Abstract
The Zap1 transcription factor of Saccharomyces cerevisiae and the Loz1 transcription factor of Schizosaccharomyces pombe both play a central role in zinc homeostasis by controlling the expression of genes necessary for zinc metabolism. Zap1 activates gene expression when cells are limited for zinc, while Loz1 is required for gene repression when zinc is in excess. In this review we highlight what is known about the underlying mechanisms by which these factors are regulated by zinc, and how transcriptional activation and repression in eukaryotic cells can be finely tuned according to intracellular zinc availability.
Keywords: Zap1, Loz1, Zinc homeostasis, Yeast, Transcription, Nutrient
1. Introduction
Zinc is an essential cofactor in a large number of enzymes and transcription factors, but is toxic when present in excess. The tight regulation of intracellular zinc levels is therefore a basic cellular process that must occur in all eukaryotic cells. Much of what is currently known about the basic mechanisms by which eukaryotic cells regulate zinc levels is derived from studies using yeast model systems. Yeast are easy to use, genetically tractable models, which can survive as haploids or diploids [1]. As yeast can survive as haploids, they can be easily used in genetic screens to identify recessive mutant alleles that lead to a desired phenotype. Many genes involved in zinc transport and zinc homeostasis are also robustly regulated at a transcriptional level in yeast [2, 3]. These large changes in gene expression have made yeast a powerful system to identify genes that are important for zinc homeostasis, and to study how zinc-dependent changes in transcription occur. Here we review what is known about the regulatory factors that facilitate zinc-dependent changes in gene expression in the budding yeast Saccharomyces cerevisiae, and in its distant cousin, the fission yeast Schizosaccharomyces pombe. Specifically, we focus on what is known about the underlying mechanisms by which two factors, Zap1 and Loz1, ‘sense’ changes in intracellular zinc levels and alter gene expression in response to zinc. Information regarding zinc sensing in other organisms can be found in the following recent reviews [4-6].
2. The Saccharomyces cerevisiae zinc sensor Zap1
The first fungal zinc-responsive transcription factor to be identified was Zap1 from S. cerevisiae [7]. Since its discovery in the late 1990s, functional homologs of Zap1 have been characterized from Aspergillus fumigatus, Cryptococcus gattii, Candida albicans, and Candida dubliniensis [8-11]. Homologs of Zap1 are also widely distributed throughout all of the major fungal phyla suggesting that the majority of the fungal kingdom may use a Zap1-like protein to maintain zinc homeostasis.
Zap1 was discovered in a genetic screen to identify genes required for the zinc-dependent regulation of ZRT1 - a gene required for high affinity zinc uptake. As further deletion analysis revealed that strains lacking ZAP1 grew poorly in zinc-limited medium, and had low levels of ZRT1 expression in zinc-limiting and zinc-replete medium, it was hypothesized that Zap1 was necessary for inducing ZRT1 expression during zinc starvation [7]. A large body of work has now demonstrated that Zap1 activates the expression of ~ 80 genes in response to zinc deficiency, and that many of these genes are required for zinc homeostasis or surviving/adapting to longer periods of zinc starvation [2, 12-14]. Zap1 induces gene expression by binding to one or more Zinc Responsive Elements (ZREs) that are located in the promoter regions of its targets [2, 15]. The consensus DNA sequence for a ZRE is 5’-ACCTTNAAGGT-3’.
2.1 Regulation of Zap1 by zinc
Zap1 plays a central role in zinc homeostasis by inducing target gene expression under zinc-limiting conditions. Both in vivo and in vitro studies addressing how Zap1 ‘senses’ zinc have shown that Zap1 is regulated at both a transcriptional level and post-translational level by zinc, and that the combination of these mechanisms allow Zap1 activity to be tightly regulated over a broad range of zinc levels.
2.2 Auto-regulation of ZAP1
At a transcriptional level, Zap1 binds to a ZRE within its own promoter and auto-regulates its own expression [15]. As Zap1 functions as an activator, auto-regulation results in the rapid amplification of ZAP1 transcripts and Zap1 protein, allowing the rapid induction of target gene expression. Although most Zap1 target genes are induced in response to zinc limitation, global transcript profiling has shown differences in expression patterns in response to zinc. Genes required for zinc homeostasis contain high affinity ZREs within their promoters and are typically induced under mildly zinc-limiting conditions. While genes that help cells to survive the oxidative stress of zinc deficiency, or survive longer periods of zinc starvation are only expressed in severely zinc-limited cells. As these latter genes usually contain low affinity ZREs within their promoters, and auto-regulation of ZAP1 leads to higher levels of Zap1 protein, this increase may facilitate binding to low affinity ZREs under severely zinc-limiting conditions [2]. Although this hypothesis has not been directly tested, ZAP1 transcript levels are regulated by zinc in other fungal species [8-10], suggesting that auto-regulation is an important component of zinc homeostasis.
2.3 Post-translational regulation of Zap1
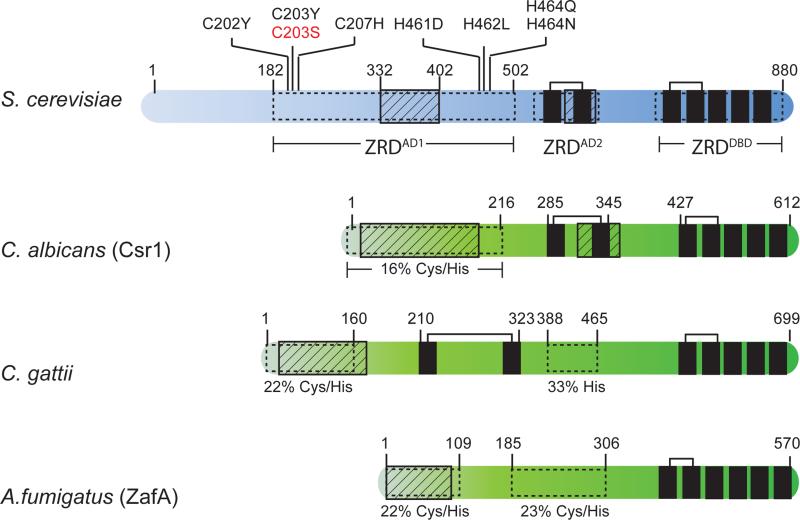
Zap1 is an 880 amino acid protein that contains two transactivation domains, designated AD1 and AD2. Zap1 also contains seven C2H2-type zinc finger domains (Figure 1). Two of the zinc finger domains (ZF1 and ZF2) overlap with AD2. The remaining five zinc fingers (ZF3-7) are located at the C-terminus and are all required for site-specific DNA binding. Combinations of mutagenesis, truncation, and deletion analyses have demonstrated that the activities of AD1, AD2, and the Zap1 DNA binding domain are all independently regulated by zinc, and that the full zinc responsiveness of Zap1 is a combination of these three independent mechanisms [16-19]. Figure 2 summarizes the contribution of each of the individual regulatory domains to Zap1 zinc-responsiveness.
Figure 1.
Domain structures of Zap1 from S. cerevisiae, C. albicans, C. gattii, and A. fumigatus. Zap1 homologs in C. albicans and A. fumigatus are named Csr1 and ZafA respectively. Features highlighted in Zap1 from S. cerevisiae include the acidic rich activation domains (striped rectangles), zinc-responsive domains (white rectangles), and zinc finger domains (black rectangles). tCWCH2 zinc fingers are connected by brackets. Amino acid substitutions that disrupt the zinc-dependent inactivation of ZRDAD1 are also indicated. The original Zap1-1up allele is shown in red. Additional features highlighted in other Zap1 homologs include regions rich in acidic residues (striped rectangles) and cysteine and histidine residues (white rectangles). Numbers above each protein represent amino acid position.
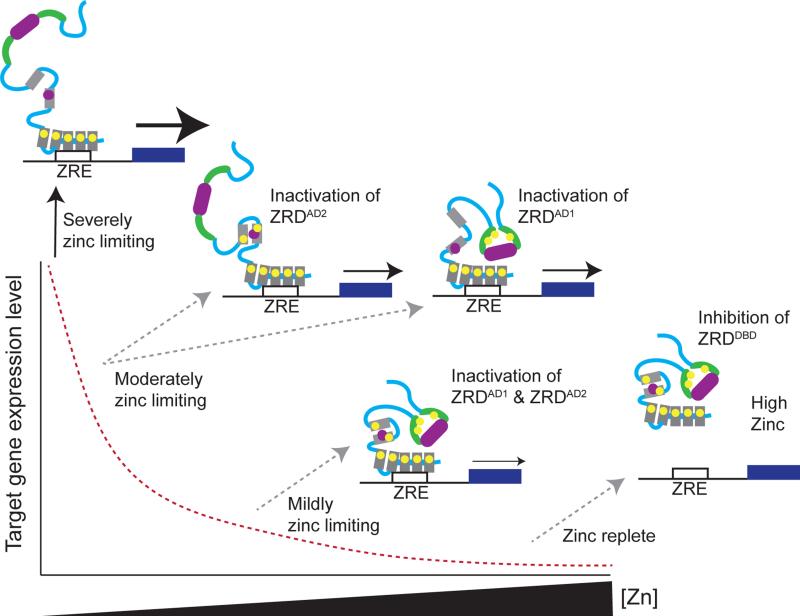
Figure 2.
Predicted model of Zap1 regulation in S. cerevisiae. Under severely zinc-limiting conditions, Zap1 target gene expression is maximally induced. Under these conditions Zap1 is bound to ZREs in target gene promoters, and AD1 and AD2 recruit coactivator complexes to drive target gene expression. Under moderately zinc-limiting conditions, the ZF1/ZF2 tCWCH2 zinc fingers bind zinc leading to the inactivation of AD2, or the cysteine/histidine rich region of ZRDAD1 binds zinc leading to the inactivation of AD1. Both of these events lead to reduced target gene expression. Under mildly zinc-limiting conditions, inactivation of ZRDAD1 and ZRDAD2 further reduces Zap1 target gene expression. When zinc is in excess, Zap1 dissociates from DNA, preventing Zap1-dependent transcription. Features shown include zinc finger domains (gray boxes), zinc ions (yellow circles), activation domains (purple rectangles with +'s), and the cysteine/histidine rich regions of AD1 (green crescents). The red dashed line represents Zap1 target gene expression in response to zinc.
2.3.1 Post-translational regulation of Zap1 – AD1
AD1 is located between amino acid residues 332-402 and is embedded within a larger region required for zinc-dependent changes in AD1 activity, designated Zinc-Responsive Domain AD1 (ZRDAD1). The zinc-dependent inactivation of AD1 is also strictly dependent upon the Zap1 DNA binding domain [17]. As discussed later, the activity of the DNA binding domain is also independently regulated by zinc. However, the inhibition of DNA binding activity occurs when cells are exposed to high zinc levels, while the inhibition of ZRDAD1 occurs over a much lower range of zinc levels [17, 18]. These differences indicate that that ZRDAD1 and the DNA binding domain form their own unique sensing domains, and their combined actions allow dynamic alterations in Zap1 activity over a much broader range of zinc levels.
Although the underlying mechanism by which AD1 function is regulated by zinc is not known, a number of observations suggest that ZRDAD1 directly binds zinc and that this may be the key to the zinc-responsiveness of this domain. ZRDAD1 does not contain any characterized zinc-binding motif, but it is enriched for the amino acids cysteine and histidine. Many of these cysteine and histidine residues are conserved in Zap1 homologs from species closely related to S. cerevisiae and are necessary for the zinc-dependent inactivation of AD1 [17]. The known amino acid substitutions that significantly alter AD1 activity are highlighted in Figure 1. As these residues surround AD1, and combinations of cysteine and histidine residues commonly coordinate zinc ions in proteins, it is thought that alterations in zinc ion levels may directly affect AD1 function. In support of this model, recombinant ZRDAD1 binds 3 mol. equivalents of zinc [17]. However, facets of this model that remain untested include experiments to determine if zinc binding to ZRDAD1 is reversible and dependent upon intracellular zinc ion levels. It is also unknown if mutations that disrupt ZRDAD1 function in vivo, alter zinc binding in vitro.
Another aspect of the ZRDAD1 regulation that remains unsolved is the role of the DNA binding domain. Studies examining the activity of the Zap1 ZRDAD1 domain from S. cerevisiae and Ashbya gossypii indicate that the robust zinc-responsiveness of ZRDAD1 is dependent upon the DNA binding domain [17]. Potential models to explain the requirement of DNA binding domain for ZRDAD1 regulation include that zinc binding to ZRDAD1 triggers an intramolecular interaction with the DNA binding domain masking AD1, or that the DNA binding domain recruits a repressor necessary for the zinc-dependent inactivation of AD1. Future studies examining the precise mechanism by ZRDAD1 are therefore still necessary to unravel its dependency on the DNA binding domain.
One remaining observation that may have broader biological significance is that the Zap1 ZRDAD1 domain from A. gossypii is modestly regulated by zinc in a manner that is independent of the DNA binding domain [17]. This result suggests that the activity of ZRDAD1 may be directly regulated by zinc in some Zap1 homologs. In more distantly related fungi, Zap1-like proteins typically contain a region rich in acidic residues at their N-terminus, which is adjacent to, or is embedded within, a region rich in cysteine and histidine residues (Figure 1). While these features resemble those of ZRDAD1 from S. cerevisiae and A. gossypii, the total number and ratios of cysteine to histidine residues, the clustering of these amino acids, and their positions relative to the acidic residues of AD1, are dependent upon the species. As the role of ZRDAD1 in overall Zap1 zinc-responsiveness has not been studied in any of these more distantly related homologs, it is unknown if the sequence differences in this domain affect zinc binding and the zinc responsiveness of Zap1. However, many fungal species have evolved unique mechanisms to acquire zinc from their own environmental niche [20]. Since Zap1 is central to the induction of these genes in response to zinc, it is possible that the differences in ZRDAD1 may also be significant.
2.3.2 Post-translational regulation of Zap1 – AD2
The most widely studied regulatory domain from Zap1 controls the activity of AD2, an acidic activation domain that directly overlaps with ZF2. In contrast to AD1, the zinc responsive domain of AD2 (ZRDAD2) is well defined and maps to ZF1 and ZF2 [16]. Another critical aspect of the regulation of ZRDAD2 by zinc is that ZF1 and ZF2 both belong to the tandem CWCH2 (tCWCH2) zinc finger family.
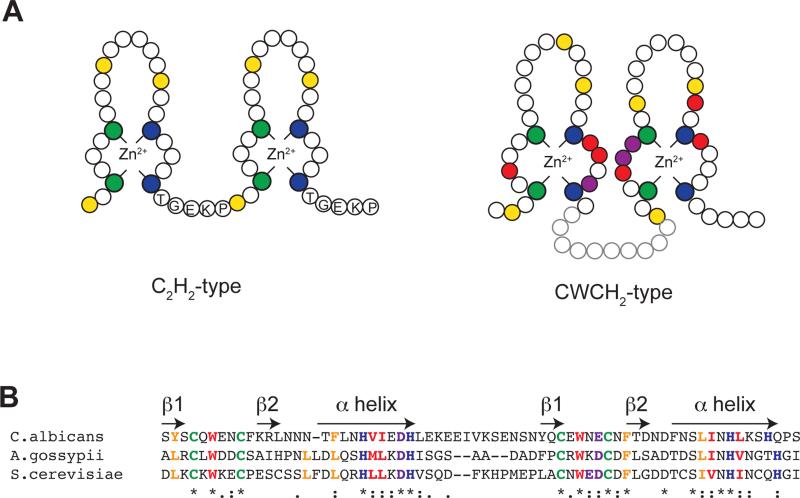
Zinc fingers are well-characterized structural domains that can have a variety of functions ranging from DNA or RNA binding, protein-protein interactions, and membrane association [21, 22]. In eukaryotes, the majority of zinc finger domains fall into a class known as the C2H2-type zinc finger. The conserved features of this class are shown in Figure 3A. Classical C2H2-type zinc fingers have a consensus of ϕ-X-Cys-X2-5-Cys-X3-ϕ-X5-ϕ-X2-His-X2-5-H, where ϕ represents a hydrophobic amino acid, X any amino acid, Cys, cysteine, and His, histidine. Within this sequence, the two conserved cysteine and histidine residues coordinate a single zinc ion, while the conserved hydrophobic residues stabilize the hydrophobic core of the individual zinc finger. Although C2H2-type zinc fingers can be solitary within proteins, in most cases two or more zinc fingers are arranged in tandem, often with a conserved linker sequence of TGEKP connecting each of the adjacent fingers [21, 22]. tCWCH2 zinc fingers also utilize two highly conserved cysteine and histidine residues to coordinate a zinc ion. However, they differ from classical C2H2-type zinc fingers in that two adjacent CWCH2 zinc fingers fold together to form a single structural unit with a shared hydrophobic core [23, 24]. Features of tCWCH2 zinc fingers that allow them to fold together include an invariable tryptophan residue that is located between the two conserved cysteine residues, slight differences in the positions of some of the core hydrophobic residues, and a slightly longer linker region [25] (also see Figure 3A). As outlined below, the ability of ZF1 and ZF2 from Zap1 to fold to form a single structural unit with a shared hydrophobic core is central to the zinc-responsiveness of ZRDAD2.
Figure 3.
Features of the ZF1/ZF2 tCWCH2 zinc fingers from Zap1. (A) A schematic diagram comparing features of two adjacent C2H2-type zinc finger and features of the ZF1/ZF2 tCWCH2 zinc fingers from Zap1. Cysteine and histidine residues that coordinate zinc ions are shown as green and blue circles respectively. The positions of hydrophobic residues involved in stabilizing the hydrophobic core of individual zinc fingers are shown in yellow, while the positions of hydrophobic residues that are involved in the formation of the inter-finger hydrophobic core are shown in red. Conserved acidic residues are shown as purple circles. The positions of highlighted residues are based of the solution structure of AD2 from Zap1 [23]. (B) An alignment showing the conservation of the above highlighted residues in Zap1 ZF1 and ZF2 from C. albicans, A. gossypii, and S. cerevisiae.
The first experiments to implicate the AD2 zinc fingers in zinc sensing were reporter assays that compared the activity of AD2, to the activities of mutated and truncated forms of AD2. These experiments revealed that: 1) AD2 is active in zinc-limited cells and is inactive in zinc-replete cells, 2) truncated forms of AD2 containing just ZF2 are constitutively active and, 3) mutated forms of AD2 containing substitutions preventing ZF1 or ZF2 from binding zinc are constitutively active [16]. Together these results indicate that AD2 is autonomously regulated by zinc and that both ZF1 and ZF2 have to be present and intact for AD2 zinc responsiveness.
Additional mutagenesis analysis highlighted the importance of the shared inter-finger hydrophobic core. Crystal and solution structures of tCWCH2 zinc fingers from GLI and Zap1 respectively, have illustrated that the side chains from the conserved tryptophan residues form the basis of the inter-finger hydrophobic core [23, 24]. As mutations targeting these tryptophan residues in ZF1/ZF2 from Zap1 lead to the constitutive activation of AD2 [16], the ability of these two zinc fingers to fold together to form a single structural unit also appears to be a critical requirement for the regulation of AD2 by zinc. Because of this requirement, it is thought that the reversible binding of zinc to ZF1 and ZF2 controls the formation of the zinc finger pair and AD2 function. Specifically, it is though that ZF1 and ZF2 are largely in an apo, unfolded form in zinc-limited cells. In this unfolded state, acidic residues that facilitate gene activation would be maximally exposed. In contrast, when zinc is in excess, ZF1 and ZF2 bind zinc and fold to form a single structural unit. In this more structured form, acidic residues necessary for activation domain function would be masked, reducing transcription.
Although no experiment has directly shown that the Zap1 ZF1/2 zinc fingers reversibly bind zinc in vivo, elegant studies using AD2-based FRET reporters have demonstrated that the conformation of AD2 is dependent upon intracellular zinc levels, and more importantly is dynamically and reversibly regulated by zinc [26]. The AD2-based FRET reporters contain ZF1 and ZF2 from Zap1 flanked by yellow fluorescent protein and cyan fluorescent protein. When expressed in S. cerevisiae, these reporters lead to higher levels of FRET in zinc-replete cells as compared to zinc-limited cells. The higher level of FRET in zinc-replete cells is also dependent upon ZF1 and ZF2 being able to fold to form a single structural domain with a shared hydrophobic core. An important difference between the AD2-based FRET reporter and other structurally related reporters is that the kinetics of zinc binding and zinc release from the AD2-based FRET reporter is significantly faster. This unique ability of the ZF1/ZF2 pair to rapidly bind and exchange zinc is consistent with the proposed model of zinc ions controlling AD2 conformation and activity. Modified forms of the AD2-based FRET sensors have also been used to monitor dynamic changes in cytosolic and organelle zinc levels in mammalian cell lines [27, 28]. The robust regulation of AD2-based FRET sensors in mammalian cells illustrates that no yeast-specific factor is required for the zinc-dependent regulation of AD2, and further supports a model in which zinc ions directly control AD2 function.
An important aspect of the regulation of ZRDAD2 by zinc is what property, or properties, of ZF1 and/or ZF2 lead to their conformation, and presumably occupancy by zinc, being dependent upon zinc status. Eleven protein families have been identified that contain tCWCH2-type zinc fingers [25]. Studies investigating the role of tCWCH2-type zinc fingers in other proteins have found that they typically have a structural role, and often facilitate binding to DNA [24, 29, 30]. To date, only the ZF1/ZF2 pair from Zap1 has a known regulatory role in zinc sensing, suggesting that some property of these zinc fingers is critical for facilitating zinc responsiveness. Although both zinc fingers bind zinc with a high affinity, zinc will readily exchange from the zinc fingers with other zinc-binding ligands in vitro [16]. The kinetically labile nature of the zinc bound to ZF1 and ZF2, therefore allows zinc to exchange from the zinc finger pair with adjacent zinc binding ligands, potentially allowing it to ‘sense’ the levels of zinc in the surrounding environment. In vitro studies examining zinc binding to the individual zinc fingers or the ZF1/ZF2 pair have shown that binding is cooperative and that ZF1 can independently form a zinc finger fold, while ZF2 cannot form a zinc finger fold without ZF1/ZF2 inter-finger interactions [23]. In vivo and in vitro analyses of chimeric zinc fingers also suggests that residues within the α-helix of ZF2 are critical for the zinc-responsiveness of the ZF1/ZF2 pair [31]. The presence of atypical amino acids within ZF1 and ZF2, particularly within the α-helix of ZF2, therefore may lead to the kinetically labile nature of zinc binding to this pair. To date, studies have shown that the ZRDAD2 from C. albicans and A. gossypii are independently regulated by zinc when expressed in S. cerevisiae [26, 31]. The amino acid residues conserved in ZF1 and ZF2 from these proteins are shown in Figure 3B.
2.3.3 Two activation domains are better than one
AD1 and AD2 are both conserved in the large majority of Zap1 homologs, suggesting that they are important for Zap1 function. So why would Zap1 need two zinc-responsive activation domains? Although AD1 and AD2 recruit similar coactivator complexes, they do not have redundant roles [32, 33]. Under zinc-limiting conditions, AD1 plays the major role in activating the expression of most Zap1 target genes, while AD2 is only required for the full activation of a few targets. In contrast, when zinc-deficient cells are subject to other environmental stresses, such as heat shock, AD2 is necessary for strong activation of Zap1 target gene expression. Outside of the laboratory yeast are most likely subject to feast or famine environments, and are more likely to be subjected to multiple stresses [32]. Two activation domains may therefore make Zap1 a more versatile activator able to induce gene expression even when many other stress pathways are being triggered.
2.3.4 Post-translational regulation of Zap1 – DNA binding domain
The Zap1 DNA binding domain is located between amino acids 687 and 880 and contains of 5 zinc fingers (ZF3-7) that are all necessary for site-specific binding to ZREs [19]. ZF3 and ZF4 are tCWCH2-type zinc fingers, while ZF5-7 are all C2H2-type zinc fingers. In most proteins that use multiple C2H2-type zinc finger domains to contact DNA, individual zinc fingers recognize a 3-4 bp sub-site [21]. This simple rule raises the question of why Zap1 requires 5 zinc fingers to bind to an 11 bp ZRE? In vitro DNA binding studies indicate that ZF4 and ZF7 are critical for high affinity binding to a ZRE and likely make bp contacts with the ACC-GGT ends of the ZRE, while ZF5 and ZF6 have a less important role in DNA recognition and likely make contacts with the 5 bp core of the ZRE [34]. Although ZF3 is not thought to directly contact DNA, structural studies of the DNA binding from the GLI oncogene have provided a potential explanation for its requirement. GLI utilizes two tCWCH2-type zinc fingers and 3 C2H2-type zinc fingers to bind to a 9 bp DNA element. In GLI, zinc fingers 2-5 make direct DNA contacts while zinc finger 1 makes extensive inter-finger contacts with zinc finger 2 [24]. Based on the similarities between the Zap1 and GLI DNA binding domains, ZF3 and ZF4 from Zap1 most likely fold together to form a single structural unit, which may optimize ZF4 DNA interactions. Thus, all 5 zinc fingers of the Zap1 DNA binding domain have structural roles and are critical for site-specific binding to ZREs.
In vivo, protein fusions consisting of the Zap1 DNA binding domain (amino acids 687-880) and the Gal4 activation domain are able to confer zinc-dependent changes in gene expression [18, 19]. These observations indicate that the Zap1 DNA binding domain forms its own unique zinc-responsive domain (ZRDDBD). Importantly, in vivo Zap1 directly binds to ZREs in zinc-limited medium, but not when zinc is in excess. However, in vitro, Zap1 DNA binding activity is not directly regulated by zinc ions [18]. Together these results suggest that another zinc-regulated post-translational mechanism controls Zap1 DNA binding function. As little is known about this regulation, the studies with the ZRDDBD illustrate that there is still much more to learned about zinc sensing in S. cerevisiae.
3. The Schizosaccharomyces pombe zinc sensor Loz1
While homologs of Zap1 are widely dispersed throughout the fungal kingdom, a notable exception is that a homolog of Zap1 is not present in the genome of the 4 known fission yeasts, which include the widely used model system, S. pombe. Despite the absence of a Zap1 homolog in S. pombe, RNA blot, reporter gene, and microarray analyses have demonstrated that genes required for zinc acquisition (zrt1 and fet4) or zinc conservation (adh4) are highly expressed in zinc-limited cells, while genes that are necessary for zinc storage (zym1) are highly expressed in zinc-replete cells [3, 35]. These robust zinc-dependent changes in gene expression suggest that fission yeasts use an alternative factor or regulatory pathway to maintain zinc homeostasis.
Although genetic screens were set up to identify this alternative zinc regulatory factor [3], the first clue to the mechanism by which S. pombe senses zinc was found in some of our work examining zinc-dependent changes in alcohol dehydrogenase gene expression. As in S. cerevisiae, the expression of adh1 and adh4 is tightly regulated by zinc availability [36, 37]. However, in S. pombe the inhibition of adh1 gene expression under zinc-limiting conditions is dependent upon the increased expression of an antisense transcript (adh1AS) that traverses the entire adh1 gene locus [35]. Whilst generating an adh1 knockout strain to further characterize the adh1AS transcript, we isolated an adh1 deletion, which expressed adh4 in zinc-replete cells. Further analysis of this strain revealed that it has obtained a second site mutation that resulted in many genes that are typically expressed in zinc-limiting cell, including adh4, zrt1, and the adh1AS transcript, being expressed in zinc-replete cells. The allele leading to impaired zinc homeostasis was named loz1-1 (for Loss of Zinc sensing allele 1), and mapped to a gene encoding a 522 amino acid protein on chromosome 1 [38]. As deletion of loz1 also results in the expression of adh4, zrt1 and the adh1AS transcripts in zinc-replete cells, we propose that Loz1's primary role in a cell is to facilitate gene repression when zinc is in excess.
Loz1 contains two C2H2-type zinc fingers at it extreme C-terminus. These two zinc fingers and an adjacent accessory domain are required for high affinity binding to a GNMGATC DNA element, which is present in multiple copies in the adh4 and zrt1 promoters [37, 38]. The zinc fingers and adjacent accessory domain are also conserved in Loz1 homologs within the Schizosaccharomyces genus, consistent these domains being important for Loz1 activity (Figure 4). Three other regions of Loz1 are also conserved within most members of the Schizosaccharomyces genus; two of which have multiple conserved cysteine and histidine residues. The significance of these conserved regions is unknown. Outside of the Schizosaccharomyces genus, many other fungi express proteins containing two zinc finger domains that share significant sequence homology with the Loz1 zinc fingers [38]. However, these proteins share no additional sequence similarity with Loz1, and so far have no known role in zinc homeostasis.
Figure 4.
Conserved amino acids in Loz1 homologs from Schizosaccharomyces sp. A schematic diagram showing conserved amino acids in Schizosaccharomyces pombe, Schizosaccharomyces cryophilus, Schizosaccharomyces japonicus, and Schizosaccharomyces octosporus. One region is absent from the S. cryophilus Loz1. Conserved cysteine and histidine residues have been highlighted by green and blue rectangles, respectively.
3.1 Regulation of Loz1 by zinc
Although it is not yet clear if Loz1 directly senses zinc ions, Loz1 activity is regulated at a transcriptional and post-translational level [37, 38]. Loz1 negatively auto-regulates its own expression leading to lower levels of loz1 transcripts in zinc-replete cells [38]. Studies with other transcriptional repressors have found that negative auto-regulation can lead to faster response rates of transcriptional networks [39], minimal transcriptional noise [40], and/or provide enhanced stability against naturally occurring mutations [41]. Thus, auto-regulation of loz1 may be important for optimizing target gene expression in response to changes in intracellular zinc, and/or buffering against naturally occurring sequence changes that alter zinc homeostasis.
When Loz1 is expressed at a constant level within a cell, it accumulates in the nucleus irrespective of zinc status. Under these conditions, the expression of zrt1 and adh4 is robustly regulated by zinc, indicating that Loz1 activity is also regulated at a post-translational level [37]. In vivo analysis of truncated forms of Loz1 suggest that the last 96 amino acids of Loz1, which include the double zinc finger domain and adjacent accessory domain, are sufficient for partial zinc-dependent repression of zrt1 and adh4 [37]. Studies examining the activity of chimeras containing domains from Loz1 and MtfA, a transcription factor from Aspergillus nidulans, also suggest that this region is important for zinc responsiveness [37]. As the zinc-responsiveness of Loz1 maps to the DNA binding domain, it is possible that zinc binding directly to these zinc fingers or accessory domain may control DNA binding activity. However, it is not known if Loz1 DNA binding is regulated by zinc in vivo or in vitro, and if the GNMGATC DNA element within the promoters of Loz1-regulated genes is sufficient for zinc-dependent repression. These critical questions are fundamental to determining the mechanism by which Loz1 affects gene expression.
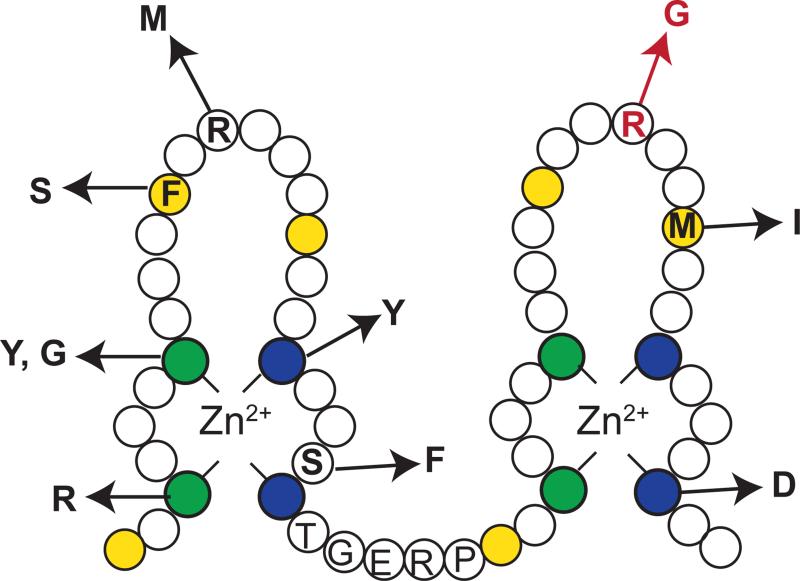
To identify other factors affecting zinc homeostasis, Ehrensberger et al. performed a genetic screen to isolate mutations that allowed adh1Δ cells to grow in the presence of respiratory inhibitor Antimycin A [37]. The rationale for this screen was that adh1Δ cells cannot survive in the presence of Antimycin A in nutrient-rich medium, however, genetic mutations that leads to increased expression of adh4 suppress this lethality [38, 42]. 36 mutations were identified in screen that led to the growth of adh1Δ cells in the presence of Antimycin A. 27 of these mutations resulted in the truncation of Loz1, while the remaining 9 led to amino acid substitutions within the Loz1 zinc finger domain that disrupted Loz1 function, most likely by impairing DNA binding activity. The amino acid substitutions that were identified in this screen are shown in Figure 5. Although this screen did not identify any new factors involved in the zinc-dependent changes of adh4 gene expression, the screen reemphasized the importance of Loz1 in zinc homeostasis.
Figure 5.
Amino acids residues critical for Loz1-dependent repression. A schematic diagram of the double zinc finger domain from Loz1, highlighting known amino acid substitutions that disrupt Loz1 function. The amino acid substitution in the original Loz1-1 allele is shown in red.
A new report suggests that the Extender of Chronological Lifespan (Ecl) proteins are required for the induction of zrt1 and SPBC1348.06c gene expression under zinc-limiting conditions [43]. The Ecl family consists of three small nuclear localized proteins named Ecl1, Ecl2, and Ecl3, all of which have a conserved cysteine rich motif at their N-terminus [44]. However, the role of Loz1 was not investigated in these studies. It therefore is currently not clear if Ecl proteins are involved in the inactivation of Loz1, or if they are simply required for the activation of these genes. Thus, future studies are necessary to determine whether the Ecl proteins have any role in zinc sensing in S. pombe.
4. Conclusions
Studies with yeast model systems have greatly facilitated our knowledge of zinc homeostasis and how a single cell can detect changes in intracellular zinc levels. Studies with Zap1 have demonstrated how tCWCH2 zinc fingers can be used as zinc sensing domains, and how multiple mechanisms can allow a single factor to sense a broad range of zinc levels. Studies with Loz1 reveal how the activity of a transcriptional repressor can be regulated by zinc, and raise the possibility that other types of zinc finger domains may also be involved in zinc sensing. Despite these discoveries, key details are still missing in our understanding of the mechanisms by which these factors sense zinc. It is also largely unknown what molecules/proteins are involved in buffering zinc ions within the cytosol. As Zap1 and possibly Loz1 sense changes in the labile pool of zinc within cells, this information is critical to understanding the regulation of the zinc sensing domains. Uncovering these details will provide a more complete picture of the mechanisms of zinc sensing in yeast, and likely insight into the types of molecules and proteins involved in zinc sensing in other organisms.
Highlights.
Zap1 is a transcription factor from S. cerevisiae that activates gene expression in zinc-limited cells.
Zap1 is regulated at both a transcriptional and post-translational level by zinc.
Zap1 contains three zinc-responsive domains that independently detect changes in intracellular zinc levels.
Loz1 is a transcription factor from S. pombe that represses gene expression in zinc-replete cells.
Acknowledgements
We would like to thank Dr. R. Michael Townsend for critical reading of the manuscript. This work was supported by grant from the National Institutes of Health Grant GM105695 to A.J.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hoffman CS, Wood V, Fantes PA. An Ancient Yeast for Young Geneticists: A Primer on the Schizosaccharomyces pombe Model System. Genetics. 2015;201:403–23. doi: 10.1534/genetics.115.181503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu CY, Bird AJ, Chung LM, Newton MA, Winge DR, Eide DJ. Differential control of Zap1-regulated genes in response to zinc deficiency in Saccharomyces cerevisiae. BMC Genomics. 2008;9:370. doi: 10.1186/1471-2164-9-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dainty SJ, Kennedy CA, Watt S, Bahler J, Whitehall SK. Response of Schizosaccharomyces pombe to zinc deficiency. Eukaryot Cell. 2008;7:454–64. doi: 10.1128/EC.00408-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi S, Bird AJ. Zinc'ing sensibly: controlling zinc homeostasis at the transcriptional level. Metallomics. 2014;6:1198–215. doi: 10.1039/c4mt00064a. [DOI] [PubMed] [Google Scholar]
- 5.Gunther V, Lindert U, Schaffner W. The taste of heavy metals: gene regulation by MTF-1. Biochim Biophys Acta. 2012;1823:1416–25. doi: 10.1016/j.bbamcr.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem Rev. 2009;109:4644–81. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao H, Eide DJ. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:5044–52. doi: 10.1128/mcb.17.9.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreno MA, Ibrahim-Granet O, Vicentefranqueira R, Amich J, Ave P, Leal F, et al. The regulation of zinc homeostasis by the ZafA transcriptional activator is essential for Aspergillus fumigatus virulence. Mol Microbiol. 2007;64:1182–97. doi: 10.1111/j.1365-2958.2007.05726.x. [DOI] [PubMed] [Google Scholar]
- 9.Schneider Rde O, Fogaca Nde S, Kmetzsch L, Schrank A, Vainstein MH, Staats CC. Zap1 regulates zinc homeostasis and modulates virulence in Cryptococcus gattii. PLoS One. 2012;7:e43773. doi: 10.1371/journal.pone.0043773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bottcher B, Palige K, Jacobsen ID, Hube B, Brunke S. Csr1/Zap1 Maintains Zinc Homeostasis and Influences Virulence in Candida dubliniensis but Is Not Coupled to Morphogenesis. Eukaryot Cell. 2015;14:661–70. doi: 10.1128/EC.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim MJ, Kil M, Jung JH, Kim J. Roles of Zinc-responsive transcription factor Csr1 in filamentous growth of the pathogenic Yeast Candida albicans. J Microbiol Biotechnol. 2008;18:242–7. [PubMed] [Google Scholar]
- 12.De Nicola R, Hazelwood LA, De Hulster EA, Walsh MC, Knijnenburg TA, Reinders MJ, et al. Physiological and transcriptional responses of Saccharomyces cerevisiae to zinc limitation in chemostat cultures. Appl Environ Microbiol. 2007;73:7680–92. doi: 10.1128/AEM.01445-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacDiarmid CW, Taggart J, Kerdsomboon K, Kubisiak M, Panascharoen S, Schelble K, et al. Peroxiredoxin chaperone activity is critical for protein homeostasis in zinc-deficient yeast. J Biol Chem. 2013;288:31313–27. doi: 10.1074/jbc.M113.512384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.North M, Steffen J, Loguinov AV, Zimmerman GR, Vulpe CD, Eide DJ. Genome-wide functional profiling identifies genes and processes important for zinc-limited growth of Saccharomyces cerevisiae. PLoS Genet. 2012;8:e1002699. doi: 10.1371/journal.pgen.1002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao H, Butler E, Rodgers J, Spizzo T, Duesterhoeft S, Eide D. Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J Biol Chem. 1998;273:28713–20. doi: 10.1074/jbc.273.44.28713. [DOI] [PubMed] [Google Scholar]
- 16.Bird AJ, McCall K, Kramer M, Blankman E, Winge DR, Eide DJ. Zinc fingers can act as Zn2+ sensors to regulate transcriptional activation domain function. EMBO J. 2003;22:5137–46. doi: 10.1093/emboj/cdg484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herbig A, Bird AJ, Swierczek S, McCall K, Mooney M, Wu CY, et al. Zap1 activation domain 1 and its role in controlling gene expression in response to cellular zinc status. Mol Microbiol. 2005;57:834–46. doi: 10.1111/j.1365-2958.2005.04734.x. [DOI] [PubMed] [Google Scholar]
- 18.Frey AG, Bird AJ, Evans-Galea MV, Blankman E, Winge DR, Eide DJ. Zinc-regulated DNA binding of the yeast Zap1 zinc-responsive activator. PLoS One. 2011;6:e22535. doi: 10.1371/journal.pone.0022535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bird A, Evans-Galea MV, Blankman E, Zhao H, Luo H, Winge DR, et al. Mapping the DNA binding domain of the Zap1 zinc-responsive transcriptional activator. J Biol Chem. 2000;275:16160–6. doi: 10.1074/jbc.M000664200. [DOI] [PubMed] [Google Scholar]
- 20.Wilson D. An evolutionary perspective on zinc uptake by human fungal pathogens. Metallomics. 2015;7:979–85. doi: 10.1039/c4mt00331d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q Rev Biophys. 2010;43:1–21. doi: 10.1017/S0033583510000089. [DOI] [PubMed] [Google Scholar]
- 22.Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Feng LS, Matskevich V, Venkataraman K, Parasuram P, Laity JH. Solution structure of a Zap1 zinc-responsive domain provides insights into metalloregulatory transcriptional repression in Saccharomyces cerevisiae. J Mol Biol. 2006;357:1167–83. doi: 10.1016/j.jmb.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Pavletich NP, Pabo CO. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993;261:1701–7. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- 25.Hatayama M, Aruga J. Characterization of the tandem CWCH2 sequence motif: a hallmark of inter-zinc finger interactions. BMC Evol Biol. 2010;10:53. doi: 10.1186/1471-2148-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiao W, Mooney M, Bird AJ, Winge DR, Eide DJ. Zinc binding to a regulatory zinc-sensing domain monitored in vivo by using FRET. Proc Natl Acad Sci U S A. 2006;103:8674–9. doi: 10.1073/pnas.0600928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin Y, Miranda JG, Stoddard CI, Dean KM, Galati DF, Palmer AE. Direct comparison of a genetically encoded sensor and small molecule indicator: implications for quantification of cytosolic Zn(2+). ACS Chem Biol. 2013;8:2366–71. doi: 10.1021/cb4003859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin Y, Dittmer PJ, Park JG, Jansen KB, Palmer AE. Measuring steady-state and dynamic endoplasmic reticulum and Golgi Zn2+ with genetically encoded sensors. Proc Natl Acad Sci U S A. 2011;108:7351–6. doi: 10.1073/pnas.1015686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekimata M, Homma Y. Sequence-specific transcriptional repression by an MBD2-interacting zinc finger protein MIZF. Nucleic Acids Res. 2004;32:590–7. doi: 10.1093/nar/gkh249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez-Martinez J, Brown CV, Diez E, Tilburn J, Arst HN, Jr., Penalva MA, et al. Overlap of nuclear localisation signal and specific DNA-binding residues within the zinc finger domain of PacC. J Mol Biol. 2003;334:667–84. doi: 10.1016/j.jmb.2003.09.072. [DOI] [PubMed] [Google Scholar]
- 31.Bird AJ, Swierczek S, Qiao W, Eide DJ, Winge DR. Zinc metalloregulation of the zinc finger pair domain. J Biol Chem. 2006;281:25326–35. doi: 10.1074/jbc.M600655200. [DOI] [PubMed] [Google Scholar]
- 32.Frey AG, Eide DJ. Roles of two activation domains in Zap1 in the response to zinc deficiency in Saccharomyces cerevisiae. J Biol Chem. 2011;286:6844–54. doi: 10.1074/jbc.M110.203927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frey AG, Eide DJ. Zinc-responsive coactivator recruitment by the yeast Zap1 transcription factor. Microbiologyopen. 2012;1:105–14. doi: 10.1002/mbo3.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans-Galea MV, Blankman E, Myszka DG, Bird AJ, Eide DJ, Winge DR. Two of the five zinc fingers in the Zap1 transcription factor DNA binding domain dominate site-specific DNA binding. Biochemistry. 2003;42:1053–61. doi: 10.1021/bi0263199. [DOI] [PubMed] [Google Scholar]
- 35.Ehrensberger KM, Mason C, Corkins ME, Anderson C, Dutrow N, Cairns BR, et al. Zinc-dependent regulation of the Adh1 antisense transcript in fission yeast. J Biol Chem. 2013;288:759–69. doi: 10.1074/jbc.M112.406165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bird AJ, Gordon M, Eide DJ, Winge DR. Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts. EMBO J. 2006;25:5726–34. doi: 10.1038/sj.emboj.7601453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ehrensberger KM, Corkins ME, Choi S, Bird AJ. The double zinc finger domain and adjacent accessory domain from the transcription factor loss of zinc sensing 1 (loz1) are necessary for DNA binding and zinc sensing. J Biol Chem. 2014;289:18087–96. doi: 10.1074/jbc.M114.551333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corkins ME, May M, Ehrensberger KM, Hu YM, Liu YH, Bloor SD, et al. Zinc finger protein Loz1 is required for zinc-responsive regulation of gene expression in fission yeast. Proc Natl Acad Sci U S A. 2013;110:15371–6. doi: 10.1073/pnas.1300853110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenfeld N, Elowitz MB, Alon U. Negative autoregulation speeds the response times of transcription networks. J Mol Biol. 2002;323:785–93. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- 40.Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–3. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- 41.Denby CM, Im JH, Yu RC, Pesce CG, Brem RB. Negative feedback confers mutational robustness in yeast transcription factor regulation. Proc Natl Acad Sci U S A. 2012;109:3874–8. doi: 10.1073/pnas.1116360109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paquin CE, Williamson VM. Ty insertions at two loci account for most of the spontaneous antimycin A resistance mutations during growth at 15 degrees C of Saccharomyces cerevisiae strains lacking ADH1. Mol Cell Biol. 1986;6:70–9. doi: 10.1128/mcb.6.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohtsuka H, Ishida M, Naito C, Murakami H, Aiba H. Sexual development of Schizosaccharomyces pombe is induced by zinc or iron limitation through Ecl1 family genes. Mol Genet Genomics. 2015;290:173–85. doi: 10.1007/s00438-014-0911-8. [DOI] [PubMed] [Google Scholar]
- 44.Ohtsuka H, Ogawa Y, Mizuno H, Mita S, Aiba H. Identification of Ecl family genes that extend chronological lifespan in fission yeast. Biosci Biotechnol Biochem. 2009;73:885–9. doi: 10.1271/bbb.80804. [DOI] [PubMed] [Google Scholar]