Abstract
Recently, a new Chlamydia-related organism, Protochlamydia naegleriophila KNic, was discovered within a Naegleria amoeba. To decipher the mechanisms at play in the modeling of genomes from the Protochlamydia genus, we sequenced the full genome of Pr. naegleriophila, which includes a 2,885,090 bp chromosome and a 145,285 bp megaplasmid. For the first time within the Chlamydiales order, we describe the presence of a clustered regularly interspaced short palindromic repeats (CRISPR) system, the immune system of bacteria, located on the chromosome. It is composed of a small CRISPR locus comprising eight repeats and associated cas-cse genes of the subtype I-E. A CRISPR locus is also present within Chlamydia sp. Diamant, another Pr. naegleriophila strain, suggesting that the CRISPR system was acquired by a common ancestor of Pr. naegleriophila, after its divergence from Pr. amoebophila. Both nucleotide bias and comparative genomics approaches identified probable horizontal gene acquisitions within two and four genomic islands in Pr. naegleriophila KNic and Diamant genomes, respectively. The plasmid encodes an F-type conjugative system highly similar to 1) that found in the Pam100G genomic island of Pr. amoebophila UWE25 chromosome, as well as on the plasmid of Rubidus massiliensis and 2) to the three genes remaining in the chromosome of Parachlamydia acanthamoebae strains. Therefore, this conjugative system was likely acquired on an ancestral plasmid before the divergence of Parachlamydiaceae. Overall, this new complete Pr. naegleriophila genome sequence enables further investigation of the dynamic processes shaping the genomes of the family Parachlamydiaceae and the genus Protochlamydia.
Keywords: comparative genomics, CRISPR, T4SS, Chlamydiales, plasmid, genomic island
Introduction
The order Chlamydiales is very diverse, as suggested by the discovery of a large number of Chlamydia and Chlamydia-related bacteria belonging to nine different families (Everett et al. 1999; Greub 2010; Horn 2011) and further broadened by cross examination of metagenomics data (Lagkouvardos et al. 2014). The family Parachlamydiaceae comprises five genera that are each represented by a small number of isolated strains. The genus Protochlamydia was enriched by the isolation of a Naegleria endosymbiont (Michel et al. 2000) that presented 97.6% identity in the 16S rRNA with Pr. amoebophila UWE25 and was thus named Pr. naegleriophila strain KNic (Casson et al. 2008). Because other members of the Parachlamydiaceae family are suspected to be associated with lung infections (Greub 2009), a diagnostic PCR specific for Pr. naegleriophila was then developed and applied to bronchoalveolar lavages. Pr. naegleriophila DNA was detected in the bronchoalveolar lavage of an immunocompromised patient with pneumonia by two PCRs targeting different genomic regions and the presence of the bacterium in the sample was confirmed by direct immunofluorescence (Casson et al. 2008). These results indicate a potential role of Pr. naegleriophila in lower respiratory tract infections.
A recent study including Chlamydia genomes and other members of the Planctomycetes-Verrucomicrobia-Chlamydia superphylum suggested that the branch leading to the order Chlamydiales is shaped mainly by genome reduction and displayed limited occurrence of gene birth, duplication, and transfer within the chlamydial clades (Kamneva et al. 2012), as is the case in other strict intracellular pathogens (Darby et al. 2007). On the contrary, the occurrence of large families of paralogs in the genome of various families within the order Chlamydiales, and particularly in Parachlamydiaceae, suggested evolution by extensive gene duplication (Eugster et al. 2007; Domman et al. 2014). The chromosome sequence of Pr. amoebophila UWE25 exhibits little evidence for the occurrence of lateral gene transfer (Horn et al. 2004). However, a number of probable lateral gene transfers were identified between Parachlamydia and other amoeba-infecting bacteria such as Legionella (Gimenez et al. 2011), a process that may take place within the amoeba itself (Bertelli and Greub 2012). The Pr. amoebophila genome has a genomic island (Pam100G) that encodes a type IV secretion system of the F-type that might be involved in conjugative DNA transfer (Greub et al. 2004). A similar system is also found on the plasmid of Simkania negevensis (Collingro et al. 2011) and a partial operon was described in Parachlamydia acanthamoebae (Greub et al. 2009), suggesting active DNA transfer capabilities in the ancestor of the Chlamydiales and some of its descendants.
Small interspaced repetitions were initially observed in Escherichia coli (Ishino et al. 1987) and they were then named CRISPR, an acronym for clustered regularly interspaced short palindromic repeats (Jansen et al. 2002). Although found in 50% of bacteria and in 90% of archaea (Weinberger et al. 2012), a CRISPR system has never been reported before in a member of the order Chlamydiales (Makarova et al. 2011). The CRISPR locus usually consists of a variable number of 23–47 bp repeats (up to 587) with some dyad symmetry, but not truly palindromic, interspaced by 21–72 bp spacers (Horvath and Barrangou 2010). Associated with these repeats are two core cas genes and additional subtype-specific genes putatively providing mechanistic specificity (Koonin and Makarova 2013). Similarity between spacers and extrachromosomal elements first suggested a role in immunity against phage infection and more generally against conjugation or transformation by acquisition of external DNA (Bolotin et al. 2005). The CRISPR-Cas system was shown to mediate an antiviral response thus inducing resistance to phage infection (Deveau et al. 2010), notably in E. coli (Brouns et al. 2008). More recently, CRISPR-Cas systems were shown to regulate stress-related response, changing gene expression and virulence traits in several pathogens, including the intracellular bacteria Francisella novicida (Louwen et al. 2014; Sampson and Weiss 2014).
In this study, we sequenced and analyzed the complete genome of Pr. naegleriophila strain KNic and discovered two potentially antagonistic systems, a type IV secretion system likely implicated in conjugative DNA transfer and a CRISPR system that generally controls foreign DNA acquisition. Furthermore, the complete genome sequence of a new species within the genus Protochlamydia offered the possibility to look into the genome dynamics throughout evolution by comparing Pr. naegleriophila KNic gene content and genome architecture to its closest relatives within the family Parachlamydiaceae.
Results
Chromosome Features
Pr. naegleriophila KNic possesses a 2,885,090 bp circular chromosome with a mean Guanine-Cytosine (GC) content of 42.7%. The genome size and the GC content are surprisingly high compared with the most closely related species, Pr. amoebophila (table 1), but it is consistent with its closest relative Chlamydia sp. Diamant, another so far unpublished Pr. naegleriophila strain (hereafter referred to as Pr. naegleriophila Diamant). The chromosome of Pr. naegleriophila strain KNic was predicted to encode 2,415 proteins and exhibited four ribosomal operons and 43 tRNAs, more than any other Chlamydiales (table 1). Two types of spacers were found between the 16S and the 23S rRNA: Either a simple intergenic spacer or a spacer containing two tRNAs for Ala and Ile.
Table 1.
Genomics characteristics of bacteria belonging to the family Parachlamydiaceae
| Species | Strain | Status | Scaffolds | Genome Size | GC Content | CDS | tRNAs | rRNA Genes | Plasmid Size | Plasmid CDS | Plasmid GC Content |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pr. amoebophila | UWE25 | Complete | 1 | 2,414,465 | 34.7 | 1,855 | 35 | 7 | — | — | — |
| Pr. amoebophila | EI2 | Draft | 178 | 2,397,675 | 34.8 | 1,797 | 36 | 3 | NA | — | — |
| Pr. amoebophila | R18 | Draft | 795 | 2,881,499 | 34.8 | 2,025 | 41 | 13 | NA | — | — |
| Pr. naegleriophila | KNic | Complete | 1 | 2,885,090 | 42.7 | 2,415 | 43 | 12 | 145,285 | 160 | 37.2 |
| Pr. naegleriophila | Diamant | Draft | 4a | 2,864,073 | 42.8 | 2,424 | 39 | 7 | 91,928 | 98 | 40.9 |
| P. acanthamoebae | UV7 | Complete | 1 | 3,072,383 | 39 | 2,531 | 40 | 10 | — | — | — |
| P. acanthamoebae | Hall’s coccus | Draft | 95 | 2,971,261 | 39 | 2,474 | 35 | 3 | NA | — | — |
| P. acanthamoebae | OEW1 | Draft | 162 | 3,008,885 | 39 | 2,321 | 38 | 4 | NA | — | — |
| P. acanthamoebae | Bn9 | Draft | 72 | 2,999,361 | 38.9 | 2,498 | NA | NA | NA | — | — |
| Parachlamydiaceae bacterium | HS-T3 | Draft | 34 | 2,307,885 | 38.7 | 2,003 | 39 | 3 | NA | — | — |
| R. massiliensis | Rubis | Draft | 3a | 2,701,449 | 32.4 | 2,446 | 36 | 5 | 80,697 | 107 | 40.2 |
| — | — | — | — | — | — | — | — | — | 39,075 | 40 | 29.8 |
| Neochlamydia sp. | EPS4 | Draft | 112 | 2,530,677 | 38.1 | 1,843 | 36 | 4 | NA | — | — |
| Neochlamydia sp. | TUME1 | Draft | 254 | 2,546,323 | 38 | 1,834 | 36 | 4 | NA | — | — |
| Neochlamydia sp. | S13 | Draft | 1342 | 3,187,074 | 38 | 2,175 | 42 | 10 | NA | — | — |
Note.—As available on NCBI database on September 22, 2015, all genomes except KNic have been reannotated by the NCBI Prokaryotic Genome Annotation Pipeline.
aFollowing removal of the plasmid(s) present, according to our analyses. NA: information not available.
The cumulative G versus C nucleotide bias (GC skew) presented a typical pyramidal shape (supplementary fig. S1, Supplementary Material online) that is expected in the absence of particular large genomic islands and confirmed the assembly accuracy. The GC skew of Pr. naegleriophila was smoother than that of Pr. amoebophila, and did not present the small inversion in the slope that is caused by the Pr. amoebophila genomic island (supplementary fig. S1, Supplementary Material online) (Greub et al. 2004). The origin of replication (ori) and the terminus of replication (ter), at the minimum and maximum of the curve (supplementary fig. S1, Supplementary Material online), respectively, showed an almost perfectly balanced chromosome with 49.8% of the base on one arm, that is, between ori and ter, and 50.2% on the other arm, that is, between ter and ori.
Genomic Rearrangements, Genomic Islands, and Indels
The alignment of available complete and nearly complete (<5 contigs) genomes of the family Parachlamydiaceae (fig. 1A), showed that the two strains of Pr. naegleriophila are highly collinear and presented no rearrangement, except for a small unplaced contig in the Pr. naegleriophila Diamant sequence. Within genus comparison of Pr. naegleriophila and Pr. amoebophila showed the occurrence of 24 recombination and inversion events. As expected, further distantly related organisms from a different genus exhibited less collinearity and an increasing number of recombination events (>180). The number of rearrangements was positively correlated (Rho = 0.96; P-value 0.04) to the cophenetic distance (fig. 1B).
Fig. 1.—
Genomic rearrangements in the Parachlamydiaceae family. (A) Left side, the phylogenetic branching of bacterial strains as inferred by a neighbor-joining tree reconstruction based on five conserved proteins (DnaA, FtsK, HemL, FabI, and SucA). Right side, visualization of genomic rearrangements in the family Parachlamydiaceae. The two strains of the species Pr. naegleriophila are highly collinear, with no apparent rearrangement except for differences in the choice of the genome start. (B) With increasing cophenetic distances between organisms, the genomes show increasing number of rearrangements.
Genomic islands are generally defined as large regions (>10 kb) that were likely acquired by horizontal gene transfer. Nucleotide bias-based methods predicted three and four genomic islands in Pr. naegleriophila strain KNic and strain Diamant, respectively (supplementary table S1, Supplementary Material online), that generally exhibited particularly high- or low-GC content (fig. 2 and supplementary fig. S1, Supplementary Material online). A comparative genomics approach identified two large genomic islands of 37 kb and 15 kb, respectively, in Pr. naegleriophila KNic (table 2). The first and largest PnaK_GI1 contained hallmarks of genomic islands: an integrase, seven transposases, as well as many hypothetical proteins and genes with poorly determined function, such as short chain dehydrogenases (supplementary table S2, Supplementary Material online). A deoxyribodipyrimidine photolyase-like protein was also present and could play a role in DNA damage repair, as was described for other bacteria (Oberpichler et al. 2011). PnaK_GI1 encompassed two of the three regions predicted as genomic islands by IslandViewer, the third probably being a false positive due to particular codon usage in two close U-box domain containing proteins. PnaK_GI2 is situated directly downstream of tRNA-Thr—tRNAs, which are preferential sites for genomic island integration—and encoded hypothetical proteins, a probable transporter for potassium, as well as a putative phage terminase large subunit. Interestingly, some genes had best BLAST hits to genes with similar broad functions in other bacteria of the Chlamydiales order, raising the question of their origin.
Fig. 2.—
Pr. naegleriophila genomic islands. Probable genomic islands identified in both Pr. naegleriophila by IslandViewer (green) and by comparative genomics (blue) are located in regions with low- or high-GC content compared with the genomic mean GC content. The similarity between the two strains is indicated by red shading, and regions differing between the two strains appear in white. Only one region in Pr. naegleriophila strain KNic and two in Pr. naegleriophila strain Diamant were identified by both methods.
Table 2.
Genomic islands identified in Pr. naegleriophila
| Region_ID | Genome | Contig | Orientation | Start | Stop | Length | Predicted | tRNA |
|---|---|---|---|---|---|---|---|---|
| PnaK_GI1 | Pr. naegleriophila KNic | LN879502 | 1 | 1,493,284 | 1,530,803 | 37,519 | Y | |
| PnaK_GI2 | Pr. naegleriophila KNic | LN879502 | 1 | 2,793,793 | 2,808,968 | 15,175 | tRNA-Thr | |
| PnaD_GI1 | Pr. naegleriophila Diamant | NZ_CCJF01000005 | 1 | 984,976 | 992,323 | 7,347 | Y | tRNA_Leu |
| PnaD_GI2 | Pr. naegleriophila Diamant | NZ_CCJF01000001 | 1 | 4,231 | 13,743 | 9,512 | ||
| PnaD_GI3 | Pr. naegleriophila Diamant | NZ_CCJF01000005 | 1 | 1,796,680 | 1,802,137 | 5,457 | ||
| PnaD_GI4 | Pr. naegleriophila Diamant | NZ_CCJF01000004 | −1 | 314,516 | 325,842 | 11,326 | tRNA_Met | |
| PnaD_GI5 | Pr. naegleriophila Diamant | NZ_CCJF01000004 | −1 | 117,214 | 125,110 | 7,896 | Y |
Pr. naegleriophila Diamant possessed five regions absent from strain KNic ranging from 5.5 to 11.3 kb (table 2). All of them contained mobility genes: Three harbored one or two integrases, one had a number of transposases and another one encoded a recombinase (supplementary table S3, Supplementary Material online). Several mobility genes seemed to have evolved toward pseudogenisation as they harbored frameshifts. PnaD_GI1 and PnaD_GI5 were also predicted as genomic islands by IslandViewer (Dhillon et al. 2015). PnaD_GI1 and PnaD_GI4 were found close to tRNAs-Leu and -Met, respectively. PnaD_GI4 presented an interesting case as it is partially conserved with Pr. naegleriophila KNic, from the integrase to gene BN1093_RS01990 (supplementary table S3, Supplementary Material online). The region unique to Pr. naegleriophila Diamant included a putative chloramphenicol acetyltransferase which is an antibiotic resistance gene, as well as an OMP-like protein—a member of a large and diverse family of Chlamydial outer membrane proteins that were not expected to be found in genomic islands.
The pairwise alignment of both Pr. naegleriophila strains also enabled the identification of multiple smaller gaps (supplementary tables S4 and S5, Supplementary Material online) representing other events of insertions, deletions, or possibly gene acquisitions. Some of these gaps might also reflect errors in sequencing, such as homopolymers or misassemblies in repetitive regions as well as poorly aligned regions. Respectively, 30% and 25% of gaps identified in strains KNic and Diamant fell within or included a coding region. Four tandem duplications that arose after the divergence of KNic and Diamant strains were identified, all of which involved hypothetical proteins (supplementary table S6, Supplementary Material online). The frequency of gaps differed along the chromosome, suggesting the existence of hot spots for genome evolution by insertion or deletion. Pr. naegleriophila KNic harbored two hot spots between 1.46 Mb and 1.58 Mb as well as between 1.99 Mb and 2.01 Mb, which include a genomic island each. Pr. naegleriophila Diamant contained multiple regions with a slightly higher frequency of gaps, the most prominent being located between 1.93 Mb and 1.99 Mb where one of the genomic islands was found. In both cases, gap size seemed randomly distributed along the chromosome, with no observable pattern.
pPNK Is an F-Type Conjugative Megaplasmid
The bacterial chromosome was circularized, leaving behind several contigs with a 23-fold coverage, 1.4 times higher than the 16× average chromosomal coverage. These contigs formed a 145,285 bp large plasmid—the largest known plasmid in the order Chlamydiales. The plasmid pPNK presented a GC content of 37.2% and included 160 genes among which are several transposase and integrase remnants, doc proteins, and systems for the maintenance of the plasmid (parA and PNK_p0119) that are all characteristic of extrachromosomal elements.
The plasmid also encoded a type IV secretion system with highest similarity to the F-type system found in the genomic island of Pr. amoebophila UWE25 chromosome (Greub et al. 2004), the plasmids of S. negevensis (Collingro et al. 2011) and Rubidus massiliensis, and the remnants traU, traN and traF present in members of the family Parachlamydiaceae (Greub et al. 2009; Collingro et al. 2011) (fig. 3). The type IV secretion system of R. massiliensis is located on plasmid pRm1 that contains almost exclusively the tra operon as well as core genes for plasmid replication, such as parA. R. massiliensis and KNic tra operons shared a striking collinearity. The comparison of gene conservation showed that traN has undergone different rearrangements in both Pr. amoebophila strains, and traC was split in strain Pr. amoebophila R18. On the other hand, R. massiliensis, S. negevensis, and Pr. naegleriophila KNic, the three bacteria that possess the tra operon on a plasmid, retained intact genes. Moreover, these bacteria presented Ti-type traA and traD genes downstream that shared similarity to other amoeba-infecting bacteria, such as Rickettsia bellii and Legionella spp.
Fig. 3.—
Chlamydiales order, plasmids, and type IV secretion system. The left panel represents a neighbor joining tree of bacteria belonging to the order Chlamydiales, whose genome sequences are available, based on four conserved proteins (DnaA, FtsK, HemL, and FabI). The presence of a plasmid in each strain is represented by a small circular DNA molecule and the draft genomes with no described plasmid are indicated by a question mark as plasmids may be hidden among the numerous contigs. Orange ovals indicate the presence of a conjugative tra operon on the plasmid or in the bacterial chromosome. The right panel shows the conservation of the type IV secretion system tra operon and the surrounding genes. Pac: P. acanthamoebae, Nsp: Neochlamydia sp., Rma: R. massiliensis, Pna: Pr. naegleriophila, Pam: Pr. amoebophila, Pac HS-T3: Parachlamydiaceae bacterium HS-T3, Wch: Waddlia chondrophila, Cse: Criblamydia sequanensis, Ela: Estrella lausannensis, and Sne: Simkania negevensis.
Gene Content of the Parachlamydiaceae Family
Orthologous groups of proteins were reconstructed to investigate the gene content of members of the Parachlamydiaceae, both for chromosomes and plasmids (fig. 4 and supplementary table S7, Supplementary Material online). All members of the family Parachlamydiaceae shared 753 groups of orthologs encoded on their chromosomes, some groups including more than one paralog. The genus Protochlamydia shared 1,265 groups of orthologs, whereas at the species level Neochlamydia sp., Pr. naegleriophila, Pr. amoebophila, and P. acanthamoebae shared 1,382, 2,109, 1,476, and 2,032 groups of orthologs, respectively. The number of groups of orthologs shared among different subgroups of bacteria was significantly correlated to the cophenetic distance between the subgroups (Pearson coefficient −0.82, P-value = 0.0001).
Fig. 4.—
Core and accessory genome of the Parachlamydiaceae family. Venn diagram representing the number of orthologous groups of proteins shared by selected representative species of the family Parachlamydiaceae encoded on the chromosome (A) and on the plasmid (B).
Only one orthologous protein was shared by all four plasmids, ParA, an essential component for plasmid partitioning. The two plasmids of R. massiliensis carried each a copy of ParA, further strengthening the presence of two separate plasmids. The small R. massiliensis pRm1 39 kb plasmid had no other protein than ParA in common with Pr. naegleriophila Diamant plasmid, but it was highly similar to Pr. naegleriophila KNic plasmid, sharing 72% of groups of orthologs (28/39), mainly driven by the tra operon. In contrast, the R. massiliensis pRm2 80 kb plasmid shared most of its orthologs with the Pr. naegleriophila Diamant plasmid (fig. 4) and Criblamydia sequanensis plasmid (Bou Khalil et al. 2016).
A CRISPR—Cas System for the First Time within Chlamydiales
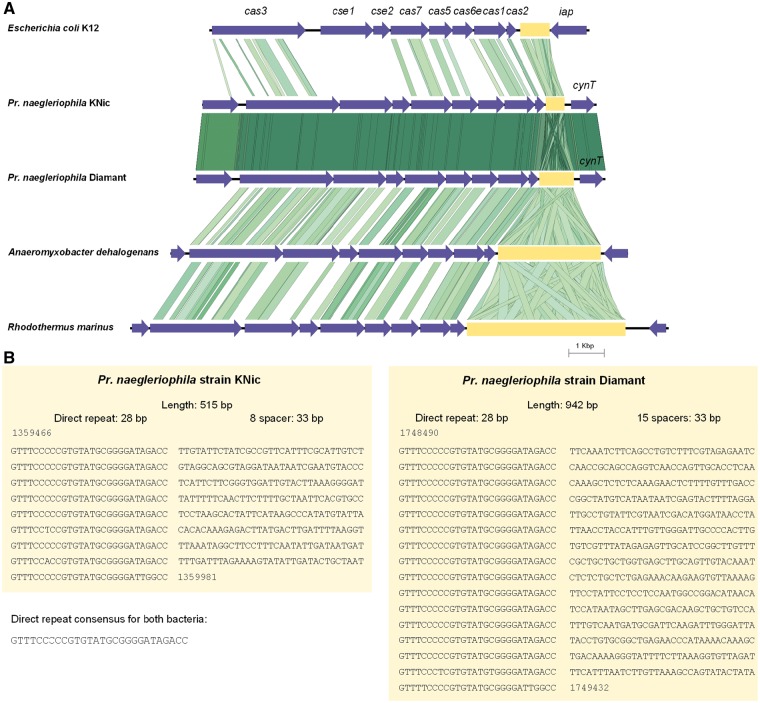
In Pr. naegleriophila, the CRISPR locus comprised eight 28 bp-long repeats separated by 33 bp-long spacers. The upstream operon of CRISPR-associated genes from the E. coli subtype I-E consists of the core genes cas1-2, the type I gene cas3 and subtype-specific genes cse1-2, cas5, cas6e, and cas7 (fig. 5). An almost identical cas operon and a CRISPR locus were identified in Pr. naegleriophila Diamant (fig. 5). This system is absent from other Parachlamydiaceae, such as strains Pr. amoebophila UWE25, EI2, and R18. Although a confirmed CRISPR locus was predicted by CRISPRfinder (Grissa et al. 2007) in the recently released genomes of Neochlamydia sp. (Domman et al. 2014; Ishida et al. 2014), no cas genes could be identified and the repeats were found to be due to a highly repeated protein sequence.
Fig. 5.—
CRISPR locus and its associated genes. (A) CRISPR associated genes consist of eight CDS, cas3, cse1, cse2, cas7, cas5, cas6e, cas1, and cas2, shown in blue within their genomic environment. Green lines connecting the genes in different organisms represent BLAST sequence homology with a gradient from light green to dark green for low to high percentage sequence identity, respectively. Genes neighboring the CRISPR locus present homology in Pr. naegleriophila genomes, but not to other genomes showing that the site of CRISPR locus insertion in Pr. naegleriophila genomes is different than in other bacteria. CRISPR repeats are found directly downstream of the cas operon, as highlighted by the yellow box. (B) Direct repeats and spacer sequences are detailed in the panel B.
The CRISPR spacers could give an interesting imprint of recent invasions by extrachromosomal elements, but unfortunately no significant homology was found by BLASTN against the nonredundant nucleotide database (nt) for strains KNic and Diamant (supplementary tables S8 and S9, Supplementary Material online). Genes surrounding this locus were found in conserved order in all Protochlamydia species, indicating that this CRISPR region was most likely acquired by horizontal gene transfer after the divergence of Pr. naegleriophila and Pr. amoebophila. The gene operon structure is commonly found in bacteria and two species present particular homology to Pr. naegleriophila KNic CRISPR locus: Anaeromyxobacter dehalogenans, a Deltaproteobacteria from soil and Rhodothermus marinus, a Bacteroidetes (fig. 5).
Discussion
The analysis of the complete genome sequence of Pr. naegleriophila enabled us to investigate the genome evolution of this recently described bacterial genus, and more broadly, the family Parachlamydiaceae. We described, for the first time, the presence of a CRISPR-locus in the order Chlamydiales, in the chromosome of two Pr. naegleriophila strains. In addition, Pr. naegleriophila harbors the largest known chlamydial plasmid that encodes a conjugative type IV secretion system similar to that found in the genomic island Pam100G of Pr. amoebophila UWE25 chromosome (Greub et al. 2004). We discuss here the current evolutionary scenario in light of these new key findings.
Based on the complete genome sequence of P. acanthamoebae UV-7 and Pr. amoebophila UWE25 as well as four draft genomes, Domman et al. (2014) suggested the occurrence of few rearrangements between strains of the same species and extensive rearrangements between the different genera of the family Parachlamydiaceae. The addition of new species, with the complete genome of Pr. naegleriophila strain KNic and the almost complete genome of strain Diamant now permits the identification of rearrangements within genera, that is, between species. Our comparison showed the absence of rearrangements between the two strains Pr. naegleriophila KNic and Diamant, but an increasing number of genome rearrangements with more distantly related organisms. In addition, comparison of complete genomes is essential to accurately infer rearrangements because highly fragmented genome sequences tend to show perfect collinearity when reordered according to a highly similar reference (supplementary fig. S2, Supplementary Material online).
Amoebae were proposed to act as a reservoir of different amoebae-resisting bacteria where horizontal gene transfer may preferentially take place (Moliner et al. 2010). The presence of an F-like conjugation plasmid putatively involved in DNA transfer in Pr. naegleriophila stresses the likelihood of gene exchange with other bacteria or with the eukaryotic host. The maintenance of intact tra genes in bacteria possessing the tra operon on a plasmid leads us to hypothesize that the system has retained functionality, whereas it has evolved toward pseudogenisation and deletion after being integrated into the genome of Pr. amoebophila strains and P. acanthamoebae strains.
The presence of an F-type conjugative operon in the plasmid or in the chromosome of various strains, combined with the lack of conjugative operon in the plasmid or in the chromosome of the Waddliaceae, Criblamydiaceae (Bertelli et al. 2014, 2015), and some Parachlamydiaceae (fig. 3) challenges the most parsimonious scenario proposed by Collingro et al. (2011) that plasmids evolved from a single conjugative plasmid acquired by an ancestor of the Parachlamydiaceae, Waddliaceae, and Simkaniaceae. In favor of this hypothesis is the shared presence of the Ti-type traA and traD in the paraphyletic R. massiliensis, Pr. naegleriophila KNic, as well as S. negevensis. However, if this hypothesis is correct, the plasmid and its tra operon were integrated within the chromosome at least twice in the genera Parachlamydia and Protochlamydia. In addition, the tra operon was completely lost several times, in the families Waddliaceae and Criblamydiaceae, in the genus Neochlamydia, and in some strains of Protochlamydia and Parachlamydia (fig. 3). Furthermore, it was partially lost in the Parachlamydia genus, where only a few genes remain. Alternative parsimonious scenarios could involve 1) the separate acquisition of the tra operon by an ancestor of Simkania and an ancestor of the family Parachlamydiaceae, after the divergence from the Parachlamydiaceae sp HS-T3 or 2) a transfer between an ancestor of Simkania and an ancestor of the family Parachlamydiaceae, which likely shared similar ecological niches or even sympatric intracellular lifestyles in light of their ability to infect the same hosts. In addition, the striking pattern of sequence similarity between R. massiliensis plasmids and Pr. naegleriophila KNic and Diamant plasmids suggest that the common ancestor of Parachlamydiaceae may have harbored at least two different large plasmids. In any case, this highlights the highly dynamic nature of the genomes of Chlamydia-related bacteria and the potential of the tra operon to be readily acquired, and lost among these bacteria.
A CRISPR-Cas system has been reported in approximately 50% of bacteria with sporadic distribution patterns suggesting that CRISPR loci are subject to frequent horizontal gene transfer, a hypothesis supported by the presence of CRISPR loci on plasmids (Haft et al. 2005). The CRISPR locus of Pr. naegleriophila and its associated genes have most probably been acquired horizontally but the proteins have insufficient homology to infer a direct transfer from a given organism. This CRISPR-Cas system is of a different subtype than that of another intracellular amoeba-resisting bacteria F. novicida ruling out the possibility of intra-amoebal transfer between these organisms. The functionality and the exact role of this CRISPR-Cas system in Pr. naegleriophila remain to be phenotypically determined, but by similarity to the type I-E locus present in E. coli, we can hypothesize that it plays a role in preventing DNA acquisition or protecting the bacteria against phages.
Although Pr. naegleriophila is an obligate intracellular bacterium, it may still be exposed to phages. Indeed, several phages of the genus Chlamydiamicrovius were isolated from classical Chlamydia and shown to grow in various species, including C. psitacci, C. abortus, C. felis, C. caviae, and C. pneumoniae (Śliwa-Dominiak et al. 2013). As a bacterium thriving in amoebal cells, and mostly found in water, Pr. naegleriophila could even be exposed to more diverse phages than the classical Chlamydia. Little is known about the diversity of phages able to infect amoeba-resisting bacteria and most reports concern the discovery of prophages, that is, phages integrated into the host genome, and phages remnants. The analysis of Legionella pneumophila pan genome revealed the presence of seven genomic islands harboring phage-like genes (D’Auria et al. 2010). The genome of Candidatus Amoebophilus asiaticus harbors genomic loci with similarity to the antifeeding prophage (afp), an essential virulence factor of the insect pathogen Serratia entomophila (Penz et al. 2010). In addition, two different coevolution experiments using Pseudomonas or Serratia strains with Tetrahymena thermophila and Acanthamoeba had diverging results: in one study protist selection did not increase resistance to phages (Friman and Buckling 2013), whereas in the second study the bacteria having coevolved with the amoebae were less susceptible to phage infection than those grown alone (Ormala-Odegrip 2015). The difference in CRISPR spacers between Pr. naegleriophila strains KNic and Diamant clearly highlights the dynamic and likely functional status of the system, as well as the exposure of such obligate intracellular bacteria to DNA of foreign origin. The absence of similarity between CRISPR spacers and sequences of the non-redundant nucleotide database underlines the currently limited knowledge on phages and extrachromosomal DNA circulating in amoebae-resisting bacteria, especially those growing in the ubiquitous amoeba Naegleria. The presence of genomic islands in both Pr. naegleriophila strains KNic and Diamant underlines the current ability of both bacteria to acquire DNA, despite the presence of a CRISPR system. Indeed, as shown in numerous examples, the CRISPR systems do not completely inhibit the acquisition of all exogenous DNA but are a component of the ongoing coevolution of mobile elements (Croucher et al. 2016). Notably, some bacteriophages have evolved genes to counter CRISPR activity and can infect cells with a CRISPR system (Nozawa et al. 2011; Lopez-Sanchez et al. 2012; Bondy-Denomy et al. 2013). Finally, the limited number of complete Parachlamydiaceae genomes available in public databases does not yet allow testing if the rate of gene and genomic island acquisition is lower in Pr. naegleriophila that harbor a CRISPR system compared with its sister phylum Pr. amoebophila that does not have such a system.
The complete genome sequence of Pr. naegleriophila represents a first step toward the understanding of mechanisms triggering genome evolution and evolutionary pressures at play in the Parachlamydiaceae family.
Materials and Methods
Culture and Purification of Pr. Naegleriophila
Pr. naegleriophila strain KNic was grown in Acanthamoeba castellanii ATCC 30010 at 32 °C using 75 cm2 cell culture flasks (Becton Dickinson, Franklin Lakes, USA) with 30 ml of peptone-yeast extract glucose broth. Pr. naegleriophila were purified from amoebae by a first centrifugation step at 120 × g for 10 min. Then, remnants from amoebae were removed from the resuspended bacterial pellet by centrifugation at 6500 × g for 30 min onto 25% sucrose (Sigma Aldrich, St Louis, USA) and finally at 32000 × g for 70 min onto a discontinuous Gastrographin (Bayer Schering Pharma, Zurich, Switzerland) gradient (48%/36%/28%).
Genome Sequencing, Assembly and Gap Closure
Pr. naegleriophila genomic DNA was isolated with the Wizard Genomic DNA purification kit (Promega Corporation, Madison, USA). Reads obtained with Genome Sequencer 20TM by Roche Applied Science (Penzberg, Germany) were assembled de-novo using Newbler V1.1.02.15 yielding 93 large contigs with a mean 16× coverage. Scaffolding on Pr. amoebophila strain UWE25 and PCR-based techniques were used to close the gaps between those contigs. Solexa 35 bp reads obtained from sequencing with Genome Analyzer GaIIx (Illumina, San Diego, USA) by Fasteris (Plan les Ouates, Switzerland) were then mapped to the final assembly with BWA (Li and Durbin 2009) and visualized with Consed (Gordon and Green 2013). Homopolymer errors were corrected in the plasmid and the chromosome sequence after manual inspection of discrepancies covered by >2 reads with a Phred base quality score of >10. Sequence start was placed in an intergenic region closest to the minimum of the GC skew, as determined with a sliding window of 100 nt.
Genome Annotation
GenDB 2.4 pipeline (Meyer et al. 2003) was used for a first automatic annotation of the genome that was followed by manual curation of annotation. Coding sequence (CDS) prediction was performed using Prodigal (Hyatt et al. 2010). All predicted CDS were submitted to similarity searches against nr, Swissprot, InterPro, Pfam, TIGRfam, and KEGG databases. Putative signal peptides, transmembrane helices, and nucleic acid binding domains were predicted using SignalP (Petersen et al. 2011), TMHMM (Krogh et al. 2001), and Helix-Turn-Helix (Dodd and Egan 1990), respectively. Protein domain identification was used to manually curate genome annotations with a scheme as proposed in Bertelli et al. (2015). The complete and annotated genome sequences have been deposited in the European Nucleic Archive under the project PRJEB7990 with accession numbers LN879502 and LN879503.
Genome Analysis
To identify CRISPR repeats, the genome sequences were submitted to CRISPRFinder (Grissa et al. 2007). The spacers within the CRISPR locus of Pr. naegleriophila strains KNic and Diamant were submitted to BLASTN (Altschul et al. 1997) homology searches against the nonredundant nucleotide database.
For phylogenetic reconstruction, multiple sequence alignments were performed with Muscle V3.7 (Edgar 2004), and a neighbor-joining tree was reconstructed using Mega 6 (Tamura et al. 2013) with 1000 bootstrap, Poisson distribution, and gamma equal to 1.
The two nearly complete genomes of R. massiliensis and Pr. naegleriophia Diamant were reordered with Mauve (Darling et al. 2004) by similarity to the closest available complete genome sequence P. acanthamoebae UV7 and Pr. naegleriophila KNic, respectively. These genomes and the complete genomes were aligned using Mauve and the alignment was represented using GenoPlotR (Guy et al. 2010). To investigate the number of rearrangements between genomes, a list of collinear block permutations was exported using Mauve (Darling et al. 2004). Each rearrangement arising after the divergence of two bacterial strains should lead to two pairs of subsequent collinear blocks being separated. Therefore, the number of rearrangements is counted as the number of two subsequent collinear blocks in the reference that were separated in the query genome, divided by two.
To investigate the occurrence of horizontal acquisition of genetic material, a prediction of genomic islands in the genomes of Pr. naegleriophila strains KNic and Diamant and Pr. amoebophila UWE25 was performed using IslandViewer (Dhillon et al. 2015). Also, regions unique to each Pr. naegleriophila strains were retrieved from a pairwise alignment using Mauve (Darling et al. 2004). Regions larger than 4000 bp were considered as potential genomic islands and were manually inspected. Smaller regions unique to each strain were not manually curated to remove spurious indels caused by contig borders or unplaced contigs in the unfinished genome of strain Diamant. Pr. amoebophila and Pr. naegleriophila strains were too distantly related to infer the presence of genomic islands accurately by comparative genomics.
Groups of orthologous proteins were computed using OrthoFinder (Emms and Kelly 2015). Home-made scripts for data analysis and visualization were written in R (Cran 2010).
Supplementary Material
Supplementary figures S1–S2 and tables S1–S9 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We are grateful to Sébastien Aeby (University of Lausanne, Switzerland) for his technical help during the gap closure stage. We would like to thank Burkhard Linke (Justus-Liebig-University Giessen, Germany) for his assistance in maintaining this GenDB project. We thank Bhavjinder K. Dhillon for editing the final version of the manuscript to correct the remaining English errors. Part of the computations was performed at the Vital-IT (http://www.vital-it.ch) Center for high-performance computing of the SIB Swiss Institute of Bioinformatics.
We acknowledge technical assistance by the Bioinformatics Core Facility at JLU Giessen and access to resources financially supported by the BMBF grant FKZ 031A533 within the de.NBI network. CB is supported by fellowships from the Société Académique Vaudoise and the Swiss National Science Foundation (P300PA_164673). The research performed in Greub’s group is supported by funding from various agencies including the Swiss National Science foundation (no 310030_141050, PDFMP3_127302 & 310030_162603)
Literature Cited
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C, et al. 2015. Sequencing and characterizing the genome of Estrella lausannensis as an undergraduate project: training students and biological insights. Front Microbiol. 6:101 doi: 10.3389/fmicb.2015.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C, Goesmann A, Greub G. 2014. Criblamydia sequanensis harbors a megaplasmid encoding arsenite resistance. Genome Announc. 2:5. doi: 10.1128/genomeA.00949-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelli C, Greub G. 2012. Lateral gene exchanges shape the genomes of amoeba-resisting microorganisms. Front Cell Infect Microbiol. 2:110. doi: 10.3389/fcimb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151:2551–2561. [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. 2013. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493:429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou Khalil JY, et al. 2016. Developmental cycle and genome analysis of “Rubidus massiliensis,” a new vermamoeba vermiformis pathogen. Front Cell Infect Microbiol. 6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJJ, et al. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson N, Michel R, Müller K-D, Aubert JD, Greub G. 2008. Protochlamydia naegleriophila as etiologic agent of pneumonia. Emerg Infect Dis. 14:168–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingro A, et al. 2011. Unity in variety—the pan-genome of the chlamydiae. Mol Biol Evol. 28:3253–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cran 2010. The comprehensive R archive network. Wiley Interdiscip. Rev. Comput. Stat. doi: 10.1002/wics.1212. [Google Scholar]
- Croucher NJ, et al. 2016. Horizontal DNA transfer mechanisms of bacteria as weapons of intragenomic conflict barton, NH, editor. PLoS Biol. 14:e1002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Auria G, Jiménez-Hernández N, Peris-Bondia F, Moya A, Latorre A. 2010. Legionella pneumophila pangenome reveals strain-specific virulence factors. BMC Genomics 11:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby AC, Cho N-H, Fuxelius H-H, Westberg J, Andersson SGE. 2007. Intracellular pathogens go extreme: genome evolution in the Rickettsiales. Trends Genet. 23:511–520. [DOI] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Garneau JE, Moineau S. 2010. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 64:475–493. [DOI] [PubMed] [Google Scholar]
- Dhillon BK, et al. 2015. IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res. 43:W104–W108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd IB, Egan JB. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domman D, et al. 2014. Massive expansion of ubiquitination-related gene families within the Chlamydiae. Mol Biol Evol. 31:2890–2904. doi: 10.1093/molbev/msu227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugster M, Roten C-AH, Greub G. 2007. Analyses of six homologous proteins of Protochlamydia amoebophila UWE25 encoded by large GC-rich genes (lgr): a model of evolution and concatenation of leucine-rich repeats. BMC Evol Biol. 7:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett KDE, Bush RM, Andersen AA. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards. Int J Syst Bacteriol. 49:415–440. [DOI] [PubMed] [Google Scholar]
- Friman V-P, Buckling A. 2013. Effects of predation on real-time host-parasite coevolutionary dynamics. Ecol Lett. 16:39–46. [DOI] [PubMed] [Google Scholar]
- Gimenez G, et al. 2011. Insight into cross-talk between intra-amoebal pathogens. BMC Genomics 12:542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Green P. 2013. Consed: a graphical editor for next-generation sequencing. Bioinformatics 29:2936–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greub G. 2009. Parachlamydia acanthamoebae, an emerging agent of pneumonia. Clin Microbiol Infect. 15:18–28. [DOI] [PubMed] [Google Scholar]
- Greub G. 2010. International Committee on Systematics of Prokaryotes * Subcommittee on the taxonomy of the Chlamydiae: minutes of the inaugural closed meeting, 21 March 2009, Little Rock, AR, USA. Int J Syst Evol Microbiol. 60:2691–2693. [DOI] [PubMed] [Google Scholar]
- Greub G, et al. 2004. A genomic island present along the bacterial chromosome of the Parachlamydiaceae UWE25, an obligate amoebal endosymbiont, encodes a potentially functional F-like conjugative DNA transfer system. BMC Microbiol. 4:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greub G, et al. 2009. High throughput sequencing and proteomics to identify immunogenic proteins of a new pathogen: the dirty genome approach. PLoS One 4:e8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35:W52–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy L, Kultima JR, Andersson SGE. 2010. genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26:2334–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 1:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn M. 2011. Phylum XXIV. Chlamydiae In: Krieg NR. editor. Bergey’s Manual of Systematic Bacteriology. Vol. 4 New York: Springer Berlin Heidelberg. [Google Scholar]
- Horn M, et al. 2004. Illuminating the evolutionary history of chlamydiae. Science 304:728–730. [DOI] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170. [DOI] [PubMed] [Google Scholar]
- Hyatt D, et al. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida K, et al. 2014. Amoebal endosymbiont Neochlamydia genome sequence illuminates the bacterial role in the defense of the host amoebae against Legionella pneumophila. PLoS One 9:4 doi: 10.1371/journal.pone.0095166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. 1987. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 169:5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Embden JDA, van, Gaastra W, Schouls LM. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 43:1565–1575. [DOI] [PubMed] [Google Scholar]
- Kamneva OK, Knight SJ, Liberles DA, Ward NL. 2012. Analysis of genome content evolution in PVC bacterial super-phylum: assessment of candidate genes associated with cellular organization and lifestyle. Genome Biol Evol. 4:1375–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS. 2013. CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 10:679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305:567–580. [DOI] [PubMed] [Google Scholar]
- Lagkouvardos I, et al. 2014. Integrating metagenomic and amplicon databases to resolve the phylogenetic and ecological diversity of the Chlamydiae. ISME J. 8:115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sanchez M-J, et al. 2012. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol Microbiol. 85:1057–1071. [DOI] [PubMed] [Google Scholar]
- Louwen R, Staals RHJ, Endtz HP, van Baarlen P, van der Oost J. 2014. The role of CRISPR-Cas systems in virulence of pathogenic bacteria. Microbiol Mol Biol Rev. 78:74–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, et al. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 9:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, et al. 2003. GenDB–an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 31:2187–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel R, Müller K-D, Hauröder B, Zöller L. 2000. A Coccoid bacterial parasite of Naegleria sp. (Schizopyrenida: Vahlkampfiidae) inhibits cyst formation of its host but not transformation to the flagellate stage. Acta Protozool. 39:199–207. [Google Scholar]
- Moliner C, Fournier P-E, Raoult D. 2010. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol Rev. 34:281–294. [DOI] [PubMed] [Google Scholar]
- Nozawa T, et al. 2011. CRISPR inhibition of prophage acquisition in streptococcus pyogenes. PLoS One 6:e19543.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberpichler I, et al. 2011. A photolyase-like protein from Agrobacterium tumefaciens with an iron-sulfur cluster. PLoS One 6:e26775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Örmälä-Odegrip A-M, et al. 2015. Protist predation can select for bacteria with lowered susceptibility to infection by lytic phages. BMC Evolutionary Biology 15:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penz T, Horn M, Schmitz-Esser S. 2010. The genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” encodes an afp-like prophage possibly used for protein secretion. Virulence 1:541–545. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 8:785–786. [DOI] [PubMed] [Google Scholar]
- Sampson TR, Weiss DS. 2014. CRISPR-Cas systems: new players in gene regulation and bacterial physiology. Front Cell Infect Microbiol. 4:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śliwa-Dominiak J, Suszyńska E, Pawlikowska M, Deptuła W. 2013. Chlamydia bacteriophages. Arch Microbiol. 195:765–771. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger AD, Wolf YI, Lobkovsky AE, Gilmore MS, Koonin EV. 2012. Viral diversity threshold for adaptive immunity in prokaryotes. MBio. 3:e00456–e00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.