Abstract
Centromere organization has evolved dramatically in one clade of fungi, the Saccharomycotina. These yeasts have lost the ability to make normal eukaryotic heterochromatin with histone H3K9 methylation, which is a major component of pericentromeric regions in other eukaryotes. Following this loss, several different types of centromere emerged, including two types of sequence-defined (“point”) centromeres, and the epigenetically defined “small regional” centromeres of Candida albicans. Here we report that centromeres of the methylotrophic yeast Komagataella phaffii (formerly called Pichia pastoris) are structurally defined. Each of its four centromeres consists of a 2-kb inverted repeat (IR) flanking a 1-kb central core (mid) region. The four centromeres are unrelated in sequence. CenH3 (Cse4) binds strongly to the cores, with a decreasing gradient along the IRs. This mode of organization resembles Schizosaccharomyces pombe centromeres but is much more compact and lacks the extensive flanking heterochromatic otr repeats. Different isolates of K. phaffii show polymorphism for the orientation of the mid regions, due to recombination in the IRs. CEN4 is located within a 138-kb region that changes orientation during mating-type switching, but switching does not induce recombination of centromeric IRs. Our results demonstrate that evolutionary transitions in centromere organization have occurred in multiple yeast clades.
Keywords: yeast, centromeres, biotechnology, inversion polymorphism, Pichia pastoris
Introduction
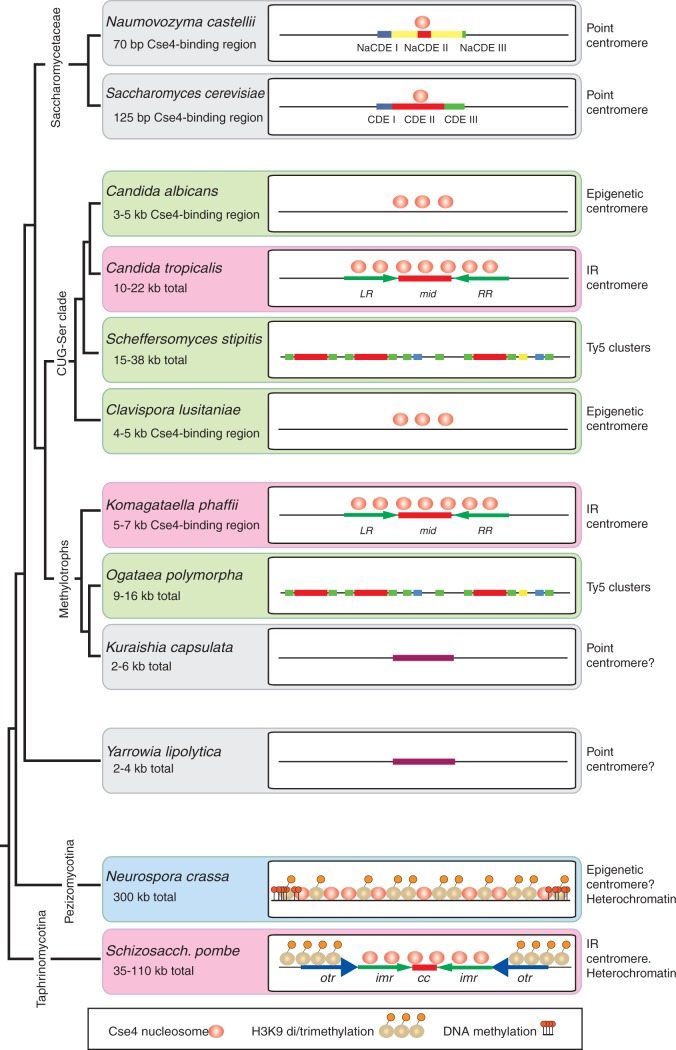
Centromeres are the connection point between the genome and the cytoskeleton during cell division. In most eukaryotes the centromere has a core region that is wrapped around nucleosomes containing the centromeric variant of histone H3 (called CenH3, or Cse4 in yeasts). A large protein complex, the kinetochore, connects these nucleosomes to the spindle microtubules that move the chromosomes during mitosis and meiosis. In most eukaryotes, the core region of the centromere is flanked by extensive arrays of heterochromatin that are repetitive in sequence and largely transcriptionally inert (Pluta et al. 1995; Volpe et al. 2002; Chan and Wong 2012; Hall et al. 2012). The heterochromatin has a characteristic modification, di- or tri-methylation of lysine 9 of histone H3 (H3K9me2/3) that makes it compact. This typical eukaryotic organization is seen in most fungi including basidiomycetes (phylum Basidiomycota), filamentous ascomycetes (subphylum Pezizomycotina) such as Neurospora crassa, and ascomycete yeasts related to Schizosaccharomyces pombe (subphylum Taphrinomycotina) (Allshire 2004; Smith et al. 2012; Janbon et al. 2014). However, an early ancestor of the subphylum Saccharomycotina lost the enzymatic machinery (Swi6/Clr4/Epe1) for making and maintaining H3K9me2/3 heterochromatin (Malik and Henikoff 2009; Allshire and Ekwall 2015; Riley et al. 2016). Yeasts that are descended from this ancestor have centromeres that are very different from other eukaryotes, being smaller and also very variable among species (summarized in fig. 1). Their centromeres are usually categorized into two types, “point centromeres” and “small regional centromeres” (Roy and Sanyal 2011), whereas the term “large regional centromeres” is used for those present in H3K9me2/3-containing fungi and other eukaryotes.
Fig. 1.—
Heterogeneity of ascomycete fungal centromere organization. Schematic (not to scale) showing the structure and epigenetic modification of centromeres across clades of ascomycete fungi. Pink backgrounds indicate IR-containing centromeres; green backgrounds indicate other types of “small regional” centromere. Cse4 nucleosome occupancy is unknown for some species. The species phylogeny is based on Morales et al. (2013) and Riley et al. (2016).
All studied species of the yeast family Saccharomycetaceae have point centromeres. As first identified in Saccharomyces cerevisiae (Fitzgerald-Hayes et al. 1982; Hieter et al. 1985; Hegemann and Fleig 1993), point centromeres are small and have a clear consensus sequence consisting of two short sequence motifs (CDE I and CDE III) to which kinetochore proteins bind, separated by an A + T-rich region (CDE II). Mutations in the conserved (CDE) motifs of point centromeres can result in chromosome instability, and plasmids containing a cloned centromere (CEN) are mitotically stable (Hegemann and Fleig 1993; Kobayashi et al. 2015). In other words, a point centromere’s sequence is necessary and sufficient for centromeric function. Each point centromere of S. cerevisiae is only 125-bp long and binds only a single CenH3-containing nucleosome (Henikoff and Henikoff 2012). As well as being well conserved across all S. cerevisiae chromosomes (Fleig et al. 1995), the CDE I–CDE III consensus is also well conserved among most species of the family Saccharomycetaceae (Gordon et al. 2011) but a notable exception occurs in the genus Naumovozyma (Kobayashi et al. 2015). Although Naumovozyma lies within the family Saccharomycetaceae (fig. 1), and therefore originally had point centromeres similar to those of S. cerevisiae, the two examined species in this genus (N. castellii and N. dairenensis) now have unorthodox point centromeres with a consensus sequence completely unlike that in the rest of the family. As the Naumovozyma centromeres are also not at absolutely conserved chromosomal locations relative to centromeres in other Saccharomycetaceae, it is probable that this new type of point centromere invaded Naumovozyma genomes and displaced the old type (Kobayashi et al. 2015).
The CUG-Ser clade shows wide variation in centromere structures (fig. 1). “Small regional” centromeres were first identified as Cse4-binding regions in Candida albicans and its close relative C. dubliniensis. These centromeres show no consensus sequence or structural features (Sanyal et al. 2004; Padmanabhan et al. 2008). They range from 4 to 18 kb of gene-free DNA, and are thus intermediate in size between point centromeres and “large regional” centromeres. About 3–5 kb of each C. albicans centromere is occupied by Cse4 (Sanyal et al. 2004; Roy and Sanyal 2011). The sequence of a C. albicans centromere appears to be neither necessary nor sufficient for centromeric function. It is not necessary, because if a centromere is deleted a neocentromere can form spontaneously elsewhere on the chromosome (Ketel et al. 2009; Thakur and Sanyal 2013). It is not sufficient, because plasmids or chromosome fragments containing CEN regions do not show mitotic stability when transformed into C. albicans (Baum et al. 2006). Thus, the mechanism that has kept centromeres at a conserved location, both among strains of C. albicans and between C. albicans and C. dubliniensis, is not well understood (Padmanabhan et al. 2008; Thakur and Sanyal 2013). Clavispora (Candida) lusitaniae has similarly nonrepetitive “small regional” centromeres, located in deep troughs of G + C content, with 4–5 kb occupied by Cse4 (Kapoor et al. 2015). However C. tropicalis was recently shown to have 3–6 kb inverted repeat (IR) structures at its centromeres, flanking a central 2–10 kb region that binds Cse4 (Chatterjee et al. 2016). Also in the CUG-Ser clade, the putative centromere regions of Debaryomyces hansenii and Scheffersomyces stipitis contain large clusters of Ty5-like retrotransposon elements within which the precise location and structure of the centromere remains unknown (Lynch et al. 2010).
The Saccharomycetaceae, CUG-Ser and methylotroph clades are the three major clades of Saccharomycotina (fig. 1) (Riley et al. 2016). Outside these but still in subphylum Saccharomycotina, Yarrowia lipolytica has small centromeres with a weak dyad consensus sequence (Vernis et al. 2001; Yamane et al. 2008), located in G + C troughs (Lynch et al. 2010). These centromeres lie in 2–4 kb intergenic regions and are often described as point centromeres, although the consensus sequence is much more poorly conserved among chromosomes than that in Saccharomycetaceae.
The Pezizomycotina and Taphrinomycotina have “large regional” centromeres with H3K9me2/3 heterochromatin, resembling typical eukaryotic centromeres. The best-characterized species in these subphyla are Schizosaccharomyces pombe and Neurospora crassa. The centromeres of Sch. pombe’s three chromosomes are 35–110 kb and contain two types of sequence repeat (Takahashi et al. 1992; Baum et al. 1994; Wood et al. 2002; Allshire and Ekwall 2015). Each centromere has a Cse4-binding domain of ∼15 kb comprising a nonrepetitive central core (cc) flanked by two inverted copies of an innermost repeat (imr) sequence which is chromosome-specific (fig. 1). The pericentromeric regions, marked by H3K9 methylation, consist of 1–9 tandem copies of an outer repeat (otr) sequence, arranged in opposite orientations on the two chromosome arms and therefore extending the overall IR organization of the whole centromere. Whereas the imr repeats are different on each chromosome, all the otr repeats are similar. Variation in the number of otr repeats is the main cause of size difference among the Sch. pombe centromeres. Neurospora centromeres consist of 150–300 kb of A + T-rich DNA, rich in heterogeneous repeat sequences and inactivated transposons (Smith et al. 2011). Unlike Sch. pombe, they contain no large IR structures (Roy and Sanyal 2011). H3K9 trimethylation is found throughout the entire N. crassa centromeric region, and is required for appropriate CenH3 occupancy (Smith et al. 2011).
In this study, we characterize the centromeres of Komagataella phaffii, a haploid species in the methylotrophs clade (fig. 1). The only previous studies of methylotroph centromeres have been in Kuraishia capsulata which has a 200-bp conserved consensus sequence on five of its seven chromosomes (Morales et al. 2013), and Ogataea (Hansenula) polymorpha which has Ty5-like retrotransposon clusters without other obvious conservation of centromere sequence or structure (Ravin et al. 2013; Hanson et al. 2014). Komagataella phaffii is better known by an obsolete name, Pichia pastoris, and is widely used in biotechnology (Kurtzman 2009; Dikicioglu et al. 2014). Komagataella phaffii has a compact genome of only 9 Mb and four chromosomes, with a ribosomal DNA array located at one end of each chromosome, and no known transposable elements (De Schutter et al. 2009; Kuberl et al. 2011). Its centromeres were recently mapped with low resolution by a Hi-C method (Varoquaux et al. 2015). Here, we localize the centromeres with high-resolution ChIP-seq and identify an IR structure at each K. phaffii centromere, reminiscent of Sch. pombe and C. tropicalis but with some significant differences. Thus, following the loss of H3K9me2/3, centromere structure has continued to evolve in diverse directions within each clade of Saccharomycotina, as well as between the clades.
Results
One Transcription Coldspot Per Chromosome in K. phaffii
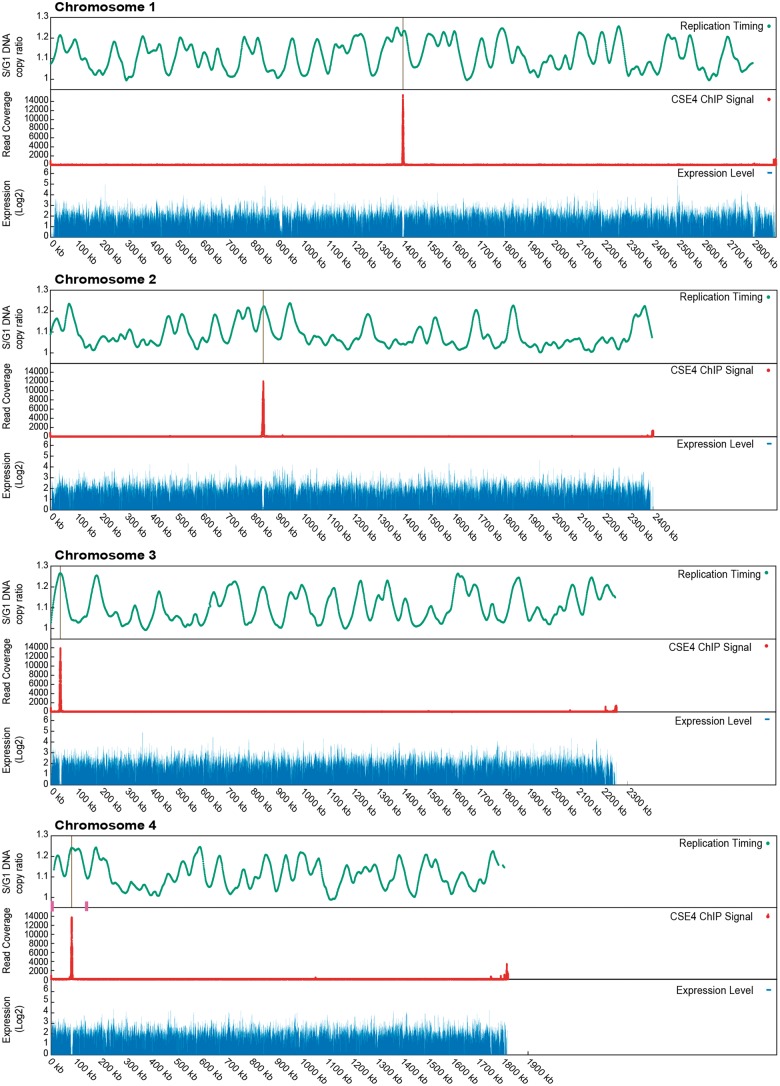
An extensive RNAseq data set for transcriptomes of K. phaffii cells was published by Liang et al. (2012). We mapped these reads to the K. phaffii genome, which in general is very compact with little noncoding DNA (Kuberl et al. 2011), and noticed that there is one large nontranscribed region on each chromosome (figs. 2 and 3). These nontranscribed regions are 6–9 kb long. They were present in both of the growth conditions studied by Liang et al.—glycerol (figs. 2 and 3) and methanol (not shown).
Fig. 2.—
Cse4 occupancy, replication timing and transcription density along the 4 Komagataella phaffii chromosomes. Red, ChIP-seq signal from HA-tagged Cse4. The single large peak on each chromosome identifies the centromere. Weaker signals at the right end of each chromosome coincide with ribosomal DNA repeats. Green, replication data from Liachko et al. (2014) re-mapped to the genome of strain CBS7435. Peaks indicate early replicating regions. Blue, transcription data from RNA-seq experiment of Liang et al. (2012) for cultures grown in glycerol. The gaps in transcription at 900 and 2800 kb on chromosome 1 are due to the very large gene MDN1 and a gap (poly-N region) in the genome sequence, respectively. Magenta boxes indicate the positions of the two MAT loci on chromosome 4 (Hanson et al. 2014).
Fig. 3.—
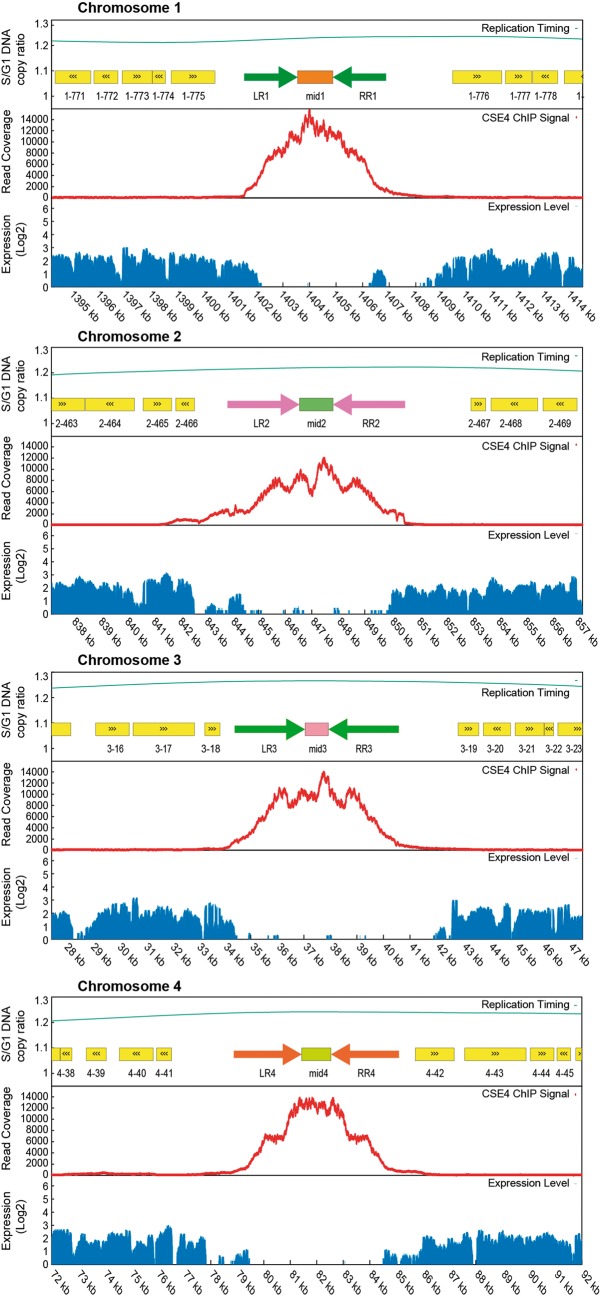
Inverted repeat structure and Cse4 occupancy in centromeric regions. A 20-kb region centered on the highest Cse4 peak is shown for each chromosome. Cse4 occupancy, transcription and replication data are as in figure 2. Paired arrows show the locations of the IRs flanking each mid region. The sequences of LR and RR are virtually identical on each chromosome, but different between chromosomes; mid sequences are different between chromosomes. Yellow boxes indicate annotated protein-coding genes.
Each nontranscribed region contains a simple IR structure, with two almost identical copies of a 2-kb sequence separated by ∼1 kb (fig. 3 and table 1). We hypothesized that these IR-containing regions could be centromeres. They are reasonably close to the previous estimates of centromere locations by Hi-C (Varoquaux et al. 2015), ranging from 0 to 19 kb away (table 1). Independently, Sturmberger et al. (2016) also proposed these IR regions to be candidate centromeres.
Table 1.
Chromosomal Coordinates and Sizes of the Components of the Centromeres of Komagataella phaffii Strain CBS7435
| Chr. | IR Length (bp) | Diffs.a | Mid Length (bp) | Total CEN Length (bp) | Left Repeat (LR) Coordinatesb | Right Repeat (RR) Coordinatesb | Varoquaux et al. |
|
|---|---|---|---|---|---|---|---|---|
| 20 kb callc | 40 kb callc | |||||||
| 1 | 2,012 | 1 | 1,330 | 5,354 | 1,401,537–1,403,548 | 1,404,879–1,406,890 | 1,408,922 | 1,404,619 |
| 2 | 2,699 | 1 | 1,257 | 6,655 | 843,845–846,543 | 847,801–850,499 | 838,077 | 837,858 |
| 3 | 2,650 | 1 | 883 | 6,182 | 34,396–37,045 | 37,929–40,578 | 18,617 | 35,592 |
| 4 | 2,559 | 0 | 1,111 | 6,229 | 78,869–81,427 | 82,539–85,097 | 76,386 | 69,315 |
aNumber of nucleotide differences between the LR and RR.
bCBS7435 genome coordinates of Kuberl et al. (2011). Accession numbers FR839628.1 to FR839631.1.
cHi-C estimates of centromere position by Varoquaux et al. (2015) using two methods. GS115 coordinates were converted to CBS7435 coordinates.
Cse4 ChIP-Seq Confirms IR Regions as Centromeres
Chromatin immunoprecipitation is a technique that has been used widely to locate centromeres in other yeasts (Sanyal et al. 2004; Lefrancois et al. 2013; Hanson et al. 2014; Kobayashi et al. 2015; Chatterjee et al. 2016). To locate the K. phaffii centromeres, we used ChIP-seq to identify sequences associated with centromeric nucleosomes containing CenH3 (Cse4). We constructed a strain of K. phaffii with a haemagglutinin (3xHA) tag at an internal site (Stoler et al. 1995; Wisniewski et al. 2014) of the endogenous CSE4 gene (supplementary fig. S1, Supplementary Material online).
We obtained a single robust ChIP-seq signal for a centromere on each of the four chromosomes (fig. 2). The signal is highest in the nonrepetitive central (mid) region between the two IRs of each chromosome, and declines along the left and right arms of the IR (LR and RR in fig. 3). The region of Cse4 binding coincides almost exactly with the region of no transcription.
The LR and RR of each centromere are virtually identical to each other (table 1), but completely unrelated on each chromosome despite being similar in size. The mid regions of each centromere are also unrelated. There is, however, a small region of similarity between the mid region of CEN1 and the IRs of CEN4 which is visible in a dot matrix plot (supplementary fig. S2, Supplementary Material online). The aligned region has 65% sequence identity over 518 bp. Other than this region, we could not find any conserved sequence motifs in the K. phaffii centromeres using MEME (Tanaka et al. 2014). The base composition of the centromeres is not exceptional: the mid regions range from 37.4–40.7% G + C, and the IRs from 37.3–38.2% G + C; for comparison the whole genome averages 41.1% G + C.
Interestingly, CEN4 is located within a 138-kb section of chromosome 4 whose orientation becomes inverted during mating-type switching (Hanson et al. 2014). In this process, recombination occurs between two identical 2.6 kb sequences (the “outer IR”) that lie in inverted orientations, one near the expressed copy of the MAT genes and the other near the silenced copy. The centromere is in the approximate center of the invertible region, 58 and 79 kb from the two MAT locus outer IRs (fig. 2).
It is also notable that CEN3 and CEN4 are both located close to one end of their chromosomes (37 and 82 kb from the telomere, respectively), making the right arm 60 times longer than the left on chromosome 3, and 22 times longer on chromosome 4. In both cases the centromere is at the opposite end of the chromosome from the rDNA array.
Centromeres Are Early Replicating
Because cell division can only proceed after the centromeres have replicated and kinetochores have been assembled on them, eukaryotic centromeres tend to be early replicating (McCarroll and Fangman 1988; Kim et al. 2003; Koren et al. 2010; Pohl et al. 2012). Liachko et al. (2014) previously mapped replication origins throughout the K. phaffii genome, using a high-throughput sequencing method to compare the copy number of 1-kb windows of the genome in S versus G1 phase of the cell cycle in K. phaffii strain GS115. We re-mapped their data to coordinates for strain CBS7435, whose genome is almost identical to GS115 (Kuberl et al. 2011). The four centromeres are all seen to be located at peaks of early replication (figs. 2 and 3, green lines), though each chromosome contains multiple similarly early peaks, confirming the result from the lower resolution Hi-C study (Varoquaux et al. 2015). The two MAT locus regions on chromosome 4 are both late-replicating (fig. 2).
Polymorphic Inversions at Centromeres
We searched for identical DNA sequence repeats in the K. phaffii genome, and found that there are only seven sequences >1 kb that occur in multiple identical copies. These are the four centromeric IRs; the inner and outer IRs flanking the MAT locus (Hanson et al. 2014); and the ribosomal DNA unit which occurs as an array at the right telomere of each chromosome. We previously found that natural isolates of K. phaffii are polymorphic for the orientation of the 138-kb region of chromosome 4, due to recombination between the MAT locus IRs. Recombination in the 2.6-kb outer IR of the MAT locus is induced during mating-type switching, whereas recombination in the 5.7-kb inner IR is not inducible but appears to occur in nature (Hanson et al. 2014).
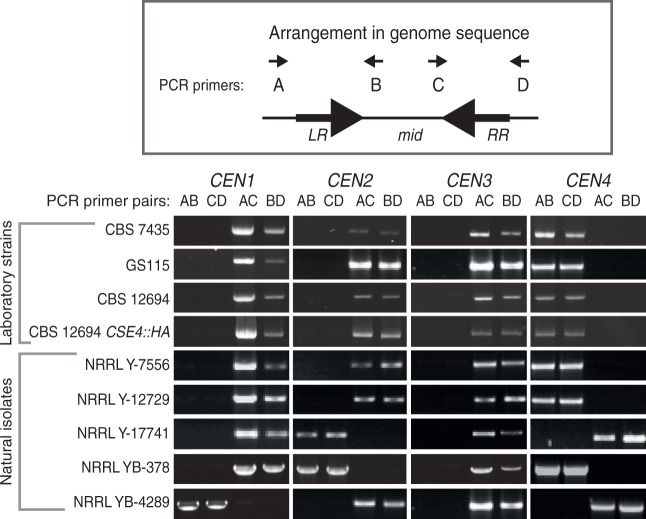
To test whether the orientation of centromeres is polymorphic due to recombination in their IRs, we used orientation-specific PCR assays to determine the arrangement of each mid region relative to the flanking chromosome arms. Among five natural isolates of K. phaffii that we tested, the orientations of three centromeres (CEN1, CEN2 and CEN4) are variable (fig. 4), indicating that ectopic recombination between the centromeric IRs also occurs in nature.
Fig. 4.—
Polymorphism of centromere orientation among Komagataella phaffii strains. Genomic DNAs from four haploid laboratory strains and five natural isolates of K. phaffii from the USDA Agricultural Research Service NRRL collection (Kurtzman 2009) were amplified using orientation-specific PCR assays. For each centromere, two primers (B and C) bind to the mid region and two (A and D) bind to the chromosome arms outside the IR regions. The 16 primer sequences are listed in supplementary table S1, Supplementary Material online. Amplification with the AB and CD primer pairs for a centromere indicates that its mid region is in the same orientation as the reference genome sequence (Kuberl et al. 2011), whereas amplification with the AC and BD pairs indicates inversion of the mid region relative to the chromosome arms. PCR products are the size of the IRs (2.0–2.7 kb; table 1). PCR amplifications used GoTaq (Promega) polymerase for 30 cycles with an annealing temperature of 55 °C.
We found that centromere orientation is conserved between the laboratory strain CBS7435 and three derivatives of this strain (fig. 4). These derivatives are the biotech strain GS115, the ku70 knockout strain CBS12694 (Naatsaari et al. 2012), and the ku70 CSE4::HA strain we used for the ChIP-seq experiment. However, the PCR-determined orientation of CEN1, CEN2, and CEN3 in all the laboratory strains is opposite to that reported in the genome sequences of CBS7435 (Kuberl et al. 2011) and GS115 (De Schutter et al. 2009), probably due to errors in genome assembly.
Because the centromeric IRs are similar in size to the MAT locus IRs, and because CEN4 is located within the 138-kb region that inverts during mating-type switching, we were curious whether induction of mating-type switching might also induce inversion of some centromeres. We induced mating-type switching by growing strain GS115 (MATalpha) in liquid NaKG media (Hanson et al. 2014), then plating for single colonies on YPD, and isolating genomic DNA from multiple cultures each grown from a single colony. Among these cultures, we identified four switched MATa clones by MAT locus orientation-specific PCR (Hanson et al. 2014). We then assayed their centromere orientations. However, the orientations of all four centromeres remained unchanged in the four MATa clones (supplementary fig. S3, Supplementary Material online).
Discussion
Komagataella phaffii, C. tropicalis and Sch. pombe are the only three fungi known to have centromeres with an IR organization, but there are significant differences among these species. They are compared in dotplots in supplementary figures S2–S5, Supplementary Material online. All seven centromeres of C. tropicalis are similar to one another (>60% sequence identity in both their mid and IR regions; supplementary fig. S4, Supplementary Material online) suggesting that they have been homogenized by gene conversion (Chatterjee et al. 2016). In contrast, each centromere of K. phaffii has a unique sequence, except for a small patch of gene conversion between CEN1 and CEN4 (supplementary fig. S2, Supplementary Material online). Similarly, each imr-cc region of Sch. pombe is different, except for a patch of gene conversion (tm region) between cc1 and cc3 (supplementary fig. S5, Supplementary Material online; Takahashi et al. 1992). The centromeres of K. phaffii therefore strongly resemble the structure of the imr-cc regions at the heart of Sch. pombe centromeres (and other Schizosaccharomyces species; Rhind et al. 2012) but they lack the additional nested arrays of otr repeats that extend the IRs and are similar across all Sch. pombe chromosomes (supplementary fig. S5, Supplementary Material online). In both K. phaffii (this study) and Sch. pombe (Steiner et al. 1993), natural isolates are polymorphic for the orientation of the central region relative to the chromosome arms, for at least some centromeres.
In both K. phaffii and Sch. pombe, the CenH3 ChIP-seq signal is maximal in the unique region (mid or cc) and decays along the flanking IRs (imr) (fig. 3; Thakur et al. 2015). In contrast, the signal in C. tropicalis is primarily in the mid regions and to a lesser extent the proximal 1 kb of the IRs (Chatterjee et al. 2016). A possible evolutionary role of the IRs is to specify where CenH3 nucleosomes are placed, preventing drift of the centromere position onto neighboring genes, affecting their transcription. This idea is supported by studies in C. albicans, which showed that neocentromeres are capable of moving locally along DNA under certain conditions (Ketel et al. 2009).
The major difference between the Sch. pombe centromeres and those of K. phaffii and C. tropicalis is the presence of the outer repeats (otr) and their associated heterochromatin. H3K9 methylation at otr is established by bidirectional transcription. The RNAi machinery processes these transcripts to direct the H3K9 methyltransferase (Clr4) to this region of the centromere (Volpe et al. 2002). In contrast, the Saccharomycotina yeasts lack most of the components of this system: there are no outer repeats and no H3K9 methylation proteins (Clr4/Swi6/Epe1) in either K. phaffii or C. tropicalis, and no RNAi apparatus (Ago1/Dcr1) in K. phaffii. As heterochromatic centromeres are the norm in most eukaryotes, we must presume that the Saccharomycotina IR-containing centromeres are derived from heterochromatic ancestors and have become simplified. What is the function of otr? Assays in Sch. pombe found that a partial otr repeat and most of the central domain is sufficient for establishing a functional centromere on naive DNA (Baum et al. 1994; Steiner and Clarke 1994; Ngan and Clarke 1997), but otr and H3K9me2/3 heterochromatin are not required for maintenance of an already active centromere (Baum et al. 1994; Folco et al. 2008). CenH3 when overexpressed can bind specifically to a central core (cc2) region entirely lacking both otr and imr (Catania et al. 2015). These results indicate the central cores of Sch. pombe centromeres have some feature that gives them an innate ability to attract CenH3, but that otr enhances centromere establishment. Once CenH3 chromatin is established, it is capable of self-maintenance and thus is the primary epigenetic mark for kinetochore formation.
The diversity of eukaryotic centromere structures is a consequence of rapid coevolution between centromere DNA and the kinetochore proteins (Padmanabhan et al. 2008; Malik and Henikoff 2009; Bensasson 2011; Kobayashi et al. 2015). Ascomycete centromeres have more structural diversity than can be conveyed by a simple binary distinction between point and regional centromeres. We suggest that it might instead be useful to consider yeast centromeres in terms of whether their CenH3-binding region is sequence-defined, repeat-defined or epigenetically defined. Point centromeres are sequence-defined, meaning that a specific DNA sequence is required for them to function. Repeat-defined centromeres appear to require a particular arrangement of repeat sequences in the CenH3-bound region, such as the IRs in K. phaffii, C. tropicalis and Sch. pombe, but the actual sequence of the repeating units may not be important. Epigenetically defined centromeres are those such as in C. albicans and Cl. lusitaniae, where the only factor determining where new CenH3 nucleosomes are deposited appears to be the presence of CenH3 in the previous generation. It is unclear whether species with Ty5-like retrotransposon clusters at their centromeres constitute a different (fourth) group; detailed CenH3 mapping has not been carried out in any yeasts with this structure, and it is possible that the kinetochore is localized to specific discrete sites and not across the whole retrotransposon cluster. The IR-containing centromere structure may be ancestral in ascomycetes, because it is present in two subphyla of Ascomycota and two families of Saccharomycotina, in which case the epigenetic and Ty5-cluster categories would represent degenerated forms of IR-containing centromeres. In this regard, it is interesting that one centromere (CEN5) of C. albicans has a large IR (Sanyal et al. 2004; Chatterjee et al. 2016) and that neocentromeres in this species frequently contain repeats (Ketel et al. 2009).
Further experiments will be needed to identify the components required for IR centromere establishment and maintenance in K. phaffii, and to determine the role of the IRs in these processes. It will be of interest to know what happens if the IRs are deleted, and whether the current IRs can be replaced by other pairs of identical sequences. In C. tropicalis, the IRs were found to enhance the activity of CEN8 in a plasmid stability assay compared with a mid8 sequence alone, and the IRs of CEN8 could not be replaced by several alternative sequences that were tested (Chatterjee et al. 2016). Komagataella phaffii has several features that could make it an attractive system for further dissection of IR-containing centromeres: it has only four chromosomes; their centromere sequences are all different; and powerful tools for genetic analysis and genome manipulation have already been developed for this species (Naatsaari et al. 2012; Sturmberger et al. 2016; Weninger et al. 2016).
Materials and Methods
Tagging the Endogenous K. phaffii CSE4 Gene
We synthesized a DNA fragment containing the K. phaffii CBS7435 CSE4 gene with a 3xHA (haemagglutinin) tag inserted between amino acids 41 and 42 (supplementary fig. S1, Supplementary Material online). We chose this tagging site based on a multiple alignment of Cse4 proteins that had previously been tagged successfully, after our initial attempt to tag CSE4 at the C-terminus yielded no viable clones. Fusion PCR was used to fuse the synthetic fragment to a kanamycin resistance (kanMX) marker and a short region downstream of the endogenous K. phaffii CSE4 gene (supplementary fig. S1, Supplementary Material online). The cassette was transformed into a ku70 mutant strain (strain CBS12694, a derivative of CBS7435; Naatsaari et al. 2012) by electroporation (Lin-Cereghino et al. 2005). Integrants were selected on YPD containing 200 μg/ml G418. Colonies were screened for correct integration by PCR and verified by sequencing.
ChIP-seq
Chromatin immunoprecipitation followed Hanson et al. (2014). Yeast cells were cultured in 200 ml YPD to log phase and then crosslinked with 1% formaldehyde at room temperature. Crosslinking was stopped by addition of 2.5 M glycine. Crosslinked cells were washed with TBS (100 mM Tris–HCl, pH 7.5, 150 mM NaCl) and resuspended in FA lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% v/v Triton X-100, 0.1% w/v sodium deoxycholate, 0.1% w/v SDS) containing 1 mM PMSF. Cells were lysed with glass beads and chromatin was fragmented by sonication with a Bioruptor Standard (Diagenode). Immunoprecipitation of chromatin fragments was performed with EZview Red Anti-HA Affinity Gel (Sigma–Aldrich) or mouse IgG1 (Cell Signaling Technology) as isotype control. After washes, bound DNA was eluted with HA peptide (Sigma–Aldrich), crosslinks were reversed, and the samples were purified by phenol–chloroform extraction before sequencing. Unpaired Illumina ChIP-seq reads (51 bp) were mapped to the 2011 version of the K. phaffii CBS7435 genome (Kuberl et al. 2011) including the mitochondrial genome, using BWA v0.7.9a-r786 (aln/samse algorithm) with default parameters (Li and Durbin 2009). Samtools v0.1.12a (r862) was used to create sorted and indexed BAM files of the results. Bedtools v2.19.0 was used to create genome coverage Bedgraph files to produce figures 2 and 3.
Replication Data
We converted the normalized replication ratio data from table S5 of Liachko et al. (2014), which were reported for 1-kb bins in the genome sequence of K. phaffii strain GS115, into equivalent coordinates for strain CBS7435. GS115 is a laboratory derivative of CBS7435 and therefore has an almost identical genome, but the published genome sequences of GS115 (De Schutter et al. 2009) and CBS7435 (Kuberl et al. 2011) differ in some details including the orientations of chromosomes 2, 3 and 4, as well as the locations of gaps and the completeness of telomeric regions. To map the replication data, we aligned the sequences of each chromosome between the two strains using MUMMER (Delcher et al. 1999) and calculated a nucleotide-to-nucleotide coordinate conversion table. In figures 2 and 3, the normalized replication ratio for each 1-kb bin in GS115 is plotted at the CBS7435 coordinate corresponding to the center of that bin.
Supplementary Material
Supplementary figures S1–S5 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This study was supported by Science Foundation Ireland (13/IA/1910) and the European Research Council (268893). High-throughput sequencing services were performed at the University of Missouri DNA Core Facility.
Literature Cited
- Allshire RC. 2004. Centromere and kinetochore structure and function In: Egel R, editor. The molecular biology of Schizosaccharomyces pombe. Berlin: Springer; p. 149–169. [Google Scholar]
- Allshire RC, Ekwall K. 2015. Epigenetic regulation of chromatin states in Schizosaccharomyces pombe. Cold Spring Harb Perspect Biol. 7:a018770.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M, Ngan VK, Clarke L. 1994. The centromeric K-type repeat and the central core are together sufficient to establish a functional Schizosaccharomyces pombe centromere. Mol Biol Cell. 5:747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M, Sanyal K, Mishra PK, Thaler N, Carbon J. 2006. Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc Natl Acad Sci U S A. 103:14877–14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensasson D. 2011. Evidence for a high mutation rate at rapidly evolving yeast centromeres. BMC Evol Biol. 11:211.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catania S, Pidoux AL, Allshire RC. 2015. Sequence features and transcriptional stalling within centromere DNA promote establishment of CENP-A chromatin. PLoS Genet. 11:e1004986.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan FL, Wong LH. 2012. Transcription in the maintenance of centromere chromatin identity. Nucleic Acids Res. 40:11178–11188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee G, et al. 2016. Repeat-associated fission yeast-like regional centromeres in the ascomycetous budding yeast Candida tropicalis. PLoS Genet. 12:e1005839.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter K, et al. 2009. Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol. 27:561–566. [DOI] [PubMed] [Google Scholar]
- Delcher AL, et al. 1999. Alignment of whole genomes. Nucleic Acids Res. 27:2369–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikicioglu D, Wood V, Rutherford KM, McDowall MD, Oliver SG. 2014. Improving functional annotation for industrial microbes: a case study with Pichia pastoris. Trends Biotechnol. 32:396–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M, Clarke L, Carbon J. 1982. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell 29:235–244. [DOI] [PubMed] [Google Scholar]
- Fleig U, Beinhauer JD, Hegemann JH. 1995. Functional selection for the centromere DNA from yeast chromosome VIII. Nucleic Acids Res. 23:922–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco HD, Pidoux AL, Urano T, Allshire RC. 2008. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319:94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JL, Byrne KP, Wolfe KH. 2011. Mechanisms of chromosome number evolution in yeast. PLoS Genet. 7:e1002190.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LE, Mitchell SE, O’Neill RJ. 2012. Pericentric and centromeric transcription: a perfect balance required. Chromosome Res. 20:535–546. [DOI] [PubMed] [Google Scholar]
- Hanson SJ, Byrne KP, Wolfe KH. 2014. Mating-type switching by chromosomal inversion in methylotrophic yeasts suggests an origin for the three-locus Saccharomyces cerevisiae system. Proc Natl Acad Sci U S A. 111:E4851–E4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann JH, Fleig UN. 1993. The centromere of budding yeast. Bioessays 15:451–460. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. 2012. “Point” centromeres of Saccharomyces harbor single centromere-specific nucleosomes. Genetics 190:1575–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter P, et al. 1985. Functional selection and analysis of yeast centromeric DNA. Cell 42:913–921. [DOI] [PubMed] [Google Scholar]
- Janbon G, et al. 2014. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 10:e1004261.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor S, Zhu L, Froyd C, Liu T, Rusche LN. 2015. Regional centromeres in the yeast Candida lusitaniae lack pericentromeric heterochromatin. Proc Natl Acad Sci U S A. 112:12139–12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketel C, et al. 2009. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 5:e1000400.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM, Dubey DD, Huberman JA. 2003. Early-replicating heterochromatin. Genes Dev. 17:330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, et al. 2015. Discovery of an unconventional centromere in budding yeast redefines evolution of point centromeres. Curr Biol. 25:2026–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A, et al. 2010. Epigenetically-inherited centromere and neocentromere DNA replicates earliest in S-phase. PLoS Genet. 6:e1001068.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuberl A, et al. 2011. High-quality genome sequence of Pichia pastoris CBS7435. J Biotechnol. 154:312–320. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP. 2009. Biotechnological strains of Komagataella (Pichia) pastoris are Komagataella phaffii as determined from multigene sequence analysis. J Ind Microbiol Biotechnol. 36:1435–1438. [DOI] [PubMed] [Google Scholar]
- Lefrancois P, Auerbach RK, Yellman CM, Roeder GS, Snyder M. 2013. Centromere-like regions in the budding yeast genome. PLoS Genet. 9:e1003209.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko I, et al. 2014. GC-rich DNA elements enable replication origin activity in the methylotrophic yeast Pichia pastoris. PLoS Genet. 10:e1004169.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, et al. 2012. Comprehensive structural annotation of Pichia pastoris transcriptome and the response to various carbon sources using deep paired-end RNA sequencing. BMC Genomics 13:738.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Cereghino J, et al. 2005. Condensed protocol for competent cell preparation and transformation of the methylotrophic yeast Pichia pastoris. BioTechniques 38:44, 46, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch DB, Logue ME, Butler G, Wolfe KH. 2010. Chromosomal G + C content evolution in yeasts: systematic interspecies differences, and GC-poor troughs at centromeres. Genome Biol Evol. 2:572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. 2009. Major evolutionary transitions in centromere complexity. Cell 138:1067–1082. [DOI] [PubMed] [Google Scholar]
- McCarroll RM, Fangman WL. 1988. Time of replication of yeast centromeres and telomeres. Cell 54:505–513. [DOI] [PubMed] [Google Scholar]
- Morales L, et al. 2013. Complete DNA sequence of Kuraishia capsulata illustrates novel genomic features among budding yeasts (Saccharomycotina). Genome Biol Evol. 5:2524–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naatsaari L, et al. 2012. Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS One 7:e39720.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngan VK, Clarke L. 1997. The centromere enhancer mediates centromere activation in Schizosaccharomyces pombe. Mol Cell Biol. 17:3305–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan S, Thakur J, Siddharthan R, Sanyal K. 2008. Rapid evolution of Cse4p-rich centromeric DNA sequences in closely related pathogenic yeasts, Candida albicans and Candida dubliniensis. Proc Natl Acad Sci U S A. 105:19797–19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG, Earnshaw WC. 1995. The centromere: hub of chromosomal activities. Science 270:1591–1594. [DOI] [PubMed] [Google Scholar]
- Pohl TJ, Brewer BJ, Raghuraman MK. 2012. Functional centromeres determine the activation time of pericentric origins of DNA replication in Saccharomyces cerevisiae. PLoS Genet. 8:e1002677.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravin NV, et al. 2013. Genome sequence and analysis of methylotrophic yeast Hansenula polymorpha DL1. BMC Genomics 14:837.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, et al. 2012. Comparative functional genomics of the fission yeasts. Science 332:930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley R, et al. 2016. Comparative genomics of biotechnologically important yeasts. Proc Natl Acad Sci U S A. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Sanyal K. 2011. Diversity in requirement of genetic and epigenetic factors for centromere function in fungi. Eukaryot Cell 10:1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal K, Baum M, Carbon J. 2004. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc Natl Acad Sci U S A. 101:11374–11379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Galazka JM, Phatale PA, Connolly LR, Freitag M. 2012. Centromeres of filamentous fungi. Chromosome Res. 20:635–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Phatale PA, Sullivan CM, Pomraning KR, Freitag M. 2011. Heterochromatin is required for normal distribution of Neurospora crassa CenH3. Mol Cell Biol. 31:2528–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner NC, Clarke L. 1994. A novel epigenetic effect can alter centromere function in fission yeast. Cell 79:865–874. [DOI] [PubMed] [Google Scholar]
- Steiner NC, Hahnenberger KM, Clarke L. 1993. Centromeres of the fission yeast Schizosaccharomyces pombe are highly variable genetic loci. Mol Cell Biol. 13:4578–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9:573–586. [DOI] [PubMed] [Google Scholar]
- Sturmberger L, et al. 2016. Refined Pichia pastoris reference genome sequence. J Biotechnol. doi: 10.1016/j.jbiotec.2016.1004.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, et al. 1992. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell 3:819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka E, Bailey TL, Keich U. 2014. Improving MEME via a two-tiered significance analysis. Bioinformatics 30:1965–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J, Sanyal K. 2013. Efficient neocentromere formation is suppressed by gene conversion to maintain centromere function at native physical chromosomal loci in Candida albicans. Genome Res. 23:638–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J, Talbert PB, Henikoff S. 2015. Inner kinetochore protein interactions with regional centromeres of fission yeast. Genetics 201:543–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoquaux N, et al. 2015. Accurate identification of centromere locations in yeast genomes using Hi-C. Nucleic Acids Res. 43:5331–5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernis L, et al. 2001. Only centromeres can supply the partition system required for ARS function in the yeast Yarrowia lipolytica. J Mol Biol. 305:203–217. [DOI] [PubMed] [Google Scholar]
- Volpe TA, et al. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833–1837. [DOI] [PubMed] [Google Scholar]
- Weninger A, Hatzl AM, Schmid C, Vogl T, Glieder A. 2016. Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris. J Biotechnol. doi: 10.1016/j.jbiotec.2016.1003.1027. [DOI] [PubMed] [Google Scholar]
- Wisniewski J, et al. 2014. Imaging the fate of histone Cse4 reveals de novo replacement in S phase and subsequent stable residence at centromeres. Elife 3:e02203.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871–880. [DOI] [PubMed] [Google Scholar]
- Yamane T, Ogawa T, Matsuoka M. 2008. Derivation of consensus sequence for protein binding site in Yarrowia lipolytica centromere. J Biosci Bioeng. 105:671–674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.