Abstract
Patients with pancreatic ductal adenocarcinoma (PDA) have a poor prognosis—in spite of new treatments, approximately 7% survive for 5 years. Although there have been advances in systemic, primarily cytotoxic, therapies, it has been a challenge to treat patients with PDA using targeted therapies. Sequence analyses have provided a wealth of information about the genetic features of PDA and identified potential therapeutic targets. Preclinical and early-phase clinical studies have found specific pathways could be rationally targeted; it might also be possible to take advantage of the genetic diversity of PDAs to develop therapeutic agents. The genetic diversity and instability of PDA cells have long been thought of as obstacles to treatment, but now are considered exploitable features. We review the latest findings in pancreatic cancer genetics and the promise of targeted-approaches in pancreatic ductal adenocarcinoma therapy.
Pancreatic ductal adenocarcinoma (PDA) is the most common type of pancreatic cancer1. The disease encompasses multiple histological subtypes, which affect patients’ prognoses2. For example, patients with adenosquamous cancers have particularly poor outcomes, whereas mucinous neoplasms are generally lower grade and are considered to be a less aggressive form of the disease 3, 4. Irrespective, most cases of PDA are a challenge to treat, with 5 year rates of survival lower than 10% for patients with cancers of all stages1. To put this into perspective, it has been estimated that by 2020 that PDA will become the 2nd leading cause of cancer-related death in the United States 5.
Most PDA is identified at a late stage, when surgical intervention is not possible. Even with complete resection and negative results from analyses of tumor margins, long-term survival after surgery is poor—tumors recur in virtually all patients 6. Presumably, this is because micrometastases are present, even in patients whose disease appears confined to the pancreas. These features of the disease have driven the need for systemic treatments to control disseminated disease. Recently approved therapies for treatment of metastatic PDA include combination chemotherapy regimens, such as fluorouracil, leucovorin, irinotecan, and oxaliplatin (FOLFIRINOX) or gemcitabine and albumin-bound paclitaxel protocols7–9. The only targeted agent approved in treatment of PDA is the epidermal growth factor receptor (EGFR) inhibitor erlotinib, which given in combination with gemcitabine, only slightly increases overall survival time compared with gemcitabine alone10. These treatment approaches increase survival times of patients with metastatic PDA; however, the average increase in overall survival is measured in weeks to months11. Although many other types of cancer are now treated based on selective features of the disease (e.g. HER2-targeted therapies for HER2+ breast cancer, crizotinib for ALK-rearranged lung cancer), there are no validated marker-based therapies for PDA.
Deregulated Pathways
In recognition that a targeted approach could be particularly important for the treatment of PDA there have been extensive genetic analyses of the disease.
Genetics
Initially, genetic characterization of PDA was directed at evaluating known oncogenic and tumor suppressive pathways12—essentially searching for genetic variants frequently associated with tumors, such as bi-allelic loss of tumor suppressor genes or activating mutations in oncogenes. Performed before the high-throughput era, these experiments involved classic gene-cloning, single-stranded conformational polymorphism, PCR, and Sanger-sequencing methodologies. In many cases, these efforts explored genes that had been functionally defined in other tumor systems. For example the KRAS gene was originally discovered in mouse oncogenic retroviruses and found to be mutated in human bladder cancer cell lines13–15. Subsequently, targeted genetic approaches demonstrated that KRAS mutations occur in more than 90% of PDA tumors 13–15.
Through functional studies of cultured cells and mouse models, KRAS mutations were found to be required for tumor initiation and maintenance, regulating a range of cell activities, from proliferation to metabolic reprogramming16–20. PDAs and other tumors were also found to have frequent mutation of the tumor suppressor TP5321, 22, which synergizes with KRAS mutations to facilitate tumor development. This synergy was used to develop the long-accepted mouse model of PDA: the K-rasLSL.G12D/+; p53R172H/+; PdxCre (KPC) mouse23. Similarly the CDK4/6 inhibitor gene CDKN2A is often deleted, mutated, or epigenetically silenced, in PDA24. Individuals with mutations in CDKN2A that define familial melanoma syndrome have a 20-fold increase in risk of pancreatic cancer, compared to individuals without these mutations12, 25.
The aforementioned combinations of genetic features are detected in many other tumor types, including colon cancer and lung adenocarcinoma. A relatively unique event in gastrointestinal malignancies (e.g. colorectal cancer) is loss of SMAD4, also referred to as DPC4 (deleted in pancreatic cancer), which mediates transforming growth factor (TGF)-b signaling 26, 27. The frequent loss at of SMAD4 from 18q in PDAs is a marker of increased metastatic potential and indicates a poor prognosis 28, 29. Linking genetic events with the histologic features of PDA precursor lesions, such as pancreatic ductal intraepithelial neoplasia (PanIN), has provided an important model of pancreatic cancer progression 30. For instance, the high-frequency of KRAS mutations in PanIN lesions suggests that this event drives disease initiation with subsequent mutations/genetic events necessary for tumor progression.
Findings from next-generation sequencing analyses
To date approximately 300 PDA genomes/exomes have been sequenced (TABLE 1). This represents a limited collection of cases when compared against lung or breast cancer where greater than 1000 cases have been subjected to exome and whole genome sequencing 31, 32. PDA is often dominated by desmoplastic stroma, which can constitute up to 90% of the tumor mass33; this makes analyses of tumor epithelial cells difficult. This is one reason that the molecular characterization of PDA has lagged behind that of other tumor types.
Table 1.
Summary of whole exome and genome sequencing studies in PDA.
| Study | Clinical Cases |
Xenograft Cell Lines |
Method |
|---|---|---|---|
| Jones et al., 2008 | 24 | Exome | |
| Yachida et al., 20102 | 7 | Exome | |
| Campbell et al., 20102 | 3 | 10 | Genome |
| Liang et al., 2012 | 3 | Genome | |
| Biankin et al., 2012 | 99 | Exome | |
| Wang et al., 2012 | 15 | Exome | |
| Murphy et al., 20141,3 | 10 | Exome | |
| Waddell et al., 2015 | 75 | 25 | Genome |
| Witkiewicz et al., 20151 | 109 | Exome | |
| Dal Molin et al., 2015 | 8 | Exome |
Denotes microdissected cases.
Denotes models from matched primary metastatic cases.
Denotes studies inclusive of PANIN lesions
Researchers have used several approaches to circumvent issues of tumor cellularity. Initially, sequence analyses were performed using tumor xenografts and cell lines, to limit contamination by non-neoplastic human cells, which alter calculations of mutant allele frequencies and copy number alterations34. However, this approach potentially selects for specific genetic events required for proliferation of cells in culture or growth of tumors in immune-compromised mice, which might not occur in patients’ tumors (or the multiple clones within human tumors). Now that sequencing is more affordable, it is possible to sequence to great depth (e.g. 1000x reads for each nucleotide) in order to computationally enrich for the presence of tumor selective variants. This approach is clearly feasible and yields important insight into PDA genetic features 35, 36, albeit using such an approach could limit the sensitivity of detection and therefore under-represent the mutational burden in relation low frequency alleles to sub-clonal features of disease. The Cancer Genome Atlas and other sequencing efforts require the presence of at least 50% of tumor cells in samples analyzed37. Several groups have used microdissection approaches to select tumor cells from tissues 38, 39.
Combining resultant genetic characterization of PDA with knowledge of the clinical features of the disease has further allowed investigators to study the evolution of cancer from the primary to the metastatic lesion, the genetic features of precursor lesions, and select PDA subtypes (e.g. adenosquamous) 38–42. In spite of the varied approaches employed, a consensus view on the landscape of pancreatic cancer genetics is emerging, wherein there are a plethora of genetic alterations beyond the canonical KRAS, TP53, CDKN2A and SMAD4 spectrum. In fact, some of these non-canonical pathways and genes may become the most ‘targetable’ in PDA.
Deregulated pathways
The earliest exome sequence analyses of PDA identified core signaling pathways that were altered at the genetic level 34. However, due to the relatively limited number of tumor genomes sequenced, the prevalence of these core alterations was not clear. Subsequent genetic analysis has reinforced specific features of the core-signaling concept and characterized important additional features of PDA (FIGURE 1, TABLE 2).
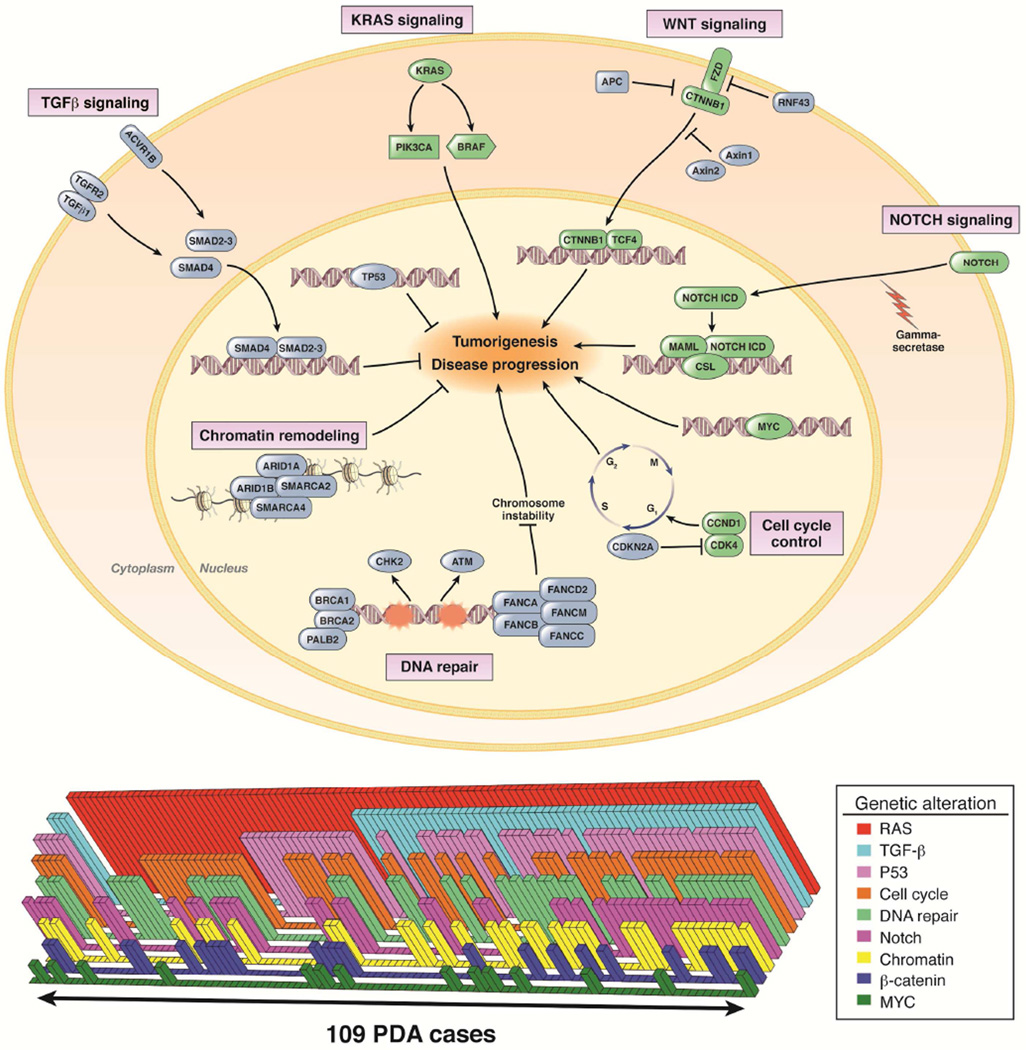
Figure 1. Diversity of oncogenic and tumor suppressor pathways in PDA.
A diagramatic representation of pathways that contribute to the etiology or progression of pancreatic cancer. Green denotes oncogenic and blue denotes tumor suppressive activities that are genetically altered in PDA. The lower bars represent alterations in the indicated pathway across 109 cases from Wiktiewicz et al., 2015. This graph illustrates the multi-fold complexity of pathway alterations between individual cases.
Table 2.
Approximate frequency of selected genetic alterations in PDA.
| Pathway | Gene | Approximate % Altered |
Predominant Genetic Events |
Potential Therapeutic Targets |
|---|---|---|---|---|
| KRAS | KRAS | 90 | Mutation | MEK, PI3K |
| BRAF | 3 | Mutation | BRAF | |
| PIK3CA | 3 | Mutation | PI3K | |
| TGF-β | SMAD4 | 30–40 | Mutation/Deletion | NA |
| TGFBR2 | 5–10 | Mutation | ||
| ACVR1B | 6 | Mutation | ||
| P53 | TP53 | 50–70 | Mutation/Deletion | P53 reactivation |
| MYC | MYC | 10 | Amplification | CDK9, BET domain |
| CELL CYCLE | CDKN2A | 40–60 | Deletion/Mutation | CDK4/6 |
| CCND1 | 10 | Amplification | CDK4/6 | |
| CDK4 | 10 | Amplification | CDK4/6 | |
| WNT | RNF43 | 10–15 | Mutation/Deletion | WNT, Tankyrase, Porcupine |
| AXIN1 | 5–10 | Mutation/Deletion | WNT, Tankyrase, Porcupine | |
| APC | 2 | Mutation/Deletion | WNT, Tankyrase, Porcupine | |
| NOTCH | NOTCH1 | 10 | Amplification | Gamma Secretase, Notch |
| NOTCH2 | 6 | Amplification | Gamma Secretase, Notch | |
| NOTCH3 | 6 | Amplification | Gamma Secretase, Notch | |
| CHROMATIN | ARID1A | 10–25 | Mutation/Deletion | EZH2, PI3K |
| ARID1B | 5–20 | Mutation/Deletion | ||
| SMARCA4 | 5 | Mutation/Deletion | ||
| SMARCA2 | 5–20 | Mutation/Deletion | ||
| DNA Damage | BRCA1 | 5 | Mutation | DNA cross link, PARP |
| BRCA2 | 7 | Mutation | DNA cross link, PARP | |
| PALB2 | 3 | Mutation | DNA cross link, PARP | |
| ATM | 6 | Mutation | ||
| FANC genes (A-M) | 10–15 | Mutation | DNA cross link |
Like many cancers, while the established mutations (e.g. KRAS and TP53) occur at high-frequency many other genetic events occur less frequently (TABLE 2). Only by analyzing many tumor samples can such genes be defined as “significantly mutated in cancer”; in general, it is viewed that more than 1000 different tumors will need to be sequenced to reach saturation43, 44. The current definition of significantly mutated is based on the observed frequency of mutations in coding regions of a tumor gene, compared with the chance of random mutations in the gene. Sequence analyses of PDAs have identified significant mutations in TGFBR2, KDM6A, AXIN1, ACVR1B, PIK3CA RNF43, GNAS, ATM, GLI3, ARID1A, RBM10 35, 39. Some of these mutations have been identified in select subtypes of PDA, but could also represent a feature of PDA cases as a whole. For example, mutations in GNAS, RNF43, and RBM10 were initially identified in intraductal papillary mucinous neoplasms (IPMNs), but subsequently found in PDAs that did not appear to arise from IPMNs. 45–47. Additionally, many of these genes are mutated in other cancers, lending credence to their significance in PDA 44.
In addition to mutations within genes, many cancer cells contain copy number alterations, which support the biological significance of a given genetic alteration 48. For example, approximately 10% of PDAs contain point mutations in CDKN2A; however, homozygous deletions are a more common event targeting this gene 35, 39. Computational approaches can identify regions of significant deletion or amplification in tumor cells 49. In the case of PDA, this includes many known tumor suppressors (e.g. CDKN2A and SMAD4) and oncogenes (e.g. MYC and CCND1) 35, 39
In spite of the emerging depth of information, the low frequency of many genetic events identified has called into question their ultimate clinical utility. However, many of the mutations identified are in genes whose products participate in the same pathways. For example, AXIN1, RNF43, APC are all mutated in PDAs and are members of the WNT pathway (TABLE 2)50. Similarly CDKN2A, CCND1, and RB function in a single pathway 51. Therefore, from a therapeutic perspective, although single genetic variants may be too rare to be viable targets, the pathways they alter might be targeted therapeutically. Sequencing studies have identified the KRAS, TGFB, TP53, MYC, chromatin remodeling, DNA repair, cell cycle, WNT–β-catenin, and NOTCH signaling pathways, among others, as those that are disrupted at the genetic level in PDAs and might be targeted (TABLE 2, FIGURE 1). Interactions among these pathways are complex; and most PDAs have genetic alterations that alter distinct subsets of these pathways (FIGURE 1)
Chromosome instability
In addition to gene specific genetic alterations, the overall genetic landscape of PDA could be an important and therapeutically prognostic feature of this disease. Analyses of chromosome architecture and copy number alterations by whole genome and exome sequencing have indicated that there are distinct subtypes of PDA, related to chromosomal instability. Using whole-exome sequencing, it is possible to capture features of amplification and deletion, and it is apparent that some PDA cases have relatively stable chromosome architecture whereas others have many amplifications and deletions 35, 39, 48. A caveat of exome sequencing is that it cannot be used to identify variants in intragenic regions, which constitute the bulk of translocations and other structural alterations. However, whole-genome sequencing can identify variants in intragenic regions; these types of studies have shown that PDAs contain a wide-spectrum of chromosome alterations36. Importantly, there appears to be a correlation between the extent of chromosome instability and mutations in genes involved in DNA break repair by homologous recombination, but not related to p53 36, 39. Chromosome instability is a feature of BRCA-deficient cancers and, multiple genes involved in DNA break repair are disrupted in PDAs, including BRCA1, BRCA2, and PALB2 (TABLE 2). These genetic variants appear to be directly involved in the etiology of pancreatic cancer, as germline mutations in these genes have been associated with familial predisposition to PDA 52–54.
Additional studies have focused on determining the frequency of the microsatellite instability (MSI) genotype of PDA. Some studies have reported that PDA is more likely to arise in families with Lynch Syndrome 55. A study performed more than 15 years ago found that fewer than 5% of PDAs could be classified as having MSI 56. Other studies supported this finding, and correlated MSI genotype with PDA with medullary histology 57. Nonetheless there has been debate over the frequency of the MSI genotype in all PDAs; one study found MSI frequency to be “irrelevant”, in that it was detected in only 0.3% of 338 consecutive surgically resected sporadic PDA cases 58. From recent sequencing studies there do appear to be hypermutated cases that harbor a mutation burden consistent with deficiency in mismatch repair occurring in ∼2% of cases36, 39. Similarly in a computational analysis of PDA mutational spectra, a contribution of mismatch repair deficiency was observed in tumor specimens 59.
Collectively, there appear to be distinct forms of PDA that can be identified based on the extent of mutation burden or chromosomal instability. These factors are likely to be associated with the etiology and/or progression of PDA, as well as patient outcomes and responses to treatment.
Genetic Alterations as Therapeutic Targets
The genetics of PDA could provide a roadmap to targeted therapy. Specifically, multiple pathways that are genetically dysregulated in PDA could serve as targets of therapy (TABLE 2). In general, the genetic features of disease provide the basis for considering two relatively simple approaches for targeted treatment of cancer. Conventionally, it is easy to envision how a specific activating genetic event can be targeted. Classic examples include targeting HER2 amplification in breast tumors or BCR–ABL in chronic myelogenous leukemia with kinase inhibitors. In these cases, the genetics of the tumor yield the direct target for pharmaceutical inhibition. The alternative approach is to exploit the biological or functional features of the genetic event therapeutically. For example, PARP inhibitors are effective against BRCA1/2-defective tumors due to impact of BRCA loss on DNA repair 60. In the case of PDA, historically neither approach to targeting the disease has been routinely employed in directing treatment. However, recent findings from genetic studies have identified new targets that can be tested in trials 61. Below select genetic features that are targetable with agents in clinical development are discussed.
KRAS–BRAF–MEK
The KRAS pathway is one of the best characterized signaling pathways in cancer 62. Because most PDAs (∼90%) have activating KRAS mutations, the pathway is an obvious choice for targeting. To date, no inhibitor of KRAS has been brought to clinical application, although the National Cancer Institute has a new program specifically directed toward developing KRAS inhibitors 63. Therefore, whether specific targeting of KRAS in PDA will represent a successful treatment approach remains unknown. PDA cell lines have variable responses to KRAS knockdown64, 65. Importantly, in genetically engineered mouse models of PDA, selective deletion of KRAS in established tumors led to a dormant population of cells that could ultimately recover from the ablation of KRAS and were driven by alternative signaling pathways 66, 67. Thus, even disruption of a key driver of PDA may not produce a durable therapeutic effect.
In recognition of the challenge of targeting KRAS directly, there have been multiple attempts to target effector pathways downstream of KRAS. In particular, MEK signaling is often required for the viability and proliferation of KRAS-driven tumors. Multiple potent MEK inhibitors have been developed, and have activity in models of PDA 68, 69. In a series of trials, the MEK inhibitors CI-1040A and AZD6244 as single agents were not effective in patients whose disease progressed on prior therapy70, 71. AZD6244 did not increase patient survival time, compared with capecitabine therapy, in a randomized phase 2 trial70. Trametinib in combination with gemcitabine therapy was not found to be superior to gemcitabine as a single agent in a randomized phase 2 trial 72.
These findings reveal the challenges of targeting a single pathway in PDA. In fact, multiple studies have shown MEK inhibitors to be particularly effective in combination with PI3K inhibitors, due to simultaneous effects of targeting 2 effectors of KRAS signaling 68, 69. This approach is being tested in a phase 1b trial with the MEK inhibitor MEK162 in combination with the PI3K inhibitor BYL719 in patients with solid tumors, including pancreatic cancer (NCT01449058) (TABLE 5). In addition, the effects of the combination of a MEK and AKT inhibitor, compared to FOLFOX (5-FU, oxaliplatin, leucovorin) as a second-line therapy for PDA, are to be presented in the near future—this strategy is evaluating a combined targeted approach to try to overcome the limitations of single pathway inhibition. A number of mutant KRAS directed trials are underway to test various MEK-targeted combinations in patient with PDAs (TABLE 5)
Table 5.
Selected marker targeted studies that could enroll PDA patients for targeted interventions
| NCT | Trial Design | N | Target | Sponsor |
|---|---|---|---|---|
| 02079740 | Trametinib and Navitoclax in Treating Patients With Advanced or Metastatic Solid Tumors (KRAS mutant tumors) |
130 | MEK BCL2 |
GSK |
| 02230553 | Lapatinib Plus Trametinib in KRAS Mutant Malignancies (M14LTK) |
30 | MEK EGFR |
Netherlands Cancer Institute |
| 02039336 | Dacomitinib Plus PD-0325901 in Advanced KRAS Mutant Malignancies |
35 | MEK EGFR |
Netherlands Cancer Institute |
| 01986166 | A Study of MEHD7945A and Cobimetinib (GDC-0973) in Patients With Locally Advanced or Metastatic Cancers With Mutant KRAS |
50 | MEK EGFR |
Genentech |
| 01449058 | A Phase Ib Study of MEK162 Plus BYL719 in Adult Patients With Selected Advanced Solid Tumors |
138 | MEK PI3K |
Novartis |
| 02187783 | LEE-011 for tumors with pathway defects (loss of CDKN2A amplification of CCND1/3 or CDK4/6) SIGNATURE |
90 | CDK4/6 | Novartis |
| 02065063 | Palbociclib in combination with trametinib for solid tumors (Phase 1/2) |
100 | CDK4/6 MEK |
GSK |
| 02022982 | Palbociclib in combination with PD- 0325901 for KRAS mutant tumors (Phase 1) |
30 | CDK4/6 MEK |
Pfizer |
| 01959139 | LGK974 in Patients With Malignancies Dependent on Wnt Ligands |
100 | WNT | novartis |
| 02152254 | IMPACT 2: Randomized Study Evaluating Molecular Profiling and Targeted Agents in Metastatic Cancer |
1362 | Multiple | MD Anderson |
| 01827384 | NCI-MPACT: Molecular Profiling-Based Assignment of Cancer Therapy for Patients With Advanced Solid Tumors |
700 | DNA repair KRAS PI3K/MTOR |
NCI |
| Pending | NCI-MATCH: Molecular Analysis for Therapy Choice |
1,000 | 20–25 Agents |
NCI |
Although patients with PDA containing KRAS mutations are a challenge to treat, little is known about the behavior of PDA without mutation in KRAS. From recent sequencing studies several potential oncogenic drivers have emerged for this subset of PDA. Activating mutations in the GNAS gene, which encodes a G-protein subunit, were identified in IPMN-derived PDAs 45, 46. Mutations in BRAF that activate kinase activity (such as V600E) have been identified and are mutually exclusive with KRAS mutations 39, 73. Cells from a tumor with mutant BRAF had selective sensitivity to vemurafenib, a BRAF inhibitor. Correspondingly, BRAF promotes development of PDA in mice 69. There is anecdotal evidence that patients with PDAs with the BRAF V600E mutation respond to an approved BRAF inhibitor; patients with metastatic pancreatic cancer may be eligible to receive a BRAF inhibitor based on the genetic profile of the tumor. Thus, simple genetic screening of the conventional KRAS/BRAF pathway could elicit a new therapeutic avenue for a minor subset of patients with PDA. The Individualized Molecular Pancreatic Cancer Therapy (IMPACT) trial is identifying patients with PDAs without mutations in KRAS for testing of specific therapeutic agents 74.
Activating mutations in PIK3CA have been identified in PDAs, but it is not clear how they promote disease progression or whether mutant PIK3CA is a good therapeutic target. In mice, activating mutations in PIK3CA are not sufficient to cause tumor development, and oncogenic mutations in PIK3CA are found in both mutant and non-mutant KRAS tumors 39, 69. In the context of mouse models, PIK3CA could augment the activity of KRAS in promoting tumor development, and these tumors might be more reliant on PI3K signaling. However, even in the case of breast cancer where PIK3CA mutation contributes to disease initiation/progression, it is unclear whether this event yields selective sensitivity to PI3 kinase inhibitors in the clinic. This is an active area of investigation, as PI3K inhibitors can augment the activity of MEK inhibitors.
DNA repair and chromosome instability
Many PDA cases contain genetic alterations that affect DNA damage repair pathways. Before the advent of next-generation sequencing, a proportion of PDAs were known to contain either germline or somatic mutations in BRCA1, BRCA2, or Fanconi anemia genes (e.g. FANCC, FANCG, and FANCN/PALB2)75–77. These genes function in a complex fashion to mediate homologous recombination mediated DNA repair that is required for the maintenance of chromosome stability, and could be hypersensitive to established and new DNA damaging agents75, 78.
The frequency of BRCA deficiency is estimated to be 5%–8% in unselected patient populations and 12%–15% in certain populations (such as Ashkenazi patients with a family history of breast or ovarian cancer). Recent sequencing studies identified subtypes of PDA characterized by chromosomal instability, probably due to BRCA deficiency or similar deficits in DNA repair36, 39. Such deficits in BRCA function have been shown to increase the sensitivity of tumor cells to platinum agents, in multiple models. Consistent with this concept, platinum-based therapy was shown to be effective, in retrospective studies of BRCA-deficient PDAs 79–81. These observations contradict the concept that BRCA is a biomarker for sensitivity to chemotherapy, as opposed to platinum agents. Trials are underway to evaluate the efficacy of the combination of cisplatin and gemcitabine in patients with locally advanced or untreated BRCA-deficient PDA. A study recently reported that some patients with chromosomal instability indicative of BRCA deficiency have exceptional responses to platinum-based regimens 36. Many of these PDAs contained genetic alterations in BRCA1, BRCA2, or PALB2. However, there were cases for which a specific genetic event was not identified.
In addition to platinum agents, work in breast and ovarian cancer have shown that BRCA-deficient cancers are selectively sensitive to poly-ADP-ribose polymerase (PARP) inhibitors82–84 (Table 3). Ongoing clinical trials are further investigating whether addition of the PARP inhibitor, veliparib, increases patients’ response to platinum agents (NCT01585805). Researchers recently presented data from a phase IB trial evaluating the triple combination of cisplatin, gemcitabine, and veliparib in newly diagnosed, untreated patients with PDA and germline mutations in BRCA or PALB2 85. These data have defined the safety and tolerability of cisplatin, gemcitabine, and veliparib and indicate the efficacy of the 3-drug combination in these individuals. Significantly higher rates and duration of response and survival were observed in this subgroup, compared in a non-randomized manner to a subgroup of patients with sporadic disease. These observations will be clarified in a prospective randomized phase 2 trial of cisplatin and gemcitabine, with or without veliparib, in patients with newly diagnosed, locally advanced, or metastatic pancreas adenocarcinoma and germline mutations in BRCA or PALB2 (NCT01585805). Results from the first part of phase 1 and 2 trials of 5-FU, oxaliplatin, and veliparib were presented at the American Society for Clinical Oncology-Gastrointestinal Cancers Conference in 201386.
Table 3.
Selected trials evaluating PARP inhibitors in advanced pancreatic adenocarcinoma
| NCT | Trial Description | N | Sponsor |
|---|---|---|---|
| 01489865 | FOLFOX + Veliparib Wild-type + germline BRCA Untreated, previously treated Phase I-II |
48 | AbbVie |
| 01585805 | Cisplatin, Gemcitabine +/− Veliparib Germline BRCA, PALB2 Randomized phase II |
50 | MSKCC/NCI Lustgarten |
| 01585805 | Veliparib Germline BRCA, PALB2 (previously treated) Phase II |
15 | MSKCC/NCI Lustgarten |
| 01296763 | Irinotecan, Cisplatin, Mitomycin C +/− Olaparib Wild-type + germline BRCA Phase I-II Results awaited |
18 | John Hopkin’s Cancer Center |
| 01482715 | Rucaparib Germline, somatic BRCA (previously treated) Phase II |
100 | Clovis |
| 01286987 | BMN-673 Germline BRCA (any solid tumor) Phase I |
BioMarin | |
| 02184195 | Platinum therapy followed by Olaparib/Placebo Germline BRCA Phase III maintenance |
145 | Astra-Zeneca POLO Trial |
PARP inhibitors have activity as single agents and in combination therapies for patients with advanced pancreas adenocarcinoma87. Findings have been reported from 2 studies. Kaufmann, et al88 reported that 5/23 (22%) previously treated patients (23 with gemcitabine and 14 with platinum therapy) with pancreas adenocarcinoma and germline mutations in BRCA responded to olaparib as a single agent. Their median survival time was 9.8 months and 41% survived for 1 year. More recently, Lowery et al89 evaluated velaparib in 16 previously heavily pre-treated patients with PDA and germline mutations in BRCA. Although no objective responses were observed, 4 patients (25%) had stable disease ranging from 4 months to 1 year. These studies indicate that a subset of patients with BRCA deficiencies and advanced pancreas cancer can benefit from a PARP-targeted agent. Analogous to the development and registration strategy of olaparib for patients with ovarian cancer, a phase 3 trial (the POLO trial, NCT02184195), is evaluating the maintenance value of olaparib in a 3:2 randomization to placebo following initial treatment with platinum-based therapy in germline BRCA-mutated PDA. This trial has a number of distinctions in that it stands alone as the only phase III trial that is underway in metastatic PDA, and is the first trial to evaluate the role of maintenance therapy in this disease. Table 3 summarizes other related PARP studies underway in patients with pancreatic cancer.
The limits with regard to patient subgroups in targeting tumors with homologous repair defects remain under study. Theoretically, DNA-damaging agents and PARP inhibitors may benefit patients with mutations in ATM, ATR, CHEK, mismatch repair genes, and other genes with similar functions 90. Targeting the mitotic checkpoint inhibitor WEE1 in cancer cells would ostensibly further sensitize chromosomally unstable PDA cells to chemotherapeutic agents91–93. Trials are underway (NCT01748825 and NCT0182734) to determine the efficacy of this approach in select populations (TABLE 3 and TABLE 5).
Collectively, data support the concept that germline or somatic mutations in BRCA could predict which patients with PDA are most likely to respond to platinum- and PARP-based therapies. Further studies are needed to determine whether mutations in BRCA can also be used as prognostic factors for patients with pancreas cancer.
Loss of CDKN2A and CDK4/6 inhibitors
One of the most frequently detected genetic alterations in PDA is disruption or silencing of the tumor suppressor gene CDKN2A 94,95. CDKN2A encodes the p16ink4a protein, which inhibits the kinase activity of CDK4 and CDK651, 96. In normal tissue, oncogenic activation of KRAS elicits a stress response that leads to activation of p16ink4a and oncogene-induced senescence 97, 98 Therefore, in many cancers driven by KRAS there is potent selection for the loss of p16ink4a. In PDA this appears to be the preferred mechanism of cell cycle deregulation, consequently PDA may be particularly sensitive to agents that recapitulate the activity of p16ink4a—i.e. the suppression of CDK4/6 activity. Highly potent CDK4/6 inhibitors have been developed, including LEE-011 (ribociclib), PD-0332991 (palbociclib), and LY2835219 (abemaciclib)51. These drugs are given orally and are being evaluated in multiple clinical trials.
Preclinical models of PDA have shown mixed responses to these agents. Although a subset of cell lines and patient-derived xenografts appear sensitive to CDK4/6 inhibitors, other PDA cell lines that lack p16ink4a are either intrinsically resistant or rapidly develop resistance in culture 99–101. These data indicate that loss of CDKN2A/p16ink4a does not, per se, predict response, and that combination approaches will be the most effective means for the use of CDK4/6 inhibitors in PDA. This supposition is consistent with the strong activity of CDK4/6 inhibitors in combination with endocrine therapy in patients with breast cancer 102. Drug screens have identified mTOR, IGF1R, and MEK inhibitors as effective agents, in combination with CDK4/6 inhibitors, in models of PDA100, 101.
CDK4/6 inhibitors are not currently in clinical trials for treatment of PDA, specifically. However, there are several ongoing trials of CDK4/6 inhibitors that are germane to PDA (Table 5). Since loss of CDKN2A is common in PDAs, it is likely that patients with this cancer who are enrolled in the Novartis-sponsored SIGNATURE trial will receive the single agent LEE-011 (NCT02187783). Similarly, the trial of palbociclib with the MEK inhibitors PD-0325901 (NCT02022982) or trametinib (NCT02065063) will likely include patients with PDA, given the targeted scope toward tumors with RAS mutations. With the recent Food and Drug Administration approval of palbociclib in combination with letrozole for estrogen receptor-positive breast cancer, it is likely that the number trials investigating the effects of the CDK4/6 combination in patients with PDA will increase.
The WNT pathway
The WNT pathway is altered in many types of gastrointestinal malignancies, such as via APC mutation in colorectal tumors 103. It has come to be recognized that this pathway is deregulated via multiple distinct genetic events in PDA and is functionally important for disease 104, 105. Interestingly, unlike colon cancer, in which APC mutations are particularly common, pancreatic tumors contain a wide spectrum of mutations. RNF43 and AXIN1 are more frequently disrupted in PDA, whereas APC is less-frequently lost36, 39, 73. RNF43 alterations are observed in IPMN as well as PDAs and associated cell lines 45, 47, 106. WNT signals to the β-catenin/TCF4 transcription factor that represents the downstream target of the pathway. RNF43 expression is induced by TCF4 in order to attenuate deregulated WNT signaling50; therefore, mutation of RNF43 leads to constitutive signaling through the pathway. AXIN and APC participate directly in the degradation of β-catenin. Although WNT signaling has been considered a therapeutic target for many years, only recently have agents specific for the pathway emerged reflecting difficulty in developing therapeutic agents that act on tumor suppressors and transcription factors50. The most advanced of these are WNT-974 (also known as LGK974). This agent functions by suppressing porcupine, which is required for secretion of WNT ligands; it has selective activity in pancreatic cancer cell lines deficient in RNF43, as well as in xenograft tumors106. Based on these data there is a clinical trial testing LGK974 in patients with tumors with dysregulated WNT signaling (NCT01351103). Criteria for inclusion in the study include the loss of RNF43 or other mediators of WNT signaling that are deregulated in pancreatic cancer.
NOTCH
The NOTCH pathway is also deregulated in multiple tumor types. NOTCH mediates self-renewal and proliferation of cancer stem cells and its activity is associated with chemoresistance and metastasis107, 108. Based on genetic analyses, mutations in NOTCH are relatively rare, but multiple components of the pathway appear to be amplified, consistent with the overexpression and observed deregulation of the pathway in PDA 39, 109, 110. Overexpression of NOTCH signaling components in pancreatic tumors has been associated with poor outcomes of patients. NOTCH signaling is required for pancreatic tumor progression and metastasis in mouse models 111, so pancreatic cancer is considered to be relatively dependent on NOTCH signaling—either in parallel with KRAS signaling or independently. Irrespective of the mechanism, studies of cell lines, xenograft tumors, and genetically engineered models have demonstrated that suppression of NOTCH has potential for therapeutic efficacy112–115.
The NOTCH signaling pathway can be inhibited pharmacologically, with inhibitors of γ-secretase, antibodies, and other mechanisms108. The γ-secretase is required to transmit NOTCH signals from the membrane to the nucleus. Inhibitors of γ-secretase have been developed by multiple pharmaceutical companies and have been tested in clinical trials, including those of patients with pancreatic cancer. BMS-906024 and PF-03084014 inhibitors are in clinical development (TABLE 4). There are only a few results from studies of single agents in patients with PDA. The agent RO4929097 was evaluated in a single-arm phase 2 trial of patients with previously treated metastatic PDA, the trial was closed for accrual with discontinuation of the agent by the sponsor.
Table 4.
Selected novel agents in development in pancreatic adenocarcinoma in the metastatic setting
| NCT | Trial Design | N | Target | Sponsor |
|---|---|---|---|---|
| 02428270 | A Study of GSK2256098 and Trametinib in Advanced Pancreatic Cancer |
24 | FAK MEK |
University Health Network, Toronto |
| 01839487 | Gem + nab-P ± PEGPH20 Rand phase II |
132 | Hyaluronan | Halozyme |
| 01959139 | mFOLFIRINOX ± PEGPH20 Rand phase II |
172 | Hyaluronan | SWOG/ S1313 |
| 01621243 | Gem + nab-P ± Necuparanib Rand phase II |
148 | Anti-stromal Heparin mimetic |
Momenta |
| 01647828 | Gem + nab-P ± Tarextumab (Alpine) Rand phase II |
140 | Notch, stem cell | OncoMed |
| 01844817 | Gem + nab-P ± OGX-427 (Rainier) Rand phase II |
132 | HSP27 | OncoGenix |
| 02101021 | Gem + nab-P ± Momelotinib Phase IB - rand ph II |
336 | JAK 1/JAK2 | Gilead |
| 02289898 | Gem + nab-P ± Demcizumab (Yosemite) Rand phase II |
201 | Anti-DLL4, stem cell |
OncoMed |
| 02077881 | Gem + nab-P ± Indoximod Rand phase II |
80 | IDO | NewLink Genetics |
| 02109445 | Gem + nab-P ± PF-03084014 Rand phase II |
193 | γ-Secretase inhibitor, Notch |
Pfizer |
| 02194829 | Gem + nab-P ± MK-1775 Phase IB - rand ph II |
133 | Wee-1 inhibitor | Merck |
| 02050178 | Gem + nab-P + OMP-54F28 Phase IB |
20 | Frizzled | OncoMed |
| 02101580 | Gem + nab-P + ADIPEG 20 Phase IB |
21 | Arginine depletion |
Polaris |
Monoclonal antibody-based therapies have also been evaluated. Tarextumab (OMP-59R5) is a fully human antibody against NOTCH2 and NOTCH3 that slows growth of xenograft tumors in mice in combination with cytotoxic agents 116. It is currently being tested in a randomized, placebo-controlled, phase 2 trial (NCT01647828), in combination with gemcitabine and nab-paclitaxel in untreated patients with metastatic pancreas adenocarcinoma (TABLE 4). In addition to evaluating the treatment signal in all patients, the trial will evaluate specifically the activity of Tarextumab in a biomarker selected subgroup of patients with high levels of NOTCH 3 expression, a particularly unfavorable prognostic subgroup. Preliminary results from the phase 1B trial demonstrated significant activity of a 15 mg/kg dose of tarextumab combined with standard doses of gemcitabine and nab-paclitaxel. The reported median time of progression-free survival was 5.6 months, and median time of overall survival was 11.6 months; 38% of this patient population had a response to the 3-drug combination, and response rate was even higher in the small number of patients whose tumors expressed high levels of NOTCH3 117.
SWI/SNF chromatin remodeling complex
Members of the family of SWI/SNF chromatin remodeling factors are mutated in many tumor types 118, 119. All sequence studies of pancreatic tumors have reported disruptions in genes encoding these factors 34, 35, 38, 39. Loss of ARID1A is the most common single event, but loss of other subunits, including ARID1B and SMARCA4, has been observed (TABLE 2). These molecules function in a large complex to facilitate the fluidity of chromatin between activated and repressed states. The specific effects of loss of these factors are hard to determine, because disruption of this complex affects chromatin stability as well as transcription of many genes. As for many tumor suppressors, it is unclear whether inhibitors of chromatin remodeling factors could be effective, although such agents are under development.
Rather, in response to the loss of ARID1A and other factors that control chromatin remodeling, compensatory pathways could be activated. RNA-intereference screens demonstrated that ARID1A–deficient cells were particularly sensitive to the selective depletion of ARID1B120. Although this finding is important for our understanding of the mechanisms of chromatin remodeling, it might not have much clinical application. A complementary drug screen found EZH2 activity to be required for the viability of ARID1A–deficient ovarian cancer cells121. Multiple agents have been developed that target EZH2. Several EZH2 inhibitors are being tested in clinical trials. E7438, GSK281612, and CPI-1205 are in phase 1 dose-finding studies, mostly comprising patients with lymphoid malignancies. Given the recent nature of the preclinical findings, no trials are underway to study the efficacy of EZH2 in patients with tumors that have lost ARID1A.
Canonically untargetable pathways
In addition to the pathways described above, there are multiple additional genetic alterations that could in principle be used as the basis for rational treatment. However, these pathways are not routinely targeted pharmaceutically.
Amplifications in MYC are frequently observed in PDA (∼15% of cases), and overexpression of MYC has been shown to promote tumor development in mice. While generally considered untargetable (as is the case with many oncogenic transcription factors), recent studies have suggested unique vulnerabilities that could be exploited in the context of MYC-driven disease. These include CDK9 and BET-bromodomain inhibitors which are potently selective for MYC tumors in preclinical models122, 123. However, whether this strategy could be effective in MYC-amplified PDA remains unknown.
Approaches to therapeutically target the loss of TGFB signaling in PDA cells could be useful given the frequent loss of SMAD4, TGFBR2, and other elements of the pathway. In spite of the clear importance of targeting loss of this pathway, there has been no definition of synthetic lethal or other approaches that could selectively target this subset of tumors. However, SMAD4 is emerging as a biomarker for a poor prognostic phenotype in PDA 28. It’s utility as a biomarker for clinical decision making is also being prospectively evaluated in a randomized phase II trial in locally advanced pancreas adenocarcinoma where SMAD4 loss and intact status will be evaluated and correlated with an intensively focused loco-regional treatment approach of combination cytotoxic therapy and high dose intensity modulated radiation versus a systemic therapy based approach (NCT01921751).
TP53 is mutated in most human tumor types, and multiple drug development programs have been initiated to exploit this event in a targeted manner. Although there have been many promising results from preclinical studies, clinical development of agents designed to reactivate TP53 has been slow. APR-246 is the only agent in this category that is being evaluated in clinical trial (NCT02098343).
Targeting Genetic Diversity in PDAs
PDA contains many genetic alterations that could be targeted therapeutically, based on studies from other tumor types or preclinical investigations. However, there are several important factors to consider in leveraging such information to improve clinical outcomes.
Importance of additional preclinical studies
Preclinical studies are needed to determine the functional effects of the genetic alterations observed in PDA cells. Many agents are known to have strong potency against a select target; at the same time there is an ever-emerging sense that genetic activation of a target does not universally predict response. For example, colon cancer cells with BRAF mutations rarely respond to BRAF inhibitors, due to compensatory EGFR signaling125.
Preclinical models that recapitulate the genetic diversity of PDA are of paramount importance. Genetically engineered mouse models provide one approach that could be complemented by studies of patient-derived xenografts and/or new, sophisticated, patient-derived in vitro models 126–128.
Pancreatic tumors in genetically engineered mouse models develop along the same pathways as human PDAs (e.g., in the context of KRAS activation) and in an immune-competent host. However, studies from other systems suggested that the genetic factors that contribute to development of tumors in these mice differ from those of humans 129, 130. Although there have been no formal investigations, it is unlikely that tumors from mouse models have the same level of genetic diversity and pathway activation observed in tumors from patients.
In contrast with genetically engineered mouse models, patient derived xenografts should harbor the genetic features of the parental tumor and therefore better recapitulate the biology of an individual tumor. However, these tumors are grown in a different environment, which could select for additional features distinct from the primary tumor and affect their response to test agents. An emerging model for PDA is that of the organoid culture model system, which has attractive features of providing a model system for interrogation in a proximate time period and providing the opportunity for genetic evaluation, pre-clinical modeling, and other considerations126.. Importantly, biotechnology companies and academic centers routinely generate models from resected tumors or biopsies in real-time, and the sensitivity of such models to therapeutic agents can be determined and provided to the physician. The extent to which these approaches will translate into patient care remains unclear; however, clinical trials testing this concept are being initiated.
Defining surrogates of response
In contrast with other diseases, it is a challenge to evaluate responses of PDA to drugs via window, neoadjuvant, or serial biopsy analyses. Although there is increasing use of systemic therapy in the neoadjuvant setting, there is no consistent use of pre-operative therapy to determine the ability of agents to impinge on tumors or their target 131, 132. Additionally tissues are not routinely collected pre and during treatment hampering discovery of potential response markers. In the context of breast cancer, these studies proved that acute suppression of Ki67 to endocrine therapy pre-operatively largely predicted durable response in ER-positive breast cancer. This type of approach and trials which are exploratory in relation to genetics and determinants of response to investigational drugs, are limited in patients with PDA, although several examples are extant (e.g., NCT02241187). Functional imaging or other non-invasive approaches, including evaluation of cell-free DNA and other liquid biopsy approaches to measure tumor response, will be particularly important in evaluating targeted treatment approaches. The only routinely used surrogate in the clinic is the serum marker of tumor burden, CA19–9133, 134.
Beyond conceived genetic sensitivities
There are a number of promising therapeutic modalities for which there is not a clear concept as to what genetic events could be associated with sensitivity. For example, immunotherapeutic strategies are providing some impressive, landmark outcomes in other tumor systems (e.g., melanoma) 135, 136 To date, such successes have not been matched in PDA, yet still there is hope that development of a vaccine or checkpoint-based approaches will work in some patients137. However, it has recently emerged that mismatch repair deficiency, as is observed in a small set of PDA cancers likely represent a predictive marker138. Similarly, approaches to target the PDA microenvironment, such as hyaluronic acid, are being tested in early-phase trials139.
The NCI Match trial is a large-scale, phase 2, disease-agnostic trial design that proposes to evaluate a series of targeted agents in parallel arms140. Eligibility will be determined based on results of next-generation sequence analyses; it is anticipated that approximately 3000 patients will be screened and approximately 1000 patients will be selected based on targetable genomic features of their tumors. The study will evaluate approved and investigational agents. Tumor response will be the primary endpoint, along with progression-free survival. This trial will present a broad opportunity for a percentage of patients with PDA to participate in a precision medicine-based approach.
Doublet and Combination Therapies
With a few exceptions, it seems unlikely that targeting a single genetic feature of pancreatic cancer will produce durable or transformative effects. This concept is reinforced when reviewing the pathway landscape of PDA (FIGURE 1), wherein most case exhibit a range of deregulated pathways. Studies of MEK inhibitors in PDA have provided insight into lack of efficacy—most PDAs have genetic deregulation of KRAS, so MEK should be an ideal target. Therefore, it seems to be necessary to target multiple pathways to improve treatment outcomes. This concept has been particularly well developed for estrogen receptor-positive breast tumors, in that addition of an active agent to endocrine therapy improved the durability of response to therapy (as shown with mTOR and CDK4/6 inhibitors). Against this backdrop, trials such as the NCI MATCH and Novartis SIGNATURE are largely matching single agents with single genetic features of tumors. Given that most pancreatic tumors have multiple genetic features that promote their progression, it will be important to move beyond single agent approaches, based on genetics, and consider strategies to target two or more key signaling pathways in parallel.
How can we perform multi-genetic and combination studies? Ostensibly, patients with tumors that contain two genetic variants frequently detected in pancreatic cancer could be treated with a particular drug combination. For example, patients whose tumors have a combination of KRAS mutation and CDKN2A loss could receive a combination of MEK and CDK4/6 inhibitors. Although there have already been a number of combination trials directed specifically against tumors with mutant KRAS (TABLE 5), specification of the drug combination has not been rationally directed. An alternative approach would be to use a standardized backbone therapy, which would be combined with a pathway selective inhibitor (e.g. NOTCH amplification specifies a g-secretase inhibitor, or an RNF43 mutation specifies a porcupine inhibitor). Such a trial design would be completely dependent on a better understanding of the effects of drug combinations, not only in relation to their potential benefit but the toxicity profile and optimized dosing schedules.
Future Directions
Progress in the treatment of PDA has been incremental. Arguably, combination cytotoxic therapies such as FOLFIRINOX, along with gemcitabine and albumin-bound paclitaxel, have provided meaningful gains, but there is room for improvement. Our understanding of the PDA genome has increased and provides insight into focused therapeutic approaches; there is emerging consensus that subsets of patients with PDA may benefit from targeted approaches. Agents designed to exploit DNA repair pathways and NOTCH signaling are in late stages of clinical development. It will be important to identify subgroups of patients with tumors most likely to benefit from agents designed to target specific pathways or genomic features.
The next generation of clinical trials needs to be thoughtfully designed and based on optimal preclinical results. It is important to select rationally tailored approaches for each study participant, and produce detailed results that provide insight into mechanisms of sensitivity and resistance. Yielding a transformative impact on survival rates for PDA will require a multi-fold approach. Fundamental research that provides a better understanding of the pathways/genes driving PDA singly and in the complex patterns observed in human disease will be required to define key drug targets and therapeutic vulnerabilities that can be exploited in the clinic. Well-designed biomarker-driven clinical trials that acknowledge the genetic complexity and challenges of treating PDA will be seminal for a targeted approach to treatment of PDA. Iterative learning from mis-steps, exceptional responses, and selected subgroup analyses will support the ultimate development of guided treatment for progressively more patients with PDA. Hopefully, such a concerted effort will yield the critical advances that have long proved elusive in this therapy recalcitrant disease.
Acknowledgments
The authors thank their colleagues and collaborators for thought-provoking discussions related to the use of genetic features of disease to provide clues to treatment. Any oversight of citations is unintended, and all efforts were made to provide a comprehensive and unbiased review of the field and to consider paths forward. The authors thank Uthra Balaji (UTSW) and Sarah Williamson (Gastroenterology) for informatic and graphical assistance respectively.
ACKNOWLEDGEMENTS OF SUPPORT:
EK: NIH-CA142543-05S2
EO: Andrea J. Will Foundation
JB: AACR-PANCAN RAN grant, NIH-CA182692
AW: NIH-CA142543-05S2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES:
EK: Research funding and advisory: Pfizer, Eli Lilly
EO: Reseach funding: OncoMed, Celgene, Sanofi-Aventis, Astra-Zeneca, Bristol Myers Squibb, Incyte Pharmaceuticals
JB: Advisory: Perthera
AW: N/A
REFERENCES
- 1.Vincent A, Herman J, Schulick R, et al. Pancreatic cancer. Lancet. 2011;378:607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yeo CJ, Cameron JL, Sohn TA, et al. Six hundred fifty consecutive pancreaticoduodenectomies in the 1990s: pathology, complications, and outcomes. Ann Surg. 1997;226:248–257. doi: 10.1097/00000658-199709000-00004. discussion 257–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kardon DE, Thompson LD, Przygodzki RM, et al. Adenosquamous carcinoma of the pancreas: a clinicopathologic series of 25 cases. Mod Pathol. 2001;14:443–451. doi: 10.1038/modpathol.3880332. [DOI] [PubMed] [Google Scholar]
- 4.Wilentz RE, Albores-Saavedra J, Zahurak M, et al. Pathologic examination accurately predicts prognosis in mucinous cystic neoplasms of the pancreas. Am J Surg Pathol. 1999;23:1320–1327. doi: 10.1097/00000478-199911000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 6.Yeo CJ, Cameron JL, Lillemoe KD, et al. Pancreaticoduodenectomy for cancer of the head of the pancreas. 201 patients. Ann Surg. 1995;221:721–731. doi: 10.1097/00000658-199506000-00011. discussion 731–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conroy T, Desseigne F, Ychou M, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 8.Saif MW. Advancements in the management of pancreatic cancer: 2013. JOP. 2013;14:112–118. doi: 10.6092/1590-8577/1481. [DOI] [PubMed] [Google Scholar]
- 9.Von Hoff DD, Ervin T, Arena FP, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369:1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore MJ, Goldstein D, Hamm J, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 11.Paulson AS, Tran Cao HS, Tempero MA, et al. Therapeutic advances in pancreatic cancer. Gastroenterology. 2013;144:1316–1326. doi: 10.1053/j.gastro.2013.01.078. [DOI] [PubMed] [Google Scholar]
- 12.Maitra A, Hruban RH. Pancreatic cancer. Annu Rev Pathol. 2008;3:157–188. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forrester K, Almoguera C, Han K, et al. Detection of high incidence of K-ras oncogenes during human colon tumorigenesis. Nature. 1987;327:298–303. doi: 10.1038/327298a0. [DOI] [PubMed] [Google Scholar]
- 14.Herzig KH, Schaffer M, Rosewicz S, et al. [Detection of mutated ras-oncogene in pancreatic cancers: improved diagnostic possibilities using molecular biology techniques?] Z Gastroenterol. 1992;30:436–439. [PubMed] [Google Scholar]
- 15.Lemoine NR. Molecular advances in pancreatic cancer. Digestion. 1997;58:550–556. doi: 10.1159/000201500. [DOI] [PubMed] [Google Scholar]
- 16.Ying H, Kimmelman AC, Lyssiotis CA, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson L, Mercer K, Greenbaum D, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 18.Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- 19.Tuveson DA, Hingorani SR. Ductal pancreatic cancer in humans and mice. Cold Spring Harb Symp Quant Biol. 2005;70:65–72. doi: 10.1101/sqb.2005.70.040. [DOI] [PubMed] [Google Scholar]
- 20.Aguirre AJ, Bardeesy N, Sinha M, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scarpa A, Capelli P, Mukai K, et al. Pancreatic adenocarcinomas frequently show p53 gene mutations. Am J Pathol. 1993;142:1534–1543. [PMC free article] [PubMed] [Google Scholar]
- 22.Kalthoff H, Schmiegel W, Roeder C, et al. p53 and K-RAS alterations in pancreatic epithelial cell lesions. Oncogene. 1993;8:289–298. [PubMed] [Google Scholar]
- 23.Hingorani SR, Wang L, Multani AS, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 24.Schutte M, Hruban RH, Geradts J, et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126–3130. [PubMed] [Google Scholar]
- 25.Bartsch DK, Sina-Frey M, Lang S, et al. CDKN2A germline mutations in familial pancreatic cancer. Ann Surg. 2002;236:730–737. doi: 10.1097/00000658-200212000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahn SA, Schutte M, Hoque AT, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 27.Liu F, Hata A, Baker JC, et al. A human Mad protein acting as a BMP-regulated transcriptional activator. Nature. 1996;381:620–623. doi: 10.1038/381620a0. [DOI] [PubMed] [Google Scholar]
- 28.Blackford A, Serrano OK, Wolfgang CL, et al. SMAD4 gene mutations are associated with poor prognosis in pancreatic cancer. Clin Cancer Res. 2009;15:4674–4679. doi: 10.1158/1078-0432.CCR-09-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iacobuzio-Donahue CA, Fu B, Yachida S, et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol. 2009;27:1806–1813. doi: 10.1200/JCO.2008.17.7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hruban RH, Goggins M, Parsons J, et al. Progression model for pancreatic cancer. Clin Cancer Res. 2000;6:2969–2972. [PubMed] [Google Scholar]
- 31.Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neesse A, Michl P, Frese KK, et al. Stromal biology and therapy in pancreatic cancer. Gut. 2010;60:861–868. doi: 10.1136/gut.2010.226092. [DOI] [PubMed] [Google Scholar]
- 34.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy SJ, Hart SN, Lima JF, et al. Genetic alterations associated with progression from pancreatic intraepithelial neoplasia to invasive pancreatic tumor. Gastroenterology. 2013;145:1098–1109. doi: 10.1053/j.gastro.2013.07.049. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Witkiewicz AK, McMillan EA, Balaji U, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iacobuzio-Donahue CA, Velculescu VE, Wolfgang CL, et al. Genetic basis of pancreas cancer development and progression: insights from whole-exome and whole-genome sequencing. Clin Cancer Res. 18:4257–4265. doi: 10.1158/1078-0432.CCR-12-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yachida S, Iacobuzio-Donahue CA. Evolution and dynamics of pancreatic cancer progression. Oncogene. 2013;32:5253–5260. doi: 10.1038/onc.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yachida S, Jones S, Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–248. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Furukawa T, Kuboki Y, Tanji E, et al. Whole-exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Sci Rep. 2011;1:161. doi: 10.1038/srep00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu J, Matthaei H, Maitra A, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med. 2011;3:92ra66. doi: 10.1126/scitranslmed.3002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amato E, Molin MD, Mafficini A, et al. Targeted next-generation sequencing of cancer genes dissects the molecular profiles of intraductal papillary neoplasms of the pancreas. J Pathol. 2014;233:217–227. doi: 10.1002/path.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calhoun ES, Hucl T, Gallmeier E, et al. Identifying allelic loss and homozygous deletions in pancreatic cancer without matched normals using high-density single-nucleotide polymorphism arrays. Cancer Res. 2006;66:7920–7928. doi: 10.1158/0008-5472.CAN-06-0721. [DOI] [PubMed] [Google Scholar]
- 49.Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 51.Asghar U, Witkiewicz AK, Turner NC, et al. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14:130–146. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones S, Hruban RH, Kamiyama M, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klein AP. Genetic susceptibility to pancreatic cancer. Mol Carcinog. 2012;51:14–24. doi: 10.1002/mc.20855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhen DB, Rabe KG, Gallinger S, et al. BRCA1, BRCA2, PALB2, and CDKN2A mutations in familial pancreatic cancer: a PACGENE study. Genet Med. 2014 doi: 10.1038/gim.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kastrinos F, Mukherjee B, Tayob N, et al. Risk of pancreatic cancer in families with Lynch syndrome. JAMA. 2009;302:1790–1795. doi: 10.1001/jama.2009.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goggins M, Offerhaus GJ, Hilgers W, et al. Pancreatic adenocarcinomas with DNA replication errors (RER+) are associated with wild-type K-ras, characteristic histopathology. Poor differentiation, a syncytial growth pattern, and pushing borders suggest RER+ Am J Pathol. 1998;152:1501–1507. [PMC free article] [PubMed] [Google Scholar]
- 57.Wilentz RE, Goggins M, Redston M, et al. Genetic, immunohistochemical, and clinical features of medullary carcinoma of the pancreas: A newly described and characterized entity. Am J Pathol. 2000;156:1641–16451. doi: 10.1016/S0002-9440(10)65035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laghi L, Beghelli S, Spinelli A, et al. Irrelevance of microsatellite instability in the epidemiology of sporadic pancreatic ductal adenocarcinoma. PLoS One. 2012;7:e46002. doi: 10.1371/journal.pone.0046002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lord CJ, Ashworth A. Targeted therapy for cancer using PARP inhibitors. Curr Opin Pharmacol. 2008;8:363–369. doi: 10.1016/j.coph.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 61.Chantrill LA, Nagrial AM, Watson C, et al. Precision Medicine for Advanced Pancreas Cancer: The Individualized Molecular Pancreatic Cancer Therapy (IMPaCT) Trial. Clin Cancer Res. 2015;21:2029–2037. doi: 10.1158/1078-0432.CCR-15-0426. [DOI] [PubMed] [Google Scholar]
- 62.McCormick F. KRAS as a Therapeutic Target. Clin Cancer Res. 2015;21:1797–1801. doi: 10.1158/1078-0432.CCR-14-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson H. US National Cancer Institute’s new Ras project targets an old foe. Nat Med. 2013;19:949–950. doi: 10.1038/nm0813-949. [DOI] [PubMed] [Google Scholar]
- 64.Hamidi H, Lu M, Chau K, et al. KRAS mutational subtype and copy number predict in vitro response of human pancreatic cancer cell lines to MEK inhibition. Br J Cancer. 2014;111:1788–1801. doi: 10.1038/bjc.2014.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zorde Khvalevsky E, Gabai R, Rachmut IH, et al. Mutant KRAS is a druggable target for pancreatic cancer. Proc Natl Acad Sci U S A. 2013;110:20723–20728. doi: 10.1073/pnas.1314307110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kapoor A, Yao W, Ying H, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158:185–197. doi: 10.1016/j.cell.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Viale A, Pettazzoni P, Lyssiotis CA, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alagesan B, Contino G, Guimaraes AR, et al. Combined MEK and PI3K inhibition in a mouse model of pancreatic cancer. Clin Cancer Res. 2015;21:396–404. doi: 10.1158/1078-0432.CCR-14-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Collisson EA, Trejo CL, Silva JM, et al. A central role for RAF-->MEK-->ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov. 2012;2:685–693. doi: 10.1158/2159-8290.CD-11-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bodoky G, Timcheva C, Spigel DR, et al. A phase II open-label randomized study to assess the efficacy and safety of selumetinib (AZD6244 [ARRY-142886]) versus capecitabine in patients with advanced or metastatic pancreatic cancer who have failed first-line gemcitabine therapy. Invest New Drugs. 2012;30:1216–1223. doi: 10.1007/s10637-011-9687-4. [DOI] [PubMed] [Google Scholar]
- 71.Rinehart J, Adjei AA, Lorusso PM, et al. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J Clin Oncol. 2004;22:4456–4462. doi: 10.1200/JCO.2004.01.185. [DOI] [PubMed] [Google Scholar]
- 72.Infante JR, Somer BG, Park JO, et al. A randomised, double-blind, placebo-controlled trial of trametinib, an oral MEK inhibitor, in combination with gemcitabine for patients with untreated metastatic adenocarcinoma of the pancreas. Eur J Cancer. 2014;50:2072–2081. doi: 10.1016/j.ejca.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 73.Dal Molin M, Zhang M, de Wilde RF, et al. Very Long-term Survival Following Resection for Pancreatic Cancer Is Not Explained by Commonly Mutated Genes: Results of Whole-Exome Sequencing Analysis. Clin Cancer Res. 2015;21:1944–1950. doi: 10.1158/1078-0432.CCR-14-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chantrill LA, Nagrial AM, Watson C, et al. Precision Medicine for Advanced Pancreas Cancer: The Individualized Molecular Pancreatic Cancer Therapy (IMPaCT) Trial. Clin Cancer Res. 2015 doi: 10.1158/1078-0432.CCR-15-0426. [DOI] [PubMed] [Google Scholar]
- 75.van der Heijden MS, Brody JR, Dezentje DA, et al. In vivo therapeutic responses contingent on Fanconi anemia/BRCA2 status of the tumor. Clin Cancer Res. 2005;11:7508–7515. doi: 10.1158/1078-0432.CCR-05-1048. [DOI] [PubMed] [Google Scholar]
- 76.van der Heijden MS, Brody JR, Gallmeier E, et al. Functional defects in the fanconi anemia pathway in pancreatic cancer cells. Am J Pathol. 2004;165:651–657. doi: 10.1016/S0002-9440(10)63329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van der Heijden MS, Yeo CJ, Hruban RH, et al. Fanconi anemia gene mutations in young-onset pancreatic cancer. Cancer Res. 2003;63:2585–2588. [PubMed] [Google Scholar]
- 78.Gallmeier E, Calhoun ES, Rago C, et al. Targeted disruption of FANCC and FANCG in human cancer provides a preclinical model for specific therapeutic options. Gastroenterology. 2006;130:2145–2154. doi: 10.1053/j.gastro.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 79.Lowery MA, Kelsen DP, Stadler ZK, et al. An emerging entity: pancreatic adenocarcinoma associated with a known BRCA mutation: clinical descriptors, treatment implications, and future directions. Oncologist. 2011;16:1397–1402. doi: 10.1634/theoncologist.2011-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lowery M, Shah MA, Smyth E, et al. A 67-Year-Old Woman with BRCA 1 Mutation Associated with Pancreatic Adenocarcinoma. J Gastrointest Cancer. 2011;42:160–164. doi: 10.1007/s12029-010-9197-1. [DOI] [PubMed] [Google Scholar]
- 81.Golan T, Kanji ZS, Epelbaum R, et al. Overall survival and clinical characteristics of pancreatic cancer in BRCA mutation carriers. Br J Cancer. 2014;111:1132–1138. doi: 10.1038/bjc.2014.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ashworth A. Drug resistance caused by reversion mutation. Cancer Res. 2008;68:10021–10023. doi: 10.1158/0008-5472.CAN-08-2287. [DOI] [PubMed] [Google Scholar]
- 83.Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol. 2008;26:3785–3790. doi: 10.1200/JCO.2008.16.0812. [DOI] [PubMed] [Google Scholar]
- 84.Hucl T, Rago C, Gallmeier E, et al. A syngeneic variance library for functional annotation of human variation: application to BRCA2. Cancer Res. 2008;68:5023–5030. doi: 10.1158/0008-5472.CAN-07-6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.O’Reilly EM, Lowery MA, Segal MF, et al. Phase IB trial of cisplatin (C), gemcitabine (G), and veliparib (V) in patients with known or potential BRCA or PALB2-mutated pancreas adenocarcinoma (PC) ASCO Meeting Abstracts. 2014;32:4023. [Google Scholar]
- 86.Michael J, Pishvaian HW, Tingting Zhuang, Aiwu Ruth He, Hwang Jimmy J, Amy Hankin, Lisa Ley, Keisha White, Susan Joy Littman, Weiner Louis M, John Marshall, Robert Brody Jonathan. A phase I/II study of ABT-888 in combination with 5-fluorouracil (5-FU) and oxaliplatin (Ox) in patients with metastatic pancreatic cancer (MPC) J Clin Oncol. 2013:31. [Google Scholar]
- 87.Bendell J, O’Reilly EM, Middleton MR, et al. Phase I study of olaparib plus gemcitabine in patients with advanced solid tumours and comparison with gemcitabine alone in patients with locally advanced/metastatic pancreatic cancer. Ann Oncol. 2015;26:804–811. doi: 10.1093/annonc/mdu581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol. 2015;33:244–250. doi: 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lowery MA, Kelsen DP, Smith SC, et al. Phase II trial of veliparib (V) in patients (pts) with previously treated BRCA or PALB2-mutated (mut) pancreas adenocarcinoma (PC) ASCO Meeting Abstracts. 2015;33:358. [Google Scholar]
- 90.Do K, Chen AP. Molecular pathways: targeting PARP in cancer treatment. Clin Cancer Res. 2013;19:977–984. doi: 10.1158/1078-0432.CCR-12-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aarts M, Sharpe R, Garcia-Murillas I, et al. Forced mitotic entry of S-phase cells as a therapeutic strategy induced by inhibition of WEE1. Cancer Discov. 2012;2:524–539. doi: 10.1158/2159-8290.CD-11-0320. [DOI] [PubMed] [Google Scholar]
- 92.Bridges KA, Hirai H, Buser CA, et al. MK-1775, a novel Wee1 kinase inhibitor, radiosensitizes p53-defective human tumor cells. Clin Cancer Res. 2011;17:5638–5648. doi: 10.1158/1078-0432.CCR-11-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rajeshkumar NV, De Oliveira E, Ottenhof N, et al. MK-1775, a potent Wee1 inhibitor, synergizes with gemcitabine to achieve tumor regressions, selectively in p53-deficient pancreatic cancer xenografts. Clin Cancer Res. 2011;17:2799–2806. doi: 10.1158/1078-0432.CCR-10-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Caldas C, Hahn SA, da Costa LT, et al. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- 95.Fukushima N, Sato N, Ueki T, et al. Aberrant methylation of preproenkephalin and p16 genes in pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Am J Pathol. 2002;160:1573–1581. doi: 10.1016/S0002-9440(10)61104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Witkiewicz AK, Knudsen KE, Dicker AP, et al. The meaning of p16(ink4a) expression in tumors: functional significance, clinical associations and future developments. Cell Cycle. 2011;10:2497–2503. doi: 10.4161/cc.10.15.16776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Serrano M, Gomez-Lahoz E, DePinho RA, et al. Inhibition of ras-induced proliferation and cellular transformation by p16INK4. Science. 1995;267:249–252. doi: 10.1126/science.7809631. [DOI] [PubMed] [Google Scholar]
- 98.Serrano M, Lin AW, McCurrach ME, et al. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 99.Liu F, Korc M. Cdk4/6 inhibition induces epithelial-mesenchymal transition and enhances invasiveness in pancreatic cancer cells. Mol Cancer Ther. 2012;11:2138–2148. doi: 10.1158/1535-7163.MCT-12-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Heilmann AM, Perera RM, Ecker V, et al. CDK4/6 and IGF1 Receptor Inhibitors Synergize to Suppress the Growth of p16INK4A–Deficient Pancreatic Cancers. Cancer Res. 2014;74:3947–3958. doi: 10.1158/0008-5472.CAN-13-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Franco J, Witkiewicz AK, Knudsen ES. CDK4/6 inhibitors have potent activity in combination with pathway selective therapeutic agents in models of pancreatic cancer. Oncotarget. 2014;5:6512–6525. doi: 10.18632/oncotarget.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Richard S, Finn JPC, Istvan Lang, Katalin Boer, Igor MBondarenko, Kulyk Sergey O, Johannes Ettl, Ravindranath Patel, Tamas Pinter, Marcus Schmidt, Shparyk Yaroslav V, Thummala Anu R, Voytko Nataliya L, Xin Huang, Kim Sindy T, Randolph Sophia S, Slamon Dennis J. Final results of a randomized Phase II study of PD 0332991, a cyclin-dependent kinase (CDK)-4/6 inhibitor, in combination with letrozole vs letrozole alone for first-line treatment of ER+/HER2- advanced breast cancer (PALOMA-1; TRIO-18) Cancer Res, AACR Supplement. 2014 [Google Scholar]
- 103.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 104.Pasca di Magliano M, Biankin AV, Heiser PW, et al. Common activation of canonical Wnt signaling in pancreatic adenocarcinoma. PLoS One. 2007;2:e1155. doi: 10.1371/journal.pone.0001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Heiser PW, Cano DA, Landsman L, et al. Stabilization of beta-catenin induces pancreas tumor formation. Gastroenterology. 2008;135:1288–1300. doi: 10.1053/j.gastro.2008.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jiang X, Hao HX, Growney JD, et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A. 2013;110:12649–12654. doi: 10.1073/pnas.1307218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer. 2011;11:338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 108.Andersson ER, Lendahl U. Therapeutic modulation of Notch signalling--are we there yet? Nat Rev Drug Discov. 2014;13:357–378. doi: 10.1038/nrd4252. [DOI] [PubMed] [Google Scholar]
- 109.Doucas H, Mann CD, Sutton CD, et al. Expression of nuclear Notch3 in pancreatic adenocarcinomas is associated with adverse clinical features, and correlates with the expression of STAT3 and phosphorylated Akt. J Surg Oncol. 2008;97:63–68. doi: 10.1002/jso.20894. [DOI] [PubMed] [Google Scholar]
- 110.Mann CD, Bastianpillai C, Neal CP, et al. Notch3 and HEY-1 as prognostic biomarkers in pancreatic adenocarcinoma. PLoS One. 2012;7:e51119. doi: 10.1371/journal.pone.0051119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thomas MM, Zhang Y, Mathew E, et al. Epithelial Notch signaling is a limiting step for pancreatic carcinogenesis. BMC Cancer. 2014;14:862. doi: 10.1186/1471-2407-14-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Palagani V, Bozko P, El Khatib M, et al. Combined inhibition of Notch and JAK/STAT is superior to monotherapies and impairs pancreatic cancer progression. Carcinogenesis. 2014;35:859–866. doi: 10.1093/carcin/bgt394. [DOI] [PubMed] [Google Scholar]
- 113.Mizuma M, Rasheed ZA, Yabuuchi S, et al. The gamma secretase inhibitor MRK-003 attenuates pancreatic cancer growth in preclinical models. Mol Cancer Ther. 2012;11:1999–2009. doi: 10.1158/1535-7163.MCT-12-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yabuuchi S, Pai SG, Campbell NR, et al. Notch signaling pathway targeted therapy suppresses tumor progression and metastatic spread in pancreatic cancer. Cancer Lett. 2013;335:41–51. doi: 10.1016/j.canlet.2013.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cook N, Frese KK, Bapiro TE, et al. Gamma secretase inhibition promotes hypoxic necrosis in mouse pancreatic ductal adenocarcinoma. J Exp Med. 209:437–444. doi: 10.1084/jem.20111923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yen WC, Fischer MM, Axelrod F, et al. Targeting notch signaling with a notch2/notch3 antagonist (tarextumab) inhibits tumor growth and decreases tumor-initiating cell frequency. Clin Cancer Res. 2015;21:2084–2095. doi: 10.1158/1078-0432.CCR-14-2808. [DOI] [PubMed] [Google Scholar]
- 117.O’Reilly EM, Smith LS, Bendell JC, et al. Final results of phase Ib of anticancer stem cell antibody tarextumab (OMP-59R5, TRXT, anti-Notch 2/3) in combination with nab-paclitaxel and gemcitabine (Nab-P+Gem) in patients (pts) with untreated metastatic pancreatic cancer (mPC) ASCO Meeting Abstracts. 2015;33:278. [Google Scholar]
- 118.Kadoch C, Hargreaves DC, Hodges C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 120.Helming KC, Wang X, Wilson BG, et al. ARID1B is a specific vulnerability in ARID1A–mutant cancers. Nat Med. 2014;20:251–254. doi: 10.1038/nm.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bitler BG, Aird KM, Garipov A, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A–mutated cancers. Nat Med. 2015;21:231–238. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Huang CH, Lujambio A, Zuber J, et al. CDK9-mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma. Genes Dev. 2014;28:1800–1814. doi: 10.1101/gad.244368.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hirai H, Arai T, Okada M, et al. MK-1775, a small molecule Wee1 inhibitor, enhances anti-tumor efficacy of various DNA-damaging agents, including 5-fluorouracil. Cancer Biol Ther. 2010;9:514–522. doi: 10.4161/cbt.9.7.11115. [DOI] [PubMed] [Google Scholar]
- 125.Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 126.Boj SF, Hwang CI, Baker LA, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu X, Ory V, Chapman S, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yuan H, Myers S, Wang J, et al. Use of reprogrammed cells to identify therapy for respiratory papillomatosis. N Engl J Med. 2012;367:1220–1227. doi: 10.1056/NEJMoa1203055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Francis JC, Melchor L, Campbell J, et al. Whole exome DNA sequence analysis of Brca2 and Trp53 deficient mouse mammary gland tumours. J Pathol. 2015 doi: 10.1002/path.4517. [DOI] [PubMed] [Google Scholar]
- 130.Westcott PM, Halliwill KD, To MD, et al. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature. 2015;517:489–492. doi: 10.1038/nature13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Brody JR, Gallmeier E, Yoshimura K, et al. A proposed clinical test for monitoring fluoropyrimidine therapy: detection and stability of thymidylate synthase ternary complexes. Cancer Biol Ther. 2006;5:923–927. doi: 10.4161/cbt.5.8.2976. [DOI] [PubMed] [Google Scholar]
- 132.Patel K, Iacobuzio-Donahue CA, Gormley PE, et al. Are we systematically under-dosing patients with fluorouracil? J Clin Oncol. 2015;33:e36–e37. doi: 10.1200/JCO.2013.49.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ko AH, Hwang J, Venook AP, et al. Serum CA19-9 response as a surrogate for clinical outcome in patients receiving fixed-dose rate gemcitabine for advanced pancreatic cancer. Br J Cancer. 2005;93:195–199. doi: 10.1038/sj.bjc.6602687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chan A, Prassas I, Dimitromanolakis A, et al. Validation of biomarkers that complement CA19.9 in detecting early pancreatic cancer. Clin Cancer Res. 2014;20:5787–5795. doi: 10.1158/1078-0432.CCR-14-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Topalian SL, Sznol M, McDermott DF, et al. Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. J Clin Oncol. 2014;32:1020–1030. doi: 10.1200/JCO.2013.53.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62–68. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Soares KC, Rucki AA, Wu AA, et al. PD-1/PD-L1 blockade together with vaccine therapy facilitates effector T-cell infiltration into pancreatic tumors. J Immunother. 2015;38:1–11. doi: 10.1097/CJI.0000000000000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Provenzano PP, Cuevas C, Chang AE, et al. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:418–429. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Conley BA, Doroshow JH. Molecular analysis for therapy choice: NCI MATCH. Semin Oncol. 2014;41:297–299. doi: 10.1053/j.seminoncol.2014.05.002. [DOI] [PubMed] [Google Scholar]