Summary
Synaptic adhesion molecules regulate synapse development and plasticity through mechanisms including trans-synaptic adhesion and recruitment of diverse synaptic proteins. We report here that the immunoglobulin superfamily member 11 (IgSF11), a homophilic adhesion molecule preferentially expressed in the brain, is a novel and dual-binding partner of the postsynaptic scaffolding protein PSD-95 and AMPAR glutamate receptors (AMPARs). IgSF11 requires PSD-95 binding for its excitatory synaptic localization. In addition, IgSF11 stabilizes synaptic AMPARs, as shown by IgSF11 knockdown-induced suppression of AMPAR-mediated synaptic transmission and increased surface mobility of AMPARs, measured by high-throughput, single-molecule tracking. IgSF11 deletion in mice leads to suppression of AMPAR-mediated synaptic transmission in the dentate gyrus and long-term potentiation in the CA1 region of the hippocampus. IgSF11 does not regulate the functional characteristics of AMPARs, including desensitization, deactivation, or recovery. These results suggest that IgSF11 regulates excitatory synaptic transmission and plasticity through its tripartite interactions with PSD-95 and AMPARs.
Introduction
Synaptic adhesion molecules regulate diverse aspects of synapse development and plasticity 1, 2, 3, 4, 5, 6, 7, 8, 9. Mechanistically, trans-synaptic adhesions are thought to trigger the recruitment of a large number of pre- and postsynaptic cytoplasmic and membrane proteins, contributing to the formation of synapses. Some other synaptic adhesion molecules are thought to be targeted to synapses and contribute to the maintenance of synapses, by interacting with and stabilizing synaptic proteins. Given that postsynaptic receptors play key roles in the establishment, maintenance, and plasticity of synaptic strength, synaptic adhesion molecules are expected to interact with postsynaptic receptors, but the molecular details are largely unclear.
Recent studies have described several interactions between synaptic adhesion molecules and postsynaptic receptors. One example is that between N-cadherin and AMPA receptors (AMPARs) 10, 11, in which N-cadherin recruits AMPARs to excitatory synapses, decreases the surface mobility of AMPARs by immobilizing them at the synapse, and promotes GluA2 (a subunit of AMPARs)-dependent spine morphogenesis and excitatory synaptic transmission 11. N-cadherin also forms a complex with β-catenin and AMPARs in a calcium-dependent manner, and enhances the surface expression of AMPARs 10. Another example is neuroligin-1, which interacts with the GluN1 subunit of NMDA receptors (NMDARs) through extracellular domains 12. In addition, several synaptic adhesion molecules, such as neuroligin-2, NGLs, SALMs, LRRTMs, TrkC, and neuroplastin-65, have been shown to associate with NMDARs, AMPARs, and GABA receptors 1, 2, 3, 4, 5, 6, 7, 8, 9, 13. Although the mechanistic details of these interactions remain to be further clarified, these results suggest that adhesion molecules and postsynaptic receptors may act in concert to regulate diverse steps of synapse development.
IgSF11 (also known as BT-IgSF) was originally identified as a novel member of the immunoglobulin superfamily, and termed BT-IgSF for its preferential expression in the brain and testis 14. IgSF11 mediates homophilic adhesion in a calcium-independent manner 15. In zebrafish, IgSF11 regulates the formation of adult pigment patterns by controlling the migration and survival of melanophores 16. Intriguingly, IgSF11 expression levels are decreased in neurons derived from induced pluripotent stem cells of schizophrenic individuals, which also show decreased neurite number and neuronal connectivity 17. These findings suggest that IgSF11 may regulate aspects of neuronal development and synapse formation.
In the present study, we found that IgSF11 interacts with PSD-95 and AMPARs in a tripartite manner. In addition, knockdown, dominant-negative inhibition, single-molecule tracking, in vivo mouse deletion, and virus rescue experiments provide evidence that IgSF11 is targeted to excitatory synapses through its interaction with PSD-95 and regulates AMPAR-mediated synaptic transmission and plasticity.
Results
IgSF11 interacts with PSD-95
Using a yeast two-hybrid screen, we identified IgSF11 as a novel binding partner of PSD-95. IgSF11, which is conserved among various vertebrate species (Supplementary Fig. 1), contains two extracellular Ig domains, a trans-membrane domain, and a C-terminal PDZ-binding motif (Fig. 1a). IgSF11, along with CAR (coxsackievirus and adenovirus receptor), ESAM (endothelial cell-selective adhesion molecule) and CLMP (coxsackie- and adenovirus receptor-like membrane protein), belongs to the CAR subgroup of the CTX family of Ig-like cell-adhesion molecules (Supplementary Fig. 2a) 18. Data in the Allen Brain Atlas indicate that IgSF11, CAR, ESAM, and CLMP mRNAs are abundantly expressed in various mouse brain regions (Supplementary Fig. 2b).
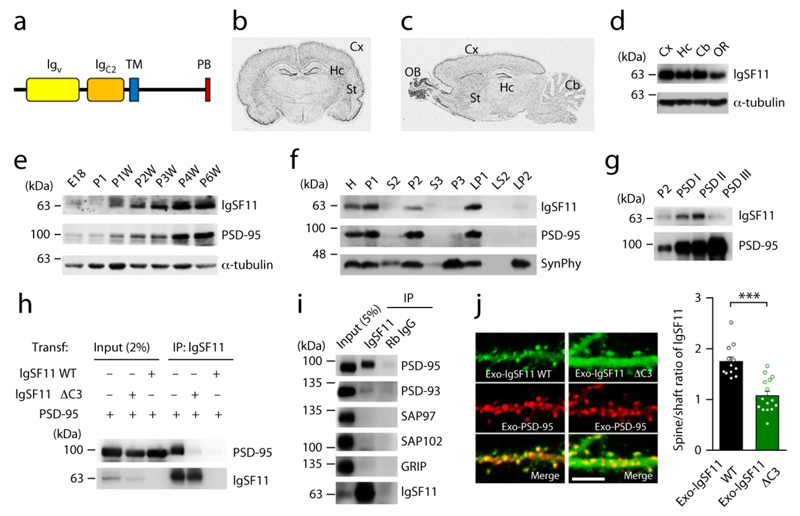
Figure 1. IgSF11 interacts with PSD-95 and is targeted to excitatory synapses in a PDZ interaction-dependent manner.
(a) Domain structure of IgSF11. Igv/C2, immunoglobulin V/C2-like; TM, transmembrane; PB, PDZ-binding.
(b and c) Distribution of IgSF11 mRNA in rat brains (6 weeks) revealed by in situ hybridization. OB, olfactory bulb; Cx, cerebral cortex; St, striatum; Hc, hippocampus; Cb, cerebellar cortex.
(d) Expression of IgSF11 protein in different rat brain regions, by immunoblot analysis. OR, other region.
(e) Expression of IgSF11 protein during rat brain development. E, embryonic; P, postnatal; W, week; PSD-95 and α-tubulin, controls.
(f) Distribution of IgSF11 protein in rat brain fractions. H, homogenates; P1, cells and nuclei-enriched pellet; P2, crude synaptosomes; S2, supernatant after P2 precipitation; S3, cytosol; P3, light membranes; LP1, synaptosomal membranes; LS2, synaptosomal cytosol; LP2, synaptic vesicle-enriched fraction. PSD-95 and synaptophysin (SynPhy; a presynaptic protein) are controls. Three independent experiments were performed.
(g) Detection of IgSF11 in PSD fractions (2 μg of proteins loaded), extracted with Triton X-100 once (PSD I), twice (PSD II), or Triton X-100 and sarcosyl (PSD III). PSD-95 is a control.
(h) IgSF11 forms a complex with PSD-95 in HEK293T cells. IgSF11 ΔC3, a mutant IgSF11 that lacks the last three aa residues and PSD-95 binding; Transf, Transfection; IP, immunoprecipitation. Three independent experiments were performed.
(i) IgSF11 coimmunoprecipitates with PSD-95 in vivo. Deoxycholate (1%) extracts of rat brain crude synaptosomes (6 weeks) were immunoprecipitated and immunoblotted. Gp, ginea pig. Three independent experiments were performed.
(j) IgSF11 is targeted dendritic spines in a PDZ binding-dependent manner. In cultured rat hippocampal neurons transfected with PSD-95 + IgSF11 WT/ΔC3 (DIV14–17), IgSF11 WT shows a greater spine localization (spine/dendritic shaft ratio) than IgSF11 ΔC3. n = 12 (WT) and 15 (ΔC3) neurons, ***p < 0.001, Student’s t-test. Scale bar: 5 μm.
In situ hybridization detected IgSF11 mRNA in various regions of the rat brain, including the olfactory bulb, cortex, striatum, hippocampus and cerebellum (Fig. 1b,c), consistent with previous Northern and in situ hybridization data 14. IgSF11 protein (~52 kDa; 428 amino acids in rats) was detected in several rat brain regions, and their levels steadily increased during brain development (Fig. 1d,e). IgSF11 protein was detected in synaptic fractions, including crude synaptosomal and postsynaptic density fractions (Fig. 1f,g).
The interaction between IgSF11 and PSD-95 involved the C-terminal PDZ-binding motif of IgSF11 and the first two PDZ domains of PSD-95 (Fig. 1h and Supplementary Fig. 3). In the rat brain, IgSF11 formed a complex with PSD-95, but not with other PSD-95 family proteins (PSD-93, SAP97, and SAP102) (Fig. 1i). In cultured hippocampal neurons, a mutant IgSF11 that lacks the PSD-95-interacting C-terminal tail (IgSF11 ΔC3) showed decreased spine localization relative to wild-type IgSF11 (Fig. 1j), suggesting that IgSF11 is targeted to excitatory synapses in a PDZ binding-dependent manner.
IgSF11 lacks synaptogenic activity
Because PSD-95-associated synaptic adhesion molecules often display synaptogenic activities, we tested whether IgSF11 was capable of inducing synaptic protein clustering in contacting axons or dendrites of cocultured neurons. We found that HEK293T cells expressing IgSF11 did not induce clustering of the presynaptic marker synapsin in contacting axons, or the postsynaptic scaffolds PSD-95 (excitatory) and gephyrin (inhibitory) in contacting dendrites (Supplementary Fig. 4a-c). In line with this, IgSF11 lacked interactions with known synaptic adhesion molecules, including neuroligins, LRRTMs, NGLs, SALMs, and ADAM22, except for the homophilic interaction with IgSF11 (Supplementary Fig. 4d).
IgSF11 interacts with AMPARs
IgSF11 binding to PSD-95 could bring IgSF11 close to other PSD-95-associated membrane proteins such as AMPARs. We thus tested if IgSF11 associates with AMPARs. Intriguingly, IgSF11 formed a coimmunoprecipitable complex with the GluA1 and GluA2 subunits of AMPARs, but not with the GluN1 subunit of NMDARs, in rat brains (Fig. 2a). In COS7 cells, IgSF11 also formed complexes with GluA1 and GluA2 (Fig. 2b). Experiments with deletion variants of IgSF11 revealed that the transmembrane domain of IgSF11 is important for the interaction with GluA1 (Supplementary Fig. 5).
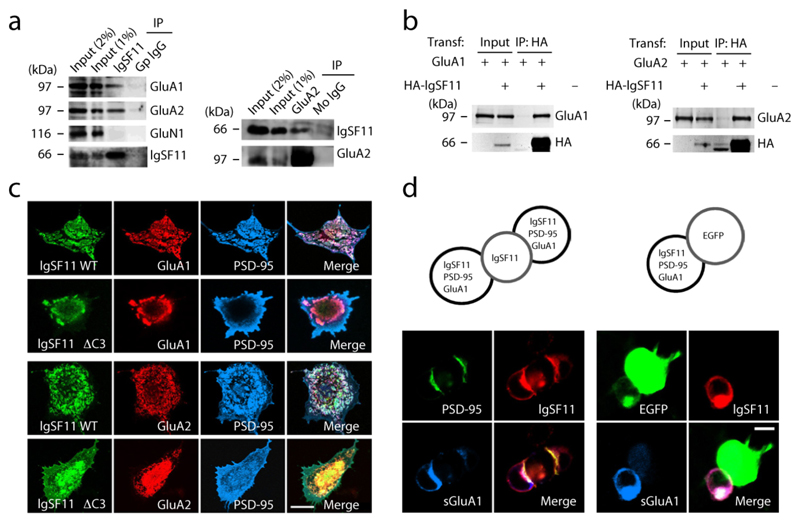
Figure 2. IgSF11 interacts with AMPARs and recruits both PSD-95 and AMPARs to sites of homophilic IgSF11 adhesion.
(a) IgSF11 coimmunoprecipitates with AMPARs, but not with NMDARs, in the rat brain. Deoxycholate (1%) extracts of the crude synaptosomal fraction of adult rat brain (6 weeks) were immunoprecipitated with IgSF11 (top) or GluA2 (bottom) antibodies and immunoblotted. GluA1 and GluA2, AMPAR subunits; GluN1, an NMDAR subunit; Gp, guinea pig; Mo, mouse. The two weak bands appearing in the mouse IgG control lane represent IgG heavy chains. Three independent experiments were performed.
(b) IgSF11 forms a complex with AMPAR subunits in HEK293T cells. HEK293T cells transfected with HA-IgSF11 and GluA1/2, or GluA1/2 alone, were subjected to immunoprecipitation with HA antibodies and immunoblotting. HA-IgSF11, N-terminally HA-tagged IgSF11. Three independent experiments were performed.
(c) IgSF11 colocalizes with PSD-95 and AMPARs in discrete clusters in heterologous cells, whereas IgSF11 ΔC3 fails to colocalize with PSD-95, but retains its colocalization with AMPARs. COS7 cells triply transfected with PSD-95 + IgSF11 (WT/ΔC3) + GluA1/2 were visualized by specific antibodies. Scale bar, 20 μm. Three independent experiments were performed.
(d) GluA1 and PSD-95 are recruited to sites of homophilic IgSF11 adhesion. HEK293T cells triply transfected with IgSF11 (untagged) + PSD-95-EGFP + HA-GluA1 were mixed with the cells singly transfected with IgSF11 or EGFP (control), followed by staining for IgSF11, EGFP, and HA (surface GluA1). Scale bar, 10 μm. Three independent experiments were performed.
In triply transfected heterologous cells, IgSF11 colocalized with both GluA1 and PSD-95 in discrete clusters (Fig. 2c). In contrast, IgSF11 ΔC3 failed to colocalize with PSD-95 but maintained its association with GluA1. Similar results were obtained with the GluA2 subunit. We next found that IgSF11 proteins expressed in two neighboring cells accumulated at sites of cell-to-cell contacts, forming a zipper-like line where GluA1 and PSD-95 co-accumulated (Fig. 2d). These results suggest that IgSF11 forms a tripartite complex with PSD-95 and AMPARs, and recruits them to sites of IgSF11 adhesion on the cell surface.
IgSF11 knockdown suppresses surface AMPAR clustering
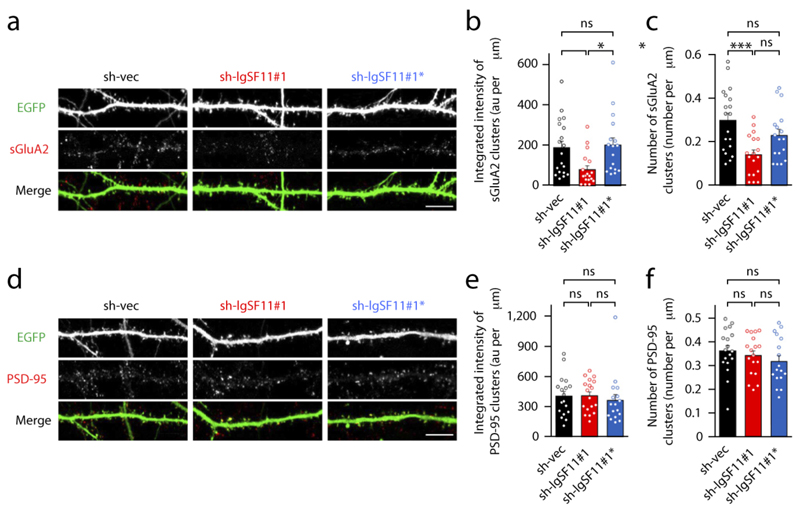
In order to explore IgSF11 function, we acutely knocked down IgSF11 using shRNA. In cultured hippocampal neurons, IgSF11 knockdown (sh-IgSF11#1) reduced the integrated intensity and number of surface GluA2 clusters, whereas a mutant shRNA construct (sh-IgSF11#1*) had no such effect (Fig. 3a-c; Supplementary Fig. 6a). Similar results were obtained for GluA1 (Supplementary Fig. 7a-c). In contrast, sh-IgSF11#1 had no effect on the number or intensity of PSD-95 clusters (Fig. 3d-f) or excitatory synapses (synapsin I-positive PSD-95) (Supplementary Fig. 7d-g). An independent knockdown construct for IgSF11 (sh-IgSF11#2) yielded similar results for surface GluA2 and PSD-95 (Supplementary Figs. 6b and 7h-k).
Figure 3. IgSF11 knockdown reduces surface clustering of AMPARs without affecting PSD-95 clustering.
(a-c) Acute knockdown of IgSF11 decreases surface clustering of GluA2 in cultured neurons. Cultured hippocampal neurons were transfected with sh-vec (empty vector), sh-IgSF11#1, or sh-IgSF11#1* (a mutant incapable of knockdown) (DIV 14–17), and stained for surface GluA2 and EGFP (for transfected neurons). au, arbitrary unit; n = 19 for sh-vec, 18 for sh-IgSF11#1, and 17 for sh-IgSF11#1*, *p < 0.05, ***p < 0.001, ns, not significant, one-way ANOVA. Scale bar: 10 μm.
(d-f) IgSF11 knockdown has no effect on dendritic PSD-95 clusters. Cultured neurons were transfected with sh-vec, sh-IgSF11#1, or sh-IgSF11#1* (DIV14–17), and immunostained for EGFP and PSD-95. n = 18 for sh-vec, 18 for sh-IgSF11#1, and 17 for sh-IgSF11#1*, ns, not significant, one-way ANOVA. Scale bar: 10 μm.
IgSF11 knockdown suppresses AMPAR synaptic transmission
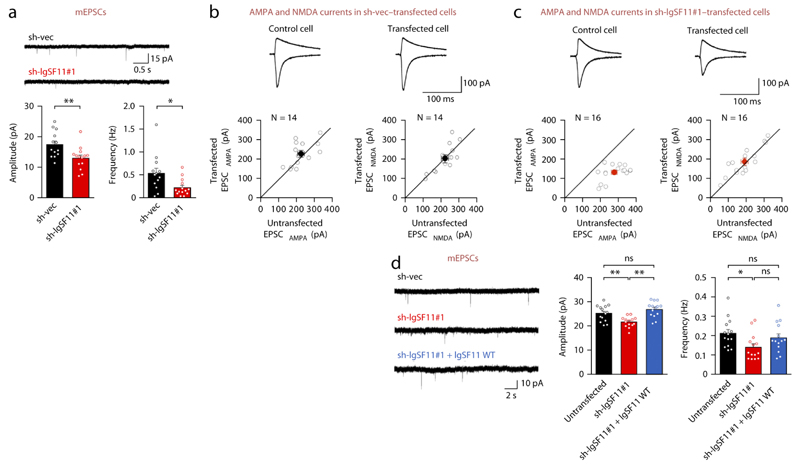
Functionally, IgSF11 knockdown in cultured rat neurons reduced both the frequency and amplitude of miniature excitatory postsynaptic currents (mEPSCs), causing a greater reduction in the frequency (Fig. 4a). Similarly, IgSF11 knockdown in hippocampal CA1 neurons in rat slice culture caused a reduction in AMPAR-mediated synaptic transmission (EPSCAMPA), but not NMDA-mediated transmission (EPSCNMDA), at Schaffer collateral (SC)-CA1 pyramidal synapses (Fig. 4b,c). Control slices transfected with empty knockdown vector (sh-vec) had no effect on either EPSCAMPA or EPSCNMDA (Fig. 4b,c).
Figure 4. IgSF11 knockdown suppresses mEPSCs and evoked EPSCs.
(a) IgSF11 knockdown reduces mEPSC frequency and amplitude in cultured hippocampal neurons, exerting a stronger influence on frequency. Hippocampal neurons were transfected with sh-IgSF11#1, or empty vector (sh-vec) (DIV 13–16). n = 13 (sh-vec) and 14 (sh-IgSF11#1), *p < 0.05, **p < 0.01, Student’s t-test.
(b and c) IgSF11 knockdown selectively suppresses AMPAR-EPSCs (EPSCAMPA) but not NMDAR-EPSCs (EPSCNMDA) at SC-CA1 synapses in slice culture. Organotypic hippocampal slice culture (DIV 3–4) were transfected with sh-IgSF11#1, or empty vector (sh-vec), and measured of EPSCA or EPSCN in transfected and neighboring untransfected (control) neurons (DIV 6–8). sh-vec transfected cells vs. neighbouring untransfected neurons (EPSCA transfected, 224 ± 17 pA; EPSCA untransfected, 227 ± 17 pA, n = 14 pairs, p > 0.05; EPSCN transfected, 202 ± 19 pA; EPSCN untransfected, 215 ± 16 pA, n = 14 pairs, p > 0.05). sh-IgSF11#1 transfected cells vs. neighbouring untransfected neurons (EPSCA transfected, 135 ± 10 pA; EPSCA untransfected, 282 ± 15 pA, n = 16 pairs, p < 0.001; EPSCN transfected, 183 ± 18 pA; EPSCN untransfected, 190 ± 20 pA, n = 16 pairs, p > 0.05). White circles indicate pairs of synaptic transmissions in transfected and untransfected cells. Black/red filled circles with error bars indicate mean ± SEM.
(d) IgSF11 knockdown reduces mEPSC frequency and amplitude in slice culture CA1 pyramidal neurons. Hippocampal organotypic slice culture (DIV 3–4) was transfected with sh-IgSF11#1, or sh-IgSF11#1 + IgSF11-res (rescue), and mEPSCs in CA1 pyramidal neurons (DIV 6–8) were measured. n = 14 for untransfected, sh-IgSF11#1, and rescue, *p < 0.05, **p < 0.01, ns, not significant, ANOVA.
IgSF11 knockdown in slice culture CA1 pyramidal neurons caused decreases in mEPSC frequency and amplitude, with a greater effect on the frequency (Fig. 4d), similar to the results of IgSF11 knockdown in dissociated hippocampal neurons (Fig. 4a). In addition, these decreases were rescued by coexpressed sh-RNA-resistant IgSF11 to levels comparable to untransfected control neurons (Fig. 4d; Supplementary Fig. 6c,d), pointing to the specificity of the knockdown construct. These results suggest that IgSF11 is important for AMPAR-mediated synaptic transmission.
IgSF11 knockdown increases surface mobility of AMPARs
The results to this point suggest that IgSF11 is localized at excitatory synapses via its interaction with PSD-95 and enhances the synaptic stability of AMPARs. If this is the case, the lack of IgSF11 should enhance the diffusion of AMPARs out of synapses and increase AMPAR mobility on the dendritic surface, as previously suggested 19. To test this idea, we performed high-throughput, single-molecule tracking of surface AMPARs in live neurons using the uPAINT method, which monitors the diffusion dynamics of a large number (tens of thousands) of surface proteins 20.
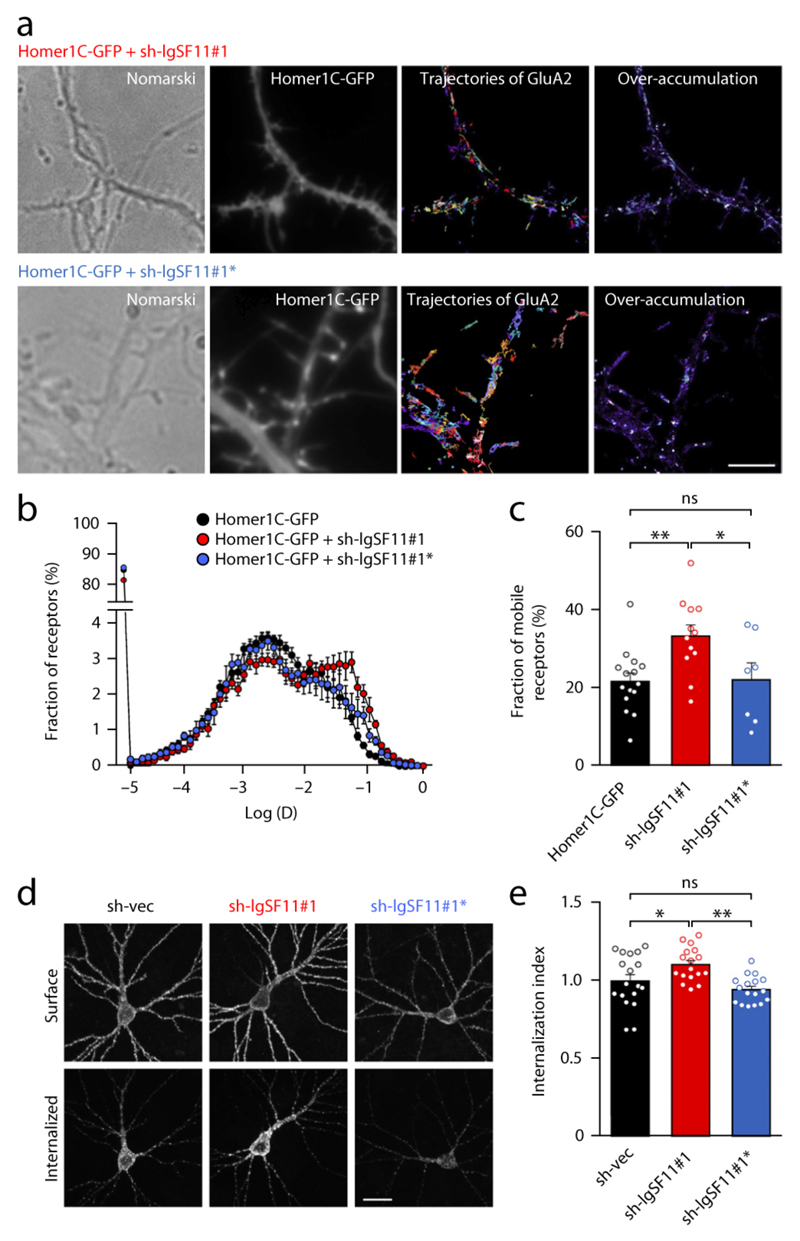
Analyses of single-molecule trajectories of approximately 31,000 to 45,000 surface GluA2 molecules in live cultured hippocampal neurons indicated that IgSF11 knockdown (sh-IgSF11#1) increased the mobile fraction of AMPARs by ~50%, compared with control neurons transfected with sh-IgSF11#1*, or Homer1C::GFP alone, which was expressed in all neurons to mark excitatory synapses/dendritic spines (Fig. 5a-c). These results suggest that IgSF11 promotes synaptic retention of AMPARs.
Figure 5. IgSF11 knockdown increases surface mobility and endocytosis of AMPARs.
(a) Representative images of single-molecule dynamics of endogenous AMPARs on live neurons obtained using uPAINT. Different single molecules are indicated by different colours. Cultured neurons were cotransfected with Homer1C-GFP (to visualize excitatory synapses/dendritic spines) and sh-IgSF11#1/sh-IgSF11#1*. Panels represent, from left to right, images for a wide-field Nomarski image, Homer1C-GFP, trajectories of GluA2-containing AMPARs labelled with anti GluA2-AT647Ns, and the super-resolved image resulting from the over-accumulation of all detections. Scale bar: 5 μm.
(b) Distribution pattern of instantaneous diffusion constants of GluA2-containing AMPARs.
(c) Sh-IgSF11#1, but not sh-IgSF11#1*, increases the mobile fraction of GluA2-containing AMPARs, reflected in instantaneous diffusion constants greater than 0.01 μm2/s. n = 15 for Homer only, 12 for sh-IgSF11#1, and 7 for sh-IgSF11#1*, one-way ANOVA.
(d) IgSF11 knockdown enhances endocytosis of GluA2. Cultured neurons transfected with HA-GluA2 and sh-vec, sh-IgSF11#1, or IgSF11#1* (DIV15–18) were subjected to antibody feeding assays. The internalization index (the ratio of internalized to internalized + surface receptors) of sh-IgSF11#1 or sh-IgSF11#1* was normalized to sh-vec control. n = 18 for sh-vec, 17 for sh-IgSF11#1, 17 for sh-IgSF11#1*, *p < 0.05, **p < 0.01, one-way ANOVA. Scale bar: 30 μm.
AMPARs that are not stable at synapses should also be more likely to undergo endocytosis. To test this idea, we performed antibody feeding assays in which N-terminally HA-tagged GluA2 on the surface of live neurons is labeled with HA to monitor GluA2 endocytosis. IgSF11 knockdown in cultured neurons enhanced internalization of GluA2, as compared with control neurons transfected with empty knockdown vector (Fig. 5d), and also decreased the steady-state levels of surface GluA2 (Supplementary Fig. 8). These results suggest that IgSF11 knockdown reduces the synaptic and surface stability of AMPARs.
IgSF11 inhibition suppresses AMPAR transmission
We next tested whether overexpression of IgSF11 affected AMPAR-mediated synaptic transmission, using herpes simplex virus (HSV)-mediated delivery of the IgSF11 gene into DG granule cells. The amplitude and frequency of mEPSCs in IgSF11-transduced DG granule cells were not significantly different from those in control neurons expressing enhanced green fluorescence protein (EGFP) (Fig. 6a-d), which suggests that the endogenous levels of IgSF11 proteins are sufficient to support synaptic stabilization of AMPARs. Notably, the frequency of mEPSCs in EGFP-expressing neurons showed a decreasing tendency relative to uninfected controls, although it did not reach statistical significance, which may be attributable to HSV infection.
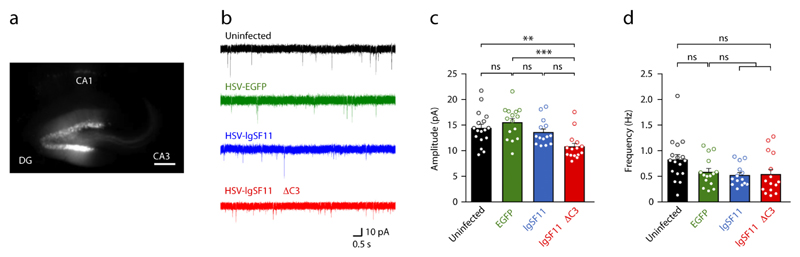
Figure 6. Dominant-negative inhibition of IgSF11 suppresses mEPSC amplitude in DG granule cells.
(a) Representative image of DG granule cells transduced with herpes simplex virus (HSV) carrying IgSF11, IgSF11 ∆C3, or EGFP only (control) at P21 and measured of mEPSCs at 60–72 hrs post-infection. DG, CA3, and CA1, subregions of the hippocampus. Scale bar, 200 μm.
(b) Examples of mEPSCs from uninfected neurons, and neurons expressing EGFP (control), IgSF11, and IgSF11 ∆C3.
(c and d) Quantification of the mEPSC results in (b). n = 17 cells (7 mice) for uninfected, n = 15 (9) for EGFP control, n = 14 (7) for IgSF11 WT, and n = 15 (8) for IgSF11 ∆C3, **p < 0.01, ns, not significant, one-way ANOVA.
In contrast to wild-type IgSF11, PSD-95 binding-defective IgSF11 (IgSF11 ΔC3) induced a significant reduction in the amplitude, but not frequency, of mEPSCs in DG granule cells, relative to EGFP-infected control neurons (Fig. 6a-d). This suggests that IgSF11 ΔC3 suppresses AMPAR-mediated transmission in a dominant-negative manner.
IgSF11 does not affect functional properties of AMPARs
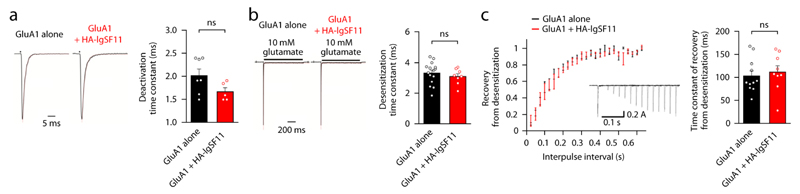
We next tested whether IgSF11 regulates the functional properties of AMPARs, by coexpressing IgSF11 with GluA1 in heterologous cells. IgSF11 coexpressed with GluA1 in HEK293FT cells had no effect on the kinetic properties of AMPARs, including deactivation, desensitization, and recovery from desensitization (Fig. 7a-c). These results suggest that IgSF11 regulates synaptic strength without changing functional properties of AMPARs.
Figure 7. IgSF11 has no effect on the functional properties of AMPARs.
(a) IgSF11 coexpression does not affect the deactivation time constant of GluA1 currents evoked by brief applications (1 ms) of glutamate (10 mM). Outside-out patches from HEK293FT cells expressing GluA1i (flip version) alone, or GluA1i + HA-IgSF11, were recorded of GluA1 currents using ultrafast perfusion. Currents were normalized for the analysis of time constants. n = 7 for GluA1i, and 5 for GluA1i + HA-IgSF11, ns, not significant. Student’s t-test.
(b) IgSF11 coexpression does not affect the desensitization time constant of GluA1 currents evoked by a longer application (1 s) of glutamate (10 mM). HEK293FT cells expressing GluA1i alone, or GluA1i + HA-IgSF11, were recorded of GluA1 currents, followed by quantitative analysis of desensitization time constants. n = 15 for GluA1i, and 8 for GluA1i + HA-IgSF11, ns, not significant, Student’s t-test.
(c) IgSF11 coexpression does not affect recovery of GluA1 currents from desensitization. HEK293FT cells expressing GluA1i alone, or GluA1i + HA-IgSF11, were recorded and superimposed of the GluA1-mediated currents for responses to brief applications of glutamate with varying time intervals, followed by quantitative analysis of recovery from desensitization (inset, examples of current traces) and time constant of recovery. n = 11 for GluA1i, and 9 for GluA1i + HA-IgSF11, ns, not significant, Student’s t-test.
Reduced mEPSC amplitude in Igsf11−/− DG granule cells
In order to explore the in vivo functions of IgSF11, we generated two lines of IgSF11-deficient mice, one using gene trap and the other using conventional gene knockout methods (Igsf11−/− (LacZ) and Igsf11−/−, respectively), both of which lacked detectable IgSF11 proteins in the brain (Supplementary Fig. 9; Fig. 8a,b).
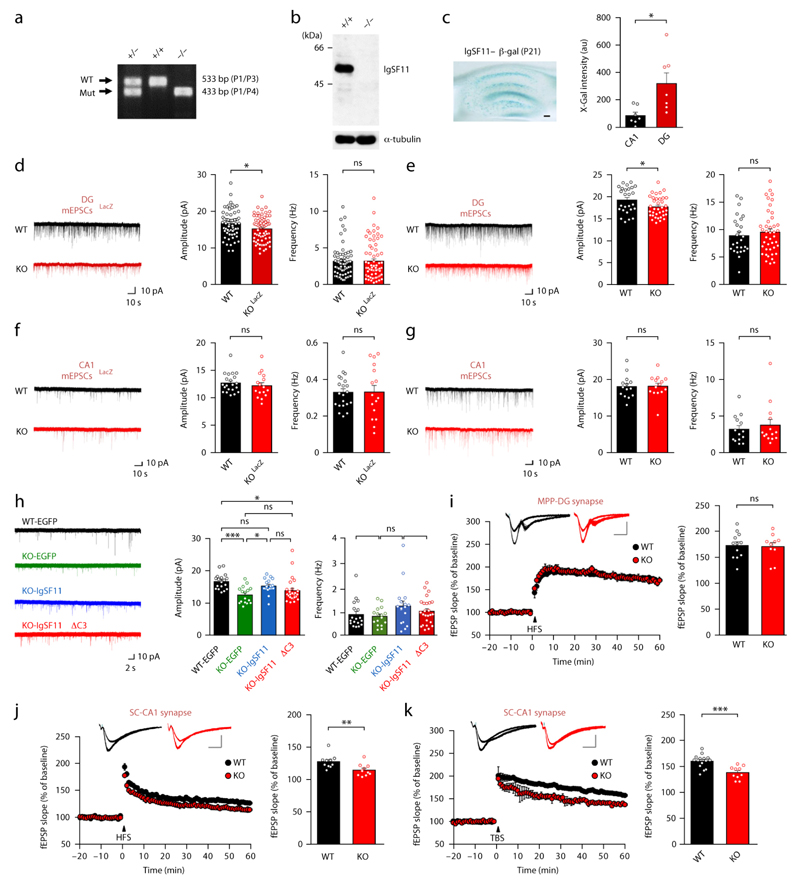
Figure 8. Reduced mEPSC amplitude in DG cells and suppressed LTP at SC-CA1 synapses in the Igsf11−/− hippocampus.
(a) PCR genotyping of WT, Igsf11+/−, and Igsf11−/− mice.
(b) Igsf11−/− brains lack detectable IgSF11 proteins. α-tubulin immunoblot, control.
(c) Strong expression of IgSF11 in the hippocampal DG (P21), revealed by X-Gal staining of brain slices expressing IgSF11 (first 50 aa)-β-galactosidase fusion proteins. n = 7, *p < 0.05, Student’s t-test.
(d,e) DG granule cells from Igsf11−/− (LacZ) and Igsf11−/− mice (2–3 weeks) show similar decreases in the amplitude, but not frequency, of mEPSCs. Igsf11−/− (LacZ), n = 63 cells (4 mice) for WT and 69 (4) for KO; Igsf11−/−, n = 26 (6) for WT and 36 (8) for KO; *p < 0.05, ns, not significant, Student’s t-test.
(f,g) CA1 pyramidal cells from Igsf11−/− (LacZ) and Igsf11−/− mice (3 weeks) show normal mEPSC amplitude and frequency. Igsf11−/− (LacZ), n = 21 cells (5) for WT and 18 (4) for KO; Igsf11−/−, n = 14 (7) for WT and KO, ns, not significant, Student’s t-test.
(h) Rescue of the reduced mEPSC amplitude in Igsf11−/− (LacZ) DG granule cells by HSV-mediated re-expression of IgSF11 but not IgSF11 ∆C3 (P20/21–23/24). n = 18 (5) for WT-EGFP, 16 (5) for KO-EGFP, 17 (5) for KO-IgSF11, and 25 (7) for KO-IgSF11 ∆C3, *p < 0.05, **p < 0.01, ***p < 0.001, ns, not significant, one-way ANOVA.
(i) MPP-DG synapses in the Igsf11−/− (LacZ) hippocampus (9–10 weeks) show normal LTP induced by four trains of high frequency stimulation. n = 12 slices from 6 mice for WT and 10, 4 for KO, ns, not significant, Student’s t-test.
(j and k) SC-CA1 synapses in the Igsf11−/− (LacZ) hippocampus (6–13 weeks) show suppressed LTP induced by high frequency stimulation (j) and theta burst stimulation (k). n = 10, 9 for WT and KO (HFS), 15, 8 for WT and 10, 7 for KO (TBS), **p < 0.01, ***p < 0.001, Student’s t-test.
IgSF11 protein expression, as determined by X-Gal staining of Igsf11−/− (LacZ) brain slices, was barely detectable at postnatal day or P5, but was clearly detected in a number of subcortical regions by postnatal week 2 (Supplementary Fig. 10). IgSF11 expression gradually increased and largely concentrated in the hippocampus over the postnatal 3–12-week period. Within the hippocampus, IgSF11 expression was ~3-times stronger in the DG than in the CA1 region at 3 weeks (Fig. 8c), consistent with in situ hybridization results. Expression levels of synaptic proteins, including synaptic scaffolding proteins, signaling molecules, and glutamate receptor subunits were not changed in Igsf11−/− (LacZ) mice (Supplementary Fig. 11).
Functionally, DG granule cells from Igsf11−/− (LacZ) and Igsf11−/− mice showed a similar reduction in the amplitude of mEPSCs (to ~90% of wild-type/WT levels), with no change in the frequency (Fig. 8d,e). In contrast, hippocampal CA1 pyramidal neurons from both Igsf11−/− (LacZ) and Igsf11−/− mice (in which IgSF11 expression is relatively weak) showed no significant changes in mEPSC amplitude or frequency compared with wild-type mice (Fig. 8f,g).
When DG granule cells in acute slices were incubated with AMPA (1 μM), a condition known to induce mainly extrasynaptic AMPAR currents 21, there was no difference in AMPAR currents between genotypes (Supplementary Fig. 12).
Re-expression of IgSF11 rescues reduced mEPSC amplitude
In the rescue experiment, re-expression of IgSF11 in Igsf11−/− DG granule cells (P20/21–23/24) by HSV-mediated gene delivery significantly improved the reduced amplitude of mEPSCs, as shown by the levels of mEPSC amplitude comparable to those of EGFP-expressing WT neurons but significantly different from those of EGFP-expressing KO neurons (Fig. 8h).
In contrast, IgSF11 ΔC3, which lacks PSD-95 binding, expressed in KO neurons yielded levels of mEPSC amplitude that are not significantly different from those of EGFP-expressing KO neurons (Fig. 8h). Notably, mEPSC amplitudes in KO neurons expressing IgSF11 and IgSF11 ΔC3 were not significantly different, indicative of a partial rescue. These results suggest PSD-95 binding is important for, but partly contributes to, IgSF11-dependent regulation of AMPARs.
Suppressed LTP at Igsf11−/− SC-CA1 synapses
When synaptic plasticity was measured, Igsf11−/− MPP (medial perforant pathway)-DG synapses showed normal long-term potentiation (LTP) induced by four trains of high frequency stimulation (Fig. 8i). Basal transmission (input-output relationship) and the paired pulse ratio at these synapses were not different between genotypes (Supplementary Fig. 13a,b).
Intriguingly, Igsf11−/− SC-CA1 synapses, which showed normal mEPSCs (Fig. 8f,g), showed suppressed LTP induced by high frequency stimulation and theta burst stimulation (Fig. 8j,k). Basal transmission and paired pulse ratio were normal at these synapses, although basal transmission showed a strong tendency for a decrease (Supplementary Fig. 13c,d). These results collectively suggest that IgSF11 deletion leads to distinct changes synaptic transmission and plasticity in two different hippocampal regions.
Normal levels of immature AMPARs in the Igsf11−/− brain
Lastly, we sought to test whether IgSF11 associates with AMPARs early in their secretory pathways to the plasma membrane such as the endoplasmic reticulum and cis-Golgi complex (ER-CG) and thus promotes the maturation and forward trafficking of AMPARs. If this is the case, the lack of IgSF11 would hold AMPARs largely in ER-CG, and increase the levels of immature AMPARs enriched with N-linked high-mannose carbohydrates, which can be detected by their sensitivity to endoglycosidase H (Endo H). We found that the Endo H-sensitive population of GluA2 in the Igsf11−/− (LacZ) hippocampus was comparable to that in the wild-type hippocampus (Supplementary Fig. 14). This suggests that IgSF11 does not associate with AMPARs early in their biosynthetic pathways.
Discussion
In the present study, we found that IgSF11 interacts with PSD-95 and AMPARs to promote synaptic stabilization of AMPARs. IgSF11 deletion in mice leads to two distinct functional changes in the hippocampus; decreased excitatory synaptic strength in DG granule cells and decreased LTP at SC-CA1 synapses.
The CAR family of immunoglobulin superfamily of proteins contains three close relatives of IgSF11 (CAR, ESAM, and CLMP), all of which are expressed in the brain and share a C-terminal PDZ-binding motif (Supplementary Fig. 2). Some of these proteins have already been shown to interact with PDZ domain-containing proteins in non-neural cells 22. Specifically, CAR interacts with PSD-95, PICK-1, ZO-1, MUPP-1, and MAGI-1b, and CLMP interacts with ZO-1. Therefore, similar PDZ interactions between the CAR family proteins and diverse synaptic PDZ proteins are expected to occur and play important roles in neuronal cells.
IgSF11 associates with AMPARs, as supported by the coimmunoprecipitation of IgSF11 with AMPARs both in vitro and in vivo (Fig. 2a,b). The apparently weak coimmunoprecipitation between the two proteins may be attributable to the restricted expression of IgSF11 in the DG region, the weak nature of the cis-interaction involving the transmembrane domain of IgSF11, or the restricted subcellular site of the interaction, occurring mainly at synapses but not in biosynthetic pathways. Interestingly, this interaction appears to promote the synaptic stabilization of AMPARs and AMPAR-mediated synaptic transmission. In support of this hypothesis, we found that (i) IgSF11 knockdown suppresses mEPSCs in cultured neurons and mEPSCs and EPSCAMPA (but not EPSCNMDA) in slice culture (Fig. 4), (ii) IgSF11 knockdown increases surface mobility and endocytosis of AMPARs (Fig. 5), (iii) dominant-negative inhibition of IgSF11 decreases mEPSC amplitude (Fig. 6), and (iv) in vivo knockout of IgSF11 by two different methods (gene trap and conventional knockout) decreases mEPSC amplitude but not frequency (Fig. 8d,e).
The significant reductions in both mEPSC frequency and amplitude in dissociated hippocampal neurons and slice culture CA1 pyramidal neurons (Fig. 4a,d) contrasts with the small decrease in mEPSC amplitude observed in Igsf11−/− DG granule cells and normal mEPSCs in CA1 pyramidal neurons (Fig. 8d-g). Whether this represents compensatory changes is unclear, given that we do not have any molecular evidence to support this possibility. However, if it is the case, compensating for the loss of IgSF11 might be easier in CA1 with a weaker (~3 times) IgSF11 expression (Fig. 8c). Notably, IgSF11 knockdown in slice culture CA1 neurons leads to a bigger decrease in mEPSC frequency than in amplitude (Fig. 4d). This, together with the normal mEPSCs in Igsf11−/− CA1 neurons (Fig. 8f,g), suggests the possibility that mEPSC frequency is rescued to a greater extent than the amplitude. If this reflects presynaptic changes, again, the stronger IgSF11 expression in the CA1 region relative to CA3 (Fig. 8c, Supplementary Fig. 2b), might be a possible explanation.
Field recordings revealed that Igsf11−/− MPP-DG synapses display unaltered basal transmission (input-output relationship) (Supplementary Fig. 13a), which contrasts with the reduced mEPSC amplitude in these cells (Fig. 8d,e). These results, although obtained using two different methods of measuring synaptic strength (synaptic currents vs. field potentials), may reflect a weak impact of chronic IgSF11 deletion on synaptic strength. In addition, the normal basal transmission at Igsf11−/− SC-CA1 synapses (Supplementary Fig. 13c) contrasts with the strong reduction in EPSCAMPA at these synapses induced by acute IgSF11 knockdown (Fig. 4b,c). A surprising result was the reduced LTP (both high frequency stimulation and theta burst stimulation) at Igsf11−/− SC-CA1 synapses, contrasting sharply with the normal LTP at Igsf11−/− MPP-DG synapses (Fig. 8i-k). This difference may reflect again the stronger IgSF11 expression in DG, or different nature of DG and CA1 synapses. Although further details remain to be studied, the dual role of IgSF11 in the regulation of synaptic strength and plasticity is reminiscent of the dual role of LRRTM4 in the regulation of synaptic strength and activity-dependent synaptic AMPAR targeting in DG granule cells 23.
IgSF11 interacts with PSD-95 and AMPARs, and recruits them to sites of IgSF11 adhesion at the cell surface in heterologous cells (Figs. 1 and 2). In cultured neurons, however, IgSF11 requires PSD-95 binding for its excitatory synaptic localization (Fig. 1j). Assuming that the results from neurons are more physiological than those from heterologous cells, these results suggest that PSD-95 binding drives IgSF11 to excitatory synapses, where IgSF11 interacts with and regulates AMPARs. Because PSD-95 associates with both IgSF11 and TARPs (transmembrane AMPAR regulatory proteins) 24, PSD-95 may be able to bring the two proteins (IgSF11 and AMPARs) close together and facilitate their association. In line with this hypothesis, re-expression of IgSF11, but not IgSF11 ΔC3 that lacks PSD-95 binding, in Igsf11−/− DG granule cells rescues the reduced mEPSC amplitude (Fig. 8h). In addition, IgSF11 ΔC3 suppresses synaptic AMPAR currents in a dominant-negative manner, suggesting that IgSF11 ΔC3 proteins that are mislocalized to extrasynaptic sites may interact with AMPARs and inhibit their synaptic targeting. It is possible that the molar abundance of IgSF11 proteins may not match that of PSD-95 and AMPARs. However, this does not rule out certain levels of contribution of IgSF11 to the synaptic regulation of AMPARs.
IgSF11-dependent synaptic stabilization of AMPARs is reminiscent of the role of neuroligin-1 in synaptic stabilization of NMDARs and AMPARs. Neuroligin-1 directly interacts with the GluN1 subunit of NMDARs through extracellular domains and promotes synaptic retention of NMDARs 12, consistent with the neuroligin-1-dependent synaptic clustering of NMDARs 25 and promotion of NMDAR-mediated synaptic transmission 26, 27, 28, 29, 30, 31. Neuroligin-1 also promotes synaptic localization of AMPARs and AMPAR-mediated transmission, through its cytoplasmic domains 32, 33. An apparent difference between IgSF11 and neuroligin-1 is that IgSF11 strongly associates with AMPARs relative to NMDARs, as shown by the weak biochemical association of IgSF11 with NMDARs (Fig. 2a) and a selective reduction in EPSCAMPA but not EPSCNMDA by IgSF11 knockdown (Fig. 4b,c). In this respect, IgSF11 is similar to neuroligin-3 and LRRTM1/2, which preferentially regulates AMPARs relative to NMDARs 26, 33.
Another examples of synaptic adhesion molecules that interact with AMPARs include N-cadherin and β3-integrin. N-cadherin associates with AMPARs and promotes the surface expression and synaptic localization of AMPARs, and AMPAR-dependent spine morphogenesis and synaptic transmission 10, 11. β3 integrin interacts with the GluA2 subunit of AMPARs through cytoplasmic domains and regulates AMPAR-mediated synaptic transmission and homeostatic synaptic plasticity 34, 35. Some of these functions are also shared by IgSF11, including synaptic AMPAR stabilization and AMPAR-mediated transmission and plasticity. It should be noted, however, that N-cadherin requires calcium for its homophilic adhesion 36, whereas IgSF11 does not 15. In addition, while IgSF11 directly interacts with PSD-95 through its C-terminal tail, N-cadherin interacts with β-catenin in a neuronal activity- and calcium-dependent manner 10, 36. The extracellular region of β3 integrin interacts with the extracellular matrix and other synaptic adhesion molecules, and the intracellular region interacts with the actin cytoskeleton and diverse signaling molecules 43,44. Therefore, extracellular and intracellular domains of N-cadherin, β3 integrin, and IgSF11 may have distinct influences on AMPARs.
Synaptic localization, function, and plasticity of AMPARs are regulated by transmembrane AMPAR auxiliary proteins 24. Specific examples include TARPs, CNIHs, CKAMP44, SynDIG1, and GSG1L 24, 37, 38, 39, 40, 41, 42. TARPs are unique among these proteins in that they regulate both the trafficking and functional properties of AMPARs 24. Is IgSF11 one of the AMPAR auxiliary proteins? We suspect not because (i) IgSF11 was not isolated in proteomic analyses of AMPAR complexes 37, 38, 39, 42, (ii) IgSF11 does not regulate the functional properties of AMPARs (Fig. 7), (iii) IgSF11 does not regulate the forward trafficking of AMPARs (Supplementary Fig. 14), and (iv) synaptic stabilization of AMPARs by IgSF11, which we propose is one of the primary functions of IgSF11, predicts their short-term interactions at the synapse.
A growing body of evidence indicates that synaptic adhesion molecules interact with neighboring receptors and adhesion molecules in a cis manner. Neuroplastin-65 cis-interacts with a small fraction of GABAA receptors and regulates their synaptic localization and subunit compositions 13. Slitrk5 cis-interacts with TrkB receptors, in addition to its trans interaction with PTPδ, in a manner requiring BDNF stimulation to promote TrkB targeting to Rab11-positive recycling endosomes 43. Neuroligin-1 interacts in cis with neurexin-1β on dendrites and inhibits the trans-synaptic interaction between neuroligin-1 and presynaptic neurexins, suppressing neuroligins-1-dependent excitatory synapse formation 44. Similarly, postsynaptic MDGA1 interacts in cis with neuroligin-2 and suppresses neuroligin-2-dependent inhibitory synapse formation 45, 46. Postsynaptic SynCAM1 cis-interacts with each other to modulate adhesive properties, promote synapse formation, and modulate synapse structure 47. In addition, NGL-1, an excitatory postsynaptic adhesion molecule, binds to presynaptic netrin-G1, and enhances the cis interaction between netrin-G1 and LAR, a presynaptic receptor tyrosine phosphatase, to promote presynaptic differentiation 48. These results, together with the present and other results, suggest that synaptic adhesion molecules can interact in cis with adjacent receptor proteins as well as adhesion molecules.
It is unclear whether IgSF11 mediates homophilic or heterophilic trans-synaptic adhesion at excitatory synaptic cleft. We favor the possibility of homophilic adhesion for the following reasons. Our results indicate that IgSF11 interacts homophilically but not heterophilically with the synaptic adhesion molecules that we tested (Supplementary Fig. 4d). In addition, our molecular modeling based on the X-ray crystal structure of CAR 49, a close relative of IgSF11, predicts the width of the synaptic cleft, known to be about ~20–24 nm long 50, may be covered by the extracellular adhesion of IgSF11 (~15–21 nm) (Supplementary Fig. 15).
In conclusion, our study identifies a novel tripartite interaction among IgSF11, PSD-95, and AMPARs, and suggests a concerted role for IgSF11 and PSD-95 in the regulation of AMPAR-mediated synaptic transmission and plasticity. Our findings add to the growing evidence that synaptic adhesion molecules regulate synaptic strength and plasticity, in addition to synapse development.
Online Methods
cDNA constructs
Full-length human IgSF11 (aa 1-430), rat IgSF11 (aa 1-428), and human IgSF11ΔC3 (a mutant that lacks the last three residues, aa 1-427) were amplified by PCR from brain cDNA libraries (Clontech) and subcloned into GW1 (British Biotechnology). For Myc-IgSF11 and HA-IgSF11, the Myc and HA epitopes were added to the N-terminus of human IgSF11 in GW1. Deletion variants of IgSF11 were generated by inverse PCRs on HA-IgSF11 (human) deleting the following regions; aa 30-222 (IgSF11ΔEcto), aa 30-156 (IgSF11ΔIgv), aa 156-222 (IgSF11ΔIgC2), and aa 263-430 (IgSF11ΔCyto). For yeast two-hybrid screen and assays, the PDZ2 domain of PSD-95 (aa 89-299) in the pBHA bait vector was used to screen a human brain yeast two-hybrid cDNA library contained in the pACT2 prey vector (Clontech). For yeast two-hybrid experiments, human IgSF11 [WT, aa 424-430; and VA, aa 424-430] were subcloned into pBHA. For shRNA knockdown of IgSF11, two independent construct of pSuper IgSF11 were generated by annealing oligonucleotides containing nt 534-552 (GGAGAAGTTAGATAATACA, sh-IgSF11#1) and nt 218-236 (TCATTCTCTATCAGGGCGG, sh-IgSF11#2) of rat IgSF11 cDNA (NM_001013120) and subcloning into pSUPER.gfp/neo (OligoEngine). shRNA mutant construct against sh-IgSF11#1 (sh-IgSF11#1*, GGAGACGTGCGATAATACA; underline, changed sequence) was generated using QuickChange kit. For the rescue of the effects of sh-IgSF11#1 in rat slice culture, sh-RNA-resistant untagged rat IgSF11 expression constructs (WT and ΔC3) were generated by PCR-based mutagenesis to introduce the following mutant target sequence (GGAGAAGCTAGACAATACA). The following constructs have been described: GW1-PSD-95 51, PDZ domains in pGAD10 (a prey vector; Clontech) 52. HA-GluA1 and HA-GluA2 were kindly provided by Dr. Maria Passafaro. Human IgSF11 was purchased (Origene).
HSV production and infection
We used herpes simplex virus (HSV)-mediated gene delivery to overexpress IgSF11-WT, IgSF11 ΔC3 or EGFP in the dentate gyrus (DG) of mouse hippocampus. The HSV vector we used, p1005(+), was a kind gift from Dr. Rachael Neve at MIT. IgSF11 WT or IgSF11 ΔC3 was subcloned into the HSV vector that express EGFP. Three different viruses (HSV-GFP-IgSF11 WT, HSV-GFP- IgSF11 ΔC3 and HSV-GFP) were produced and packaged using 2-2 host cell line and replication-defective helper virus (5dl1.2), as previously described 53, 54. The purified virus was resuspended in 10% sucrose. The titer of each virus stock was 4.6 x 107 infectious units/ml for HSV-GFP-IgSF11-WT, 1.2 x 108 for HSV-GFP-IgSF11 ΔC3, and 1.2 x 108 for HSV-GFP. Virus was stereotaxically injected into the DG (-1.6 AP/anteroposterior, ±1.6 ML/medial lateral, -1.6 DV/dorsal ventral) of C57/B6 mouse for overexpression experiments, or IgSF11−/− mice for rescue experiments, at P21. Electrophysiological experiments were performed 60-72 hours after stereotaxic injection.
Antibodies
MBP fusion proteins containing human IgSF11 (aa 262-430) were used for immunization of guinea pigs (2003), and peptides containing mouse IgSF11 (aa 399-427) were used to immunize guinea pigs (2067) and rabbits (2068 and 2070). The specificity of anti-IgSF11 antibodies (2003) was confirmed by immunoblot experiments using Igsf11−/− brain lysates. The following antibodies have been described: PSD-95 (1688) 55, SAP102 (1145) 52, CaMKIIα (1299) 55, CASK (1640) 56, GluN2B (21266) 57, GluA1 (1193) 56, GluA2 (1195) 56, and pan-GRIP (1756) 74. The following antibodies were purchased: Myc and HA rabbit polyclonal, synaptophysin (Santa Cruz sc-789, sc-805, sc-9116), HA mouse monoclonal (Boehringer Mannheim 12CA5), GluA2 (75-002), PSD-95 (75-028), PSD-93 (75-057), SAP97 (75-030), SAP102 (75-058) (NeuroMab), GluA1 (Calbiochem PC246), GluA1, GluA2, synapsin I (Millipore MAB2263, MAB397, AB1543), GluN1 (BD Pharmingen 556308), GluN2B (Invitrogen 320600), GluN2A (AGC-002), GluN2B (AGC-003) (Alomone labs), SNAP-25 (BD Transduction Laboratories 610366), MAP2, and α-tubulin (Sigma M1406, T5168).
In situ hybridization
In situ hybridization was performed as described previously 58. Adult (6 weeks) rat brain sections (12 μm thick) were prepared with a cryostat (Leica CM 1950). The hybridization probes were prepared from pGEM7zf plasmids containing nucleotides 279-1444 of rat IgSF11 (XM_221443) using a Riboprobe System (Promega) and α-[35S]UTP.
Subcellular and PSD fractions
Subcellular and PSD fractions of rat brains were prepared as described previously 59, 60. Immunoblot analysis of these fractions was performed using IgSF11 (2003), PSD-95 (1688), synaptophysin and α-tubulin antibodies.
Dissociated neuron culture, transfection and immunocytochemistry
Cultured hippocampal neurons were prepared from embryonic day 18 Sprague-Dawley rat brains. Dissociated neurons on poly-L-lysine coated (1 mg/ml) coverslips were placed in neurobasal medium supplemented with B27 (Invitrogen), 0.5 mM L-glutamine, and penicillin-streptomycin. Cultured neurons were transfected using mammalian transfection kit (Invitrogen) and fixed with 4% paraformaldehyde/sucrose, permeabilized with 0.2% Triton X-100, and incubated with primary and dye-conjugated secondary antibodies. For surface staining of GluA1 or GluA2, live neurons were incubated with GluA1 or GluA2 antibodies for 20 min at 37°C, followed by fixation and incubation with primary and secondary antibodies.
Antibody feeding assay
Antibody feeding assay was performed as described 61. Live neurons expressing HA-GluA2 were incubated with mouse HA antibodies (10 μg/ml) for 10 min at 37°C. After DMEM washing, neurons were returned to conditioned medium for 10 min. Neurons were incubated with Cy3 antibodies for surface GluA2, permeabilized, and labeled with Cy5 and FITC antibodies for internalized GluA2 and coexpressed EGFP, respectively.
Surface and internal GluA2 labeling
Surface and internal GluA2 labeling for the determination of steady-state levels was performed as described 61. HA-GluA2-expressing neurons were fixed and incubated with rabbit HA antibodies for surface GluA2, followed by permeabilization with 0.2% Triton X-100 and incubation with mouse HA antibodies for internal GluA2. Cy3-, Cy5-, and FITC-conjugated secondary antibodies were used to visualize surface GluA2, internal GluA2, and coexpressed EGFP, respectively.
Image acquisition and quantification
Z-stack images were acquired using a confocal microscope (LSM 510 and LSM 780; Zeiss) under the same parameter settings for all scanning. All transfected neurons, with the exception of those with obvious morphological abnormalities, were imaged in an unbiased manner. Image analyses were performed by a researcher blinded to the experimental conditions. Morphometric measurements on randomly selected images were performed using MetaMorph (Universal Imaging). Neuronal areas for surface/internal GluA2 analysis were manually selected excluding neuronal cell bodies.
Slice culture, transfection, and electrophysiology
Hippocampal slice cultures were prepared from 6–7-day-old Wistar rats, as previously described 62. At DIV 3–4, using a biolistic GeneGun (Biorad, USA), neurons in the CA1 or DG region were transfected with plasmids expressing shRNA-IgSF11#1 (sh-IgSF11#1), or sh-IgSF11#1 + HA-IgSF11-res (rescue expression construct). Whole-cell patch clamp recordings of CA1 or DG neurons were made 3–4 days following transfection. A non-effective shRNA vector was used as a negative control against IgSF11#1-shRNA. Excitatory postsynaptic currents (EPSCs) were recorded using a Multiclamp 700B amplifier (Axon Instruments, Foster City, USA), with pipettes (4–6 MΩ) filled with recording solutions (280 mOsm [pH 7.2]) comprised (mM) CsMeSO4, 130; NaCl, 8; Mg-ATP, 4; Na-GTP, 0.3; EGTA, 0.5; HEPES 10; QX-314, 6. Recordings were carried out in solution containing (mM): NaCl 119, KCl 2.5, CaCl2 4, MgCl2 4, NaHCO3 26, NaH2PO4 1, glucose 11, picrotoxin 0.02, and 2-chloroadenosine 0.01, gassed with 5 % CO2/95 % O2, at pH 7.4. Dual patch clamp recordings were made simultaneously from a pair of neighbouring CA1 pyramidal neurons, one transfected and the other untransfected. AMPAR-mediated EPSC amplitude (EPSCAMPA) was determined as the peak EPSC amplitude at a holding potential of -70 mV. NMDAR-mediated EPSC amplitude (EPSCNMDA) was determined 80 ms after the EPSCN peak at a holding potential of +40 mV. mEPSCs were measured as described below (Electrophysiology-mini and patch recordings section).
Electrophysiology for AMPAR kinetics
HEK-293 FT cells were transfected with GluA1 iQ alone, or in combinations with IgSF11 at equal amounts, via calcium-phosphate method. Two days after transfection, to measure deactivation, outside-out membrane patches were excised and positioned in front of an ultrafast perfusion system mounted on a piezoelectric translator (Physic instruments). To determine the desensitization time constant, 10 mM glutamate was applied for 1 sec. Recovery from desensitization was measured using a paired pulse protocol and an increasing interpulse interval. Data analysis was performed using axograph 4.6 (Molecular devices) and graphed using Kaleidagraph 3.5 (Synergy software) and Excel 2004 (Microsoft). Statistical analysis was done using Instat 2.03 (Synergy software).
Universal points-accumulation-for-imaging-in-nanoscale-topography (U-PAINT)
AMPAR mobility on hippocampal neurons was assessed using the U-PAINT technique as reported 63. Hippocampal neurons were cultured from E18 Sprague-Dawley rats following a method previously described 64. Briefly, hippocampal neurons from 18 days-old rat embryos were cultured on glass coverslips following the Banker method. After 10 days in vitro, neurons were co-transfected with Homer 1C-GFP (a postsynaptic marker) and sh-IgSF11#1 or sh-IgSF11#1*. 2-3 days after transfection, cells were used for mobility recordings in U-PAINT. Briefly, transfected cells were mounted on a Ludin chamber filled with 600 μL of cooled HEPES based solution. A low concentration of ATTO-647nm-coupled anti-GluA2 antibodies was then added to the chamber. Stochastic labelling of coupled antibodies on endogenous GluA2-containing AMPA receptors allowed us to record thousands of trajectories lasting over 1 sec. Analysis and representation of these single-particle tracking movies were performed with a homemade software developed under MetaMorph (kindly provided by Jean-Baptiste Sibarita).
Generation of Igsf11−/− (LacZ) and Igsf11−/− mice
A mouse ES cell line (JM8.N4, strain C57BL/6N) dual trapped and targeted in the Igsf11 gene was provided by the Knockout mouse project (KOMP) and originated from the international knockout mouse consortium (IKMC). The targeting cassette contained non-genomic region between two FRT sites including the En2 (Engrailed-2) splice acceptor upstream of the IRES-lacZ, human β-actin promoter, neomycin gene, a polyadenylation signal and floxed Igsf11 gene exon3. The gene targeting scheme has been described 65. Igsf11 mutant mice were generated by homologous recombination in embryonic stem cells and implanted into C57BL/6 albino-type blastocysts using standard procedures to generate chimera in the same B6 genetic background. Heterozygotes (N1; Igsf11−/− (LacZ)) were backcrossed to C57BL/6J for 3-5 generations. In order to produce conventional KO mice with exon 3 deletion, Igsf11−/− (LacZ) mice were crossed with a protamine-Cre mouse line (JAX 007252), which efficiently targets floxed genes in the male germline 66. Littermates derived from heterozygous parents were used for all analysis. Genotyping of the Igsf11−/− (LacZ) mice was performed using PCR and the following three primers; P1 (5’- CAC ATG CAC AGG AAG GTC CTC-3’), P2 (5’- CCA ACT GAC CTT GGG CAA GAA C -3’), P3 (5’- CGC CGT CAA ACA TTT GTC CAC-3’). The size of the PCR products for WT (P1 and P3) and mutant (Igsf11−/− (LacZ); P1 and P2) genes were 533 and 316 bp, respectively. For genotyping of IgSF11-floxed mice and exon 3-deleted (Igsf11−/−) mice, we used the following PCR primers; P1 (5’- ACA TGC ACA GGA AGG TCC TCA TT -3’), P3 (5’- GCG CCG TCA AAC ATT TGT CCA C-3’), P4 (5’- AGG ATG TCC TTA AGA GTA CAC AGG AG -3’). The size of the PCR products for Igsf11-floxed (P1 and P3) and exon 3-deleted (Igsf11−/−) (P1 and P4) genes were 533 and 433 bp, respectively. Both male and female mice were used for X-Gal staining and mEPSCs measurements. All mice were bred and maintained according to the KAIST Animal Research Requirements, and all procedures were approved by the Committees of Animal Research at KAIST. Mice were fed ad libitum by standard rodent chow and tap water, and housed under 12-h light/dark cycle (lights off at 19:00 in KAIST).
X-Gal staining
For X-Gal staining, mice at various developmental stages were transcardially perfused with heparin in phosphate buffered saline without 4% paraformaldehyde for β-galactosidase activity. Sagittal brain slices (300 μm thick) were incubated with X-Gal (1 mg/ml).
Endo H and PNGase F digestion
To digest high-mannose oligosaccharides and all N-linked carbohydrates, we tried digestion of brain lysates with Endo H and PNGase F (New England Biolabs), respectively, according to manufacturer’s protocols. Briefly, mice hippocampal lysates were denatured by heating at 100 °C for 10 min and incubated with Endo H or PNGase F at 37 °C for 1 hr, followed by immunoblotting with GluA2 antibodies.
Brain lysates preparation
Whole mouse brain lysates (3 weeks) were prepared as previously described 67. After the brain dissection, obtained brain tissues were briefly homogenized in 3 volumes of ice-cold homogenization buffer (0.32 M sucrose, 10 mM HEPES pH 7.4, 2 mM EDTA, protease inhibitors, phosphatase inhibitors). Protein concentrations were measured by the Bradford assay. The relative amount of α-tubulin was used as a loading control. The immunoblot intensity were measured using Metamorph 7.1 (Molecular Devices)
Electrophysiology-mini and patch recordings
Electrophysiological recordings for whole-cell patch were performed as previously described 56, 61, 68. Briefly, acute sagittal hippocampal slices (300-400 μm thick for whole-cell recordings) from WT and Igsf11−/− mice (2–3- week-old littermates for whole-cell recordings) were prepared using a vibratome (Leica VT1200s) in ice-cold high sucrose cutting solution containing (in mM) 212 sucrose, 25 NaHCO3, 5 KCl, 1.25 NaH2PO4, 0.5 CaCl2, 3.5 MgCl2, 10 D-glucose, 1.25 L-ascorbic acid, 2 Na-pyruvate equilibrated with 95% O2/5% CO2. The slices were then allowed to recover at 32 °C for 30 min in artificial cerebral spinal fluid (aCSF) containing (in mM): 125 NaCl, 25 NaHCO3, 2.5 KCl, 1.25 NaH2PO4, 2.5 CaCl2, 1.3 MgCl2, 10 D-glucose) with pH 7.3-7.4 and osmolarity 296-300 mOsm and maintained at room temperature before recordings (0.5 ~ 1 hr).
Whole-cell patch recordings were performed with recording pipettes pulled from borosilicate glass capillaries (Harvard Apparatus, 1.5 mm OD, GC150T-7.5) with a micropipette puller (Narishige PC-10). For whole-cell recordings, CA1 pyramidal cells or DG granule cells were held at –70 mV with recording pipettes (3–4 MΩ) at 30.5 °C (400 μm slice thickness) using Multiclamp 700B amplifier (Axon Instruments) except for that DG and CA1 whole-cell recordings of Igsf11−/− (LacZ) slices (300 μm) were performed at – 60 mV at room temperature using a Axopatch 200B (Axon Instruments). For mEPSC recordings, Cs-based intracellular solution contained (in mM) 110 Cs-gluconate, 30 CsCl, 20 HEPES, 4 MgATP, 0.3 NaGTP, 4 NaVitC, 0.5 EGTA with pH 7.3 and osmolarity ~295 mOsm. Picrotoxin (100 μM) and TTX (0.5 μM) was used to inhibit inhibitory synaptic currents and sodium channel-mediated action potentials, respectively. For consistent mEPSC measurements, the baseline was monitored for 5~10 min, and mEPSCs began to be measured at the same time point after whole-cell access was established to minimize time-dependent fluctuation. Liquid junction potentials were not corrected. Series access resistance was 15-30 MΩ, and only the cells with a change in series access resistance < 20 % were included in the analysis.
For extrasynaptic AMPAR experiments, AMPA-mediated whole-cell currents were obtained by bath application of 1 μM (S)-α -amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (S-AMPA) for 5 min in the presence of 1 μM TTX and 100 uM picrotoxin after stable 5~10 min baseline holding currents were established. Reagents were purchased from Tocris (tetrodotoxin, (S)-AMPA), and Sigma (picrotoxin).
Electrophysiology-field recordings
Acute hippocampal slices (400 μm thick, sagittal slice for CA1, sagittal or coronal slice for DG) were used for extracellular field recordings at 28-30 °C (TC-324B, Warner Instruments). Field excitatory postsynaptic potentials (fEPSPs) were recorded with glass electrodes (1–5 MΩ tip resistance) filled with ACSF or 3 M NaCl, and evoked every 20 s with a stimulating glass electrode filled with ACSF. The recording electrode was also placed in the medial perforant path (MPP) for DG or in stratum radiatum for CA1 but ~200 μm away from the stimulating electrode. fEPSPs in the DG were recorded in the presence of 100 μM picrotoxin (Sigma).
For input-output experiments measuring basal transmission, input was the peak amplitude of the fiber volley, and the output was the initial slope of the fEPSP from averages of five individual traces. For each slice, we established an initial input-output (I-O) curve, by measuring fiber volley amplitudes and initial slopes of the fEPSPs in the range of stimulation from 5 to 50 μA with 5 μA increments. To assess statistical difference for the Input-output relationships, linear regression was performed on the individual slices to calculate the linear fit slope as the slope of the linear input–output relationship 69. Then, stimulus strength was adjusted to yield ~30-40 % of the maximal fEPSP slope. Paired pulse ratios were determined by evoking two fEPSPs (averages of three individual traces) that are 20-3000 ms apart and dividing the initial slope of the second fEPSP by that of the first (fEPSP2/fEPSP1). The paired pulses were delivered every 10 s.
For synaptic plasticity experiments, CA3 was removed to minimize epileptic activities. LTP at SC-CA1 synapses was induced by a single tetanus of 100 pulses at 100 Hz, or theta burst stimulation consisting of 10 trains of 4 pulses at 100 Hz, delivered at an inter-train interval of 200 ms, repeated 4 times at 20 s interval. LTP at MPP-DG synapses was induced by four trains of tetanus of 100 pulses at 100 Hz with 20 sec intervals.
Electrophysiology-data acquisition and analysis
All the recording data were filtered at 1 or 2 kHz and digitized at 10 kHz. Analog to digital conversion was performed using Digidata 1320A or 1440A (Molecular Devices). Data were acquired using Clampex 9.2 or 10 (Molecular Devices), and analyzed using custom macros written in Igor Pro 4.0 (Wavemetrics), Minianalysis 6.0.7 (Synaptosoft), and Clampfit 10 (Axon Instruments). The experimenters were blind to the genotypes of the mice.
Molecular modeling of IgSF11 homophilic adhesion
The structure of the extracellular domain from mouse IgSF11 was modelled by homology modelling, using as a template the crystal structure of the extracellular domain of CAR (a close relative of IgSF11) [PDB ID: 3JZ7; http://www.rcsb.org/pdb/home/home.do] 70, which shows 34% aa sequence identity with mouse IgSF11, and SWISS-MODEL server (http://swissmodel.expasy.org) 71. The homophilic dimer of IgSF11 was modelled also using the homodimeric structure of mouse CAR 70, which yielded an initial dimeric structure. Rigid body docking simulation on this initial dimer (an input model) was performed using RosettaDock server 72. This simulation generated 1000 structures and returned 10 structures with best scores, of which three with different orientations and lowest energy levels were selected as final models. All structural images were generated using PyMOL molecular visualization software (version 1.3; http://www,pymol.org) 73.
Experimental designs
The experiments involving representative figures such as Western blot analysis and immunofluorescence/X-Gal staining were performed at least three times. Experiments, data collection, and analyses were performed by researchers blinded to the experimental conditions. For proper exclusion of data points, the following criteria were established before data collection. Animals, in which the virus expression could not be observed, were excluded. Cells that are morphologically unhealthy were excluded. In input-output experiments, slices, in which the slopes that did not achieve a fit of R square > 0.80, were discarded from the analysis. In whole-cell experiments, cells, which did not fulfill the standard criteria, were excluded by monitoring the electrophysiological properties, including cell capacitance, input resistance, series resistance, resting membrane potential, and baseline holding current.
Statistics
No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those generally employed in the field. All the data were randomly collected. Data distribution was assumed to be normal but this was not formally tested. Statistical tests and data point plotting were performed using GraphPad Prism 6.
Supplementary methods checklist
A supplementary methods checklist is available.
Acknowledgements
This work was supported by the Institute for Basic Science (to E.K.), the Brain Research Program through the National Research Foundation of Korea(NRF) funded by the Ministry of Science, ICT & Future Planning (2013-056732 to H.K.), and Wellcome Trust and UK MRC Neurodegenerative Disease Initiative Programme (to K.C.).
Footnotes
Competing interest declaration
The authors declare that they have no competing financial interests.
References
- 1.Yuzaki M. Cbln1 and its family proteins in synapse formation and maintenance. Current opinion in neurobiology. 2011;21:215–220. doi: 10.1016/j.conb.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi H, Craig AM. Protein tyrosine phosphatases PTPdelta, PTPsigma, and LAR: presynaptic hubs for synapse organization. Trends in neurosciences. 2013;36:522–534. doi: 10.1016/j.tins.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krueger DD, Tuffy LP, Papadopoulos T, Brose N. The role of neurexins and neuroligins in the formation, maturation, and function of vertebrate synapses. Current opinion in neurobiology. 2012;22:412–422. doi: 10.1016/j.conb.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 4.de Wit J, Hong W, Luo L, Ghosh A. Role of leucine-rich repeat proteins in the development and function of neural circuits. Annual review of cell and developmental biology. 2011;27:697–729. doi: 10.1146/annurev-cellbio-092910-154111. [DOI] [PubMed] [Google Scholar]
- 5.Shen K, Scheiffele P. Genetics and Cell Biology of Building Specific Synapse Connectivity. Annual review of neuroscience. 2010 doi: 10.1146/annurev.neuro.051508.135302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalva MB, McClelland AC, Kayser MS. Cell adhesion molecules: signalling functions at the synapse. Nature reviews Neuroscience. 2007;8:206–220. doi: 10.1038/nrn2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biederer T, Stagi M. Signaling by synaptogenic molecules. Current opinion in neurobiology. 2008;18:261–269. doi: 10.1016/j.conb.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Um JW, Ko J. LAR-RPTPs: synaptic adhesion molecules that shape synapse development. Trends in cell biology. 2013;23:465–475. doi: 10.1016/j.tcb.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nuriya M, Huganir RL. Regulation of AMPA receptor trafficking by N-cadherin. Journal of neurochemistry. 2006;97:652–661. doi: 10.1111/j.1471-4159.2006.03740.x. [DOI] [PubMed] [Google Scholar]
- 11.Saglietti L, et al. Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron. 2007;54:461–477. doi: 10.1016/j.neuron.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Budreck EC, et al. Neuroligin-1 controls synaptic abundance of NMDA-type glutamate receptors through extracellular coupling. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:725–730. doi: 10.1073/pnas.1214718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrera-Molina R, et al. Structure of excitatory synapses and GABAA receptor localization at inhibitory synapses are regulated by neuroplastin-65. The Journal of biological chemistry. 2014;289:8973–8988. doi: 10.1074/jbc.M113.514992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzu S, et al. Molecular cloning of a novel immunoglobulin superfamily gene preferentially expressed by brain and testis. Biochemical and biophysical research communications. 2002;296:1215–1221. doi: 10.1016/s0006-291x(02)02025-9. [DOI] [PubMed] [Google Scholar]
- 15.Harada H, Suzu S, Hayashi Y, Okada S. BT-IgSF, a novel immunoglobulin superfamily protein, functions as a cell adhesion molecule. Journal of cellular physiology. 2005;204:919–926. doi: 10.1002/jcp.20361. [DOI] [PubMed] [Google Scholar]
- 16.Eom DS, et al. Melanophore migration and survival during zebrafish adult pigment stripe development require the immunoglobulin superfamily adhesion molecule Igsf11. PLoS genetics. 2012;8:e1002899. doi: 10.1371/journal.pgen.1002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brennand KJ, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raschperger E, Engstrom U, Pettersson RF, Fuxe J. CLMP, a novel member of the CTX family and a new component of epithelial tight junctions. The Journal of biological chemistry. 2004;279:796–804. doi: 10.1074/jbc.M308249200. [DOI] [PubMed] [Google Scholar]
- 19.Opazo P, Sainlos M, Choquet D. Regulation of AMPA receptor surface diffusion by PSD-95 slots. Current opinion in neurobiology. 2012;22:453–460. doi: 10.1016/j.conb.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Giannone G, et al. Dynamic superresolution imaging of endogenous proteins on living cells at ultra-high density. Biophysical journal. 2010;99:1303–1310. doi: 10.1016/j.bpj.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnell E, Sizemore M, Karimzadegan S, Chen L, Bredt DS, Nicoll RA. Direct interactions between PSD-95 and stargazin control synaptic AMPA receptor number. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:13902–13907. doi: 10.1073/pnas.172511199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schreiber J, Langhorst H, Jüttner R, Rathjen FG. The IgCAMs CAR, BT-IgSF, and CLMP: Structure, Function, and Diseases. In: Berezin V, Walmod PS, editors. Cell adhesion molecules: implications in neurological diseases. Springer; 2014. [Google Scholar]
- 23.Siddiqui TJ, et al. An LRRTM4-HSPG complex mediates excitatory synapse development on dentate gyrus granule cells. Neuron. 2013 doi: 10.1016/j.neuron.2013.06.029. in press. [DOI] [PubMed] [Google Scholar]
- 24.Jackson AC, Nicoll RA. The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits. Neuron. 2011;70:178–199. doi: 10.1016/j.neuron.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chih B, Engelman H, Scheiffele P. Control of excitatory and inhibitory synapse formation by neuroligins. Science. 2005;307:1324–1328. doi: 10.1126/science.1107470. [DOI] [PubMed] [Google Scholar]
- 26.Soler-Llavina GJ, Fuccillo MV, Ko J, Sudhof TC, Malenka RC. The neurexin ligands, neuroligins and leucine-rich repeat transmembrane proteins, perform convergent and divergent synaptic functions in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16502–16509. doi: 10.1073/pnas.1114028108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chubykin AA, et al. Activity-Dependent Validation of Excitatory versus Inhibitory Synapses by Neuroligin-1 versus Neuroligin-2. Neuron. 2007;54:919–931. doi: 10.1016/j.neuron.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung SY, et al. Input-specific synaptic plasticity in the amygdala is regulated by neuroligin-1 via postsynaptic NMDA receptors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4710–4715. doi: 10.1073/pnas.1001084107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blundell J, et al. Increased Anxiety-like Behavior in Mice Lacking the Inhibitory Synapse Cell Adhesion Molecule Neuroligin 2. Genes, brain, and behavior. 2008 doi: 10.1111/j.1601-183X.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shipman SL, Nicoll RA. A subtype-specific function for the extracellular domain of neuroligin 1 in hippocampal LTP. Neuron. 2012;76:309–316. doi: 10.1016/j.neuron.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J, et al. Neuroligin-1 is required for normal expression of LTP and associative fear memory in the amygdala of adult animals. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9087–9092. doi: 10.1073/pnas.0803448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mondin M, et al. Neurexin-neuroligin adhesions capture surface-diffusing AMPA receptors through PSD-95 scaffolds. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:13500–13515. doi: 10.1523/JNEUROSCI.6439-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shipman SL, Schnell E, Hirai T, Chen BS, Roche KW, Nicoll RA. Functional dependence of neuroligin on a new non-PDZ intracellular domain. Nature neuroscience. 2011 doi: 10.1038/nn.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cingolani LA, et al. Activity-dependent regulation of synaptic AMPA receptor composition and abundance by beta3 integrins. Neuron. 2008;58:749–762. doi: 10.1016/j.neuron.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pozo K, Cingolani LA, Bassani S, Laurent F, Passafaro M, Goda Y. beta3 integrin interacts directly with GluA2 AMPA receptor subunit and regulates AMPA receptor expression in hippocampal neurons. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1323–1328. doi: 10.1073/pnas.1113736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tai CY, Kim SA, Schuman EM. Cadherins and synaptic plasticity. Current opinion in cell biology. 2008;20:567–575. doi: 10.1016/j.ceb.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 37.Schwenk J, et al. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science. 2009;323:1313–1319. doi: 10.1126/science.1167852. [DOI] [PubMed] [Google Scholar]
- 38.Shanks NF, et al. Differences in AMPA and kainate receptor interactomes facilitate identification of AMPA receptor auxiliary subunit GSG1L. Cell reports. 2012;1:590–598. doi: 10.1016/j.celrep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Engelhardt J, et al. CKAMP44: a brain-specific protein attenuating short-term synaptic plasticity in the dentate gyrus. Science. 2010;327:1518–1522. doi: 10.1126/science.1184178. [DOI] [PubMed] [Google Scholar]
- 40.Kalashnikova E, et al. SynDIG1: an activity-regulated, AMPA- receptor-interacting transmembrane protein that regulates excitatory synapse development. Neuron. 2010;65:80–93. doi: 10.1016/j.neuron.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, et al. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408:936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- 42.Schwenk J, et al. High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron. 2012;74:621–633. doi: 10.1016/j.neuron.2012.03.034. [DOI] [PubMed] [Google Scholar]
- 43.Song M, et al. Slitrk5 Mediates BDNF-Dependent TrkB Receptor Trafficking and Signaling. Developmental cell. 2015 doi: 10.1016/j.devcel.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taniguchi H, et al. Silencing of neuroligin function by postsynaptic neurexins. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:2815–2824. doi: 10.1523/JNEUROSCI.0032-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee K, et al. MDGAs interact selectively with neuroligin-2 but not other neuroligins to regulate inhibitory synapse development. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:336–341. doi: 10.1073/pnas.1219987110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pettem KL, Yokomaku D, Takahashi H, Ge Y, Craig AM. Interaction between autism-linked MDGAs and neuroligins suppresses inhibitory synapse development. The Journal of cell biology. 2013;200:321–336. doi: 10.1083/jcb.201206028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fogel AI, Stagi M, Perez de Arce K, Biederer T. Lateral assembly of the immunoglobulin protein SynCAM 1 controls its adhesive function and instructs synapse formation. The EMBO journal. 2011;30:4728–4738. doi: 10.1038/emboj.2011.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song YS, Lee HJ, Prosselkov P, Itohara S, Kim E. Trans-induced cis interaction in the tripartite NGL-1, netrin-G1 and LAR adhesion complex promotes development of excitatory synapses. Journal of cell science. 2013;126:4926–4938. doi: 10.1242/jcs.129718. [DOI] [PubMed] [Google Scholar]
- 49.Patzke C, et al. The coxsackievirus-adenovirus receptor reveals complex homophilic and heterophilic interactions on neural cells. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:2897–2910. doi: 10.1523/JNEUROSCI.5725-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lucic V, Yang T, Schweikert G, Forster F, Baumeister W. Morphological characterization of molecular complexes present in the synaptic cleft. Structure. 2005;13:423–434. doi: 10.1016/j.str.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 51.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 52.Choi J, et al. Regulation of dendritic spine morphogenesis by insulin receptor substrate 53, a downstream effector of Rac1 and Cdc42 small GTPases. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:869–879. doi: 10.1523/JNEUROSCI.3212-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neve RL, Neve KA, Nestler EJ, Carlezon WA., Jr Use of herpes virus amplicon vectors to study brain disorders. BioTechniques. 2005;39:381–391. doi: 10.2144/05393PS01. [DOI] [PubMed] [Google Scholar]
- 54.Kim J, Kwon JT, Kim HS, Josselyn SA, Han JH. Memory recall and modifications by activating neurons with elevated CREB. Nature neuroscience. 2014;17:65–72. doi: 10.1038/nn.3592. [DOI] [PubMed] [Google Scholar]
- 55.Yang J, et al. DGKiota regulates presynaptic release during mGluR-dependent LTD. The EMBO journal. 2011;30:165–180. doi: 10.1038/emboj.2010.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim MH, et al. Enhanced NMDA receptor-mediated synaptic transmission, enhanced long-term potentiation, and impaired learning and memory in mice lacking IRSp53. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:1586–1595. doi: 10.1523/JNEUROSCI.4306-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature. 1994;368:144–147. doi: 10.1038/368144a0. [DOI] [PubMed] [Google Scholar]
- 58.Han S, et al. Altered expression of synaptotagmin 13 mRNA in adult mouse brain after contextual fear conditioning. Biochemical and biophysical research communications. 2012;425:880–885. doi: 10.1016/j.bbrc.2012.07.166. [DOI] [PubMed] [Google Scholar]
- 59.Huttner WB, Schiebler W, Greengard P, De Camilli P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. The Journal of cell biology. 1983;96:1374–1388. doi: 10.1083/jcb.96.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cho KO, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- 61.Han K, et al. Regulated RalBP1 binding to RalA and PSD-95 controls AMPA receptor endocytosis and LTD. PLoS biology. 2009;7:e1000187. doi: 10.1371/journal.pbio.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jo J, et al. Abeta(1-42) inhibition of LTP is mediated by a signaling pathway involving caspase-3, Akt1 and GSK-3beta. Nature neuroscience. 2011;14:545–547. doi: 10.1038/nn.2785. [DOI] [PubMed] [Google Scholar]
- 63.Giannone G, et al. Dynamic superresolution imaging of endogenous proteins on living cells at ultra-high density. Biophysical journal. 2010;99:1303–1310. doi: 10.1016/j.bpj.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Groc L, et al. Differential activity-dependent regulation of the lateral mobilities of AMPA and NMDA receptors. Nature neuroscience. 2004;7:695–696. doi: 10.1038/nn1270. [DOI] [PubMed] [Google Scholar]
- 65.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O'Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carlin RK, Grab DJ, Cohen RS, Siekevitz P. Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. The Journal of cell biology. 1980;86:831–845. doi: 10.1083/jcb.86.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Won H, et al. GIT1 is associated with ADHD in humans and ADHD-like behaviors in mice. Nature medicine. 2011;17:566–572. doi: 10.1038/nm.2330. [DOI] [PubMed] [Google Scholar]
- 69.Schoch S, et al. RIM1alpha forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature. 2002;415:321–326. doi: 10.1038/415321a. [DOI] [PubMed] [Google Scholar]
- 70.Patzke C, et al. The coxsackievirus-adenovirus receptor reveals complex homophilic and heterophilic interactions on neural cells. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:2897–2910. doi: 10.1523/JNEUROSCI.5725-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 72.Lyskov S, Gray JJ. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 2008;36:W233–238. doi: 10.1093/nar/gkn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010 [Google Scholar]
- 74.Wyszynski M, Kim E, Yang FC, Sheng M. Biochemical and immunocytochemical characterization of GRIP, a putative AMPA receptor anchoring protein, in rat brain. Neuropharmacology. 1998;37:1335–1344. doi: 10.1016/s0028-3908(98)00120-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.