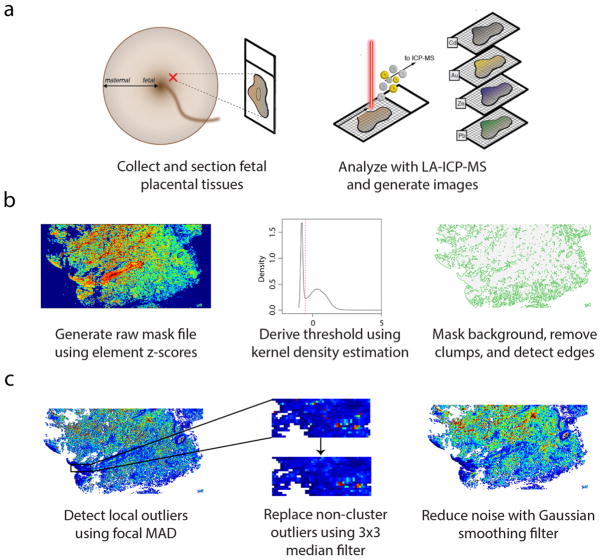
Fig. 1. Overview of LA-ICP-MS imaging workflow.
(a) Formalin-fixed, paraffin-embedded placental tissues, sampled from the fetal aspect, were sectioned (5-μm thick) and affixed to glass slides. Spatial distribution of multiple elements was determined using LA-ICP-MS, and data was read into R as a grid stack for image processing. (b) Background pixels were masked using a global thresholding algorithm. A raw mask file was created by converting the elements in the grid stack to z-scores, removing elements from the stack with high background noise, and finding the mean pixel z-scores across all elements in the stack, weighted on the squared gas-blank z-score means for each element. A threshold cutoff value was determined from the first-occurring local minimum value from the kernel density estimate of the weighted mean grid, and pixels with values below the cutoff were set to null values. Then, the algorithm removed small clusters of remaining background pixels and identified tissue edges. (c) Extreme outliers were detected using a focal median absolute deviation algorithm that ignored outliers in biologically-relevant clusters, and these outliers were replaced using a 3×3 median filter. After outlier replacement, images were smoothed using a Gaussian filter (σ = 2/3, r = 2).