Abstract
Intralocus sexual conflict (IASC) prevents males and females from reaching their disparate phenotypic optima and is widespread, but little is known about its genetic underpinnings. In Rhizoglyphus robini, a mite species with alternative male morphs, elevated sexual dimorphism of the armored fighter males (compared to more feminized scramblers males) was previously reported to be associated with increased IASC. Because IASC persists if gene expression patterns are correlated between sexes, we compared gene expression patterns of males and females from the replicate lines selected for increased proportion of fighter or scrambler males (F- and S-lines, respectively). Specifically, we tested the prediction that selection for fighter morph caused correlated changes in gene expression patterns in females. We identified 532 differentially expressed genes (FDR < 0.05) between the F-line and S-line males. Consistent with the prediction, expression levels of these genes also differed between females from respective lines. Thus, significant proportion of genes differentially expressed between sexually selected male phenotypes showed correlated expression levels in females, likely contributing to elevated IASC in F-lines reported in a previous study.
Keywords: ontogenetic conflict, sexual selection, alternative reproductive tactics (ARTs), polyphenism, differential gene expression
Introduction
Sexes often have distinct phenotypic optima for many traits that are expressed in both males and females. If these sex-specific optima cannot be achieved simultaneously due to the constraint of a shared genome, intralocus sexual conflict (IASC) results, characterized by negative fitness correlation between the sexes (Lande 1980). Although the extent to which IASC occurs in natural populations is still debated, it has been observed in multiple species across several taxonomic groups (reviewed in Bonduriansky and Chenoweth 2009; van Doorn GS 2009). IASC has been described to have potential consequences for various evolutionary processes such as speciation, evolution of sex chromosomes, sex determination, regulation of gene expression, sexual selection, sex allocation, and aging (reviewed in Bonduriansky and Chenoweth 2009; van Doorn GS 2009). Recently it was also suggested that IASC may facilitate the maintenance of genetically determined alternative reproductive tactics (ARTs) (Plesnar-Bielak et al. 2014).
Species that exhibit ARTs provide an excellent system to study IASC as they show discontinuous male phenotypes with differing degrees of sexual dimorphism. Typically, the male types that exhibit a dominant and aggressive phenotype are more phenotypically distant from females than are subordinate male phenotypes. Comparative evidence shows that sexually-selected dimorphism is associated with elevated IASC (Cox and Calsbeek 2009). Occurrence of ARTs makes it possible to investigate this association more directly, by manipulating the degree of IASC through changing frequencies of ARTs within populations. Indeed, in horned beetles (Harano et al. 2010) and bulb mites (Plesnar-Bielak et al. 2014), artificial selection for armored and aggressive morphs resulted in decreased female fitness, confirming that sexually selected dimorphism can act as a driver of sexual conflict.
Elevated sexual conflict should select for various mechanisms of its resolution (reviewed in Stewart et al. 2010), including gene duplication and subfunctionalization (Wyman et al. 2012), sex-dependent regulation of gene expression (McIntyre et al. 2006) or genomic imprinting (Day and Bonduriansky 2004). Studies of ARTs in turkeys (Pointer et al. 2013), horned beetles (Snell-Rood et al. 2011), and bulb-mites (Stuglik et al. 2014) have shown that higher phenotypic dimorphism is indeed correlated with higher magnitude of sex bias in gene expression. Yet, elevated IASC associated with more sexually dimorphic male phenotypes suggests that sex bias does not fully resolve sexual conflict (Harano et al. 2010; Plesnar-Bielak et al. 2014). This is likely to result from constraints resulting from males and females sharing most of their genomes. Such constraints were inferred by Griffin et al. (2013), who documented that evolution of sex-bias in gene expression among Drosophilids can be predicted by intersexual correlations of gene expression within species. Thus, while sex bias apparently evolved for some genes expressed in armored morphs (Snell-Rood et al. 2011; Stuglik et al. 2014), many other genes might be constrained, so that the sex bias cannot evolve. Based on this reasoning, it was predicted that genes which changed expression in males in response to selection on male morphs would undergo a correlated expression change in females.
This prediction was tested in the present study in the bulb mite, Rhizoglyphus robini. As in several other species within mite family Acaridae (Radwan et al. 2009), two male morphs are observed in R. robini: aggressive fighters and benign scramblers. The fighter males possess a heavily sclerotized, thickened and sharply terminated third pair of legs used during male–male competition to stab other males, whereas the scrambler males have unmodified legs, similar to those in females. While fighters outcompete scramblers in direct competition for mates (Radwan and Klimas 2001), Plesnar-Bielak et al. (2014) hypothesized that scramblers are still maintained in populations because IASC leads to lower fitness of daughters of fighter males. The present study took advantage of the replicate lines selected by Plesnar-Bielak et al. (2014) for fighter males (F-lines) and scrambler males (S-lines) to compare how they differ in gene expression patterns using RNAseq. It was hypothesized that increased IASC in the F-lines might be a result of correlated changes in gene expression in males and females as a response to selection on male morph. The first aim here was to identify genes showing significant expression difference between males from the F-lines and S-lines. The second aim was to test whether changes in male gene expression resulting from selection on morph cause correlated changes in gene expression in females. Specifically, we tested if the genes showing differential expression between the F-line and S-line males differ in expression significantly between females from respective directions of selection. Finally, we explored functions of these genes.
Materials and Methods
Samples and Sequencing
Samples came from lines selected for increased proportions of either fighter males (F-lines) or scrambler males (S-lines) in four replicates in each direction, with 50 males of the desired morph and 50 females giving rise to each generation (see Plesnar-Bielak et al. 2014 for further details). After about 40 generations, all lines maintained >90% of the desired morph (Plesnar-Bielak et al. 2014).
At generation 96, tritonymphs (last juvenile stage of mites) were isolated from the selection lines (300 per line), adults were collected as they emerged and separated by sex and morph. Selection line females and males (either morph) were respectively paired each with a single scrambler male and a single female from the stock population, for 24 h to allow mating. This was done because these mites typically mate right after emergence, and preventing mating might cause unnatural modification of gene expression profiles. For each replicate line of the two selection regimes, RNA was extracted from samples of females and males, each sample containing approximately 100 individuals of the same sex and age. Samples were collected in a randomized order with respect to selection regimes. RNA was also extracted from pooled samples of females, fighter male morphs and scrambler male morphs from the stock population (100 individuals pooled for each sex and morph), which was used for transcriptome assembly. RNA extractions were done using RNAzol®RT kit (Chomczynski et al. 2010). RNA samples were purified using DNA-free™ kit (Ambion) to eliminate DNA contamination. Quality of extracted RNA was checked using Agilent 2100 Bioanalyzer System. Library and RNA-Sequence samples were prepared using NEXTflex™ Rapid Illumina RNA-Seq library prep kit (Bioo Scientific) and Illumina sequencing was performed at the Medical University of Warsaw producing single end (SE) 100 bp reads.
Quality assessment for the resulting data was done using FastQC (Andrews 2015; http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc) prior to the transcriptome assemblies and analysis. Reads with low quality bases or short read length were removed from subsequent analyses using Skewer (Jiang et al. 2014). Although the bulb mite genome has been sequenced and assembled (M. Konczal, F.A. Kondrashov, F. Camara, A. Vlasova, J. Radwan, unpublished), its annotation is still in progress. Therefore a comprehensive transcriptome database was built using genome-guided and de novo transcriptome assembly with Program to assemble spliced alignments (PASA) (http://pasapipeline.github.io/#A_ComprehensiveTranscriptome).
Assembly and Gene Models
De novo assembly of transcriptome was carried out using reads from selection lines (377.7 mln SE reads) and unselected populations (137.3 mln paired-end reads) sequenced earlier (Stuglik et al. 2014) (data deposited in BioProject portal (PRJNA213807)). Transcripts were reconstructed with the Trinity transcriptome assembler (Grabherr et al. 2011, release 2014-07-17; Haas et al. 2013) with an extra parameter, -PasaFly, which reduced the number of reported isoforms. Additionally, a genome guided assembly of transcriptome was made with Trinity (http://trinityrnaseq.github.io/#genome_guided), using draft genome consisting of 6,599 scaffolds with N50 of 345 kb and half of the genome assembled in 174 largest scaffolds (Konczal et al. in prep.). Reads were first aligned to the draft genome with Genomic Short-read Nucleotide Alignment Program (gsnap) (Wu and Nacu 2010) and separated according to putative loci followed by de novo transcriptome assembly at each genome-defined locus. Transcripts expressed at very low levels are likely to be artefacts (Bhargava et al. 2014); therefore transcripts below 0.1 FPKM were removed from the assemblies before building the comprehensive transcriptome database.
Using the de novo and genome-guided transcriptome assemblies as input, a comprehensive transcriptome database was generated within PASA, a program that infers and annotates gene models for assembled transcripts. The program identifies clusters of aligned transcripts that are likely to represent transcribed genes and the longest transcript from each cluster is selected as a representative of gene model. Protein coding regions for each transcript are inferred by TransDecoder, software bundled with PASA. The comprehensive transcriptome database, containing coding as well as noncoding gene models, was used as the reference for further analysis.
Gene Expression Analysis
Gene expression analysis was performed with the pipeline included in Trinity. For each sample, reads were aligned to the reference comprehensive transcriptome database with Bowtie (Langmead et al. 2009), followed by estimation of transcript abundance using RSEM (Li and Dewey 2011). A minimum expression filter of 5 transcripts per kilobase million (TPM) in at least one sample was used to remove lowly expressed genes from further analysis (https://github.com/trinityrnaseq/trinityrnaseq/wiki/Trinity-Transcript-Quantification#counting-numbers-of-expressed-transcripts-or-genes). This threshold resulted in retention of 25,519 genes, which is close to the number of genes predicted to be protein coding (31,100) based on the ongoing annotation of the R. robini genome (Konczal et al. in prep). Next, edgeR (Robinson et al. 2010) was used to identify and analyze differentially expressed genes in pairwise comparisons between the F-line and S-line males, and between the F-line and S-line females, treating the replicate lines within each selection regime as biological replicates. Fighters and scramblers do not differ in body size and do not appear to invest differentially in testes (Radwan; 1997), and females do not express fighter phenotypes at all, hence differences in expression between the lines (we considered only within-sex differences) were unlikely to be due to differences in body size allometries (Harrison et al. 2015). Nevertheless, to minimize any effects of allometry, we considered only genes with at least a 2-fold change in expression (counted as FPKM) between F-lines and S-lines. False discovery rate (FDR) ≤ 0.05 was used to identify genes showing significant expression difference.
For those genes with significant expression difference between F-line and S-line males, the differences in mean expression levels in F-line and S-line females were also analyzed. If selection on male morph caused correlated expression changes in females, the F-male-biased genes would show higher mean expression in the F-line females than in the S-line females, and vice versa for the S-male-biased genes. Means over four replicate lines per selection regime for log-transformed FPKM values of gene expression in females were analyzed with paired Wilcoxon signed rank test (with means for selection regimes paired across genes) in R Core Team (2015 stats package, version 3.1.3); the nonparametric test was used due to non-normality of distribution.
In addition to the analysis of differential gene expression between F-lines and S-lines described above, considering all genes in females (which should identify both genes with correlated expression between the sexes, and those which evolved in females in response to a difference in their social environment, i.e. the type of males they interacted with), we carried out a second differential expression analysis between F-line and S-line females, taking into account the expression levels in females of only the genes identified above as differentially expressed between F-line and S-line males. Differential expression in females of the genes identified in this second analysis should be due to their strong genetic correlation with expression in respective male morphs. We thus considered these genes as potential candidates for IASC associated with the evolution of fighter morph.
Functional Annotation, Gene Ontology and Enrichment Analysis
For the genes that exhibited significant differential expression between F-lines and S-lines in both males and females, gene ontology (GO) analyses were performed using Blast2GO (ver.3.0) (Conesa et al. 2005). A command-line BLAST+ blastx (Camacho et al. 2009) was performed for the gene model reference transcriptome using the Swissprot database on NCBI FTP site (ftp://ftp.ncbi.nlm.nih.gov/blast/db/) (version 21-03-2016) and the results were imported into Blast2GO (Version 3.0) (Conesa et al. 2005), where mapping and functional annotation were done using default settings. Enrichment analyses were performed in R (Version 3.2.3) (R Core Team 2015) using Bioconductor packages GOstats (Falcon and Gentleman 2007), GSEABase (Morgan et al. 2008), and GOstatsPlus (https://github.com/davfre/GOstatsPlus/blob/master/vignettes/GOstatsPlus.Rmd#use-gostats-to-test-for-over-represented-go-terms). For the genes differentially expressed in the F-line and S-line females, NCBI conserved domain search (Marchler-Bauer and Bryant 2004; http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was performed using the database CDD v3.14-47363 PSSMs (Marchler-Bauer et al. 2015) and the GO categories for these domains were identified with EMBL-EBI InterPro (http://www.ebi.ac.uk/interpro/; Mitchell et al. 2015).
Results
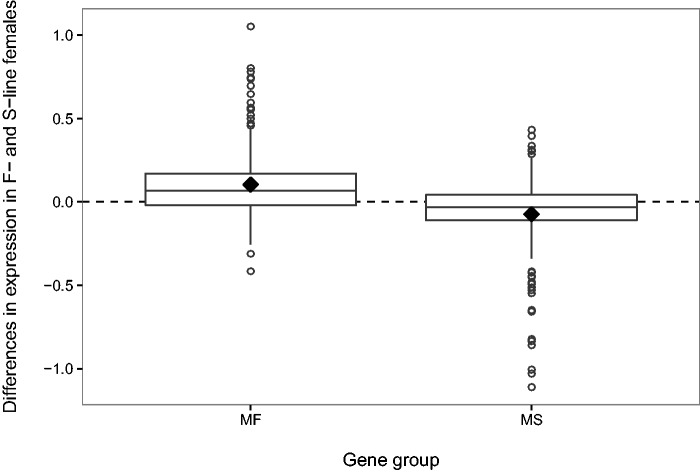
Of the total 54,475 gene models in the reference comprehensive transcriptome database (henceforth referred to as genes for simplicity), 28,956 were excluded from analysis as a result of the minimum expression filter of 5 TPM. 25,519 genes were used for further analysis, of which 532 genes showed significant differential gene expression between males from F-lines and S-lines (Fold change ≥ 2, FDR < 0.05, P < 0.005). Consistent with our hypothesis, expression patterns in females changed in response to selection on male morph. Of the 295 genes upregulated in F-line males, only 267 were expressed in females in at least one replicate line. The mean expression levels for these genes were significantly higher in the females from F-lines than S-lines (V = 27,899, P < 0.001, Wilcoxon signed rank test) (fig. 1). Similarly, for 219 genes (out of the 237 genes upregulated in S-line males) expressed in females in at least one replicate line, the mean expression levels were higher in the females from S-lines than F-lines (V = 8,178, P < 0.001, Wilcoxon signed rank test) (fig. 1).
Fig. 1.—
Boxplot for differences in expression between F-line and S-line females for genes biased in males (positive difference represents higher expression in F-line females). Y-axis depicts difference in mean over replicate lines for log transformed expression values (FPKM) in F-line and S-line females. Diamonds represent means. F-male-biased genes (MF), S-male-biased genes (MS).
When the 532 genes differentially expressed in the F-line and S-line males were analyzed separately for differential expression in F-line and S-line females, 11 genes with significant differential expression (Fold change ≥ 2, FDR < 0.05, P < 0.005) were identified. The five genes with higher relative expression in F-line females were also upregulated in F-line males, and similarly the six genes with higher relative expression in S-line females were also upregulated in S-line males.
Taking into account all 25,519 genes, differential gene expression analysis between F-line and S-line females identified only two genes with a significant expression difference (Fold change ≥ 2, FDR < 0.05, P < 0.005). The gene showing higher relative expression in the F-line females was also present among the 295 genes with upregulated expression in the F-line males. Similarly, the gene with a higher relative expression in S-line females showed a significant upregulation in S-line males. These two genes were also present in the group of 11 genes from the analysis mentioned above.
The enrichment analysis showed that the genes with a higher relative expression in F-line and S-line males were neither over- nor under-represented for any GO categories. The 11 genes differentially expressed between the F-line and S-line males, which also showed significant expression difference between the F-line and S-line females, were not annotated, but conserved domains and GO categories could be identified for 4 of these genes (table 1). One of the genes with a higher relative expression in F-line females showed a conserved domain (Oxidored_q3 super family NADH-ubiquinone/plastoquinone oxidoreductase chain 6) involved in oxidation-reduction process, while another showed a conserved domain (DDE superfamily endonuclease/transposase) associated with DNA cleavage followed by strand transfer reaction. One of the genes upregulated in S-line females showed conserved domains associated with PMT_2 super family, specifically the enzyme dolichyl-phosphate-mannose-protein mannosyltransferase involved in protein glycosylation. Another gene upregulated in S-line females showed a conserved protein kinase domain, which catalyzes protein phosphorylation.
Table 1.
Conserved Domains and GO Categories Identified for Genes Showing Expression Difference Between F-Line and S-Line Females
| Bias | Conserved Domain | GO-ID | Term | Category |
|---|---|---|---|---|
| F-f | Oxidored_q3 super family | GO:0055114 | oxidation-reduction process | P |
| GO:0008137 | NADH dehydrogenase (ubiquinone) activity | F | ||
| F-f | DDE_Tnp_4 | GO:0003676 | Nucleic acid binding | F |
| S-f | PMT_2 super family | GO:0004169 | Dolichyl-phosphate-mannose-protein mannosyltransferase activity | F |
| S-f | Pkinase | GO:0006468 | Protein phosphorylation | P |
| GO:0005524 | ATP binding | F | ||
| GO:0004672 | Protein kinase activity | F |
NOTE.—Genes upregulated in F-line females (F-f), Genes upregulated in S-line females (S-f), Biological process (P) and Molecular function (F).
Discussion
The results from this study showed that divergent selection on male phenotype in a species with alternative male morphs causes changes in patterns of gene expression in both males and females. Thus, genes differing between sexually selected male phenotypes showed correlated expression in females, suggesting that these genes underlie IASC reported to be associated with the fighter phenotype (Plesnar-Bielak et al. 2014). While sex bias in gene expression is considered a way of resolution of this conflict (McIntyre et al. 2006; Stewart et al. 2010), our data indicate that such resolution is not necessarily easily achieved for genes associated with sexually selected male phenotypes, even though these phenotypes are not expressed in females. These results confirm earlier suggestions that evolution of exaggerated sexual traits affects a number of other characters, selecting for gene variants which may cause negative intersexual fitness correlations (Harano et al. 2010; Plesnar-Bielak et al. 2014).
Probably not all of detected differences in gene expression we observed between males of different morphs represented changes in allele frequencies of regulatory sequences or transacting expressed genes resulting from selection we applied. Some of these differentially expressed genes might reflect downstream consequences of morphological and behavioral differences between morphs. However, this argument does not apply to females, which were phenotypically indistinguishable between lines (Plesnar-Bielak et al. 2014). Thus, the fact that these genes showed a correlated difference in expression between F-line and S-line females suggests that a substantial proportion of them reflected repeatable differentiation in allele frequencies between the lines.
In order to understand why sexual conflict persists in populations, sexually antagonistic genes need to be identified, yet it has been done in only few studies. The 11 genes which were upregulated in males and also in females from the same selection regimes can be considered candidates for loci underlying sexual conflict. Although not all 11 could be annotated, conserved domains could be identified for 4 of these genes such that biological functions may be inferred for them. One of the genes with a higher relative expression in both males and females from F-lines showed conserved domains for NADH dehydrogenase enzyme involved in the oxidation–reduction process, while the other for a DNA endonuclease/transposase enzyme. One of the genes upregulated in both males and females from S-lines showed conserved domains for a glycosyltransferase enzyme involved in protein modification while another showed conserved domains associated with protein kinase activity. The nonspecific nature of functions associated with these genes may indicate that these genes are expressed in many tissues, rather than being tissue-specific, highlighting that sexually selected male phenotypes may affect expression levels in both sexes of genes with diverse functions.
Similarly, a previous study in Drosophila melanogaster (Innocenti and Morrow 2010) had demonstrated that sexually antagonistic genes were highly expressed in most tissues, not only in sex-limited reproductive tissues but also in neural tissues and tissues associated with metabolism, nutrient uptake and transport, such as crop, midgut, hindgut fat body, and heart. Many of these sexually antagonistic genes were also associated with diverse categories of metabolic processes (Innocenti and Morrow 2010).
Apart from changes in female expression profiles resulting from correlated response to changes in male expression profiles, there might be changes in expression of other genes responsible for adjusting female physiology to direct effects of changed proportion of male morphs. However, gene expression analysis across all 25,519 genes revealed only two genes with significantly different expression between S and F-line females, and these genes were also upregulated in males from respective lines. This does not necessarily imply that changes in female expression profiles other than those resulting from correlated response to selection on males did not occur; but they may be subtle or tissue-specific.
Overall, the results were consistent with the hypothesis that the higher IASC in the lines selected for the more sexually dimorphic male phenotype is attributable to correlated changes in gene expression patterns in sexually selected male phenotypes and females in response to selection. Eleven candidate genes underlying the conflict were identified. With improved annotation of the R. robini genome in the future, the functions of these genes may be inferred with more certitude which would facilitate further elucidation of the genetic basis of IASC in this species.
Acknowledgments
This work was supported by the Foundation for Polish Science, International PhD Projects Program co-financed by the European Regional Development Fund within the project MPD/2009-3/5, “Environmental stress, population viability and adaptation”, KNOW RNA Research Centre in Poznań 01/KNOW2/2014, and the Jagiellonian University (DS/WBiNoZ/INoS/762/14). The bulb mite genome analyses were supported by HHMI International Early Career Scientist Program (55007424), the MINECO (BFU2012-31329 and BFU2015-68723-P), Spanish Ministry of Economy and Competitiveness Centro de Excelencia Severo Ochoa 2013-2017 grant (SEV-2012-0208), Secretaria d'Universitats i Recerca del Departament d'Economia i Coneixement de la Generalitat’s AGAUR program (2014 SGR 0974), and the European Research Council under the European Union's Seventh Framework Program (FP7/2007-2013, ERC grant agreement (335980_EinME).
Literature Cited
- Andrews S. FastQC a quality control tool for high throughput sequence data [cited 2015 Jul 4]. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bhargava V, Head SR, Ordoukhanian P, Mercola M, Subramaniam S. 2014. Technical variations in low-input RNA-seq methodologies. Sci Rep. 4:3678.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonduriansky R, Chenoweth SF. 2009. Intralocus sexual conflict. Trends Ecol Evol. 24:280–288. [DOI] [PubMed] [Google Scholar]
- Build a Comprehensive Transcriptome Database Using Genome-guided and De novo RNA-Seq Assembly [cited 2015 Jan 7]. Available from: http://pasapipeline.github.io/#A_ComprehensiveTranscriptome.
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Wilfinger W, Kennedy A, Rymaszewski M, Mackey K. 2010. RNAzol®RT: a new single-step method for isolation of RNA. Nat Methods 7:4–5. [Google Scholar]
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Cox RM, Calsbeek R. 2009. Sexually antagonistic selection, sexual dimorphism, and the resolution of intralocus sexual conflict. Am Nat. 173:176–187. [DOI] [PubMed] [Google Scholar]
- Day T, Bonduriansky R. 2004. Intralocus sexual conflict can drive the evolution of genomic imprinting. Genetics 167:1537–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon S, Gentleman R. 2007. Using GOstats to test gene lists for GO term association. Bioinformatics 23:257–258. [DOI] [PubMed] [Google Scholar]
- FTP: BLAST Databases [cited 2016 Mar 21]. Available from: ftp://ftp.ncbi.nlm.nih.gov/blast/db/.
- GOstatsPlus [cited 2016 April 20]. Available from: https://github.com/davfre/GOstatsPlus/blob/master/vignettes/GOstatsPlus.Rmd#use-gostats-to-test-for-over-represented-go-terms.
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin RM, Dean R, Grace JL, Rydén P, Friberg U. 2013. The shared genome is a pervasive constraint on the evolution of sex-biased gene expression. Mol Biol Evol. 30:2168–2176. [DOI] [PubMed] [Google Scholar]
- Haas B. Counting numbers of expressed transcripts or genes [cited 2016 March 3] Available from: https://github.com/trinityrnaseq/trinityrnaseq/wiki/Trinity-Transcript-Quantification#counting-numbers-of-expressed-transcripts-or-genes.
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. 8:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harano T, Okada K, Nakayama S, Miyatake T, Hosken DJ. 2010. Intralocus sexual conflict unresolved by sex-limited trait expression. Curr Biol. 20:2036–2039. [DOI] [PubMed] [Google Scholar]
- Harrison PW, et al. 2015. Sexual selection drives evolution and rapid turnover of male gene expression. Proc Natl Acad Sci U S A. 112:4393–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti P, Morrow EH. 2010. The sexually antagonistic genes of Drosophila melanogaster. PLoS Biol. 8:e1000335.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- InterPro protein sequence analysis & classification.—European Bioinformatics Institute [cited 2016 Mar 18]. Available from: http://www.ebi.ac.uk/interpro/.
- Jiang H, Lei R, Ding SW, Zhu S. 2014. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15:182.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. 1980. Sexual dimorphism, sexual selection, and adaptation in polygenic characters. Evolution 34:292–305. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327–W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res. 43:D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre LM, et al. 2006. Sex-specific expression of alternative transcripts in Drosophila. Genome Biol. 7:R79.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A, et al. 2015. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43:D213–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M, Falcon S, Gentleman R. 2008. GSEABase: Gene set enrichment data structures and methods. R package version 1.32.0. Available from: http://bioconductor.org/packages/release/bioc/html/GSEABase.html.
- NCBI Conserved Domain Search [cited 2016 Mar 18]. Available from: http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi.
- Plesnar-Bielak A, Skrzynecka AM, Miler K, Radwan J. 2014. Selection for alternative male reproductive tactics alters intralocus sexual conflict. Evolution 68:2137–2144. [DOI] [PubMed] [Google Scholar]
- Pointer MA, Harrison PW, Wright AE, Mank JE. 2013. Masculinization of gene expression is associated with exaggeration of male sexual dimorphism. PLoS Genetics 9:e1003697.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- Radwan J. 1997. Sperm precedence in the bulb mite, Rhizoglyphus robini: context-dependent variation. Ethol. Ecol. Evol. 9:373–383. [Google Scholar]
- Radwan J. 2009. Alternative mating tactics in acarid mites. Adv Stud Behav. 39:185–208. [Google Scholar]
- Radwan J, Klimas M. 2001. Male dimorphism in the bulb mite, Rhizoglyphus robini: fighters survive better. Ethol Ecol Evol. 13:69–79. doi: 10.1080/08927014.2001.9522788. [Google Scholar]
- RNA-Seq De novo Assembly Using Trinity [cited 2014 Dec 20]. Available from: http://trinityrnaseq.github.io/#genome_guided.
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell-Rood EC, et al. 2011. Developmental decoupling of alternative phenotypes: insights from the transcriptomes of horn-polyphenic beetles. Evolution 65:231–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart AD, Pischedda A, Rice WR. 2010. Resolving intralocus sexual conflict: genetic mechanisms and time frame. J Hered. 101:S94–S99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuglik MT, Babik W, Prokop Z, Radwan J. 2014. Alternative reproductive tactics and sex‐biased gene expression: the study of the bulb mite transcriptome. Ecol Evol. 4:623–632. [Google Scholar]
- van Doorn GS. 2009. Intralocus sexual conflict. Ann N Y Acad Sci. 1168:52–71. [DOI] [PubMed] [Google Scholar]
- Wu TD, Nacu S. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26:873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman MJ, Cutter AD, Rowe L. 2012. Gene duplication in the evolution of sexual dimorphism. Evolution 66:1556–1566. [DOI] [PubMed] [Google Scholar]