Abstract
Only few studies on snake venoms were dedicated to deeply characterize the toxin secretion of animals from the Colubridae family, despite the fact that they represent the majority of snake diversity. As a consequence, some evolutionary trends observed in venom proteins that underpinned the evolutionary histories of snake toxins were based on data from a minor parcel of the clade. Here, we investigated the proteins of the totally unknown venom from Phalotris mertensi (Dipsadinae subfamily), in order to obtain a detailed profile of its toxins and to appreciate evolutionary tendencies occurring in colubrid venoms. By means of integrated omics and functional approaches, including RNAseq, Sanger sequencing, high-resolution proteomics, recombinant protein production, and enzymatic tests, we verified an active toxic secretion containing up to 21 types of proteins. A high content of Kunitz-type proteins and C-type lectins were observed, although several enzymatic components such as metalloproteinases and an L-amino acid oxidase were also present in the venom. Interestingly, an arguable venom component of other species was demonstrated as a true venom protein and named svLIPA (snake venom acid lipase). This finding indicates the importance of checking the actual protein occurrence across species before rejecting genes suggested to code for toxins, which are relevant for the discussion about the early evolution of reptile venoms. Moreover, trends in the evolution of some toxin classes, such as simplification of metalloproteinases and rearrangements of Kunitz and Wap domains, parallel similar phenomena observed in other venomous snake families and provide a broader picture of toxin evolution.
Keywords: snake, venom, transcriptome, proteome, acid lipase, molecular evolution
Introduction
The protein and peptide composition of the venoms from a number of dangerous snake species have been extensively studied by means of high-throughput technologies such as transcriptomics, proteomics, and peptidomics (Durban et al. 2011; Tashima et al. 2012; Aird et al. 2013; Rokyta et al. 2013; Hargreaves et al. 2014; Lomonte et al. 2014; Margres et al. 2014; Mcgivern et al. 2014, 2015; Chapeaurouge et al. 2015; Junqueira-de-Azevedo et al. 2015; Petras et al. 2015; Viala et al. 2015; Reeks et al. 2016; Tan et al. 2016).
By extension, the compositions and general activities of venoms of snakes belonging to the same genus or to the same families of the most medically relevant ones become more predictable. Most of the recent efforts to characterize these venoms resulted in the identification of species-specific versions of proteins already known and rare are the recent examples where new types of venom molecules are described in these groups, such as the case of a novel Kazal-type protein in the venom of Bothriechis schlegelii (Lomonte et al. 2008; Fernández et al. 2016). However, an unknown universe of toxins may be hidden in the venomous secretions of snakes more distantly related to the medically important species.
Although the families comprising species hazardous to humans, that is, Viperidae, Elapidae, and Atractaspididae, represent only about 30% of snake species (The Reptile Database 2016), the majority of snake biodiversity in the World (∼65% of species) is spread within a group generally called “colubrid.” This group can be considered paraphyletic or monophyletic, according to the (sub)families included within the clade, but we will adopt the recent classification proposed by Pyron et al. (2013), who considered Colubridae as a monophyletic family that includes Dipsadinae, Colubrinae, Natricinae, among other subfamilies. Colubridae species are highly heterogeneous, however a ubiquitous feature of them is the presence of cephalic glands (venom gland, Duvernoy’s gland, supra-, and infralabial glands), which may produce toxin secretions used to capture and kill preys. Their bites are, with few exceptions, nonlethal to humans mainly due to the inability to deeply inject the venom, once they have rear fangs (opistoglyph dentition) or no specialized fangs (aglyph). Nevertheless, human injuries have been reported (Mackinstry 1983; Minton 1990; Datta and Tu 1993; Sawai et al. 2002). Particularly, the Dipsadinae subfamily, which comprises some of the most commonly observed colubrids in South America, has been reported in a large number of epidemiological studies related to snake bites (Prado-Franceschi and Hyslop 2002; Puorto and França 2003; Salomão et al. 2003).
Over the past years, the venom proteomes (and venom gland transcriptomes) of a few colubrid species have been reported (Fry et al. 2003; Ching et al. 2006; Mackessy et al. 2006; OmPraba et al. 2010; Peichoto et al. 2012; McGivern et al. 2014), bringing important contributions to the knowledge of venom composition in the group. These studies also provided insights into the molecular evolution of snake toxins, including the recruitment of new toxin types (OmPraba et al. 2010; Ching et al. 2012), and into the adoption of different venom strategies in different subfamilies, paralleling the different specializations observed in traditionally venomous snakes of Elapide and Viperidae families (McGivern et al. 2014). However, the specific examples provided by these works may not reflect the full diversity of venom compositions and protein types existing in colubrid snakes. Consequently, the trends in snake venom evolution largely discussed in the literature are mostly based on observations from a minority of species, though of high medical relevance.
In order to obtain a comprehensive profile of an unknown colubrid venom from the Dipsadinae subfamily and to evaluate whether known trends in the evolution of snake toxins occur in the group, we investigated the venom activities, the proteome and the venom gland transcriptome of the species Phalotris mertensi in an integrated way. The genus Phalotris (Dipsadinae) occurs from Central Brazil down to the Patagonia region. The singular pattern of body colors of P. mertensi resembles that verified in some members of the Elapidae family (e.g., coral snakes belonging to Micrurus genus) and it is likely an evolutionary mimicry strategy adopted in order to avoid predation (Brodie 1993). Phalotris mertensi is a fossorial snake, with diurnal and nocturnal activity. The diet of this particular species is poorly known, however, due to its fossorial habit, it is believed that it feeds mainly on amphisbenids and other elongated vertebrates (Sawaya et al. 2008). To date, there are no data concerning venom characterization of any member of Phalotris genus, although there is an interesting report of human envenomation (Lema 1978) by P. trilineatus. The symptoms observed were cephalea, local and oral mucosa hemorrhage, edema, and severe renal insufficiency, suggesting the presence of various classes of toxins in the venomous secretion of the snake.
Our investigation shows that P. mertensi venom, besides harboring toxin types commonly observed in other snakes, contains an unusual acid lipase similar to mammalian lysosomal lipases. It also revealed particular structural features of P. mertensi toxins that evidence more general trends in the molecular evolution of snake toxins. These findings reinforce previous examples in the literature about the potential of unexplored venoms to reveal novel biomolecules and important information for understanding the evolution of venoms.
Experimental Section
Venom
Venom was extracted from specimens of P. mertensi after injection of pilocarpin (10 mg/kg in 0.15 M NaCl), which was used to stimulate venom secretion, since colubrids produce minimum amounts of venom. The venom was collected (∼2 mg of protein) with the aid of capillary tubes, lyophilized, and stored at −20 °C (Ferlan et al. 1983).
2-D Electrophoresis
Phalotris mertensi venom sample (350 µg of proteins) was dissolved in MilliQ® water and mixed with DeStreak rehydration solution (GE Healthcare) containing 1% IPG buffer to a final volume of 450 µl (for 24 cm precast IPG strip; pH 3–10, linear) (GE Healthcare). First, dimension was carried out in an Ettan IPGphor Isoeletric Focusing System (GE Healthcare) as described by the manufacturer, at 20 °C, using a three-phase electrophoresis program: 30 V for 6 h, 150 V for 2 h, 350 V for 1 h, 500 V for 1 h, 1,000 V for 1 h, 3,000V for 1 h, and 5,000 V for 13 h. Prior to running the second dimension, the IPG strip was placed in a rehydration tray and the proteins were reduced and alkylated by sequential incubation in the following solutions: 0.05 M Tris-HCl, pH 8.4, 1% sodium dodecyl sulfate (SDS); 30% glycerol (equilibration buffer-EB), 20 mg/ml dithiothreitol in EB for 12 min; and then a solution of 30 mg/ml iodoacetamide in EB, for 10 min. Afterwards, it was directly applied onto a 12% SDS–polyacrylamide gel (20 cm × 26 cm × 1.5 mm) for the second dimension electrophoresis. The gel was stained with silver according to Mortz et al. (2001).
In-Gel Protein Digestion and Mass Spectrometric Protein Identification
Protein spots were excised and in-gel trypsin digestion was performed according to Hanna et al. (2000). An aliquot (4.5 μl) of the resulting peptide mixture was injected into a trap column (180 μm i.d.×20 mm) packed with C18 chromatographic medium (Waters) for desalting with 100% solvent A (0.1% formic acid) at 5 μl/min for 3 min. Peptides were then eluted onto an analytical C18 column (75 μm i.d.×100 mm) (Waters) using a 20 min gradient at a flow rate of 600 nl/min where solvent A was 0.1% formic acid and solvent B was 0.1% formic acid in acetonitrile. The gradient was 0–80% acetonitrile in 0.1% formic acid over 20 min. A QTOF Ultima mass spectrometer (Waters) was used to acquire spectra. Spray voltage was set at 3.1 kV and the instrument was operated in data-dependent mode in which one full mass spectrometry (MS) scan was acquired in the m/z range of 200–2,000 followed by in tandem mass spectrometry (MS/MS) acquisition using collision induced dissociation of the three most intense ions from the MS scan. A dynamic peak exclusion was applied to avoid the same m/z being selected for the next 90 s. Raw data were processed using ProteinLynx™ software (Waters) and the resulting .pkl files were created and searched through Mascot software version 2.4.1 (Matrix Science) against the P. mertensi database (5,320 protein sequences derived from the translation of cDNA sequences obtained from P. mertensi venom gland transcriptome) with a parent and fragment tolerance of 0.5 Da. Iodoacetamide derivatives of cysteine and oxidation of methionine were specified in Mascot software version 2.4.1 (Matrix Science) as fixed and variable modifications, respectively. Only peptides sequences with expect values (e-values) ≤0.05 were considered.
In-Solution Protein Digestion and Mass Spectrometric Protein Identification
Phalotris mertensi venom sample (100 μg of proteins) was dissolved in MilliQ® water and the (in-solution) trypsin digestion procedure was carried out according to Kleifeld et al. (2011) with slight modifications. Briefly, guanidine hydrochloride (GuHCl) was added to the protein solution to a final concentration of 4 M, followed by the addition of dithiothreitol (in MilliQ® water) to a final concentration of 5 mM and the mixture was incubated at 65 °C, for 1 h. Iodoacetamide was added to the solution to a final concentration of 15 mM and the mixture was incubated at room temperature (RT), in the dark, for 1 h. Excess of iodoacetamide was quenched by the addition of dithiothreitol to a final concentration of 15 mM. Proteins were then precipitated with 8 volumes of ice cold acetone and 1 volume of ice cold methanol and the mixture was incubated for 3 h at −80 °C. After centrifugation at 14,000 × g for 10 min, 4 °C, the protein pellet was washed three times with ice-cold methanol and resuspended with the addition of a minimal volume of a 100 mM NaOH solution followed by the addition of MilliQ® water to a final protein concentration of 1 μg/μl. Trypsin (Sigma, Proteomics grade) was added to the solution at 1:100 enzyme:substrate ratio (w/w) and the mixture was incubated for 18 h at 37 °C.Tryptic peptides were desalted using Sep Pak tC18 cartridge (Waters) according to manufacturer’s instructions, dried, and dissolved in 50 µl of 0.1% formic acid. The peptide mixture (5 μl) was injected into a 2 cm C18 trap column (100 μm i.d. × 360 µm o.d.) using an EASY II-nanoLC system (Proxeon) coupled to an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). The chromatographic separation of tryptic peptides was performed on a 10-cm long column (75 µm i.d. × 350 µm e.d.) packed in-house with 5 µm Aqua® C-18 beads (Phenomenex). Peptides eluted with a linear gradient of 3–35% acetonitrile in 0.1% formic acid in 60 min at 200 nl/min. Spray voltage was set at 2.0 kV and the mass spectrometer was operated in data-dependent mode, in which one full MS scan was acquired in the m/z range of 300–1,800 followed by MS/MS acquisition using collision induced dissociation of the ten most intense ions from the MS scan. The MS spectra were acquired in the Orbitrap analyzer at 60,000 resolution (at 400 m/z) whereas the MS/MS scans were acquired in the linear ion trap. Isolation window, activation time, and normalized collision energy were set to 3 m/z, 30 ms, and 35%, respectively. A dynamic peak exclusion was applied to avoid the same m/z being selected for the next 90 s.
LTQ-Orbitrap Velos raw data were converted to MGF format using the MS convert tool (ProteoWizard 2015; version 3.0.3535) for database searching using Mascot server version 2.4.1 (Matrix Science) against the P. mertensi database (5,320 sequences derived from the translation of cDNA sequences obtained from P. mertensi venom gland transcriptome) with a parent tolerance of 10 ppm and fragment tolerance of 0.5 Da. Iodoacetamide derivatives of cysteine and oxidation of methionine were specified in Mascot as fixed and variable modifications, respectively.
Label-Free Quantitation
The data from Mascot search were analyzed by Scaffold version 4.4.4 (Proteome Software) and a false discovery rate of 1% was required for both protein and peptide identifications. Spectra count values for each protein were used to calculate the relative abundance of proteins using the normalized spectral abundance factor (NSAF) as described by Zybailov et al. (2006). For the calculations of protein coverage and NSAF, the length of mature proteins was considered.
Amidolytic Activity
Amidolytic activity was determined on D-Val-Leu-Arg-pNA and D-Arg-Gly-Arg-pNA at 37 °C in a system containing 0.1M Tris HCl buffer (buffer A), pH 8.0, substrate, and venom (8.25 µg and 16.5 µg) diluted in buffer A. Substrate assay concentrations were D-Val-Leu-Arg-pNA, 0.5 mM and D-Arg-Gly-Arg-pNA, 0.25 mM (Chromogenix). Reactions were stopped by adding 3% acetic acid, and release of p-nitroaniline was monitored at 405 nm. Amidolytic activity was expressed as nanomoles of substrate hydrolysed per minute per mg of protein.
Coagulant Activity
The minimum coagulant dose (MCD) is defined as the minimum amount of venom resulting in clot formation within 60 s at 37 °C (Theakston and Reid 1983). Coagulation activity was determined on human citrated plasma (200 μl) using serial dilutions of venom samples in a final volume of 100 μl (dissolved in150 mM NaCl).
Caseinolytic Activity
Caseinolytic activity was determined by the method of Lomonte and Gutiérrez (1983) using casein as substrate. Venom samples were dissolved in 150 mM NaCl to a final concentration of 50 μg/ml and were incubated at 37 °C for 30 min with 1 ml of 1% casein solution in phosphate buffered saline, pH 7.5. Then, 3 ml of 5% trichloroacetic acid were added, and after 30 min at 22–25 °C, the tubes were centrifuged. The absorbance of supernatants was determined at 280 nm. Proteolytic activity was expressed in Units/mg venom, calculated as change in absorbance at 280 nm divided by venom concentration (mg) and multiplied by 100.
Gelatinolytic Activity
2D-Polyacrylamide gel electrophoresis (PAGE) gelatin zymography of venom was performed as described elsewhere (Paes-Leme et al. 2009). Gelatin (Sigma) was copolymerized with 12% polyacrylamide. Samples containing 25 μg of venom were incubated in the presence and absence of inhibitors ethylenediaminetetraacetic acid (EDTA) (20 mM) and phenylmethylsulfonyl fluoride (PMSF) (10 mM) for 1 h at RT and then dissolved in sample buffer in the absence of reducing agent. After electrophoresis, gels were incubated for 1 h at RT in 50 mM Tris-HCl, pH 7.4 containing 2.5% Triton X-100 to remove traces of SDS and incubated in zymography incubation buffer (50 mM Tris-HCl, pH 8.0, containing 200 mM NaCl, 10 mM CaCl2, and 0.02% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) at 37 °C for 12 h. Gels were stained with Coomassie Blue and destained. Gelatin digestion was identified as clear zones of lysis against a blue background.
Fibrinogenolytic Activity
A mixture of human fibrinogen (Calbiochem) and venom samples (1:100 venom:fibrinogen ratio) was incubated at 37 °C for 2 h in presence or absence of inhibitors 1,10-phenantroline (10 mM), EDTA (10 mM), and PMSF (5 mM). A sample of fibrinogen was incubated without enzyme under identical conditions. Reactions were stopped by adding Laemmli sample buffer and then subjected to SDS–PAGE using a 10% polyacrylamide gel (Laemmli 1970) and stained with silver.
Hyaluronidase Activity
Hyaluronidase activity was determined turbidimetrically by the method of Ferrante (1956) modified by Pukrittayakamee (1988). Venom samples (50 μg) were dissolved in assay mixture (200 mM acetate buffer, pH 6.0, containing 150 mM NaCl), and the substrate (hyaluronic acid 0.5 mg/ml) was added. The mixture was incubated at 37 °C for 15 min and reaction was stopped by the addition of 1 ml of 2.5% cetylmethylammonium bromide in 2% NaOH. Absorbance was determined at 400 nm against a blank in which no venom was added. Turbidity reducing activity was expressed as a percentage of the remaining hyaluronic acid, taking the absorbance of a tube in which no venom as added as 100%. Specific activity was expressed as turbidity reducing units (TRU)/milligram of venom, in which one TRU was defined as the amount of venom able to promote reduction of 50% of turbidity.
Phospholipase A2 Activity
The activity was measured following the method described by Holzer and Mackessy (1996). Samples of 200 μg of venom, dissolved in 150 mM NaCl, were added in 10 mM Tris HCl buffer containing 10 mM CaCl2 and 100 mM NaCl. Then, 100 μl of substrate (0.3 mM 4-nitro-3-octanoloixy acid in acetonitrile) was added. The mixture was incubated at 37 °C for 15 min. The reaction was stopped by adding 2.5% Triton X-100 and the tubes were placed on ice bath. Absorbance was determined at 425 nm. Specific activity was expressed as nanomoles of substrate hydrolysed per minute per mg of protein.
Myotoxic Activity
Male Swiss mice (18–22 g) were obtained from Instituto Butantan and received water and food ad libitum. All the procedures involving mice were in accordance with the ethical principles in animal research adopted by the Brazilian Society of Animal Science and the National Brazilian Legislation. The protocol was approved by Institutional Animal Care and Use Committee (No 485/08) in 2008/11/06. Creatine kinase (CK) assay was performed as described by Gutiérrez et al. (1980). Animals were injected intramuscularly with 50 μg of venom in a volume of 50 μl (n = 6). The control animals (n = 6) were injected with sterile saline. After 3 h, blood samples were collected via ophthalmic plexus. Plasma was collected after centrifugation and CK levels were determined by CPK-520 kit (Sigma). The CK levels were determined according to CK standard in a calibration curve.
cDNA Preparation
A pair of venom (Duvernoy’s) glands of one specimen of P. mertensi kept in Laboratorio de Herpetologia, Instituto Butantan, was removed after 4 days of manual extraction of the venom. Total RNA was isolated using TRIzol reagent (LifeSciences) and messenger RNA purification was performed using a column of oligo-dT cellulose (GE). The cDNA was synthesized from 5 µg of mRNA using the Superscript Plasmid System for cDNA Synthesis and Cloning (Invitrogen-Thermo). The cDNA was ligated with the adaptors included in the kit, size selected in two ranges (250–600 bp and over 600 bp) and used to downstream transcriptomic analyses.
Expressed Sequence Tag Sequencing
Part of the unamplified cDNAs of both size ranges were directionally cloned in the pSPORT-1 plasmid and transformed in Escherichia coli DH5α electrocompetent cells. Plasmid DNA was isolated using alkaline lysis from randomly chosen clones as described before (Junqueira-de-Azevedo et al. 2006). DNA was sequenced on an ABI 3100 sequencer using the BigDye 3.1 kit (Applied Biosystems) with a standard 5′ primer (M13R). The electropherogram files were analyzed in a semiautomatic way as described elsewhere (Junqueira-de-Azevedo et al. 2006). The Phred program (www.phrap.com) was used to remove poor quality sequences (window length of 75 bases with 75% of standard quality < 25). Adapter and vector sequences were further removed by the CrossMatch program. An examination was carried out manually and sequences below 150 bp were discarded. The remaining Expressed Sequence Tags (ESTs) were then assembled in clusters of contiguous sequences using the CAP3 program (Huang and Madan 1999) set for 98% or more of base identity in a high-quality region. The relative representation of each cluster was given by the number of ESTs used in its assembly.
RNAseq
One nanogram of the cDNA from each range was fragmented using the Nextera XT DNA Library Prep Kit (Illumina), adding Illumina-specific oligonucleotide adapters for sequencing by synthesis procedure. The two libraries were sequenced on an Illumina HiSeq 1500 system, in Rapid Run mode, using standard manufacturer procedures to produce paired end reads of 150 bp. All Illumina reads were processed for removing adaptors and low quality ends of the sequences. The paired-end reads were first merged using CLC Genomics Workbench (Qiagen) to generate longer continuous reads. These merged reads were further assembled by the software seqMan NGen (DNAstar) using De Novo assembly module for transcriptomic data, with standard parameter except for “Match Window Length: 40; Min Match Percent: 98; Mismatch Penalty: 25; Match Repeat Percent: 400.” For a relative quantification of the gene expression, all passed filter sequencing reads were mapped back to the assembled contigs using CLC Genomics suite (Qiagen), RNAseq module, allowing the mapping of more than 80% of the read length with 97% identity. The RPKM (Reads Per Kilobase of exon model per Million mapped reads, Mortazavi et al. 2008) was calculated in order to allow a relative comparison of the expression level among genes of different sizes. The raw sequence data are available on GenBank SRA database under accession numbers (SRR3141929, SRR3141934, SRR3141937, SRR3141938).
Sequence Annotation and Curation
The de novo assembled contigs generated from Illumina reads were first automatically annotated by performing different BLAST searches against: 1) UniProt database with the algorithm BLASTX to identify similar sequences, 2) an in-house compiled set of representative sequences of snake toxin classes obtained from GenBank, Uniprot, and transcriptome shotgun assembly (TSA); 3) GenBank nt database, using BLASTN, for the sequences not annotated in the previous steps. Putative toxin categories were attributed to the sequences showing relevant (e-value < 105) matches to known snake toxins. The contig sequences in each category were further inspected and reanalyzed to improve the quality and extension of the informatically assembled cDNAs. Depending on the case, one or more contigs were fused in a single referential transcript, or the contig sequence was trimmed to the CDS (coding DNA sequence) to avoid assembly errors outside of the verified protein-coding region, or the contigs were extended by remapping the reads to their ends using CLC Genomics suite (Qiagen). Additionally, the ESTs and the ESTs assembled clusters were used to guide the curation process and to validate reassembled transcripts. In some cases, as described in the Results, full-length clones were resequenced by primer-walking to confirm and obtain their full cDNAs. The sequences were deposited in GenBank TSA database linked to Bioproject PRJNA310618).
Cloning, Expression, Purification, and Refolding of Acid Lipase
Based on the svLIPA (snake venom acid lipase) sequence obtained from the transcriptomic analysis, the primers (svLIPA-F: 5′- CTCGAGTCAGTGCTTGAGAGAAGA -3′; and svLIPA-R: 5′- GGTACCGCAGCACTACATGATGTTTGG -3′) were designed with restriction endonuclease sites for enzymes XhoI and KnpI, in the forward and reverse primers, respectively, and used to amplify by polymerase chain reaction (PCR) a mature svLIPA sequence from a complete EST clone. The amplified gene with the expected size was gel-purified, cloned into pGEM®-T Easy Vector Systems (Promega) and transformed into E. coli DH5α. This amplicon was further subcloned into the plasmid pAE, which express the protein in the bacteria cytoplasm with a N-terminal 6 × His residue tag (Ramos et al. 2004). Chemically competent E. coli BL21 Star™ (DE3) pLysS (Invitrogen) cells were transformed with the construction and plated on lysogeny broth (LB) agarose plates containing 100 μg/ml ampicillin and 34 mg/ml chloramphenicol and grown overnight at 37 °C. A single colony was inoculated in the same media and grown overnight at 37 °C. In the next day, it was diluted 1:100 into 250 ml of fresh LB (ampicillin/chloramphenicol) and at an optical density (600 nm) of 0.6, isopropyl-β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM to induce recombinant svLIPA (svLIPAr) expression. After 4 h the cells were harvested by centrifugation at 5,000 × g for 10 min at 4 °C and the supernatant discarded. The inclusion bodies were solubilized with 8M urea in binding buffer (100 mM Tris-HCl pH 8.0, 300 mM NaCl, and 5 mM of imidazole) and intermittently sonicated on ice for 60 s with intervals of 4 min for cooling, with total sonication time of 6 min. Cell debris were then harvested by centrifugation (13,700 × g, 10 min, 4 °C), and the recombinant protein was purified from the supernatant by affinity chromatography using Ni-NTA resin (Qiagen). The unbound proteins were removed by washing the column with binding buffer until no protein was detected and the bound svLIPAr was eluted with elution buffer (100 mM Tris-HCl, 1 M imidazole, 300 mM NaCl, 6 M urea, pH 8.0). The urea-denatured protein was refolded by dialysis with a gradient of decreasing concentration of urea at pH 8.0. The purity and concentration were analyzed in a 12% SDS–PAGE under reducing conditions (Laemmli 1970) and Bradford assays (Bradford 1976) using bovine serum albumin (BSA) as standard.
Immune Detection of svLIPA by Western Blot
Standard protocols were followed to raise mouse polyclonal antiserum (Harlow and Lane 1988). Briefly, purified svLIPAr was emulsified with Al(OH)3 adjuvant and 5 μg was injected into Swiss mice (n = 5). Three boosts of 5 μg each of the svLIPAr emulsified with Al(OH)3 adjuvant were given at an interval of 15 days. Seven days after the last injection, serum from the mice was collected. Immune reactivity was evaluated by indirect ELISA: Maxisorp plates (Nunc) were coated overnight at 4 °C with a solution of 10 μg/ml of the svLIPAr in 50 mM Na2CO3, pH 9.6, and blocked with PBS-T (PBS/0,1% Tween 20) containing 1% BSA. Antibody binding was detected by horseradish peroxidase conjugated anti-mouse antibody (Calbiochem) followed by addition of TMB (3,3′,5,5′-Tetramethylbenzidine) solution (Bio-Rad). Absorbance values were determined at 450 nm with a FlexStation microplate reader. For the western blot experiments, the samples were separated by electrophoresis on a 12% polyacrylamide gel and transferred in nitrocellulose membranes using a semidry western transfer system (Bio-Rad) at a constant voltage for 60 min. The membranes were blocked with 5% skimmed milk in phosphate buffered saline containing 0.1% Tween-20 for 2 h. The blots were treated with the mouse anti-svLIPAr serum (1:1,000 dilution) and conjugated horseradish peroxidase anti-mouse IgG (GE) was used as secondary antibodies (1:1,000 dilution). After three washes with PBS-T, binding of antibodies was detected using a chemiluminescent kit (Millipore) according to the manufacturer’s instructions.
Lipase Activity Assays
Lipase activity was measured using lipase assay kit with glycerol standard (Sigma) by adding 100 μl of the reaction mix (Lipase Assay Buffer, Peroxidase Substrate, and Lipase Substrate Enzyme Mix) with 10 μg of svLIPAr or 5 μg of P. mertensi venom or 5 μl of lipase positive control solution in microtiter plate. The plate was incubated at 37 °C and the absorbance (570 nm) was measured every 10 min. The lipase activity is expressed as nmol/min/ml = milliunit/ml, in which one lipase unit is the amount of enzyme that generated 1 μmol of glycerol from the triglyceride per minute at 37 °C. Lipase activity was also measured by using 4-methylumbelliferyl oleate (4-MUO) substrate (Modified Yan et al. 2006). The 4-MUO was resuspended in hexane (100 mg/ml) and diluted 1:100 in 4% Triton X-100. For assay was used 50 µl diluted 4-MUO and 100 µl (10 µg) svLIPAr in 50 μl buffer (0.2 mol/l Na2Ac added with 0.01% Tween 80, pH 5.5) at 37 °C for 30 min. The resulting fluorescence signal was detected at an excitation 360 nm and emission at 430 nm and the readings were deduced from the absorbance values emitted only in the presence of the substrate.
Results and Discussion
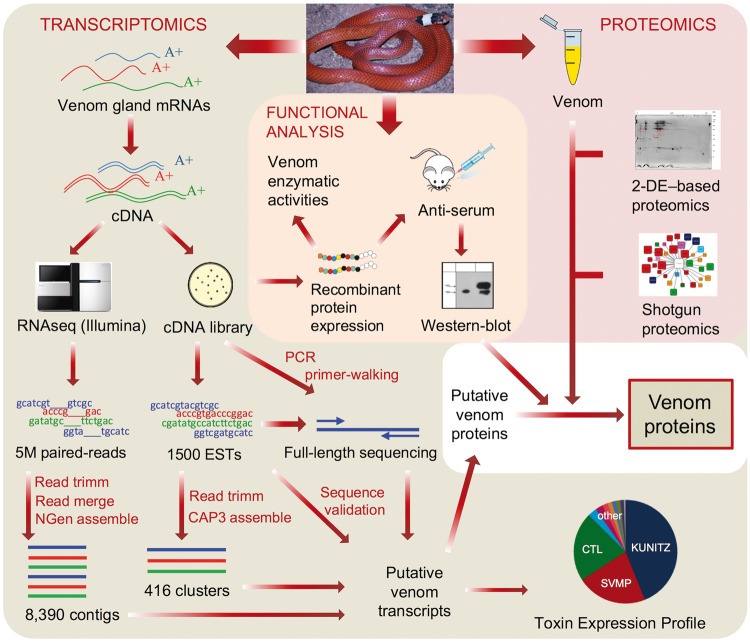
To achieve a reliable profiling of the toxin composition and activities of a venom from a species belonging to a poorly studied group of snakes and for which we lack any sequence background, it is required the use of multiple approaches, preferably in an integrated fashion. Enzymatic characterization may provide initial clues regarding the functional role of toxins, whereas a shotgun proteomic characterization is a powerful approach for a more descriptive analysis. However, given the phylogenetic distance of P. mertensi to other species and the lack of a comprehensive public database regarding colubrid proteins/toxins, the generation of a species-specific venom gland transcriptomic data set is a step required for the accurate identification of proteins by mass spectrometric analysis, besides a way of knowing the primary sequences of putative toxins. Since most of the assemblers for Next Generation Sequencing (NGS) data tend to fail in resolving the multiple isoforms of abundantly expressed toxins, as extensively discussed in the literature (Wagstaff et al. 2009; Rokyta et al. 2015), the correctness of these transcriptomic sequences could be checked by the use of cloning-based approaches such as EST survey or RT-PCR. On the other hand, the proteomic analysis of the venom secretion is important to confirm if the possible venom proteins predicted from the cDNAs are present or not in the venom. Finally, due to the very low amounts of venom produced by colubrids, a more detailed characterization of particularly interesting proteins could be only feasible by obtaining recombinant proteins. And this is facilitated by the availability of physical clones from the cloning-based approaches, which is not obtainable in NGS-only transcriptomics. In this study, we applied this combined strategy, as illustrated in figure 1, to characterize a totally unknown venom from a rear-fanged snake.
Fig. 1.—
Overview of the experimental approach adopted to investigate the venom of Phalotris mertensi. Transcriptomics, proteomics, and functional analysis were combined to provide corroborating evidences of the proteins present in the venom.
SDS–PAGE Profile and Enzymatic Activities of P. mertensi Venom
In order to get a comprehensive picture of snake venom, both reducing and nonreducing SDS–PAGE can be used as complementary approaches to examine the proteome complexity. Under both conditions the general profile of P. mertensi venom showed less complexity as compared with viperid venoms (supplementary fig. S1A, Supplementary Material online), and this pattern seems to be shared by other Colubridae venoms, such as Boiga irregularis, Philodryas olfersii, and Thamnodynastes strigatus (Ching et al. 2006, 2012; Mackessy et al. 2006). Under nonreducing conditions, its main protein bands showed molecular masses ranging from 75 to 20 kDa. Some differences between the reducing and nonreducing profiles suggest that the venom may contain polypeptide chains linked by disulfide bonds.
Gelatin zymography of P. mertensi venom was performed to visualize its proteinase proteome, since gelatin is degraded by both metalloproteinases and serine proteinases in snake venoms. Phalotris mertensi venom proteinases showed gelatinolytic activity, as illustrated by intense clear zones on the gel at the molecular mass range of around 48–38 kDa (supplementary fig. S1B, Supplementary Material online). Gelatinolytic activity was not inhibited by incubation of venom with EDTA, however it was affected by the treatment with PMSF, suggesting the presence of serine proteinases with gelatinolytic activity.
The evaluation of other proteolytic activities showed that P. mertensi venom displays lower amidolytic activity and higher caseinolytic activity compared with Bothrops jararaca venom (Menezes et al. 2006). Moreover, the venom showed hyaluronidase activity, which may be related to the process of venom spreading among prey tissues, by the hydrolysis of hyaluronic acid, an important component of the connective tissue (Pukrittayakamee et al. 1988). The phospholipase A2 activity of the venom was similar to that of B. jararaca, however, and its myotoxic activity was approximately 3 times higher (supplementary table S1, Supplementary Material online) (Furtado et al. 2006). The coagulopathy observed upon envenomation with viperid venoms is contributed by both serine proteinases and metalloproteinases. The determination of P. mertensi venom coagulant activity on human plasma showed an MCD approximately 20 times higher than that of adult B. jararaca venom (Furtado et al. 2006), being similar to values found for newborn viperid venoms, which are known by their intense procoagulant activity (Furtado et al. 1991; Zelanis et al. 2010). The venom also showed hydrolysis of the fibrinogen alpha chain, which was inhibited by metal-chelating agents (o-phenanthroline and EDTA) but not by PMSF, suggesting the activity of fibrinogenolytic metalloproteinases (supplementary fig. S1C, Supplementary Material online). Taken together, these results suggest a role of metalloproteinases in the procoagulant activity of P. mertensi venom.
De Novo RNAseq Profile of P. mertensi Venom Gland Validated by Sanger Sequencing
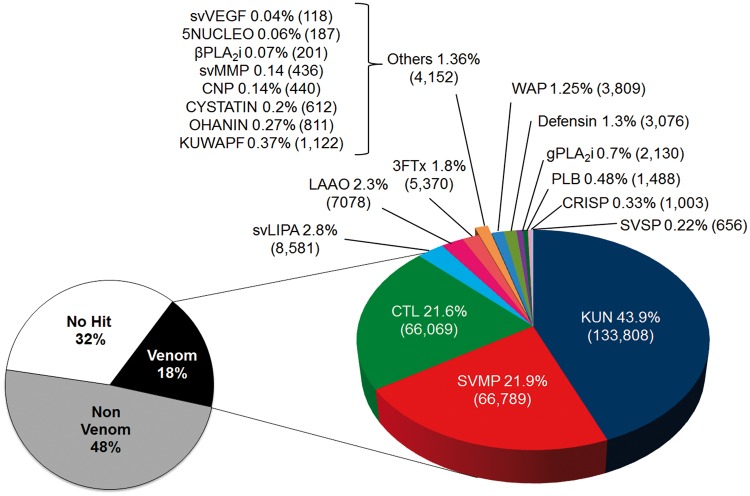
The transcriptome of P. mertensi venom gland was first analyzed by de novo RNAseq based on Illumina technology. Sequencing of the cDNA library resulted in around 3 million raw paired-end reads of 150 nt, which were filtered for high quality sequences yielding a final data set of 2,470,539 reads. The paired-end reads were merged for elongating the sequenced fragments and then assembled using NGen software under stringent parameters to allow the distinction between toxin isoforms, resulting in 8,390 contigs. According to the similarity searches using a BLASTX algorithm against Uniprot database and against a compiled data set of representative sequences of known snake toxin classes, these contigs were initially separated into three categories: transcripts coding for putative venom components, transcripts coding for nonvenom proteins, and transcripts not identified. The putative venom components were subcategorized in protein classes according to the most similar protein they match, which could be classical snake toxins or proteins suggested but not stablished as venom components. By using downstream analytical steps, the contigs identified as coding for putative toxins had their sequences curated and validated in order to reduce the redundancy, to improve the completeness and to correct the protein prediction. Figure 2 shows the RNAseq transcriptomic profile of the venom glands, considering the relative expression of each transcript in the category.
Fig. 2.—
Transcriptomic profile of the venom glands of P. mertensi by RNAseq. The “venom” category corresponds to transcripts coding for known or putative venom components, classified based on their sequence similarities to known toxin superfamilies. The relative expression is indicated by the percentage of expression values (RPKM–reads per kilobase per million of mapped reads) of each category among all transcripts (left chart) or among venom-related transcripts (right chart). In parentheses are the total RPKM in each category. 3FTx: three finger toxin; 5NUCLEO: 5′nucleotidase; CYST, cystatin; CRISP: cysteine-rich secretory protein; CNP: C-type natriuretic peptide; CTL: C-type lectin; KUN: Kunitz-type proteins; KUWAPF: Ku-wap-fusin-like protein; LAAO: l-amino acid oxidase; svLIPA: snake venom acid lipase; svMMP: snake venom matrix metalloproteinase; OHANIN: Ohanin-like protein; PLB: phospholipase B; bPLA2i: beta type PLA2 inhibitor; gPLA2i: gamma type PLA2 inhibitor; WAP: wap domain containing protein.
We also generated ESTs based on Sanger sequencing method, using the same cDNA sample of the RNAseq as source material, mainly to curate and refine the Illumina-based contigs (supplementary fig. S2, Supplementary Material online). It was interesting to note that both EST and RNAseq profiles were similar regarding the proportion of majorly expressed toxins. However, some relevant but less expressed classes of toxins, such as SVSP (snake venom serine proteinase) and CRISP (cysteine-rich secretory protein), were absent in the EST-based analysis due to the expected low coverage of the transcriptome. This was also evidenced by the high number of singlet sequences, most of them corresponding to transcripts involved in cellular functions. For the most expressed toxin genes, however, there was a reasonable correspondence and a quantitative equivalence in both profiles, that is, the most abundant venom genes were the same in both analyses. As far as we know, this is the only comparison between the results of the two methodological approaches for snake venom glands, especially considering that they were based on the exact same RNA sample. Moreover, it indicates that the EST-based profiles of many snake venom glands performed in the early 2000's may have provided a fair quantification of the majorly expressed venom genes and pointed out the most relevant transcripts, however, they may have missed less represented toxin categories, as expected. This might be one of the explanations as to why previous venom gland transcriptomic analyses based on ESTs sometimes provided very simple toxin profiles, whereas venom proteomic analyses, including two-dimensional gel electrophoresis and mass spectrometric identification, suggested a rather complex picture (e.g., Ching et al. 2006). Regardless of the sequencing technology employed and sequence coverage achieved, other levels of protein modifications, such as protein processing, glycosylation, complex formation, etc., probably contribute significantly to the differences observed between proteome and transcriptome profiles.
Proteomic Characterization of P. mertensi Venom
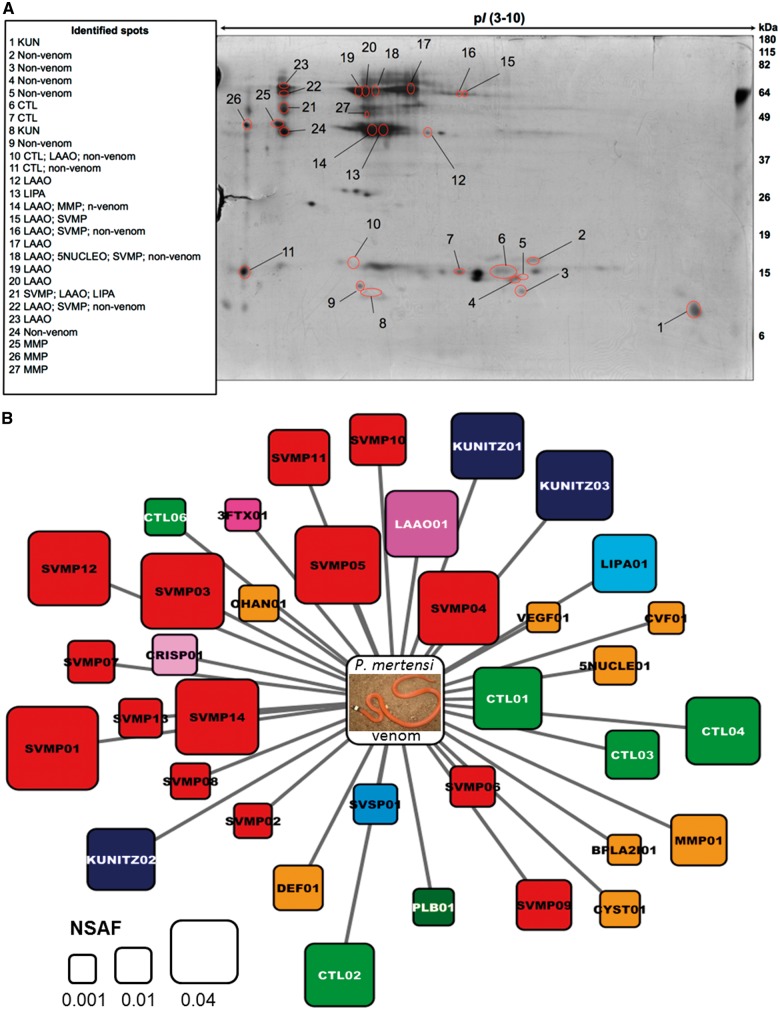
The first approach used for the proteomic characterization of the venom was 2-D electrophoresis (2-DE) and in-gel trypsin digestion coupled to mass spectrometric protein identification based on searches against a database of proteins predicted from the P. mertensi venom gland transcriptome generated in this study. As shown in figure 3, the 2-DE image of P. mertensi venom is composed of spots within a broad range of isoelectric points (pI from 3 to 10) and molecular masses of ∼10–70 kDa. The total number of spots, however, is much lower than that observed with viperid venoms under similar electrophoretic conditions. The overall proteomic pattern showed that high molecular mass spots are composed of groups of approximately 45 and 70 kDa with pI ranging from 4 to 5.5. On the other hand, low molecular mass spots (of ∼15 kDa and below) seem to have widespread isoelectric points, ranging from 4 to 10. The identification of proteins present in 27 selected spots (supplementary table S2, Supplementary Material online) revealed that P. mertensi venom contains some of the typical toxin classes of viperid venoms (Kunitz type protein [KUN]; L-amino acid oxidase [LAAO], C-type lectin [CTL], snake venom metalloproteinase [SVMP]) and also snake venom matrix metalloproteinases (svMMP), as the venom of T. strigatus (Ching et al. 2012). Among the identified proteins there were some cellular and plasma components, which were likely contaminants from the milking process, and, interestingly, a peptide corresponding to an acid lipase was identified in spot #21 (fig. 3A).
Fig. 3.—
Proteomic analysis of P. mertensi venom. (A) 2-DE profile of P. mertensi venom. Venom (350 µg) was applied to IPG strips (24 cm, pI 3-10) followed by electrophoresis on 12% SDS–polyacrylamide gels (20 cm × 26 cm). Proteins were stained with silver. Protein spots that were selected for in-gel trypsin digestion and LC-MS/MS analysis are indicated with red circles. Left side panel shows identified protein classes. (B) Graphical overview of toxin classes identified in P. mertensi venom by in-solution trypsin digestion followed by LC-MS/MS and label-free quantitative analysis. The area of each square is proportional to the NSAF. Graphical view generated with Cytoscape v. 2.8.1 (Cline et al. 2007).
In order to further explore the complexity of P. mertensi venom proteome, we performed an “in-solution” trypsin digestion followed by high-resolution LC-MS/MS analysis (supplementary table S3, Supplementary Material online). This approach allowed a comprehensive qualitative analysis of the venom proteome, whereas the values for spectral count for each protein were used to calculate their approximate relative abundance (fig. 3B and supplementary table S3, Supplementary Material online). The MS/MS spectra resulted in the identification of 67 proteins of which 37 are annotated as putative toxins. As it was observed in the protein identification of 2-DE spots, the presence of various nonvenom proteins, derived from cellular components and plasma, suggests the damage of venom gland tissue upon venom extraction. Besides, it may be relevant to consider that, in order to improve the yield of venom production, the animals used in this study were treated with pilocarpine, which is an agonist of the muscarinic acetylcholine receptor (Fox et al. 2001), and may have enhanced the production of other buccal fluids, possibly contributing to the presence of such proteins, though the venom was collected using capillaries attached to the rear fags.
More importantly, toxin classes that had not been detected by spot identification, such as SVSP, CRISP, svVEGF (snake venom Vascular Endothelial Growth Factor), Ohanin-like, CVF, phospholipase B, and defensin, were identified in the shotgun analysis. According to the relative abundance evaluation, SVMPs were the most abundant toxins in P. mertensi venom (fig. 3B).
An Integrated View of P. mertensi Venom Composition
The curated set of possible venom components of P. mertensi is summarized in table 1, and, in general, it revealed a composition of typical toxin classes found in other snake venoms. It includes some toxin types more abundantly found in the Viperidae family, such as SVMP and SVSP, other more typical of the Elapidae family, such as 3FTx, CVF, WAP, and KUN (Kunitz-type peptide), and one exclusive from Colubridae: svMMP. Some other identified toxin types are relatively abundant in both families (Viperidae and Elapidae), such as CTL, LAAO, and CRISP. The transcriptomic and proteomic analyses also revealed some other known venom constituents that occur in minor amounts in many snake venoms, such as 5NUCLEO (5′ nucleosidase), defensin, cystatins, svVEGF, Ohanin-like protein, PLB, and inhibitors of PLA2, whose occurrences in P. mertensi venom transcriptome were also at low transcriptional levels. Almost all the potential toxin classes identified in the transcriptome were detected in the venom proteomic analysis (except for WAP and KUWAFUS [ku-wap-fusin], which are potentially small proteins and whose corresponding tryptic fragments may have fallen out of the lower mass limit of the mass spectrometric analysis conditions used herein), confirming a very rich composition of this venom.
Table 1.
Summarization of the Transcriptomic and Proteomic Analyses of P. mertensi Venom
| Transcriptome |
Proteome |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Venom Protein Family | ORF Size | Expression Level |
ESTs Coverage | Peptides Identified |
MS/MS Coverage | Spectral Count | Relative Abundance | |||||||||
| ID | bp | Full/ Partial | reads | RPKM | % of venom | % of all | % of ORF | in gel | In solution | % | No of spectra | NSAF | ||||
| Kunitz type protein (KUN) | 25340 | 133808.5 | 43.89 | 12.120 | ||||||||||||
| PMERREF_KUNITZ01 | 258 | Full | 9332 | 49578.4 | 16.26 | 4.491 | 100 | 1 | 5 | 52.9 | 10 | 0.0320 | ||||
| PMERREF_KUNITZ02 | 261 | Full | 13365 | 70188.5 | 23.02 | 6.358 | 100 | 1 | 4 | 45.3 | 9 | 0.0284 | ||||
| PMERREF_KUNITZ03 | 258 | Full | 2643 | 14041.6 | 4.61 | 1.272 | 100 | 2 | 5 | 55.3 | 11 | 0.0352 | ||||
| Snake venom metalloproteinase (SVMP) | 84619 | 66786.7 | 21.90 | 6.050 | ||||||||||||
| PMERREF_SVMP01 | 1356 | Full | 14277 | 14431.6 | 4.73 | 0.013 | 96 | 11 | 31.3 | 60 | 0.0473 | |||||
| PMERREF_SVMP02 | 1623 | Part 5' | 1147 | 968.7 | 0.32 | 0.001 | 21 | 3 | 7.8 | 9 | 0.0053 | |||||
| PMERREF_SVMP03 | 1872 | Full | 3809 | 2789.0 | 0.91 | 0.003 | 68 | 1 | 26 | 45.5 | 85 | 0.0404 | ||||
| PMERREF_SVMP04 | 1875 | Full | 3698 | 2703.4 | 0.89 | 0.002 | 51 | 12 | 26 | 46.3 | 81 | 0.0385 | ||||
| PMERREF_SVMP05 | 1872 | Full | 10780 | 7893.2 | 2.59 | 0.007 | 100 | 24 | 44.4 | 89 | 0.0424 | |||||
| PMERREF_SVMP06 | 1455 | Part 5' | 2560 | 2411.6 | 0.79 | 0.002 | 0 | 7 | 18.0 | 15 | 0.0105 | |||||
| PMERREF_SVMP07 | 1875 | Full | 7726 | 5648.0 | 1.85 | 0.005 | 95 | 1 | 17 | 30.3 | 25 | 0.0119 | ||||
| PMERREF_SVMP08 | 1851 | Full | 2210 | 1636.5 | 0.54 | 0.001 | 73 | 1 | 9 | 15.7 | 13 | 0.0063 | ||||
| PMERREF_SVMP09 | 1875 | Full | 3356 | 2453.3 | 0.80 | 0.002 | 29 | 18 | 30.0 | 39 | 0.0186 | |||||
| PMERREF_SVMP10 | 1848 | Full | 10401 | 7714.6 | 2.53 | 0.007 | 82 | 20 | 40.8 | 40 | 0.0194 | |||||
| PMERREF_SVMP11 | 1714 | Part 3' | 1504 | 1202.8 | 0.39 | 0.001 | 18 | 15 | 23.3 | 43 | 0.0233 | |||||
| PMERREF_SVMP12 | 1875 | Full | 13591 | 9935.5 | 3.26 | 0.009 | 98 | 1 | 22 | 38.0 | 83 | 0.0395 | ||||
| PMERREF_SVMP13 | 1863 | Full | 2113 | 1554.6 | 0.51 | 0.001 | 29 | 11 | 23.2 | 19 | 0.0091 | |||||
| PMERREF_SVMP14 | 1875 | Full | 7447 | 5444.0 | 1.79 | 0.005 | 51 | 26 | 44.9 | 86 | 0.0404 | |||||
| C-type lectin (CTL) | 22870 | 66069.7 | 21.67 | 5.985 | ||||||||||||
| PMERREF_CTL01 | 453 | Full | 7741 | 23422.7 | 7.68 | 2.122 | 100.0 | 9 | 7 | 68.7 | 18 | 0.0288 | ||||
| PMERREF_CTL02 | 483 | Full | 4604 | 13065.5 | 4.29 | 1.183 | 100.0 | 12 | 68.1 | 20 | 0.0297 | |||||
| PMERREF_CTL03 | 498 | Full | 2257 | 6212.1 | 2.04 | 0.563 | 100.0 | 9 | 49.1 | 12 | 0.0172 | |||||
| PMERREF_CTL04 | 474 | Full | 5446 | 15748.4 | 5.17 | 1.426 | 100.0 | 12 | 11 | 73.9 | 22 | 0.0334 | ||||
| PMERREF_CTL05 | 513 | Full | 938 | 2506.2 | 0.82 | 0.227 | 96.7 | 0 | – | 0 | 0 | |||||
| PMERREF_CTL06 | 498 | Full | 705 | 1940.4 | 0.64 | 0.176 | 76.9 | 5 | 30.9 | 5 | 0.0072 | |||||
| PMERREF_CTL07 | 525 | Full | 382 | 997.3 | 0.33 | 0.090 | 100.0 | 0 | – | 0 | 0 | |||||
| PMERREF_CTL08 | 483 | Full | 1 | 2.8 | 0.00 | 0.000 | 43.7 | 0 | – | 0 | 0 | |||||
| PMERREF_CTL09 | 528 | Full | 213 | 553.0 | 0.18 | 0.050 | 0.0 | 0 | – | 0 | 0 | |||||
| PMERREF_CTL10 | 489 | Full | 232 | 650.3 | 0.21 | 0.059 | 93.3 | 0 | – | 0 | 0 | |||||
| PMERREF_CTL11 | 495 | Full | 174 | 481.8 | 0.16 | 0.044 | 0.0 | 0 | – | 0 | 0 | |||||
| PMERREF_CTL12 | 495 | Full | 113 | 312.9 | 0.10 | 0.028 | 0.0 | 0 | – | 0 | 0 | |||||
| PMERREF_CTL13 | 498 | Full | 64 | 176.1 | 0.06 | 0.016 | 66.9 | 0 | – | 0 | 0 | |||||
| Acid lipase (svLIPA) | ||||||||||||||||
| PMERREF_LIPA01 | 1200 | Full | 6931 | 8582.0 | 2.81 | 0.777 | 100.0 | 2 | 18 | 70.9 | 42 | 0.0251 | ||||
| L-amino acid oxidase (LAAO) | ||||||||||||||||
| PMERREF_LAAO01 | 1460 | Full | 7565 | 7068.3 | 2.32 | 0.640 | 62.6 | 54 | 33 | 63.4 | 73 | 0.0324 | ||||
| Three-finger toxin (3FTx) | 1013 | 5370.9 | 1.76 | 48.650 | ||||||||||||
| PMERREF_3FTX01 | 361 | Full | 902 | 4737.0 | 1.55 | 0.429 | 100 | 1 | 12.8 | 1 | 0.0032 | |||||
| PMERREF_3FTX02 | 241 | Full | 111 | 633.9 | 0.21 | 0.057 | 0 | 0 | – | 0 | 0 | |||||
| Waprin (WAP) | ||||||||||||||||
| PMERREF_WAP01 | 240 | Full | 667 | 3809.4 | 1.25 | 0.345 | 100 | 0 | - | 0 | 0 | |||||
| Defensin | 563 | 3976.1 | 1.30 | 36.015 | ||||||||||||
| PMERREF_DEF01 | 195 | Full | 515 | 3638.7 | 1.19 | 0.330 | 100 | 4 | 46.9 | 3 | 0.0142 | |||||
| PMERREF_DEF02 | 195 | Full | 48 | 337.4 | 0.11 | 0.031 | 0 | 0 | – | 0 | 0 | |||||
| Gamma PLA2 inhibitor (gPLA2i) | ||||||||||||||||
| PMERREF_GPLA2I01 | 384 | Full | 597 | 2131.0 | 0.70 | 0.193 | 0 | 1 | 12.6 | 0 | – | |||||
| Phospholipase B (PLB) | ||||||||||||||||
| PMERREF_PLB01 | 1662 | Full | 135 | 1478.72 | 0.48 | 0.134 | 0 | 18 | 39.8 | 19 | 0.0074 | |||||
| Ku-wap-fusin (KUWAPF) | ||||||||||||||||
| PMERREF_KUWAPF01 | 387 | Full | 317 | 1122.8 | 0.37 | 0.102 | 0 | 0 | – | 0 | 0 | |||||
| Cysteine-rich secretory protein (CRISP) | ||||||||||||||||
| PMERREF_CRISP01 | 780 | Full | 527 | 1003.3 | 0.33 | 0.091 | 0 | 8 | 42.3 | 11 | 0.0104 | |||||
| Ohanin-like (OHANIN) | ||||||||||||||||
| PMERREF_OHAN01 | 642 | Full | 380 | 811.3 | 0.27 | 0.073 | 69.3 | 5 | 32.4 | 6 | 0.0065 | |||||
| Serine proteinase (SVSP) | ||||||||||||||||
| PMERREF_SVSP01 | 771 | Full | 369 | 656.0 | 0.22 | 0.059 | 0 | 9 | 43.8 | 11 | 0.0099 | |||||
| Cystatin | 191 | 618.4 | 0.20 | 5.602 | ||||||||||||
| PMERREF_CYST01 | 426 | Full | 158 | 508.4 | 0.17 | 0.046 | 0 | 3 | 30.5 | 4 | 0.0069 | |||||
| PMERREF_CYST02 | 411 | Full | 33 | 110.1 | 0.04 | 0.010 | 0 | 0 | – | 0 | 0 | |||||
| Matrix metalloproteinase (svMMP) | ||||||||||||||||
| PMERREF_MMP01 | 1986 | Part | 632 | 436.2 | 0.14 | 0.040 | 0 | 24 | 34 | 61.3 | 61 | 0.0192 | ||||
| C-type natriuretic peptide (CNP) | ||||||||||||||||
| PMERREF_CNP01 | 861 | Full | 277 | 441.0 | 0.14 | 0.040 | 0 | 1 | – | 1 | – | |||||
| Cobra venom Factor-like (CVF) | ||||||||||||||||
| PMERREF_CVF01 | 1242 | Part 5' | 195 | 215.2 | 0.07 | 0.019 | 0 | 6 | 16.7 | 6 | 0.0030 | |||||
| Beta PLA2 inhibitor (bPLA2i) | ||||||||||||||||
| PMERREF_BPLA2I01 | 918 | Part 5' | 135 | 201.6 | 0.07 | 0.018 | 0 | 2 | 6.9 | 2 | 0.0015 | |||||
| 5' nucleotidase (5NUCLEO) | ||||||||||||||||
| PMERREF_5NUCLE01 | 1575 | Part 5' | 216 | 188.0 | 0.06 | 0.017 | 59.4 | 1 | 18 | 42.4 | 21 | 0.0087 | ||||
| Vascular endothelial groth factor (svVEGF) | ||||||||||||||||
| PMERREF_VEGF01 | 579 | Full | 50 | 118.4 | 0.04 | 0.011 | 0 | 1 | 15.1 | 1 | 0.0013 | |||||
Note.—The assembled transcripts coding for known snake venom proteins are listed according to their expression levels (RPKM), showing the corresponding sequence features and proteomic identification in the venom.
A recent investigation on the venom and venom glands of two colubrids from different subfamilies showed that each species had a trend to be more similar to one of the families of front-fanged snakes (McGivern et al. 2014). Specifically, while the venom of Boiga irregularis (Colubrinae) shows a marked Elapidae-like profile due to the prevalence of 3FTx, Hypsiglena sp, which is within the same subfamily of the P. mertensi (Dipsadidae), has a Viperidae-like venom profile (McGivern et al. 2014). For the present study, the overall predominance of SVMPs and diverse CTLs, and the low expression of 3FTx in P. mertensi venom suggest a more Viperidae-like venom, though the high content of Kunitz-type inhibitors indicates a rather mixed toxin pattern.
Other previous investigations on the venoms of nonfront fanged snakes have shown that despite being rich in classical snake toxins found in venoms of Viperidae and Elapidae families, some of them contain new and unexpected molecules that make them unique. For example, veficolins and svMMPs were proposed as new venom classes from Cerberus rynchops OmPraba et al. (2010) and T. strigatus (Ching et al. 2012), respectively. In the present work, a particular finding was the presence in the venom gland transcriptome of an abundant transcript coding for an enzyme similar to lysosomal acid lipases from vertebrates, which was previously argued to be a possible new toxin (Casewell et al. 2009) without further confirmation. Here, however, the presence of the protein in the venom was confirmed by proteomic identification and further characterization, as presented in detail later.
Some other transcripts coding for secreted proteins, such as lysozyme and phospholemman, were also found as highly represented in P. mertensi venom gland transcriptome, but they were not detected in the venom proteome. It is particularly interesting to mention two novel full-length transcripts for lysozyme highly expressed in the venom glands. In humans, the major expression sites of this enzyme are the parotid and submandibular salivary glands (EMBL-EBI. 2015), which are homologous to venom glands. The production of lysozyme could be easily associated with a protective function of the secretory apparatus against microbial infection transmitted by preys, but its fate in the snake oral cavity remains an interesting point to be studied, since it is not apparently secreted to the venom.
Kunitz-Type Proteins: Key Proteinase Inhibitors Evolving Alone or Combined with WAP Domains
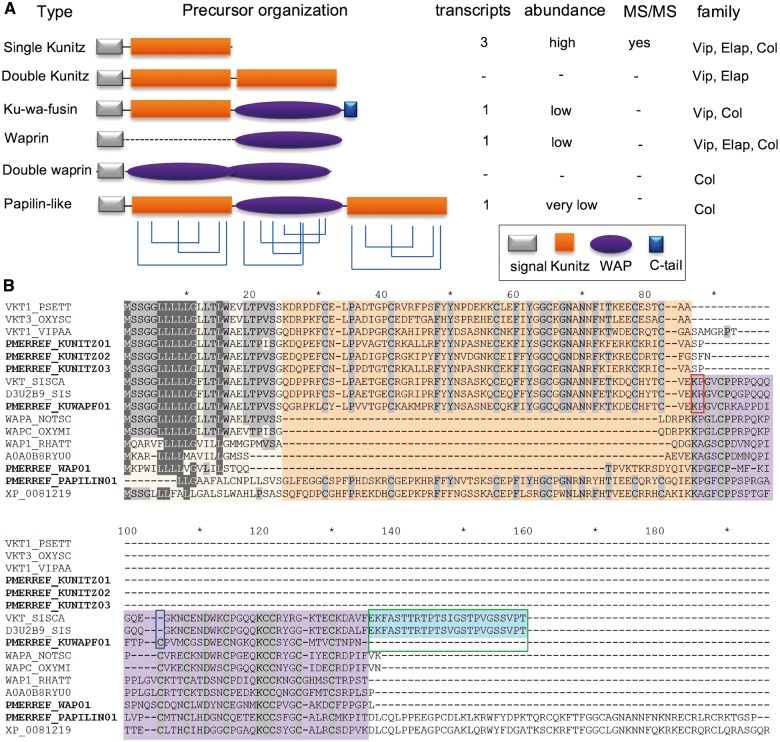
The most expressed group of transcripts in the P. mertensi venom gland, considering the RPKM value, encodes Kunitz-type proteins (table 1), which usually function as serine protease inhibitors. There were three distinct transcripts encoding classical venom Kunitz-type protein precursors, which have a signal peptide and one copy of the BPTI/Kunitz inhibitor domain containing an Arg residue after the second Cys (fig. 4). This residue was shown to be the S1 site necessary for plasmin and trypsin inhibition in the Textilinins-1 and -2 from the Australian elapid Pseudonaja textilis. Its substitution by nonpositively charged residues in naturally occurring isoforms of many toxin is a marker of chymotrypsin inhibitors or noninhibitory Kunitz-type proteins that act as neurotoxin active on Ca2+ channels (Schweitz et al. 1994; Filippovich et al. 2002; Millers et al. 2009). This fact indicates that these P. mertensi proteins are more likely to function as true protease inhibitors than to act as neurotoxins.
Fig. 4.—
Proteins containing Kunitz-type and Wap-type domains in P. mertensi and in other snakes. (A) Schematic organization of domains arrangement in known snake proteins suggested as toxins and their occurrence in the present work. (B) Alignment of the proteins found in P. mertensi (PMERREF_) with related proteins from public databases, emphasizing the different domains by colored backgrounds. The red box indicates potential cleavage site between kunitz and wap in ku-wap-fusin, the blue box indicates the cysteine residue present in P. mertensi ku-wap-fusin and absent in other species and the green box indicates the C-terminal tail of ku-wap-fusin from other species that is absent in P. mertensi.
There were also other transcripts coding for larger proteins containing Kunitz domains in addition to a WAP (whey acidic protein-type four-disulfide core) domain in the P. mertensi venom gland transcriptome. One of them is similar to a papilin-like protein, which is conserved in reptiles, and contains a WAP domain surrounded by two Kunitz domains (Fessler et al. 2004); the other is similar to ku-wap-fusin toxins described in some viper venoms (Pahari et al. 2007) and more recently in a colubrid (McGivern et al. 2014), in which a N-terminal Kunitz domain is followed by a C-terminal WAP domain and a short C-terminal tail. Nevertheless, we also found waprin-like toxins (Fry et al. 2008) in the transcriptome of P. mertensi venom glands, in which WAP domains occur solely (without the Kunitz domain). Figure 4 illustrates a comparison of the structural arrangement of these domains in proteins expressed in other snake venom glands with those of the proteins found in this work. It is interesting to observe that differently from the original viperid ku-wap-fusin proteins, the P. mertensi one retains the second Cys residues of the WAP domain (fig. 4B), while it lacks the acquired extra C-terminal tail and the dibasic residues in the linker region, which is believed to provide cleavage between the two domains (Doley et al. 2010). It thus seems that this colubrid sequence is a more bona fide combination of the two domains, which may occur together in the mature protein. Nevertheless, this protein was not detected in the venom proteome.
The evolutionary relationship between the toxins having these two domain types (Kunitz-type and WAP), alone or conjugated, was investigated by St Pierre et al. (2008) and Doley et al. (2010), who proposed that they arose from a common ancestral gene (Kunitz-like) with subsequent duplication and diversification through a recombination of the flanking introns. This process led to the insertion of WAP domain in ku-wap-fusin followed by the loss of the Kunitz domain in the generation of the waprin gene (Doley et al. 2010). From the transcriptomic analysis of P. mertensi venom glands it is now possible to add two players that may improve this story: a nonviper ku-wap-fusin, holding more conserved features from the WAP domains, and a papilin-like protein, containing a Kunitz domain after the WAP domain. At this point, without any genomic information about the gene structure of these transcripts, it is not possible to adequately revisit the evolutionary process that resulted in the emergence of the venom proteins, however, our findings illustrate the plasticity of domain recruitments and rearrangements in snake venom toxins.
A Trend in Metalloproteinases Evolution toward Structure Simplification
SVMPs are the most representative class of toxin, in terms of diversity, and are also highly abundant in P. mertensi venom. We were able to identify 12 different SVMP transcripts, which were all confirmed as present in the venom (table 1). All but one was identified as P-III class SVMP, that is, they possess a catalytic domain followed by disintegrin-like and cysteine-rich domains. This is in agreement with the current hypothesis for the evolution of this toxin class, which assumes that the acquisition of P-I and P-II types of enzymes occurred within the Viperidae family, after its differentiation from other snake clades (Fry et al. 2008; Casewell et al. 2011). This process is supposed to have happened through mutations in key residues and losses of some introns (Bazaa et al. 2007), which occurred in several independent events on different species within the Viperidae family (Casewell et al. 2011).
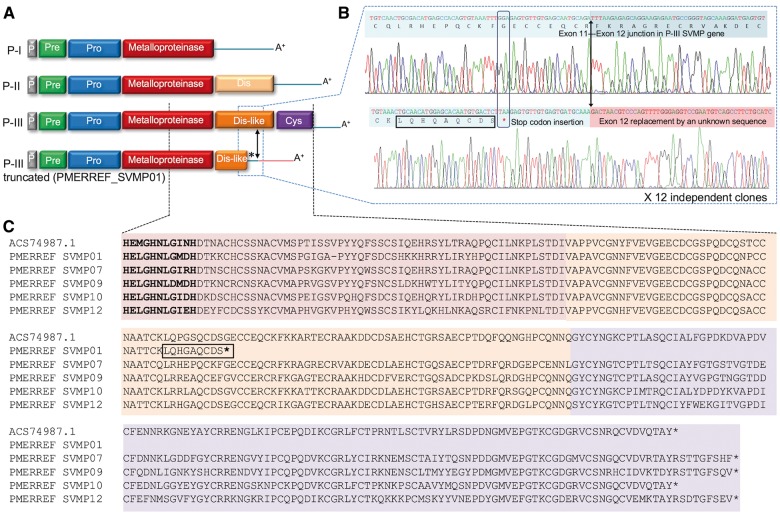
Our results, however, showed a notable exception to the canonical domain organization of the P-III class SVMPs, since one transcript (PMERREF_SVMP01) encodes a P-III SVMP truncated at the middle of the disintegrin-like domain (fig. 5). This truncation was caused by the existence of a stop codon after the 58th residue of the disintegrin-like domain, and the replacement of the remaining 3′-UTR portion of the cDNA, shortly after this stop codon, by a sequence of unknown origin lacking similarity to any sequence in the databases (fig. 5B). This unknown sequence begins shortly after the position of the 11th intron–exon boundary described for a P-III SVMP gene (Sanz et al. 2012), thus indicating that the exon 11 of P-III SVMP gene was substituted with a new sequence that generated a new 3′-UTR.
Fig. 5.—
SVMPs in P. mertensi venom. (A) Schematic organization of domains in SVMPs precursor types, emphasizing the P-III type commonly observed in colubrids and the truncated P-III type found in this work. (B) Sanger sequencing electropherograms of a normal and a truncated P-III SVMPs found in this work. Note: 1) a mutation in a coding codon (in vertical box) generating a stop codon (marked with *) in the truncated precursor; 2) the differences in the sequences of the truncated precursor (denoted by red background) when compared to a nontruncated precursor, exactly after the position known to be the Exon11-Exon 12 junction in P-III SVMP gene (marked by a vertical arrow); 3) the LQHGAQCDS peptide (in horizontal box) detected in the venom proteome, which is only present in the truncated precursor. (C) Alignment of part of the catalytic domain and disintegrin-like domain of some P. mertensi SVMPs and with the first described colubrid SVMP (ACS74987.1, from Philodryas olfersi).
Since our finding of the truncated P-III transcript could initially be interpreted as a possible nonfunctional mRNA, we carried out a more detailed investigation to evaluate its significance. Initially, we verified that the contig containing this feature (PMERREF_SVMP01) was highly expressed and the region containing the truncation was highly covered by the Illumina reads (data not shown), which is a good indication that it is a truly expressed transcript rather than a rare one or an artifact of the assembly. To confirm this, we checked for Sanger-based ESTs covering this region and identified 15 independent clones (from the primary and unamplified library) confirming this sequence (one clone exemplified in fig. 5B). We also designed specific primers and PCR amplified the primary cDNA pool to obtain a single full-length cDNA clone coding for this product, which again confirmed the originally assembled contig. In order to check if the specific protein product of this transcript existed in the venom, we carefully analyzed the peptides sequenced by MS/MS and found seven unique peptides matching only this predicted protein. Particularly, one of these peptides (LQHGAQCDS) is a specific marker of the existence of this protein, since it ends in the last residue of the shortened protein encoded by transcript PMERREF_SVMP01 (before the inserted stop codon) (fig. 5C and supplementary table S3, Supplementary Material online). Indeed, since trypsin was used during sample preparation, this semitryptic peptide could only be generated from the C-terminal portion of this protein (whose precursor terminates with this unusual stop codon position). None of the normal (nontruncated) SVMPs of P. mertensi could generate a tryptic peptide ending at this position since they do not have Arg or Lys residues in this region. Taken together, the confirmation of the sequence of this cDNA, its the high expression level in both the transcriptome and the proteome (fig. 3B) and the sequencing of its unique peptides, including the unusual C-terminal region, indicate a functional gene encoding a new type of SVMP in this venom.
An evolutionary significance of this finding is that it suggests a convergent evolution of multidomain SVMP genes in two different snake families toward a similar simplified form. Although in viperids it may have occurred through a domain loss process associated with intron losses (Bazaa et al. 2007), in the P. mertesi case it seems to be caused by the insertion of a new exon of an unknown origin, shortening the original protein sequence. Since viperid P-I SVMPs, which lack noncatalytic domains, are generally less hemorrhagic than P-III SVMPs (Moura-Da-Silva et al. 2009), it was hypothesized that, in viperids, evolution is convergently producing multiple independent generations of P-I class SVMPs, perhaps as a response to dietary selection pressures for different prey types (Casewell et al. 2011). The extrapolation of this idea to colubrids depends first on knowing if this is an apotypic character of this species or a synapotypic character shared with more colubrids, which remains to be elucidated when data from related species become available.
It is interesting to observe that a similar case of a trend toward a simplification in a colubrid protease structure was noted with the svMMPs, a different type of metalloproteinase recently shown to be a new toxin component exclusive of colubrid venoms (Ching et al. 2012). They can occur as a MMP-9 like structure (including hemopexin and fibronectin domains) or as a derived form such as those from T. strigatus venom where the ancillary domains were lost but the functional activity of the shortened protein was kept (Ching et al. 2012). In this study, we found a svMMP transcript encoding a protein with the classical MMP-9 arrangement, that is, including one hemopexin and three fibronectin domains, whose sequence was confirmed by proteomic analysis with a high spectral count in the venom. (PMERREF_MMP01, table 1) The gene for this cDNA thus represents the presimplification state, whereas the T. strigatus one represents the apotypic (derived) character, extending the notion that the evolutionary trend through simplification of metalloproteinases (svMMP or SVMP) may happen in a species-specific manner.
An Acid Lipase as True Venom Component
The transcriptomic analysis revealed an unexpected and highly expressed transcript coding for a lysosomal acid lipase-like protein. The expression level dictated by 6,931 reads (RPKM = 8,582) was comparable to that of the most abundant SVMPs and Kunitz-type toxins, and far above the major venom gland house-keeping genes, such as protein disulfide-isomerase and calreticulin, which use to be the most expressed nonvenom transcripts in venom gland transcriptomes (Junqueira-de-Azevedo and Ho 2002). The EST analysis also indicated the high expression of the transcript coding for the acid lipase-like protein and confirmed the sequence of the assembled contig with an overlap of 100% and an average coverage of five times. One of the EST clones was resequenced showing a full-length cDNA of 2,055 bp with a 1,200 bp open reading frame (ORF) and a 3′-UTR containing a classical poly adenylation signal and a poly A+ tail (supplementary fig. S3, Supplementary Material online). The encoded protein precursor, named PMERREF_LIPA01, contains 400 amino acids, with a predicted signal peptide of 17 residues followed by a mature protein chain of a 383 amino acids. The molecular mass of the mature protein was predicted to be 39.9 kDa but it includes three potential N-glycosylation sites that may increase the observed protein molecular mass.
Comparison of the PMERREF_LIPA01 precursor sequence against confirmed and curated proteins form the UniProt database revealed best similarity (∼66% identity) to mammalian lysosomal acid lipases (cholesteryl ester hydrolase—EC: 3.1.1.13). However, a broader search against the TSA_nr database (containing predicted proteins derived from TSA) revealed several predicted Acid Lipases from Squamate reptiles, including snakes, which share around 80% of sequence identity. In fact, an acid lipase transcript was firstly detected in the EST based transcriptome of the African viper Echis coloratus (Casewell et al. 2009), summing 2% of the venom-related transcripts, whereas it was not detected in the three other species of the same genus investigated in that study. The authors proposed that it could represent a new toxin type, however, since the work did not involve protein analysis, the protein was not found in the venom. In the venom gland transcriptome of the South American Elapidae Micrurus altirostris, a partial transcript for an acid lipase representing 0.4% of venom-related transcripts was also identified and, in this case, one single peptide was detected by MS/MS analysis of the HPLC-fractionated venom (Correa-Netto et al. 2011). Some of the other snake species, including two colubrids, for which transcripts coding for acid lipase were available in TSA also had their crude venoms analyzed by MS/MS but none of them revealed this protein as a venom component. The absence of protein detection in these species suggests that the acid lipase may not be regularly secreted to their venoms or it is secreted at very low levels, making difficult to identify it at the protein level.
The existence of this protein, among many other protein types, in venoms was in fact questioned by Hargreaves et al. (2014), when they put in check the recruitment of several proposed minor snake toxins because of their relative widespread expression in nonvenom tissues. The authors concluded that the acid lipase should not be a toxin due to its coexpression in the other tissues of Echis colloratus tested in the same study, although its expression level in the venom gland of that species was much higher than in the other tissues. They preferred to associate the higher expression of acid lipase in the venom glands to a possible role in the high cell turnover of this tissue than to a potential function as a toxin. They also ignored other previous evidences in favor of this protein as a venom component, such as its elevated expression levels in M. altirostris venom gland transcriptome and its detection in the venom proteome of this species and they did not look further for the presence of this protein in the venoms of the species studied. Thus, the arguable proposition of acid lipase as a new toxin would benefit from a clear demonstration of its occurrence in a snake venom secretion.
Here, the venom proteomic characterization by in-solution trypsin digestion and MS/MS analysis provided the identification of 18 unique peptides matching the mature sequence of the acid lipase, corresponding to a coverage of approximately 71% of the protein sequence (table 1 and supplementary table S3, Supplementary Material online). The spectral count of 42 for unique peptides of this protein indicates that it is relatively abundant in the venom (table 1, fig. 3B). The identification of a 2-DE spot of approximately 55 kDa provided another evidence of the presence of the acid lipase in the venom (fig. 3A).
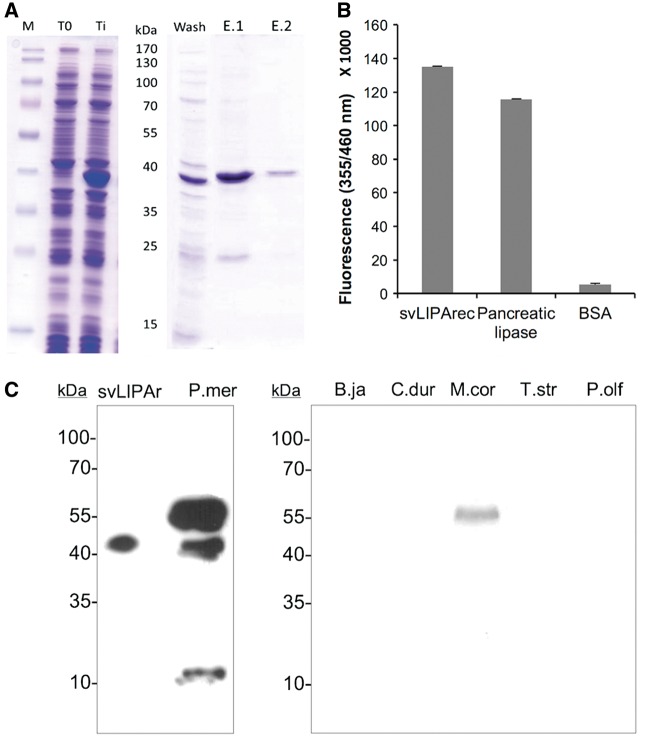
In order to get a proteomic-independent confirmation of its occurrence in the venom of P. mertensi (and other snakes) and to further characterize it, we first obtained a recombinant acid lipase in E. coli (fig. 6A). The purified protein was refolded in solution and its enzymatic activity was tested. The 4-MUO assay showed the recombinant protein to be active, producing a fluorescence emission comparable to that of control human acid lipase (fig. 6B). Using a colorimetric assay to measure glycerol release, the activity was determined as 0.165 milliunits/ml, corresponding to a specific activity of 1.65 nmol.min − 1.−1.mg − 1. The recombinant protein was also used to immunize mice to produce a specific antiserum. After the immunization scheme, the mouse serum was used to immunostain the protein in a pooled preparation of venom from P. mertensi individuals and of other snake species. A marked band of 55 kDa was observed in the venom of P. mertensi (the same molecular mass of the spot identified as an acid lipase in the 2-DE analysis), followed by a less reactive one of approximately 39 kDa (the expected mass for the nonglycosylated protein) and a weakly immunostained band of approximately 10 kDa that may be the result of partial protein degradation (fig. 6). Interestingly a protein band of the same mass (∼55 kDa) was detected in the venom of Micrurus corallinus (Elapidae), a species closely related to M. altitrostris, the one we first obtained proteomic evidence of the occurrence of acid lipase in the venom (Correa-Netto et al. 2011).
Fig. 6.—
Production and analysis of a recombinant acid lipase. (A) The mature portion of the cDNA of LIPA was subcloned in a bacterial expression vector, transformed in E. coli BL21(DE3) pLys and induced by IPTG. M: molecular marker, T0: bacterial extract previously to induction, Ti: bacterial extract after 3h induction of expression by IPTG. After solubilizing the inclusion bodies in the presence of 6M urea, it was bound into Ni2+ affinity resin, washed with 5 mM imidazole (Wash) and eluted in two volumes of 1M imidazole (E.1) and (E.2). (B) Enzymatic activity of recombinant LIPA, compared to a commercial bovine pancreatic lipase and BSA as a negative control. (C) After immunizing mice with the purified recombinant LIPA, the raised antiserum was able to recognize the recombinant protein (svLIPAr) and the wild protein in the venom of P. mertensi (P.mer) and also in the venom of Micrurus corallinus (M.cor) but not in other venoms such as Bothrops jararaca (B.ja), Crotalus durissus terrificus (C.dur), Thamnodynastes strigatus (T.str), and Philodryas olfersi (P.olf).
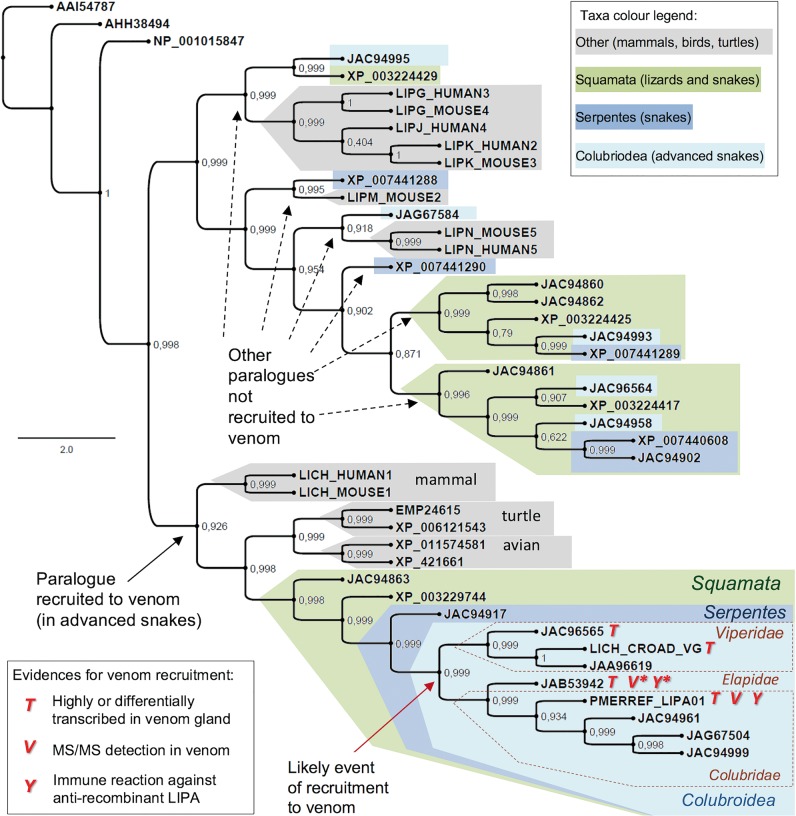
Considering the existence of various new predicted acid lipase genes (or cDNAs, or proteins) in many squamate reptiles, including the occurrence of multiple “isoforms” (possible paralogues) in the same species with different tissue expression (Hargreaves et al. 2014), it is interesting to predict the evolutionary relationship between all of them and to hypothesize about the origin of venom acid lipases. We thus performed a phylogenetic analysis with acid lipase-related sequences from all reptiles in the context of similar orthologues from other vertebrates. The phylogenetic tree (fig. 7) indicates that there are up to four paralogues of acid lipase in the squamata reptiles. Although the snake paralogue genes recognized as equally expressed in nonvenom and venom producing tissues (Hargreaves et al. 2014) form several independent clades containing orthologues from Anolis lizard and other vertebrates, the snake paralogue transcripts with a differential (but not exclusive) gene expression in the venom glands form a single robust clade. In other words, all the acid lipases evidenced or suggested as venom components, as previously discussed, form a unique monophyletic group distinct from the physiological (nonvenom) sequences. This includes P. mertensi (this work), Ech. colloratus (Casewell et al. 2009), and Micrurus spp sequences (note: since M. altirostris had only a partial sequence available, we included in the phylogeny a Micrurus fulvius sequence (Margres et al. 2013). The tree topology was actually in a general agreement with that elaborated by Hargreaves et al. (2014), though we recognize the clade containing the venom gland differentially expressed transcripts as a result of a recruitment to the venom. A calculation of rates of nonsynonymous to synonymous substitutions among nucleotide sequences in the “venom” clade of lipases indicated that these proteins are not under diversifying selection (average dn/ds = 0.4274), as observed in some but not all toxin classes.
Fig. 7.—
Bayesian tree of acid lipase proteins from vertebrates focusing on the existence of various paralogues in squamata reptiles with a possible recruitment of one paralogue to venom in advanced snakes. The protein sequences are referred by their accession numbers. *JAB53942 is from M. fulvius but the proteomic evidence was obtained from similar sequence from M. altirostris (Correa-Netto et al. 2011) and the immunochemical evidence is from M. corallinus venom (this work). Numbers on the nodes represent Bayesian posterior probabilities. Bayesian inferences was obtained by MrBayes software version 3.1.1 30, utilizing 1 × 106 numbers of generations for the Markov chain Monte Carlo algorithm in four chains, sampled every 100 and using the aamodelpr prior set to mixed models.
We thus conclude that acid lipase is most likely a venom constituent considering: 1) the reliable identification of this protein in the venom P. mertensi by proteomic and immunochemical analyses (this work); 2) the presence of lipase activity in the venom of P. mertensi (this work); 3) its previous suggestive identification in the venom of the Elapidae M. corallinus (Correa-Netto et al. 2011); 4) its high representation in P. mertensi venom gland transcriptome (this work); 5) its wide transcription in the oral glands of other snakes (including a Viperidae, Elapidae, and Colubridae), where expression levels are generally high or higher than in other tissues (Casewell et al. 2009; Hargreaves et al. 2014); and 6) the predicted common origin of the venom gene in all the suggested species (this work). Thus we propose the name svLIPA for this venom component, avoiding the use of the term “lysosomal” because of the evidences of its secretion to the venom solution rather than its sorting to the organelle. The confirmation of this venom component as a toxin, and not only a venom component, depends on more complex tests about its biological function and pathophysiological effects, especially in ecological context of the species. Unfortunately, a strong limitation for such investigations is the low amount of venom obtained from colubrid specimens, making the use of the recombinant protein a useful alternative for future investigations.
Conclusion
In order to obtain a complete picture of the venom from P. mertensi, we adopted complementary omics and functional approaches involving: 1) the evaluation of some of the enzymatic activities commonly observed in snake venoms, 2) the generation of a de novo NGS transcriptome based on Illumina technology, 3) the cloning and sequencing of random ESTs to validate transcript assemblies, 4) the primer-walking sequencing of specific full-length toxin cDNAs, 5) the proteomic analysis of the venom using the venom gland transcriptome as a reference database, and 6) the production of a recombinant protein to characterize a possible new toxin.
This combined approach allowed us to trace the components of this venom, observing an enzyme-rich secretion resembling a Viperidae venom, however with an unusual high content of Kunitz-type inhibitors that may play a role in the hemostatic disturbance of the prey. Nevertheless, the presence in the venom of three-finger toxins, which are usually found in the venom of Elapidae snakes, the presence of svMMPs, a colubrid-specific protein type, and the identification of a novel acid lipase that may be a new toxin with an unknown role, underscore the complexity of P. mertensi venom and evidence its unique characteristics.
The finding of the acid lipase in the venom could also be considered in the context of the debate about a single versus a multiple origin of reptilian venom systems (Fry et al. 2006; Hargreaves et al. 2014). Although far from providing a considerable contribution to solve this question, we demonstrated that one minor putative component (acid lipase) previously suggested (and contested) in some species could actually be a true venom component in other species. Thus, some care should be taken in assuming a nonvenom related function of a certain protein simply because of its (or its complement) coexpression in other tissues. The investigation of the presence of the protein in the actual secretion, claimed to be venom, seems to be a critical requirement to foster this discussion.
Moreover, a detailed view of the structural features of the toxins investigated here suggested that some evolutionary tendencies observed in more studied venoms from medically relevant families also occur in colubrids. As an example of protein simplification, domain losses were observed in the two types of colubrid metalloproteinases (SVMP and svMMPs), paralleling the evolution of SVMPs in Viperidae. As an example of protein rearrangement, distinct domains such as wap and kunitz-type were found rearranged in multiple configurations across the snake families. These findings reinforce the importance of investigating colubrid venoms to achieve a more complete picture of the evolutionary history of snake toxins.
Supplementary Material
Supplementary figures S1–S3 and tables S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to Dr Maria de Fátima D. Furtado for calling our attention to the venom of P. mertensi and helping in obtaining the first samples used. This work was supported by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (13/07467-1) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (fellowship 387422).
Literature Cited
- Aird SD, et al. 2013. Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophis okinavensis and Protobothrops flavoviridis). BMC Genomics 14:790.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aird SD, et al. 2015. Snake venoms are integrated systems, but abundant venom proteins evolve more rapidly. BMC Genomics 16:647.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazaa A, et al. 2007. Loss of introns along the evolutionary diversification pathway of snake venom disintegrins evidenced by sequence analysis of genomic DNA from Macrovipera lebetina transmediterranea and Echis ocellatus. J Mol Evol. 64:261–271. [DOI] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 72:248–254. [DOI] [PubMed] [Google Scholar]
- Brodie ED., 3rd 1993. Differential avoidance of coral snake banded patterns by free-ranging avian predators in Costa Rica. Evolution 47:227–235. [DOI] [PubMed] [Google Scholar]
- Casewell NR, Harrison RA, Wüster W, Wagstaff SC. 2009. Comparative venom gland transcriptome surveys of the saw-scaled vipers (Viperidae: Echis) reveal substantial intra-family gene diversity and novel venom transcripts. BMC Genomics 10:564.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casewell NR, Wagstaff SC, Harrison RA, Renjifo C, Wuster W. 2011. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol Biol Evol. 28:2637–2649. [DOI] [PubMed] [Google Scholar]
- Chapeaurouge A, et al. 2015. Interrogating the Venom of the Viperid Snake Sistrurus catenatus edwardsii by a combined approach of electrospray and MALDI mass spectrometry. PLoS One 10(5):e0092091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching AT, et al. 2006. Some aspects of the venom proteome of the Colubridae snake Philodryas olfersii revealed from a Duvernoy's (venom) gland transcriptome. FEBS Lett. 580:4417–4422. [DOI] [PubMed] [Google Scholar]
- Ching AT, et al. 2012. Venomics profiling of Thamnodynastes strigatus unveils matrix metalloproteinases and other novel proteins recruited to the toxin arsenal of rear-fanged snakes. J Proteome Res. 11:1152–1162. [DOI] [PubMed] [Google Scholar]
- Cline M, et al. 2007. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2:2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa-Netto C, et al. 2011. Snake venomics and venom gland transcriptomic analysis of Brazilian coral snakes, Micrurus altirostris and M. corallinus. J Proteomics. 74:1795–1809. [DOI] [PubMed] [Google Scholar]
- Datta G, Tu AT. 1993. Toxicology and Biochemistry of Colubridae Venom. J Toxicol. 12:63–89. [Google Scholar]
- Doley R, Pahari S, Md Abu R, Mackessy SP, Manjunatha K. 2010. The gene structure and evolution of ku-wap-fusin (Kunitz Waprin Fusion Protein), a novel evolutionary intermediate of the kunitz serine protease inhibitors and waprins from Sistrurus catenatus (Massasauga Rattlesnake) venom glands. Open Evol J. 4:31–41. [Google Scholar]
- Durban J, et al. 2011. Profiling the venom gland transcriptomes of Costa Rican snakes by 454 pyrosequencing. BMC Genomics 12:259.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMBL-EBI. 2015. Expression atlas. [cited 2015 Nov] Available from: http://www.ebi.ac.uk/gxa/home.
- Fessler JH, Kramerova I, Kramerov A, Chen Y, Fessler LI. 2004. Papilin, a novel component of basement membranes, in relation to ADAMTS metalloproteases and ECM development. Int J Biochem Cell Biol. 36:1079–1084. [DOI] [PubMed] [Google Scholar]
- Ferlan I, Ferlan A, King T, Russell FE. 1983. Preliminary studies on the venom of colubrid snake Rhabdophis subminatus (Red-necked Keelback). Toxicon 21:570–574. [DOI] [PubMed] [Google Scholar]
- Fernández J, Gutiérrez JM, Calvete JJ, Sanz L, Lomonte B. 2016. Characterization of a novel snake venom component: kazal-type inhibitor-like protein from the arboreal pitviper Bothriechis schlegelii. Biochimie. 125:83–90. [DOI] [PubMed] [Google Scholar]
- Ferrante N. 1956. Turbidimetric measurement of acid mucopolysaccharides and hyaluronidase activity. J Biol Chem. 220:303–306. [PubMed] [Google Scholar]
- Filippovich I, et al. 2002. A family of textilinin genes, two of which encode proteins with antihaemorrhagic properties. Br J Haematol. 119:376–384. [DOI] [PubMed] [Google Scholar]
- Fox RI, Konttinen Y, Fisher A. 2001. Use of muscarinic agonists in the treatment of Sjögren's syndrome. Clin Immunol. 101(3):249–263. [DOI] [PubMed] [Google Scholar]
- Fry BG, et al. 2003. Isolation of a neurotoxin (alpha-colubritoxin) from a nonvenomous colubrid: evidence for early origin of venom in snakes. J Mol Evol. 57:446–452. [DOI] [PubMed] [Google Scholar]
- Fry BG, et al. 2006. Early evolution of the venom system in lizards and snakes. Nature 439:584–588. [DOI] [PubMed] [Google Scholar]
- Fry BG, et al. 2008. Evolution of an arsenal: structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Mol Cell Proteomics. 7:215–246. [DOI] [PubMed] [Google Scholar]
- Furtado MF, Maruyama M, Kamiguti AS, Antonio LC. 1991. Comparative study of nine Bothrops snake venoms from adult female snakes and their offspring. Toxicon 29:219–226. [DOI] [PubMed] [Google Scholar]
- Furtado MF, Travaglia-Cardoso SR, Rocha MM. 2006. Sexual dimorphism in venom of Bothrops jararaca (Serpentes: Viperidae). Toxicon 48:401–410. [DOI] [PubMed] [Google Scholar]
- Gutiérrez JM, Arroyo O, Bolaños R. 1980. Myonecrosis, hemorragia y edema inducidos por el veneno de Bothrops asper en ratón blanco. Toxicon 18:603–610. [DOI] [PubMed] [Google Scholar]
- Hanna SL, Sherman NE, Kinter MT, Goldberg JB. 2000. Comparison of proteins expressed by Pseudomonas aeruginosa strains representing initial and chronic isolates from a cystic fibrosis patient: an analysis by 2-D gel electrophoresis and capillary column liquid chromatography-tandem mass spectrometry. Microbiology 146:2495–2508. [DOI] [PubMed] [Google Scholar]
- Hargreaves AD, Swain MT, Logan DW, Mulley JF. 2014. Testing the Toxicofera: comparative transcriptomics casts doubt on the single, early evolution of the reptile venom system. Toxicon 92:140–156. [DOI] [PubMed] [Google Scholar]
- Greenfield AE, 1988. Immunizing Animals. In: Harlow E, Lane D, editors. Antibodies a laboratory manual. New York: Cold Spring Harbor Laboratory Publications; p. 139–243. [Google Scholar]
- Holzer M, Mackessy SP. 1996. An aqueous endpoint assay of snake venom phospholipase A2. Toxicon 34:1149–1155. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junqueira-de-Azevedo IL, et al. 2006. Lachesis muta (Viperidae) cDNAs reveal diverging pit viper molecules and scaffolds typical of cobra (Elapidae) venoms: implications for snake toxin repertoire evolution. Genetics 173:877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junqueira-de-Azevedo IL, et al. 2015. Venom-related transcripts from Bothrops jararaca tissues provide novel molecular insights into the production and evolution of snake venom. Mol Biol Evol. 32(3):754–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junqueira-de-Azevedo IL, Ho PL. 2002. A survey of gene expression and diversity in the venom glands of the pitviper snake Bothrops insularis through the generation of expressed sequence tags (ESTs). Gene 299:279–291. [DOI] [PubMed] [Google Scholar]
- Kleifeld O, et al. 2011. Identifying and quantifying proteolytic events and the natural N terminome by terminal amine isotopic labeling of substrates. Nat Protoc. 6:1578–1611. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- Lema T. 1978. Relato de um envenenamento por uma cobra não venenosa. Nat Em Revista. 4:62–63. [Google Scholar]
- Lomonte B, et al. 2008. Snake venomics and antivenomics of the arboreal neotropical pitvipers Bothriechis lateralis and Bothriechis schlegelii. J Proteome Res. 7(6):2445–2457. [DOI] [PubMed] [Google Scholar]
- Lomonte B, et al. 2014. Venomics of new world pit vipers: genus-wide comparisons of venom proteomes across Agkistrodon. J Proteomics. 96:103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonte B, Gutiérrez JM. 1983. Proteolytic activity of snake venoms of Costa Rica on casein. Nat Protoc. 6:1578–1611. [PubMed] [Google Scholar]
- Mackessy SP, Sixberry NM, Heyborne WH, Fritts T. 2006. Venom of the Brown Treesnake, Boiga irregularis: ontogenetic shifts and taxa-specific toxicity. Toxicon 5:537–548. [DOI] [PubMed] [Google Scholar]
- Mackinstry DM. 1983. Morphologic evidence of toxic saliva in Colubrid snakes: a checklist of world genera. Herpetol Rev. 14:12–15. [Google Scholar]
- Margres MJ, Aronow K, Loyacano J, Rokyta DR. 2013. The venom-gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC Genomics 14:531.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margres MJ, et al. 2014. Linking the transcriptome and proteome to characterize the venom of the eastern diamondback rattlesnake (Crotalus adamanteus). J Proteomics. 96:145–158. [DOI] [PubMed] [Google Scholar]
- McGivern JJ, et al. 2014. RNA-seq and high-definition mass spectrometry reveal the complex and divergent venoms of two rear-fanged colubrid snakes. BMC Genomics 15:1061.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes MC, Furtado MF, Travaglia-Cardoso SR, Camargo AC, Serrano SM. 2006. Sex-based individual variation of snake venom proteome among eighteen Bothrops jararaca siblings. Toxicon 47:304–312. [DOI] [PubMed] [Google Scholar]
- Millers EK, et al. 2009. Crystal structure of textilinin-1, a Kunitz-type serine protease inhibitor from the venom of the Australian common brown snake (Pseudonaja textilis). FEBS J. 276:3163–3175. [DOI] [PubMed] [Google Scholar]
- Minton SA. 1990. Venomous bites non venomous snakes: a bibliography of Colubrid envenomation. J Wild Med. 1:119–127. [Google Scholar]
- Mortz E, Krogh TN, Vorum H, Gorg A. 2001. Improved silver staining protocols for high sensitivity protein identification using matrix assisted laser desorption/ionization-time of flight analysis. Proteomics 1:1359–1363. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 5:621–628. [DOI] [PubMed] [Google Scholar]
- Moura-Da-Silva AM, Serrano SMT, Fox JW, Gutierrez JM. 2009. Snake venom metalloproteinases. Structure, function and effects on snake bite pathology In: Maria Elena DL, et al. , editors. Animal toxins: state of the art. perspectives in health and biotechnology. Belo Horizonte: UFMG Press; p. 525-546. [Google Scholar]
- OmPraba G, et al. 2010. Identification of a novel family of snake venom proteins Veficolins from Cerberus rynchops using a venom gland transcriptomics and proteomics approach. J Proteome Res. 9:1882–1893. [DOI] [PubMed] [Google Scholar]
- Paes-Leme AF, et al. 2009. Analysis of the subproteomes of proteinases and heparin-binding toxins of eight Bothrops venoms. Proteomics 9:733–745. [DOI] [PubMed] [Google Scholar]
- Pahari S, Mackessy SP, Kini RM. 2007. The venom gland transcriptome of the Desert Massasauga rattlesnake (Sistrurus catenatus edwardsii): towards an understanding of venom composition among advanced snakes (Superfamily Colubroidea). BMC Mol Biol. 8:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichoto ME, Tavares FL, Santoro ML, Mackessy SP. 2012. Venom proteomes of South and North American opisthoglyphous (Colubridae and Dipsadidae) snake species: a preliminary approach to understanding their biological roles. Comp Biochem Physiol Part D Genomics Proteomics. 7:361–369. [DOI] [PubMed] [Google Scholar]
- Petras D, Heiss P, Süssmuth RD, Calvete JJ. 2015. Venom proteomics of indonesian king cobra, Ophiophagus hannah: integrating top-down and bottom-up approaches. J Proteome Res. 14(6):2539–2556. [DOI] [PubMed] [Google Scholar]
- Prado-Franceschi J, Hyslop S. 2002. South American colubrid envenomations. J Toxicol. 21:117–158. [Google Scholar]
- ProteoWizard. 2015. [cited 2015 Nov]. Available from: http://proteowizard.sourceforge.net/.
- Pukrittayakamee S, et al. 1988. The hyaluronidase activities of some Southeast Asian snake venoms. Toxicon 26:629–637. 6. [DOI] [PubMed] [Google Scholar]
- Puorto G, França FOS. 2003. Serpentes não peçonhentas e aspectos clínicos dos acidentes In: Cardoso JLCEA, editor. Animais peçonhentos no Brasil. Biologia, clínica e terapêutica dos acidentes, 1a ed.. São Paulo: Sarvier; p. 108-114. [Google Scholar]
- Pyron RA, Burbrink FT, Wiens JJ. 2013. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol Biol. 13:93.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos CR, Abreu PA, Nascimento AL, Ho PL. 2004. A high-copy T7 Escherichia coli expression vector for the production of recombinant proteins with a minimal N-terminal His-tagged fusion peptide. Braz J Med Biol Res. 37:1103–1109. [DOI] [PubMed] [Google Scholar]
- Reeks T, et al. 2016. Deep venomics of the Pseudonaja genus reveals inter- and intra-specific variation. J Proteomics. 133:20–32. [DOI] [PubMed] [Google Scholar]
- Rokyta DR, Margres MJ, Calvin K. 2015. Post-transcriptional mechanisms contribute little to phenotypic variation in snake venoms. G3 (Bethesda) 5:2375–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta DR, Wray KP, Margres MJ. 2013. The genesis of an exceptionally lethal venom in the timber rattlesnake (Crotalus horridus) revealed through comparative venom-gland transcriptomics. BMC Genomics 14:394.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomão MG, Albolea ABP, Santos SMA. 2003. Colubrid snakebite: a public health problem in Brazil. Herpetolog Rev. 34:307–312. [Google Scholar]
- Sanz L, Harrison RA, Calvete JJ. 2012. First draft of the genomic organization of a PIII-SVMP gene. Toxicon 60:455–469. [DOI] [PubMed] [Google Scholar]
- Sawai Y, Honma M, Kawamura Y, Saki A, Hatsuse M. 2002. Rhabdophis tigrinus in Japan: pathogenesis of envenomation and production of antivenom. J Toxicol. 21:181–201. [Google Scholar]
- Sawaya RJ, Marques OAV, Martins M. 2008. Composição e história natural das serpentes do cerrado de Itirapina, São Paulo, Sudeste do Brasil. Biota Neotrop. 8:127–149. [Google Scholar]
- Schweitz H, et al. 1994. Calcicludine, a venom peptide of the kunitz-type protease inhibitor family, is a potent blocker of high-threshold Ca2+ channels with a high affinity for L-type channels in cerebellar granule neurons. Proc Natl Acad Sci U S A. 91:878–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Pierre L, et al. 2008. Common evolution of waprin and kunitz-like toxin families in Australian venomous snakes. Cell Mol Life Sci. 65:4039–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CH, et al. 2016. Unveiling the elusive and exotic: venomics of the malayan blue coral snake (Calliophis bivirgata flaviceps). J Proteomics. 132:1–12. [DOI] [PubMed] [Google Scholar]
- Tashima AK, et al. 2012. Peptidomics of three Bothrops snake venoms: insights into the molecular diversification of proteomes and peptidomes. Mol Cell Proteomics. 11(11):1245–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Reptile Database. 2016. [cited 2016 Feb]. Available from: www.reptile-database.org.
- Theakston RDG, Reid HA. 1983. Development of simple standard assay procedures for the characterization of snake venoms. Bull World Health Org. 61:949–956. [PMC free article] [PubMed] [Google Scholar]
- Viala VL, et al. 2015. Venomics of the Australian eastern brown snake (Pseudonaja textilis): detection of new venom proteins and splicing variants. Toxicon 107(Pt B):252–265. [DOI] [PubMed] [Google Scholar]
- Wagstaff SC, Sanz L, Juárez P, Harrison RA, Calvete JJ. 2009. Combined snake venomics and venom gland transcriptomic analysis of the ocellated carpet viper, Echis ocellatus. J Proteomics. 71:609–623. [DOI] [PubMed] [Google Scholar]
- Yan C, et al. 2006. Macrophage-specific expression of human lysosomal acid lipase corrects inflammation and pathogenic phenotypes in lal-/- mice. Am J Pathol. 169:916–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelanis A, et al. 2010. Analysis of the ontogenetic variation in the venom proteome/peptidome of Bothrops jararaca reveals different strategies to deal with prey. J Proteome Res. 9:2278–2291. [DOI] [PubMed] [Google Scholar]
- Zybailov B, et al. 2006. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 5:2339–2347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.