Abstract
Within each individual, a highly diverse T cell receptor (TCR) repertoire interacts with peptides presented by major histocompatibility complex (MHC) molecules. Despite extensive research, it remains controversial whether germline-encoded TCR-MHC contacts promote TCR-MHC specificity and if so, whether there exist differences in TCR V-gene compatibilities with different MHC alleles. We applied eQTL mapping to test for associations between genetic variation and TCR V-gene usage in a large human cohort. We report strong trans associations between variation in the MHC locus and TCR V-gene usage. Fine mapping of the association signals reveals specific amino acids in MHC genes that bias V-gene usage, many of which contact or are spatially proximal to the TCR or peptide. Hence, these MHC variants, several of which are linked to autoimmune diseases, can directly affect TCR-MHC interaction. These results provide the first examples of trans-QTLs mediated by protein-protein interactions, and are consistent with intrinsic TCR-MHC specificity.
MHC proteins are an essential component of the adaptive immune system, due to their role in presenting self and foreign processed peptides for inspection by T cells1. Human MHC genes - also referred to as HLA (Human Leukocyte Antigen) genes - are extremely polymorphic, and variants of these genes have been associated with many traits, including most autoimmune diseases2,3. It has been suggested that in some cases the increased disease risk conferred by particular MHC alleles is due to differences in the peptides they present, or to differences in the intrinsic stability of the MHC variants4. However, beyond their importance in shaping the sequence repertoire of antigenic peptides presented, in most cases we still have limited understanding of the functional implications of different MHC alleles and their interplay with TCR diversity3,4.
One possible functional effect of MHC genotypes could be to impact the usage of paralogous genes that encode the TCR repertoire. Each individual has a highly diverse TCR repertoire that is able to recognize and respond to a huge variety of foreign peptides when they are presented on MHC proteins. Each TCR is a heterodimer, usually comprised of α (TRA) and β (TRB) chains (1–5% of T cells instead carry γ (TRG) and δ (TRD) chains5). Each T cell clone expresses a unique pair of TCR chains resulting from somatic V(D)J recombination of one of each of the paralogous variable (V), joining (J) and, in β and δ chains, diversity (D) genes. During this recombination, the joints are partially digested and nucleotides are randomly added to form the highly variable and non-germline-encoded Complementarity Determining Region (CDR) 3 loop that recognizes presented peptides6. Additional contacts with the MHC are formed by the CDR1 and CDR2 loops of the T cell receptor, which are encoded by the V-genes6–10. T cells subsequently undergo both positive and negative selection in the thymus to ensure specificity for foreign, but not for self, peptides11.
The TCR repertoire is reshaped in response to infection8,12 and varies between individuals13. However, little is known about the extent to which the usage of different V-genes in the TCR repertoire is shaped by host genetics, apart from limited observations of increased repertoire similarity among close relatives14,15, and a report that usage of Vα genes in response to an EBV epitope depends on HLA-B genotype16. Moreover, while it is intuitive that MHC genotype might affect the TCR repertoire, the precise nature of the MHC-TCR interaction remains controversial. In contrast to B-cell-secreted antibodies (which are also generated by V(D)J recombination), TCRs interact specifically with MHC-peptide complexes. Yet despite numerous structural, in vitro, and murine in vivo studies, there is still active debate about whether the germline-encoded TCR-MHC contacts help to promote this specificity10,17–23 or are merely bystanders24–26,23. Recent studies have reported conflicting conclusions on this point26,27. If germline-encoded contacts influence TCR-MHC interaction, then we might expect different TCR V-genes to differ in their compatibilities with different MHC alleles. Such differences might bias V-gene usage in the post-thymic repertoire, since both thymic selection and clonal expansion of T cells are dependent on TCR-MHC interactions21.
Here, we address the question of how the host genotype influences the make-up of the TCR repertoire using expression quantitative trait loci (eQTL) analysis28 of a large human cohort29 for which both RNA sequencing of peripheral blood and genotyping data are available (Fig. 1). We took an undirected approach of testing, genome-wide, for trans associations between genetic variation and expression of TCR V-genes. (We will also use the term ‘bias’ to refer to genotype-dependent shifts in V-gene usage). Our results suggest that MHC genotypes play an important role in determining the V-gene usage profiles of each individual’s TCR repertoire.
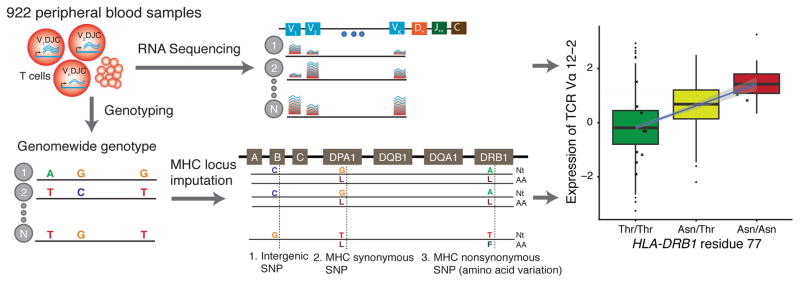
Figure 1. Illustration of our approach.
Expression of TCR V-genes in peripheral blood was estimated by mapping whole blood RNA-seq29 reads to V-genes while controlling for relevant individual level covariates and the total number of reads mapped to each TCR chain in that individual. Genotypes were measured genome-wide using Illumina HumanOmni1-Quad BeadChip29. MHC genotypes were imputed with SNP2HLA31. Associations between nucleotide or amino acid genotypes and V-gene expression were tested using Pearson correlations.
Results
Expression of TCR V-genes is associated with MHC variation
We analyzed RNA-sequencing (RNA-seq) data collected from the peripheral blood of 922 individuals29 of European ancestry. To estimate the relative expression of each V-gene, we counted the number of reads that mapped uniquely to each V-gene while controlling for the total expression of each TCR chain and other relevant covariates (Supplementary Table 1; Fig. 1, Supplementary Figs 1 and 2; see Methods). After removing genes and individuals with low numbers of mapped reads, we obtained expression measurements for 44 Vα, 40 Vβ, 11 Vγ and 3 Vδ genes in each of 895 individuals (Supplementary Tables 2 and 3; Supplementary Figs. 3–5). Since ordinarily only one functional TCR is expressed in each T cell, the estimated expression levels will be determined by the fraction of T cells expressing each TCR, as well as the expression level of the TCR in each cell. As a control, we applied a similar pipeline to analyze the V-genes from B cell-secreted antibodies (immunoglobulin; Ig), which are not expected to interact with MHC.
To test for associations between genotype and the expression of TCR V-genes, we used genome-wide genotype measurements in the same individuals29 (Fig. 1). We initially tested for short-range eQTLs, i.e., within 1Mb of each V-gene. We excluded from this analysis a small number of genes in which read mappability varies across haplotypes (Supplementary Fig. 6; see Methods). As expected, we found many short-range eQTLs - for 78% of TCR V-genes and 46% of Ig V-genes at 5% False Discovery Rate (FDR) (Supplementary Fig. 7a; Supplementary Table 4) - presumably reflecting cis-acting effects on gene regulation.
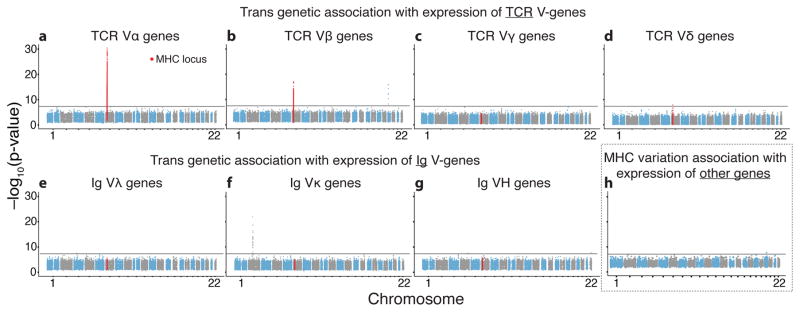
We next tested genome-wide for long-range eQTLs. Notably, we found multiple highly significant associations between the MHC locus and expression of TCR V-genes. 47.7% and 22.5% of the Vα and Vβ genes, respectively, were associated at a stringent threshold of p<5×10−8, which accounts for genome-wide significance testing (Figs. 2a and 2b, Supplementary Fig. 8; Supplementary Table S5). Restricting the analysis to the extended MHC locus reduces the multiple testing burden so that 66% and 35% of the Vα and Vβ genes, respectively, were significantly associated with variation in the MHC locus at a 5% FDR (Supplementary Fig. 7b; Supplementary Table 4; see Methods). The MHC locus stands out, as we observed just one other genome-wide significant trans association with any TCR V-gene (TRVB24-1, associated with variation near ZNF443) (Figs. 2a, 2b, and Supplementary Fig. 9). Interestingly, despite the lack of MHC restriction of γδ-TCRs30, the few significant associations with expression of Vδ-genes also mapped to the MHC locus (Figs. 2c and 2d). We speculate that these associations might be caused by the small population of δβ T cells that recognize MHC-presented peptides30. Importantly, there were no genome-wide significant associations between MHC and the Ig V-genes, which encode antibodies (Figs. 2e–2g, Supplementary Figs. 7b and 8), and just one association between MHC variants and any other gene (Fig. 2h), highlighting the specific relationship between MHC and TCR.
Figure 2. Expression of TCR Vα and Vβ genes is significantly associated with genetic variation in the MHC locus.
(a–d) Manhattan plots showing for each of 649,863 measured SNPs the most significant association across all V genes in each TCR α chain (a), β chain (b), γ chain (c) and δ chain. (e–g) Similar to (a–d) for immunoglobulin (Ig) λ chain (e), κ chain (f) and heavy chain (g). (h) A Manhattan plot showing, for each of 13,732 tested genes, the most significant association between its expression and any of the 9,967 measured SNPs in the extended MHC locus. Excluded from the plot were all Ig and TCR V-genes, genes within 1Mb of the MHC locus, and one pseudogene with high sequence similarity to MHC (RNF5P1). Black horizontal lines correspond to a p-value of 5×10−8.
Classical MHC genes drive most MHC locus-TCR associations
To localize functional elements in the MHC locus that associate with Vα or Vβ expression, we used the genotyped SNPs to impute genotype estimates for all known MHC variants31 (Fig. 1). We then performed forward stepwise regression to identify independent associations with any SNPs within the MHC locus or with amino acid polymorphisms in classical MHC genes. We iteratively added polymorphic positions to a predictive model of each gene’s expression until no additional position significantly improved the model fit (F-test, p-value threshold=0.05 under Bonferroni correction; Fig. 3a, Supplementary Fig. 10; Supplementary Table 6, see Methods).
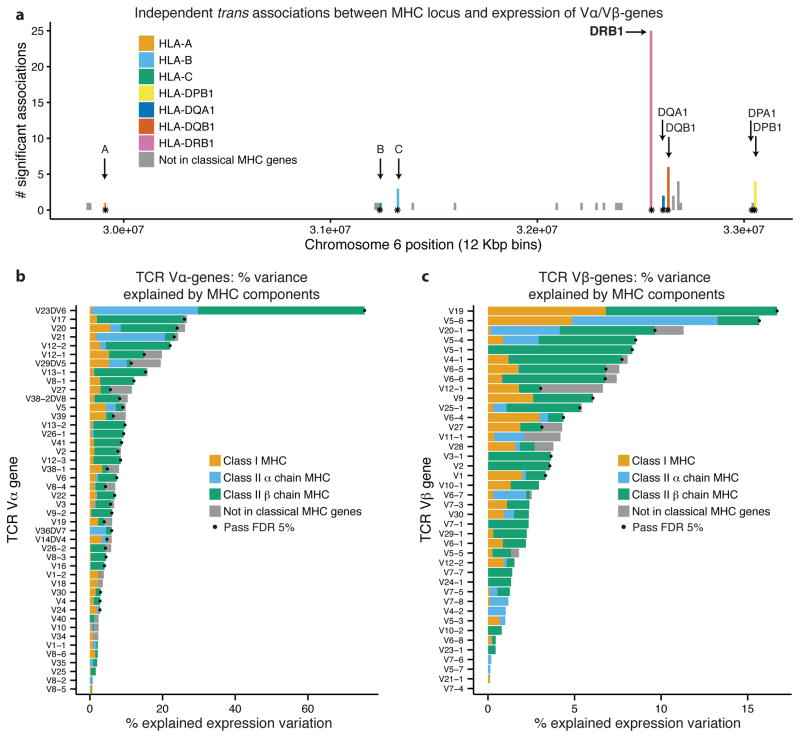
Figure 3. Expression of TCR Vα and Vβ genes is associated with amino acid variation in MHC proteins.
(a) Independent associations between Vα or Vβ expression and nucleotide or amino acid variation in the MHC locus (p<0.05 with Bonferroni correction). SNPs are binned according to their genomic position (12Kb bins); points mark centerpoints of the classical MHC genes. For binning by equal numbers of SNPs see Supplementary Fig. 10. (b) and (c) show TCR Vα and TCR Vβ expression variation explained by amino acid variation in classical MHC genes and genetic variability in the MHC locus outside the transcribed regions of classical MHC genes. Values computed using GCTA33. Dots indicate that the total fraction of variation explained by the MHC gene components was significant at 5% FDR.
Using this conservative threshold, we identified 66 independent MHC-TCR associations for 28 TCR Vα and 15 Vβ genes. These associations explained from 2.2% to 37% of the expression variation of each V-gene (Supplementary Fig. 11 and Supplementary Table 7). It is often difficult to precisely locate causal sites in QTL mapping due to linkage disequilibrium (LD - i.e., the property that genotypes at linked sites tend to be correlated). Nonetheless, despite the fact that LD tends to spread association signals, we observed strong enrichment of signals within transcribed regions of classical MHC genes, especially in HLA-DRB1. 42 of 66 associations are in MHC genes - of these 37 are in class II and 25 are in the class II gene HLA-DRB1 alone (these are 3.5–, 7.5– and 15.8–fold enrichments respectively, relative to all variants). In addition, many of the remaining 24 associations outside genes are near classical MHC proteins, and may thus be in LD with causal variants in genes. The larger number of associations with variation in MHC class II proteins than in MHC class I proteins may be biologically meaningful, but it might also reflect greater power in our data set to detect class II interactions due to the higher abundance of CD4 than CD8 T cells in peripheral blood32.
To test the robustness of our results, we conducted two further analyses. First, we tested for independent associations using classical MHC 4-digit haplotypes instead of nucleotide and amino acid variation. This analysis yielded qualitatively similar results: 75 of 92 associations were with MHC class II haplotypes; of these, 32 were with HLA-DRB1 haplotypes (these are 1.6- and 2.4-fold enrichments respectively, relative to all classical MHC haplotypes; (Supplementary Figs. 12–13, and Supplementary Table 8). HLA-DRB1*03:01 was associated with the largest number of different V-genes (12 genes). Second, we performed a joint multi-phenotype regression analysis. This analysis also indicated that MHC class II genes and especially HLA-DRB1 contribute the most signals (Supplementary Figs. 14–16, Methods).
To assess the specific contribution of coding variants to these associations, we used a variance components method designed for genomic data, GCTA33, to estimate the fraction of the expression variation of TCR V-genes that can be explained by coding variants in classical MHC genes. We fit a model with genetic components representing the amino acid variation in each MHC gene, and a component for variants in the MHC locus outside the transcribed regions of classical MHC genes (Figs. 3b and 3c, and Supplementary Figs. 17–22). We found that amino acid variants in MHC genes explain a significant fraction of expression variation for 33 out of 44 TCR Vα genes and 16 of 40 Vβ genes and 92% and 88%, respectively, of the total variance explained (Figs. 3b and 3c, Supplementary Fig. 21; 5% FDR, see Methods). As a negative control, only one Ig V-gene out of 149 was significant at a 5% FDR. For significant TCR V-genes, the fraction of variance explained ranges from 5–75% (additional variance may be due to environmental or random factors as well as measurement noise - especially for genes expressed at low levels; Supplementary Fig. 22). Thus, in summary, we conclude that the vast majority of the TCR variation explained by the MHC locus is due to amino acid variation in MHC proteins, with major contributions of MHC class II β chains and especially HLA-DRB1.
MHC residues that bias TCR expression contact the TCR
Our next goal was to infer which amino acid positions are most likely responsible for expression biases of the TCR V-genes (see Methods). Since many of the positions are in strong LD, it is often unclear which position is causal for a particular association. To quantify the uncertainty, we implemented a Bayesian Markov chain Monte Carlo (MCMC) approach that sampled over the joint distribution of potential causal positions that are consistent with the association data for each V-gene. Compared to frequentist approaches such as variable selection methods, which generally make firm choices about which sites to include, Bayesian models are generally better at quantifying and reflecting the uncertainty due to LD34–37. Our model started with a low, uniform prior probability that any given amino acid position would be causal, and incorporated the intuition that if a particular position is causal for one variant then it may be more likely causal for others as well (separately for Vα and Vβ genes). The model outputs a posterior probability that any given amino acid position is causally associated with expression of a particular V-gene or with any V-gene, while accounting for correlations across sites due to LD.
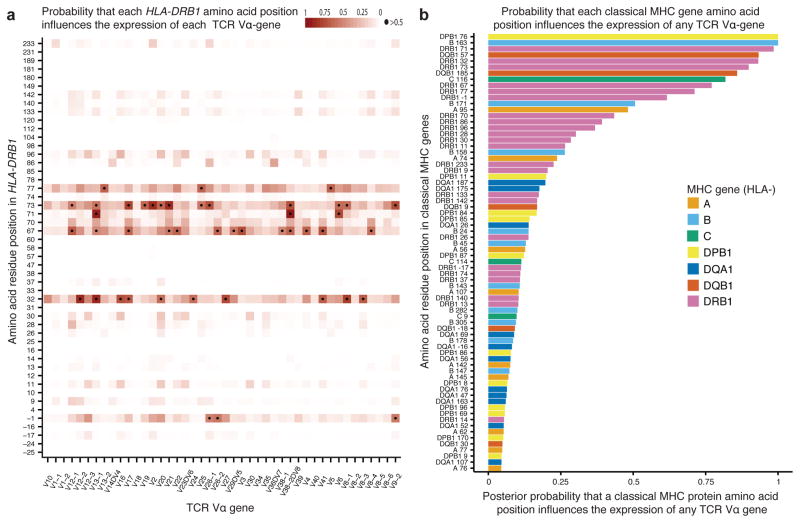
The results revealed several MHC amino acid positions with high posterior probabilities of influencing expression of TCR Vα-genes (Fig. 4 and Supplementary Table 9), though for some associations the posterior is shared across multiple potential causal positions in strong LD (Supplementary Fig. 23). Interestingly, three of the top 15 amino acid positions influencing expression of TCR Vα-genes (HLA-DRB1 71 and 86, HLA-DQB 57) are strongly associated with several autoimmune diseases. Alleles of these three amino acids are strongly correlated with expression biases of TCR Vα-genes (Supplementary Fig. 24). For example, HLA-DRB1 71, the third-ranked amino acid position, is strongly associated with seropositive rheumatoid arthritis, type 1 diabetes and multiple sclerosis38–40. Different variants of the amino acid at this position are correlated with increased expression of different V-genes. It is possible that TCR V-gene expression biases, previously shown to affect the outcome of autoimmunity and infection41,42, are related to some MHC associations with autoimmune and infectious disease risk. Consistent with the analysis above, we found fewer positions that likely influence expression of TCR Vβ-genes (Supplementary Fig. 25 and Supplementary Table 10).
Figure 4. Bayesian inference of MHC amino acid residues that influence expression of TCR Vα genes.
(a) Estimated posterior probabilities that amino acid residues in HLA-DRB1 (y-axis) influence expression of each TCR Vα gene (x-axis). Dots indicate positions with >50% probability of influencing expression of a particular V gene. (b) Posterior probabilities that particular MHC amino acid residues influence expression of any TCR Vα-gene.
If the detected associations result from MHC residues influencing the TCR-peptide-MHC interaction, then the relevant residues should cluster near the contact interface involved in TCR interaction with MHC or the presented peptide (influencing the TCR indirectly). A priori, MHC residues that affect TCR germline interactions could be direct pairwise structural contacts, or could cause indirect effects relayed through subtle conformational changes of the MHC or peptide from within the MHC groove, as seen for alloreactive MHC in graft rejection43.
To test whether the associated residues tend to be at or near the TCR-peptide-MHC interface, we first superimposed our genetic mapping results for DRB1 residues onto protein structures of class II MHC-TCR interactions (Figs. 5a and 5b). Remarkably, the residues with high posterior probability of influencing V-gene usage tend to be either physically near, or in direct contact with the TCR in structures that contain DRB1 (Fig. 5d). We next aligned structures of TCRs bound to human class II MHCs and identified all contacts between the DRB, DQB, or DPB chains and any of the TCRα chains or the presented peptides (Fig. 5e, and Supplementary Figs. 26–27; see Methods for details).
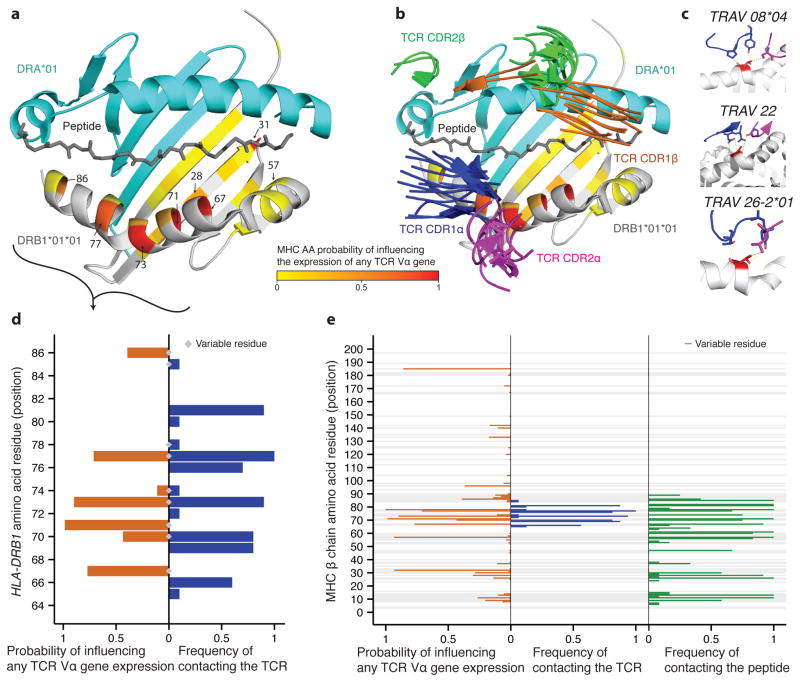
Figure 5. MHC residues that are functionally important for TCR recognition are also associated with TCR expression.
(a) Residue posterior probabilities of influencing expression of at least one TCR Vα-gene (yellow-red color scale) mapped onto a structure of HLA-DRB1 (white) and HLA-DRA1 (teal) (PDB ID: 2iam). (b) Similar to a but showing CDR1 (α blue, β green) and CDR2 (α magenta, β orange) loops of solved TCRs complexed with class II human MHC molecules. (c) Zoom-in on the consistent interaction between HLA-DRB1 residue 77 (red) and TCR CDR loops in three different TCR-MHC complexes (top to bottom PDB IDs: 1j8h, 2iam and 3t0e). (d) Comparison of probabilities that HLA-DRB1 residues influence expression of any TCR Vα-gene, and their frequency of physically contacting the TCR in ten DRB1/5 containing complexes. (e) Comparison of probabilities that MHC residues influence expression of at least one TCR Vα-gene (max over DRB1, DQB1 and DPB1), and their frequency of physically contacting the TCR and the peptide in 16 solved complexes (see also Supplementary Fig. 15 and Supplementary Table 4 lists the TCR Vα used in each structure).
Despite the large diversity in TCRα-MHCβ interaction chemistries, a small subset of MHC residues contact TCRα in a large fraction of the structures, in agreement with similar analyses of TCR-MHC class I complexes25,44. For example, residue 77, predicted by our model to influence TCR Vα expression (posterior probability of 0.71), shows diverse but consistent contact with germline TCR residues in all analyzed TCR-class-II MHC complexes (Fig. 5c and Supplementary Table 11). To quantitatively test whether MHC amino acid residues near the TCR interaction surface tend to be associated with TCR V-gene expression, we correlated our model posterior probabilities with the mean distance between each MHC residue centroid and the closest TCR (Supplementary Fig. 28) or peptide (Supplementary Fig. 29) residue centroid in all analyzed complexes. Although inter-residue proximity does not strictly correlate with energetic importance in protein-protein interactions, we found the association probabilities and TCR distances to be significantly correlated (Supplementary Fig. 30a; Pearson R= −0.28, p-value=0.022; Spearman R=−0.35, p-value=0.0044). This correlation would be higher if not for one outlier, residue 185, which may either be a false prediction or may indirectly affect the structural integrity of the MHC protein45. The results were similar when distances between amino acid Cα atoms are used instead of distances between centroids (Supplementary Fig. 30b). Hence, our structural analysis indicates that several of the residues in close proximity to the TCR or peptide affect expression of TCR V-genes.
Discussion
Our results show that MHC genotype plays a key role in shaping the TCR repertoire in a broad population sample, even in the absence of a shared immune challenge. We see an excess of signals in MHC Class II loci, and within HLA-DRB1 in particular; however it is unclear at this time whether this reflects a greater role for Class II genes in shaping the TCR repertoire, or simply differences in power due to the greater abundance of CD4 T cells in whole blood32. Many of the observed associations are linked to MHC residues, implying a direct role for protein-protein interactions in mediating these effects. We suggest that germline-encoded TCR-MHC compatibilities may bias thymic selection of some V-genes in favor of others, in an MHC-dependent manner.
Further, we were able to fine map some of these signals to specific amino acids that lie at the TCR-pMHC contact interface. Overall we find evidence for the intuitive result that positions near the interaction surface have higher probability of influencing expression of V-genes. That said, not all of the most-associated MHC positions are in direct physical contact with the TCR. More distant associations - e.g., at residue 185 - may reflect LD with other more proximal sites, or may be responsible for longer-range effects within the protein complex. It is known that amino acids distal to contact interfaces do sometimes have important effects on interaction energetics, even when they are not in direct contact45. In particular, this has been shown previously for alloreactive MHC in graft rejection43. Nonetheless, our Bayesian approach does highlight several amino acid residues positioned in the TCR-pMHC contact interface as being important for biasing V-gene usage. To the best of our knowledge, these are the first examples of trans associations mediated by protein-protein interactions.
Our observations also have implications for a long-standing debate about the basis of TCR specificity for MHC molecules, and the molecular forces responsible for MHC restriction. In 1971 Niels Jerne postulated that germline TCR and MHC genes co-evolved to be predisposed towards interacting46 - a phenomenon also referred to as ‘germline bias’. Our observation that MHC genotype has a direct association with TCR V-gene usage in the broader population implies that germline-encoded TCR-MHC contacts influence their interaction specificity. This orthogonal genetic evidence is in agreement with a variety of structural and functional data supporting a model of intrinsic specificity between TCR and MHC proteins. Other examples of supportive data include structural studies of numerous TCR-pMHC complexes that show persistent germline contacts between V-regions and MHC molecules20,47–49 and compatible residues between TCR loops and HLA-A*02:0150. Some of these contacts are necessary for functional recognition of MHC molecules and, when mutated, can lead to drastically altered outcomes of thymic selection20,21. Germline-derived Vα elements have been shown to influence MHC class I versus II selection51. Moreover, T cells have recently been shown to recognize MHC molecules independent of the MHC allele and peptide27, supplying functional evidence that the TCR has intrinsic specificity for the MHC via germline-encoded V-genes. Other studies argue against the intrinsic specificity model. For example, it has been reported that specific MHC residues are not essential for TCR recognition25 and there are no known constraints on the CDR sequences24; TCRs in mice that lack MHC I & II, CD4 and CD8 are activated by a non-MHC ligand23; and recently two TCR-pMHC structures composed of the same V-genes showed reversed polarity for MHC binding26. Our observations that TCR-MHC compatibilities exist within a large cohort of individuals likely reflect how most (though not necessarily all) TCR-MHC interactions occur. This leads us to view the studies that are discordant with the co-evolution model as representing bona fide deviations from a ‘canonical’ continuum of TCR-MHC recognition modes. Some level of ‘non-canonical’ recognition may be expected, considering the enormous repertoire of TCR sequences possible through recombination. In summary, our genetic results are in agreement with some degree of ‘hard-wired’ TCR-MHC recognition and are supportive of the Jerne Hypothesis.
One key limitation of our approach is that it is based on RNA sequencing of peripheral whole blood. Therefore, our measurements reflect average V-gene usage across different sub-populations of T cells, most notably, aggregating across the CD4 and CD8 T cell subsets. Previous work has identified several variants in the MHC region that are associated with individual-level variation in CD4:CD8 ratios52; moreover, V gene usage differs between CD4 and CD8 cells53. Therefore, one concern is whether the CD4:CD8 variants might drive the signals reported here. However, we find that those SNPs are only modestly associated with our signals (Supplementary Figs. 31a and 31b) and are not selected by our conditional analyses (Fig. 3a, and Supplementary Figs. 12 and 15). Controlling for those SNPs results in similar, highly significant associations with expression of V-genes (Supplementary Figs. 31c and 31d). While we cannot rule out some effect on the fine mapping, especially in the HLA-B gene, which reportedly harbors the strongest signal for CD4:CD8 ratio, the localization of the high posteriors to the TCR-pMHC interface suggests that the effect is not very strong. Additionally, some measurement noise may also result from the expansion of particular T cell clones on an individual-specific basis. These, and other, sources of variation are implicitly modeled by our Bayesian fine mapping approach, however, future studies with longer-read sequencing of sorted cell populations may be able to improve mapping resolution. A final concern is the possibility that we may not be able to identify causal residues that are poorly imputed. Most residues in our data have high imputation quality, and we find no correlation between imputation quality and posterior probability (Supplementary Fig. 32), however it remains possible that we may have overlooked poorly imputed causal sites.
In summary, we have found that usage of TCR Vα and Vβ-genes is associated with the genotype of MHC proteins. Our structural analysis suggests that these associations result from differences in the specificity of different TCR V-genes to different MHC variants. Our results provide a compelling example of strong trans associations that are mediated by protein-protein interaction between a receptor and its ligand, and shed light on the basis of TCR-MHC recognition.
Online Materials and Methods
Genotype and TCR V-gene expression data
We analyzed whole blood RNA sequencing and genotyping data from 922 individuals from the Depression Genes and Networks Project reported by Battle et al.29 (National Institute of Mental Health Grant 5RC2MH089916). Measured SNPs were filtered as described previously, resulting in genotype data at 649,863 SNPs29. The expression of each V-gene relative to the total chain expression was estimated from peripheral blood RNA-sequencing (~70M 51bp reads per individual). Sequencing reads were mapped as in Battle et al. (using Bowtie254 with Tophat55 default parameters) and the number of unique reads that mapped to each V/J/C-TCR/Ig gene was counted using a modified version of HTSeq56 which allow reads to map to a sequence of more than one V/D/J/C-gene (see Supplementary Fig. 1 for the average number of reads mapped in an individual to each V-gene). Individuals and genes with low read-counts were removed: specifically, we removed from our analysis individuals with < 80,000 reads mapping uniquely to TCR/Ig V/D/J/C genes (31% of the median). For analysis, we required that each V-gene have on average >1 read per individual, at most 100 samples with zero reads, and at least 1% of the individuals with at least 20 reads. Next, the read counts were log transformed (0.1 pseudo reads were added to avoid zeroes); and regressed on known technical and biological confounding factors as in Battle et al.29 and on the log total number of reads mapped to each TCR or Ig chain (including reads that mapped to J and C genes; see Supplementary Table 1, and Supplementary Fig. 3). The median coefficient of variation (CV) of each V-gene was 6% (Supplementary Fig. 3). Finally, to avoid the effect of outliers, the residuals were quantile-normalized to a normal distribution (though in practice this made little difference to the detected associations). Similar methodology was applied to Ig V-genes. The final dataset contains measurements of 44 TCR Vα, 40 TCR Vβ, 11 TCR Vγ, 3 TCR Vδ, 31 Ig Vκ, 38 Ig Vλ and 41 Ig VH genes in 895 individuals.
Since read mappability may vary by genotype, we analyzed the mappability of 25-mers from the reference and imputed alternative haplotypes (Supplementary Fig. 6). We found that in 9 V-genes >0% and up to 10% of 25-mers have better mappability to the reference sequence than to alternative haplotypes. We thus excluded all of these genes from the short-range (cis) association analysis (thus dropping 6, 1 and 3 TCR Vα, Vβ and Vγ-genes, respectively). We note that mapping differences may increase measurement noise and reduce power for detecting long range (trans) associations, but should not create false positives, and therefore these genes were not removed from that analysis. Moreover, we avoid comparing expression of different V-genes, since it is difficult to estimate the mappable lengths of the V-genes due to possible differences between the partial digestion of V-genes and J-gene pairing during V(D)J recombination.
eQTL detection
Associations between genotypes and TCR V-gene-normalized expression values were tested using Pearson correlations. Similarly, in testing for trans associations between MHC and other genes, we used Pearson correlations to test all genes at least 1Mb away from the MHC locus against all variants genotyped in the extended MHC locus. The extended MHC locus is defined as the region from the SLC17A2 gene at the telomeric end to the DAXX gene at the centromeric end of chromosome 6 (hg19 coordinates 25,912,984–33,290,793)57. For genome-wide trans-QTL mapping we used a significance threshold of p<5×10−8, which accounts for genome-wide significance testing.
Identification of V-genes that are significantly associated with genotypes in cis or in the MHC locus
The empirical significance of the association between the expression of each V-gene and short-range (cis) or MHC locus genotype was evaluated by comparing the p-value of the most significantly associated SNP to the most significant p-values in each of 10,000 random permutations of expression values across individuals. 5% FDR was then used to control for the testing of multiple V-genes (Supplementary Fig. 7). The SNPs used for the cis-analysis were imputed using SHAPEIT58 (for pre-phasing) and IMPUTE259. The 1000 Genomes Phase 160 panel was used for imputation with a EUR MAF > 0.01 filter and genotype likelihood > 0.9. In testing for trans associations with MHC genotypes, we used genotyped SNPs within the extended MHC locus57.
Imputation of MHC locus genotype
SNP2HLA31 was used to impute the MHC locus genotypes, using a reference panel of 5,225 individuals of European descent collected by the Type 1 Diabetes Genetics Consortium31. The software imputes 2 and 4-digit classical alleles for HLA-A, HLA-C, HLA-B, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1 and HLA-DPB1, and their corresponding amino acid (AA) polymorphisms and single-nucleotide polymorphisms (SNPs) (Fig. 1, bottom part). SNPs and polymorphic amino acids with maximum allele frequency > 97.5 and alleles with frequency lower than 0.5% were removed. The imputation quality (Beagle R2 with predicted true genotypes) is larger than 0.95 and 0.99 for 97% and 80% of the imputed amino acids, respectively (excluding alleles with MAF<2.5%). There is no significant correlation between the imputation quality of amino acid residues, and the posterior probability that they influence expression of TCR V-genes (Supplementary Fig. 32).
Conditional analysis of associations between expression of TCR Vα and Vβ-genes and genetic variation in the MHC locus
The imputed genotype of each variable amino acid position or nucleotide position was encoded by the allelic dosage variables while omitting the most common allele (a similar encoding was used by Raychaudhuri et al.38). The conditional analysis was performed separately for each V-gene using forward stepwise linear regression. In each step of the regression, and for every amino acid residue or nucleotide position, we considered an expanded model that included the allelic dosage variables for that position. The significance was tested using F-tests. If the most significant position had a p-value < 0.05/n, where n was the number of tested positions, then this position was added to the model as a covariate. Regression stopped when the p-value of the most significant position was greater than 0.05/n (this threshold was p~10−5). We performed two additional variants of this type of analysis. First, instead of testing for associations with nucleotide or amino acids positions in the MHC locus, we tested for associations with classical MHC gene 4-digit haplotypes. Second, instead of testing for associations of MHC genetic variability (at the position or 4-digit haplotype level) with expression of each V-gene separately, we tested for associations with joint expression of all V-genes from a single chain. Since each gene expression was quantile normalized to a normal distribution, using the joint expression of V-genes gives equal weight to explaining the expression of each gene in the chain.
Estimating the fraction of expression variation for each TCR V-gene that is explained by genetic variation in the MHC proteins
We used the Genomewide Complex Trait Analysis tool (GCTA) developed by Yang et al.33. We fit a model with a component for each variable classical MHC protein (HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1, HLA-DPB1) representing the variability at the amino acid level and a component for all SNPs in the MHC locus outside the transcribed regions of the classical MHC genes. Amino acid level variation is used in place of genotype because the annotation of transcribed SNPs in the MHC genes is often ambiguous and allele-dependent. To adapt GCTA to multi-allelic positions, we calculated the genetic relationship between individuals in a multi-allelic position by averaging the relationship over dosage variables of all possible allele. To estimate the significance of the total variation explained by the MHC protein components, we ran GCTA on 100 permutations of the expression data for each TCR V-gene. The fraction of explained variation of these permuted expression vectors follows a truncated normal distribution61. We estimated the variance of this distribution and used it to compute a p-value for the fraction of variation of the expression of each TCR V-gene that is explained by genetic variation in MHC proteins. We corrected for multiple hypothesis testing using 5% FDR62.
Inferring which MHC protein amino acid residues are associated with expression biases of TCR V-genes
Identifying which amino acid residues drive the association of the MHC proteins with the expression of TCR V-genes is challenging due to the linkage disequilibrium (LD) between nearby variants. To test the association of each residue while accounting for the genotype of other residues, we used a Bayesian variable selection approach with a spike and slab prior63,64. The Bayesian modeling framework has a number of practical advantages for this problem: in particular, it allows us to appropriately account for the joint uncertainty when there are multiple causal positions and extensive LD. We used priors that reflect our expectation that most residues do not directly influence expression, and that a residue that influences expression of one V-gene is more likely to be relevant for expression of other V-genes. The priors were set such that the model required strong evidence to return high posterior probabilities of association.
Specifically, we modeled the expression of each Vα or Vβ gene as a linear combination of the imputed alleles of amino acid residues of classical MHC genes. Each residue’s allelic dosage variable can either be included in the model or excluded (coefficient=0) from the model of expression of each V-gene. We sampled the space of possible models to estimate the posterior probability that each residue has at least one of its allelic dosage variables in the model of a specific V-gene. Assuming that the associations are caused by the residues’ effects on V-gene expression, we refer to this posterior as the probability that a residue influences expression of a TCR V-gene.
In detail, our response vectors are the relative expression of each V-gene from a specific chain across the N=895 individuals. For a TCR chain with T paralog V-genes, we define a multiple response matrix YN×T with yi,t being the standardized relative expression of TCR V-gene t∈{1,2,…,T} in individual i∈{1,2,…,N}. Our features are the imputed amino acid residues in each individual for each of the classical MHC genes (HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1 and HLA-DPB1). The allelic dosage of a specific amino acid variant (a∈ {1,2,…,Ar}) of a residue (r) of an MHC gene (g) in individual (i) is represented by a continuous variable xi,(g,r,a) )∈[0,2]. For clarity, we also use below h∈{1,2,…,H} as the index for all possible combinations of (g,r,a). Our genotype matrix is therefore XN×H. We filtered out residues in which the maximal allele frequency was larger than 97.5% and dropped alleles with frequency lower than 0.5%. We define zt,h∈{0,1} to be an indicator of whether genotype xh is included (has a coefficient βt,h~ N(0,σβ2) or excluded (has a zero coefficient) from the model of yt. Together, the expression of gene t in individual i is modeled by
| (1) |
where
| (2) |
independently, and
| (3) |
Previous structural analysis of TCR-peptide-MHC complexes have found that a limited and consistent set of MHC residues interacts with the TCR and the docking orientation of the TCR is semi-conserved. This is reflected by the choice of the prior probability of zt,h=1 to be small and shared across V-genes (P(zt,h=1)= Ph, for t∈{1,2,…,T}) and the modeling of α and β chain genes separately.
We consider that residue r of gene g influences the expression of TCR V-gene t, if at least one of the allelic dosage variables that represent it is in the model (i.e. its indicator variable zt,(g,r,a) is equal to one). We denote this event F(Z,t,g,r):
| (4) |
where
| (5) |
To avoid an a priori preference for residues with different numbers of possible alleles, we modified the prior beta distribution parameters of the indicator prior πh in (3) such that the a priori expectation and variance of F(Z,t,g,r) are equal for residues with different number of possible variants and match the prior in (3) for a residue with two possible alleles.
| (6) |
| (7) |
To find suitable values for the prior parameters, we assume that xh is an allelic dosage variable of a residue r that has only two possible alleles and therefore is represented by a single dosage variable. We define πh~ Beta(a,b) to be the prior of zh~Ber(πh). Now let xh* be one of A dosage variables that represent a residue r*, and πh*~ Beta(a*,b* ) be the prior of zh*~Ber(πh*). We seek a*,b* such that the a priori expectation and variance of F over residue r* are equal to the expectation and variance of F over residue r.
| (8) |
| (9) |
Equation (8) is equal to
| (9) |
| (10) |
| (11) |
Where B is the Beta function. In a similar manner equation (9) is equal to:
| (12) |
We then solved (11) and (12) numerically using R ‘nleqslv’ package to achieve (a*,b*) for various values of A.
In our analyses, the prior probability of any MHC residue to interact with a specific TCR V-gene was set at 0.1% and the prior probability of each position of the MHC to interact with any of the 44 TCR α V-genes was 4.3%, and 3.9% for any of the 40 TCR β chain V-genes.
To compute the posterior probability the posterior probabilities with which each MHC residue influences TCR V-gene expression that (i.e. Eθ|X,Y (F(Z,t,g,r)) ) for all r) we sampled the space of possible models using an efficient Gibbs sampler65 where the likelihood is integrated over the coefficients (β)
| (13) |
Altogether, we sampled 100,000 samples from 10 different starting points (excluding the first 2000 samples).
Structural analysis
A full list of TCR-pMHC structures and intermolecular interactions is curated by the international ImMunoGeneTics information system (IMGT66). We used all structures that were available in the protein databank (RCSB PDB67) on June 15, 2015. In order to compare the intermolecular interactions to the genetic analysis, the IMGT numbering was converted to the RCSB PDB numbering for all residues (RCSB PDB is indexed in the same manner as proteins in the UniProt database68). For the structure 4p4k, we converted the PDB numbering for the DP residues to the DQ and DR numbering using the IMGT labeling (for example, residue 72AG in the IMGT numbering equates to PDB residue 75 for DP structures, but 77 for all DP and DQ structures.) This allows for a more consistent comparison between the different alleles. The IMGT structural contact algorithm determined the structural contacts, which were defined as polar, non-polar, and hydrogen-bonds.
Pairwise Distance Analysis
The distances between pairs of amino acid residues were calculated as the distance between the centroid of the amino acids using the PDB xyz coordinates for every atom in a given amino acid. This accounts for directionality of the side chains within the structure. For each MHC β chain residue, its minimum distance to any TCRα chain residue and its minimum distance to every peptide residue were calculated. For comparison, we also calculated the distance between the Cα of each residue using the PDB xyz coordinates of the Cα’s (Supplementary Fig. 28b). The results using the two distance measurements are similar. A list of PDB files used can be found in Supplementary Table 10.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants HG0070736, 1R01GM097171-01A1, RO1 AI03867, U19 AI057229, the Howard Hughes Medical Institute, the EMBO Long-Term Fellowship and a National Science Foundation Graduate Research Fellowship. We thank David Golan, David Knowles, Audrey Fu, Michael Birnbaum, Marvin Gee, Juan Mendoza, Anand Bhaskar and Towfique Raj for helpful discussions, and the anonymous referees for valuable comments.
Footnotes
Author Contributions
E.S and J.K.P. conceived the project. E.S performed genetic analyses with input from A.B., H.B.F. and J.K.P. E.S. and L.V.S. performed structural analyses with input from K.C.G. E.S., L.V.S., K.C.G. and J.K.P. wrote the manuscript. The work was supervised by K.C.G. and J.K.P. All authors reviewed, revised, and provided feedback on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
URLs
IMGT, the international ImMunoGeneTics information system, http://www.imgt.org; RCSB PDB, protein data bank, http://www.rcsb.org.
References for main text
- 1.Neefjes J, Jongsma MLM, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11:823–36. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 2.McDevitt HO, Bodmer WF. HL-A, immune-response genes, and disease. Lancet (London, England) 1974;1:1269–75. doi: 10.1016/s0140-6736(74)90021-x. [DOI] [PubMed] [Google Scholar]
- 3.Gutierrez-Arcelus M, Rich SS, Raychaudhuri S. Autoimmune diseases - connecting risk alleles with molecular traits of the immune system. Nat Rev Genet. 2016;17:160–74. doi: 10.1038/nrg.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyadera H, Tokunaga K. Associations of human leukocyte antigens with autoimmune diseases: challenges in identifying the mechanism. J Hum Genet. 2015;60:697–702. doi: 10.1038/jhg.2015.100. [DOI] [PubMed] [Google Scholar]
- 5.Vantourout P, Hayday A. Six-of-the-best: unique contributions of γδ T cells to immunology. Nat Rev Immunol. 2013;13:88–100. doi: 10.1038/nri3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossjohn J, et al. T Cell Antigen Receptor Recognition of Antigen-Presenting Molecules. Annu Rev Immunol. 2014;33:141210135520002. doi: 10.1146/annurev-immunol-032414-112334. [DOI] [PubMed] [Google Scholar]
- 7.Rudolph MG, Stanfield RL, Wilson Ia. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 8.Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of T-cell receptor bias in immunity. Nat Rev Immunol. 2006;6:883–94. doi: 10.1038/nri1977. [DOI] [PubMed] [Google Scholar]
- 9.Housset D, Malissen B. What do TCR-pMHC crystal structures teach us about MHC restriction and alloreactivity? Trends Immunol. 2003;24:429–37. doi: 10.1016/s1471-4906(03)00180-7. [DOI] [PubMed] [Google Scholar]
- 10.Garcia KC, et al. A closer look at TCR germline recognition. Immunity. 2012;36 doi: 10.1016/j.immuni.2012.05.018. 887-8-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see) Nat Rev Immunol. 2014;14:377–91. doi: 10.1038/nri3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roudier J. Association of MHC and rheumatoid arthritis. Association of RA with HLA-DR4: the role of repertoire selection. Arthritis Res. 2000;2:217–20. doi: 10.1186/ar91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robins HS, et al. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med. 2010;2:47ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zvyagin IV, et al. Distinctive properties of identical twins’ TCR repertoires revealed by high-throughput sequencing. Proc Natl Acad Sci U S A. 2014;111:5980–5. doi: 10.1073/pnas.1319389111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gulwani-Akolkar B, et al. Do HLA genes play a prominent role in determining T cell receptor V alpha segment usage in humans? J Immunol. 1995;154:3843–51. [PubMed] [Google Scholar]
- 16.Miles JJ, et al. TCR alpha genes direct MHC restriction in the potent human T cell response to a class I-bound viral epitope. J Immunol. 2006;177:6804–14. doi: 10.4049/jimmunol.177.10.6804. [DOI] [PubMed] [Google Scholar]
- 17.Garcia KC. Reconciling views on T cell receptor germline bias for MHC. Trends Immunol. 2012;33:429–36. doi: 10.1016/j.it.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10:143–7. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castro CD, Luoma AM, Adams EJ. Coevolution of T-cell receptors with MHC and non-MHC ligands. Immunol Rev. 2015;267:30–55. doi: 10.1111/imr.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P. Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature. 2009;458:1043–6. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Laethem F, et al. Lck availability during thymic selection determines the recognition specificity of the T cell repertoire. Cell. 2013;154:1326–41. doi: 10.1016/j.cell.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Laethem F, et al. Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC. Immunity. 2007;27:735–50. doi: 10.1016/j.immuni.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 24.Holland SJ, et al. The T-cell receptor is not hardwired to engage MHC ligands. Proc Natl Acad Sci U S A. 2012;109:E3111–8. doi: 10.1073/pnas.1210882109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burrows SR, et al. Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability. Proc Natl Acad Sci U S A. 2010;107:10608–13. doi: 10.1073/pnas.1004926107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beringer DX, et al. T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex. Nat Immunol. 2015;16:1153–61. doi: 10.1038/ni.3271. [DOI] [PubMed] [Google Scholar]
- 27.Parrish HL, Deshpande NR, Vasic J, Kuhns MS. Functional evidence for TCR-intrinsic specificity for MHCII. Proc Natl Acad Sci U S A. 2016:1–6. doi: 10.1073/pnas.1518499113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rockman MV, Kruglyak L. Genetics of global gene expression. Nat Rev Genet. 2006;7:862–72. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 29.Battle A, et al. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sottini A, Imberti L, Fiordalisi G, Primi D. Use of variable human V delta genes to create functional T cell receptor alpha chain transcripts. Eur J Immunol. 1991;21:2455–9. doi: 10.1002/eji.1830211023. [DOI] [PubMed] [Google Scholar]
- 31.Jia X, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinclair C, Bains I, Yates AJ, Seddon B. Asymmetric thymocyte death underlies the CD4:CD8 T-cell ratio in the adaptive immune system. Proc Natl Acad Sci U S A. 2013;110:E2905–14. doi: 10.1073/pnas.1304859110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kichaev G, et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 2014;10:e1004722. doi: 10.1371/journal.pgen.1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wallace C, et al. Dissection of a Complex Disease Susceptibility Region Using a Bayesian Stochastic Search Approach to Fine Mapping. PLoS Genet. 2015;11:e1005272. doi: 10.1371/journal.pgen.1005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wellcome Trust Case Control Consortium et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raychaudhuri S, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–6. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu X, et al. Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat Genet. 2015 doi: 10.1038/ng.3353. advance on. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patsopoulos NA, et al. Fine-mapping the genetic association of the major histocompatibility complex in multiple sclerosis: HLA and non-HLA effects. PLoS Genet. 2013;9:e1003926. doi: 10.1371/journal.pgen.1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Messaoudi I, Guevara Patiño JA, Dyall R, LeMaoult J, Nikolich-Zugich J. Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science. 2002;298:1797–800. doi: 10.1126/science.1076064. [DOI] [PubMed] [Google Scholar]
- 42.Price DA, et al. Public clonotype usage identifies protective Gag-specific CD8+ T cell responses in SIV infection. J Exp Med. 2009;206:923–936. doi: 10.1084/jem.20081127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luz JG. Structural Comparison of Allogeneic and Syngeneic T Cell Receptor-Peptide-Major Histocompatibility Complex Complexes: A Buried Alloreactive Mutation Subtly Alters Peptide Presentation Substantially Increasing Vbeta Interactions. J Exp Med. 2002;195:1175–1186. doi: 10.1084/jem.20011644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murray JS. An old Twist in HLA-A: CDR3α Hook up at an R65-joint. Front Immunol. 2015;6 doi: 10.3389/fimmu.2015.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levin AM, et al. Exploiting a natural conformational switch to engineer an interleukin-2 ‘superkine’. Nature. 2012;484:529–33. doi: 10.1038/nature10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jerne NK. The somatic generation of immune recognition. Eur J Immunol. 1971;1:1–9. doi: 10.1002/eji.1830010102. [DOI] [PubMed] [Google Scholar]
- 47.Dai S, et al. Crossreactive T Cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 2008;28:324–34. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams JJ, et al. Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCR-peptide-MHC cross-reactivity. Nat Immunol. 2016;17:87–94. doi: 10.1038/ni.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol. 2007;8:975–83. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 50.Blevins SJ, et al. How structural adaptability exists alongside HLA-A2 bias in the human αβ TCR repertoire. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1522069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sim BC, Zerva L, Greene MI, Gascoigne NR. Control of MHC restriction by TCR Valpha CDR1 and CDR2. Science. 1996;273:963–6. doi: 10.1126/science.273.5277.963. [DOI] [PubMed] [Google Scholar]
- 52.Ferreira MAR, et al. Quantitative trait loci for CD4:CD8 lymphocyte ratio are associated with risk of type 1 diabetes and HIV-1 immune control. Am J Hum Genet. 2010;86:88–92. doi: 10.1016/j.ajhg.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klarenbeek PL, et al. Somatic Variation of T-Cell Receptor Genes Strongly Associate with HLA Class Restriction. PLoS One. 2015;10:e0140815. doi: 10.1371/journal.pone.0140815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anders S, Pyl PT, Huber W. HTSeq - A Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Bakker PIW, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–72. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Delaneau O, Marchini J. Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat Commun. 2014;5:3934. doi: 10.1038/ncomms4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Visscher PM, Yang J, Goddard ME. A commentary on ‘common SNPs explain a large proportion of the heritability for human height’ by Yang et al (2010) Twin Res Hum Genet. 2010;13:517–24. doi: 10.1375/twin.13.6.517. [DOI] [PubMed] [Google Scholar]
- 62.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B …. 1995 [Google Scholar]
- 63.Mitchell T, Beauchamp J. Bayesian variable selection in linear regression. J Am Stat …. 1988 [Google Scholar]
- 64.Ishwaran H, Rao J. Spike and slab gene selection for multigroup microarray data. J Am Stat …. 2005 [Google Scholar]
- 65.Geman S, Geman D. Stochastic Relaxation, Gibbs Distributions, and the Bayesian Restoration of Images. IEEE Trans Pattern Anal Mach Intell. 1984:721–741. doi: 10.1109/tpami.1984.4767596. PAMI-6. [DOI] [PubMed] [Google Scholar]
- 66.Lefranc MP, et al. IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 2015;43:D413–22. doi: 10.1093/nar/gku1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.The UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2014;43:D204–212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.