Abstract
Misfolding of the prion protein (PrP) is the key step in the transmission of spongiform pathologies in humans and several animals. Although PrP is highly conserved in mammals, a few changes in the sequence of endogenous PrP are proposed to confer protection to dogs, which were highly exposed to prion during the mad-cow epidemics. D159 is a unique amino acid found in PrP from dogs and other canines that was shown to alter surface charge, but its functional relevance has never been tested in vivo. Here, we show in transgenic Drosophila that introducing the N159D substitution on mouse PrP decreases its turnover. Additionally, mouse PrP-N159D demonstrates no toxicity and accumulates no pathogenic conformations, suggesting that a single D159 substitution is sufficient to prevent PrP conformational change and pathogenesis. Understanding the mechanisms mediating the protective activity of D159 is likely to lessen the burden of prion diseases in humans and domestic animals.
Keywords: Prion protein, neurotoxicity, dog, mouse, Drosophila, misfolding, protective substitution, sequence alignment
Introduction
Prion diseases are aggressive neurodegenerative disorders affecting humans and other mammals. These conditions are characterized by spongiform brain pathology and deposition of misfolded isoforms of the prion protein (PrP) (Colby and Prusiner, 2011; Prusiner, 1998). Protease resistant conformations of PrP termed scrapie PrP (PrPSc) are transmissible, but transmission requires conformational conversion of the host cellular PrP (PrPC) (Bueler et al., 1993; Prusiner et al., 1993; Telling et al., 1995). Despite dedicated efforts to uncover the intrinsic and extrinsic factors regulating PrP conversion, we still have a limited understanding of the molecular mechanisms mediating pathogenesis. One avenue to shed light on these complex processes consists on exploiting the natural resistance of some animals to prion diseases. Scrapie is an endemic disease in sheep and goat that is not transmissible to humans, but can be transmitted experimentally to rodents (mouse, rat, hamster, bank vole) (Chandler, 1971; Chandler and Fisher, 1963; Zlotnik and Rennie, 1963; Zlotnik and Rennie, 1965). The accidental transmission of scrapie to bovine resulted in the mad cow epidemics of the 1980’s and variant Creutzfeldt-Jacob disease in humans. Contaminated bone meal subsequently spread prions to several domestic and zoo animals, demonstrating the susceptibility of many mammals to prion diseases, even felines (cat, tigers) and mustelids (mink, ferret) in which the disease is not endemic (Kirkwood and Cunningham, 1994; Kretzschmar et al., 1992). The exposure of dogs to prions was comparable to that of other domestic and zoo animals, but despite the best veterinary care not a single case of prion disease has been described among dogs (Kirkwood and Cunningham, 1994). Transmission experiments in MDCK dog kidney cells have shown that they do not replicate human and mouse prions despite normal biogenesis of endogenous PrP, further supporting the resistance of dogs to prions (Polymenidou et al., 2008). Since endogenous PrP is critical for prion transmission and prion disease, the identification of animals resistant to prion diseases suggest that either their endogenous PrPC is conformationally stable or their cells express co-factors that prevent PrP misfolding. Identifying the factors promoting dog PrP stability can reveal key insight to understand PrP misfolding and pathogenesis.
The C-terminal region of PrP contains a globular domain comprising three helices and a short β-sheet (James et al., 1997; Riek et al., 1996). NMR studies of dog (Canis genus) PrP (CaPrP) found that the structure of its globular domain is highly conserved compared to that of rodent or human PrP (Lysek et al., 2005). Sequence alignment identified a few amino acid substitutions in dogs not present in susceptible animals and humans. Interestingly, only four amino acids differentiate the globular domain of dog and cat PrP, resistant and susceptible animals respectively. D159 and R177 (human PrP numbering, see Fig. 1A) are unique substitutions in CaPrP (Lysek et al., 2005) that substitute the highly conserved N159 and H177 in cat PrP and in most mammals. D159 is on the α1-β2 loop and exposed on the surface of CaPrP, resulting in increased negative charge. This amino acid is polymorphic in dogs, with 68% carrying D and 32% carrying E at 159 (Stewart et al., 2012), both acidic residues that introduce the same surface alterations. Other members of the dog family, canines like wolfs and foxes, also carry D159, suggesting that they enjoy the same protection to prion diseases as dogs. It has been proposed that this change in surface charge will result in altered interactions with other proteins, possibly proteins that contribute to PrP conversion. Additionally, in vitro studies have demonstrated the high stability of CaPrP under strong denaturing conditions, more so than its rabbit and horse cognates, two other prion resistant animals (Khan et al., 2010). However, the functional relevance of the D159 residue has not been probed in vivo.
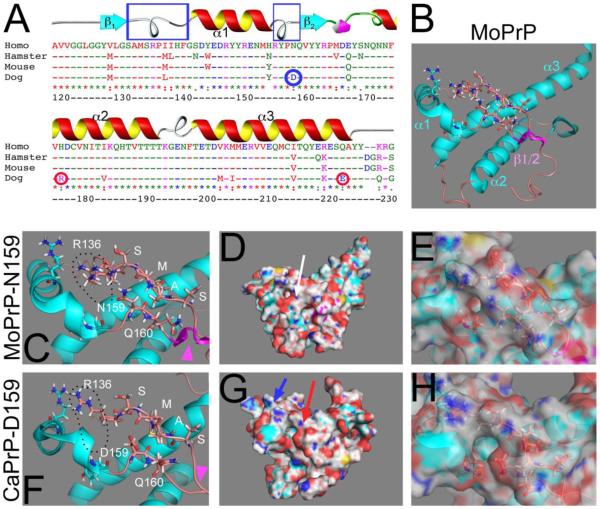
Figure 1. D159 is unique in dog PrP and affects surface charge.
A, Sequence alignment (ClustalW) for the globular domain of PrP from human, Chinese hamster, mouse, and dog. The numbering corresponds to human PrP. CaPrP has three non-conserved substitutions, N159 (blue circle) in α1-β2 loop, R177 in helix 2 and E223 (red circles). The two loops that are close in the 3D structure are indicated by blue boxes. B, 3D structure of MoPrP in Cartoon view indicating the secondary structures. C, Position of N159 and regional effects in the Cartoon view of MoPrP. R136 in the α1-β1 loop is open, creating a negative charge. D and E, Surface view of MoPrP showing the mostly neutral central region (D, arrow). In the semi-transparent view, the position of N159 is shown with respect to the charge (E). F, Position of D159 and regional effects in the Cartoon view of CaPrP. D159 causes a new negative surface charge that also affects distant residues, like the displacement of R136 and its negative charge. G and F, Surface view of CaPrP. New positive (blue arrow) and negative (red arrow) charges appear on the surface of CaPrP (G). The position of D159 with respect of charge is shown (F).
Here, we describe the generation of transgenic flies expressing mutant PrP from mouse carrying the D159 substitution from CaPrP to examine the ability of a single amino acid change to confer conformational stability and suppress toxicity. We and others showed previously that mouse PrP (MoPrP)-WT induces progressive toxicity in locomotor neurons associated with the accumulation of pathogenic conformations (Fernandez-Funez et al., 2010; Gavin et al., 2006). In contrast, MoPrP-N159D has lower turnover than MoPrP-WT, suggesting that Drosophila cells can detect aberrant structural features on MoPrP-WT and promote its degradation. Moreover, flies expressing MoPrP-N159D show no progressive locomotor dysfunction and no accumulation of pathogenic conformations. Overall, these studies identify D159 as a key residue responsible for preventing PrP conformational change and disease. This knowledge can be leveraged to search for genetic or pharmacologic agents that stabilize these domains and prevent PrP pathogenesis in humans and farm animals.
Materials and Methods
Sequence alignment and protein modeling
The alignment of the globular domain of human, Syrian hamster, mouse, and dog PrP spanning positions 120-231 was done on ClustalW2 (ebi.ac.uk/Tools/clustalw2) using human PrP as reference. Amino acid sequences were obtained from the NCBI protein database with the following accession numbers: AAH22532 (human), B34759 (Syrian hamster), AAA39996 (mouse) and ACO71291 (dog). The color- coded amino acids indicate properties relevant for protein structure (polarity and charge). To generate 3D views of mouse and dog PrP, we opened in PyMOL (pymol.org) the NMR structures for MoPrP (2L1H) and CaPrP (1XYK) from the RSCB Protein Data Bank (rcsb.org/pdb). We visualized the proteins in Cartoon and Surface formats, and showed only relevant amino acids to optimize their recognition. In the Surface format, we created an opaque representation and another with 40% transparency to visualize the underlying amino acids.
Molecular and Drosophila genetics
The cDNA for mouse PrP carrying the N159D substitutions (MoPrP-N159D) was kindly provided by J. Castilla. We amplified the construct with specific primers containing EcoRI and NotI sites to facilitate cloning into the pUAST vector to generate transgenic flies (18). The construct was injected into yw embryos at Rainbow Transgenics following standard procedures (37) to generate multiple independent transgenic lines. MoPrP-WT flies were kindly provided by S. Supatappone (Gavin et al., 2006). The reporter line UAS-CD8-GFP, expressed GFP attached to the membrane (Lee et al., 1999) and the driver lines da-Gal4 (ubiquitous expression) and OK107-Gal4 (mushroom bodies) were obtained from the Bloomington Drosophila Stock Center. The BG380-Gal4 (motor neuron) driver was a gift from B. Zhang (University of Missouri).
Quantitative RT-PCR
To quantify the levels of MoPrP-WT and MoPrP-N159D mRNA transcripts, we performed real time RT- PCR assays, using Taqman probes labeled with FAM. Total RNA was isolated (RNAqueous, Ambion) from ten whole flies expressing MoPrP-WT or MoPrP-N159D ubiquitously under the control of da-Gal4. Real-time PCRs reactions were done using Taqman Universal PCR Master Mix (Applied Biosystems) and were run in the ABI PRISM 7500 system (Applied Biosystems). RNA polymerase II (RpII) mRNA was used as control to normalize the expression for the two groups. The following primers and probes were used: MoPrP-Fw: GAGACCGATGTGAAGATGATGGA; MoPrP-Rev: TAATAGGCCTGGGACTCCTTCTG; MoPrP probe: (FAM)-CGTGGTGGAGCAGAT; dRpII-Fw: CAGAGTCCGCGTAACACCTA; dRpII-Rev: CAGGTACTCCATGGAACGTG; and dRpII probe (FAM)-TCCGATGAAGCCACTTGTGACCA. Primers and probes were designed using the GeneScript® TaqMan real-time PCR design software and the experiment was done in six technical replicates. The experiment was done in six technical replicates. The graphs showing normalized levels of PrP transcripts were made in excel.
Tissue homogenates and western blot
One whole fly expressing MoPrP-WT or –N159D ubiquitously under the control of da-Gal4 were homogenized in 30 μl of RIPA buffer (Thermo Scientific) and Complete protease inhibitors (Roche Applied Science) using a motorized pestle. Protein extracts were fractionated by sodium dodecyl sulfate- polyacrylamide gel electrophoresis (SDS-PAGE) in 4-12 % Bis–Tris gels (Invitrogen) under reducing conditions and electroblotted into nitrocellulose membranes. Membranes were blocked in TBS-T containing 5% non-fat milk, and probed against the 6H4 anti-PrP antibody (1: 10,000, Prionics) and the anti-α-tubulin (1:200,000, Sigma) antibody as loading control, followed by HRP-conjugated secondary antibodies at 1:2,500. The 6H4 antibody detects a conserved epitope in helix 1 (amino acids 144–152) not present in hamster PrP that is sensitive to the state of the disulfide bond. Immunoreactive bands were visualized by enhanced chemiluminescence (ECL, Amersham). For quantification of MoPrP-WT and – N159D, we performed three independent western blots and quantified the PrP bands by normalizing with respect to the Tubulin bands.
Immunofluorescence and image processing
For subcellular localization, we co-expressed the MoPrP and CD8-GFP as a cell membrane marker in interneurons of the larval ventral ganglion under the control of OK107-Gal4. Whole-mount immunohistochemistry of fixed larval brains was conducted by fixing in 4% formaldehyde, washing with PBT, and blocking with 3% bovine serum albumin before incubating with the primary antibody as described previously (Fernandez-Funez et al., 2010). We incubated first with the 6H4 anti-PrP antibody (1: 1,000) followed by the secondary antibody anti-mouse-Cy3 (Molecular Probes) at 1:600. We mounted the stained larval brain in Vectashield antifade (Vector) mounting medium for microscopic observation and documentation. We collected two-channel fluorescent images with AxioVision (Zeiss) in an Axio- Observer Z1 microscope (Zeiss) by optical sectioning using ApoTome (structured light microscopy) with a 63x NA: 1.4 (oil) objective. Representative single plane images of local interneurons were extracted from Z-stacks. Final images were combined using Adobe Photoshop and processing included brightness/contrast adjustment to whole images for optimal viewing and printing.
Locomotor Assay
Flies expressing MoPrP or control (LacZ) constructs were crossed with the BG380-Gal4 driver, raised at 27°C, and the progeny was collected at day 1 post-eclosion, and subjected to climbing assays with the genotypes blinded to the experimenter as previously described (Le Bourg and Lints, 1992). Briefly, 25 newborn adult females were placed in empty vials and forced to the bottom by firmly tapping against the bottom. After 8 seconds, the number of flies that climb above 5 cm was recorded. This was repeated 8 times every day for the first 10 days and every other day until all the flies stopped climbing. The climbing index was calculated as flies above line / total flies × 100 and plotted as a function of age in Microsoft Excel. Data points are presented as mean values ± standard deviation.
Immunoprecipitation Assays
We detected pathogenic PrP conformations in Drosophila homogenates using the 15B3 immunoprecipitation kit (Prionics AG, Switzerland) (Korth et al., 1997). The monoclonal 15B3 conformational antibody was bound to Dynabeads M-280 Tosylactivated as specified by the manufacturer (Invitrogen). We incubated homogenates from 5 whole flies expressing MoPrP with 15B3 and the immunoprecipitated proteins were subjected to western blot using anti-PrP 6H4 (1: 10,000, Prionics). We used controls with beads not bound to antibody or with homogenates not expressing MoPrP.
Results
D159 has local and regional effects on the surface of PrP
Sequence alignment of the globular domain of human, Syrian hamster, mouse, and dog PrP shows that these are highly conserved proteins, with most changes resulting in conservative substitutions (Fig. 1A). CaPrP carries three prominent non-conservative changes with respect to the three susceptible species shown: D159, R177, and E223. Since cat PrP also carries the E223 substitution (Lysek et al., 2005), this residue should have no role in the stability of CaPrP. Both D159 and R177 are unique substitutions on CaPrP not conserved in cats, making them prime suspects to modulate PrP conformation. R177 and H177 are positively charged residues that should have minor effects on PrP structure and surface charge. Thus, the only prominent substitution in CaPrP is an asparagine (N) to aspartic acid (D) at 159 in the α1-β2 loop (Fig. 1A and B). Since the residue at 159 is exposed to the surface, D159 has been proposed previously to change the surface charge (Lysek et al., 2005).
The α1-β2 loop is in close proximity to the β1-α1 loop in the tertiary structure of MoPrP (Fig. 1B) with potential local effects on helix 1 and the β-sheet. In fact, the D159 substitution seem to perturb the β-sheet because it appears less defined by NMR (Lysek et al., 2005) (Fig. 1C and F, arrowheads). In addition, the change in surface charge in one loop could potentially affect the orientation of nearby charged residues. To test this, we visualized the 3D models of mouse and dog PrP by PyMOL. The most obvious effect of D159 is the negative charge of the side chain sticking out and causing a change in the surface (Fig. 1C, D, F and G). Although R136 and R151 maintained their general position, we noticed that in MoPrP, R136 occupied a region closer to N159 and the α1-β2 loop (Fig. 1C). In CaPrP, R136 moved slightly towards α2, thus displacing its negative charge. Although the increase in negative charge by D159 would allow a closer interaction with R136, the charges of the backbone chains may preclude it, instead displacing R136 (Fig. 1F). N159 creates a neutral surface in MoPrP that extends over the surface of the two loops and helix 2 (Fig. 1D and E). In contrast, D159 creates a negative surface charge and a displacement of positively charged residues that result in a positively charged region of the surface of the β1-α1 loop and the α2 helix (Fig. 1G and H) [see also (Lysek et al., 2005)]. These sequence and structural studies support the key role of D159 on CaPrP, but in vivo validation is still missing.
N159D decreases MoPrP turnover
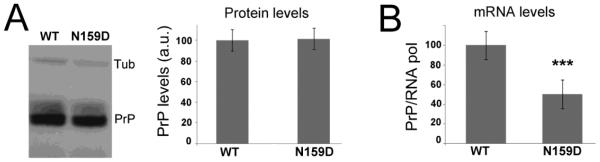
To examine the protective effect of D159, we introduced the N159D on the MoPrP backbone. Then, we created transgenic flies to determine how N159D alters the biological and biochemical properties of MoPrP. Taking advantage of existing flies expressing high levels of MoPrP-WT (Gavin et al., 2006), we selected for transgenic MoPrP-N159D lines expressing comparable levels from western blots. We prepared homogenates of whole flies expressing MoPrP ubiquitously and used the 6H4 antibody to detect MoPrP. We identified WT and N159D lines expressing PrP at almost identical levels following quantitation of three independent western blots (Fig. 2A), providing key tools to interpret the differences caused by a single amino acid replacement. We also examined the expression of the WT and N159D lines at the messenger RNA (mRNA) level expecting to find close to identical amounts as indicated by the protein levels. For this, we extracted total RNA from whole fly homogenates and performed quantitative PCR (qPCR). Surprisingly, we found that MoPrP-WT mRNA was twice as abundant as MoPrP-N159D mRNA (Fig. 2B). These results indicate that the turnover of MoPrP-WT is twice as fast as that of MoPrP- N159D, suggesting that Drosophila cells can detect abnormal biogenesis and/or conformations in MoPrP- WT that accelerate its degradation. In contrast, MoPrP-N159D may acquire more stable tertiary structures that promote its stability.
Figure 2. Expression and stability of MoPrP-N159D in flies.
A, Western blot shows comparable expression levels of MoPrP-WT and MoPrP-N159D strains detected with the 6H4 antibody. The quantification of three independent experiments indicates that the expression levels are highly comparable. B, Determination of mRNA levels for MoPrP-WT and MoPrP-N159D using qPCR. Using the same flies as in A, we extracted total RNA and performed qPCR using the same primers for both genotypes. The MoPrP-N159D shows significantly lower levels of PrP transcripts. *** p<0.001
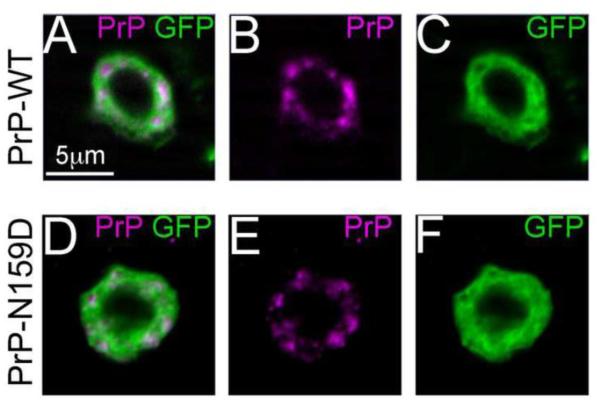
Next, we examined if N159D substitution had additional effects on MoPrP biogenesis and sub-cellular localization by detecting PrP by immunofluorescence in interneurons of the larval ventral nerve cord. For this, we co-expressed MoPrP-WT and -N159D with the reporter CD8-GFP under the control of the OK107-Gal4 driver line. MoPrP-WT accumulated mainly in the Golgi apparatus (Fig. 3A-C) as we reported previously (Fernandez-Funez et al., 2010). MoPrP-N159D displayed a similar sub-cellular distribution with large clumps in the Golgi (Fig. 3D-F). Thus, we observed no obvious effects of the N159D substitution on PrP distribution.
Figure 3. N159D does not alter the subcellular distribution of MoPrP.
A-F, MoPrP-WT and MoPrP-N159D were co-expressed with GFP in interneurons of the ventral ganglion and detected with the 6H4 antibody in whole-mount brains. Both MoPrP-WT (A-C) and -N159D (D-F) showed a mixed distribution in the cell, with some aggregates and some diffuse localization.
N159D protects against motor neuron dysfunction
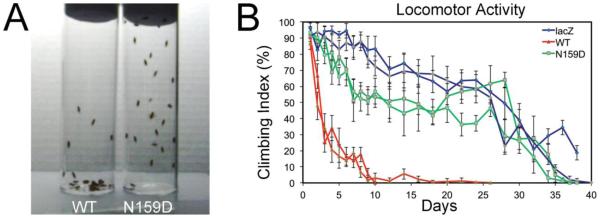
To determine the biological consequences of the N159D substitution on MoPrP, we performed locomotor assays. To test for progressive neuronal dysfunction, we expressed MoPrP-WT, -N159D, and the reporter transgene LacZ (control) in motor neurons using the strong BG380-Gal4 driver. We assayed motor coordination of females in climbing assays, which reflect the ability of flies to climb the walls of an empty vial as a function of age. We tested the flies every day for the first ten days to identify early phenotypes and then every other day until control flies stopped climbing. A snapshot comparing flies expressing MoPrP-WT or -N159D at day ten illustrates the locomotor difference between WT and mutant PrP (Fig 4A). Control flies expressing LacZ continued to climb until day 35 and reached 50% climbing index by day 27 (Fig. 4B). As we reported previously, MoPrP-WT induced severe locomotor dysfunction, with 50% climbing index by day 3 (Fig. 4A and B) (Fernandez-Funez et al., 2010). In contrast, flies expressing MoPrP-N159D continued to climb until day 35 (similar to LacZ flies), reached 50% climbing index around day 27, and hovered around 50% for two weeks (Fig. 4A and B). Although flies expressing MoPrP-N159D did not perform as well as flies expressing LacZ, they performed much better that flies expressing MoPrP-WT, indicating that a single amino acid replacement is sufficient to suppress PrP toxicity.
Figure 4. N159D prevents progressive locomotor dysfunction.
A, Flies expressing either MoPrP-WT or MoPrP-N159D show different locomotor abilities by day 10, with most of the MoPrP-WT flies in the bottom of the vial, whereas the MoPrP-N159D were on the walls and climbed to the top of the vial. B, MoPrP-WT flies show a very dramatic locomotor dysfunction in 5 days and almost stop climbing after day 10. In contrast, MoPrPN159D flies show a mild decrease in climbing activity and retain their locomotor activity for 30 days, similar to control flies expressing LacZ. Climbing graphs are shown in duplicates to illustrate robustness of the experiment.
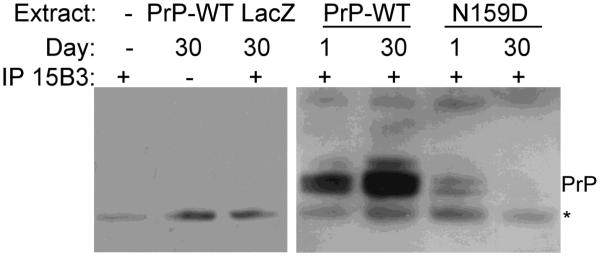
N159D prevents the accumulation of pathogenic conformations
After showing that N159D has a protective activity on MoPrP, we next examined if the change in biological properties was accompanied by changes in biochemical features. For this, we analyzed the accumulation of pathogenic PrP isoforms recognized by the 15B3 conformational antibody. 15B3 specifically recognizes pathogenic PrP conformations including PrPSc regardless of species, but does not recognize PrPC from any species (Korth et al., 1997). We have shown previously that 15B3 specifically recognizes pathogenic conformations of mouse and hamster PrP that accumulate over time (Fernandez- Funez et al., 2009; Fernandez-Funez et al., 2010). First, we demonstrated the specificity of the antibody with several controls (Fig. 5, left panel). A column with 15B3 and no fly extract produced a small unspecific band. A column loaded with flies expressing MoPrP-WT incubated in the absence of 15B3 also produced a small unspecific band. Finally, non-PrP flies (LacZ) incubated with 15B3 produced the same pattern, demonstrating the lack of immunoreactivity of 15B3 with Drosophila tissues. Then, we prepared homogenates from day 1 and day 30 flies expressing MoPrP-WT or -N159D, and followed the same procedures for immunoprecipitation with 15B3 and detection by western blot (Fig. 5, right panel). Young flies expressing MoPrP-WT produced two weak bands positive for 15B3 at the correct molecular weight for PrP, indicating the accumulation of small amounts of pathogenic conformations. Flies expressing the MoPrP-WT and aged for 30 days accumulated larger amounts of 15B3-positive conformations, consistent with the progressive misfolding of PrP into pathogenic conformations. In contrast, both young and old flies expressing MoPrP-N159D accumulated very small amounts of 15B3- positive conformations, indicating the inability of this mutant to undergo conversion into pathogenic conformations.
Figure 5. N159D prevents the accumulation of pathogenic conformations.
Detection of pathogenic PrP conformations using the 15B3 antibody. MoPrP-WT is immunoprecipitated by 15B3 in both young (day 1) and old flies (day 30), with the older flies accumulating higher amounts of PrPSc-like conformers. However, very small amounts of MoPrP-N159D are immunoprecipitated by 15B3. Controls on the left show the specificity of the antibody for MoPrP. * indicates a non-specific band.
Discussion
Few animals seem to be resistant to prion diseases and these exceptions provide unique opportunities to uncover the intrinsic factors (amino acid sequence) mediating the structural stability of PrPC and its resistance to conversion. Since expression of endogenous PrPC and direct interaction with PrPSc are critical for conformational conversion and pathogenesis (Telling et al., 1995), the intriguing resistance to disease of dogs has to reside on unique features of its endogenous sequence. Additionally, in vitro studies have confirmed the high stability of CaPrP under strong denaturing conditions (Khan et al., 2010), while dog MDCK cells and coyote brain extract do not support prion replication (Kurt et al., 2009; Polymenidou et al., 2008). This study demonstrates that introducing a single amino acid change in mouse PrP that is only present in canids is enough to suppress toxicity and conformational change. These are strong arguments supporting the key role of endogenous CaPrP in the resistance to prion disease, although a secondary role for the protein quality control machinery, including chaperones, cannot be ignored. D159 is the most salient difference in the sequence of CaPrP compared to animals susceptible to prion diseases. Previous studies have shown that D159 alters the surface charge of CaPrP, which should affect the interactions of PrP with other proteins (Lysek et al., 2005). Here we provide the first in vivo evidence for the relevance of D159 by introducing the N159D substitution into the MoPrP backbone. This single substitution made MoPrP more stable based on the protein to mRNA ratio. These results suggest that the WT allele is targeted for degradation and the N159D substitution is sufficient to prevent its high turnover. Although we do not know the exact reasons for the high turnover of MoPrP-WT, it is likely that partially misfolded conformations interact with chaperones that target them for degradation. This is confirmed by the presence of 15B3-reactive conformations at day 1 in adult flies. More importantly, MoPrP-N159D induces no progressive dysfunction of locomotor neurons and no conversion into 15B3- positive conformations, supporting the biological relevance of D159. Although dogs carry another unique substitution in helix 2 – R177 – that is not conserved in the close sequence of cat PrP (Lysek et al., 2005), our studies demonstrate that D159 is sufficient to provide resistance into murine PrP. Overall, a single amino acid substitution on the α1-β2 loop is sufficient to alter MoPrP biogenesis, prevent conformational changes, and suppress toxicity in vivo.
This work conclusively demonstrates the relevance of D159 in regulating PrP structure and pathogenesis in a transgenic animal model sensitive to PrP misfolding and toxicity. However, the short α1-β2 loop has received little attention for its role on PrP structure. Most of the focus is on the three helices because PrP misfolding requires the loss of some helicity and many pathogenic mutations are located on helices 2 and 3. The β2-α2 loop also seems critical because it harbors four pathogenic mutations while natural variations contribute to disease susceptibility in sheep and cervids (Kurt et al., 2009; Sigurdson et al., 2010). In contrast, the α1-β2 loop only harbors a nonsense pathogenic mutation (Q160X) and its sequence is highly conserved among mammals, except in canids (dogs, wolfs, foxes). D159 changes the surface charge due to its acidic side chain, but the effects of D159 spread to nearby amino acids creating new negatively charged areas (Lysek et al., 2005). A recent report comparing several PrP structures by NMR suggests that the β1-α1 and α1-β2 loops interact more closely in CaPrP than in susceptible animals (Hasegawa et al., 2013). The subtle changes in the orientation of the side chains and the closer loops may affect the stability of the β-sheet, explaining its poor resolution by NMR (Lysek et al., 2005). All these changes are expected to alter protein-protein interactions and our in vivo results suggest that these interactions are critical regulators of PrP conformation and toxicity. Thus, this critical domain should be investigated in more detail, because the identification of α1-β2 loop-binding proteins are expected to reveal clues about the molecular mechanisms and the extrinsic factors mediating PrP conversion.
Combined, in vitro and in vivo evidence support the intrinsic resistance of the canine family to prion disease associated with D/E159 (Stewart et al., 2012). Although this prion resistance is highly relevant in a physiological in vivo context, in vitro studies demonstrate that recombinant CaPrP is amyloidogenic and can accumulate in the β-state (misfolded conformation) under strong denaturing conditions (Khan et al., 2010; Nystrom and Hammarstrom, 2015). Additionally, CaPrP can be converted into a protease-resistant conformation when seeded with high concentrations of bovine PrPSc in brain homogenates, and this material can induce pathogenesis in mice expressing bovine PrP, but not in humanized mice (Vidal et al., 2013). These are extreme laboratory conditions and CaPrP is unlikely to adopt these conformations naturally or under physiological conditions, thus diminishing the risk of canines acquiring prion diseases naturally to virtually negligible. The fact remains that dogs and wild canids have been exposed to bovine, sheep, and cervid prions for decades and shown no susceptibility to disease, supporting the protective role of CaPrP and the critical role of D159.
Interestingly, two other mammals resistant to prion diseases, horses and rabbits, carry key substitutions in the β2-α2 loop and the first turn of helix 2 (Khan et al., 2010; Perez et al., 2010). The presence of three protective substitutions in two different domains indicates that several intrinsic and extrinsic factors contribute to the stability of PrPC. Similarly, multiple mutations along the globular domain are pathogenic in humans (Lloyd et al., 2011), indicating that different perturbations favor PrP misfolding and pathogenesis. The available evidence suggests that PrPC is a protein in a precarious folded balance that is highly prone to misfolding, but several intrinsic and extrinsic factors seem to contribute to its conformational stability. Exploiting the natural protective mechanisms identified in prion-resistant animals can contribute to alleviate these devastating diseases in humans and domestic animals.
Highlights.
Dogs and other canines are rare animals resistant to prion diseases
D159 is a unique residue in dogs and canines that substitutes the conserved N159
Introducing the N159D substitution in the sequence of mouse PrP suppresses its conformational change and toxicity in Drosophila
D159 is the key protective residue in dog PrP that provides conformational stability and confers protection against prions
Acknowledgements
We thank S. Supatappone and the Bloomington Drosophila Stock Center (NIH P40OD018537) for transgenic flies and J. Castilla for the MoPrP-N159D construct. This work was supported by the NIH grant DP2 OD002721-01 to PF-F. JS-G was supported by a postdoctoral fellowship from the Basque Government.
Abbreviations
- α1-3
(helices 1-3)
- CaPrP
(Canis PrP)
- MoPrP
(mouse PrP)
- PrP
(Prion protein)
- WT
(wild type)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests: The authors declare that no competing interests exist.
References
- Bueler H, et al. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–47. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- Chandler RL. Experimental transmission of scrapie to voles and Chinese hamsters. Lancet. 1971;1:232–3. doi: 10.1016/s0140-6736(71)90966-4. [DOI] [PubMed] [Google Scholar]
- Chandler RL, Fisher J. Experimental Transmission of Scrapie to Rats. Lancet. 1963;2:1165. [PubMed] [Google Scholar]
- Colby DW, Prusiner SB. Prions. Cold Spring Harb Perspect Biol. 2011;3:a006833. doi: 10.1101/cshperspect.a006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Funez P, et al. In vivo generation of neurotoxic prion protein: role for hsp70 in accumulation of misfolded isoforms. PLoS Genet. 2009;5:e1000507. doi: 10.1371/journal.pgen.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Funez P, et al. Sequence-dependent prion protein misfolding and neurotoxicity. J Biol Chem. 2010;285:36897–36908. doi: 10.1074/jbc.M110.174391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin BA, et al. Accelerated accumulation of misfolded prion protein and spongiform degeneration in a Drosophila model of Gerstmann-Straussler-Scheinker syndrome. J Neurosci. 2006;26:12408–14. doi: 10.1523/JNEUROSCI.3372-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K, et al. Comparison of the local structural stabilities of mammalian prion protein (PrP) by fragment molecular orbital calculations. Prion. 2013;7:185–91. doi: 10.4161/pri.23122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TL, et al. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc Natl Acad Sci U S A. 1997;94:10086–91. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MQ, et al. Prion disease susceptibility is affected by beta-structure folding propensity and local side-chain interactions in PrP. Proc Natl Acad Sci U S A. 2010;107:19808–13. doi: 10.1073/pnas.1005267107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood JK, Cunningham AA. Epidemiological observations on spongiform encephalopathies in captive wild animals in the British Isles. Vet Rec. 1994;135:296–303. doi: 10.1136/vr.135.13.296. [DOI] [PubMed] [Google Scholar]
- Korth C, et al. Prion (PrPSc)-specific epitope defined by a monoclonal antibody. Nature. 1997;390:74–7. doi: 10.1038/36337. [DOI] [PubMed] [Google Scholar]
- Kretzschmar HA, et al. Molecular cloning of a mink prion protein gene. J Gen Virol. 1992;73:2757–61. doi: 10.1099/0022-1317-73-10-2757. Pt 10. [DOI] [PubMed] [Google Scholar]
- Kurt TD, et al. Trans-species amplification of PrP(CWD) and correlation with rigid loop 170N. Virology. 2009;387:235–43. doi: 10.1016/j.virol.2009.02.025. [DOI] [PubMed] [Google Scholar]
- Le Bourg E, Lints FA. Hypergravity and aging in Drosophila melanogaster. 4. Climbing activity. Gerontology. 1992;38:59–64. doi: 10.1159/000213307. [DOI] [PubMed] [Google Scholar]
- Lee T, et al. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–76. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Lloyd S, et al. Genetics of prion disease. Top Curr Chem. 2011;305:1–22. doi: 10.1007/128_2011_157. [DOI] [PubMed] [Google Scholar]
- Lysek DA, et al. Prion protein NMR structures of cats, dogs, pigs, and sheep. Proc Natl Acad Sci U S A. 2005;102:640–5. doi: 10.1073/pnas.0408937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom S, Hammarstrom P. Generic amyloidogenicity of mammalian prion proteins from species susceptible and resistant to prions. Sci Rep. 2015;5:10101. doi: 10.1038/srep10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez DR, et al. Horse prion protein NMR structure and comparisons with related variants of the mouse prion protein. J Mol Biol. 2010;400:121–8. doi: 10.1016/j.jmb.2010.04.066. [DOI] [PubMed] [Google Scholar]
- Polymenidou M, et al. Canine MDCK cell lines are refractory to infection with human and mouse prions. Vaccine. 2008;26:2601–14. doi: 10.1016/j.vaccine.2008.03.035. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–83. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB, et al. Ablation of the prion protein (PrP) gene in mice prevents scrapie and facilitates production of anti-PrP antibodies. Proc Natl Acad Sci U S A. 1993;90:10608–12. doi: 10.1073/pnas.90.22.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riek R, et al. NMR structure of the mouse prion protein domain PrP(121-321) Nature. 1996;382:180–2. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- Sigurdson CJ, et al. A molecular switch controls interspecies prion disease transmission in mice. J Clin Invest. 2010;120:2590–9. doi: 10.1172/JCI42051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart P, et al. Genetic predictions of prion disease susceptibility in carnivore species based on variability of the prion gene coding region. PLoS One. 2012;7:e50623. doi: 10.1371/journal.pone.0050623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telling GC, et al. Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- Vidal E, et al. Bovine spongiform encephalopathy induces misfolding of alleged prion-resistant species cellular prion protein without altering its pathobiological features. J Neurosci. 2013;33:7778–86. doi: 10.1523/JNEUROSCI.0244-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnik I, Rennie JC. Further observations on the experimental transmission of scrapie from sheep and goats to laboratory mice. J Comp Pathol. 1963;73:150–62. doi: 10.1016/s0368-1742(63)80018-1. [DOI] [PubMed] [Google Scholar]
- Zlotnik I, Rennie JC. Experimental Transmission of Mouse Passaged Scrapie to Goats, Sheep, Rats and Hamsters. J Comp Pathol. 1965;75:147–57. doi: 10.1016/0021-9975(65)90005-8. [DOI] [PubMed] [Google Scholar]