Abstract
Heliconius possess a unique ability among butterflies to feed on pollen. Pollen feeding significantly extends their lifespan, and is thought to have been important to the diversification of the genus. We used RNA sequencing to examine feeding-related gene expression in the mouthparts of four species of Heliconius and one nonpollen feeding species, Eueides isabella. We hypothesized that genes involved in morphology and protein metabolism might be upregulated in Heliconius because they have longer proboscides than Eueides, and because pollen contains more protein than nectar. Using de novo transcriptome assemblies, we tested these hypotheses by comparing gene expression in mouthparts against antennae and legs. We first looked for genes upregulated in mouthparts across all five species and discovered several hundred genes, many of which had functional annotations involving metabolism of proteins (cocoonase), lipids, and carbohydrates. We then looked specifically within Heliconius where we found eleven common upregulated genes with roles in morphology (CPR cuticle proteins), behavior (takeout-like), and metabolism (luciferase-like). Closer examination of these candidates revealed that cocoonase underwent several duplications along the lineage leading to heliconiine butterflies, including two Heliconius-specific duplications. Luciferase-like genes also underwent duplication within lepidopterans, and upregulation in Heliconius mouthparts. Reverse-transcription PCR confirmed that three cocoonases, a peptidase, and one luciferase-like gene are expressed in the proboscis with little to no expression in labial palps and salivary glands. Our results suggest pollen feeding, like other dietary specializations, was likely facilitated by adaptive expansions of preexisting genes—and that the butterfly proboscis is involved in digestive enzyme production.
Keywords: heliconiinae, proteolysis; trypsin-like serine protease; luciferin 4-monooxygenase; lipase; proboscipedia
Introduction
Adaptations to novel food sources are found in numerous species from flies to humans. During dietary specialization, modifications to gene expression regulation are likely to have played an important role (Blekhman et al. 2008; Luca et al. 2010; Blekhman et al. 2014). Gene expression evolution may occur through adjustments in transcriptional and posttranscriptional regulation, and gene duplication events (Zhang 2003; Chen and Rajewsky 2007; Haygood et al. 2007; Wray 2007). For example, the continuous production of lactase into late human development allowed for adult digestion of lactose in milk, and is thought to be due to cis-regulatory changes in an enhancer region upstream of the lactase gene (Ingram et al. 2009). Similarly, copy number variation has been observed in the amylase genes of humans, with increased expression and corresponding protein levels being associated with a high starch diet (Perry et al. 2007).
Pollen feeding is one such dietary specialization that occurs at different developmental stages across herbivorous insect species (Boggs 1986; Wackers et al. 2007). Some pollen feeding species feed only as larvae, laying down fat reserves later used for adult nutrition. Others feed as adults, and some as both larvae and adults, to provide a source of nutrition for sexual maturation and body maintenance (Cook et al. 2004). Female honey bees (Apis mellifera; Imdorf et al. 1998) and hoverflies, for example, require pollen for successful reproduction (Gilbert 1985). Among butterflies, Heliconius are the only species known to supplement their adult diet by feeding on pollen (Roulston and Cane 2000). Heliconius females do not require pollen for reproduction but, when ingested by females, pollen provides essential amino acids that are directly provisioned into the eggs (Gilbert 1972; Dunlap-pianka et al. 1977). Additionally, adult pollen feeding for both Heliconius sexes allows for the extension of lifespan from around 3–5 weeks to 6 months, and increased reproductive output (Gilbert 1972; Boggs 1979; O'Brien et al. 2003).
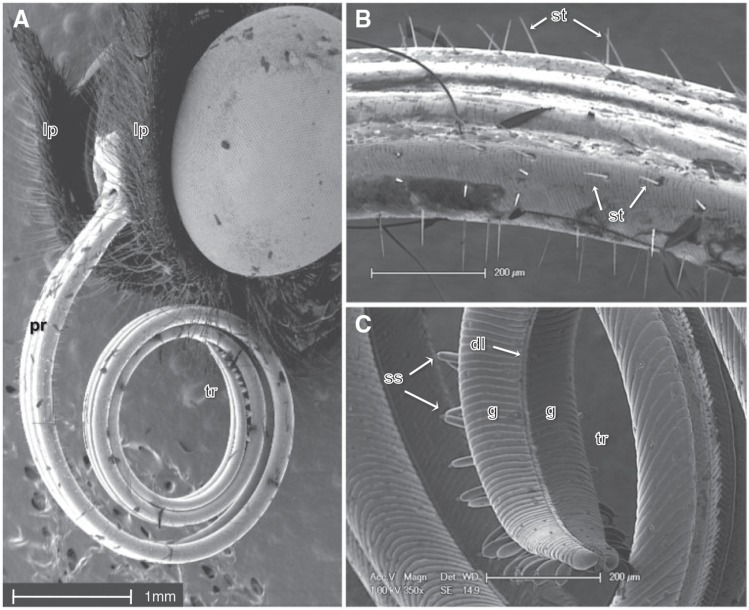
Pollen feeding in Heliconius involves a suite of adaptive phenotypes that differentiate them from nonpollen feeding species, including morphological, behavioral and metabolic differences (Gilbert 1972). Pollen feeding Heliconius species have a significantly longer proboscis, with longer and more numerous sensilla trichodea, which may aid in the collection and retention of pollen grains (fig. 1; Krenn and Penz 1998). Once collected, pollen is masticated on the proboscis through a unique pollen processing behavior that involves the coiling and uncoiling and side-to-side movements of the proboscis combined with salivary discharge (Gilbert 1972; Krenn and Penz 1998; Eberhard and Krenn 2003; Krenn et al. 2009). The saliva of pollen feeding species is known to contain proteolytically active proteases, and some evidence suggests increased proteolytic activity in response to the presence of pollen (Eberhard et al. 2007). The salivary glands of Heliconius are larger than those of nonpollen feeders, although differences in anatomical features that control saliva secretion in the proboscis do not appear to differ (Eberhard et al. 2009). Intriguingly, although the salivary glands are thought to produce the majority of digestive proteins in lepidopterans, in moths the proboscis produces at least one enzyme directly: the trypsin-like serine protease cocoonase (Kafatos et al. 1967).
Fig. 1.—.
Scanning electron micrograph of the head and mouthparts of Heliconius melpomene. (A) Labial palps (lp), proboscis (pr) and proboscis tip region (tr). (B) Sensilla trichodea (st) found on the proboscis, (C) Magnified view of the proboscis which is comprised of dorsally and ventrally linked galeae (g) linked by dorsal ligulae (dl). The tip region contains sensilla styloconica (ss) that are club shaped and flattened.
It has been suggested that the evolution of adult pollen feeding in Heliconius allowed for a greater investment of adult resources in reproduction, which freed larval resource investment for defensive purposes (Cardoso and Gilbert 2013). This suggests that pollen feeding could have played an important role in the evolution of Müllerian mimicry within the genus (Müller 1879; Gilbert 1991). However, little is known about the genetic basis of pollen feeding or the pathway by which pollen feeding evolved.
Although RNA sequencing has been used to identify the expression of gustatory, olfactory, and ionotropic receptors in mouthparts of individual Heliconius butterflies (Briscoe et al. 2013, van Schooten et al. 2016), no studies have quantified genome-wide expression in the butterfly proboscis. The proboscis is of particular interest because it harbors morphological features associated with pollen feeding, and, in moths, directly secretes at least one digestive protein. Thus, we used high-throughput RNA sequencing to characterize gene expression and identify candidate pollen feeding genes in the mouthparts of four Heliconius species and one out-group species, Eueides isabella, that does not feed on pollen. We hypothesized that due to morphological differences and because pollen has a much higher amino acid content compared with nectar, we might observe an upregulation of genes annotated with morphological and proteolysis functions in our Heliconius mouthparts transcriptomes compared with the nonpollen feeding Eueides. Our aims were to: 1) identify common upregulated genes and their functions in the mouthparts of all five species, and 2) identify Heliconius-specific mouthparts-upregulated genes that might have specific functions in pollen feeding adaptations.
Materials and Methods
Sampling, mRNA Extraction and Sequencing
Butterfly samples were obtained from a Costa Rican butterfly farm, Suministros Entomológicos Costarricenses, S.A. Butterflies were shipped as pupae and hung within a humidified chamber at room temperature until eclosion. Adult butterflies were fed on diluted ∼1:10 100% honey and were then fresh frozen in −80 °C freezer 1–2 days after eclosion. mRNA was extracted from the antennae, legs (all six legs) and mouthparts (proboscis and labial palps) of one male and one female H. erato, H. doris, H. sara, and a nonpollen feeding sister species to Heliconius, Eueides isabella, using TRIzol (Life Technologies, Inc.). The labial palps were included due to their potential for a role in feeding. Illumina sequencing resulted in 24 sequenced libraries of 100 bp paired-end reads (see Supplementary Methods for details). Raw read data were also obtained for H. melpomene rosina from Briscoe et al. (2013), which includes three biological replicate males and three females for each tissue type (ArrayExpress accession: E-TAB-1500). These data include resequenced samples from each tissue type of one male and one female comprising of 100 bp single-end reads. See supplementary table S1, Supplementary Material online for details of samples used in this study. Illumina read libraries are available as fastq files from the ArrayExpress database (www.ebi.ac.uk/arrayexpress), accession number E-MTAB-3446.
Trinity Assembly and Mapping
De novo assemblies were performed for each of the five species using Trinity (Grabherr et al. 2011), assembling reads into contiguous sequences (>300 bp) representing mRNA transcripts. For each species, libraries were mapped back to each final reference assembly using RSEM (Li and Dewey 2011) and expression levels of each contig were quantified. FPKM (fragments per kilobase of exon per million reads mapped) was calculated using RSEM, and normalized between libraries with the trimmed mean of M-values (TMM) method in the R package NOISeq (Tarazona et al. 2011). Fasta files of all five Trinity transcriptome assemblies and expression level counts of each Trinity “gene” are available from Dryad under the data identifier doi: 10.5061/dryad.8d724.
Differential Expression Analysis
Differential expression analysis was performed on raw count data using the R package edgeR (Robinson et al. 2010). Count data from single-end libraries were merged with the counts for their respective paired-end sequenced samples for analysis. The H. melpomene dataset thus comprised of three tissues—mouthparts, antennae and legs—from males and females, and three biological replicates of each. The remaining four species included six libraries per species, comprised of the three tissue types, each from one male and one female. In order to obtain contigs that demonstrated signals of strong upregulation in the mouthparts, differential expression analyses were performed to compare mouthparts gene expression to antennal and leg tissues, the latter tissues being used as “control” tissues due to their similarity to the proboscis in terms of developmental specification. Data from each species were analyzed separately. Each data set was filtered to remove contigs with low expression, retaining those contigs with > 1 count per million in at least three groups for the H. melpomene data, and at least two groups for the other four species. Data were normalized between samples using the default TMM normalization method (Robinson and Oshlack 2010).
The H. melpomene count data were analyzed using a generalized linear model in edgeR, modeled as ∼tissue + sex. The remaining four species were modeled using the classic edgeR approach for single factor designs, with male and female samples grouped as biological replicates of each tissue type. In order to control for performing multiple tests, a false discovery rate (FDR; adjusted P-values) was calculated for each contig as per Storey and Tibshirani (2003). Contigs were defined as showing evidence of significant upregulation in the mouthparts when the FDR was < 0.05 and contig expression increased by > 1 log fold change (logFC) compared with the other tissues. Heatmaps were constructed with the R package Heatplus (Ploner 2012). The expression of several candidate genes in the proboscis, labial palps, and salivary glands was verified using reverse transcription polymerase chain reaction (RT-PCR; see Supplementary Methods and supplementary table S2, Supplementary Material online for details).
Functional Assignment and Cross-Species Orthology Determination
Open reading frames (ORFs) for Trinity assembled contigs were predicted from the longest contig for each Trinity “gene” using the Trinity Transdecoder program, which produces the most likely nonoverlapping ORF(s). Instances of multiple ORFs per contig were filtered by choosing the ORF with the top BLAST hit (e-value) to a protein in the UniProt database (see below for BLAST details). Functional assignment of significant mouthparts-upregulated contigs was performed in Blast2GO (Conesa et al. 2005), using default parameters. Blast2GO-assigned gene ontology (GO) functions were summarized by quantifying the number of contigs within level 2 biological process GO terms, and dissected to more specific terms, using the node information score to identify “hot spots” of GO terms within a network of GO term associations (Conesa and Gotz 2008). All GO terms were for biological processes, the parameter α was set at the default 0.6 and GO terms were only considered if they had scores of > 5.
Additionally, functions for each Trinity assembled contig were obtained through a separate BLAST of each species’ ORFs to the UniProt (Apweiler et al. 2004) invertebrate database using Transcriptome Computational Workbench (e-value < 1 × 10 − 10; TCW v1.2; Soderlund et al. 2013). TCW obtains the best top hit and assigns a gene description to that ORF. TCW also utilizes OrthoMCL (Li et al. 2003), which runs a Markov Cluster algorithm to group putative ortholog and recent paralog sequences into ortholog clusters (orthoclusters) across species. To identify orthoclusters, a joint all-against-all BLAST of Trinity-assembled contig ORFs across species was performed using BLAST+, filtering out contigs with >99% pairwise identity. A database of cross-species sequence associations was created, with each orthocluster assigned a consensus functional description from the top UniProt BLAST hits. The filtering of highly similar sequences between species and the BLAST to the UniProt database ensured that contaminant sequences were not included in the final set of orthoclusters. Orthoclusters were then used to compare the orthology of genes upregulated in the mouthparts.
Orthoclusters that contained mouthparts-upregulated genes were compared for their presence in each Heliconius species, and absence in E. isabella. A five-way Venn diagram was created using web tools from the Bioinformatics and Systems Biology group, Ghent University (http://bioinformatics.psb.ugent.be/webtools/Venn/). Additional comparisons were made to identify contigs in at least three of the four Heliconius species and not in E. isabella. A text file containing the predicted ORF functional annotations (best BLAST hits) and orthocluster membership from each of the five Trinity assemblies are available from Dryad under the data identifier doi:10.5061/dryad.8d724.
Results
De Novo Trinity Assembly
Several assembly protocols were explored (see Supplementary Methods). Trimmed reads without digital normalization produced the most optimal de novo assemblies and had a high mapping efficiency (supplementary tables S3 and S4, Supplementary Material online). The final assemblies included one male and one female from each tissue type for each species. Trinity de novo assemblies across species averaged 40,524 Trinity “genes” (nonredundant mRNA contigs) and 80,144 total contigs (averaging ∼2 isoforms per gene; table 1). The N50 of transcriptome assemblies across species averaged 2,617 bp (table 1) and mapping efficiencies across libraries and species ranged from 80–97%, with the exception of one library that had an efficiency of 75% (supplementary table S4, Supplementary Material online).
Table 1.
Summaries of De Novo Assemblies Using Trinity
| Species | Number of Trinity “Genes” | Total Number of Trinity Contigs | Contig N50 (bp) |
|---|---|---|---|
| H. melpomene | 57047 | 121904 | 3244 |
| H. erato | 42628 | 86726 | 2497 |
| H. sara | 32211 | 60789 | 2334 |
| H. doris | 36180 | 70081 | 2715 |
| E. isabella | 34555 | 61221 | 2294 |
| Average | 40524 | 80144 | 2617 |
Functions of Genes Upregulated in Labial Palps and Proboscis
Differential expression analysis revealed an average of 463 contigs upregulated in the mouthparts across species (FDR < 0.05, logFC > 1), ranging from 236 in H. erato to 618 in H. melpomene (table 2; fig. 2). Blast2GO annotations of these contigs suggested they had similar broad functions, the most common of which involved the term metabolic process (supplementary fig. S1, Supplementary Material online). Multi-level GO term annotations coupled with an enrichment score provided more detailed functions, including terms for proteolysis, carbohydrate and lipid metabolism (all Blast2GO node information scores > 5; supplementary figs. S2–S6, Supplementary Material online). Proteolysis terms had the second highest score for all species except H. erato where its score was < 5. Carbohydrate and lipid metabolism GO terms were present in each species except for H. erato and E. isabella where carbohydrate metabolism had a score of < 5.
Table 2.
Number of Significantly Upregulated Contigs in the Mouthparts of Each Species
| Species | Number of Significant Contigs* |
|---|---|
| H. melpomene | 618 |
| H. erato | 236 |
| H. sara | 616 |
| H. doris | 555 |
| E. isabella | 289 |
*Significance threshold of < 0.05 FDR (adjusted P-value), and > 1 log fold change in expression.
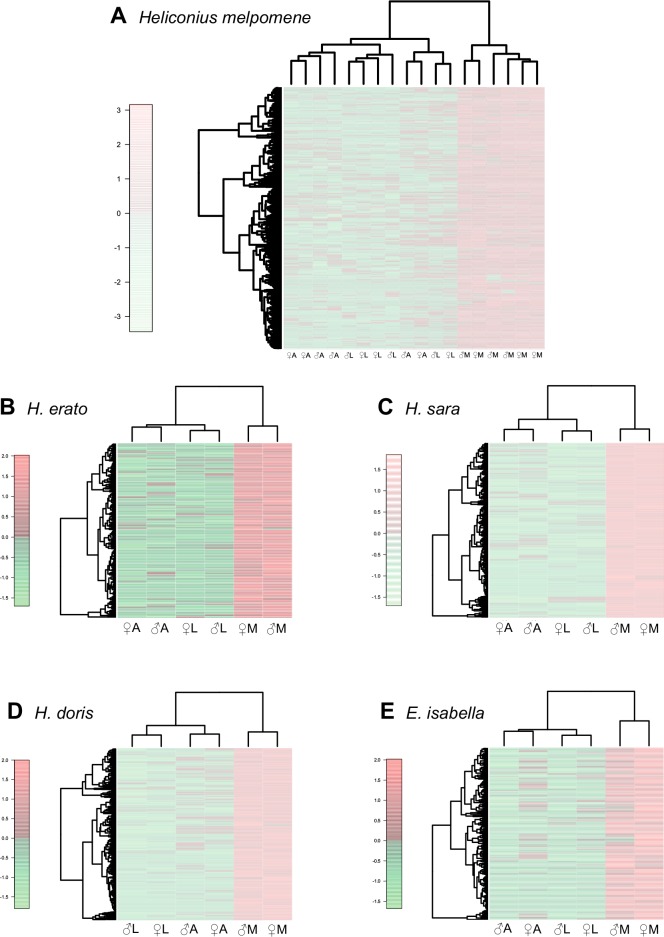
Fig. 2.—.
Heatmaps of significantly upregulated contigs in the mouthparts of each species (FDR < 0.05, logFC of > 1). Color scale indicates the scaled log counts-per-million expression difference between libraries, with each row being a contig. (A) H. melpomene, (B) H. erato, (C) H. sara, (D) H. doris, (E) E. isabella. A = antennae, L = legs, M = mouthparts.
Transcriptome Computational Workbench was used to examine the orthology of genes upregulated in the mouthparts by determining cross-species orthoclusters (putative ortholog and recent paralog sequences). In total, 69,022 amino acid sequences were obtained from the five assembled transcriptomes (including multiple instances of ORFs within a contig from Transdecoder), which clustered into 10,102 orthoclusters containing at least two amino acid sequences per cluster. In total, mouthparts-upregulated genes fell into 230, 116, 252, 219, and 154 different orthoclusters in H. melpomene, H. erato, H. sara, H. doris and E. isabella, respectively (supplementary table S5, Supplementary Material online). Orthologs were compared to determine those upregulated in the mouthparts of all species (fig. 3).
Fig. 3.—.
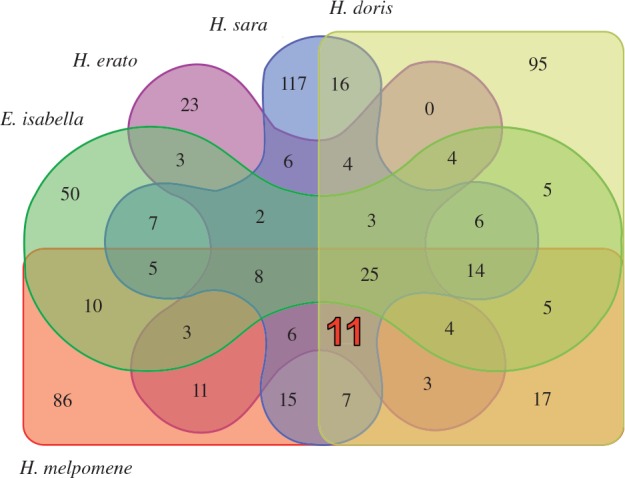
Venn diagram of overlapping orthoclusters containing mouthparts-upregulated contigs. Highlighted is the overlap containing Heliconius-specific upregulated contigs (see table 4 for details).
Twenty-five orthoclusters were common to all species, including genes annotated with morphology and behavior (table 3). Two of these 25 clusters encoded homeobox proteins, Pox-neuro (OM001839) and proboscipedia (OM001622), which are involved in neurological (Nottebohm et al. 1992) and proboscis development (Abzhanov et al. 2001), respectively. Dopa decarboxylase (Ddc; OM000102) plays a role in learning (Tempel et al. 1984) and longevity (De Luca et al. 2003) in Drosophila. Chemosensory proteins (CSPs; OM000015) were also upregulated, including two in H. melpomene orthologous to genome annotations HMEL010989 and HMEL010990 (CSP3; Dasmahapatra et al. 2012). Three of the 25 clusters were cuticle protein genes containing RR motifs (CPRs; Rebers and Riddiford 1988), one contained a juvenile hormone binding protein (OM000238), and one was orthologous to the Drosophila buffy gene, which functions in the cellular response to starvation (Hou et al. 2008).
Table 3.
Orthoclusters of Mouthparts-Upregulated Genes Common to All Five Heliconiine Species
| Ortholog Cluster ID | Gene Name | Functional Description |
Number of Paralogs Demonstrating Significant Upregulation in the Mouthparts of Each Species |
Contigs in Each Cluster | ||||
|---|---|---|---|---|---|---|---|---|
| E. isabella | H. melpomene | H. erato | H. sara | H. doris | ||||
| OM000011 | l(2)03659 | Probable multidrug resistance-associated protein lethal(2) | *1* | *2*** | **1* | **1*** | **1** | 79 |
| OM000014 | FACR1 | Fatty-acyl-CoA reductase (alcohol-forming) activity, ether lipid biosynthetic process | **3** | **6* | *5** | *3*** | *6** | 76 |
| OM000015 | CSP | Chemosensory protein | *2** | **2** | *3* | *6* | *5** | 77 |
| OM000022 | Wwox | WW domain-containing oxidoreductase | *2* | ***1*** | **1* | ***1** | *1* | 66 |
| OM000026 | - | Aldo-keto reductase | *1** | *4* | *2** | *4*** | *4* | 61 |
| OM000035 | - | UDP-glucuronosyltransferase | *2* | *1** | **1** | *2* | *1** | 50 |
| OM000040 | - | Carbonyl reductase, NADPH activity | *1* | ***1*** | *1** | *1** | **1** | 46 |
| OM000051 | CPR | Cuticular protein RR-1 family, chitin-based cuticle development | *1* | **2*** | ***2* | *6*** | **5** | 44 |
| OM000062 | - | Elongase, elongation of very long chain fatty acids | *4* | **4** | *4* | *4** | *3* | 41 |
| OM000102 | Ddc | Aromatic-L-amino-acid decarboxylase, response to stimulus, behavior, growth, pigmentation, cognition | *1* | **1** | *1* | *1* | **1*** | 25 |
| OM000188 | - | Lipase, lipid metabolism | ***2** | **2** | *2* | ***1*** | *2** | 18 |
| OM000193 | - | C2 domain transmembrane protein | *1* | ***1*** | **1** | **1*** | **1*** | 15 |
| OM000233a | cocoonase | Trypsin-like serine protease, proteolysis | ***3** | **4** | ***1*** | **3* | ***2*** | 17 |
| OM000238 | - | Juvenile hormone binding protein | *2* | *2* | *2** | *2*** | *2** | 16 |
| OM000325 | - | ABC transporter family G member | *3* | ***1*** | *2* | *2** | *1** | 14 |
| OM000556 | CPR | Cuticular protein RR-1 family, chitin-based cuticle development | **1** | *3* | ***2*** | **3* | ***3*** | 11 |
| OM000832 | - | Seminal fluid protein HACP060 | *1* | **1** | *1* | *1* | *1* | 6 |
| OM001210 | - | Cellular retinaldehyde binding protein | *1* | ***1*** | *1** | **1*** | *1* | 8 |
| OM001622 | proboscipedia | Homeobox protein, proboscis development | ***1*** | ***1*** | ***1*** | ***1*** | ***1*** | 6 |
| OM001839 | Pox neuro | Homeobox protein, nervous sytem development, adult feeding behavior, sensory organ development | ***1*** | ***1*** | ***1*** | ***1*** | ***1*** | 6 |
| OM002337 | - | Monocarboxylic acid transmembrane transporter activity | *1** | ***1*** | ***1*** | ***1*** | **1*** | 6 |
| OM003749 | - | Steroid dehydrogenase activity | **1** | ***1*** | ***1*** | ***1*** | ***1** | 5 |
| OM004900 | CPR | Cuticular protein RR-3 family, chitin-based cuticle development | **1*** | **1*** | *1*** | *1*** | **1*** | 5 |
| OM005185 | buffy | Regulation of cell death, cellular response to starvation | *1* | **1*** | **1* | *1* | **1* | 5 |
| OM007063 | GPAT | Glycerol-3-phosphate acyltransferase, phospholipid metabolism | *1* | **1*** | **1** | **1*** | **1** | 5 |
Note.—At least one gene per orthocluster was significantly upregulated in the mouthparts (FDR < 0.05, logFC >1) of all species. The number of significantly upregulated mouthparts genes belonging to each cluster is presented alongside cluster totals. Genes were named according to their closest Drosophila or insect homologs.
a Paralog counts obtained from manual annotation, all other clusters from automated annotation (supplementary table S5, Supplementary Material online). Asterisks denote FDR (adjusted P-value) strength of upregulated genes in mouthparts to legs (left of number) and mouthparts to antennae (right of number) comparisons. Where more than one paralog is upregulated, the least significant FDRs are presented: *< 0.05, **< 1 × 10 − 5, ***< 1 × 10 − 10.
The majority of the remaining common orthoclusters was annotated with metabolic functions. One was annotated with proteolysis: cocoonase (OM000233, see below and fig. 4), and several with lipid metabolism. Lipase (OM000188) is involved in several key roles in insects, including embryogenesis, flight and metamorphosis (Gilbert 1967; Horne et al. 2009). Fatty-acyl-CoA reductase (OM000014) and GPAT (glycerol-3-phosphate acyltransferase; OM007063) play a role in lipid metabolism, specifically GPAT is associated with phospholipid metabolism, using glycerol-3-phosphate and acyl-CoA as substrates (Gimeno and Cao 2008; Wendel et al. 2009). Common clusters also included drug detoxification genes such as UDP-glucuronosyltransferase, which is responsible for a large proportion of drug metabolism (Oda et al. 2015). Lastly, the cellular retinaldehyde binding protein cluster (OM001210) belongs to the CRAL-TRIO domain containing protein family that underwent a recent large expansion in Lepidoptera (Smith and Briscoe 2015).
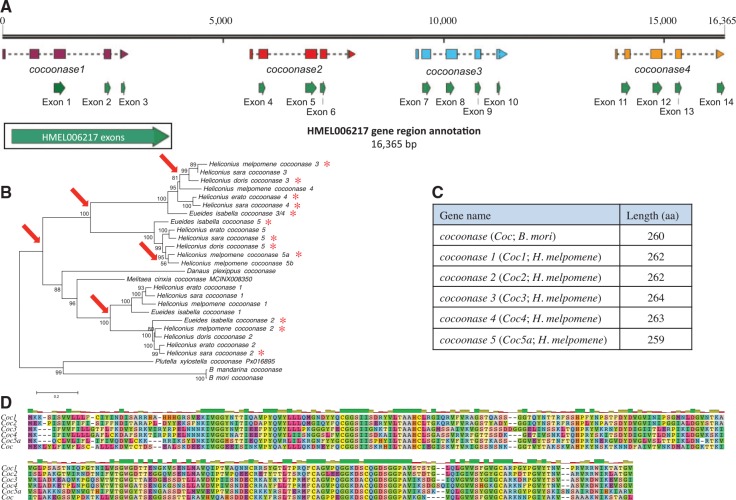
Fig. 4.—.
Genomic annotation of Heliconius melpomene cocoonase 1–4. (A) Previous annotation of gene HMEL006217 (green) and new annotations of each cocoonase (purple, red, blue, orange). (B) A maximum-likelihood tree of cocoonase nucleotide sequences with bootstrap support values (above branches; < 50% values not shown) and branch length scale bar (substitutions per site). Red asterisks denote mouthparts-upregulated contigs and red arrows are inferred duplication events. (C) Amino acid (aa) lengths of representative cocoonase genes. (D) Amino acid alignment of cocoonases.
Genes that were uniquely upregulated in the mouthparts of each species were annotated with numerous functions, including metabolism of different nutrient classes (supplementary table S6, Supplementary Material online).
Candidate Pollen-Feeding Genes
Genes belonging to eleven orthoclusters demonstrated Heliconius-specific upregulation in the mouthparts (FDR < 0.05, logFC > 1; table 4; fig. 5A). Six were annotated with metabolism related functions. These included two single-copy clusters related to proteolysis: coagulation factor X (OM002911), a trypsin-like serine protease, and Serpin100A (OM001769), a serine protease inhibitor. Also present were carbohydrate metabolism (maltose phosphorylase; OM001951), luciferase-like (luciferin 4-monooxygenase; OM000048), and Adh-like (alcohol dehydrogenase; OM000265) genes. Three clusters contained CPR cuticle proteins (see Dittmer et al. 2015 for a recent classification of these proteins), and two contained genes that impact behavior: orthologs of egon/eagle genes (embryonic gonad; OM003483) and takeout-like genes (OM000111).
Table 4.
Orthoclusters of Mouthparts-Upregulated Genes in Heliconius but not Eueides
| Ortholog CLuster ID | Gene Name | Functional Description |
Number of Paralogs Demonstrating Significant Upregulation in the Mouthparts of Each Species |
Contigs in Each Cluster | ||||
|---|---|---|---|---|---|---|---|---|
| E. isabella | H. melpomene | H. erato | H. sara | H. doris | ||||
| OM000048 | luciferase-like | Luciferin 4-monooxygenase, ATP-dependent monooxygenase/long chain fatty acyl-CoA synthetase | 0 | *1*** | *1* | ***1*** | ***2* | 40 |
| OM000111 | takeout-like | Circadian clock-controlled, adult feeding behavior | 0 | *2** | **1* | ***2** | *2** | 28 |
| OM000264 | CPR | Cuticular protein RR1 family, chitin-based cuticle development | 0 | ***1*** | *1* | ***1** | **2** | 16 |
| OM000265 | Adh-like | Alcohol dehydrogenase, 15-hydroxyprostaglandin dehydrogenase | 0 | *2** | ***1*** | *2*** | ***1*** | 16 |
| OM001769 | Serpin100A | Serine protease inhibitor 100A, regulation of proteolysis | 0 | ***1** | *1* | ***1* | **1* | 5 |
| OM001951 | - | Maltose phosphorylase, carbohydrate metabolism | 0 | *1* | *1* | **1** | *1** | 5 |
| OM002911 | coagulation factor X | Trypsin-like serine endopeptidase, proteolysis | 0 | *1** | **1* | *1** | *1* | 5 |
| OM003483 | egon/eagle | Zinc finger transcription factor | 0 | **1** | *1* | ***1** | **1** | 5 |
| OM003746 | CPR | Cuticular protein RR1 motif, chitin-based cuticle development | 0 | *1* | **1** | **1** | *1** | 5 |
| OM004221 | - | ATP-citrate synthase, TCA cycle | 0 | *1** | *1** | *1** | **1*** | 5 |
| OM005658a | CPR | Cuticular protein RR1 motif, chitin-based cuticle development | - | ***1* | **1* | ***1** | ***1*** | 4 |
Note.—At least one gene per orthocluster was significantly upregulated in the mouthparts (FDR < 0.05, logFC >1) of all four Heliconius species, but not E. isabella. The number of significantly upregulated mouthparts genes belonging to each cluster is presented alongside cluster totals. Genes were named according to their closest Drosophila or insect homologs.
a Eueides isabella sequence not found in cluster. Asterisks denote FDR (adjusted P-value) strength of upregulated genes in mouthparts to legs (left of number) and mouthparts to antennae (right of number) comparisons. Where more one than paralog is upregulated, the least significant FDRs are presented: *< 0.05, **< 1 × 10 − 5, ***< 1 × 10 − 10.
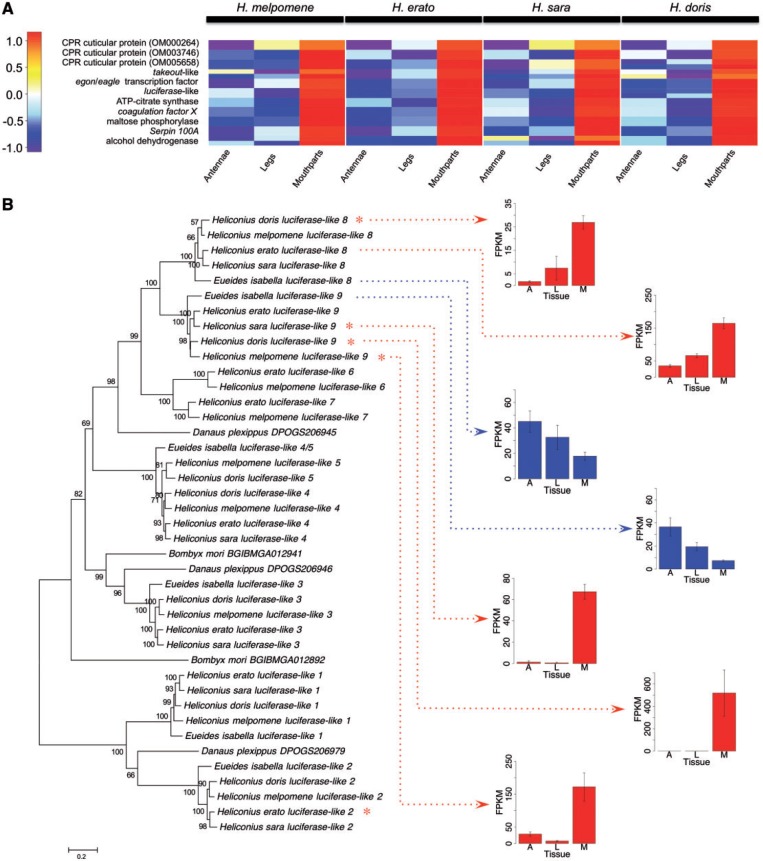
Fig. 5.—
Average expression levels of genes upregulated in Heliconius mouthparts and luciferase-like gene family phylogeny. (A) Color bar is the scaled log counts-per-million difference between tissues. Multiple homologs from a single orthocluster are presented as half-size boxes in the same row. Genes are also listed in table 4. (B) Maximum-likelihood tree of luciferase-like genes including bootstrap support values (above branches; < 50% values not shown) and branch length scale bar (substitutions per site). Red asterisks indicate genes significantly upregulated in mouthparts (FDR < 0.05, logFC > 1). H. erato luciferase-like 8 did not reach this threshold. Mean and standard error of gene expression (FPKM) for luciferase-like 8 and 9 are shown (Heliconius genes in red, E. isabella in blue). FPKM was normalized between species (to H. melpomene) using a normalization factor derived from a highly and stably expressed reference gene (Drosophila ortholog cryptocephal). A = antennae, L = legs and M = mouthparts.
We also examined genes that were upregulated in the mouthparts in any three Heliconius species, but not upregulated in E. isabella mouthparts (supplementary table S7, Supplementary Material online). This group consisted of 20 additional orthoclusters that had similar functions to the 11 Heliconius-specific clusters. Functions included one proteolysis-related peptidase cluster (OM001715), four cuticle-related clusters, including cuticle pigmentation (yellow-c; OM000067) and development, and nine clusters involved in metabolism including a glycoside hydrolase (myrosinase 1; OM000005).
Signal Peptide Analysis
To assess whether putative proteins upregulated in the mouthparts were destined for the secretory pathway, signal peptide analysis was performed on ORFs common to all species (25), and Heliconius alone (11) (see Supplementary Methods for details). Of these 36 proteins, 19 contained a signal peptide region in their N-terminal domain, suggesting they are secreted (supplementary table S8, Supplementary Material online). Seventeen of the 36 had digestion-related annotations, and of these 10 contained ORFs with signal peptides. These included cocoonase, coagulation factor X, Serpin100A, luciferase-like, lipase and GPAT. Additionally, all cuticular proteins contained signal peptides, as well as takeout-like, chemosensory, and juvenile hormone binding proteins. It should be noted that this method detects classically secreted proteins and thus the remaining ORFs could include nonclassically secreted proteins with currently unknown sequence motifs.
Cocoonase and Luciferase Gene Duplications
Cocoonase appears to be present as a single copy in the silkmoth Bombyx mori, the diamond backed moth Plutella xylostella, the monarch butterfly Danaus plexippus, and the Glanville fritillary Melitaea cinxia genomes (ISGC 2008; Zhan et al. 2011; You et al. 2013; Ahola et al. 2014). However, we uncovered a more complex evolutionary history of cocoonase within heliconiine butterflies. We discovered four tandem duplicates of cocoonase on genomic scaffold HE672036 of the H. melpomene genome, originally annotated as one large predicted gene containing 14 exons (HMEL006217; Dasmahapatra et al. 2012; fig. 4). Two additional cocoonases were discovered on an unscaffolded genomic contig (HMEL017107). These additional copies are considered here as different versions of cocoonase 5 due to the close similarity of their nucleotide sequences (named cocoonase 5a and 5b; fig. 4A). Each of the five cocoonase genes was present in the de novo transcriptomes of the other Heliconius species, except for H. erato cocoonase 3, and H. doris cocoonase 1 and 4. Only four cocoonase genes were present in the E. isabella transcriptome assembly. Phylogenetic analysis indicates that cocoonase 3 and 4 arose via a gene duplication event specific to Heliconius (fig. 4B). Most cocoonase paralogs were upregulated in the mouthparts across all five species (fig. 4B); however, while cocoonase paralogs 2–5 were upregulated across species, cocoonase 1 was not in any species, although it was still expressed in the proboscis (see below). Lengths of predicted amino acid sequences and their alignments are shown in figure 4C and D.
Maximum-likelihood trees of genes upregulated in Heliconius mouthparts also revealed other gene duplication events (fig. 5 and supplementary fig. S6, Supplementary Material online). For example, luciferase-like genes appear to have undergone 3–4 rounds of duplication in heliconiines, and a unique duplication event occurred in Heliconius producing luciferase-like 4 and 5, and resulting in nine Heliconius paralogous groups in total. Three luciferase-like paralogs showed mouthparts-upregulation, most notably paralogs 8 and 9; orthologs of these three genes in E. isabella did not show upregulation (fig. 5B).
Confirmation of Tissue-Specific Expression of Candidate Genes
To clarify whether mouthparts-upregulated genes related to metabolism are indeed expressed in the proboscis, we performed separate RT-PCRs using mRNA extracted from H. melpomene male and female proboscides, labial palps, and salivary glands (fig. 6). Overall, we found no difference in expression between the sexes. Peptidase (OM001715) and cocoonase 4 were exclusively expressed in the proboscis tissue, and coagulation factor X, cocoonase 1, cocoonase 5, and luciferase 9 displayed high expression levels in the proboscis with only faint bands amplifying from one or both of the other two tissues. Cocoonase 3 displayed similar expression levels in the proboscis and labial palps, with lower expression in the salivary glands. The remaining luciferases were either expressed across all tissues or had reduced expression in the proboscis. Serpin 100A and positive control EF1alpha were expressed across all tissue types.
Fig. 6.—
RT-PCR results of candidate genes involved in proteolysis and lipid metabolism in H. melpomene female and male proboscis, labial palps and salivary glands. Each row is a separate gene and columns are male and female samples from each tissue type, with the final column being the negative control (water). EF1alpha is a positive control and is expressed in every sample.
Discussion
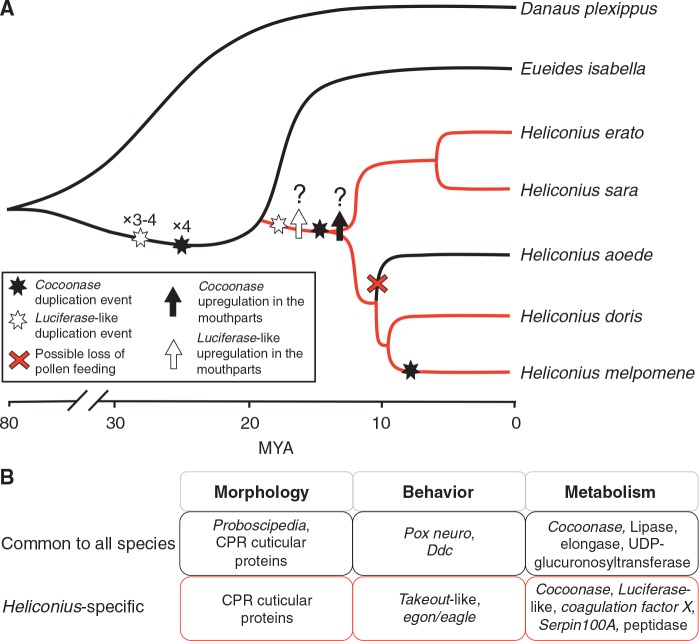
Pollen feeding in Heliconius is unique amongst butterflies and may have played an important role in their evolutionary history. We used RNA-Seq data from four species of Heliconius and one nonpollen feeding species, E. isabella, across three tissue types to identify genes that demonstrated upregulation in the proboscis and labial palps. We identified several hundred mouthparts-upregulated genes in each species, which were primarily involved in metabolism. Twenty-five of these genes were upregulated in all five species, and 11 displayed Heliconius-specific upregulation. We find evidence for both gene duplication and expression regulation changes during the evolution of mouthpart-specific gene expression, and these candidate genes provide a starting point from which to examine the genetic basis of pollen feeding (fig. 7).
Fig. 7.—
Cartoon of cocoonase and luciferase-like evolution within heliconiine butterflies. (A) Gene duplication events and potential regulatory changes mapped on to a species tree. Heliconiinae divergence time estimates obtained from Kozak et al. (2015), and D. plexippus divergence from Heliconiinae from Kozak et al. (2015) and Pohl et al. (2009) (range of 50–80 Ma). Red branches indicate pollen feeders and black branches nonpollen feeders. (B) Examples of mouthparts-upregulated genes common to heliconiine species and those specific to Heliconius, across three broad classes of phenotypes (morphology, behavior and metabolism).
Gene Expression in Butterfly Proboscis and Labial Palps
The majority of genes upregulated in the mouthparts of all five species was annotated with metabolic functions, some of which might be linked to their role in feeding. Digestion involves the three broad nutrient classes: proteins, carbohydrates and lipids (Terra and Ferreira 2012). Genes encoding enzymes involved in processing these three classes were seen in the Blast2GO annotations, particularly genes involved in proteolysis (supplementary figs. S2–S6, Supplementary Material online).
The broad functions of the Blast2GO annotations for genes upregulated in the mouthparts (supplementary fig. S1, Supplementary Material online) were similar to those of genes expressed in the saliva and salivary glands of several insect species. Transcriptional profiles of the salivary glands from the potato leaf hopper (Empoasca fabae), the white fly (Bemisia tabaci) and the western flower thrip (Frankliniella occidentalis) show the majority of genes to fall into the GO term metabolic process, closely followed by cellular process, localization or biological regulation (DeLay et al. 2012; Su et al. 2012). Some specific classes of genes are thought to be particularly important to insect digestion, including lipases, trypsin-like serine proteases and glycoside hydrolases (Valenzuela et al. 2003; Tunaz and Stanley 2004; Eberhard et al. 2007; Hosseininaveh et al. 2009; Shukle et al. 2009), all of which were detected as being upregulated in the mouthparts of the five species examined (table 3 and supplementary table S5, Supplementary Material online). However, despite the broad functional overlap between species, ortholog comparisons of mouthparts-upregulated genes revealed only twenty-five clusters common to all species.
One such cluster was annotated with proteolysis and contained five cocoonase homologs (OM000233) and a potential copy number variant in the reference genome of H. melpomene. Cocoonase aids in eclosure in moths by degrading sericin in silk that holds together the fibroin silk fibers; it is produced during the pupal-adult transformation by specialized cells in the galeae tissue of the proboscis (Kafatos and Williams 1964; Kafatos and Feder 1968; Law et al. 1977; Krenn 2010; Fukumori et al. 2014). Cocoonase is deposited as a dry enzyme on the proboscis and immediately mixed with a solvent produced by the labial glands, hydrolyzing the proteinaceous matrix of the cocoon (Kafatos et al. 1967). We found evidence for five duplication events of cocoonase genes within Lepidoptera, which led to four paralogous clades in heliconiine butterflies, a fourth duplication specific to Heliconius creating cocoonases 3 and 4, and a more recent duplication event leading to cocoonases 5a and 5b in H. melpomene (fig. 4). Given the digestive properties of cocoonase, the absence of a silk cocoon in butterflies, and the expression of duplicates in adult butterfly mouthparts, it is possible that the retention of heliconiine-specific duplications is linked to feeding in adults.
The presence of mRNA in adult mouthparts from genes encoding digestive enzymes further suggests that the proboscis might play a more direct role in digestion—and possibly, nutrient uptake (see below)—than previously thought. Our RT-PCR results indicate that several metabolism genes were highly expressed in the proboscis tissue but not in the labial palps or salivary glands (fig. 6). Further, many of the digestion annotated genes also contained signals of secretion, including cocoonase, suggesting that the proboscis might directly secrete digestive enzymes. However, few studies have examined the protein content of insect saliva directly (rather than salivary glands). Interestingly, at least one protein directly secreted by the proboscis, cocoonase (Kafatos et al. 1967), has also been isolated from butterfly saliva (Harpel et al. 2015). Harpel et al. (2015) isolated 31 proteins from H. melpomene aglaope saliva that were annotated with carbohydrate hydrolysis, proteolysis, and immunity functions. Of these 31 saliva proteins, 19 were expressed in the mouthparts of our H. melpomene individuals with > 1 FPKM average expression level, although only two, matching the previous annotations of cocoonase genes (HMEL006217 and HMEL017107), were upregulated in the mouthparts. The presence of key digestive enzyme transcripts in the mouthparts of butterflies suggests that the proboscis, in conjunction with the salivary glands, could be a source of enzymes involved in extraoral digestion. Intriguingly, we found amino acid transporters upregulated in the mouthparts of most species, suggesting that the proboscis might also play a role in nutrient absorption (supplementary table S5, Supplementary Material online).
Gene Expression in Heliconius Proboscis and Labial Palps
Several morphological, behavioral and metabolic phenotypes have been associated with pollen feeding that could potentially be the result of adaptation. These include proboscis size and shape, pollen processing behavior, and the secretion of digestive enzymes, particularly proteases. We discovered genes upregulated in the mouthparts of Heliconius that were annotated with functions for each of these three categories (fig. 7).
Morphology related expression included three clusters of CPR cuticle genes. The precise role of cuticle proteins in the formation of insect cuticles is still unclear, however amino acid motifs common to all CPR proteins, RR consensus motifs (Rebers and Riddiford 1988), are known to play a key role in cuticle structure. Lepidopterans, such as Manduca sexta, have an enormous repertoire of these genes, more than any other insect for which these proteins have been characterized (Dittmer et al. 2015). Proteins containing RR motifs can be split into three groups. The RR1 motif is predominantly found in soft (flexible) tissues, the RR2 in hard (rigid) tissues and the tissue specificity of a third form, RR3, has yet to be elucidated (Andersen 1998, 2000; Iconomidou et al. 2005). Both RR1 and RR3 families of CPRs were upregulated in both Eueides and Heliconius mouthparts (table 3). Three additional clusters of CPRs with RR1 motifs were upregulated only in Heliconius mouthparts (table 4), suggesting a critical role for flexible tissue cuticle genes in Heliconius proboscis morphology.
Two genes annotated with specific functions for feeding behavior and development, takeout-like (OM000111) and egon/eagle (OM003483) respectively, were upregulated in Heliconius mouthparts (supplementary fig. S7, Supplementary Material online, table 4). The circadian clock-regulated gene takeout is a transcription factor expressed in the head, proventriculus, crop, and antennae in Drosophila and appears to be a direct molecular link between the circadian clock and the feeding/starvation response (Sarov-Blat et al. 2000). Further, takeout has been associated with the aging process and overexpression of this gene in the adult nervous system, head fat body or abdominal fat body of Drosophila can lead to around a 20% increase in longevity (Bauer et al. 2010). In Drosophila, the embryonic gonad gene egon/eagle plays a role in neurological differentiation during development, specifying the fate of serotonin neurons (Lundell and Hirsh 1998). Serotonin is a neurotransmitter that is conserved across both vertebrates and invertebrates (Lundell and Hirsh 1998), and plays a general role in locomotion, and in feeding behavior in ants (Falibene et al. 2012), bees (French et al. 2014) and flies (Dacks et al. 2003; Neckameyer 2010). The upregulation of gene family members of these two feeding behavior genes in the mouthparts of Heliconius species implicates them as potential candidate genes for the unique feeding behavior seen in this group.
Finally, the nutritional content of pollen can vary greatly, however the general consensus for the composition of pollen collected by bees is 10–40% protein, 1–13% lipid, and 12–55% carbohydrate (Campos et al. 2008). Orthologs of the firefly luciferase were upregulated in Heliconius mouthparts and can have a dual function, one as an ATP-dependent monooxygenase in bioluminescence pathways, and another in lipid metabolism due to its structural similarity to fatty acyl-CoA synthetase (Oba et al. 2003). One key predication of this study was that, due to the high protein content of pollen, genes relating to protein digestion would show signals of strong upregulation in the mouthparts of Heliconius species and not the nonpollen feeding Eueides species. Indeed we found proteases and a protease-inhibitor (Serpin100A) with Heliconius-specific mouthparts upregulation. However, we also found proteolysis genes to be upregulated in the mouthparts of E. isabella (supplementary fig. S6, Supplementary Material online) suggesting that nonpollen feeders may have the capability to digest pollen-derived amino acids. Terra and Ferreira (1994) note that the presence of complex digestive enzymes in nectar-feeding moths, and the ability of adult Lepidoptera to feed on very different nutritional sources (e.g., pollen, blood, feces, and more), suggests there might be no enzymatic constraint other than perhaps protein expression levels to the evolution of feeding on additional food sources (Banziger 1970; Gilbert 1972). Nectar can contain trace amounts of proteins and free amino acids either naturally, or released from pollen grains (Linskens and Schrauwen 1969; Gilbert 1972). Thus some nonpollen feeders might be exposed to, and be able to metabolize, amino acids derived from pollen. It has been hypothesized that feeding on amino acids in nectar coupled with the proboscis grooming behavior initiated by pollen grains caught on the proboscis might have been the starting point towards the evolution of pollen feeding in Heliconius (Erhardt and Baker 1990; Hikl and Krenn 2011). This in turn suggests that the evolution of pollen feeding might have begun with behavioral and morphological changes involving the collection, retention, and processing of pollen.
Conclusions
We find evidence of gene duplication events and regulatory changes in genes upregulated in the Heliconius proboscis and labial palps, and identify several candidate genes for pollen feeding behavior. For example, luciferase-like genes demonstrated a recent gene expansion event followed by the evolution of tissue-specific expression in Heliconius species (figs. 5B and 7). This suggests that the upregulation of luciferase-like genes could have played a role in the Heliconius pollen-feeding adaptation. One way to test this hypothesis would be to examine the levels of luciferase-like expression in Neruda spp. Neruda consists of several species that have been reclassified as members of the genus Heliconius (Beltrán et al. 2007; Kozak et al. 2015) and which appear to have secondarily lost the pollen-feeding trait (Gilbert 1972; Brown 1981; Neil Rosser, pers. comm.). We predict that levels of luciferase-like expression in Neruda would resemble those of Eueides. Future genetic studies examining pollen feeding should quantify gene expression in the Heliconius salivary glands, investigate enzyme production by the proboscis, and examine gene expression linked to the fitness benefits of pollen feeding.
Supplementary Material
Acknowledgments
We thank Paola Vargas of the Entomological Supply Company in Costa Rica for going out of her way to provide us with butterfly pupae. Also Ana Catalan, Kyle McCulloch, Donovan German, and Susan Finkbeiner for comments on the manuscript, and three anonymous reviewers for their help with improving the manuscript. Nucleotide sequences have been deposited in GenBank under accession numbers KU925713–KU925777. This work was partially supported by National Science Foundation cooperative agreement DBI-0939454 to A.D.B.
Supplementary Material
Supplementary figures S1–S7 and tables S1–S8 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Literature Cited
- Abzhanov A, Holtzman S, Kaufman TC. 2001. The Drosophila proboscis is specified by two Hox genes, proboscipedia and Sex combs reduced, via repression of leg and antennal appendage genes. Development 128:2803–2814. [DOI] [PubMed] [Google Scholar]
- Ahola V, et al. 2014. The Glanville fritillary genome retains an ancient karyotype and reveals selective chromosomal fusions in Lepidoptera. Nat Commun.. 5:4737.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SO. 1998. Amino acid sequence studies on endocuticular proteins from the desert locust, Schistocera gregaria. Insect Biochem Mol Biol.. 28:421–434. [DOI] [PubMed] [Google Scholar]
- Andersen SO. 2000. Studies on proteins in post-ecdysial nymphal cuticle of locust, Locusta migratoria, and cockroach, Blaberus craniifer. Insect Biochem Mol Biol.. 30:569–577. [DOI] [PubMed] [Google Scholar]
- Apweiler R, et al. 2004. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res.. 32:D115–D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banziger H. 1970. The piercing mechanism of the fruit-piercing moth Calpe (calyptra) thalictri Bkh. (Noctuidae) with reference to the skin-piercing blood-sucking moth C. eustrigata Hmps. Acta Trop. 27:54–88. [PubMed] [Google Scholar]
- Bauer J, et al. 2010. Comparative transcriptional profiling identifies takeout as a gene that regulates life span. Aging 2:298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrán M, Jiggins CD, Brower AVZ, Bermingham E, Mallett J. 2007. Do pollen feeding, pupal-mating and larval gregariousness have a single origin in Heliconius butterflies? Inferences from multilocus DNA sequence data. Biol J Linnean Soc. 92:221–239. [Google Scholar]
- Blekhman R, et al. 2014. Comparative metabolomics in primates reveals the effects of diet and gene regulatory variation on metabolic divergence. Sci Rep. 4:5809.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman R, Oshlack A, Chabot AE, Smyth GK, Gilad Y. 2008. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet. 4:e1000271.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs CL. 1979. Resource allocation and reproductive strategies in several heliconiine butterfly species. PhD thesis. Austin, USA: University of Texas.
- Boggs CL. 1986. Reproductive strategies of female butterflies: variation in and constraints on fecundity. Ecol Entomol. 11:7–15. [Google Scholar]
- Briscoe AD, et al. 2013. Female behaviour drives expression and evolution of gustatory receptors in butterflies. PLoS Genet. 9:e1003620.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KS. 1981. The biology of Heliconius and related genera. Annu Rev Entomol. 36:427–456. [Google Scholar]
- Campos MGR, et al. 2008. Pollen composition and standardisation of analytical methods. J Apic Res. 47:154–161. [Google Scholar]
- Cardoso MZ, Gilbert LE. 2013. Pollen feeding, resource allocation and the evolution of chemical defence in passion vine butterflies. J Evol Biol. 26:1254–1260. [DOI] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. 2007. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 8:93–103. [DOI] [PubMed] [Google Scholar]
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S. 2008. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook SM, Murray DA, Williams IH. 2004. Do pollen beetles need pollen? The effect of pollen on oviposition, survival, and development of a flower-feeding herbivore. Ecol Entomol. 29:164–173. [Google Scholar]
- Dacks AM, Nickel T, Mitchell BK. 2003. An examination of serotonin and feeding in the flesh fly Neobellieria bullata (Sarcophagidae: Diptera). J Insect Behav. 16:1–21. [Google Scholar]
- Dasmahapatra KK, et al. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLay B, et al. 2012. Transcriptome analysis of the salivary glands of potato leafhopper, Empoasca fabae. J Insect Physiol. 58:1626–1634. [DOI] [PubMed] [Google Scholar]
- De Luca M, et al. 2003. Dopa decarboxylase (Ddc) affects variation in Drosophila longevity. Nat Genet. 34:429–433. [DOI] [PubMed] [Google Scholar]
- Dittmer NT, et al. 2015. Annotation and expression analysis of cuticular proteins from the tobacco hornworm, Manduca sexta. Insect Biochem Mol. 62:100–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap-pianka H, Boggs CL, Gilbert LE. 1977. Ovarian dynamics in heliconiine butterflies: programmed senescence versus eternal youth. Science 197:487–490. [DOI] [PubMed] [Google Scholar]
- Eberhard SH, Krenn HW. 2003. Salivary glands and salivary pumps in adult Nymphalidae (Lepidoptera). Zoomorphology 122:161–167. [Google Scholar]
- Eberhard SH, Hrassnigg N, Crailsheim K, Krenn HW. 2007. Evidence of protease in the saliva of the butterfly Heliconius melpomene (L.) (Nymphalidae, Lepidoptera). J Insect Physiol. 53:126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard SH, Nemeschkal HL, Krenn HW. 2009. Biometrical evidence for adaptations of the salivary glands to pollen feeding in Heliconius butterflies (Lepidoptera: Nymphalidae). Biol J Linnean Soc. 97:604–612. [Google Scholar]
- Erhardt A, Baker I. 1990. Pollen amino acids - an additional diet for a nectar feeding butterfly. Plant Syst Evol. 169:111–121. [Google Scholar]
- Falibene A, Rossler W, Josens R. 2012. Serotonin depresses feeding behaviour in ants. J Insect Physiol. 58:7–17. [DOI] [PubMed] [Google Scholar]
- French AS, et al. 2014. The role of serotonin in feeding and gut contractions in the honeybee. J Insect Physiol. 61:8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumori H, et al. 2014. Purification and characterization of cocoonase from the silkworm Bombyx mori. Biosci Biotechnol Biochem. 78:202–211. [DOI] [PubMed] [Google Scholar]
- Gilbert FS. 1985. Ecomorphological relationships in hoverflies (Diptera, Syrphidae). Proc R Soc Lond B Biol Sci. 224:91–105. [Google Scholar]
- Gilbert LE. 1972. Pollen feeding and reproductive biology of Heliconius butterflies. Proc Natl Acad Sci U S A. 69:1403–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LE. 1991. Biodiversity of a Central American Heliconius community: pattern, process, and problems In: Price PW, Lewinsohn TM, Fernandes TW, and Benson WW. Plant-animal interactions: evolutionary ecology in tropical and temperate regions. New York: John Wiley; p. 403–427. [Google Scholar]
- Gilbert LI. 1967. Lipid metabolism and function in insects. Adv Insect Physiol. 4:69–211. [Google Scholar]
- Gimeno RE, Cao J. 2008. Mammalian glycerol-3-phosphate acyltransferases: new genes for an old activity. J Lipid Res. 49:2079–2088. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpel D, Cullen DA, Ott SR, Jiggins CD, Walters JR. 2015. Pollen feeding proteomics: salivary proteins of the passion flower butterfly, Heliconius melpomene. Insect Biochem Mol. 63:7–13. [DOI] [PubMed] [Google Scholar]
- Haygood R, Fedrigo O, Hanson B, Yokoyama KD, Wray GA. 2007. Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nat Genet. 39:1140–1144. [DOI] [PubMed] [Google Scholar]
- Hikl AL, Krenn HW. 2011. Pollen processing behavior of Heliconius butterflies: a derived grooming behavior. J Insect Sci. 11:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne I, Haritos VS, Oakeshott JG. 2009. Comparative and functional genomics of lipases in holometabolous insects. Insect Biochem Mol Biol. 39:547–567. [DOI] [PubMed] [Google Scholar]
- Hosseininaveh V, Bandani A, Hosseininaveh F. 2009. Digestive proteolytic activity in the Sunn pest, Eurygaster integriceps. J Insect Sci. 9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou YCC, Chittaranjan S, Barbosa SG, McCall K, Gorski SM. 2008. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J Cell Biol. 182:1127–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iconomidou VA, Willis JH, Hamodrakas SJ. 2005. Unique features of the structural model of 'hard' cuticle proteins: implications for chitin-protein interactions and cross-linking in cuticle. Insect Biochem Mol Biol. 35:553–560. [DOI] [PubMed] [Google Scholar]
- Imdorf A, Rickli M, Kilchenmann V, Bogdanov S, Wille H. 1998. Nitrogen and mineral constituents of honey bee worker brood during pollen shortage. Apidologie 29:315–325. [Google Scholar]
- Ingram CJ, Mulcare CA, Itan Y, Thomas MG, Swallow DM. 2009. Lactose digestion and the evolutionary genetics of lactase persistence. Hum Genet. 124:579–591. [DOI] [PubMed] [Google Scholar]
- ISGC (International Silkworm Genome Consortium). 2008. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. 38:1036–1045. [DOI] [PubMed] [Google Scholar]
- Kafatos FC, Feder N. 1968. Cytodifferentiation during insect metamorphosis: the galea of silkmoths. Science 161:470–472. [DOI] [PubMed] [Google Scholar]
- Kafatos FC, Tartakoff AM, Law JH. 1967. Cocoonase: I. Preliminary characterization of a proteoloytic enzyme from silk moths. J Biol Chem. 242:1477–1487. [PubMed] [Google Scholar]
- Kafatos FC, Williams CM. 1964. Enzymatic mechanism for the escape of certain moths from their cocoons. Science 146:538–540. [DOI] [PubMed] [Google Scholar]
- Kozak KM, et al. 2015. Multilocus species trees show the recent adaptive radiation of the mimetic Heliconius butterflies. Syst Biol. 64:505–524. doi:10.1093/sysbio/syv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenn HW. 2010. Feeding mechanisms of adult Lepidoptera: structure, function, and evolution of the mouthparts. Annu Rev Entomol. 55:307–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenn HW, et al. 2009. Mechanical damage to pollen aids nutrient acquisition in Heliconius butterflies (Nymphalidae). Arthropod Plant Interact. 3:203–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenn HW, Penz CM. 1998. Mouthparts of Heliconius butterflies (Lepidoptera: Nymphalidae): a search for anatomical adaptations to pollen-feeding behavior. Int J Insect Morphol. 27:301–309. [Google Scholar]
- Law JH, Dunn PE, Kramer KJ. 1977. Insect proteases and peptidases In: Meister A, editor. Advances in enzymology and related areas of molecular biology. New York: John Wiley; p. 389–425. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens HF, Schrauwen J. 1969. Release of free amino acids from germinating pollen. Acta Bot Neerl. 18:47–53. [Google Scholar]
- Luca F, Perry GH, Di Rienzo A. 2010. Evolutionary adaptations to dietary changes. Annu Rev Nutr. 30:291–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundell MJ, Hirsh J. 1998. eagle is required for the specification of serotonin neurons and other neuroblast 7-3 progeny in the Drosophila CNS. Development 125:463–472. [DOI] [PubMed] [Google Scholar]
- Müller F. 1879. Ituna and Thyridia: a remarkable case of mimicry in butterflies. Trans Entomol Soc Lond. xx–xxix. [Google Scholar]
- Neckameyer WS. 2010. A trophic role for serotonin in the development of a simple feeding circuit. Dev Neurosci. 32:217–237. [DOI] [PubMed] [Google Scholar]
- Nottebohm E, Damblychaudiere C, Ghysen A. 1992. Connectivity of chemosensory neurons is controlled by the gene Poxn in Drosophila. Nature 359:829–832. [DOI] [PubMed] [Google Scholar]
- Oba Y, Ojika M, Inouye S. 2003. Firefly luciferase is a bifunctional enzyme: ATP-dependent monooxygenase and a long chain fatty acyl-CoA synthetase. FEBS Lett. 540:251–254. [DOI] [PubMed] [Google Scholar]
- Oda S, Fukami T, Yokoi T, Nakajima M. 2015. A comprehensive review of UDP-glucuronosyltransferase and esterases for drug development. Drug Metab Pharmacokinet. 30:30–51. [DOI] [PubMed] [Google Scholar]
- O'Brien DM, Boggs CL, Fogel ML. 2003. Pollen feeding in the butterfly Heliconius charitonia: isotopic evidence for essential amino acid transfer from pollen to eggs. Proc R Soc Lond B Biol Sci. 270:2631–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, et al. 2007. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 39:1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploner A. 2012. Heatplus: heatmaps with row and/or column covariates and colored clusters. R package version 2.0. Available at: http://bioconductor.org/packages/release/bioc/html/Heatplus.html
- Pohl N, Sison-Mangus MP, Yee EN, Liswi SW, Briscoe AD 2009. Impact of duplicate gene copies on phylogenetic analysis and divergence time estimates in butterflies. BMC Evol Biol. 9:99. [DOI] [PMC free article] [PubMed]
- Rebers JE, Riddiford LM. 1988. Structure and expression of a Manduca sexta larval cuticle gene homologous to Drosophila cuticle genes. J Mol Biol. 203:411–423. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulston TH, Cane JH. 2000. Pollen nutritional content and digestibility for animals. Plant Syst Evol. 222:187–209. [Google Scholar]
- Sarov-Blat L, So WV, Liu L, Rosbash M. 2000. The Drosophila takeout gene is a novel molecular link between circadian rhythms and feeding behavior. Cell 101:647–656. [DOI] [PubMed] [Google Scholar]
- Shukle RH, Mittapalli O, Morton PK, Chen MS. 2009. Characterization and expression analysis of a gene encoding a secreted lipase-like protein expressed in the salivary glands of the larval Hessian fly, Mayetiola destructor (Say). J Insect Physiol. 55:104–111. [DOI] [PubMed] [Google Scholar]
- Smith G, Briscoe AD. 2015. Molecular evolution and expression of the CRAL_TRIO protein family in insects. Insect Bio Mol Bio. 62:168–173. [DOI] [PubMed] [Google Scholar]
- Soderlund C, Nelson W, Willer M, Gang DR. 2013. TCW: transcriptome computational workbench. PLoS One 8:e69401.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 100:9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YL, et al. 2012. Transcriptomic analysis of the salivary glands of an invasive whitefly. PLoS One 7:e39303.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A. 2011. Differential expression in RNA-seq: a matter of depth. Genome Res. 21:2213–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel BL, Livingstone MS, Quinn WG. 1984. Mutations in the dopa decarboxylase gene affect learning in Drosophila. Proc Natl Acad Sci U S A. 81:3577–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terra WR, Ferreira C. 1994. Insect digestive enzymes: properties, compartmentalization and function. Comp Biochem Physiol. 109:1–62. [Google Scholar]
- Terra WR, Ferreira C. 2012. Molecular and evolutionary physiology of insect digestion In: Panizzi AR, Parra JRP, editors. Insect bioecology and nutrition for integrated pest management. Boca Raton, FL: CRC Press; p. 93–119. [Google Scholar]
- Tunaz H, Stanley DW. 2004. Phospholipase A(2) in salivary glands isolated from tobacco hornworms, Manduca sexta. Comp Biochem Phys B. 139:27–33. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IMB, Pham VM, Garfield MK, Ribeiro JMC. 2003. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem Mol Biol. 33:717–732. [DOI] [PubMed] [Google Scholar]
- van Schooten B, Jiggins C, Briscoe AD, Papa R. 2016. Genome-wide analysis of ionotropic receptors provides insight into their evolution in Heliconius. BMC Genomics 17:254.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackers FL, Romeis J, Van Rijn P. 2007. Nectar and pollen feeding by insect herbivores and implications for multitrophic interactions. Annu Rev Entomol. 52:301–323. [DOI] [PubMed] [Google Scholar]
- Wendel AA, Lewin TM, Coleman RA. 2009. Glycerol-3-phosphate acyltransferases: rate limiting enzymes of triacylglycerol biosynthesis. BBA Mol Cell Biol Lipids. 1791:501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 8:206–216. [DOI] [PubMed] [Google Scholar]
- You M, et al. 2013. A heterozygous moth genome provides insights into herbivory and detoxification. Nat Genet. 45:220–225. [DOI] [PubMed] [Google Scholar]
- Zhan S, Merlin C, Boore JL, Reppert SM. 2011. The monarch butterfly genome yields insights into long-distance migration. Cell 147:1171–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. 2003. Evolution by gene duplication: an update. Trends Ecol Evol. 18:292–298. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.