Van Domselaar has active research programs in metagenomics, infectious disease genomic epidemiology, diagnostic antibody development, and the development of novel bioinformatics algorithms for infectious sequence analysis and investigation. His lab leads several large-scale national and international genomics projects, including two Canadian Genomics Research and Development Initiative Interdepartmental Shared Priority Projects: one on Food and Water Safety and a second project investigating the accumulation and transmission of antimicrobial resistance genes throughout Canada. He is also a principal investigator on the Genome Canada Integrated Rapid Infectious Disease Analysis (IRIDA) project to develop an end-to-end computational platform for the storage, management, analysis, and sharing of genomic epidemiological information for infectious disease outbreak investigations.
REFERENCES
- 1.Portny SE, Austin J. 12 July 2002. Project management for scientists. American Association for the Advancement of Science, Washington, DC: http://www.sciencemag.org/careers/2002/07/project-management-scientists. [Google Scholar]
- 2.Sandve GK, Nekrutenko A, Taylor J, Hovig E. 2013. Ten simple rules for reproducible computational research. PLoS Comput Biol 9:e1003285. doi: 10.1371/journal.pcbi.1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vos RA. 2016. Ten simple rules for managing high-throughput nucleotide sequencing data. bioRxiv doi: 10.1101/049338. [DOI] [Google Scholar]
- 4.Liolios K, Schriml L, Hirschman L, Pagani I, Nosrat B, Sterk P, White O, Rocca-Serra P, Sansone SA, Taylor C, Kyrpides NC, Field D. 2012. The Metadata Coverage Index (MCI): a standardized metric for quantifying database metadata richness. Stand Genomic Sci 6:438–447. doi: 10.4056/sigs.2675953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gargis AS, Kalman L, Berry MW, Bick DP, Dimmock DP, Hambuch T, Lu F, Lyon E, Voelkerding KV, Zehnbauer BA, Agarwala R, Bennett SF, Chen B, Chin EL, Compton JG, Das S, Farkas DH, Ferber MJ, Funke BH, Furtado MR, Ganova-Raeva LM, Geigenmuller U, Gunselman SJ, Hegde MR, Johnson PL, Kasarskis A, Kulkarni S, Lenk T, Liu CS, Manion M, Manolio TA, Mardis ER, Merker JD, Rajeevan MS, Reese MG, Rehm HL, Simen BB, Yeakley JM, Zook JM, Lubin IM. 2012. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol 30:1033–1036. doi: 10.1038/nbt.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budowle B, Connell ND, Bielecka-Oder A, Colwell RR, Corbett CR, Fletcher J, Forsman M, Kadavy DR, Markotic A, Morse SA, Murch RS, Sajantila A, Schmedes SE, Ternus KL, Turner SD, Minot S. 2014. Validation of high throughput sequencing and microbial forensics applications. Investig Genet 5:9. doi: 10.1186/2041-2223-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kell DB, Oliver SG. 2004. Here is the evidence, now what is the hypothesis? The complementary roles of inductive and hypothesis-driven science in the post-genomic era. Bioessays 26:99–105. doi: 10.1002/bies.10385. [DOI] [PubMed] [Google Scholar]
- 8.Junemann S, Prior K, Albersmeier A, Albaum S, Kalinowski J, Goesmann A, Stoye J, Harmsen D. 2014. GABenchToB: a genome assembly benchmark tuned on bacteria and benchtop sequencers. PLoS One 9:e107014. doi: 10.1371/journal.pone.0107014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kodama Y, Shumway M, Leinonen R. 2012. The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res 40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cock PJA, Fields CJ, Goto N, Heuer ML, Rice PM. 2010. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res 38:1767–1771. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database Collaboration. 2011. The sequence read archive. Nucleic Acids Res 39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ewing B, Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194. [PubMed] [Google Scholar]
- 14.Fusaro VA, Patil P, Gafni E, Wall DP, Tonellato PJ. 2011. Biomedical cloud computing with Amazon Web Services. PLoS Comput Biol 7:e1002147. doi: 10.1371/journal.pcbi.1002147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angiuoli SV, White JR, Matalka M, White O, Fricke WF. 2011. Resources and costs for microbial sequence analysis evaluated using virtual machines and cloud computing. PLoS One 6:e26624. doi: 10.1371/journal.pone.0026624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shanahan HP, Owen AM, Harrison AP. 2014. Bioinformatics on the cloud computing platform Azure. PLoS One 9:e102642. doi: 10.1371/journal.pone.0102642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nosowsky R, Giordano TJ. 2006. The Health Insurance Portability and Accountability Act of 1996 (HIPAA) privacy rule: implications for clinical research. Annu Rev Med 57:575–590. doi: 10.1146/annurev.med.57.121304.131257. [DOI] [PubMed] [Google Scholar]
- 18.Griebel L, Prokosch HU, Kopcke F, Toddenroth D, Christoph J, Leb I, Engel I, Sedlmayr M. 2015. A scoping review of cloud computing in healthcare. BMC Med Inform Decis Mak 15:17. doi: 10.1186/s12911-015-0145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez LL, Brooks LD, Greenberg JH, Green ED. 2013. Research ethics. the complexities of genomic identifiability. Science 339:275–276. doi: 10.1126/science.1234593. [DOI] [PubMed] [Google Scholar]
- 20.Luheshi L, Raza S, Moorthie S, Hall A, Blackburn L, Rands C, Sagoo G, Chowdhury S, Kroese M, Burton H. 2015. Pathogen genomics into practice. PHG Foundation, Cambridge, United Kingdom. [Google Scholar]
- 21.Allard MW, Strain E, Melka D, Bunning K, Musser SM, Brown EW, Timme R. 2016. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J Clin Microbiol 54:1975–1983. doi: 10.1128/JCM.00081-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dove ES, Joly Y, Tasse AM, Public Population Project in Genomics and Society (P3G) International Steering Committee, International Cancer Genome Consortium (ICGC) Ethics and Policy Committee, Knoppers BM. 2015. Genomic cloud computing: legal and ethical points to consider. Eur J Hum Genet 23:1271–1278. doi: 10.1038/ejhg.2014.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyres KL, Conway TC, Garg S, Queiroz C, Reumann M, Holt K, Rusu LI. 2014. WGS analysis and interpretation in clinical and public health microbiology laboratories: what are the requirements and how do existing tools compare? Pathogens 3:437–458. doi: 10.3390/pathogens3020437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goecks J, Nekrutenko A, Taylor J, Galaxy Team. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blankenberg D, Von Kuster G, Bouvier E, Baker D, Afgan E, Stoler N Galaxy Team, Taylor J, Nekrutenko A. 2014. Dissemination of scientific software with Galaxy ToolShed. Genome Biol 15:403. doi: 10.1186/gb4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Afgan E, Chapman B, Taylor J. 2012. CloudMan as a platform for tool, data, and analysis distribution. BMC Bioinformatics 13:315. doi: 10.1186/1471-2105-13-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Afgan E, Sloggett C, Goonasekera N, Makunin I, Benson D, Crowe M, Gladman S, Kowsar Y, Pheasant M, Horst R, Lonie A. 2015. Genomics virtual laboratory: a practical bioinformatics workbench for the cloud. PLoS One 10:e0140829. doi: 10.1371/journal.pone.0140829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu B, Madduri RK, Sotomayor B, Chard K, Lacinski L, Dave UJ, Li J, Liu C, Foster IT. 2014. Cloud-based bioinformatics workflow platform for large-scale next-generation sequencing analyses. J Biomed Inform 49:119–133. doi: 10.1016/j.jbi.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. 2014. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jasny BR, Chin G, Chong L, Vignieri S. 2011. Data replication & reproducibility. Again, and again, and again …. Introduction. Science 334:1225. doi: 10.1126/science.334.6060.1225. [DOI] [PubMed] [Google Scholar]
- 34.Peng RD. 2011. Reproducible research in computational science. Science 334:1226–1227. doi: 10.1126/science.1213847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nekrutenko A, Taylor J. 2012. Next-generation sequencing data interpretation: enhancing reproducibility and accessibility. Nat Rev Genet 13:667–672. doi: 10.1038/nrg3305. [DOI] [PubMed] [Google Scholar]
- 36.Morrison SS, Pyzh R, Jeon MS, Amaro C, Roig FJ, Baker-Austin C, Oliver JD, Gibas CJ. 2014. Impact of analytic provenance in genome analysis. BMC Genomics 15(Suppl 8):S1. doi: 10.1186/1471-2164-15-S8-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pightling AW, Petronella N, Pagotto F. 2014. Choice of reference sequence and assembler for alignment of Listeria monocytogenes short-read sequence data greatly influences rates of error in SNP analyses. PLoS One 9:e104579. doi: 10.1371/journal.pone.0104579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mardis ER. 2013. Next-generation sequencing platforms. Annu Rev Anal Chem 6:287–303. doi: 10.1146/annurev-anchem-062012-092628. [DOI] [PubMed] [Google Scholar]
- 39.Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M. 2012. Comparison of next-generation sequencing systems. J Biomed Biotechnol 2012:251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. 2012. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 41.Kwong JC, McCallum N, Sintchenko V, Howden BP. 2015. Whole genome sequencing in clinical and public health microbiology. Pathology 47:199–210. doi: 10.1097/PAT.0000000000000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glenn TC. 2011. Field guide to next-generation DNA sequencers. Mol Ecol Resour 11:759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- 43.Koren S, Phillippy AM. 2015. One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Curr Opin Microbiol 23:110–120. doi: 10.1016/j.mib.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 44.Loman NJ, Quick J, Simpson JT. 2015. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat Methods 12:733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- 45.Sit CS, Ruzzini AC, Van Arnam EB, Ramadhar TR, Currie CR, Clardy J. 2015. Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants. Proc Natl Acad Sci U S A 112:13150–13154. doi: 10.1073/pnas.1515348112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greninger AL, Naccache SN, Federman S, Yu G, Mbala P, Bres V, Stryke D, Bouquet J, Somasekar S, Linnen JM, Dodd R, Mulembakani P, Schneider BS, Muyembe-Tamfum JJ, Stramer SL, Chiu CY. 2015. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med 7:99. doi: 10.1186/s13073-015-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCoy RC, Taylor RW, Blauwkamp TA, Kelley JL, Kertesz M, Pushkarev D, Petrov DA, Fiston-Lavier AS. 2014. Illumina TruSeq synthetic long-reads empower de novo assembly and resolve complex, highly-repetitive transposable elements. PLoS One 9:e106689. doi: 10.1371/journal.pone.0106689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagarajan N, Pop M. 2010. Sequencing and genome assembly using next-generation technologies. Methods Mol Biol 673:1–17. doi: 10.1007/978-1-60761-842-3_1. [DOI] [PubMed] [Google Scholar]
- 49.Fox EJ, Reid-Bayliss KS, Emond MJ, Loeb LA. 2014. Accuracy of next generation sequencing platforms. Next Gener Seq Appl 1:1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. 2012. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. 2014. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet 15:121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- 52.Robasky K, Lewis NE, Church GM. 2014. The role of replicates for error mitigation in next-generation sequencing. Nat Rev Genet 15:56–62. doi: 10.1038/nrg3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lander ES, Waterman MS. 1988. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics 2:231–239. [DOI] [PubMed] [Google Scholar]
- 54.Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual, 4th ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 55.Centers for Disease Control and Prevention. 14 June 2013. Next generation sequencing: standardization of clinical testing (Nex-StoCT) working groups. Division of Laboratory Science and Standards, Office of Public Health Scientific Services, Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/ophss/csels/dls/glp_genetic_testing/nex-stoct.html. [Google Scholar]
- 56.Tong W, Ostroff S, Blais B, Silva P, Dubuc M, Healy M, Slikker W. 2015. Genomics in the land of regulatory science. Regul Toxicol Pharmacol 72:102–106. doi: 10.1016/j.yrtph.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 57.Global Microbial Identifer. 27 June 2016. Global Microbial Identifier consortium work group 4: ring trials and quality assurance. Global Microbial Identifier. http://www.globalmicrobialidentifier.org/Workgroups#work-group-4.
- 58.Moran-Gilad J, Sintchenko V, Pedersen SK, Wolfgang WJ, Pettengill J, Strain E, Hendriksen RS, Global Microbial Identifier Initiative's Working Group 4 (GMI-WG4). 2015. Proficiency testing for bacterial whole genome sequencing: an end-user survey of current capabilities, requirements and priorities. BMC Infect Dis 15:174. doi: 10.1186/s12879-015-0902-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paszkiewicz KH, Farbos A, O'Neill P, Moore K. 2014. Quality control on the frontier. Front Genet 5:157. doi: 10.3389/fgene.2014.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li YL, Weng JC, Hsiao CC, Chou MT, Tseng CW, Hung JH. 2015. PEAT: an intelligent and efficient paired-end sequencing adapter trimming algorithm. BMC Bioinformatics 16(Suppl 1):S2. doi: 10.1186/1471-2105-16-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM. 2013. An extensive evaluation of read trimming effects on Illumina NGS data analysis. PLoS One 8:e85024. doi: 10.1371/journal.pone.0085024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yun S, Yun S. 2014. Masking as an effective quality control method for next-generation sequencing data analysis. BMC Bioinformatics 15:382. doi: 10.1186/s12859-014-0382-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Q, Guo Y, Li J, Long J, Zhang B, Shyr Y. 2012. Steps to ensure accuracy in genotype and SNP calling from Illumina sequencing data. BMC Genomics 13(Suppl 8):S8. doi: 10.1186/1471-2164-13-S8-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen T, Gan R, Chang Y, Liao W, Wu TH, Lee C, Huang P, Lee C, Chen YM, Chiu C, Tang P. 2015. Is the whole greater than the sum of its parts? De novo assembly strategies for bacterial genomes based on paired-end sequencing. BMC Genomics 16:648. doi: 10.3389/fgene.2014.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 67.Mukherjee S, Huntemann M, Ivanova N, Kyrpides NC, Pati A. 2015. Large-scale contamination of microbial isolate genomes by Illumina PhiX control. Stand Genomic Sci 10:18. doi: 10.1186/1944-3277-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmieder R, Edwards R. 2011. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS One 6:e17288. doi: 10.1371/journal.pone.0017288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hadfield J, Eldridge MD. 2014. Multi-genome alignment for quality control and contamination screening of next-generation sequencing data. Front Genet 5:31. doi: 10.3389/fgene.2014.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou Q, Su X, Wang A, Xu J, Ning K. 2013. QC-chain: fast and holistic quality control method for next-generation sequencing data. PLoS One 8:e60234. doi: 10.1371/journal.pone.0060234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McNair K, Edwards RA. 2015. GenomePeek—an online tool for prokaryotic genome and metagenome analysis. PeerJ 3:e1025. doi: 10.7717/peerj.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, 1000 Genomes Project Analysis Group. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pabinger S, Dander A, Fischer M, Snajder R, Sperk M, Efremova M, Krabichler B, Speicher MR, Zschocke J, Trajanoski Z. 2014. A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform 15:256–278. doi: 10.1093/bib/bbs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Caboche S, Audebert C, Lemoine Y, Hot D. 2014. Comparison of mapping algorithms used in high-throughput sequencing: application to Ion Torrent data. BMC Genomics 15:264. doi: 10.1186/1471-2164-15-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H. 2014. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30:2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Treangen TJ, Salzberg SL. 2011. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet 13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Periwal V, Scaria V. 2015. Insights into structural variations and genome rearrangements in prokaryotic genomes. Bioinformatics 31:1–9. doi: 10.1093/bioinformatics/btu600. [DOI] [PubMed] [Google Scholar]
- 81.Wendl MC, Wilson RK. 2009. Statistical aspects of discerning indel-type structural variation via DNA sequence alignment. BMC Genomics 10:359. doi: 10.1186/1471-2164-10-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fonseca NA, Rung J, Brazma A, Marioni JC. 2012. Tools for mapping high-throughput sequencing data. Bioinformatics 28:3169–3177. doi: 10.1093/bioinformatics/bts605. [DOI] [PubMed] [Google Scholar]
- 83.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, Shi X, Fulton RS, Ley TJ, Wilson RK, Ding L, Mardis ER. 2009. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods 6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Olson ND, Lund SP, Colman RE, Foster JT, Sahl JW, Schupp JM, Keim P, Morrow JB, Salit ML, Zook JM. 2015. Best practices for evaluating single nucleotide variant calling methods for microbial genomics. Front Genet 6:235. doi: 10.3389/fgene.2015.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reumerman RA, Tucker NP, Herron PR, Hoskisson PA, Sangal V. 2013. Tool for rapid annotation of microbial SNPs (TRAMS): a simple program for rapid annotation of genomic variation in prokaryotes. Antonie Van Leeuwenhoek 104:431–434. doi: 10.1007/s10482-013-9953-x. [DOI] [PubMed] [Google Scholar]
- 90.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Simpson JT, Pop M. 2015. The theory and practice of genome sequence assembly. Annu Rev Genomics Hum Genet 16:153–172. doi: 10.1146/annurev-genom-090314-050032. [DOI] [PubMed] [Google Scholar]
- 92.Miller JR, Koren S, Sutton G. 2010. Assembly algorithms for next-generation sequencing data. Genomics 95:315–327. doi: 10.1016/j.ygeno.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ekblom R, Wolf JB. 2014. A field guide to whole-genome sequencing, assembly and annotation. Evol Appl 7:1026–1042. doi: 10.1111/eva.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nagarajan N, Pop M. 2013. Sequence assembly demystified. Nat Rev Genet 14:157–167. doi: 10.1038/nrg3367. [DOI] [PubMed] [Google Scholar]
- 95.Sutton GG, White O, Adams MD, Kerlavage AR. 1995. TIGR assembler: a new tool for assembling large shotgun sequencing projects. Genome Sci Technol 1:9–19. doi: 10.1089/gst.1995.1.9. [DOI] [Google Scholar]
- 96.Treangen TJ, Sommer DD, Angly FE, Koren S, Pop M. 2011. Next generation sequence assembly with AMOS. Curr Protoc Bioinformatics Chapter 11:Unit 11.8. doi: 10.1002/0471250953.bi1108s33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chaisson MJ, Pevzner PA. 2008. Short read fragment assembly of bacterial genomes. Genome Res 18:324–330. doi: 10.1101/gr.7088808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Simpson JT, Durbin R. 2012. Efficient de novo assembly of large genomes using compressed data structures. Genome Res 22:549–556. doi: 10.1101/gr.126953.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hernandez D, Francois P, Farinelli L, Osteras M, Schrenzel J. 2008. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res 18:802–809. doi: 10.1101/gr.072033.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 102.Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chitsaz H, Yee-Greenbaum JL, Tesler G, Lombardo MJ, Dupont CL, Badger JH, Novotny M, Rusch DB, Fraser LJ, Gormley NA, Schulz-Trieglaff O, Smith GP, Evers DJ, Pevzner PA, Lasken RS. 2011. Efficient de novo assembly of single-cell bacterial genomes from short-read data sets. Nat Biotechnol 29:915–921. doi: 10.1038/nbt.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang W, Chen J, Yang Y, Tang Y, Shang J, Shen B. 2011. A practical comparison of de novo genome assembly software tools for next-generation sequencing technologies. PLoS One 6:e17915. doi: 10.1371/journal.pone.0017915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Earl D, Bradnam K, St. John J, Darling A, Lin D, Fass J, Yu HO, Buffalo V, Zerbino DR, Diekhans M, Nguyen N, Ariyaratne PN, Sung WK, Ning Z, Haimel M, Simpson JT, Fonseca NA, Birol I, Docking TR, Ho IY, Rokhsar DS, Chikhi R, Lavenier D, Chapuis G, Naquin D, Maillet N, Schatz MC, Kelley DR, Phillippy AM, Koren S, Yang SP, Wu W, Chou WC, Srivastava A, Shaw TI, Ruby JG, Skewes-Cox P, Betegon M, Dimon MT, Solovyev V, Seledtsov I, Kosarev P, Vorobyev D, Ramirez-Gonzalez R, Leggett R, MacLean D, Xia F, Luo R, Li Z, Xie Y, Liu B, Gnerre S, MacCallum I, Przybylski D, Ribeiro FJ, Yin S, Sharpe T, Hall G, Kersey PJ, Durbin R, Jackman SD, Chapman JA, Huang X, DeRisi JL, Caccamo M, Li Y, Jaffe DB, Green RE, Haussler D, Korf I, Paten B. 2011. Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome Res 21:2224–2241. doi: 10.1101/gr.126599.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bradnam KR, Fass JN, Alexandrov A, Baranay P, Bechner M, Birol I, Boisvert S, Chapman JA, Chapuis G, Chikhi R, Chitsaz H, Chou WC, Corbeil J, Del Fabbro C, Docking TR, Durbin R, Earl D, Emrich S, Fedotov P, Fonseca NA, Ganapathy G, Gibbs RA, Gnerre S, Godzaridis E, Goldstein S, Haimel M, Hall G, Haussler D, Hiatt JB, Ho IY, Howard J, Hunt M, Jackman SD, Jaffe DB, Jarvis ED, Jiang H, Kazakov S, Kersey PJ, Kitzman JO, Knight JR, Koren S, Lam TW, Lavenier D, Laviolette F, Li Y, Li Z, Liu B, Liu Y, Luo R, Maccallum I, Macmanes MD, Maillet N, Melnikov S, Naquin D, Ning Z, Otto TD, Paten B, Paulo OS, Phillippy AM, Pina-Martins F, Place M, Przybylski D, Qin X, Qu C, Ribeiro FJ, Richards S, Rokhsar DS, Ruby JG, Scalabrin S, Schatz MC, Schwartz DC, Sergushichev A, Sharpe T, Shaw TI, Shendure J, Shi Y, Simpson JT, Song H, Tsarev F, Vezzi F, Vicedomini R, Vieira BM, Wang J, Worley KC, Yin S, Yiu SM, Yuan J, Zhang G, Zhang H, Zhou S, Korf IF. 2013. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience 2:10. doi: 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Salzberg SL, Phillippy AM, Zimin A, Puiu D, Magoc T, Koren S, Treangen TJ, Schatz MC, Delcher AL, Roberts M, Mar̃A§ais G, Pop M, Yorke JA. 2012. GAGE: a critical evaluation of genome assemblies and assembly algorithms. Genome Res 22:557–567. doi: 10.1101/gr.131383.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Magoc T, Pabinger S, Canzar S, Liu X, Su Q, Puiu D, Tallon LJ, Salzberg SL. 2013. GAGE-B: an evaluation of genome assemblers for bacterial organisms. Bioinformatics 29:1718–1725. doi: 10.1093/bioinformatics/btt273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Phillippy AM, Schatz MC, Pop M. 2008. Genome assembly forensics: finding the elusive mis-assembly. Genome Biol 9:R55. doi: 10.1186/gb-2008-9-3-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schatz MC, Phillippy AM, Sommer DD, Delcher AL, Puiu D, Narzisi G, Salzberg SL, Pop M. 2013. Hawkeye and AMOS: visualizing and assessing the quality of genome assemblies. Brief Bioinform 14:213–224. doi: 10.1093/bib/bbr074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hunt M, Kikuchi T, Sanders M, Newbold C, Berriman M, Otto TD. 2013. REAPR: a universal tool for genome assembly evaluation. Genome Biol 14:R47. doi: 10.1186/gb-2013-14-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Richardson EJ, Watson M. 2013. The automatic annotation of bacterial genomes. Brief Bioinform 14:1–12. doi: 10.1093/bib/bbs007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Soh S, Gordon P, Sensen C. 2012. Genome annotation. Chapman and Hall/CRC, Boca Raton, FL. [Google Scholar]
- 116.Van Domselaar GH, Stothard P, Shrivastava S, Cruz JA, Guo A, Dong X, Lu P, Szafron D, Greiner R, Wishart DS. 2005. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res 33:W455–W459. doi: 10.1093/nar/gki593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Frishman D, Mironov A, Mewes HW, Gelfand M. 1998. Combining diverse evidence for gene recognition in completely sequenced bacterial genomes. Nucleic Acids Res 26:2941–2947. doi: 10.1093/nar/26.12.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Badger JH, Olsen GJ. 1999. CRITICA: coding region identification tool invoking comparative analysis. Mol Biol Evol 16:512–524. doi: 10.1093/oxfordjournals.molbev.a026133. [DOI] [PubMed] [Google Scholar]
- 119.Lukashin AV, Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Van Domselaar G, Graham MR, Stothard PM. 2014. Prokaryotic genome annotation, p 25–49. In Bishop TO. (ed), Bioinformatics and data analysis in microbiology, 1st ed Horizon Press, Poole, United Kingdom. [Google Scholar]
- 123.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nawrocki EP, Kolbe DL, Eddy SR. 2009. Infernal 1.0: inference of RNA alignments. Bioinformatics 25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pedruzzi I, Rivoire C, Auchincloss AH, Coudert E, Keller G, de Castro E, Baratin D, Cuche BA, Bougueleret L, Poux S, Redaschi N, Xenarios I, Bridge A, UniProt Consortium . 2013. HAMAP in 2013, new developments in the protein family classification and annotation system. Nucleic Acids Res 41:D584–D589. doi: 10.1093/nar/gks1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kulikova T, Aldebert P, Althorpe N, Baker W, Bates K, Browne P, van den Broek A, Cochrane G, Duggan K, Eberhardt R, Faruque N, Garcia-Pastor M, Harte N, Kanz C, Leinonen R, Lin Q, Lombard V, Lopez R, Mancuso R, McHale M, Nardone F, Silventoinen V, Stoehr P, Stoesser G, Tuli MA, Tzouvara K, Vaughan R, Wu D, Zhu W, Apweiler R. 2004. The EMBL nucleotide sequence database. Nucleic Acids Res 32:D27–D30. doi: 10.1093/nar/gkh120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Skovgaard M, Jensen LJ, Brunak S, Ussery D, Krogh A. 2001. On the total number of genes and their length distribution in complete microbial genomes. Trends Genet 17:425–428. doi: 10.1016/S0168-9525(01)02372-1. [DOI] [PubMed] [Google Scholar]
- 129.Pruitt KD, Tatusova T, Maglott DR. 2005. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 33:D501–D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Haft DH, Selengut JD, White O. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res 31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Meyer F, Overbeek R, Rodriguez A. 2009. FIGfams: yet another set of protein families. Nucleic Acids Res 37:6643–6654. doi: 10.1093/nar/gkp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sigrist CJ, Cerutti L, Hulo N, Gattiker A, Falquet L, Pagni M, Bairoch A, Bucher P. 2002. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform 3:265–274. doi: 10.1093/bib/3.3.265. [DOI] [PubMed] [Google Scholar]
- 134.Gattiker A, Gasteiger E, Bairoch A. 2002. ScanProsite: a reference implementation of a PROSITE scanning tool. Appl Bioinformatics 1:107–108. [PubMed] [Google Scholar]
- 135.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 136.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gaasterland T, Sensen CW. 1996. MAGPIE: automated genome interpretation. Trends Genet 12:76–78. doi: 10.1016/0168-9525(96)81406-5. [DOI] [PubMed] [Google Scholar]
- 138.Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J, Kalinowski J, Linke B, Rupp O, Giegerich R, Puhler A. 2003. GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res 31:2187–2195. doi: 10.1093/nar/gkg312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Vallenet D, Labarre L, Rouy Z, Barbe V, Bocs S, Cruveiller S, Lajus A, Pascal G, Scarpelli C, Medigue C. 2006. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res 34:53–65. doi: 10.1093/nar/gkj406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Henson J, Tischler G, Ning Z. 2012. Next-generation sequencing and large genome assemblies. Pharmacogenomics 13:901–915. doi: 10.2217/pgs.12.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. 2009. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25:2271–2278. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- 142.Stewart AC, Osborne B, Read TD. 2009. DIYA: a bacterial annotation pipeline for any genomics lab. Bioinformatics 25:962–963. doi: 10.1093/bioinformatics/btp097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 144.Croucher NJ, Finkelstein JA, Pelton SI, Mitchell PK, Lee GM, Parkhill J, Bentley SD, Hanage WP, Lipsitch M. 2013. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet 45:656–663. doi: 10.1038/ng.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Grad YH, Kirkcaldy RD, Trees D, Dordel J, Harris SR, Goldstein E, Weinstock H, Parkhill J, Hanage WP, Bentley S, Lipsitch M. 2014. Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: a retrospective observational study. Lancet Infect Dis 14:220–226. doi: 10.1016/S1473-3099(13)70693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Didelot X, Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Didelot X, Wilson DJ. 2015. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Marttinen P, Hanage WP, Croucher NJ, Connor TR, Harris SR, Bentley SD, Corander J. 2012. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res 40:e6. doi: 10.1093/nar/gkr928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hawkey J, Hamidian M, Wick RR, Edwards DJ, Billman-Jacobe H, Hall RM, Holt KE. 2015. ISMapper: identifying transposase insertion sites in bacterial genomes from short read sequence data. BMC Genomics 16:667. doi: 10.1186/s12864-015-1860-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Varani AM, Siguier P, Gourbeyre E, Charneau V, Chandler M. 2011. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol 12:R30. doi: 10.1186/gb-2011-12-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dhillon BK, Chiu TA, Laird MR, Langille MG, Brinkman FS. 2013. IslandViewer update: improved genomic island discovery and visualization. Nucleic Acids Res 41:W129–W132. doi: 10.1093/nar/gkt394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, Tsai YC, Boitano M, Dayal J, Brooks SY, Schmidt B, Young AC, Thomas JW, Bouffard GG, Blakesley RW, NISC Comparative Sequencing Program, Mullikin JC, Korlach J, Henderson DK, Frank KM, Palmore TN, Segre JA. 2014. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 6:254ra126. doi: 10.1126/scitranslmed.3009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bose M, Barber RD. 2006. Prophage Finder: a prophage loci prediction tool for prokaryotic genome sequences. In Silico Biol 6:223–227. [PubMed] [Google Scholar]
- 156.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Felsenstein J. 1996. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol 266:418–427. doi: 10.1016/S0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 158.Gregory TR. 2008. Understanding evolutionary trees. Evol Educ Outreach 1:121–137. doi: 10.1007/s12052-008-0035-x. [DOI] [Google Scholar]
- 159.Yang Z, Rannala B. 2012. Molecular phylogenetics: Principles and practice. Nat Rev Genet 13:303–314. doi: 10.1038/nrg3186. [DOI] [PubMed] [Google Scholar]
- 160.Baldauf SL. 2003. Phylogeny for the faint of heart: a tutorial. Trends Genet 19:345–351. doi: 10.1016/S0168-9525(03)00112-4. [DOI] [PubMed] [Google Scholar]
- 161.Croucher NJ, Harris SR, Grad YH, Hanage WP. 2013. Bacterial genomes in epidemiology—present and future. Philos Trans R Soc Lond B Biol Sci 368:20120202. doi: 10.1098/rstb.2012.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chan CX, Ragan MA. 2013. Next-generation phylogenomics. Biol Direct 8:3. doi: 10.1186/1745-6150-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dewey CN. 2012. Whole-genome alignment, p 237–257. In Anisimova M. (ed), Evolutionary genomics: statistical and computational methods, vol 1 Humana Press, Totowa, NJ. [Google Scholar]
- 164.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gadagkar SR, Rosenberg MS, Kumar S. 2005. Inferring species phylogenies from multiple genes: concatenated sequence tree versus consensus gene tree. J Exp Zool B Mol Dev Evol 304:64–74. [DOI] [PubMed] [Google Scholar]
- 168.Altenhoff AM, Dessimoz C. 2012. Inferring orthology and paralogy. Methods Mol Biol 855:259–279. doi: 10.1007/978-1-61779-582-4_9. [DOI] [PubMed] [Google Scholar]
- 169.Li L, Stoeckert CJ Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Leache AD, Banbury BL, Felsenstein J, de Oca AN, Stamatakis A. 2015. Short tree, long tree, right tree, wrong tree: new acquisition bias corrections for inferring SNP phylogenies. Syst Biol 64:1032–1047. doi: 10.1093/sysbio/syv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kuhner MK, Beerli P, Yamato J, Felsenstein J. 2000. Usefulness of single nucleotide polymorphism data for estimating population parameters. Genetics 156:439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Davis S, Pettengill JB, Luo Y, Payne J, Shpuntoff A, Rand H, Strain E. 2015. CFSAN SNP pipeline: an automated method for constructing SNP matrices from next-generation sequence data. PeerJ Comput Sci 1:e20. doi: 10.7717/peerj-cs.20. [DOI] [Google Scholar]
- 175.Jackson BR, Tarr C, Strain E, Jackson KA, Conrad A, Carleton H, Katz LS, Stroika S, Gould LH, Mody RK, Silk BJ, Beal J, Chen Y, Timme R, Doyle M, Fields A, Wise M, Tillman G, Defibaugh-Chavez S, Kucerova Z, Sabol A, Roache K, Trees E, Simmons M, Wasilenko J, Kubota K, Pouseele H, Klimke W, Besser J, Brown E, Allard M, Gerner-Smidt P. 2016. Implementation of nationwide real-time whole-genome sequencing to enhance listeriosis outbreak detection and investigation. Clin Infect Dis 63:380–386. doi: 10.1093/cid/ciw242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bertels F, Silander OK, Pachkov M, Rainey PB, van Nimwegen E. 2014. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol Biol Evol 31:1077–1088. doi: 10.1093/molbev/msu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pettengill JB, Luo Y, Davis S, Chen Y, Gonzalez-Escalona N, Ottesen A, Rand H, Allard MW, Strain E. 2014. An evaluation of alternative methods for constructing phylogenies from whole genome sequence data: a case study with Salmonella. PeerJ 2:e620. doi: 10.7717/peerj.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gardner SN, Hall BG. 2013. When whole-genome alignments just won't work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One 8:e81760. doi: 10.1371/journal.pone.0081760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gardner SN, Slezak T, Hall BG. 2015. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- 180.Schwartz RS, Harkins KM, Stone AC, Cartwright RA. 2015. A composite genome approach to identify phylogenetically informative data from next-generation sequencing. BMC Bioinformatics 16:193. doi: 10.1186/s12859-015-0632-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Henz SR, Huson DH, Auch AF, Nieselt-Struwe K, Schuster SC. 2005. Whole-genome prokaryotic phylogeny. Bioinformatics 21:2329–2335. doi: 10.1093/bioinformatics/bth324. [DOI] [PubMed] [Google Scholar]
- 182.Sims GE, Jun SR, Wu GA, Kim SH. 2009. Alignment-free genome comparison with feature frequency profiles (FFP) and optimal resolutions. Proc Natl Acad Sci U S A 106:2677–2682. doi: 10.1073/pnas.0813249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sims GE, Kim SH. 2011. Whole-genome phylogeny of Escherichia coli/Shigella group by feature frequency profiles (FFPs). Proc Natl Acad Sci U S A 108:8329–8334. doi: 10.1073/pnas.1105168108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Haubold B, Klotzl F, Pfaffelhuber P. 2015. Andi: fast and accurate estimation of evolutionary distances between closely related genomes. Bioinformatics 31:1169–1175. doi: 10.1093/bioinformatics/btu815. [DOI] [PubMed] [Google Scholar]
- 185.Haubold B. 2014. Alignment-free phylogenetics and population genetics. Brief Bioinform 15:407–418. doi: 10.1093/bib/bbt083. [DOI] [PubMed] [Google Scholar]
- 186.Bonham-Carter O, Steele J, Bastola D. 2014. Alignment-free genetic sequence comparisons: a review of recent approaches by word analysis. Brief Bioinform 15:890–905. doi: 10.1093/bib/bbt052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Maiden MC, Jansen van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jolley KA, Bliss CM, Bennett JS, Bratcher HB, Brehony C, Colles FM, Wimalarathna H, Harrison OB, Sheppard SK, Cody AJ, Maiden MC. 2012. Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiology 158:1005–1015. doi: 10.1099/mic.0.055459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cody AJ, McCarthy ND, Jansen van Rensburg M, Isinkaye T, Bentley SD, Parkhill J, Dingle KE, Bowler IC, Jolley KA, Maiden MC. 2013. Real-time genomic epidemiological evaluation of human campylobacter isolates by use of whole-genome multilocus sequence typing. J Clin Microbiol 51:2526–2534. doi: 10.1128/JCM.00066-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bryant D, Moulton V. 2004. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol 21:255–265. [DOI] [PubMed] [Google Scholar]
- 191.Huson DH. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 192.Francisco AP, Vaz C, Monteiro PT, Melo-Cristino J, Ramirez M, Carrico JA. 2012. PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics 13:87. doi: 10.1186/1471-2105-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kohl TA, Diel R, Harmsen D, Rothganger J, Walter KM, Merker M, Weniger T, Niemann S. 2014. Whole-genome-based Mycobacterium tuberculosis surveillance: a standardized, portable, and expandable approach. J Clin Microbiol 52:2479–2486. doi: 10.1128/JCM.00567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bratcher HB, Corton C, Jolley KA, Parkhill J, Maiden MC. 2014. A gene-by-gene population genomics platform: de novo assembly, annotation and genealogical analysis of 108 representative Neisseria meningitidis genomes. BMC Genomics 15:1138. doi: 10.1186/1471-2164-15-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ruppitsch W, Pietzka A, Prior K, Bletz S, Fernandez HL, Allerberger F, Harmsen D, Mellmann A. 2015. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of listeria monocytogenes. J Clin Microbiol 53:2869–2876. doi: 10.1128/JCM.01193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Katz LS, Petkau A, Beaulaurier J, Tyler S, Antonova ES, Turnsek MA, Guo Y, Wang S, Paxinos EE, Orata F, Gladney LM, Stroika S, Folster JP, Rowe L, Freeman MM, Knox N, Frace M, Boncy J, Graham M, Hammer BK, Boucher Y, Bashir A, Hanage WP, Van Domselaar G, Tarr CL. 2013. Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. mBio 4:e00398-13. doi: 10.1128/mBio.00398-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hendriksen RS, Price LB, Schupp JM, Gillece JD, Kaas RS, Engelthaler DM, Bortolaia V, Pearson T, Waters AE, Upadhyay BP, Shrestha SD, Adhikari S, Shakya G, Keim PS, Aarestrup FM. 2011. Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak. mBio 2:e00157-11. doi: 10.1128/mBio.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VP, Nash JH, Taboada EN. 2016. The Salmonella in silico typing resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 202.Cosentino S, Voldby Larsen M, Moller Aarestrup F, Lund O. 2013. PathogenFinder—distinguishing friend from foe using bacterial whole genome sequence data. PLoS One 8:e77302. doi: 10.1371/journal.pone.0077302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O'Brien JS, Pawlowski AC, Piddock LJ, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lambert D, Carrillo CD, Koziol AG, Manninger P, Blais BW. 2015. GeneSippr: a rapid whole-genome approach for the identification and characterization of foodborne pathogens such as priority Shiga toxigenic Escherichia coli. PLoS One 10:e0122928. doi: 10.1371/journal.pone.0122928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Frenay HM, Bunschoten AE, Schouls LM, van Leeuwen WJ, Vandenbroucke-Grauls CM, Verhoef J, Mooi FR. 1996. Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur J Clin Microbiol Infect Dis 15:60–64. doi: 10.1007/BF01586186. [DOI] [PubMed] [Google Scholar]
- 207.Martin IM, Ison CA, Aanensen DM, Fenton KA, Spratt BG. 2004. Rapid sequence-based identification of gonococcal transmission clusters in a large metropolitan area. J Infect Dis 189:1497–1505. doi: 10.1086/383047. [DOI] [PubMed] [Google Scholar]
- 208.Huber CA, Foster NF, Riley TV, Paterson DL. 2013. Challenges for standardization of Clostridium difficile typing methods. J Clin Microbiol 51:2810–2814. doi: 10.1128/JCM.00143-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Harris SR, Clarke IN, Seth-Smith HM, Solomon AW, Cutcliffe LT, Marsh P, Skilton RJ, Holland MJ, Mabey D, Peeling RW, Lewis DA, Spratt BG, Unemo M, Persson K, Bjartling C, Brunham R, de Vries HJ, Morre SA, Speksnijder A, Bebear CM, Clerc M, de Barbeyrac B, Parkhill J, Thomson NR. 2012. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat Genet 44:413–419. doi: 10.1038/ng.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Moran-Gilad J, Prior K, Yakunin E, Harrison TG, Underwood A, Lazarovitch T, Valinsky L, Luck C, Krux F, Agmon V, Grotto I, Harmsen D. 2015. Design and application of a core genome multilocus sequence typing scheme for investigation of Legionnaires' disease incidents. Euro Surveill 20:21186. doi: 10.2807/1560-7917.ES2015.20.28.21186. [DOI] [PubMed] [Google Scholar]
- 211.Frerichs RR, Keim PS, Barrais R, Piarroux R. 2012. Nepalese origin of cholera epidemic in Haiti. Clin Microbiol Infect 18:E158–E163. doi: 10.1111/j.1469-0691.2012.03841.x. [DOI] [PubMed] [Google Scholar]
- 212.Grad YH, Lipsitch M, Feldgarden M, Arachchi HM, Cerqueira GC, Fitzgerald M, Godfrey P, Haas BJ, Murphy CI, Russ C, Sykes S, Walker BJ, Wortman JR, Young S, Zeng Q, Abouelleil A, Bochicchio J, Chauvin S, Desmet T, Gujja S, McCowan C, Montmayeur A, Steelman S, Frimodt-Moller J, Petersen AM, Struve C, Krogfelt KA, Bingen E, Weill FX, Lander ES, Nusbaum C, Birren BW, Hung DT, Hanage WP. 2012. Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011. Proc Natl Acad Sci U S A 109:3065–3070. doi: 10.1073/pnas.1121491109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wright MS, Haft DH, Harkins DM, Perez F, Hujer KM, Bajaksouzian S, Benard MF, Jacobs MR, Bonomo RA, Adams MD. 2014. New insights into dissemination and variation of the health care-associated pathogen Acinetobacter baumannii from genomic analysis. mBio 5:e00963-13. doi: 10.1128/mBio.00963-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lynch T, Chen L, Peirano G, Gregson DB, Church DL, Conly J, Kreiswirth BN, Pitout JD. 2016. Molecular evolution of a Klebsiella pneumoniae ST278 isolate harboring blaNDM-7 and involved in nosocomial transmission. J Infect Dis doi: 10.1093/infdis/jiw240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Cairns MD, Preston MD, Lawley TD, Clark TG, Stabler RA, Wren BW. 2015. Genomic epidemiology of a protracted hospital outbreak caused by a toxin A-negative Clostridium difficile sublineage PCR ribotype 017 strain in London, England. J Clin Microbiol 53:3141–3147. doi: 10.1128/JCM.00648-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Stoesser N, Giess A, Batty EM, Sheppard AE, Walker AS, Wilson DJ, Didelot X, Bashir A, Sebra R, Kasarskis A, Sthapit B, Shakya M, Kelly D, Pollard AJ, Peto TE, Crook DW, Donnelly P, Thorson S, Amatya P, Joshi S. 2014. Genome sequencing of an extended series of NDM-producing Klebsiella pneumoniae isolates from neonatal infections in a Nepali hospital characterizes the extent of community- versus hospital-associated transmission in an endemic setting. Antimicrob Agents Chemother 58:7347–7357. doi: 10.1128/AAC.03900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Perna NT, Plunkett G III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 218.Sturdevant GL, Kari L, Gardner DJ, Olivares-Zavaleta N, Randall LB, Whitmire WM, Carlson JH, Goheen MM, Selleck EM, Martens C, Caldwell HD. 2010. Frameshift mutations in a single novel virulence factor alter the in vivo pathogenicity of Chlamydia trachomatis for the female murine genital tract. Infect Immun 78:3660–3668. doi: 10.1128/IAI.00386-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Roetzer A, Diel R, Kohl TA, Ruckert C, Nubel U, Blom J, Wirth T, Jaenicke S, Schuback S, Rusch-Gerdes S, Supply P, Kalinowski J, Niemann S. 2013. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med 10:e1001387. doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Bonner C, Caldwell HD, Carlson JH, Graham MR, Kari L, Sturdevant GL, Tyler S, Zetner A, McClarty G. 2015. Chlamydia trachomatis virulence factor CT135 is stable in vivo but highly polymorphic in vitro. Pathog Dis 73:ftv043. doi: 10.1093/femspd/ftv043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lieberman TD, Michel JB, Aingaran M, Potter-Bynoe G, Roux D, Davis MR Jr, Skurnik D, Leiby N, LiPuma JJ, Goldberg JB, McAdam AJ, Priebe GP, Kishony R. 2011. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet 43:1275–1280. doi: 10.1038/ng.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Croucher NJ, Kagedan L, Thompson CM, Parkhill J, Bentley SD, Finkelstein JA, Lipsitch M, Hanage WP. 2015. Selective and genetic constraints on pneumococcal serotype switching. PLoS Genet 11:e1005095. doi: 10.1371/journal.pgen.1005095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sanchez-Buso L, Comas I, Jorques G, Gonzalez-Candelas F. 2014. Recombination drives genome evolution in outbreak-related Legionella pneumophila isolates. Nat Genet 46:1205–1211. doi: 10.1038/ng.3114. [DOI] [PubMed] [Google Scholar]
- 224.Chen L, Zheng D, Liu B, Yang J, Jin Q. 2016. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res 44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Zhou CE, Smith J, Lam M, Zemla A, Dyer MD, Slezak T. 2007. MvirDB—a microbial database of protein toxins, virulence factors and antibiotic resistance genes for bio-defence applications. Nucleic Acids Res 35:D391–D394. doi: 10.1093/nar/gkl791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Mao C, Abraham D, Wattam AR, Wilson MJ, Shukla M, Yoo HS, Sobral BW. 2015. Curation, integration and visualization of bacterial virulence factors in PATRIC. Bioinformatics 31:252–258. doi: 10.1093/bioinformatics/btu631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Meric G, Yahara K, Mageiros L, Pascoe B, Maiden MC, Jolley KA, Sheppard SK. 2014. A reference pan-genome approach to comparative bacterial genomics: identification of novel epidemiological markers in pathogenic campylobacter. PLoS One 9:e92798. doi: 10.1371/journal.pone.0092798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ren CP, Chaudhuri RR, Fivian A, Bailey CM, Antonio M, Barnes WM, Pallen MJ. 2004. The ETT2 gene cluster, encoding a second type III secretion system from Escherichia coli, is present in the majority of strains but has undergone widespread mutational attrition. J Bacteriol 186:3547–3560. doi: 10.1128/JB.186.11.3547-3560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, Sebaihia M, Baker S, Basham D, Brooks K, Chillingworth T, Connerton P, Cronin A, Davis P, Davies RM, Dowd L, White N, Farrar J, Feltwell T, Hamlin N, Haque A, Hien TT, Holroyd S, Jagels K, Krogh A, Larsen TS, Leather S, Moule S, O'Gaora P, Parry C, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar typhi CT18. Nature 413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 230.Holt KE, Thomson NR, Wain J, Langridge GC, Hasan R, Bhutta ZA, Quail MA, Norbertczak H, Walker D, Simmonds M, White B, Bason N, Mungall K, Dougan G, Parkhill J. 2009. Pseudogene accumulation in the evolutionary histories of Salmonella enterica serovars Paratyphi A and Typhi. BMC Genomics 10:36. doi: 10.1186/1471-2164-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ji C, Huang A, Liu W, Pan G, Wang G. 2013. Identification and bioinformatics analysis of pseudogenes from whole genome sequence of Phaeodactylum tricornutum. Chin Sci Bull 58:1010–1017. doi: 10.1007/s11434-012-5174-3. [DOI] [Google Scholar]
- 232.Dutilh BE, Backus L, Edwards RA, Wels M, Bayjanov JR, van Hijum SA. 2013. Explaining microbial phenotypes on a genomic scale: GWAS for microbes. Brief Funct Genomics 12:366–380. doi: 10.1093/bfgp/elt008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Bayjanov JR, Molenaar D, Tzeneva V, Siezen RJ, van Hijum SA. 2012. PhenoLink—a web-tool for linking phenotype to ∼omics data for bacteria: application to gene-trait matching for Lactobacillus plantarum strains. BMC Genomics 13:170. doi: 10.1186/1471-2164-13-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Marinier E, Berry C, Weedmark KA, Domaratzki M, Mabon P, Knox NC, Reimer AR, Graham MR, Van Domselaar G. 2015. Neptune: a tool for rapid genomic signature discovery. bioRxiv doi: 10.1101/032227. [DOI] [Google Scholar]
- 235.Field N, Cohen T, Struelens MJ, Palm D, Cookson B, Glynn JR, Gallo V, Ramsay M, Sonnenberg P, Maccannell D, Charlett A, Egger M, Green J, Vineis P, Abubakar I. 2014. Strengthening the reporting of molecular epidemiology for infectious diseases (STROME-ID): an extension of the STROBE statement. Lancet Infect Dis 14:341–352. doi: 10.1016/S1473-3099(13)70324-4. [DOI] [PubMed] [Google Scholar]
- 236.Aarestrup FM, Koopmans MG. 2016. Sharing data for global infectious disease surveillance and outbreak detection. Trends Microbiol 24:241–245. doi: 10.1016/j.tim.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 237.Yozwiak NL, Schaffner SF, Sabeti PC. 2015. Data sharing: make outbreak research open access. Nature 518:477–479. doi: 10.1038/518477a. [DOI] [PubMed] [Google Scholar]
- 238.Rohde H, Qin J, Cui Y, Li D, Loman NJ, Hentschke M, Chen W, Pu F, Peng Y, Li J, Xi F, Li S, Li Y, Zhang Z, Yang X, Zhao M, Wang P, Guan Y, Cen Z, Zhao X, Christner M, Kobbe R, Loos S, Oh J, Yang L, Danchin A, Gao GF, Song Y, Li Y, Yang H, Wang J, Xu J, Pallen MJ, Wang J, Aepfelbacher M, Yang R, E. coli O104:H4 Genome Analysis Crowd-Sourcing Consortium. 2011. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N Engl J Med 365:718–724. doi: 10.1056/NEJMoa1107643. [DOI] [PubMed] [Google Scholar]
- 239.Trifonov V, Khiabanian H, Greenbaum B, Rabadan R. 2009. The origin of the recent swine influenza A(H1N1) virus infecting humans. Euro Surveill 14:19193. [PubMed] [Google Scholar]
- 240.Reimer AR, Van Domselaar G, Stroika S, Walker M, Kent H, Tarr C, Talkington D, Rowe L, Olsen-Rasmussen M, Frace M, Sammons S, Dahourou GA, Boncy J, Smith AM, Mabon P, Petkau A, Graham M, Gilmour MW, Gerner-Smidt P, V. cholerae Outbreak Genomics Task Force. 2011. Comparative genomics of Vibrio cholerae from Haiti, Asia, and Africa. Emerg Infect Dis 17:2113–2121. doi: 10.3201/eid1711.110794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Orata FD, Keim PS, Boucher Y. 2014. The 2010 cholera outbreak in Haiti: how science solved a controversy. PLoS Pathog 10:e1003967. doi: 10.1371/journal.ppat.1003967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Gire SK, Goba A, Andersen KG, Sealfon RS, Park DJ, Kanneh L, Jalloh S, Momoh M, Fullah M, Dudas G, Wohl S, Moses LM, Yozwiak NL, Winnicki S, Matranga CB, Malboeuf CM, Qu J, Gladden AD, Schaffner SF, Yang X, Jiang PP, Nekoui M, Colubri A, Coomber MR, Fonnie M, Moigboi A, Gbakie M, Kamara FK, Tucker V, Konuwa E, Saffa S, Sellu J, Jalloh AA, Kovoma A, Koninga J, Mustapha I, Kargbo K, Foday M, Yillah M, Kanneh F, Robert W, Massally JL, Chapman SB, Bochicchio J, Murphy C, Nusbaum C, Young S, Birren BW, Grant DS, Scheiffelin JS, Lander ES, Happi C, Gevao SM, Gnirke A, Rambaut A, Garry RF, Khan SH, Sabeti PC. 2014. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Vayena E, Salathe M, Madoff LC, Brownstein JS. 2015. Ethical challenges of big data in public health. PLoS Comput Biol 11:e1003904. doi: 10.1371/journal.pcbi.1003904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Contreras JL, Reichman JH. 2015. Data access. Sharing by design: data and decentralized commons. Science 350:1312–1314. doi: 10.1126/science.aaa7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Mohiyuddin M, Mu JC, Li J, Bani Asadi N, Gerstein MB, Abyzov A, Wong WH, Lam HY. 2015. MetaSV: an accurate and integrative structural-variant caller for next generation sequencing. Bioinformatics 31:2741–2744. doi: 10.1093/bioinformatics/btv204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Milne I, Stephen G, Bayer M, Cock PJ, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform 14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 247.Miller CA, Qiao Y, DiSera T, D'Astous B, Marth GT. 2014. Bam.iobio: a web-based, real-time, sequence alignment file inspector. Nat Methods 11:1189. doi: 10.1038/nmeth.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Oakley T, Alexandrou M, Ngo R, Pankey M, Churchill CK, Chen W, Lopker K. 2014. Osiris: accessible and reproducible phylogenetic and phylogenomic analyses within the Galaxy workflow management system. BMC Bioinformatics 15:230. doi: 10.1186/1471-2105-15-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Zuo G, Hao B. 2015. CVTree3 web server for whole-genome-based and alignment-free prokaryotic phylogeny and taxonomy. Genomics Proteomics Bioinformatics 13:321–331. doi: 10.1016/j.gpb.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]