Abstract
Dynamin 1 is a large neuron-specific GTPase involved in the endocytosis and recycling of pre-synaptic membranes and synaptic vesicles. Mutations in the gene encoding dynamin 1 (DNM1) underlie two epileptic encephalopathy syndromes, Lennox-Gastaut Syndrome and Infantile Spasms. Mice homozygous for the Dnm1 “fitful” mutation, a non-synonymous coding variant in an alternatively spliced exon of Dnm1 (exon 10a; isoform designation: Dnm1aFtfl) have an epileptic encephalopathy-like disorder including lethal early onset seizures, locomotor and neurosensory deficits. Although fitful heterozygotes have milder recurrent seizures later in life, suggesting an additive or semi-dominant mechanism, the molecular etiology must also consider the fact that Dnm1aFtfl exerts a dominant negative effect on endocytosis in vitro. Another complication is that the fitful mutation induces alterations in the relative abundance of Dnm1 splice variants; mutants have a downregulation of Dnm1a and an upregulation of Dnm1b, changes which may contribute to the epileptic pathology. To examine whether Dnm1a loss of function, Dnm1aFtfl dominance or compensation by Dnm1b is the most critical for severe seizures, we studied alternate isoform-specific mutant mice. Mice lacking Dnm1 exon 10a or Dnm1 exon 10b have neither spontaneous seizures nor other overt abnormalities, suggesting that in normal conditions the major role of each isoform is redundant. However, in the presence of Dnm1aFtfl only exon 10a deleted mice experience severe seizures. These results reveal functional differences between Dnm1a and Dnm1b isoforms in the presence of a challenge, i.e. toxic Dnm1Ftfl, while reinforcing its effect explicitly in this model of severe pediatric epilepsy.
Keywords: Dynamin, alternative isoform, epilepsy, seizure, endocytosis, fitful
Introduction
Dnm1 encodes a large neuron-specific GTPase involved in the endocytosis and recycling of pre-synaptic membranes and synaptic vesicles. This gene undergoes alternative splicing resulting in the regulated expression of several splice variants. The first alternatively spliced site is exon 10, resulting in two mutually-exclusive variants, termed Dnm1a and Dnm1b. The fitful mutation (Dnm1aFtfl) confers a single amino acid substitution to the respective Dnm1a isoform leaving the alternative Dnm1b isoform intact (Boumil et al., 2010). Mutant Dnm1aFtfl protein does not assemble properly into the homo-oligomeric complexes necessary for proper function (Boumil et al., 2010). Complete knockout of Dnm1 (all isoforms) in the mouse demonstrated that a sub-population of neurons is more sensitive to the loss of Dnm1 and resulted in endocytic defects (Ferguson et al., 2007, Hayashi et al., 2008). Homozygous Dnm1 knockout mice display early lethality, but neither the homozygous nor heterozygous knockout mice have seizures. By comparison, the fitful mutation, while present only in the Dnm1a isoform, is associated with recurrent seizures in both heterozygous and homozygous genotypes, with the latter being severe and accompanied by behavioral comorbidities similar to human epileptic encephalopathy (EE) patients that carry de novo DNM1 variants (Boumil et al., 2010, Asinof et al., 2015, Dhindsa et al., 2015).
Expression and alternative splicing of Dnm1 is regulated during early postnatal development and synaptogenesis (Gray et al., 2003, Ferguson et al., 2007). There is a developmental shift in the expression of the Dnm1a and Dnm1b isoforms (Boumil et al., 2010). Dnm1b is expressed at the highest levels during embryonic and early postnatal development and decreases during the time of synaptogenesis. Dnm1a expression increases at this stage (Boumil et al., 2010). The Dnm1a–Dnm1b alternative exon encodes a portion of the “middle” domain of dynamin 1, previously shown to be required for multi-molecular assembly (Okamoto et al., 1999, Smirnova et al., 1999, Ramachandran et al., 2007). A paralogous middle domain exon is also present in Dnm2, but not in Dnm3 (reviewed in (Marks et al., 2001)), although the developmental expression profile of this domain is not known (Sontag et al., 1994). Interestingly, the Dnm2 alternative isoforms have Golgi- specific differential functions (Cao et al., 1998, Liu et al., 2008). In fitful animals exon 10 is spliced abnormally; Dnm1b expression persists well into the late postnatal period (Boumil et al., 2010). It is tempting to speculate that altered dynamin 1 composition during synaptic maturation may contribute to the disease phenotype – perhaps reflecting changing dynamin 1 subunit assembly requirements for endocytosis from early to late postnatal development. Indeed, the modes of endocytosis mature from basal endocytosis to stimulation-induced endocytosis as the requirement for presynaptic synaptic vesicle recycling becomes more demanding (Bonanomi et al., 2008, Clayton and Cousin, 2009). Inappropriate expression of Dnm1b in mature neurons that should express the adult Dnm1a isoform may affect the kinetics of endocytosis and exacerbate the fitful phenotype.
Dnm1b is expressed earlier than Dnm1a and, interestingly, homozygous fitful mice survive until two to three weeks of age suggesting that Dnm1b is necessary and perhaps sufficient for providing normal Dnm1 function during this stage (Boumil et al., 2010). Dnm1 null mice only survive through the first week of life (Ferguson et al., 2007). In both wild-type and fitful mutant mice, Dnm1a expression increases and becomes the more dominantly expressed isoform by two weeks of age. This is concurrent with the observed decline of fitful mutant mice, suggesting that Dnm1a is necessary for maturation and the expression of Dnm1b can no longer compensate.
In order to determine whether the Dnm1 middle domain isoform usage is critical for normal development or for the seizure phenotype in fitful mice we generated and characterized mice that lack either Dnm1a or Dnm1b. Although neither deletion alone has any obvious effect on development, in the presence of the Dnm1aFtfl mutation normal Dnm1a isoform is required to prevent lethal seizures. We conclude that subtle functional differences between the middle domain isoforms have evolved in vertebrates to specialize in differential requirements for developmental and adult brain functions.
Results
Generation of dynamin 1 isoform specific knock-out mice
It was previously demonstrated that the developmentally regulated shift in dynamin-1 isoform expression is disrupted in fitful mice carrying an isoform specific mutation (Boumil et al., 2010). Therefore, we postulated that Dnm1a and Dnm1b isoforms may have divergent functions. In order to isolate and distinguish possible altered functional requirements for the Dnm1a and Dnm1b isoforms, we generated two independent lines of knockout mice, by homologous recombination in ES cells using targeting vectors containing loxP recombination sites flanking either exon 10b or exon 10a and a Tkneo selection cassette flanked by FRT recombination sites (Fig. 1). The targeted isoform exon was deleted and the remaining isoform was left intact. Independent mouse lines were backcrossed to C57BL/6J mice for at least ten generations to establish congenic strains. The established Dnm1a and Dnm1b isoform knockout mice were viable, had a Mendelian distribution of expected genotypes and no obvious health alterations. The mice were initially observed for overt behavior and seizure phenotypes. Neither the Dnm1a nor the Dnm1b knockout mice displayed any spontaneous seizure phenotype either as young mice or as aged adults.
Fig. 1. Dynamin 1 isoform specific mice.
(A) Schematic of DNM1 with enlargement of the alternatively spliced exon structure found in the middle domain. Above the protein domains are the known disease-causing human variants (black) and the fitful mutation (red). The fitful mutation is located in exon 10a. (B) Targeting strategy. LoxP sites were created flanking either exon10b or exon10a with an adjacent FRT flanked neomycin selection cassette. Correctly targeted recombined mice were crossed with FLP mice to remove the neomycin cassette resulting in “floxed” mice. The floxed mice were crossed with germline cre recombinase mice to recombine the loxP sites and remove the targeted exon resulting in isoform specific deletion mice.
Expression levels of dynamin 1 isoforms
Analysis of RNA and protein levels demonstrated a complete loss of isoform expression in the respective deletion strain (Fig. 2). While a lack of specific isoform expression was detected, the overall level of dynamin 1 was maintained indicating upregulation of the remaining isoform. This was confirmed at the transcript and protein level, by RT-PCR and western blot respectively. Further, we confirmed that forcing isolated expression of the alternate isoform does not affect overall dynamin 1 regulation or abundance.
Fig. 2. Maintenance of dynamin 1 levels in isoform deletion mice.
(A) To determine the relative levels of Dnm1a and Dnm1b in isoform deletion mice, we used an RT-PCR restriction enzyme assay. The alternative isoform region is amplified by common primers and the two transcripts are distinguished by a diagnostic HphI restriction enzyme site specific for the Dnm1b isoform (Boumil et al., 2010). The top band is uncut Dnm1a and the bottom band is digested Dnm1b product on an agarose gel. Total Dnm1 is shown below amplified with common primers as well as control reaction. Each isoform mutant is compared to a littermate control of the same age. (B) Dnm1 protein analysis shows loss of Dnm1a (left) and loss of Dnm1b (right) in the respective deletion strains; overall Dnm1 levels remain steady. Samples were taken at two different ages to reflect the age of highest isoform expression, respectively.
Localization of isoforms in vitro
Once we established that our targeting strategy could ablate each Dnm1 isoform with high efficiency, we were interested in dissecting functional differences between Dnm1a and Dnm1b. First, we assayed whether the two isoforms were trafficked to different subcellular compartments. Previously, we have demonstrated similar generalized localization patterns of DNM1a and DNM1b isoforms when over-expressed in COS-7 cells (Boumil et al., 2010). To more closely investigate discrete isoform localization, we again expressed GFP-tagged DNM1 isoform constructs in cells and immunostained with antibodies recognizing distinct cellular markers. Both DNM1 isoforms have overlap with tubulin and with clathrin (Fig. 3A,B). DNM1a colocalizes with clathrin at the plasma membrane while DNM1b appears to have greater association with clathrin at the perinuclear region (Fig. 3B). To confirm the perinuclear localization we co-stained with giantin (a Golgi marker). DNM1b co-localizes with giantin while DNM1a does not (Fig. 3C). Dnm1a clathrin-associated plasma membrane localization and Dnm1b Golgi localization is consistent with differential localization and roles previously reported for Dnm1 and Dnm2 isoforms (Cao et al., 1998).
Fig. 3. Subcellular localization of DNM1 isoforms and endocytosis compensation.
COS-7 cells were transfected with either DNM1a or DNM1b GFP tagged constructs. (A) DNM1a (top middle, green) or DNM1b (bottom middle, green) colocalization with tubulin (red, left panels; merge, right panels). (B) DNM1a (top middle, green) or DNM1b (bottom middle, green) colocalization with clathrin (red, left panels; merge, right panels). (C) DNM1a (top middle, green) or DNM1b (bottom middle, green) localization with giantin (red, left panels; merge, right panels).
Isoform protein interactions
Next, we were interested in investigating differences in protein interactions mediated by alternative splicing of Dnm1 exon 10. We reasoned that Dnm1 from mice at age post-natal day five (p5; which have relatively high levels of Dnm1b protein) would be associated with different proteins than Dnm1 from mice at post-natal day 20 (p20; have high levels of Dnm1a protein) if protein interactions of the two isoforms differed. While there are other factors which might account for these changes, including differential expression of those Dnm1 interactors or of Dnm1 itself, co-immunoprecipitation of Dnm1 from these time points can provide an “upper bound” of biologically relevant changes in protein interactions between the Dnm1 variants. We immunoprecipitated Dnm1 from p5 and p20 brain lysates, and performed SDS-PAGE to separate all proteins which co-precipitated with Dnm1. We excised coomassie-stained bands which appeared to change between the two ages and analyzed these differences with high specificity using mass spectrometry (Supplementary Data). We discovered several proteins which appeared to interact with Dnm1 only in more mature animals, including Pi4ka, Amphiphysin 1 and Clathrin Heavy-Chain (Supplementary Data). We then immunoprecipitated Dnm1 from adult littermate wild-type (near-exclusive expression of Dnm1a) and Dnm1a deleted mice (only express Dnm1b) and performed western blot analysis to detect levels of co-immunoprecipitated proteins found to differ in our earlier study. We observed a consistent decrease in Amphiphysin 1 binding to dynamin in mice expressing Dnm1b, suggesting that it has a different affinity for each of the two isoforms (Fig. 4). In this assay, none of the other interactions that we tested had a noticeable change in affinity for different dynamin 1 isoforms (Fig. 4; data not shown).
Fig. 4. Dynamin 1 isoform specific interactions.
Immunoprecipitation of lysates from adult wild-type brains (left) or Dnm1a deleted brains (right). Input and IP lanes were hybridized with anti-Dnm1 (control) and then stripped and re-hybridized with anti-Amph1 and anti-clathrin antibodies.
Cell-type specific expression of the different Dnm1 isoforms
We were interested in cell-type specific expression of the two isoforms. Inhibitory interneurons represent a minority of neural cells and have vastly different circuit roles, functional specialization, developmental history and maturation processes than most excitatory neurons. Further, it is known that inhibitory neurons are more sensitive to the loss of dynamin 1 (Ferguson et al., 2007, Hayashi et al., 2008). We therefore wanted to determine whether these cells had a different pattern of Dnm1 splicing than in the whole brain. We used “ribotag” (Rpl22tm1.1Psam; Sanz et al., 2009) mice crossed to a Gad2 cre recombinase expressing mouse line to tag and immunoprecipitate ribosomes from all inhibitory neurons at several developmental stages. We reverse-transcribed RNA associated with those ribosomes and used a restriction digest-based assay to determine the relative levels of Dnm1a and Dnm1b (Boumil et al., 2010). In whole brain cDNA of wild-type animals, we confirmed that Dnm1b transcripts were higher at early ages but gradually shifted to Dnm1a (Fig 5A; input, p8 vs. p21). Gad2 positive cells had similar proportions of Dnm1a and Dnm1b as in total brain tissue at all ages (Fig. 5A; ribotag IP).
Fig. 5. Expression of Dynamin 1 isoforms in inhibitory neurons.
(A) Assay of Dnm1 isoform levels in inhibitory neurons. The levels of Dnm1a and Dnm1b associated with tagged, immunoprecipitated ribosomes are distinguished by a diagnostic HphI restriction enzyme site specific for the Dnm1b isoform (Boumil et al., 2010). The top band is uncut Dnm1a and the bottom band is digested Dnm1b product on an agarose gel. (B) Control RT-PCR assay demonstrating the specificity of enrichment for inhibitory neuron specific transcripts. Vglut is amplified as an excitatory neuron marker; Gad2 is inhibitory neuron specific.
Altered isoform expression in Dnm1aΔ/Ftfl mice, but not Dnm1bΔ/Ftfl mice
The fitful mutation induces a shift in developmental regulation of isoform expression such that the Dnm1b isoform is upregulated longer during development (Boumil et al., 2010). Consequently, the misregulation may contribute to the fitful phenotypes. In order to distinguish between potential confounding effects of increased persistent Dnm1b expression, we crossed the individual Dnm1a and Dnm1b deletion mice with Dnm1Ftfl heterozygous mice resulting in Dnm1aΔ/Ftfl or Dnm1bΔ/Ftfl compound heterozygous mice, respectively. This allowed us to determine whether lack of either isoform exacerbated the fitful phenotype, for example, from milder non-lethal handling-associated seizures typical of fitful heterozygotes to severe seizures observed in younger homozygotes.
We examined expression of the alternative isoforms in the compound heterozygous animals. cDNA from adult brain showed that Dnm1bΔ/Ftfl mice have the expected pattern of expression with higher levels of Dnm1a in the adult and very low levels of Dnm1b (Fig. 6A). The Dnm1aΔ/Ftfl mice have increased amount of Dnm1b comparable to the shift seen in Dnm1+/Ftfl animals (Fig. 6A,B; (Boumil et al., 2010)).
Fig. 6. Altered dynamin 1 expression in isoform deletion mice in the presence of the fitful mutation.
(A) RT-PCR followed by diagnostic enzyme restriction analysis to distinguish Dnm1a and Dnm1b levels. Overall Dnm1 amplification is shown as control. (B) Relative quantification of the levels of Dnm1a and Dnm1b. (C) Protein levels of Dnm1 alleles in heterozygous and compound heterozygous brains. (D) Relative quantification of the levels of Dnm1, Dnm1a and Dnm1b compared to loading control (tubulin).
Western blot analysis confirmed the altered isoform expression at the protein level in the compound heterozygous animals (Fig. 6C). Notably, overall amounts of Dnm1 remain steady, but isoform amounts change – with more relative Dnm1b protein in the Dnm1aΔ/Ftfl compound heterozygotes and less relative Dnm1aFtfl protein (Fig. 6C, D).
While lack of either isoform alone did not result in seizures, the presence of the Dnm1aFtfl allele in the absence of wild-type Dnm1a did result in premature lethality (Fig. 7A). Although the unpredictable timing of lethality precluded us from visibly observing seizures in most cases, we conclude this lethality is due to lethal seizures because a) we have observed individual mice found in status epilepticus and later found dead, and b) when carcasses were found intact they typically had hindlimbs and forelimbs in full extension, and forepaws clenched – tell-tale signs of a maximal seizure. In contrast, seizures of Dnm1+/Ftfl mice are neither maximal nor lethal. Also, when seizures were observed following handling, the Dnm1aΔ/Ftfl compound heterozygous mice were observed to have a significantly earlier seizure onset than the Dnm1bΔ/Ftfl or Dnm1+/Ftfl mice (Fig. 7B; p<0.05; student’s |t| test pairwise testing) with seizures beginning at approximately 6–7 weeks of age versus 11–12 weeks of age.
Fig. 7. Early seizure onset and reduced lifespan induced by Dnm1a depletion.
(A) Survival curve of the Dnm1aΔ/Ftfl, Dnm1bΔ/Ftfl and Dnm1+/Ftfl mice. Dnm1aΔ/Ftfl mice have a reduced lifespan; the Dnm1bΔ/Ftfl and Dnm1+/Ftfl mice do not. (B) Latency to first observed seizure is shown for Dnm1aΔ/Ftfl, Dnm1bΔ/Ftfl and Dnm1+/Ftfl mice; *p<0.05, Dnm1aΔ/Ftfl vs Dnm1+/Ftfl.
Lack of dynamin 1 isoforms alters seizure threshold in fitful mice
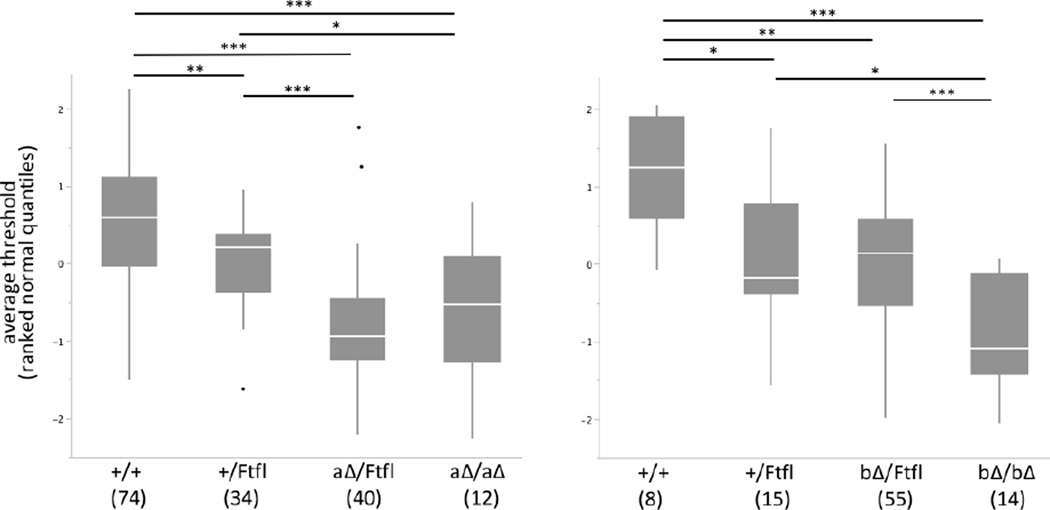
Seizure onset and severity is increased in the Dnm1aΔ/Ftfl mice. Therefore we determined whether the compound heterozygous mice had an altered seizure threshold. Electroconvulsive threshold (ECT) testing was done separately in males and females and then combined after rank and n-quantile normalization. The Dnm1aΔ/Ftfl mice had a lowered seizure threshold in comparison to both their wildtype and Dnm1+/Ftfl littermates (Fig. 8, left). The Dnm1bΔ/Ftfl mice did not show a similar decrease in threshold compared to their Dnm1+/Ftfl littermates, but did compared to wildtype littermates. Interestingly, the homozygous Dnm1bΔ/bΔ mice also had a reduced seizure threshold compared to littermates of all genotypes (Fig. 8, right).
Fig. 8. Depletion of Dnm1 isoforms lowers seizure threshold.
ECT threshold for Dnm1a deletion (left) and Dnm1b deletion (right) animals crossed with Dnm1+/Ftfl mice. Male and female data are shown together after rank and n-quantile normalization. Genotypes below indicate the alleles expressed for Dnm1: Dnm1+/+, Dnm1+/Ftfl, Dnm1aΔ/Ftfl, Dnm1aΔ/aΔ, Dnm1bΔ/Ftfl or Dnm1bΔ/bΔ. Number of animals tested is shown below genotype in parentheses. The combined data show that the reduced ECTs of the compound heterozygous mice were statistically significant (Student’s |t|-test, ***p<0.0001; **p<0.001; *p<0.01).
Altered EEG profile in Dnm1aΔ/Ftfl mice
Dnm1aΔ/aΔ and Dnm1bΔ/bΔ homozygous mice do not develop seizures and have normal electroencephalogram (EEG) recordings (data not shown). Recordings from compound heterozygous Dnm1aΔ/Ftfl mice showed abnormal interictal EEG patterns. Initially, episodes of epileptiform spikes were observed during quiet behavior patterns of the mice (Fig. 9A). Spike incidence was greatly increased in Dnm1aΔ/Ftfl mice, suggestive of a hyperexcitable network (Fig. 9B). Interictal spiking was not observed in the Dnm1bΔ/Ftfl animals in the same age range; although the Dnm1bΔ/Ftfl animals do have later onset spontaneous and routine handling-associated seizures, these are indistinguishable from seizures observed in Dnm1Ftfl/+ animals where interictal events are not observed.
Fig. 9. Hyperexcitable network in Dnm1aΔ/Ftfl mice.
(A) Differential representative EEG traces from a Dnm1aΔ/Ftfl mouse (top) depicts remarkably frequent epileptiform spikes as compared to a Dnm1bΔ/Ftfl mouse (bottom). Electrode placement: RB, right-back; RF, right-front; LF, left-front; LB, left-back. (B) Spiking activity graphed as spikes per hour. Left panel, homozygous Dnm1a deletion mice, Dnm1aΔ/Ftfl mice, homozygous Dnm1b deletion mice and Dnm1bΔ/Ftfl mice. Number of mice recorded from and scored is indicated below genotype. Right panel depicts spikes per hour for Dnm1aΔ/Ftfl mice or Dnm1bΔ/Ftfl mice scored one hour before (pre) and 2 hours after (post) PTZ injection.
We hypothesized that the interictal spikes are “pre-seizure” events that do not reach a threshold to become full-blown generalized seizures. In order to encourage the subthreshold seizure activity to reach threshold, we administered the GABA receptor antagonist pentylenetetrazole (PTZ). We recorded mice for one hour, administered PTZ at a dose of 30mg/kg, which is below the dose documented to induce tonic-clonic seizures in C57BL/6J mice (Kosobud and Crabbe, 1990, Kosobud et al., 1992), and observed for two hours further. The addition of PTZ significantly increased the frequency and rate of spike events in the Dnm1aΔ/Ftfl mice while causing only a modest increase in the Dnm1bΔ/Ftfl mice (Fig. 9B).
Endocytic compensation of isoforms in vitro
Overexpression of DNM1aFtfl protein disrupts endocytosis in COS-7 cells (Boumil et al., 2010). To determine if either dynamin 1 isoform can rescue the impaired endocytosis, we doubly transfected cells with either DNM1aFtfl and DNM1a or with DNM1aFtfl and DNM1b and assayed transferrin uptake. Both isoforms restored endocytosis levels (Fig. 10).
Fig. 10. Endocytosis compensation.
(A) Transferrin (red) endocytosis in COS-7 cells overexpressing DNM1aFtfl (left; green), overexpressing DNM1aFtfl and DNM1a (middle; green) or overexpressing DNM1aFtfl and DNM1b (right; green). (B) Quantification of transferrin localization in COS-7 cells overexpressing DNM1aFtfl alone, DNM1aFtfl and DNM1a, or DNM1aFtfl and DNM1b. Transferrin endocytosis was determined as cells that have transferrin in a perinuclear position, similar to non-transfected cells.
Isoform expression and seizure phenotype
Quantification of the isoform expression in the Dnm1 mouse models reveals an altered pattern accompanied by a seizure phenotype (Fig. 6; Fig. 11). Of the mice that live to adulthood, the Dnm1aΔ/Ftfl mice express the highest proportion of mutant DNM1a:wildtype DNM1b and also do not express wildtype DNM1A; this molecular composition corresponds to a lethal seizure predisposition and more severe phenotype.
Fig. 11. Model of Dnm1 isoform expression and composition.
This depiction of Dnm1 isoform abundance, with data taken from Figure 6D, summarizes the relative expression of Dnm1b (gray) compared to Dnm1a (wildtype;black or Ftfl; striped) in several genotypic combinations. Phenotypic outcome is summarized by premature lethality (red cross) and epileptiform activity in the EEG (thumbnail).
Discussion
Dynamin-1 is one of a growing number of proteins involved in synaptic development or function and mutated in human genetic epilepsies. Seizure disorders associated with many of these newly identified mutations manifest in early childhood and are accompanied by serious behavioral comorbidities: SYN1 (Giannandrea et al., 2013; Paemka et al., 2013), STXBP1 (Stamberger et al., 2016), Prrt2 (Liu et al., 2016; Valente et al., 2016), NAPB (Conroy et al., 2016), SV2a. (Serajee and Huq, 2015). The developmental importance of alternative exon usage is increasingly being recognized, both through specific genes and the phenotypes of individuals or animals with mutations in regulators such as RNA binding proteins and splicing factors (Eom et al., 2013; Ince-Dunn et al., 2012; Lal et al., 2013), although their respective roles in disease susceptibility remain unelaborated for many. Dnm1 undergoes regulated alternative splicing resulting in the possibility of at least eight isoforms (Cao et al., 1998). The relative expression of isoforms carrying the first alternative exon is altered during postnatal development in Dnm1aFtfl (fitful) mice, which carry a Dnm1a isoform-specific mutation. Prior studies suggested distinct roles for the alternatively spliced Dnm1 isoforms (Cao et al., 1998, Boumil et al., 2010). Here we probe the intricate, yet significant, differential requirements for Dnm1a and Dnm1b isoforms by characterizing human epileptic encephalopathy-relevant seizure phenotypes of isoform specific knockout mice, in the presence or absence of a Dnm1 variant – Dnm1Ftfl – associated with severe developmental disease in a mouse model.
Dnm1a and Dnm1b isoform specific knockout mice are overtly normal. They have a typical lifespan, do not have spontaneous seizures of any kind and phenotypically resemble wildtype littermates. Although we note that in-depth behavioral analysis has not been performed to exclude subtle behavioral abnormalities, we have observed that Dnm1aΔ/Ftfl compound heterozygotes are heavier than the other genotypes (unpublished observations) and this may be an indication of slight metabolic differences or in activity levels. Otherwise, the alternative isoforms would appear to have outwardly redundant basal roles in healthy animals.
However, subtler requirements for the alternative isoforms were revealed in the presence of a challenge, in this case a genetic challenge – the pathogenic variant Dnm1Ftfl. The increase in severe and lethal seizures in the absence of a wildtype Dnm1a isoform combined with Dnm1Ftfl reveal that Dnm1b cannot fully compensate for Dnm1a. This is further evidenced by the high levels of pre-epileptiform cortical spiking activity in the Dnm1aΔ/Ftfl mice suggestive of a hyperexcitable network.
The lowered electroconvulsive threshold observed in Dnm1bΔ/bΔ mice implies that Dnm1b is nevertheless required for normal circuitry establishment during development. Dnm1a deletion also confers a low seizure threshold compared to wildtype, however the lowered threshold in combination with Dnm1Ftfl correlates with the increased seizure severity/lethality observed in Dnm1aΔ/Ftfl compound heterozygotes.
The developmental regulation of the Dnm1a and Dnm1b isoforms may also provide insight into function. Dnm1b is expressed highest during embryonic and early postnatal development; expression decreases with synaptogenesis as Dnm1a expression increases (Boumil et al., 2010). This change in expression may reflect changing endocytic requirements over the course of development. It is appealing to speculate that an early endocytic pathway required for neuronal maturation may be lacking in the Dnm1b isoform deficient mice, leading to an increase in seizure sensitivity as indicated by the decreased seizure threshold in these mice.
Interestingly, altered isoform abundance is comparable between Dnm1+/Ftfl heterozygous and Dnm1bΔ/Ftfl mice (Fig. 6A,B; (Boumil et al., 2010)); both genotypes have increased expression of Dnm1a, which is a combination of Dnm1a and Dnm1aFtfl, and low overall amounts of Dnm1b. Conversely, the Dnm1aΔ/Ftfl animals upregulate the Dnm1b isoform. We propose a model in which Dnm1b is needed during development and Dnm1a is required for adult roles, with overlapping compensation sufficient for normal basal function. Indeed, we show that either isoform can restore DNM1aFtfl induced defective endocytosis in COS-7 cells demonstrating compensation of basal function. In adult Dnm1aΔ/Ftfl mice, there is only functional DNM1b present and the model suggests that it is not as efficient or functional as DNM1a to sustain the rapid synaptic vesicle recycling required at high-frequency firing synapses. An overlapping hypothesis suggests that the increased seizure intensity in the Dnm1aΔ/Ftfl mice (versus Dnm1bΔ/Ftfl) is the result of the higher percentage of available DNM1 being the mutant form (Fig. 11).
In addition to our genetic data suggesting that Dnm1a and Dnm1b isoforms have divergent uses, we provide direct evidence of functional differences. In COS7 cells, Dnm1a is trafficked to the plasma membrane while Dnm1b is concentrated in the perinuclear Golgi. The two isoforms also have different affinities for Amphiphysin I, as evidenced by our co-immunoprecipitation experiments. It is possible that this altered binding affinity is linked to altered subcellular localization of Dnm1a and Dnm1b. In the future, we hope to investigate protein interaction differences between the two isoforms more exhaustively.
These results suggest that there are functional differences between the two isoforms, although only 14 amino acids vary between DNM1a and DNM1b in this 46 amino acid encoding alternate exon. Since this region is involved in dynamin 1 oligomeric assembly, we hypothesize that either differential assembly kinetics or differential interactions of the isoforms underlies their specialized functions. Two scenarios may explain this. The first is that DNM1b cannot assemble efficiently on its own, requiring the interaction of DNM1a oligomers for higher order assembly. The second suggests that the DNM1b oligomers can assemble, but the enzymatic complex is not as efficient as the DNM1a complex. This could be due to self-self interactions and/or less efficient interactions with known dynamin 1 binding partners, such as amphiphysin 1, synapsin and clathrin. A recent study by Krishnan et al., demonstrates that DNM1a/DNM1b middle domain splicing can temper the allosteric regulation of the GTPase domain (Krishnan et al., 2015). Their study suggests that the middle domain may control GTPase activity by differential interactions with the SH3/PRD domains. They claim that an allosteric regulatory mechanism would explain the lack of correlation between dynamin oligomerization and GTPase activity. Indeed in our studies, we have observed DNM1a and DNM1b to have similar independent oligomerization dynamics (unpublished observations) implying that the alternative portion of the dynamin-1 middle domain may have function(s) other than regulating assembly kinetics.
It was surprising the DNM1 exon 10 splicing did not differ in Gad2+ interneurons. These cells migrate and mature much later than their radially-migrating excitatory cortical counterparts. At the developmental stage when DNM1 exon 10 shifting occurs, interneurons are still developing a mature molecular identity. Our data suggest that the exon 10 shift is regulated by a cue or combination of cues which occur in most neurons at the same time, independent of their developmental history.
Notably, RNA-protein high throughput interaction mapping studies show that the pan-neuronal splicing factor PTBP2 binds near exon 10 in DNM1, and its deletion causes inappropriate exon 10 splicing (Licatalosi et al., 2012, Li et al., 2014). PTBP2 expression is induced slightly before the change in isoform expression, and it is found in Gad2+ inhibitory neurons (unpublished observations). While we find it highly likely that PTBP2 is the primary determinant of exon 10 splicing, it remains an open question as to whether the fitful mutation induces changes in Dnm1 splicing directly through a PTBP2-dependent mechanism, or through another avenue.
In conclusion, we submit strong evidence for subtle functional differences between DNM1a and DNM1b. The two isoforms have different subcellular localizations and protein binding partners. Furthermore, they have different capacities for compensation in the presence of a genetic challenge, i.e. Dnm1Ftfl mutation; the presence of wildtype DNM1a is able to prevent fitful-induced epileptiform activity and lethal seizures through an unknown mechanism in which DNM1b is deficient. These findings have implications for human patients with epileptogenic Dnm1 mutations; one proposed therapeutic avenue for this presently untreatable disorder would be to use gene therapy to increase wildtype DNM1 levels. Our work shows that it will be important to introduce expression of a developmentally-appropriate isoform which is fully capable of negating the effects of dominant-negative DNM1 mutations.
Materials and Methods
Generation and maintenance of mouse lines
All mice were housed and procedures performed with approval of Institutional Animal Care and Use Committee (IACUC). All mice were obtained from The Jackson Laboratory, maintained in a room with a 14hour light on/10hour light off cycle, and given free access to LabChow meal and water.
The fitful mutation arose spontaneously in 2000 at The Jackson Laboratory (Bar Harbor, ME) as a spontaneous mutation on the C57BL/6J inbred strain (Boumil et al., 2010). To produce the Dnm1a and Dnm1b isoform deletion mice, targeting vectors were constructed (as described in Fig. 1) and electroporated into 129 ES cells. Clones were selected for G418 resistance (neomycin resistant gene insertion). Correctly targeted clones were confirmed by Southern analysis and were injected into C57BL/6J blastocysts for pseudopregnant mouse implantation. Chimeric mice were crossed to B6.ACT-Flpe mice to remove the Frt-neo-Frt cassette and resultant mice were either backcrossed to C57BL/6J mice to maintain the floxed allele or backcrossed to C57BL/6J Meox2-cre mice for germline removal of the floxed allele (Fig. 1 schematic). The Rpl22tm1.1Psam/J ribotag mice and Gad2tm2(cre)Zjh/J mice were obtained from the Jackson Laboratory (Bar Harbor, ME).
Genotyping
Genotypes were determined by PCR using tail DNA. Genotyping Dnm1Ftfl (fitful) was done as previously (Boumil et al., 2010). The Dnm1aΔ allele was PCR amplified with specific primers (Dnm1in10bLoxu, 5’ACTGAATAACCTACCCAGGCCA3’; Dnm1in10aLoxu, 5’CCTCTCTGTCCACTTGTAGCCATT3’; Dnm1in10aLoxd, 5’ACTGGGTGATGCTCACTAGAACCT3’). The Dnm1bΔ allele was PCR amplified with specific primers (Dnm1in9loxu, 5’GGGCATAGCACCCAGATATCATTC3’; Dnm1in10bloxu, 5’ACTGAATAACCTACCCAGGCCA3’; Dnm1in10bloxd, 5’ATCCCTTGCCTCTCCTCCTAACA3’).
Ribotag
Ribotag was performed essentially as described in (Sanz et al., 2009). Mice were sacrificed and brains were removed rapidly on ice. Each replicate was performed using pooled tissue from 2–3 animals. Brains were homogenized in ice-cold homogenization buffer (50 mM tris, 100 mM KCl, 12 mM MgCl2, 1% NP40) supplemented with 1 mM DTT, 200 U/ml RNAsin (promega), 1 mg/ml heparin, 100 µg/ml cycloheximide, and Complete Mini EDTA-free protease inhibitors (1 tablet/10 ml, Roche). After letting foam settle, samples were rocked end-over-end for 10 minutes. Homogenates were then centrifuged 10 min at 10,000 RPM. Supernatant was removed and incubated at 4 degrees (while rocking) with 10 µl mouse anti-hemagglutinin antibody (Covance; clone HA.11) for four hours. 100 µl protein G dynabeads (Thermo Fisher) were washed in homogenization buffer, then added to the supernatant and rocked at 4 degrees overnight. Beads were collected with a magnet and washed with high salt homogenization buffer (50 mM tris, 300 mM KCl, 12 mM MgCl2, 1% NP40) three times for 10 minutes apiece. RNA was eluted using the Qiagen RNEasy Mini kit.
Reverse-transcription PCR
Total RNA was prepared from brains with Trizol (Invitrogen) following the manufacturer's suggested conditions and protocol. RNA (0.5µg) was reverse transcribed with AMV reverse transcriptase (Promega). Amplification and restriction enzyme digestion was done as previously described, including use of an internal restriction site to aid with relative Dnm1 transcript quantification (Boumil et al., 2010).
Western blot
Protein extracts were made in lysis buffer (1mM HEPES, 137mMNaCl, 10% glycerol, 1% NP-40) with Complete-mini proteinase inhibitor mix (Roche) added fresh. Extracts were quantified using the Bradford reagent (Bio-Rad). Extracts (25–50 µg protein) were diluted in Laemmli buffer, incubated at 95°C for 5 minutes, resolved by SDS-PAGE and transferred to PVDF membrane. All membrane blotting steps were carried out in TBS plus Tween (TBST). Blots were blocked for 1 hour in 5% milk or 5% BSA, incubated at RT with primary antibody for 4 hours, HRP-conjugated secondary antibody (1:15000) for 1 hour and visualized with Luminata Forte (EMD Millipore). Membranes were incubated with Restore Western blot stripping buffer (Fisher) for 5 min while shaking to remove antibodies for subsequent hybridization. Primary antibodies used were rabbit anti-dynamin-1 (1:1500; Pierce Thermo Fisher, PA1-660), rabbit anti-DNM1a (1:200; Affinity BioReagents), chicken anti-DNM1b (1:100; Affinity BioReagents) and mouse anti-tubulin (1:1000; Sigma, T4026).
Image Analysis/Band Quantification
All western blots and RT-PCR/restriction digest assays were quantified using FIJI (LOCI, University of Wisconsin). Bands were selected and their intensity over space plotted using the program's Gel Analysis tool. For each intensity diagram, we acquired the area under the curve, subtracting background intensity. All data were normalized to control bands. Each technical replication was performed on a single gel with one window size to minimize intra-replication variation in background intensity.
Co-immunoprecipitation
Co-immunoprecipitation was performed in lysis buffer (see above). Briefly, fresh protein lysates (1 mg) were mixed with 7.5 µl rabbit anti-DNM1 (Pierce Thermo Fisher) in 150 µl lysis buffer and rocked at 4°C for 2.5 hours. Protein G Dynabeads (Thermo Fisher) were washed in lysis buffer, incubated with lysis buffer containing 1% BSA for one hour, then washed again. Antibody-lysate mixtures were then incubated with 50 µl beads and rocked overnight at 4°C. Beads were removed with a magnet and washed with lysis buffer; proteins were eluted for western blotting as described above.
Mass spectrometry
Coimmunoprecipitated proteins were resolved via SDS-PAGE as described above using a 4–15% TGX precast gel (BioRad). Gels were stained overnight with SYPRO Ruby (Invitrogen), then overnight with EZ-Blue (Sigma). Bands of interest were excised and processed for mass spectrometry by the Jackson Laboratory Proteomics Core, and mass spectrometry was performed by the Vermont Genetics Network at the University of Vermont using a Linear Ion Trap mass spectrometer. All identified proteins with less than 2 unique peptides were excluded from our analysis.
Assessment of handling-associated seizures
Mice were observed for age-of-onset, frequency and severity in their homecage for handling evoked seizures during weekly cage changing for at least 175 days. Our caretakers are trained to watch for generalized tonic-clonic seizures following weekly cage changes and to record the details, including animal ID, date and severity (lethal or non-lethal), onto seizure-watch cards attached to the main cage card. In related studies, we determined that sham cage changes (moving an animal into a clean cage, and back to its home cage) by laboratory personnel had a per-test penetrance of 50% in Dnm1Ftfl/+ adults, as long as the interval between tests is 3 days or longer (W. Frankel, unpublished results). Thus after 6 weeks, the cumulative penetrance for weekly observation following routine handling would be over 98%. Such observational data collected by trained caretakers has in the past been reliable and accurate enough to use as a binary phenotype for initial chromosomal mapping studies (Boumil et al., 2010).
Electroconvulsive threshold testing
For ECT, we followed previously described procedures (Frankel et al., 2001, Yang et al., 2003, Boumil et al., 2010). Briefly, mice were restrained; a drop of anesthetic containing 0.5% tetracaine and 0.9% NaCl was placed onto each eye and a fixed electrical current was applied via silver transcorneal electrodes using an electroconvulsive stimulator (Ugo Basile model 7801). For acute electroconvulsive threshold ECT, the stimulator was set to produce rectangular wave pulses with the following parameters: 299Hz, 0.2s duration, 1.6ms pulse width. The threshold to a minimal clonic forebrain seizure was determined by testing individual mice approximately daily until the endpoint was observed and group means were calculated.
EEG
Adult mice aged between 6 and 8 weeks were anesthetized with tribromoethanol (400 mg/kg i.p.). Small burr holes were drilled (1 mm anterior to the bregma and 2 mm posterior to the bregma) on both sides of the skull 2 mm lateral to the midline. Four teflon-coated silver wires were soldered onto the pins of a microconnector (Mouser electronics, Texas). The wires were placed between the dura and the brain and a dental cap was then applied. The mice were given a post-operative analgesic of carprofen (5 mg/kg subcutaneous) and allowed a minimum 48 h recovery period before recordings. Differential amplification recordings were recorded between all four electrode pairs, providing 6 channels for each subject. Mice were connected to the EEG Stellate Lamont Pro-36 programmable amplifier (Lamont Medical Instruments, Madison, WI) for a 24-hour period with accompanying video digital EEG recording. EEG data were recorded with Stellate Harmonie software (Stellate Systems, Inc., Montreal, Canada).
After baseline recording for one hour, pentylenetetrazole (PTZ; Sigma) was administered i.p. at 30mg/kg in saline. Mice were observed and recorded for two hours further after PTZ administration.
Each EEG recording was analyzed visually by scanning through the six differential traces (from four recording electrodes) on the computer. An epileptiform spike was defined as a rapid transient with both positive and negative components distinguishable from background activity, lasting between 20–70ms and occurring synchronously at the majority of electrode positions (Uchibori et al., 2002, White et al., 2010). The spikes were scored manually counting the number of spikes that met the above criteria for the recording interval, and then averaging over the time interval (# spikes/hour).
Cell culture
COS-7 cells were maintained in supplemented Dulbecco's modified Eagle's medium (DMEM; 10% fetal bovine serum, 30U/ml penicillin, 30µg/ml streptomycin) at 37°C in a 5% CO2 humidified atmosphere. Cells were split twice a week.
Transfections
Transient transfection of COS-7 cells was performed with 1–2µg of DNA/well in 6-well culture dishes using the Lipofectamine Plus Reagent (Invitrogen) according to the manufacturer's protocol.
Endocytosis assays
To assay endocytosis, cells transfected (on coverslips) with GFP tagged proteins were incubated in serum-free DMEM (Invitrogen) for 1 hour at 37°C followed by the addition of 25µg/ml AlexaFluor-555 conjugated transferrin (Invitrogen) for 15 minutes at 37°C. Cells were washed three times with PBS, and in some experiments, were incubated in an acidic solution (0.5M NaCl, 0.2M acetic acid in PBS; pH 3.0) for 4 minutes at 4°C to strip surface bound transferrin and washed immediately with PBS, fixed in 4% paraformaldehyde, 4% sucrose in PBS for 10 minutes at RT and mounted on slides with Gel/Mount (biomeda).
Supplementary Material
Highlights.
-
-
Dnm1 exon 10 is alternatively spliced to produce distinct middle domains.
-
-
The isoforms are differentially trafficked and have distinct protein interactions.
-
-
Each Dnm1 isoform can compensate for the other in normal genotypic conditions
-
-
Dnm1b cannot compensate if challenged by pathogenic Dnm1aFtfl, implying unique isoform function
-
-
The effect of Dnm1Ftfl on splice form abundance does not contribute to its pathology.
Acknowledgments
We are grateful to Rob Wilpan and The University of Vermont VGN Proteomics facility for assistance with mass spectrometry, and to Joanne Smith for exceptional animal care. This work was supported by NIH grant R01 NS073576 to RMB and R37 NS031348 to WNF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asinof SK, Sukoff Rizzo SJ, Buckley AR, Beyer BJ, Letts VA, Frankel WN, Boumil RM. Independent Neuronal Origin of Seizures and Behavioral Comorbidities in an Animal Model of a Severe Childhood Genetic Epileptic Encephalopathy. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonanomi D, Fornasiero EF, Valdez G, Halegoua S, Benfenati F, Menegon A, Valtorta F. Identification of a developmentally regulated pathway of membrane retrieval in neuronal growth cones. J Cell Sci. 2008;121:3757–3769. doi: 10.1242/jcs.033803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumil RM, Letts VA, Roberts MC, Lenz C, Mahaffey CL, Zhang ZW, Moser T, Frankel WN. A missense mutation in a highly conserved alternate exon of dynamin-1 causes epilepsy in fitful mice. PLoS Genet. 2010:6. doi: 10.1371/journal.pgen.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Garcia F, McNiven MA. Differential distribution of dynamin isoforms in mammalian cells. Mol Bio Cell. 1998;9:2595–2609. doi: 10.1091/mbc.9.9.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton EL, Cousin MA. The molecular physiology of activity-dependent bulk endocytosis of synaptic vesicles. J Neurochemistry. 2009;111:901–914. doi: 10.1111/j.1471-4159.2009.06384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy J, et al. NAPB - a novel SNARE-associated protein for early-onset epileptic encephalopathy. Clin Genet. 2016;89:E1–E3. doi: 10.1111/cge.12648. [DOI] [PubMed] [Google Scholar]
- Dhindsa RS, Bradrick SS, Yao XY, Heinzen EL, Petrovski S, Krueger BJ, Johnson MR, Frankel WN, Petrou S, Boumil RM, Goldstein DB. Epileptic encephalopathy causing DNM1 mutations impair synaptic vesicle endocytosis. Neurol Genet. 2015;1:e4. doi: 10.1212/01.NXG.0000464295.65736.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom T, et al. NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure. Elife. 2013;2:e00178. doi: 10.7554/eLife.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, Brasnjo G, Hayashi M, Wolfel M, Collesi C, Giovedi S, Raimondi A, Gong LW, Ariel P, Paradise S, O'Toole E, Flavell R, Cremona O, Miesenbock G, Ryan TA, De Camilli P. A selective activity-dependent requirement for dynamin 1 in synaptic vesicle endocytosis. Science. 2007;316:570–574. doi: 10.1126/science.1140621. [DOI] [PubMed] [Google Scholar]
- Frankel WN, Taylor L, Beyer B, Tempel BL, White HS. Electroconvulsive thresholds of inbred mouse strains. Genomics. 2001;74:306–312. doi: 10.1006/geno.2001.6564. [DOI] [PubMed] [Google Scholar]
- Giannandrea M, et al. Nonsense-mediated mRNA decay and loss-of-function of the protein underlie the X-linked epilepsy associated with the W356x mutation in synapsin I. PLoS One. 2013;8:e67724. doi: 10.1371/journal.pone.0067724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray NW, Fourgeaud L, Huang B, Chen J, Cao H, Oswald BJ, Hemar A, McNiven MA. Dynamin 3 is a component of the postsynapse, where it interacts with mGluR5 and Homer. Curr Biol. 2003;13:510–515. doi: 10.1016/s0960-9822(03)00136-2. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Raimondi A, O'Toole E, Paradise S, Collesi C, Cremona O, Ferguson SM, De Camilli P. Cell- and stimulus-dependent heterogeneity of synaptic vesicle endocytic recycling mechanisms revealed by studies of dynamin 1-null neurons. PNAS. 2008;105:2175–2180. doi: 10.1073/pnas.0712171105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince-Dunn G, et al. Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron. 2012;75:1067–1080. doi: 10.1016/j.neuron.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosobud AE, Crabbe JC. Genetic correlations among inbred strain sensitivities to convulsions induced by 9 convulsant drugs. Brain Res. 1990;526:8–16. doi: 10.1016/0006-8993(90)90243-5. [DOI] [PubMed] [Google Scholar]
- Kosobud AE, Cross SJ, Crabbe JC. Neural sensitivity to pentylenetetrazol convulsions in inbred and selectively bred mice. Brain Res. 1992;592:122–128. doi: 10.1016/0006-8993(92)91666-3. [DOI] [PubMed] [Google Scholar]
- Krishnan S, Collett M, Robinson PJ. SH3 Domains Differentially Stimulate Distinct Dynamin I Assembly Modes and G Domain Activity. PLoS One. 2015;10:e0144609. doi: 10.1371/journal.pone.0144609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal D, et al. RBFOX1 and RBFOX3 mutations in rolandic epilepsy. PLoS One. 2013;8:e73323. doi: 10.1371/journal.pone.0073323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zheng S, Han A, Lin CH, Stoilov P, Fu XD, Black DL. The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. Elife. 2014;3:e01201. doi: 10.7554/eLife.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, Darnell RB. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YW, Surka MC, Schroeter T, Lukiyanchuk V, Schmid SL. Isoform and splice-variant specific functions of dynamin-2 revealed by analysis of conditional knockout cells. Molecular Bio Cell. 2008;19:5347–5359. doi: 10.1091/mbc.E08-08-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YT, et al. PRRT2 mutations lead to neuronal dysfunction and neurodevelopmental defects. Oncotarget. 2016 May 9; doi: 10.18632/oncotarget.9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks B, Stowell MH, Vallis Y, Mills IG, Gibson A, Hopkins CR, McMahon HT. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature. 2001;410:231–235. doi: 10.1038/35065645. [DOI] [PubMed] [Google Scholar]
- Okamoto PM, Tripet B, Litowski J, Hodges RS, Vallee RB. Multiple distinct coiled-coils are involved in dynamin self-assembly. J Biol Chem. 1999;274:10277–10286. doi: 10.1074/jbc.274.15.10277. [DOI] [PubMed] [Google Scholar]
- Paemka L, et al. PRICKLE1 interaction with SYNAPSIN I reveals a role in autism spectrum disorders. PLoS One. 2013;8:e80737. doi: 10.1371/journal.pone.0080737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Surka M, Chappie JS, Fowler DM, Foss TR, Song BD, Schmid SL. The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO. 2007;26:559–566. doi: 10.1038/sj.emboj.7601491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. PNAS. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serajee FJ, Huq AM. Homozygous Mutation in Synaptic Vesicle Glycoprotein 2A Gene Results in Intractable Epilepsy, Involuntary Movements, Microcephaly, and Developmental and Growth Retardation. Pediatr Neurol. 2015;52:642 e1–646 e1. doi: 10.1016/j.pediatrneurol.2015.02.011. [DOI] [PubMed] [Google Scholar]
- Smirnova E, Shurland DL, Newman-Smith ED, Pishvaee B, van der Bliek AM. A model for dynamin self-assembly based on binding between three different protein domains. J Biol Chem. 1999;274:14942–14947. doi: 10.1074/jbc.274.21.14942. [DOI] [PubMed] [Google Scholar]
- Sontag JM, Fykse EM, Ushkaryov Y, Liu JP, Robinson PJ, Sudhof TC. Differential expression and regulation of multiple dynamins. J Biol Chem. 1994;269:4547–4554. [PubMed] [Google Scholar]
- Stamberger H, et al. STXBP1 encephalopathy: A neurodevelopmental disorder including epilepsy. Neurology. 2016;86:954–962. doi: 10.1212/WNL.0000000000002457. [DOI] [PubMed] [Google Scholar]
- Uchibori M, Saito K, Yokoyama S, Sakamoto Y, Suzuki H, Tsuji T, Suzuki K. Foci identification of spike discharges in the EEGs of sleeping El mice based on the electric field model and wavelet decomposition of multi monopolar derivations. J Neurosci Methods. 2002;117:51–63. doi: 10.1016/s0165-0270(02)00073-0. [DOI] [PubMed] [Google Scholar]
- Valente P, et al. PRRT2 Is a Key Component of the Ca(2+)-Dependent Neurotransmitter Release Machinery. Cell Rep. 2016;15:117–131. doi: 10.1016/j.celrep.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A, Williams PA, Hellier JL, Clark S, Dudek FE, Staley KJ. EEG spike activity precedes epilepsy after kainate-induced status epilepticus. Epilepsia. 2010;51:371–383. doi: 10.1111/j.1528-1167.2009.02339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Beyer BJ, Otto JF, O'Brien TP, Letts VA, White HS, Frankel WN. Spontaneous deletion of epilepsy gene orthologs in a mutant mouse with a low electroconvulsive threshold. Hum Mol Genet. 2003;12:975–984. doi: 10.1093/hmg/ddg118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.