Abstract
MicroRNAs (miRNAs) have recently come to be viewed as critical players that modulate a number of cellular features in various biological systems including the mature CNS by exerting regulatory control over the stability and translation of mRNAs. Despite considerable evidence for the regulatory functions of miRNAs, the identities of the miRNA species that are involved in the regulation of synaptic transmission and plasticity and the mechanisms by which these miRNAs exert functional roles remain largely unknown. In the present study, the expression of microRNA-188 (miR-188) was found to be upregulated by the induction of long-term potentiation (LTP). The protein level of neuropilin-2 (Nrp-2), one of the possible molecular targets for miR-188, was decreased during LTP induction. We also confirmed that the luciferase activity of the 3′-UTR of Nrp-2 was diminished by treatment with a miR-188 oligonucleotide but not with a scrambled miRNA oligonucleotide. Nrp-2 serves as a receptor for semaphorin 3F, which is a negative regulator of spine development and synaptic structure. In addition, miR-188 specifically rescued the reduction in dendritic spine density induced by Nrp-2 expression in hippocampal neurons from rat primary culture. Furthermore, miR-188 counteracted the decrease in the miniature EPSC frequency induced by Nrp-2 expression in hippocampal neurons from rat primary culture. These findings suggest that miR-188 serves to fine-tune synaptic plasticity by regulating Nrp-2 expression.
Introduction
In the CNS, microRNAs (miRNAs) have been shown to regulate development, survival, function and plasticity (Yuste and Bonhoeffer, 2001; Wayman et al., 2008; Vo et al., 2010). The recognition of targets by miRNAs generally involves the 3′-untranslated region (3′-UTR) of the mRNA target and the 5′ end of the miRNA, spanning nucleotides 2–8 of the miRNA (the seed sequence). The limited sequence complementary between a miRNA and its target allows a single miRNA to regulate many mRNA targets (Vo et al., 2010). Many miRNAs and their precursors, along with the components of the miRNA machinery, exist in synaptic fractions (Lambert et al., 2010), where they are poised to regulate neurotransmission.
Long-term potentiation (LTP) is a cellular model that mimics long-term memory, requiring protein synthesis (Kotaleski and Blackwell, 2010). The structural changes in synaptic connectivity that follow the physiological changes in synaptic strength must involve the gene regulatory networks that control synaptic development, maturation and maintenance. miRNAs rapidly and coordinately regulate the stability and translation of sets of mRNAs that mediate specific processes (Kosik, 2006; Guo et al., 2010), suggesting that miRNAs could possess an important role in homeostatic synaptic plasticity (Cohen et al., 2011). Despite considerable evidence for the regulatory functions of miRNAs, the identities of the miRNA species that are involved in the regulation of synaptic transmission and plasticity as well as the mechanisms by which these miRNAs exert their functional roles remain largely unknown (Lambert et al., 2010).
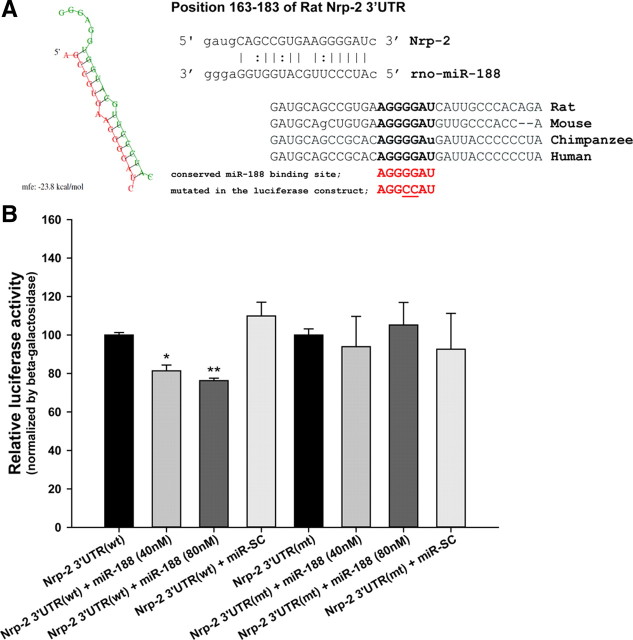
In this study, we investigated the roles and the regulatory mechanisms of miRNAs in the hippocampus during LTP. Through microarray analysis of miRNAs, we found that the expression levels of several miRNAs, including miR-188 were upregulated in rat hippocampal slices after LTP induction. The target molecules of miR-188 were, in turn, sought bioinformatically by using miRNA-target gene prediction algorithms. Neuropilin-2 (Nrp-2) has a conserved binding site for miR-188 in its 3′-UTR (positions 163–183 of the rat 3′-UTR; see Fig. 3A). In this study, we focused on the role of Nrp-2, one of the possible target molecules of miR-188, in synaptic plasticity.
Figure 3.
Nrp-2 is a target for miR-188. A, Left, the predicted hybridization of miRNAs (green) and the Nrp-2 (red) transcript using the RNAhybrid algorithm. The minimum free energy required for the hybridization is indicated. Right, a schematic diagram of rat Nrp-2 mRNA containing the predicted conserved target site of miR-188. B, HEK 293 cells were cotransfected with one of the following combinations using Lipofectamine 2000 (Invitrogen): (1) wild-type 3′-UTR of Nrp-2 in pMIR-REPORT (wt 3′UTR of Nrp-2) alone; (2) wt 3′-UTR of Nrp-2 plus 40 nm miR-188 oligonucleotide; (3) wt 3′-UTR of Nrp-2 plus 80 nm miR-188 oligonucleotide; (4) wt 3′-UTR of Nrp-2 plus miR-SC oligonucleotide; (5) mutant type 3′-UTR of Nrp-2 in pMIR-REPORT (mt 3′-UTR of Nrp-2) alone; (6) mt 3′-UTR of Nrp-2 plus 40 nm miR-188 oligonucleotide; (7) mt 3′UTR of Nrp-2 plus 80 nm miR-188 oligonucleotide; or (8) mt 3′-UTR of Nrp-2 plus miR-SC oligonucleotide. Luciferase activity assays were performed 48 h after transfection using the Luciferase Assay kit (Promega) and were measured with a Centro LB960 reader (Berthold Technologies). The β-galactosidase activity was measured to normalize the luciferase activity. The relative luciferase activity was reduced in the cells cotransfected with the wt 3′-UTR of Nrp-2 plus miR-188 (76.2822 ± 1.2642%, n = 6, **p = 0.003579), compared with the cells transfected only with the wt 3′-UTR of Nrp-2. However, in the cells cotransfected with wt 3′-UTR of Nrp-2 plus miR-SC, no significant difference was observed compared with the cells transfected only with the wt 3′-UTR of Nrp-2. The statistical analysis was performed using a nonparametric Mann–Whitney for wt 3′-UTR of Nrp-2 and independent t test for mt 3′-UTR of Nrp-2, respectively; the data are represented by the mean ± SEM, respectively.
Nrps are 130–140 kDa single-spanning transmembrane glycoproteins that function as receptors for class 3 semaphorins, polypeptides (Kolodkin et al., 1997; Chen et al., 2000) and members of the vascular endothelial growth factor (VEGF) family. Nrp-2 acts as a receptor for semaphorin-3F (Sema-3F), which induces the repulsion of Nrp-2-expressing neuronal growth cones (Kolodkin et al., 1997; Kruger et al., 2005), whereas Nrp-1 serves as a receptor for Sema-3A, which induces the collapse of the neuronal growth cone (Gu et al., 2002; Chen et al., 2005).
In this study, it was found that Nrp-2 overexpression in hippocampal neurons from rat primary culture reduced the frequency of miniature EPSCs (mEPSCs) whereas the overexpression of miR-188 prevented this reduction. Moreover, Nrp-2 overexpression diminished dendritic spine densities, but miR-188 rescued this reduction.
Together, our results suggest that a synaptic activity-regulated miRNA, miR-188, plays an important role in synaptic plasticity by downregulating Nrp-2 expression.
Materials and Methods
Hippocampal slice preparation and LTP induction.
All in vivo experiments were performed in accordance with the Guidelines for Animal Experiments set forth by the Ethics Committee of Seoul National University. Acute hippocampal slices were prepared from 4- to 5-week-old (90∼110 g) male Sprague Dawley (SD) rat brains. Briefly, brains were rapidly removed and coronal brain slices (400 μm) containing hippocampus, were cut on a Vibratome (Leica) in ice-cold artificial CSF (aCSF) [119 mm NaCl, 2.5 mm KCl, 1 mm MgSO4, 2.5 mm CaCl2, 1.25 mm NaH2PO4, 26 mm NaHCO3 and 10 mm glucose] that was bubbled with 95% O2/5% CO2 to adjust to pH 7.4. After 1.5 h recovery at 27°C, an individual slice was transferred to a submerged recording chamber and continuously superfused with oxygenated aCSF at a rate of 2.5–3 ml/min at 33 ± 1°C.
LTP was introduced by changing the bath solution to Mg2+-free aCSF solution containing 1 mm glycine, 1 μm strychnine (Sigma-Aldrich) and 100 μm picrotoxin (119 mm NaCl, 2.5 mm KCl, 1.25 mm NaH2PO4, 26 mm NaHCO3, 10 mm Glucose, 2.5 mm CaCl2, pH 7.4) for 15 min as previously described (Liu and Lachamp, 2006), then recorded in basal bath solution for 2 h. Electrical stimulation intensity was normalized to the value of basal fiber volley amplitude (FV). Then average responses (mean ± SEM) were expressed as the percentage of basal field EPSP (fEPSP) amplitude.
Electrophysiological recordings.
fEPSPs were evoked by constant stimulation of the Schaffer collaterals with a bipolar electrode and were recorded in the stratum radiatum layer of CA1 with a glass micropipette filled with 3 m NaCl (Fig. 1B). The stimulation intensities were chosen to produce a fEPSP with a slope that was 50–60% of that obtained with the maximal stimulation. The data were acquired by using an Axopatch 200A amplifier and a Digidata 1200 (Molecular Devices) interface. For AMPAR-mediated mEPSCs, the vector constructs were cotransfected into cultured primary hippocampal neurons (DIV 16–20) with one of the following combinations using CalPhos Mammalian Transfection Kit (Clontech Laboratories): (1) IRES-mGFP control vector alone; (2) Nrp-2-IRES-mGFP vector alone; (3) Nrp-2-IRES-mGFP vector plus miR-188-IRES-DsRed vector; (4) Nrp-2-IRES-mGFP vector plus 100 nm miR-124 oligonucleotide; (5) Nrp-2-IRES-mGFP vector plus 100 nm scrambled miRNA oligonucleotide (miR-SC); (6) miR-188-IRES-DsRed vector alone; or (7) Nrp-2-IRES-mGFP vector plus 100 nm 2′-O-Me miR-188 AS oligonucleotide. miR-124 is one of the most abundant miRNAs expressed in the murine brain (Lagos-Quintana et al., 2002), and the 3′-UTR of Nrp-2 does not possess binding sites for miR-124. Therefore, miR-124 was used as a negative control for miR-188 in the analyses of dendritic spine density and mEPSCs. The neurons were placed in a recording chamber and continuously superfused (1.5 ml/min) with bath solution (aCSF) containing the following: 127 mm NaCl, 5 mm KCl, 2 mm MgCl2, 2 mm CaCl2, 12 mm glucose, 10 mm HEPES, and 0.001 mm tetrodotoxin at pH 7.3–7.4 and 300–305 mOsm as described previously (Gu et al., 2009). The NMDA receptor antagonist d-aminophosphonovalerate (20 μm) and GABAA receptor antagonist picrotoxin (100 μm) were added to aCSF. The voltage-clamp of whole cells was performed with a MultiClamp 700B amplifier (Molecular Devices). The recording electrodes (8–10 MΩ) were filled with a solution containing 130 mm CsMeSO4, 8 mm NaCl, 0.5 mm EGTA, 10 mm HEPES, 2 mm MgATP, 10 mm phosphocreatine, 5 mm QX-314, and 0.1 mm NaGTP (adjusted to pH 7.2 with CsOH). The series resistance (10–30 MΩ) was monitored in all of the experiments. The membrane potential was held at −70 mV during the recording. The frequency and amplitude of the mEPSCs were analyzed with the Mini Analysis program (Synaptosoft). The noise level was <5 pA, and 10 pA was usually used as the threshold for mEPSC events. Two minutes of representative mEPSC recordings (300–400 events) were used to generate the cumulative distribution plot. The statistical comparisons of the synaptic currents were made using the Kolmogorov–Smirnov test.
Figure 1.
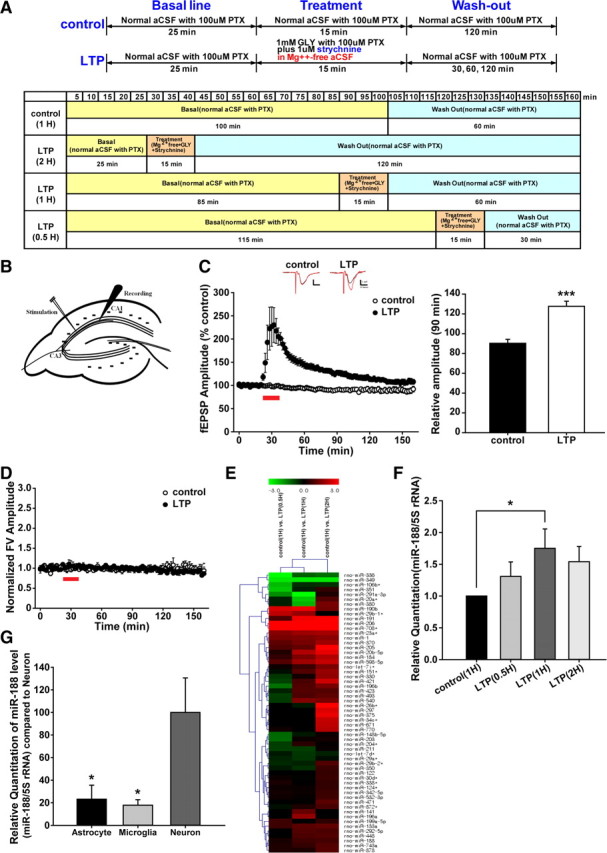
Synaptic activity upregulates miR-188 expression. LTP was introduced in acute hippocampal slices by treatment with 1 mm glycine (GLY), 1 μm strychnine and 100 μm picrotoxin (PTX) in Mg2+-free aCSF (119 mm NaCl, 2.5 mm KCl, 1.25 mm NaH2PO4, 26 mm NaHCO3, 10 mm Glucose, 2.5 mm CaCl2; pH 7.4) at 25°C. A, The experimental scheme for LTP induction is shown. The time points for collecting hippocampal slices for RNA extraction are indicated (control, LTP-0.5 h, LTP-1 h, LTP-2 h). B, A schematic illustration of the Schaffer collateral pathway in the rat hippocampal circuit that was used in the fEPSP recordings. (C, D) fEPSPs were evoked by constant stimulation in the Schaffer collaterals with a bipolar electrode and were recorded in the stratum radiatum layer of CA1 with a glass micropipette filled with 3 m NaCl. The fEPSP amplitudes were increased to 230.11 ± 37.61% (mean ± SEM, n = 7, filled circles, p = 0.0019) by LTP induction, compared with the basal aCSF solution (99.13 ± 1.54%, n = 6, open circles). The potentiated responses slowly decreased, but throughout the recording period, the cells maintained significantly potentiated responses for >60 min (126.90 ± 5.69%, n = 7 in LTP vs 90.35 ± 3.97%, n = 6 in control, ***p = 0.00017). The statistical analysis consisted of an independent t test between the control and LTP groups; the data are represented by the mean ± SEM. E, Microarray screening of miRNAs was conducted to investigate the changes in miRNA expression during LTP using an array chip with 2 sets of independent experiment [each set consisted of control (1 h), chem-LTP (0.5 h), chem-LTP (1 h), chem-LTP (2 h); the cutoff value of upregulation or downregulation is 1.5-fold] at different time points. A heatmap is shown. F, qRT-PCR was performed to examine miR-188 in the rat hippocampal slices after LTP induction. The statistical analysis used a one-way ANOVA with Fisher's least significant difference (LSD) post hoc test; the data are represented by the means ± SEM. G, qRT-PCR was performed to examine miR-188 expression in cultured primary hippocampal neurons, astrocytes, and microglia. The statistical analysis was performed using a one-way ANOVA with LSD post hoc test; the data are represented by the means ± SEM.
MicroRNA microarray analysis.
The synthesis of target miRNA probes and hybridization were performed using Agilent's miRNA Labeling Reagent and Hybridization kit (Agilent Technology) according to the manufacturer's instructions. Briefly, each 100 ng of total RNA (including small RNA fraction) was dephosphorylated with ∼15 U of calf intestine alkaline phosphatase, followed by RNA denaturation with ∼40% DMSO and 10 min incubation at 100°C. Dephosphorylated RNA were ligated with pCp-Cy3 mononucleotide and purified with MicroBioSpin 6 columns (Bio-Rad). After purification, labeled samples were resuspended with Gene Expression blocking Reagent and Hi-RPM Hybridization buffer, followed by boiling for 5 min at 100°C and chilled for 5 min on ice. Finally, denatured labeled probes were pipetted onto assembled Agilent's miRNA Microarray (15K) and hybridized for 20 h at 55°C with 20 RPM rotating in an Agilent Hybridization oven (Agilent Technology). The hybridized microarrays were washed as described in the manufacturer's washing protocol (Agilent Technology).
Data acquisition and analysis.
The hybridized images were scanned using Agilent's DNA microarray scanner and quantified with Feature Extraction Software (Agilent Technology). All data normalization and selection of fold-changed genes were performed using GeneSpringGX 7.3 (Agilent Technology). The averages of normalized ratios were calculated by dividing the average of the normalized signal channel intensity by the average of normalized control channel intensity. Functional annotation of genes was performed according to the Gene Ontology Consortium (www.geneontology.org/index.shtml) by GeneSpringGX 7.3.
MicroRNA target prediction analysis.
The target mRNAs of specific miRNAs were predicted by searching on public databases with prediction algorithms, such as MicroCosm (www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/).
Cell culture.
Human embryonic kidney 293 (HEK 293 cells) were maintained in DMEM containing 10% FBS, 50 U/ml penicillin and 50 μg/ml streptomycin at 37°C in a humidified environment of 95% O2/5% CO2. Hippocampal primary neural cultures were prepared from the hippocampi of E18–E19 pregnant SD rats by dissociation with 0.25% trypsin and plating onto coverslips coated with poly-l-lysine. The neurons were grown in Neurobasal medium (Invitrogen) supplemented with B27 (Invitrogen), 2 mm GlutaMAX-I supplement (Invitrogen), and 100 μg/ml penicillin/streptomycin (Invitrogen) at 37°C in a humidified environment of 95% O2/5% CO2.
DNA constructs.
The Nrp-2 cDNA in the IRES-mGFP vector was a generous gift from Dr. A. L. Kolodkin, The Johns Hopkins University School of Medicine, Baltimore, MD. The 3′-UTR of the rat Nrp-2 mRNA (GenBank Accession No. NM_030869) was amplified by PCR from a rat brain cDNA library and subsequently inserted downstream of the Nrp-2 open reading frame, which was confirmed by sequencing. For the luciferase activity assay, the 3′-UTR of the rat Nrp2 mRNA was amplified by PCR and then subcloned into the SpeI/MluI cloning site located downstream of the firefly luciferase gene in the pMIR-REPORT miRNA expression vector (Ambion). Point mutations were introduced into a possible miR-188 binding site located in the Nrp2 3′-UTR using an inverse PCR (iPCR)-based Site-Directed Mutagenesis Kit (KOD-Plus Mutagenesis Kit, Toyobo). The expression vectors for rno-miR-188 (miRBase Accession No. MI0006134) were prepared by introducing synthesized oligonucleotides corresponding to the precursor miRNA sequences (pre-miRNA) into the pLL3.7-DsRed2. All of the constructs were sequenced using an ABI310 Sequencer. The sequence of miR-188 mimic is 5′-CATCCCTTGCATGGTGGAGGG-3′; that of miR-SC is 5′-CCUCGUGCCGUUCCAUCAGGUAG-3′; that of miR-124 mimic is 5′-UAAGGCACGCGGUGAAUGCC-3′ (synthesized based on the sequence of rno miR-124 (miRBase Accession No. MI0000828). The sequence of the antisense 2′-O-methyl (2′-O-Me) oligonucleotide for miR-188 (2′-O-Me-188-AS) is 5′-rGrCrUrCrGrCrCrCrUrCrCrArCrCrArUrGrCmAmAmGmGmGmAmUmGrUrGrArGrA-3′ (r, RNA base; m, 2′-O-methyl base).
Luciferase activity assay.
HEK293 cells seeded at 3 × 105 cells per 6-well plate were cotransfected with one of the following combinations using Lipofectamine 2000 (Invitrogen): (1) wild-type 3′-UTR of Nrp-2 in pMIR-REPORT (wt 3′UTR of Nrp-2) alone;(2) wt 3′-UTR of Nrp-2 plus 40 nm miR-188 oligonucleotide; (3) wt 3′-UTR of Nrp-2 plus 80 nm miR-188 oligonucleotide; (4) wt 3′-UTR of Nrp-2 plus miR-SC oligonucleotide; (5) mutant type 3′-UTR of Nrp-2 in pMIR-REPORT (mt 3′-UTR of Nrp-2) alone; (6) mt 3′-UTR of Nrp-2 plus 40 nm miR-188 oligonucleotide; (7) mt 3′UTR of Nrp-2 plus 80 nm miR-188 oligonucleotide; or (8) mt 3′-UTR of Nrp-2 plus miR-SC oligonucleotide. The luciferase activity assays were performed 48 h after transfection using the Luciferase Assay kit (Promega) and were measured with a Centro LB960 reader (Berthold Technologies). The β-galactosidase activity was measured to normalize the luciferase activity.
Quantitative real-time RT-PCR.
The total RNA including small RNA fraction was extracted, and 0.5 μg was processed for cDNA synthesis using oligo(dT)20 primers and SuperScript III reverse transcriptase (Invitrogen). The cDNA was then amplified on the 7500 Fast Real-Time PCR system (Applied Biosystems), using the ΔΔCt method with SYBR Green I (Roche). The primers used were as follows: Nrp2 forward, 5′-AGGCTAACAATGATGCCACC-3′; Nrp-2 reverse, 5′-GCAACCAAAGAGCTCCAGTC-3′; GAPDH forward, 5′-ACCACCATGGAGAAGGCTGG-3′; GAPDH reverse, 5′-CTCAGTATAGCCCAGGATGC-3. To quantify the miRNA expression levels, TaqMan miRNA assay-based quantitative real-time RT-PCR (qRT-PCR; GenoExplorer miRNA qRT-PCR Kit, GenoSensor Corp., AZ) was performed on a 7500 Fast Real-Time PCR system, using the ΔΔCt method. ROX was used as an endogenous reference to standardize the miRNA expression levels. All of the data were calibrated by the universal reference data.
Dendritic spine density analysis.
Cultured primary hippocampal neurons (DIV 10–12) were transfected with one of the following combinations: (1) IRES-mGFP control vector alone; (2) Nrp-2-IRES-mGFP vector alone; (3) Nrp-2-IRES-mGFP vector plus miR-188-IRES-DsRed plasmid; (4) Nrp-2-IRES-mGFP vector plus 100 nm miR-124 oligonucleotide; (5) Nrp-2-IRES-mGFP vector plus 100 nm miR-SC; (6) miR-188-IRES-DsRed plasmid alone; or (7) IRES-mGFP control vector plus 100 nm 2′-O-Me miR-188 AS. The number of dendritic spines was evaluated at DIV 18–20. The fluorescent images were acquired with an LSM 510 confocal microscope (Carl Zeiss), using the same settings for all of the samples. The spines were counted within the 50–100 μm segments on the secondary dendrites that extended at least 40–80 μm beyond the cell body (soma).
Western blotting.
For the Western blotting, 30–50 μg of protein was loaded onto denaturing 12–15% SDS-PAGE gels and transferred to PVDF membranes (Millipore). Each membrane was then incubated in 5% skim milk or 5% bovine serum albumin for 1 h at room temperature followed by overnight incubation with appropriate primary antibodies (Nrp-2, 1:2000, Cell Signaling Technology; GAPDH, 1:5000, Santa Cruz Biotechnology). The membrane was then incubated for 1 h at room temperature with anti-rabbit or anti-mouse secondary antibodies conjugated with horse-radish peroxidase (HRP; 1:5000, Invitrogen). The HRP signals were visualized using an enhanced chemiluminescent substrate (Thermo Fisher Scientific).
Statistical analysis.
The data are expressed as the means ± SEM. values. A one-way ANOVA and independent t test (SPSS) were used for determining the statistical significance. The results were considered to be statistically significant if p < 0.05.
Results
Synaptic activity induced by LTP upregulates miR-188 expression
LTP was induced in acute hippocampal slices by treatment with 1 mm glycine, 1 μm strychnine (Sigma-Aldrich) and 100 μm picrotoxin in Mg2+ free aCSF (119 mm NaCl, 2.5 mm KCl, 1.25 mm NaH2PO4, 26 mm NaHCO3, 10 mm Glucose, 2.5 mm CaCl2; pH 7.4) at 25°C (for detailed procedures, see Materials and Methods, Hippocampal slice preparation and LTP induction, and Fig. 1A) to investigate the changes in miRNA expression profiles during LTP induction.
We first confirmed that LTP was induced by performing basal recordings at the Schaffer Collateral-CA1 synapses (Fig. 1B). The fEPSP amplitudes were substantially increased to 230.11 ± 37.61% (mean ± SEM, n = 7, **p = 0.0019) by LTP induction with the reagents (1 mm glycine, 1 μm strychnine and 100 μm picrotoxin in Mg2+-free aCSF) but not by the basal aCSF solution (99.13 ± 1.54%, n = 6; Fig. 1C). The potentiated responses slowly decreased, but throughout the recording period, the cells maintained significantly potentiated responses for >1 h (126.90 ± 5.69%, n = 7 in LTP versus 90.35 ± 3.97%, n = 6 in control, ***p = 0.00017; Fig. 1C).
We also analyzed the fiber volley amplitudes in each experimental case to normalize the electrical stimulation and to rule out the effects of any dying axons or hyperactivated axonal signals. Throughout the recording, the fiber volley amplitudes were kept as the basal values (not exceeding 10% of the basal range (Fig. 1D). In addition, the expression levels of the immediate early genes known to be induced by LTP, such as c-fos and cpg-15, were significantly increased by the LTP induction (data not shown).
Next, miRNA microarray screening was performed to investigate the changes in the miRNA expression profile during LTP induction. From the 287 screened miRNAs in the array chip containing two sets of independent experiments [each set consisted of control, LTP (0.5 h), LTP (1 h) and LTP (2 h)], we found that five miRNAs were significantly altered when assessed by microarray analysis (the cutoff value of upregulation or downregulation was 1.5-fold; Fig. 1E). We confirmed the results from the microRNA microarray analysis by performing qRT-PCR. miR-188 showed a steep increase at 1 h after LTP induction (LTP (1 h), 1.7510 ± 0.3045-fold, n = 3 compared with the control (1 h); *p = 0.045781; Fig. 1F). This result is consistent with previous reports showing the importance of protein synthesis during the induction stage of synaptic plasticity, especially within 1 h of LTP induction, as demonstrated by treatment with anisomycin, an inhibitor of protein synthesis (Fonseca et al., 2006b; Karpova et al., 2006). When anisomycin was applied to brain slices or to primary cultured neurons following LTP induction by electrophysiological or chemical methods, the level of synaptic potentiation was obscured (Fonseca et al., 2006a).
Hippocampal slice cultures should contain neurons as well as glial cells (Yokose et al., 2011). To determine which cell type was producing the miR-188, we investigated the expression of miR-188 in primary hippocampal neurons, astrocytes and microglia by qRT-PCR. As shown in Figure 1G, miR-188 was expressed approximately fourfold to fivefold more in primary neurons, compared with astrocytes and microglia.
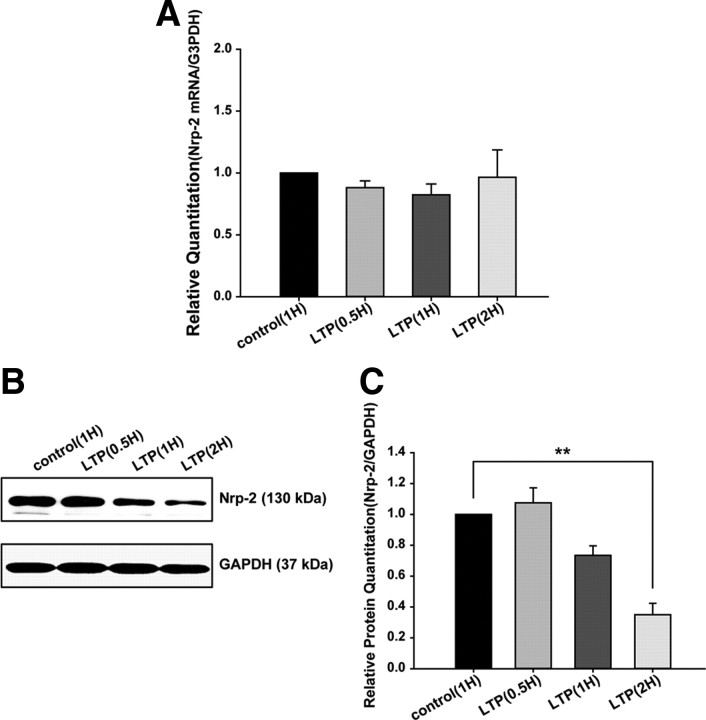
miR-188 decreases Nrp-2 protein levels, but not Nrp-2 mRNA levels
By using different miRNA-target prediction algorithms, including MicroCosm, we predicted a list of target genes for miR-188. Based on this list of target genes, we focused on Nrp-2 as a molecular target for miR-188. The qRT-PCR results showed that the mRNA level of Nrp-2 was not significantly altered at any time point (Fig. 2A), but the Nrp-2 protein level was significantly decreased 2 h after LTP induction in rat hippocampal slices (0.3500 ± 0.0740, n = 3; normalized value compared with the control; ***p = 0.000679; Fig. 2B,C). We examined the protein levels of ephrin A1, another possible target for miR-188. However, there were no significant changes in the ephrin A1 protein levels at any time point following LTP induction (data not shown).
Figure 2.
miR-188 decreases Nrp-2 protein levels, but not Nrp-2 mRNA levels. A, The mRNA level of Nrp-2 was examined in rat hippocampal slices by RT-PCR after LTP induction. B, The Nrp-2 protein level was checked by Western blotting with rat hippocampal slices after LTP induction. C, The relative densitometrical value of Nrp-2 is shown in the graph. The statistical analysis was performed by a one-way ANOVA with Bonferroni's post hoc test; the data are represented by means ± SEM.
Through the bioinformatics matching analysis, we predicted that the seed sequence of miR-188 (2–8 nt) binds to the conserved binding site, 5′-AGGGGAU-3′ (positions 176–182 of the rat 3′-UTR) on the 3′-UTR of the Nrp-2 mRNA and affects Nrp-2 protein translation (Fig. 3A). A luciferase activity assay was performed 48 h after cotransfecting pMIR-Luc-3′-UTR constructs containing either the target binding sequence of miR-188 in the wt 3′-UTR of the Nrp-2 mRNA or a mutant sequence containing the sequence (5′-AGGCCAU-3′) in the miR-188 binding site, together with 40 or 80 nm miR-188 or miR-SC oligonucleotides, in HEK 293 cells. The relative luciferase activity was reduced in the cells cotransfected with the wt 3′-UTR of Nrp-2 plus miR-188 (76.2822 ± 1.2642%, n = 6, **p = 0.003579), compared with the cells transfected only with the wt 3′-UTR of Nrp-2. However, in the cells cotransfected with the wt 3′-UTR of Nrp-2 plus miR-SC, no significant difference was observed compared with the cells transfected only with the wt 3′-UTR of Nrp-2 (109.9178 ± 7.0928%, n = 3, p = 0.432768). In case of the mt 3′-UTR, cotransfection with either miR-188 or miR-SC did not affect the luciferase activity (Fig. 3B). This result indicates that miR-188 specifically binds to the 3′-UTR of Nrp-2 and represses the translation of luciferase.
Based on this, we concluded that miR-188 binds directly to the 3′-UTR of the Nrp-2 mRNA and decreases the Nrp-2 protein level by translational repression (Fig. 2A,B,C and Fig. 3B).
miR-188 restores the Nrp-2-induced reduction in the dendritic spine density
The majority of the excitatory synapses in the mammalian CNS are formed on dendritic spines, and spine morphology and distribution are critical for synaptic transmission, synaptic integration and plasticity (Yuste and Bonhoeffer, 2001; Spruston, 2008). A previous study showed that the knock-out of Nrp-2 induces an increase in the dendritic spine number and the mEPSC frequency, especially in dentate gyrus granule cells and cortical layer V (Tran et al., 2009).
To examine the role of Nrp-2 and its regulation by miR-188 at the synapse, we transfected Nrp-2 cDNA that contained the 3′-UTR subcloned into IRES-mGFP alone or together with miR-188 (in IRES-DsRed2), miR-124 oligonucleotide, miR-SC oligonucleotide (Genolution Pharmaceuticals) into primary cultured hippocampal neurons at DIV 10–12 and analyzed the number of dendritic spines at DIV 18–20 (Fig. 4A–F). In addition, the number of dendritic spines in the neurons transfected with control vector plus 2′-O-Me-188-AS (IDT, CA) were also investigated (Fig. 4G). First, we investigated the effects of 2′-O-Me-188-AS on Nrp-2 protein levels 24 h after transfecting 2′-O-Me-188-AS into primary cultured hippocampal neurons at DIV 13. The Nrp-2 protein level was found to be increased by treatment with 2′-O-Me-188-AS (Fig. 4I), indicating that the translation of Nrp-2 is regulated by endogenous miR-188 in primary cultured hippocampal neurons. In addition, the protein level of Nrp-2 in cells cotransfected with miR-188 was found to be decreased compared with the level in cells transfected with Nrp-2 only, whereas there was no change observed when miR-SC was cotransfected with Nrp-2 (Fig. 4J).
Figure 4.
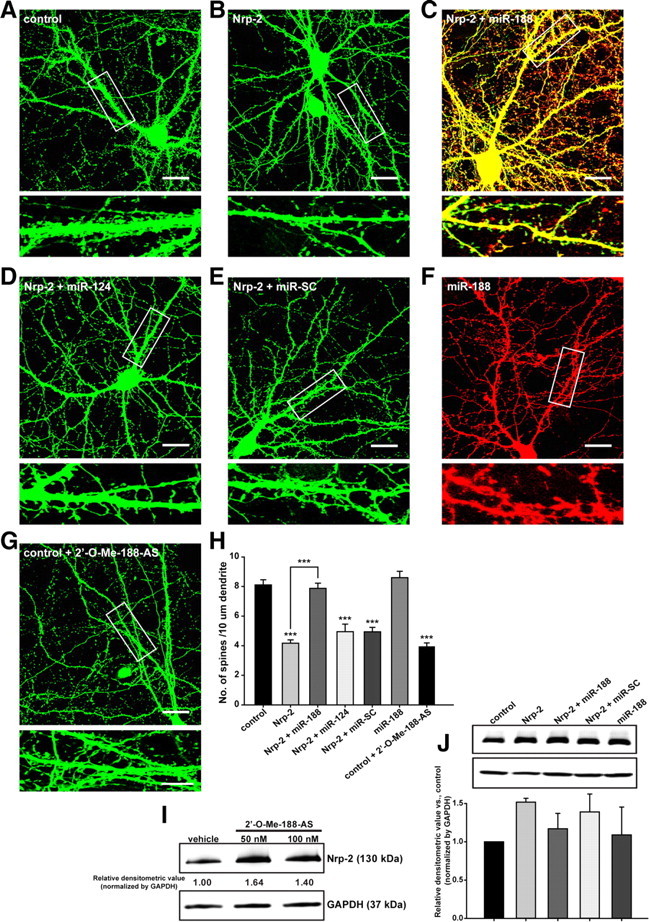
miR-188 reverses the Nrp-2-induced reduction in dendritic spine density. A–G, Representative images of dendritic spines on cultured primary hippocampal neurons at DIV 18–20 after transfecting Nrp-2 (in IRES-mGFP), either alone or with miR-188 (IRES-DsRed2), miR-124 or miR-SC oligonucleotides or transfecting with miR-188 alone or with IRES-mGFP vector plus 2′-O-Me-188-AS at DIV 10–12. The dendritic segment outlined with a white box (upper) is magnified to delineate spine morphology (bottom). Scale bars: 20 μm (low-scaled images), 5 μm (magnified images). H, A quantification of the spine density (40–80 μm of secondary dendritic spines from soma) in DIV 18–20 after transfection into cultured primary hippocampal neurons at DIV 10–12. Nrp-2 overexpression reduced the number of dendritic spines per 10 μm of dendrites by a significant 51.4% (4.1708 ± 0.2292, n = 12, ***p = 4.58 E12) compared with the mGFP-transfected control (8.1074 ± 0.3505, n = 9). Cotransfection with Nrp-2 plus miR-188 reversed the reduction in the number of dendritic spines induced by Nrp-2 (7.8778 ± 0.33497, n = 9, p = 0.637684) compared with the control (8.1074 ± 0.3505, n = 9). However, cotransfection with miR-SC or miR-124 did not reverse the reduction in the number of dendritic spines induced by Nrp-2. The statistical analysis was performed by a one-way ANOVA with least significant difference (LSD) post hoc test; the data are represented by the means ± SEM. I, The Nrp-2 protein level was assessed by Western blotting after 2′-O-Me-188-AS (50 and 100 nm) treatment for 24 h in primary hippocampal neurons. The blot is the representative of the two independent experiments. The intensities of the bands were measured relative to the amount of GAPDH in each sample. The relative densitometric value versus the vehicle-treated group is marked in numbers. J, The Nrp-2 protein level was assessed by Western blotting after transfection with Nrp-2 cDNA alone, or with Nrp-2 plus miR-188 or miR-SC or miR-188 alone in primary cultured hippocampal neurons. The blot is the representative of the three independent experiments. The intensity of the bands was measured relative to the amount of GAPDH in each sample. The relative densitometric value versus the mGFP control group is shown in graph.
Nrp-2 overexpression reduced the number of dendritic spines per 10 μm of dendrites by a significant 51.4% (4.1708 ± 0.2292, n = 12, ***p = 4.57 E12) compared with the mGFP-transfected control (8.1074 ± 0.3505, n = 9, Fig. 4B,H). However, cotransfection with Nrp-2 plus miR-188 rescued the reduction in the number of dendritic spines induced by Nrp-2 (7.8778 ± 0.33497, n = 9, p = 0.637684) compared with the control (8.1074 ± 0.3505, n = 9, Fig. 4C,H), indicating an important role for miR-188 in regulating dendritic spine density. We also confirmed that this reversal effect by miR-188 was specific by showing that cotransfection with miR-SC or miR-124 did not reverse the reduction in the number of dendritic spines induced by Nrp-2 (Fig. 4D,E,H).
When miR-188 was transfected alone, a significant change in the dendritic spine density was not observed (8.6028 ± 0.4247, n = 6, p = 0.364828) compared with the control (8.1074 ± 0.3505, n = 9, Fig. 4F,H). Although this result was somewhat inconsistent with our expectations, it is possible that endogenous miR-188 already plays a sufficiently large role in regulating Nrp-2 expression in hippocampal cultured neurons such that the addition of miR-188 does not affect the number of dendritic spines. In addition, it might be possible that the down-stream molecules of the Nrp-2 signaling pathway in dendritic spine formation, which have not been elucidated to date, are regulated by miR-188 or by other miRNAs (miR-338* and miR-29a*) found to be induced by LTP in our study.
miR-188 rescues the Nrp-2-induced reduction in basal synaptic transmission
Next, we recorded mEPSCs and analyzed the frequency and amplitude to investigate whether miR-188 plays a role in basal synaptic transmission. To investigate the relationship between Nrp-2 and miR-188 on mEPSCs, we used a dual whole-cell recording method in cultured hippocampal neurons to simultaneously record in transfected and nontransfected cells (Fig. 5A). First, we analyzed the effects of Nrp-2 overexpression on basal synaptic transmission. The mEPSC frequency of Nrp-2 transfected cultured primary hippocampal neurons was significantly decreased (2.090 ± 0.585 Hz, n = 8, **p = 0.014763; Fig. 5B,C) compared with nontransfected hippocampal neurons nearby (4.462 ± 0.471 Hz, n = 8) whereas that of control vector transfected neurons showed no difference, compared with nontransfected neurons nearby (data not shown). However, cotransfection of miR-188 with Nrp-2 completely reversed the reduction in mEPSC frequency compared with nontransfected cells (transfected, 3.118 ± 0.718 Hz vs nontransfected neurons, 3.788 ± 1.055 Hz, p = 0.608229), whereas cotransfection with miR-124 (which was used as a negative control), miR-SC or 2′-O-Me-188-AS, did not reverse the reduction in mEPSC frequency induced by Nrp-2 overexpression (Fig. 5B,C).
Figure 5.
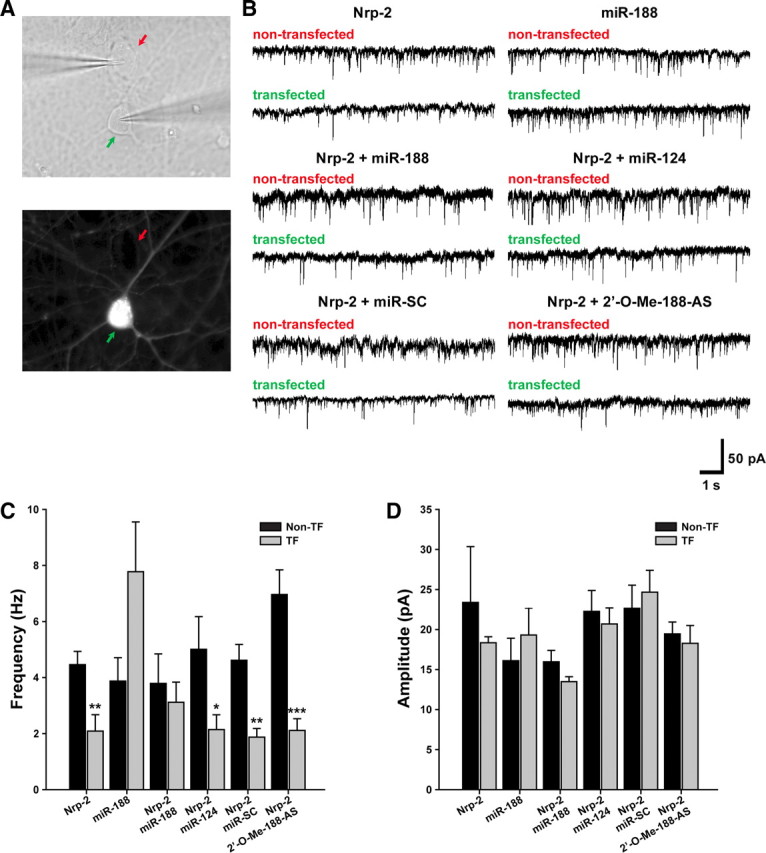
miR-188 reverses the Nrp-2-induced reduction in basal synaptic transmission. A, The configuration of the dual whole-cell recording model to simultaneously measure in transfected neuron (green arrow) and nontransfected neuron (red arrow). B, Sample traces of mEPSCs recorded in cultured primary hippocampal neurons after transfecting Nrp-2 (in IRES-mGFP) alone or Nrp-2 with miR-188 (IRES-DsRed2), miR-124 or miR-SC oligonucleotides or 2′-O-Me-188-AS at DIV 10–12. C, Bar graphs for mean values of the mEPSC frequencies of transfected (black bar) and nontransfected (gray bar) cultured primary hippocampal neurons are shown. The mEPSC frequency of Nrp-2-transfected cultured primary hippocampal neurons was significantly decreased (2.090 ± 0.585 Hz, n = 8, *p = 0.014763), compared with nontransfected hippocampal neurons nearby (4.462 ± 0.471 Hz, n = 8). However, cotransfection of miR-188 with Nrp-2 completely reversed the reduction in mEPSC frequency compared with nontransfected cells (transfected, 3.1190 ± 0.7190 Hz vs nontransfected neurons, 3.788 ± 1.055 Hz, p = 0.959130), whereas cotransfection with miR-124 (which was used as a negative control), miR-SC or 2′-O-Me-188-AS, did not reverse the reduction in mEPSC frequency induced by Nrp-2 overexpression. Statistical analysis was performed by a Mann–Whitney test; Data represents mean ± SEM. D, A bar graph of the mean values of the mEPSC amplitudes of cultured primary hippocampal neurons transfected with each construct (black bar) which were not altered in comparison with the nontransfected neurons (gray bar).
However, the mEPSC amplitude was not significantly different among all the groups (Fig. 5D). These results indicate that miR-188 modulates basal synaptic transmission by regulating Nrp-2. An increase or decrease in the number of spines is generally accompanied by a respective increase or decrease in mEPSCs frequency (but not amplitude; Hsia et al., 1998; Saglietti et al., 2007; Xie et al., 2007; Barbosa et al., 2008; Arikkath et al., 2009; Hayashi-Takagi et al., 2010).
Discussion
The high abundance of miRNAs in the mammalian brain strongly suggests that miRNAs may represent important post-transcriptional regulators of gene expression by subtle but influential changes to the genetic programs that normally occur in neural cells (Kosik, 2006; Lau and Hudson, 2010; Benoit and Tenner, 2011). One miRNA can target up to several hundred genes (Guo et al., 2010; Wang et al., 2010) to destabilize the mRNA transcripts that mediate complex cellular processes, such as development and plasticity, in a coordinated manner (Cohen et al., 2011). It is also well known that miRNAs are able to regulate the expression of at least one-third of all human genes (Bartel and Chen, 2004; Shi et al., 2011). Although miRNAs in the CNS have been studied extensively in recent years, the involvement of miRNAs in adult, neural processes, such as the fine regulation of gene expression that is essential for the adjustment of the cell and organism to the dynamic cellular and physiological conditions, is far less understood (Hébert and De Strooper, 2009; Haramati et al., 2011). The dysfunctions of miRNAs and alterations in their expression have been shown to be associated with the pathogenesis of neurodegenerative diseases such as Alzheimer's and Parkinson's diseases (Cogswell et al., 2008; Saba and Schratt, 2010).
In this study, we found that miR-188 expression was upregulated by LTP induction, and that it plays an important role in dendritic spine formation and synaptic transmission by specifically regulating the expression of Nrp-2. The 3′-UTR of Nrp-2 has possible binding sites for 29 miRNAs when predicted with MicroCosm bioinformatic program. Among the 29 miRNAs, let-7a, let-7b, let-7c, miR-124, miR-126, miR-29b, and miR-9 are known to be expressed in the hippocampus. However, none of the 7 miRNAs described above showed significant changes in expression after LTP induction in our results with miRNA microarray analysis.
We confirmed that miR-188 expression was increased 1 h after LTP induction by performing qRT-PCR (Fig. 1F) and that Nrp-2 is a target for miR-188 by luciferase activity assay (Fig. 3). miR-188 expression was detected to be increased 1 h after LTP induction and a significant reduction in Nrp-2 was observed 2 h after LTP induction. This time difference is thought to occur because a certain amount of time is required for miR-188 to repress Nrp-2 translation.
Nrp-2 is a receptor for Sema-3F, which is a negative regulator of spine development and synaptic structure. Nrp-2 and Sema-3F are expressed during synaptogenesis in the hippocampus at postnatal day (P)21. The Nrp-2 intracellular domain contains a C-terminal PDZ ligand motif that may be critical for Nrp-2/plexinA3 localization and Sema-3F/Nrp-2-mediated regulation of spine morphology and synapse structure (Tran et al., 2009). Nrp-2 is enriched in the dentate gyrus molecular layer, where the dendrites of the granule cells reside. Sema-3F is strongly expressed in the hilus, along the projection pathways of both the supra and intrapyramidal axons and the entorhinal cortex axons that innervate the dentate gyrus molecular layer. Therefore, Sema-3F and Nrp-2 are expressed in patterns consistent with the hypothesis that these proteins direct postnatal hippocampal neural circuit formation (Tran et al., 2009). Class 3 semaphorins are secreted proteins that act as potent chemorepellents for specific classes of central and peripheral nervous system neurons in vitro. Nrps are unusual in their ability to bind, with high affinity, two structurally unrelated ligands, i.e., semaphorins and VEGFs that have distinct biological functions (Tong and Buck, 2005; Zachary et al., 2009).
The VEGF family is involved in the formation of new blood vessels in both malignant and nonmalignant conditions, by binding with the Nrp-1 or -2 receptor in endothelial cells (Chen et al., 2005). However, the function of VEGF through these receptors in the CNS has not been defined, yet. Recently, it was reported that treatment with fluoxetine, an antidepressant, induces VEGF expression in the brain and that this effect is related to antidepressant behaviors. VEGF was reported to be implicated in hippocampal neurogenesis in the rodent brain (Gluzman-Poltorak et al., 2001; Lee et al., 2009). However, its exact mechanisms or action during LTP remains to be clarified. Nrp-2 may exert an effect on synaptic plasticity through other ligands such as VEGF together with Sema-3F.
Mice with null mutations in the genes encoding Sema-3F and its holoreceptor Nrp-2 exhibit increased spine number and size in the dentate gyrus granule cells and the cortical layer V pyramidal neurons as well as aberrant spine distribution. In contrast, a distinct Sema-3A-Nrp-1/PlexA4 signaling cascade controls the basal dendritic arborization in the layer V cortical neurons but does not influence spine morphogenesis or distribution (Tran et al., 2009). During brain development, the growth cones of axons navigate toward their target fields, where synapses are formed with the appropriate postsynaptic partners. It is becoming clear that pathfinding by most axons is dependent on a complex combination of secreted and membrane-bound cone attractants and repellents (Shimakawa et al., 2002). Axons are guided by both positive and negative cues that they encounter as they find their way to their targets (Naska et al., 2010).
A recent report demonstrated that neuron-glial-related-cell adhesion molecule formed a molecular complex with Nrp-2 and that the formation of this complex was required for growth cone collapse in response to Sema-3F in thalamic neuron cultures (Demyanenko et al., 2011).
In addition to the initially identified roles of miRNAs in development and cellular identity, more recently, their roles in neurodegenerative diseases have been appreciated (Hébert and De Strooper, 2009, Schonrock et al., 2011, Wang et al., 2010). In our unpublished experiments, miR-188 expression was found to be significantly diminished in hippocampi and cerebral cortices from AD patients, suggesting that the dendritic spine alterations observed in AD are at least in part due to the deregulation of miR-188 expression and the subsequent lack of homeostatic control of the target molecules.
In this study, we found that the overexpression of miR-188 reversed the Nrp-2-induced reduction in dendritic spine density in cultured primary hippocampal neurons (Fig. 4). In addition, the mEPSC frequency of Nrp-2-transfected cultured primary hippocampal neurons was significantly decreased, in comparison with nontransfected neurons nearby. However, the overexpression of miR-188 reversed the decreased frequency of mEPSCs observed in Nrp-2-transfected hippocampal neurons. In contrast, the mEPSC amplitude showed no significant difference among all of the groups (Fig. 5). The finding that the mEPSC frequency was decreased by Nrp-2 overexpression without effects on mEPSC amplitude supports the morphological findings that Nrp-2 decreases synaptic spine density but that miR-188 reverses this decrease. These results indicate that miR-188 regulates basal synaptic transmission by suppressing Nrp-2 expression.
In conclusion, our study provides additional evidence that LTP induction causes time-dependent changes in miRNA expression, suggesting the involvement of these molecules in the cellular response to LTP.
Footnotes
This work was financially supported by grants from Korea Healthcare Technology R and D Project (A080137 and A111230), by Ministry for Health, Welfare and Family Affairs, Republic of Korea, by the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology, Grant 2011-0021866, and in part by Seoul National University Bundang Hospital Research Fund (03-2010-007). We thank Dr. A. L. Kolodkin for providing the Nrp-2 cDNA in the mGFP vector.
References
- Arikkath J, Peng IF, Ng YG, Israely I, Liu X, Ullian EM, Reichardt LF. Delta-catenin regulates spine and synapse morphogenesis and function in hippocampal neurons during development. J Neurosci. 2009;29:5435–5442. doi: 10.1523/JNEUROSCI.0835-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AC, Kim MS, Ertunc M, Adachi M, Nelson ED, McAnally J, Richardson JA, Kavalali ET, Monteggia LM, Bassel-Duby R, Olson EN. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci U S A. 2008;105:9391–9396. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Benoit ME, Tenner AJ. Complement protein C1q-mediated neuroprotection is correlated with regulation of neuronal gene and microRNA expression. J Neurosci. 2011;31:3459–3469. doi: 10.1523/JNEUROSCI.3932-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Li M, Chai H, Yang H, Fisher WE, Yao Q. Roles of neuropilins in neuronal development, angiogenesis, and cancers. World J Surg. 2005;29:271–275. doi: 10.1007/s00268-004-7818-1. [DOI] [PubMed] [Google Scholar]
- Chen H, Bagri A, Zupicich JA, Zou Y, Stoeckli E, Pleasure SJ, Lowenstein DH, Skarnes WC, Chédotal A, Tessier-Lavigne M. Neuropilin-2 regulates the development of selective cranial and sensory nerves and hippocampal mossy fiber projections. Neuron. 2000;25:43–56. doi: 10.1016/s0896-6273(00)80870-3. [DOI] [PubMed] [Google Scholar]
- Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- Cohen JE, Lee PR, Chen S, Li W, Fields RD. MicroRNA regulation of homeostatic synaptic plasticity. Proc Natl Acad Sci U S A. 2011;108:11650–11655. doi: 10.1073/pnas.1017576108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demyanenko GP, Riday TT, Tran TS, Dalal J, Darnell EP, Brennaman LH, Sakurai T, Grumet M, Philpot BD, Maness PF. NrCAM deletion causes topographic mistargeting of thalamocortical axons to the visual cortex and disrupts visual acuity. J Neurosci. 2011;31:1545–1558. doi: 10.1523/JNEUROSCI.4467-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca R, Nägerl UV, Bonhoeffer T. Neuronal activity determines the protein synthesis dependence of long-term potentiation. Nat Neurosci. 2006a;9:478–480. doi: 10.1038/nn1667. [DOI] [PubMed] [Google Scholar]
- Fonseca R, Vabulas RM, Hartl FU, Bonhoeffer T, Nägerl UV. A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron. 2006b;52:239–245. doi: 10.1016/j.neuron.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Gluzman-Poltorak Z, Cohen T, Shibuya M, Neufeld G. Vascular endothelial growth factor receptor-1 and neuropilin-2 form complexes. J Biol Chem. 2001;276:18688–18694. doi: 10.1074/jbc.M006909200. [DOI] [PubMed] [Google Scholar]
- Gu C, Limberg BJ, Whitaker GB, Perman B, Leahy DJ, Rosenbaum JS, Ginty DD, Kolodkin AL. Characterization of neuropilin-1 structural features that confer binding to semaphorin 3A and vascular endothelial growth factor 165. J Biol Chem. 2002;277:18069–18076. doi: 10.1074/jbc.M201681200. [DOI] [PubMed] [Google Scholar]
- Gu Z, Liu W, Yan Z. beta-Amyloid impairs AMPA receptor trafficking and function by reducing Ca2+/calmodulin-dependent protein kinase II synaptic distribution. J Biol Chem. 2009;284:10639–10649. doi: 10.1074/jbc.M806508200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramati S, Navon I, Issler O, Ezra-Nevo G, Gil S, Zwang R, Hornstein E, Chen A. MicroRNA as repressors of stress-induced anxiety: the case of amygdalar miR-34. J Neurosci. 2011;31:14191–14203. doi: 10.1523/JNEUROSCI.1673-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ, Makino Y, Seshadri AJ, Ishizuka K, Srivastava DP, Xie Z, Baraban JM, Houslay MD, Tomoda T, Brandon NJ, Kamiya A, Yan Z, Penzes P, Sawa A. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci. 2010;13:327–332. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Hsia AY, Malenka RC, Nicoll RA. Development of excitatory circuitry in the hippocampus. J Neurophysiol. 1998;79:2013–2024. doi: 10.1152/jn.1998.79.4.2013. [DOI] [PubMed] [Google Scholar]
- Karpova A, Mikhaylova M, Thomas U, Knöpfel T, Behnisch T. Involvement of protein synthesis and degradation in long-term potentiation of Schaffer collateral CA1 synapses. J Neurosci. 2006;26:4949–4955. doi: 10.1523/JNEUROSCI.4573-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin AL, Levengood DV, Rowe EG, Tai YT, Giger RJ, Ginty DD. Neuropilin is a semaphorin III receptor. Cell. 1997;90:753–762. doi: 10.1016/s0092-8674(00)80535-8. [DOI] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Kotaleski JH, Blackwell KT. Modelling the molecular mechanisms of synaptic plasticity using systems biology approaches. Nat Rev Neurosci. 2010;11:239–251. doi: 10.1038/nrn2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger RP, Aurandt J, Guan KL. Semaphorins command cells to move. Nat Rev Mol Cell Biol. 2005;6:789–800. doi: 10.1038/nrm1740. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lambert TJ, Storm DR, Sullivan JM. MicroRNA132 modulates short-term synaptic plasticity but not basal release probability in hippocampal neurons. PLoS One. 2010;5:e15182. doi: 10.1371/journal.pone.0015182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P, Hudson LD. MicroRNAs in neural cell differentiation. Brain Res. 2010;1338:14–19. doi: 10.1016/j.brainres.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Jang DJ, Lee N, Ko HG, Kim H, Kim YS, Kim B, Son J, Kim SH, Chung H, Lee MY, Kim WR, Sun W, Zhuo M, Abel T, Kaang BK, Son H. Induction of neuronal vascular endothelial growth factor expression by cAMP in the dentate gyrus of the hippocampus is required for antidepressant-like behaviors. J Neurosci. 2009;29:8493–8505. doi: 10.1523/JNEUROSCI.1321-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Lachamp P. The activation of excitatory glutamate receptors evokes a long-lasting increase in the release of GABA from cerebellar stellate cells. J Neurosci. 2006;26:9332–9339. doi: 10.1523/JNEUROSCI.2929-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naska S, Lin DC, Miller FD, Kaplan DR. p75NTR is an obligate signaling receptor required for cues that cause sympathetic neuron growth cone collapse. Mol Cell Neurosci. 2010;45:108–120. doi: 10.1016/j.mcn.2010.05.015. [DOI] [PubMed] [Google Scholar]
- Saba R, Schratt GM. MicroRNAs in neuronal development, function and dysfunction. Brain Res. 2010;1338:3–13. doi: 10.1016/j.brainres.2010.03.107. [DOI] [PubMed] [Google Scholar]
- Saglietti L, Dequidt C, Kamieniarz K, Rousset MC, Valnegri P, Thoumine O, Beretta F, Fagni L, Choquet D, Sala C, Sheng M, Passafaro M. Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron. 2007;54:461–477. doi: 10.1016/j.neuron.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Schonrock N, Matamales M, Ittner LM, Gotz J. MicroRNA networks surrounding APP and amyloid-beta metabolism—Implications for Alzheimer's disease. Exp Neurol. 2011 doi: 10.1016/j.expneurol.2011.11.013. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Shi G, Liu Y, Liu T, Yan W, Liu X, Wang Y, Shi J, Jia L. Upregulated miR-29b promotes neuronal cell death by inhibiting Bcl2L2 after ischemic brain injury. Exp Brain Res. 2011;216:225–230. doi: 10.1007/s00221-011-2925-3. [DOI] [PubMed] [Google Scholar]
- Shimakawa S, Suzuki S, Miyamoto R, Takitani K, Tanaka K, Tanabe T, Wakamiya E, Nakamura F, Kuno M, Matsuura S, Watanabe Y, Tamai H. Neuropilin-2 is overexpressed in the rat brain after limbic seizures. Brain Res. 2002;956:67–73. doi: 10.1016/s0006-8993(02)03482-0. [DOI] [PubMed] [Google Scholar]
- Spruston N. Pyramidal neurons: dendritic structure and synaptic integration. Nat Rev Neurosci. 2008;9:206–221. doi: 10.1038/nrn2286. [DOI] [PubMed] [Google Scholar]
- Tong Y, Buck M. 1H, 15N and 13C Resonance assignments and secondary structure determination reveal that the minimal Rac1 GTPase binding domain of plexin-B1 has a ubiquitin fold [Letter] J Biomol NMR. 2005;31:369–370. doi: 10.1007/s10858-005-0943-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TS, Rubio ME, Clem RL, Johnson D, Case L, Tessier-Lavigne M, Huganir RL, Ginty DD, Kolodkin AL. Secreted semaphorins control spine distribution and morphogenesis in the postnatal CNS. Nature. 2009;462:1065–1069. doi: 10.1038/nature08628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo NK, Cambronne XA, Goodman RH. MicroRNA pathways in neural development and plasticity. Curr Opin Neurobiol. 2010;20:457–465. doi: 10.1016/j.conb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Wang WX, Wilfred BR, Madathil SK, Tang G, Hu Y, Dimayuga J, Stromberg AJ, Huang Q, Saatman KE, Nelson PT. miR-107 regulates granulin/progranulin with implications for traumatic brain injury and neurodegenerative disease. Am J Pathol. 2010;177:334–345. doi: 10.2353/ajpath.2010.091202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Srivastava DP, Photowala H, Kai L, Cahill ME, Woolfrey KM, Shum CY, Surmeier DJ, Penzes P. Kalirin-7 controls activity-dependent structural and functional plasticity of dendritic spines. Neuron. 2007;56:640–656. doi: 10.1016/j.neuron.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokose J, Ishizuka T, Yoshida T, Aoki J, Koyanagi Y, Yawo H. Lineage analysis of newly generated neurons in organotypic culture of rat hippocampus. Neurosci Res. 2011;69:223–233. doi: 10.1016/j.neures.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu Rev Neurosci. 2001;24:1071–1089. doi: 10.1146/annurev.neuro.24.1.1071. [DOI] [PubMed] [Google Scholar]
- Zachary IC, Frankel P, Evans IM, Pellet-Many C. The role of neuropilins in cell signalling. Biochem Soc Trans. 2009;37:1171–1178. doi: 10.1042/BST0371171. [DOI] [PubMed] [Google Scholar]