Abstract
A significant increase in cyclooxygenase 2 (COX2) gene expression has been shown to promote cylcooxygenase-dependent colon cancer development. Controversy associated with the role of COX2 inhibitors indicates that additional work is needed to elucidate the effects of arachidonic acid (AA) derived (cyclooxygenase and lipoxygenase) eicosanoids in cancer initiation, progression and metastasis. We have recently developed a novel Fads1 knockout mouse model, which allows for the investigation of AA-dependent eicosanoid deficiency without the complication of essential fatty acid deficiency. Interestingly, the survival rate of Fads1 null mice is severely compromised after 2 months on a semi-purified AA-free diet, which precludes long-term chemoprevention studies. Therefore, in this study, dietary AA levels were titrated to determine the minimal level required for survival, while maintaining a distinct AA-deficient phenotype. Null mice supplemented with AA (0.1, 0.4, 0.6, 2.0%, w/w) in the diet exhibited a dose-dependent increase (P < 0.05) in AA, PGE2, 6-keto PGF1α, TXB2 and EdU-positive proliferative cells in the colon. In subsequent experiments, null mice supplemented with 0.6% AA diet were injected with a colon-specific carcinogen (azoxymethane) in order to assess cancer susceptibility. Null mice exhibited significantly (P < 0.05) reduced levels/multiplicity of aberrant crypt foci (ACF) as compared to wild type sibling littermate control mice. These data indicate that (i) basal/minimal dietary AA supplementation (0.6%) expands the utility of the Fads1 Null mouse model for long-term cancer prevention studies, and (ii) that AA content in the colonic epithelium modulates colon cancer risk.
Keywords: Fads1, arachidonic acid, eicosanoids, cell proliferation, aberrant crypt foci
Introduction
Colorectal cancer is the third most common diagnosed cancer in men and the second in women (1). Elevated levels of cyclooxygenase 2 (COX2) and concomitant overproduction of prostaglandins of the 2-series, e.g., PGE2 and TXA2, are often detected in human colon adenomas and adenocarcinomas (2, 3). From a physiological perspective, AA-derived eicosanoids, including PGE2 are diverse, and have been shown to stimulate key downstream signaling cascades that regulate cell proliferation, apoptosis, angiogenesis, inflammation, and immune surveillance (4, 5).
Arachidonic acid (20:4 5,8,11,14, AA) is the preferred substrate for cyclooxygenase catalysis (6). Dietary linoleic acid (18:2 9,12, LA) is the major source of tissue dihomo-gamma-linolenic acid (20:3 8,11,14, DGLA) and AA, and its metabolism is regulated by the complementary action of fatty acid desaturases (Fads1 and Fads2) (7). Most studies have targeted prostaglandin biosynthetic and degradation enzymes in an attempt to suppress AA-derived eicosanoid–mediated inflammation and cancer initiation/promotion (8). Surprisingly, very few investigators to date have attempted to target AA substrate levels as a way of modulating prostaglandin biosynthesis and tumor development. We recently developed a novel genetic model, i.e., the Fads1 (Δ5 desaturase) knockout mouse, to delineate the role of AA-derived 2-series eicosanoids in mucosal physiology and colon cancer risk. This model allows for the specific investigation of AA deficiency without the underlying complications of essential fatty acid (LA and DGLA) deficiency (9). Unfortunately, a complication of this model is reduced viability after weaning when mice are fed an AA-free diet.
In this study, we report the usefulness of this model to address clinical questions related to colorectal cancer. We initially titrated dietary AA levels to determine the minimal AA dose capable of sustaining viability of the Fads1 knock out mice for long-term carcinogenesis studies. In a complementary proof-of-principal study, we assessed the effects of dietary AA on colonocyte cell proliferation and aberrant crypt foci (ACF) formation, early indicators of cancer initiation/progression.
Materials and Methods
Animals and diets
Mutant Fads1 null mice were originally generated using a gene-trapping technique at the Texas Institute for Genomic Medicine (TIGM) as previously described (9). Wild type (Wt), heterozygous (Het) and null (Null) mice were derived from heterozygous crosses. All procedures followed the guidelines approved by the Public Health Service and the Institutional Animal Care and Use Committee at Texas A&M University. Wt mice were fed a basal commercial 10% safflower oil diet (Research Diet, D03092902R), free of AA. Null mice were originally fed the AA-free basal diet, then switched to AA supplemented diets post-weaning (at 4–5 weeks of age). ARASCO oil (containing 42% AA, w/w) was added to the basal 10% safflower oil diet to generate 0.1, 0.4, 0.6 and 2% (w/w) AA supplemented diets. The fatty acid composition of these diets is described in Supplemental Table 1. Mice were fed the basal or AA supplemented diets for 7–11 weeks. At the end of the feeding period, mice were terminated and tissues were collected for analysis as described below.
Fatty acid analysis
Fatty acid profiles of dietary lipids and select issues (colonic mucosa, liver, tail) were extracted by the method of Folch (10) and transesterified in 6% methanolic HCl overnight, followed by gas chromatography analysis as previously described (11).
Eicosanoid analysis
Eicosanoids were extracted from colonic mucosa as previously described (12–14). Briefly, an aliquot of the 0.5 ml of PBS buffer was added to the snap frozen tissues followed with homogenization in using the Precellys tissue homogenizer (Bertin Technology). An aliquot of homogenate (400 μl) was then extracted with hexane/ethyl acetate (1:1). The upper organic layer was collected and the organic phases from three extractions were pooled and evaporated to dryness under a stream of nitrogen. All extraction procedures were performed at minimum light levels at 4°C. Samples were then reconstituted in 100 μl of methanol/0.1% acetic acid (50:50, v/v) before liquid chromatography/tandem mass spectroscopic analysis. Protein concentration was determined by the method of Bradford according to the manufacturer's instructions (Bio-Rad). Liquid chromatography/tandem mass spectroscopic analyses were performed using a Agilent 6460 triple quadrupole mass spectrometer (Agilent Technologies, Palo Alto, CA) equipped with an Agilent 1200 binary pump high-performance liquid chromatography system (Agilent) using a the method of Yang et al. (12, 14). Eicosanoids of interest were separated using a Kinetex 3 μ C-18 (4.6 × 100) mm analytic column (Phenomenex, Torrance, CA). The mobile phase consisted of 0.1% formic acid and acetonitrile with 0.1% formic acid. The flow rate was 0.4 mL/min with a column temperature of 35°C, and samples were kept at 4°C. The mass spectrometer was operated in both negative and positive electrospray ionization modes with source and gas temperatures at 350°C. Eicosanoids were detected and quantified by multiple reaction mode (MRM) monitoring the transitions of analytes and their relevant internal standards (15).
Analysis of colonic cell proliferation
In situ colonic cell proliferation was determined by immunohistochemical detection. Mice were i.p. injected with 5-ethynyl-2′-deoxyuridine (EdU, 30 mg/kg body weight) 2 h prior to termination. One cm of distal colon was removed, fixed in 4% paraformaldehyde, followed by a series of ethanol washes, and embedded in paraffin. The incorporation of EdU into DNA of actively dividing cells was determined using a commercially available kit (Click-iT EdU Alexa Fluor 647 Imaging Kit; Invitrogen). Briefly, after deparaffinization, samples were washed in 3% BSA in PBS, treated with 0.5% Triton in PBS for 20 min, washed again with 3% BSA in PBS, then incubated with Click-It reaction cocktail for 30 min. Coupling of EdU to the Alexa fluor substrate was then observed using fluorescence microscopy (9).
Characterization of immune cell populations
A single-cell suspension was produced by combining spleen, mesenteric and inguinal lymph nodes that were pushed through a sterile stainless steel wire screen (100 mesh) and resuspended in RPMI 1640 medium with 25 mmol/L HEPES (Irvine Scientific), supplemented with 10% fetal bovine serum (FBS, Irvine Scientific), 2 mmol/L GlutaMAX (Gibco), penicillin 100 U/mL and streptomycin 0.1 mg/mL (Gibco). Subsequently, a mononuclear cell suspension was produced by density gradient centrifugation using Lympholyte-M (Cedarlane Laboratories). The T cell compartment was identified by surface expression of CD3. For this purpose, 106 viable mononuclear cells were incubated for 10 min with a FcγR blocking monoclonal antibody (1 μg/mL) (2.4G2, BD Pharmingen) on ice and subsequently stained with 1 μg/mL of APC-anti-mouse CD3 (clone 145-2C11, eBioscience) antibodies for 30 min. Flow cytometric analysis was conducted using an Accuri C6 flow cytometer (Accuri Cytometers).
Carcinogen treatment
To determine whether the deletion of Fads1 impacts the formation of early preneoplastic lesions (aberrant crypt foci, ACF) in the colon, intestinal segments were collected from carcinogen treated animals. Specifically, 8–10 weeks old Wt (fed AA-free diet) and Null mice (fed 0.6% AA supplemented diet) were injected s.c. with azoxymethane (AOM) at 10 mg/kg body weight once weekly for 4 weeks. Eight weeks after the final AOM injection, mice were terminated for ACF analysis as previously described (16, 17).
Aberrant crypt evaluation
Distal colonic sections were excised and flushed with cold PBS, slit longitudinally, fixed flat between two pieces of Whatman No. 1 filter paper, and placed under a glass plate in 70% ethanol for 24 h. Fixed colons were subsequently stained with 0.2% methylene blue in PBS for 5 min and then placed on a light microscope equipped with a clear grid and the mucosal side oriented upward. Aberrant crypts, which are larger and have a thicker epithelial lining were visualized at 40× magnification and scored for total number and multiplicity (number of crypts per focus) as described previously (16, 18–20). Details related to the criteria to define aberrant crypts and representative ACF images are shown in Supplemental Figure 1.
RNA sequencing
Post AOM treatment, RNA was extracted from proximal colonic mucosa using the Ambion RNA isolation kit as per the manufacture’s protocol. RNA quantity was determined using a Nanodrop (Fisher Thermoscientific) spectrophotometer and the quality was assessed using the Nano6000 chip on a Bioanalyzer (Agilent). Only RNA with an RIN (RNA Integrity Number) of 8 or higher was used. Samples were randomized prior to RNASeq library preparation. Sequencing libraries were generated using 1 μg of RNA and the TruSeq RNA Sample Preparation kit (Illumina) as per manufacturer’s instructions. Libraries were pooled and sequenced on an Illumina HiSeq 2500 at SeqWright Genomic Services (Houston, TX). Sequencing data were demultiplexed and aligned using STAR with default parameters (21) and referenced against Mus musculus (UCSC version mm10). Differentially expressed genes were determined using EdgeR (22) based on the matrix of gene counts. Gene networks were assessed using Ingenuity Pathway Analysis (IPA) software. Upstream regulator analysis was based on prior knowledge of expected associations between transcriptional regulars and their target genes stored in the Ingenuity Knowledge database (23).
Statistical analysis
Data were analyzed using one-way ANOVA. Differences of P < 0.05 were considered significant. Data were tested for normality by D’Agostino-Pearson omnibus normality test, and are presented as means ± SE. Analyses were conducted by using the Prism 6 program (GraphPad Software, Inc., La Jolla, CA).
Results
Temporal changes in AA levels in Fads1 Null mice
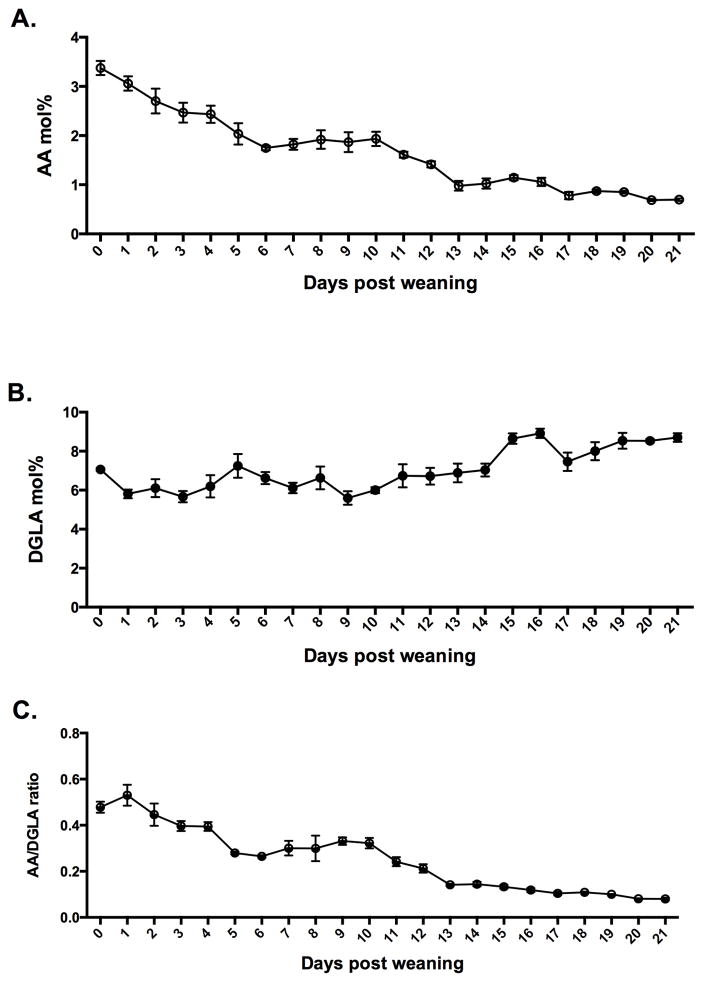
Fatty acid profiles in Fads1 Null mouse tails were assessed immediately post-weaning (~ 4 weeks old) (without AA supplementation) and were monitored daily for 3 weeks (Figure 1A&B). As Null pups transitioned from Het mother’s milk to an AA-free diet, the tail levels of AA gradually decreased, while DGLA was slightly increased due to the deletion of Fads1. Figure 1C documents tail tissue-specific changes in the AA/DGLA ratio in Null mice post-weaning.
Figure 1. Post-weaning temporal changes in AA / DGLA levels.
Total lipids were isolated from Fads1 Null mouse tails at different ages post-weaning. Profiles of (A) AA and (B) DGLA (mol%) and (C) ratios of AA/DGLA, were measured. (mean ± SE, n=3–5 animals per data point).
Dietary AA supplementation dose-dependently enhances Fads1 Null mouse viability
We have previously demonstrated that Fads1 Null mice do not survive beyond 12 weeks of age without supplementation of exogenous AA (9). To further assess their dietary AA requirement, 5 different diets containing increasing (0–2 wt %) levels of AA were administered (Supplemental Table 1). Main differences among the diets were the levels of AA and LA (precursor of DGLA via Fads2 and elongase). The Kaplan-Meier survival curves of Fads1 Null mice (Supplemental Figure 2) confirmed the essential role of AA by extending the longevity of Null mice. This effect was dose-dependent, with both 0.6 and 2% AA supplementation extending survival rates to levels comparable to Wt mice. Interestingly, compared to the slightly lower group of 0.4% AA supplementation, the 0.6% AA group exhibited a 100% survival rate. These data indicate the presence of a dietary threshold for this essential fatty acid, required to maintain basic physiological functions.
AA supplementation alters tissue fatty acid levels in a dose-dependent manner
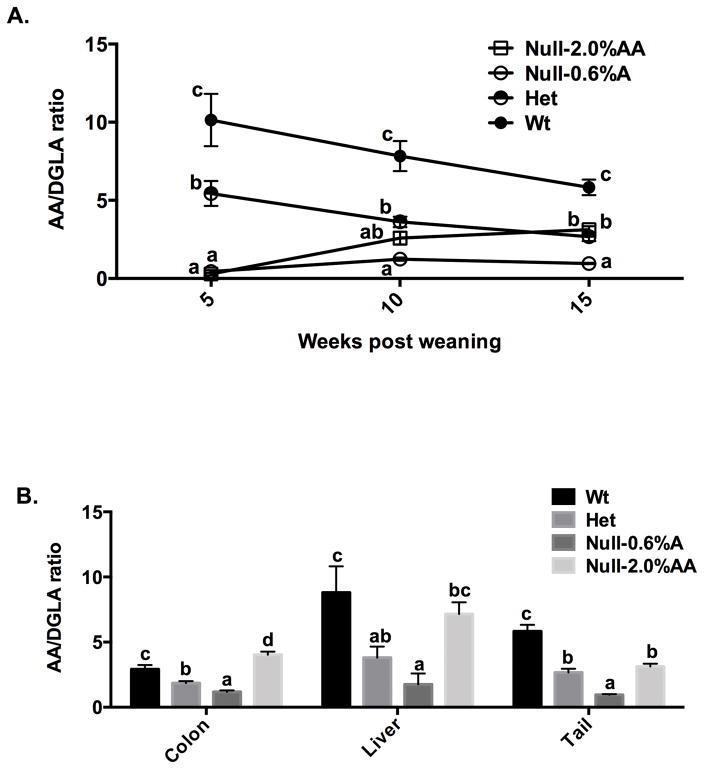
In order to compare tissue fatty acid profiles in AA-fed Null, Wt and Het mice over a 10 week feeding period, tail fatty acid profiles were assessed in the same cohort of animals at 3 time points: initial (5 wk of age), mid (10 wk of age) and end (15 wk of age). As shown in Figure 2A, the 2% AA diet normalized fatty acid levels relative to Het mice, while 0.6% AA supplementation resulted in a significantly lower AA/DGLA ratio compared to Wt and Het (P < 0.05) mice. Similar AA/DGLA profiles in Wt and 0.6% AA-fed Null were also observed in the liver and colonic mucosa at the end of the feeding period (Figures 2B).
Figure 2. AA/DGLA ratios in AA-supplemented Fads1 null mice.
Post weaning (5 wk old) Fads1 Wt, Het and Null mice were fed an AA-free diet, while Null–0.6% AA and Null–2% AA mice were fed diets supplemented with 0.6 and 2% AA (w/w), respectively, for 10 weeks. (A) Total lipids were isolated from mouse tails at the initial (5 wk of age), mid (10 wk of age) and end (15 wk of age) of the feeding period. Different letters at the same time point indicate significantly differences (P < 0.05), mean ± SE, n=4–10 mice per data point. (B) Total lipids were isolated from colonic mucosa, liver and tail, and fatty acid profiles were measured by GC-MS at the end of the feeding period (15 wk of age) (mean ± SE, n=3). Bars not sharing the same letter indicate a significant difference within respective tissues (P < 0.05).
Effects of AA supplementation on eicosanoid and cannabinoid profiles
Following AA supplementation, eicosanoid and cannabinoid levels in Null mouse colonic mucosa were significantly (P < 0.05) altered compared to Null mice fed an AA-free diet. As shown on Table 1, AA-derived eicosanoids, including PGE2, 6-keto-PGF1α, 13-hydroxyl octadecadienoic acid (13-HODE), 12-hydroxyl eicosatetraenoic acid (12-HETE), thromboxane B2 (TXB2), PGD2; and cannabinoids, including arachidonoyl ethanolamine (AEA), 2- arachidonoyl glycerol (2-AG) were significantly increased in Null mice supplemented with AA. In comparison, DGLA-derived eicosanoids, including PGE1 and 13-PGE1 were unaltered. Although 2% AA supplementation significantly (P < 0.05) increased colonic mucosal AA-derived eicosanoid levels in Null mice, nearing or surpassing the levels in Wt mice; the levels of PGE2, 6-keto-PGF1α and TXB2 in the 0.6% AA group were still significantly (P < 0.05) lower than those from Wt mice fed AA-free diet.
Table 1.
| Wt (0% AA) | Het (0% AA) | Null (0% AA) | Null (0.6% AA) | Null (2% AA) | |
|---|---|---|---|---|---|
| (ng/mg protein) | |||||
| PGE1 | 0.68±0.10a | 0.95±0.18ab | 1.19±0.13b | 1.37±0.23b | 1.15±0.19ab |
| PGE2 | 10.40±1.51c | 7.99±1.69b | 1.11±0.31a | 5.65±0.50ab | 8.39±1.13b |
| 6-keto-PGF1α | 1.45±0.24c | 1.15±0.19bc | 0.08±0.02a | 0.60±0.06ab | 1.01±0.26b |
| 13-PGE2 | 0.75±0.23b | 0.53±0.10ab | 0.00±0.00a | 1.13±0.21b | 0.71±0.18ab |
| 13-HODE | 0.62±0.30ab | 1.36±0.57b | 0.12±0.01a | 0.38±0.05ab | 3.16±0.49c |
| 12-HETE | 1.13±0.16b | 1.02±0.28ab | 0.26±0.07a | 0.84±0.14ab | 2.67±0.93c |
| TXB2 | 0.49±0.06c | 0.31±0.10b | 0.02±0.01a | 0.31±0.05b | N/A3 |
| PGD2 | 6.62±1.63b | 5.70±1.62b | 0.44±0.07a | 8.60±0.84b | N/A |
| AEA | 0.15±0.02b | 0.11±0.01b | 0.04±0.00a | 0.24±0.05c | N/A |
| 2-AG | 12.83±2.19b | 10.26±2.02b | 2.29±0.37a | 8.65±1.39b | N/A |
| 13-PGE1 | 0.10±0.03 | 0.08±0.00 | N/A | 0.10±0.03 | 0.06±0.03 |
| 15-HETE | 1.09±0.34 | 0.78±0.13 | N/A | 0.77±0.05 | 1.20±0.31 |
| 5-HETE | 0.22±0.07a | 0.18±0.06ab | N/A | 0.40±0.04b | 0.42±0.11b |
Abbreviations: PG, prostaglandin; HODE, hydroxyl octadecadienoic acid; HETE, hydroxyl eicosatetraenoic acid; TX, thromoboxane; AEA, arachidonoyl ethanolamine; AG, arachidonoyl glycerol; 13-PGE2, 13,14-dihydro 15-keto-PGE2; 13-PGE1, 13,14-dihydro 15-keto-PGE1.
Values not sharing the same letter are significantly different (p < 0.05), mean ± SE, n= 4–11.
N/A, not available.
AA supplementation promotes colonic cell proliferation
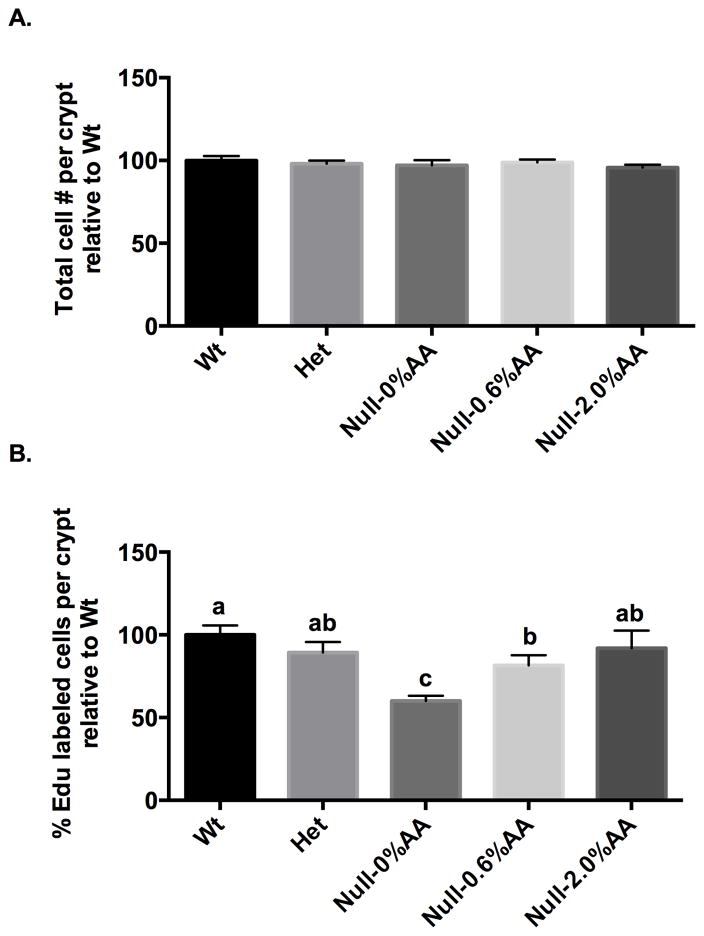
Since PGE2 has been linked to cancer progression (24), we determined the effect of dietary AA on colonic cell proliferation in Null mice. As expected, AA supplementation significantly enhanced PGE2 levels in the colonic mucosa (Table 1), and further elevated the proliferative index (Figure 3), i.e., % Edu positive cells in colonic crypts. Importantly, 2.0% AA supplementation normalized the number of cells in S-phase relative to Wt mice fed AA-free diet, similar to the eicosanoid data shown in Table 1. In comparison, supplementation with 0.6% AA only partially elevated the proliferation index, remaining significantly (P < 0.05) below Wt mice fed an AA-free diet.
Figure 3. Effect of AA supplementation on colonocyte proliferation.
Cell proliferation in mouse colonic crypts was measured by the EdU Click-It assay. Refer to Figure 2 for legend details. Data are expressed as (A) Total number of cells per crypt relative to Wt, and (B) Percentage of EdU-labeled cells per crypt relative to Wt, mean ± SE, n=3–8. Bars not sharing the same letter indicate significant differences (P < 0.05).
The percentage of CD3+ T cells is rescued by AA supplementation in Fads1 null mice
We have previously demonstrated that Null mice exhibit lower levels of CD3+ T cells (9). In this study, we determined the ability of dietary AA supplementation to normalize elements of the adaptive immune cell population, specifically the CD3+ T cells. Similar to the eicosanoid and proliferation data (Table 1, Figure 3), AA supplementation dose-dependently increased the percentage of CD3+ T cells in Null mice (Supplemental Figure 3).
Colonocyte preneoplasia histopathology is decreased in Fads1 null mice
A goal of this study was to extend longevity of the Fads1 Null mouse while maintaining their AA-depleted properties for the purpose of conducting long-term carcinogenesis studies to elucidate the role of AA in cancer susceptibility. For this purpose, we administered Null mice the lowest level of AA shown to promote viability (0.6% AA). The data in Table 2 show that 0.6% AA-fed Null mice exposed to AOM (colon carcinogen) had a significantly (P < 0.05) decreased risk of neoplasia progression, as indicated by both a lower number of ACF per cm of colon and a reduced number of high multiplicity ACF, relative to Wt siblings fed a semi-purified diet lacking AA.
Table 2.
Effect of Fads1 deficiency on AOM-induced ACF development
| Wt | Null | p-value | |
|---|---|---|---|
| # aberrant crypt/ focus | |||
| 1 | 17.27±0.31 | 14.38±2.10 | 0.385 |
| 2 | 10.46±1.43 | 7.25±0.80 | 0.096 |
| 3 | 4.27±0.97 | 1.88±0.35 | 0.060 |
| 4 | 1.18±0.23 | 0.25±0.16 | 0.007 |
| 5+ | 0.64±0.24 | 0.13±0.13 | 0.114 |
| Colon length (cm) | 3.66±0.29 | 3.76±0.14 | 0.791 |
| Total ACF#/colon | 33.82±4.28 | 23.88±2.56 | 0.088 |
| ACF#/cm colon | 9.83±1.28 | 6.38±0.66 | 0.047 |
| High Multiplicity1 ACF#/colon | 6.09±1.00 | 2.25±0.56 | 0.008 |
| % High Multiplicity2 | 18.14±1.84 | 9.21±1.71 | 0.003 |
High Multiplicity ACF: foci containing 3 or more aberrant crypts.
Percentage of High Multiplicity: number of high multiplicity ACF divided by total number of ACF.
Significantly different (P < 0.05). Data represent means ± SE, n=8–11.
Deletion of Fads1 alters tumor and cellular development related genes
Fads1 gene knock out did not significantly (FDR > 0.2) alter the expression of genes associated with lipid metabolism, including various desaturases, elongases, cyclooxygenases and lipoxygenases in colonic mucosa following AOM challenge (Supplemental Table 2). To our surprise, no significant effects on Wnt signaling related TCF/LEF family genes were observed despite the fact that PGE2 plays an important role in Wnt signaling pathway (25). Interestingly, several colon cancer markers, such as Rentib (resistin-like molecule beta) and Ceacam2 (carcinoembryonic antigen cell adhesion molecules) were significantly decreased in the Fads1 Null mice (Table 3). The resistin-like molecule beta, an intestinal goblet cell-specific protein, has been reported to be markedly overexpressed in intestinal tumors from Min (multiple intestinal neoplasia) mice as well as a human colon cancer cell line (26). In addition, members of the carcinoembryonic antigen cell adhesion molecules family are prototypical tumour markers (27). The significant 13.3-fold decrease in Ceacam2 and 1.8 fold decrease in Rentib gene expression (Table 3) are consistent with the suppressed ACF phenotype observed in Null mice (Table 2). To further explore the subtle network changes in key regulatory pathways, upstream analysis of differentially expressed genes with P<0.05 was conducted. Several cancer progression pathways involving upstream regulators, e.g., IFN, TWIST1, NOS2, PI3K, SMARCB1, HES1, S100A9, S100A8 and IRF7, were all inhibited in Null vs Wt animals, with Z scores less than −2.0. Representative signaling PI3K, NOS2 and S100A8 networks are shown in Supplemental Figure 4.
Table 3.
Significantly modulated colonic mucosal genes in Fads1 Null vs. wild type mice.
| Downregulated | Fold Δ (Null/Wt) | Upregulated | Fold Δ (Null/Wt) |
|---|---|---|---|
| Fads1 | −1489.3 | Fcer2a | 18.6 |
| Eif2s3y | −628.4 | BB123696 | 3.8 |
| Kdm5d | −585.2 | 8430408G22Rik | 3.8 |
| Uty | −281.1 | Scand1 | 3.8 |
| Ddx3y | −269.5 | S100g | 2.9 |
| Ube1y1 | −61.0 | Cldn8 | 2.1 |
| 9530053A07Rik | −14.8 | Arntl | 2.0 |
| Ceacam2 | −13.3 | Nfil3 | 2.0 |
| Gm5325 | −7.5 | Cps1 | 1.9 |
| Retnib | −1.8 | Aadac | 1.6 |
| Per3 | −1.6 | Tpt1 | 1.6 |
| Tef | −1.5 | Cyp27a1 | 1.5 |
| Atg2a | −1.4 | Cited2 | 1.4 |
| Irf2bp2 | 1.4 |
only mRNA with an FDR(Q) value less than 0.2 are reported, n=3 mice.
Discussion
The effects of non-steroidal anti-inflammatory drugs (NSAIDs) on the gastrointestinal tract are well described, however their possible propensity to cause clinical relapse in patients with inflammatory bowel disease (28) and exacerbate respiratory diseases (29), appear to limit their utility to the general public. The detrimental side effects are likely due to the inhibition of both COX1 and COX2 enzymes. We demonstrate that the Fads1 knock out mouse model can be utilized as a novel approach to suppress cell membrane AA and its derived eicosanoids (9), without compromising the metabolic status of other essential fatty acids and their metabolites, a common side effect of most NSAIDs.
Null mice can be used only for short-term studies due to the lethal effect of cellular AA depletion overtime (9). Therefore, in this study, we administered AA in the diet in order to provide Null mice with a minimum survival dose, enough to maintain normal function, while still exhibiting a significantly reduced AA/PGE2 profile compared to wild type siblings. Examination of fatty acid profiles post-weaning (Figure 1) indicated that once Null mice no longer have access to their Het mother’s milk, cellular AA mol% and AA/DGLA ratio levels decline progressively and steadily, consistent with a decrease in viability. These data reaffirm that AA is absolutely required in order to maintain normal cell function. Interestingly, excess AA in cell membranes may have a detrimental effect, as exhibited by the positive correlation between AA-derived PGE2, TXA2 and colon cancer risk (3, 30, 31).
In order to further probe the utility of the Fads1 mouse model (9), titration experiments revealed that, 0.6% AA (w/w) supplementation rescued 100% of the Null mice (Supplemental Figure 2), while maintaining a significantly lower AA/DGLA ratio in multiple tissues (Figure 2) as compared to Wt siblings. In general, tissue AA/DGLA levels and colonic mucosa AA-derived eicosanoids (PGE2, 6-keto-PGF1α, TXB2) were correlated in Null mice relative to Wt siblings (Table 1). Furthermore, the suppressed eicosanoid profile in Null mice was associated with a reduction in CD3+ T cells and colonic crypt cell proliferation (Figure 3 and Supplemental Figure 3). This finding is noteworthy because hyperproliferation of systematic T cells is associated with elevated levels of PGE2 (32) and inflammatory bowel disease (33, 34).
Collectively, these data indicate that dietary AA supplementation is essential in extending the longevity of the Null mouse. In addition, by utilizing a “minimal” survival dose level of AA (0.6 wt%), we are still able to lower membrane AA composition, AA-derived eicosanoids, and colonic cell proliferation compared to Wt animals. This affords an experimental opportunity to conduct long-term chemoprevention studies. In an initial investigation, we contrasted the effects of a colon-specific carcinogen in Null mice supplemented with 0.6% AA versus Wt (control) mice. These groups of animals were chosen because they represent the most distinct systemic AA levels (high vs. low) in this Fads1 mouse model. The use of AOM is based on its specific carcinogenic potential in colon. For example, it is well known that AOM induces colorectal adenocarcinomas at high incidence in rodents, resulting in a similar multistep progression relative to human sporadic colorectal cancer (35). This widely accepted model is commonly used to assess the chemopreventive / chemotherapeutic potency of dietary components with important bioactive properties (36). Aberrant crypt foci induced by treatment with AOM is a well-established predictive biomarker, with the formation and growth of these preneoplastic lesions representing the earliest stages of colon cancer (37, 38). Although several alternative precancerous lesions, including mucin depleted foci, beta-catenin-accumulated crypts, and flat ACF, have been described and are useful biomarkers relative to ACF (39–41), the significant correlation of high multiplicity ACF incidence and tumor outcome strongly suggest the ACF remain a useful biomarker for the initial screening of agents for chemoprevention (42, 43).
The significant reduction in cell proliferation and ACF formation in Fads1 Null mice (Figure 3, Table 2) is consistent with previous studies indicating that AA-derived eicosanoids (especially PGE2) promote aberrant Wnt signaling and malignant transformation of the colonic mucosa (25, 44). In addition, examination of colonic mucosa global transcriptional data from AOM-treated mice indicated that several tumor markers were decreased (FDR < 0.2), e.g., Ceacam2 and Retnib, suggesting that the Fads1 Null mice are more resistant to carcinogen insult as compared to Wt mice. It has been shown that Ceacem acts as a crucial factor in glioblastoma-initiating cells, a tumorigenic cell subpopulation resistant to radiotherapy, chemotherapy, by activating c-Src/STAT3 signaling (45). Up-regulation of the COX2/PGE2- JAK2/STAT3 signaling pathway has also been reported in subjects presenting with colorectal cancer invasion and metastasis (30, 46). In addition, in Fads1 null mice, the inhibition of several upstream regulator networks, including PI3K, NOS2 and S100A8, is consistent with a reduction in susceptibility to AOM induced carcinogenesis.
In summary, we propose that the Fads1 null mouse is a new model to study the role of AA-metabolites in cancer pathophysiology.
Supplementary Material
Acknowledgments
This work was supported by NIH R35CA197707 (R.S. Chapkin), P30ES02351 (R.S. Chapkin), American Institute for Cancer Research (R.S. Chapkin), and by grant RP120028 from the Cancer Prevention and Research Institute of Texas (CPRIT) (R.S. Chapkin).
We thank DSM Nutritional Products for providing the ARASCO oil and Dr. Martha Hensel for ACF related analyses.
Footnotes
The authors have no potential conflicts of interest to disclose.
References
- 1.Baena R, Salinas P. Diet and colorectal cancer. Maturitas. 2015;80:258–64. doi: 10.1016/j.maturitas.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 2.Elzagheid A, Emaetig F, Alkikhia L, Buhmeida A, Syrjanen K, El-Faitori O, et al. High cyclooxygenase-2 expression is associated with advanced stages in colorectal cancer. Anticancer Res. 2013;33:3137–43. [PubMed] [Google Scholar]
- 3.Li H, Liu K, Boardman LA, Zhao Y, Wang L, Sheng Y, et al. Circulating Prostaglandin Biosynthesis in Colorectal Cancer and Potential Clinical Significance. EBioMedicine. 2015;2:165–71. doi: 10.1016/j.ebiom.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D, Mann JR, DuBois RN. The role of prostaglandins and other eicosanoids in the gastrointestinal tract. Gastroenterology. 2005;128:1445–61. doi: 10.1053/j.gastro.2004.09.080. [DOI] [PubMed] [Google Scholar]
- 5.Schneider SL, Ross AL, Grichnik JM. Do inflammatory pathways drive melanomagenesis? Exp Dermatol. 2015;24:86–90. doi: 10.1111/exd.12502. [DOI] [PubMed] [Google Scholar]
- 6.Smith WL. Cyclooxygenases, peroxide tone and the allure of fish oil. Curr Opin Cell Biol. 2005;17:174–82. doi: 10.1016/j.ceb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Mathias RA, Vergara C, Gao L, Rafaels N, Hand T, Campbell M, et al. FADS genetic variants and omega-6 polyunsaturated fatty acid metabolism in a homogeneous island population. J Lipid Res. 2010;51:2766–74. doi: 10.1194/jlr.M008359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu WK, Sung JJ, Lee CW, Yu J, Cho CH. Cyclooxygenase-2 in tumorigenesis of gastrointestinal cancers: an update on the molecular mechanisms. Cancer Lett. 2010;295:7–16. doi: 10.1016/j.canlet.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 9.Fan YY, Monk JM, Hou TY, Callway E, Vincent L, Weeks B, et al. Characterization of an arachidonic acid-deficient (Fads1 knockout) mouse model. J Lipid Res. 2012;53:1287–95. doi: 10.1194/jlr.M024216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 11.Chapkin RS, Somers SD, Erickson KL. Dietary manipulation of macrophage phospholipid classes: selective increase of dihomogammalinolenic acid. Lipids. 1988;23:766–70. doi: 10.1007/BF02536219. [DOI] [PubMed] [Google Scholar]
- 12.Yang P, Chan D, Felix E, Madden T, Klein RD, Shureiqi I, et al. Determination of endogenous tissue inflammation profiles by LC/MS/MS: COX- and LOX-derived bioactive lipids. Prostaglandins Leukot Essent Fatty Acids. 2006;75:385–95. doi: 10.1016/j.plefa.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 13.Jia Q, Lupton JR, Smith R, Weeks BR, Callaway E, Davidson LA, et al. Reduced colitis-associated colon cancer in Fat-1 (n-3 fatty acid desaturase) transgenic mice. Cancer Res. 2008;68:3985–91. doi: 10.1158/0008-5472.CAN-07-6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pirman DA, Efuet E, Ding XP, Pan Y, Tan L, Fischer SM, et al. Changes in cancer cell metabolism revealed by direct sample analysis with MALDI mass spectrometry. PLoS One. 2013;8:e61379. doi: 10.1371/journal.pone.0061379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes D, Otani T, Yang P, Newman RA, Yantiss RK, Altorki NK, et al. NAD+-dependent 15-hydroxyprostaglandin dehydrogenase regulates levels of bioactive lipids in non-small cell lung cancer. Cancer Prev Res (Phila) 2008;1:241–9. doi: 10.1158/1940-6207.CAPR-08-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho Y, Kim H, Turner ND, Mann JC, Wei J, Taddeo SS, et al. A chemoprotective fish oil- and pectin-containing diet temporally alters gene expression profiles in exfoliated rat colonocytes throughout oncogenesis. J Nutr. 2011;141:1029–35. doi: 10.3945/jn.110.134973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma DW, Finnell RH, Davidson LA, Callaway ES, Spiegelstein O, Piedrahita JA, et al. Folate transport gene inactivation in mice increases sensitivity to colon carcinogenesis. Cancer Res. 2005;65:887–97. [PMC free article] [PubMed] [Google Scholar]
- 18.Bird RP. Observation and quantification of aberrant crypts in the murine colon treated with a colon carcinogen: preliminary findings. Cancer Lett. 1987;37:147–51. doi: 10.1016/0304-3835(87)90157-1. [DOI] [PubMed] [Google Scholar]
- 19.McLellan EA, Medline A, Bird RP. Dose response and proliferative characteristics of aberrant crypt foci: putative preneoplastic lesions in rat colon. Carcinogenesis. 1991;12:2093–8. doi: 10.1093/carcin/12.11.2093. [DOI] [PubMed] [Google Scholar]
- 20.Murray NR, Davidson LA, Chapkin RS, Clay Gustafson W, Schattenberg DG, Fields AP. Overexpression of protein kinase C betaII induces colonic hyperproliferation and increased sensitivity to colon carcinogenesis. J Cell Biol. 1999;145:699–711. doi: 10.1083/jcb.145.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Triff K, Konganti K, Gaddis S, Zhou B, Ivanov I, Chapkin RS. Genome-wide analysis of the rat colon reveals proximal-distal differences in histone modifications and proto-oncogene expression. Physiol Genomics. 2013;45:1229–43. doi: 10.1152/physiolgenomics.00136.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubois RN. Role of inflammation and inflammatory mediators in colorectal cancer. Trans Am Clin Climatol Assoc. 2014;125:358–72. discussion 72–3. [PMC free article] [PubMed] [Google Scholar]
- 25.Fan YY, Davidson LA, Callaway ES, Goldsby JS, Chapkin RS. Differential effects of 2- and 3-series E-prostaglandins on in vitro expansion of Lgr5+ colonic stem cells. Carcinogenesis. 2014;35:606–12. doi: 10.1093/carcin/bgt412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng LD, Tong QS, Weng MX, He J, Lv Q, Pu JR, et al. Enhanced expression of resistin-like molecule beta in human colon cancer and its clinical significance. Dig Dis Sci. 2009;54:274–81. doi: 10.1007/s10620-008-0355-2. [DOI] [PubMed] [Google Scholar]
- 27.Heine M, Nollau P, Masslo C, Nielsen P, Freund B, Bruns OT, et al. Investigations on the usefulness of CEACAMs as potential imaging targets for molecular imaging purposes. PLoS One. 2011;6:e28030. doi: 10.1371/journal.pone.0028030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kvasnovsky CL, Aujla U, Bjarnason I. Nonsteroidal anti-inflammatory drugs and exacerbations of inflammatory bowel disease. Scand J Gastroenterol. 2015;50:255–63. doi: 10.3109/00365521.2014.966753. [DOI] [PubMed] [Google Scholar]
- 29.Ledford DK, Wenzel SE, Lockey RF. Aspirin or other nonsteroidal inflammatory agent exacerbated asthma. J Allergy Clin Immunol Pract. 2014;2:653–7. doi: 10.1016/j.jaip.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 30.Oshima H, Oshima M. The inflammatory network in the gastrointestinal tumor microenvironment: lessons from mouse models. J Gastroenterol. 2012;47:97–106. doi: 10.1007/s00535-011-0523-6. [DOI] [PubMed] [Google Scholar]
- 31.Montrose DC, Nakanishi M, Murphy RC, Zarini S, McAleer JP, Vella AT, et al. The role of PGE2 in intestinal inflammation and tumorigenesis. Prostaglandins Other Lipid Mediat. 2015;116–117:26–36. doi: 10.1016/j.prostaglandins.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sreeramkumar V, Fresno M, Cuesta N. Prostaglandin E2 and T cells: friends or foes? Immunol Cell Biol. 2012;90:579–86. doi: 10.1038/icb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabashima K, Saji T, Murata T, Nagamachi M, Matsuoka T, Segi E, et al. The prostaglandin receptor EP4 suppresses colitis, mucosal damage and CD4 cell activation in the gut. J Clin Invest. 2002;109:883–93. doi: 10.1172/JCI14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganapathi KA, Pittaluga S, Odejide OO, Freedman AS, Jaffe ES. Early lymphoid lesions: conceptual, diagnostic and clinical challenges. Haematologica. 2014;99:1421–32. doi: 10.3324/haematol.2014.107938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luceri C, De Filippo C, Caderni G, Gambacciani L, Salvadori M, Giannini A, et al. Detection of somatic DNA alterations in azoxymethane-induced F344 rat colon tumors by random amplified polymorphic DNA analysis. Carcinogenesis. 2000;21:1753–6. doi: 10.1093/carcin/21.9.1753. [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Huang XF. The signal pathways in azoxymethane-induced colon cancer and preventive implications. Cancer Biol Ther. 2009;8:1313–7. doi: 10.4161/cbt.8.14.8983. [DOI] [PubMed] [Google Scholar]
- 37.Schmelz EM, Xu H, Sengupta R, Du J, Banerjee S, Sarkar FH, et al. Regression of early and intermediate stages of colon cancer by targeting multiple members of the EGFR family with EGFR-related protein. Cancer Res. 2007;67:5389–96. doi: 10.1158/0008-5472.CAN-07-0536. [DOI] [PubMed] [Google Scholar]
- 38.Raju J. Azoxymethane-induced rat aberrant crypt foci: relevance in studying chemoprevention of colon cancer. World J Gastroenterol. 2008;14:6632–5. doi: 10.3748/wjg.14.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hata K, Kubota M, Shimizu M, Moriwaki H, Kuno T, Tanaka T, et al. Monosodium glutamate-induced diabetic mice are susceptible to azoxymethane-induced colon tumorigenesis. Carcinogenesis. 2012;33:702–7. doi: 10.1093/carcin/bgr323. [DOI] [PubMed] [Google Scholar]
- 40.Cui C, Takamatsu R, Doguchi H, Matsuzaki A, Saio M, Yoshimi N. Pre-neoplastic lesion, mucin-depleted foci, reveals de novo high-grade dysplasia in rat colon carcinogenesis. Oncol Rep. 2012;27:1365–70. doi: 10.3892/or.2012.1657. [DOI] [PubMed] [Google Scholar]
- 41.Suzui M, Morioka T, Yoshimi N. Colon preneoplastic lesions in animal models. J Toxicol Pathol. 2013;26:335–41. doi: 10.1293/tox.2013-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corpet DE, Tache S. Most effective colon cancer chemopreventive agents in rats: a systematic review of aberrant crypt foci and tumor data, ranked by potency. Nutr Cancer. 2002;43:1–21. doi: 10.1207/S15327914NC431_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunn BK, Steele VE, Fagerstrom RM, Topp CF, Ransohoff D, Cunningham C, et al. Predictive Value Tools as an Aid in Chemopreventive Agent Development. J Natl Cancer Inst. 2015;107:djv259. doi: 10.1093/jnci/djv259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaur J, Sanyal SN. PI3-kinase/Wnt association mediates COX-2/PGE(2) pathway to inhibit apoptosis in early stages of colon carcinogenesis: chemoprevention by diclofenac. Tumour Biol. 2010;31:623–31. doi: 10.1007/s13277-010-0078-9. [DOI] [PubMed] [Google Scholar]
- 45.Kaneko S, Nakatani Y, Takezaki T, Hide T, Yamashita D, Ohtsu N, et al. Ceacam1L Modulates STAT3 Signaling to Control the Proliferation of Glioblastoma-Initiating Cells. Cancer Res. 2015;75:4224–34. doi: 10.1158/0008-5472.CAN-15-0412. [DOI] [PubMed] [Google Scholar]
- 46.Liu X, Ji Q, Ye N, Sui H, Zhou L, Zhu H, et al. Berberine Inhibits Invasion and Metastasis of Colorectal Cancer Cells via COX-2/PGE2 Mediated JAK2/STAT3 Signaling Pathway. PLoS One. 2015;10:e0123478. doi: 10.1371/journal.pone.0123478. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.