Abstract
The Wnt/β-catenin pathway is among the most frequently altered signaling networks in human cancers. Despite decades of preclinical and clinical research, efficient therapeutic targeting of Wnt/β-catenin has been elusive. RNA interference (RNAi) technology silences genes at the mRNA level, and therefore can be applied to previously-undruggable targets. Lipid nanoparticles (LNPs) represent an elegant solution for delivery of RNAi-triggering oligonucleotides to disease-relevant tissues, but have been mostly restricted to applications in the liver. In this study, we systematically tuned the composition of a prototype LNP to enable tumor-selective delivery of a Dicer-substrate siRNA (DsiRNA) targeting CTNNB1, the gene encoding β-catenin. This formulation, termed EnCore-R, demonstrated pharmacodynamic activity in subcutaneous human tumor xenografts, orthotopic patient-derived xenograft (PDx) tumors, disseminated hematopoietic tumors, genetically induced primary liver tumors, metastatic colorectal tumors, murine metastatic melanoma. DsiRNA delivery was homogeneous in tumor sections, selective over normal liver and independent of apolipoprotein-E binding. Significant tumor growth inhibition was achieved in Wnt-dependent colorectal and hepatocellular carcinoma models, but not in Wnt-independent tumors. Finally, no evidence of accelerated blood clearance or sustained liver transaminase elevation was observed after repeated dosing in nonhuman primates. These data support further investigation to gain mechanistic insight, optimize dose regimens and identify efficacious combinations with standard-of-care therapeutics.
Keywords: RNAi, DsiRNA, lipid nanoparticle, β-catenin, tumor delivery
Introduction
The Wnt/β-catenin signaling pathway promotes differentiation events during embryonic development and leads to tumor formation when aberrantly activated (1). Dysregulation of the Wnt/β-catenin pathway is observed in approximately 90% of colorectal cancers (CRC) and >50% of liver cancers (2). The two most common oncogenic mutations causing unchecked Wnt/β-catenin signaling are gain-of-function point mutations or deletions in CTNNB1, the gene encoding β-catenin, or loss-of-function mutations in APC, a tumor suppressor gene that encodes the adenomatous polyposis coli protein, which negatively regulates β-catenin function (3). These and other known genetic lesions impair the cell’s ability to degrade β-catenin via the ubiquitin-proteasome pathway, resulting in its nuclear accumulation and subsequent complexation with T-cell factor/lymphoid enhancing factor (TCF/LEF) to activate gene transcription (4). The result of such tumorigenic lesions is transcriptional up-regulation of specific genes involved in proliferation, evasion of apoptosis, deregulated metabolism and metastatic potential (2, 5, 6). Preclinical evidence in genetic tumor models suggests that inhibiting β-catenin function would yield therapeutic benefit to sizable subpopulations of cancer patients (7, 8).
Like several other well characterized oncogenes, β-catenin is very challenging to target by conventional small molecule or monoclonal antibody approaches since it is an intracellular protein with no enzymatic activity (9). Therefore, no approved therapy targeting β-catenin has emerged despite decades of research. RNA interference (RNAi) technology has enabled the inhibition of previously undruggable targets at the mRNA level and has advanced to late clinical development for several indications (10). Most RNAi triggers that have been investigated as potential therapeutics are double-stranded or partially double-stranded oligonucleotides of less than 30 base pairs, and contain varying degrees of chemical modification to confer stability and reduce potential for immunostimulatory activation (11). The mechanism of action for all RNAi triggers that leads to target degradation is well-established. After the therapeutic RNA duplex reaches the cell cytosol, the guide (antisense) strand is bound by the Ago2 endonuclease, which targets its cognate mRNA for cleavage and destruction at a well-defined position exactly ten bases from the 5′ end of the guide stand.
Dicer substrate small interfering RNAs (DsiRNAs) are a unique class of RNAi triggers that leverage the microRNA machinery found in all mammalian cells. DsiRNAs are processed by the Dicer enzyme to 21 or 22mer RNA duplexes that are loaded into Ago2 to direct the specific degradation of target mRNAs with high potency and specificity (12). DsiRNAs have higher intrinsic potency than their cognate conventional siRNAs, which bypass Dicer for many (13, 14) but not all (15) target mRNAs. The often favorable profile of DsiRNAs as a structural class has been hypothesized to involve molecular interactions between the Dicer and Ago2 endonucleases.
To achieve RNA interference in a disease-relevant tissue after systemic administration, the oligonucleotide must have sufficient biodistribution, cell internalization and cytosolic partitioning properties to enable activation of the RNA-induced silencing complex (RISC) and subsequent degradation of the target mRNA. Since RNAi duplexes are inherently unstable, rapidly cleared and cell-impermeable, engineering drug delivery solutions has been a major focus of translational RNAi research for over a decade. Recent efforts have primarily focused on one of two strategies in investigational RNAi therapeutics: ligand-mediated targeting of a liver-specific receptor, or encapsulation in a functionalized nanoparticle that drives biodistribution and cell trafficking (10). So far, systemic delivery of RNAi triggers has advanced to late clinical development only for targets in the liver, underscoring the difficulty of tumor delivery despite well-validated targets and unmet medical need. A tumor delivery strategy must overcome a unique physiology, which features a malformed vasculature responsible for heterogeneity, hypoxia, acidotic regions and increased interstitial fluid pressure (16). These features of the microenvironment have been shown to act as challenging barriers to drug delivery (17–19). In addition to overcoming complex biology, a clinically viable delivery solution must meet pharmaceutical criteria: stability, scalability, and amenability to good manufacturing practices (GMP). Classes of RNAi delivery formulations that have been investigated as cancer therapeutics either preclinically or clinically include cyclodextrin-based polymers (20), chitosan particles (21), antibody-linked conjugates (22), carbon nanotubes (23), and lipid nanoparticles (LNPs) (13, 24–26).
LNPs and liposomes represent the most clinically advanced class of delivery vehicles for oncology therapeutics (27). Liposomal doxorubicin, daunorubicin and cytarabine have been approved for indications ranging from metastatic ovarian and breast cancer to acute myelogenous leukemia (AML), and numerous other LNPs and liposomes are in late clinical development. For cytotoxic agents and other small molecules, advances in LNP-based delivery offer the potential for improved pharmacokinetic (PK) properties, tumor bioavailability and therapeutic index. For cell-impermeable oligonucleotides, specialized LNPs enable escape of siRNA payloads from the endosomal compartment of tumor cells to the cytosol, in part by undergoing pH-dependent structural transitions (28). LNP-based formulations for oncology have not yet advanced beyond early clinical evaluation, where the focus has been primarily on tumors with liver involvement (29). Consequently, there is a need for systematic development of LNPs with the ability to deliver RNAi triggers efficiently to tumors of diverse origin, with acceptable tolerability profiles.
In this paper, we describe development and characterization of Encore, an LNP system for delivery of DsiRNA to β-catenin-dependent tumors. We describe structure-activity relationship (SAR) analysis for this multi-component, modular system. EnCore LNPs represent a unique structural class of nanoparticles and are currently in clinical development for tumor delivery of RNAi triggers.(30) Encore LNPs carrying CTNNB1-targeting DsiRNA delivered pharmacology-relevant intratumoral concentrations of the oligonucleotide, yielding mRNA silencing and anti-tumor efficacy in multiple mouse models. The models evaluated include subcutaneous cell-line derived xenografts (CLDX), orthotopic patient-derived xenografts (PDX), disseminated hematological tumors, CRC liver metastases, experimental lung metastases and spontaneous liver tumors induced by specific genetic drivers. Together, these results demonstrate the feasibility of DsiRNA therapeutics using EnCore LNPs for traditionally undruggable targets. These data advance our knowledge of tumor-centric LNPs as a platform for formulation of RNAi triggers, and support clinical investigation of direct β-catenin inhibitors for cancer.
Materials and Methods
Materials
All DsiRNAs were synthesized by Integrated DNA Technologies, Inc. (IDT, Coralville, IA) or ChemGenes Inc. (Willmington, MA). Primer and probe oligonucleotides used in quantitative real-time polymerase chain reaction (qRT-PCR) detection were synthesized by IDT or Life Sciences (Carlsbad, CA).
Lipid nanoparticle (LNP) formulations
EnCore LNPs were prepared using a two-step mixing process. The first step involved mixing the DsiRNA payload with a cationic liposome to form a solid loaded Core. The cationic liposome was made by extrusion of a mixture of a cationic lipid (e.g., DL-048) and a polyethylene glycol (PEG)-lipid (e.g., DSG-PEG2K). The lipids were mixed in equal volume using either a batch process or an in-line process. The resulting loaded Core was then used in the second mixing step with the envelope lipids to form the final EnCore LNP. A typical mixture of envelope lipids comprised a cationic lipid (e.g., DL-103), DSPC, cholesterol and DSPE-PEG2K. The ethanolic solution of the envelope lipids was mixed with the solution of the loaded Core using either a batch process or an in-line process. The resulting solution was then subjected to diafiltration by KrosFlo Tangential Flow Filtration System (Spectrum Labs, Rancho Dominguez, CA). Phosphate-buffered saline (PBS) was exchanged into the formulation. The formulation was then concentrated, collected, filtered through 0.22μm membrane and stored at 4°C before vialing. The particle size of the EnCore LNPs was determined by dynamic light scattering (Zetasizer Nano ZS, Malvern, UK) and the mean diameter was in the range of 70–100 nm with a polydispersity of ~0.1–0.2. The concentration of the DsiRNA was measured using UV absorbance chromatography. Lipids quantification test was done by UPLC-CAD assay. Encapsulation efficiency was measured by RiboGreen assay and ranged from 85 to 95%.
Cell lines
Human colorectal cancer (CRC) cell lines LS411N, Ls174t and SW403, murine melanoma cell line B16F10, hepatocellular carcinoma cell line Hep3B and AML cell line KG-1 were obtained from ATCC (Manassas, VA). The acute lymphoblastic leukemia (ALL) cell lines 697 and NALM-6 were obtained from DSMZ (Braunschweig, Germany). CRC and the ALL cell lines were grown in RPMI medium supplemented with 10% fetal bovine serum (FBS). B16F10 and Hep3B cells were grown in DMEM medium supplemented with 10% FBS. KG-1 cell line was grown in IMDM medium supplemented with 20% FBS. Mice were obtained from Harlan Laboratories (Indianapolis, IN). All the Cell lines were purchased in 2014 and authenticated originally by ATCC and DSMZ using short tandem repeat analysis. Cells were expanded and frozen down at low passage within 1 month after the receipt of the original stocks. Cells were then thawed and used within 5–6 passages for this study. All the cell lines were subjected to mycoplasma testing (IMPACT ™ 1 profile by IDEXX BioResearch, Columbia, MO) before being released for use in studies.
Cell line-derived xenograft models
6–8 week old Hsd:Athymic Nude-Foxn1nu mice (hereby referred to as nude mice) were injected subcutaneously with LS411N (5×106 cells), SW403 (5×106 cells), Ls174t (5×106 cells), Hep3B (5×106 cells + matrigel) or B16F10 (2×106 cells) under the right shoulder. Tumor volume was measured twice a week to monitor tumor growth/suppression. Dosing was initiated when the tumors reached 150mm3. For tumor growth inhibition studies, animals were randomized and assigned to one of four cohorts and subjected to dosing cycles as described in Results (n=8/cohort, plus 3 additional animals for pharmacokinetic/pharmacodynamics [PK/PD] analysis after the first cycle). B16F10 experimental lung metastases were generated by injecting 7.5×105 cells intravenously in nude mice. The disseminated leukemia models were developed by injecting 5×106 697, KG-1 and NALM-6 cells intravenously into NOD.CB17-Prkdcscid/NCrHsd (NOD.SCID) mice. CRC liver metastases models were generated by surgically implanting 2×106 cells in the spleen of nude mice after midline abdominal incision. After surgery, the abdominal incision was closed with 5-0 to 6-0 absorbable, nonbraided suture and the skin was closed with a single wound clip. Mice were anesthetized with isoflurane before initiating the surgery and during surgery. Buprenorphine was given preoperatively and post-operatively at 0.1 mg/kg subcutaneously for pain relief. All intravenous dosing was performed via lateral tail vein at a total volume of 10 ml/kg. Mice were held in a pathogen-free environment and all procedures involving animals were performed according to protocols approved by Dicerna Pharmaceuticals’ Institutional Animal Care and Use Committee (Dicerna-IACUC).
Patient-derived xenograft model
Left liver lobes of 6–8-week-old male Balb/c nude mice were surgically implanted with primary human liver cancer model (LI0050) fragments (2–3 mm in diameter) for tumor development. When the orthotopic tumor size reached about 300–500 mm3 (extrapolated by serum alpha-fetoprotein [AFP] levels and satellite mice), mice were randomized and assigned to two cohorts. The protocol and any amendments or procedures involving the care and use of animals in this study were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of CrownBio (Beijing, China), and the care and use of animals were conducted in accordance with the regulations of the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC).
Quantitative RT-PCR measurements
Animal tissues were preserved by either snap-freezing or RNA-later fixative (Life Technologies, Carlsbad, CA) and were homogenized using a bead mill (TissueLyzer, Qiagen, Germantown, MD). After total RNA isolation, representative RNA samples were subjected to QC and determination of the RNA Integrity Number (RIN) score by Agilent 2100 Bioanalyzer (31). 100 ng of total RNA was used to make complementary DNA (cDNA) using high capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA). The cDNA was then diluted 4× for qRT-PCR using TaqMan Fast Advanced Master Mix (Applied Biosystems) and gene-specific primer-probe sets. Primer/probe sets were obtained from Life Technologies and included CTNNB1 (Hs00170025_m1) PP1A (4326316E), MYC (Hs00153408_m1) and AXIN2 (Hs00610344_m1). In some experiments, SuperScript III quantitative one-step RT-PCR kit (Applied Biosystems, #11732-088) was used as well. Stem loop qRT-PCR was performed as described previously, whereby cDNA was prepared from whole-tissue homogenates using a microRNA amplification protocol (Applied Biosystems #4366597) (32). Primers and probes used for detection of CTNNB1 DsiRNA included: CTNNB1 Stem Loop RT primer (5-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCAGAATA-3), CTNNB1 forward primer (5′-CGCGCGTTCTACATCATTTGTATTG-3′), universal reverse primer (5′-AGTGCAGGGTCCGAG-3′) and CTNNB1 probe (5′-FAM-TTCGCACTGGATACGACGCAG – MGB-3′). Wnt pathway profiling was performed using TaqMan Low Density Array (LDA, Applied Biosystems) technology using custom gene expression panels according to manufacturer’s protocols.
In situ hybridization and immunohistochemistry
ViewRNA technology (Affymetrix, Santa Clara, CA) was used for all in situ hybridization experiments, following vendor’s protocol with the following exceptions. Formalin-fixed/paraffin embedded tissue sections were cut to 5 μm, baked at 60°C for 60 minutes and then stored at −20°C until use. Slides were washed using the Little Dipper automated system (SciGene, Sunnyvale, CA, #1080-65-1). Histo-Clear (Electron Microscopy Sciences, Hatfield, PA, #64110-04) was used for clearing the slide after melting the paraffin. Pretreatment of tumor tissue was for 20 minutes at 95°C. Protease digestion of tumor tissue was 15–20 minutes at 40°C. Slides were mounted in Vectashield Mounting Media with DAPI (Vector Labs, Burlingame, CA, #H-1200). Analysis of slides was performed using a Nikon Microscope with a 20× objective with an NA of 0.75. Signal intensity and area of signal were measured using Nikon Elements software. β-catenin immunohistochemistry was performed as described (33), using a β-catenin antibody (Cell Signaling, Danvers, MA, 8480) at a concentration of 1:500 and Signal Stain DAB substrate kit (Cell Signaling, #8059). Ki67 immunochemistry was performed using an anti-Ki67 antibody (Abcam, Cambridge, MA, #ab16667) at a concentration of 1:100 and DAB under standard conditions.
5′RACE-Seq Methods
Total RNA was isolated using the Qiagen RNA miniprep kit (Qiagen) according to the manufacturer’s instructions. 5′RACE analysis was performed on 5 μg of total RNA using the GeneRacer kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions with the exception that RNA were directly ligated to the kit 5′ Racer Adapter. cDNA generation and RACE amplification were performed using the kit 5′ Racer Primer and CTNNB1 specific primer (5′-TGCAGCCCACCAGCTAACGCACTG-3′). A touchdown qPCR protocol was used to improve amplicon specificity. The cDNA library was constructed using the NEBNext mRNA library prep kit (NEB, Ipswich MA) according to the manufacturer’s instructions. Briefly, PCR products were purified using AMPure beads and adaptors were ligated followed by pair-end library enrichment by amplification. The cDNA library was purified and validated for size, quality and concentration using an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA). The cDNA libraries were multiplexed and sequenced on the Illumina MiSeq platform. Data were filtered and sequences with 15 nucleotides of the 3′ end of the RACE adapter were included in downstream analysis. The RACE adapter sequences were trimmed from the reads and the remaining portion was mapped to the CTNNB1 cDNA sequence. Reads that matched to CTNNB1 were reported in the final analysis.
Results
Structure-activity relationship (SAR) of EnCore LNP components
LNPs have been applied in clinical testing to achieve favorable pharmacological properties for complex biomolecules, including oligonucleotides. We investigated a novel tumor delivery platform for DsiRNA payloads termed EnCore LNPs. This modular system contains a solid pre-manufactured Core to which the oligonucleotide is complexed, and an Envelope layer that contributes to the transfection and biodistribution properties of the particle. We recently reported the activity of a CTNNB1-targeting DsiRNA formulated in a first-generation LNP, hereby termed EnCore-A, in Hep3B orthotopic hepatocellular tumors (13). The Core and Envelope of EnCore-A each contain a cationic lipid (sometimes referred to as an ionizable lipid because it is expected to be neutral at physiological pH) and a PEGylated lipid (Figure S1). The EnCore manufacturing process is fully scalable, amenable to current good manufacturing process (cGMP) standards, and subjected to analytical release criteria prior to biological evaluation. These release criteria typically include >90% RNA encapsulation efficiency and a mean diameter ranging from 70–100, depending on formulation components. The EnCore LNP system has been validated for manufacturability, efficacy and safety through formal preclinical IND-enabling studies. DCR-MYC, a first-in-class oncology therapeutic targeting the MYC oncogene, is an EnCore LNP/MYC DsiRNA-based investigational drug currently in clinical testing (30).
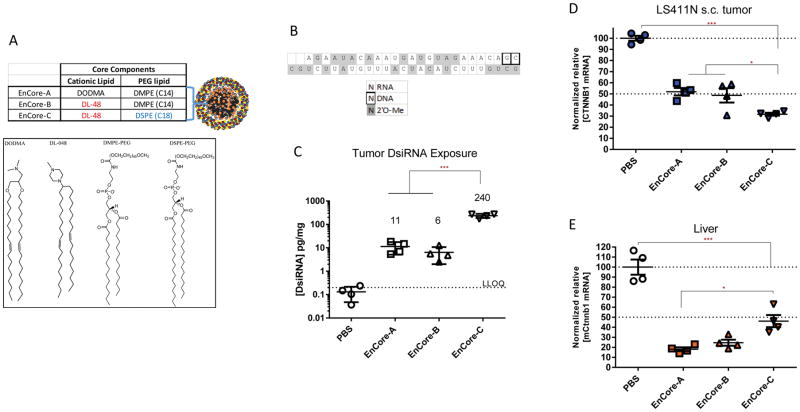
To further advance the EnCore LNP technology, we first explored the chemical space around the Core lipid components. Starting from EnCore-A, we generated EnCore-B by replacing DODMA with the novel lipid DL-48 (Figure 1A) to study potential differences in bioperformance between a charged dimethylglycine and a neutral piperizine lipid head group. Next, we generated EnCore-C by extending the acyl chain length of DMPE-PEG of the EnCore-B core from 14 to 18 carbons using DSPE-PEG. The rationale behind this investigation is that longer acyl chains on PEG-lipids can prevent diffusion of the PEG from the particle and therefore affect its PK properties (34). EnCore-A, B and C carrying CTNNB1 DsiRNA (13) (Figure 1B) were then evaluated in mice bearing subcutaneously implanted human colorectal LS411N tumors by intravenous administration. Like many colorectal tumors, LS411N cells contain an activated Wnt/β-catenin pathway since they are heterozygous for a loss of function mutation in the APC gene (35). A CTNNB1-targeting DsiRNA payload, which targets a region of 100% identity between mouse and human, was chosen so that its activity could be monitored in normal mouse tissues as well as in the human tumor.
Figure 1. Effect of the Core cationic lipid and PEG-lipid on delivery to xenografted colorectal tumors.
(A), composition of EnCore LNP Core components and key structures. (B), CTNNB1 DsiRNA payload and modification scheme legend. The top strand is the passenger (sense) strand depicted in the 5′ to 3′ orientation. The bottom strand is the guide (sense) strand depicted in the 3′ to 5′ orientation. Mice bearing LS411N subcutaneous tumors were dosed intravenously at 3 mg/kg daily for 3 days, and subjected to necropsy one day after the final dose. (C), DsiRNA guide strand concentration in tumors at time of necropsy were determined by qRT-PCR as described in Materials and Methods. The lower limit of DsiRNA quantitation (LLOQ) is indicated. CTNNB1 mRNA levels in tumor (D) and normal mouse liver (E) were determined by normalizing to a reference mRNA and are shown as relative to phosphate buffered saline (PBS)-treated animals. Data points are from individual mice. Mean and SEM (standard error of the mean) are shown, as well as p values as determined by ANOVA analysis (*, <0.05; **, <0.01; ***, <0.001). n=4 animals per group.
After intravenous (IV) administration of each of the three EnCore formulations to LS411N tumor-bearing mice, we measured the intratumoral concentration of the DsiRNA payload as well as the CTNNB1 mRNA target. While EnCore-A and EnCore-B delivered comparable DsiRNA copy numbers and yielded similar tumor CTNNB1 mRNA knockdown (KD) levels, EnCore-C delivered >30× more DsiRNA to tumors (Figure 1C) and yielded improved mRNA KD (Figure 1D). Duration of effect was dependent on the dosing regimen, but robust mRNA KD was observed for at least 5 days after treatment in both single-dose and multiple-dose regimens (Figure S2). Finally, improved duration of KD was observed when the DsiRNA payload was further modified by the incorporation of stabilizing 2′-fluoro nucleotides (36), suggesting a correlation between RNAi payload stability and tumor PD (Figure S3).
To compare the activity of Encore-A/B/C in tumors relative to normal tissue, we also measured murine Ctnnb1 (mCtnnb1) mRNA levels in the livers of the LS411N tumor bearing mice. Notably, while EnCore-C showed improved mRNA KD activity in tumors relative to its precursor, it resulted in decreased activity in the normal liver compared to the LNPs containing the shorter C14 PEG-lipid (Figure 1E). While EnCore-A and –B treatment resulted in a tumor/liver KD ratio of 0.6 to 0.7, indicating higher potency in liver than in tumor, EnCore-C treatment resulted in a tumor/liver KD ratio of 1.3, indicating tumor selectivity. These data contributed to an improved understanding of the structure-activity relationship (SAR) for the Core lipid components.
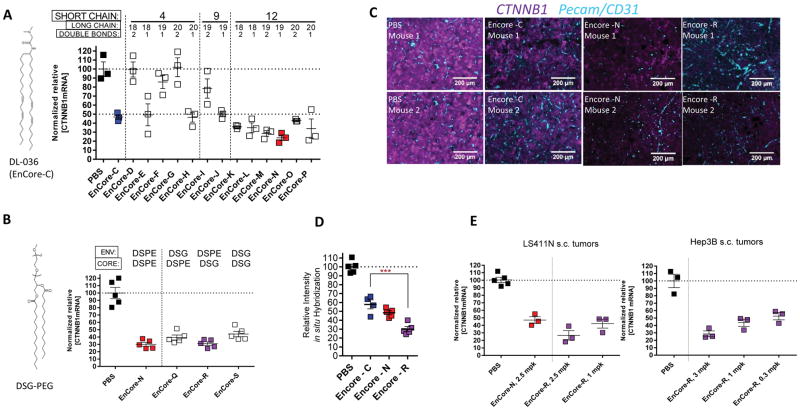
Next, we sought to investigate the SAR of the Envelope cationic lipid, which previous studies suggested is likely to be the driver of transfection efficiency (37). EnCore A-C contains DL-036, which contains a dimethylglycine head group and two symmetrical acyl chains (Figure 2A). It has been previously suggested that incorporating asymmetric acyl chains into amino lipids can facilitate pH-dependent lipid phase transitions, thereby increasing endosomal release of the payload (38–40). In a test set of otherwise identical LNPs, changing the lengths or the number of double bonds of one or both of the acyl chains significantly impacted the CTNNB1 mRNA silencing activity in tumors (Figure 2A). After three IV doses of 3 mg/kg in LS411N tumor-bearing mice, the KD of the target mRNA varied widely within the test set, from 0–76% (Figure 2A). Among the LNPs in the test set, the subset containing a short acyl chain length of 12 carbons yielded the highest potency. One of several examples of SAR discovered in this test set was that the activity of the most highly asymmetric lipids was dependent on their degree of unsaturation (see, for example, EnCore-D vs. –E and EnCore-G vs. –H, Figure 2A). When the asymmetry was reduced by increasing the length of the short acyl chain, the degree of unsaturation was less impactful. These data suggest that transfection activity is highly dependent on the geometry of the EnCore particle surface. We selected the most potent LNP in the test set, EnCore-N, for further refinement. Since electrostatic effects can influence higher-order lipid structures (41), we further modified EnCore-N by changing its negatively charged PEG-lipid, DSPE, with the neutral PEG-lipid DSG, either in the Envelope (EnCore-Q), the Core (EnCoreR), or in both (EnCore-S). Based on the higher potency of EnCore-N, we reasoned that a single IV dose, instead of the three daily doses as used in the preliminary studies, would enable differentiation between the final formulation candidates at non-saturating levels of mRNA knockdown. After a single 5 mg/kg dose in LS411N tumor-bearing mice, there was a trend toward higher potency for LNPs that contain DSPE in the Envelope, but this did not reach statistical significance. (Figure 2B). Based on these data, EnCore-R was selected as the lead formulation.
Figure 2. Multi-parameter optimization of EnCore delivery efficiency.
(A), Bioperformance of EnCore LNPs featuring cationic lipids of varying acyl chain lengths and double bond content. Note that the number of double bonds as indicated refers to long chain, while the short chains throughout the series were fully saturated. Tumor [CTNNB1 mRNA] after three daily doses of 3 mg/kg were determined as in Figure 1; n=3/group. p<0.05 for EnCore-C vs. EnCore-N. (B), EnCore-E was further derivatized by varying the Core and Envelope PEG lipids, and their potency was evaluated 24 hours after a single 5 mg/kg dose in LS411N s.c. tumors (n=5/group). (C), LS411N tumors were subjected to in situ hybridization for human CTNNB1 mRNA (magenta) and mouse Pecam/CD31 mRNA (cyan) 24 hours after a single 5 mg/kg dose of the indicated LNP (n=4/cohort, n=2 shown as images, 10× magnification). (D), quantitation of intensity of CTNNB1 mRNA signal from in situ hybridization (n=4/cohort). (E), comparison of single-dose mRNA knockdown potency in LS411N and Hep3B subcutaneous tumors, at the indicated dose levels (n=3). p<0.05 vs. PBS for all cohorts on all panels except EnCore-D, -F, -G, and –I on panel A, which are not significant vs. PBS.
To further investigate intratumoral mRNA silencing for the most potent LNPs in this test set, we performed fluorescence in situ hybridization on LS411N tumor sections (Figure 2C) 24 hours after administering a single 5 mg/kg dose. Tumor sections were stained for CTNNB1 mRNA (green), as well as mouse Pecam mRNA (also known as CD31, blue), an endothelial marker to outline the host vasculature. The EnCore-C, -N, and –R test articles all showed a marked decrease of CTNNB1 mRNA relative to the untreated control group, with EnCore-R yielding the most robust silencing according to the image intensity quantiation (Figure 2D). Importantly, mRNA silencing was generally homogeneous throughout the tumor parenchyma and not concentrated around the blood vessels, suggesting efficient extravasation of the particle. A single-dose administration in LS411N tumors as well as a second xenograft model, Hep3B hepatocellular carcinoma, suggested that the approximate in vivo tumor ED50 for EnCoreR/CTNNB1 DsiRNA was between 0.3 and 1 mg/kg in subcutaneous tumors (Fig. 2D).
Silencing of CTNNB1 in metastatic and disseminated cell line-derived tumor models
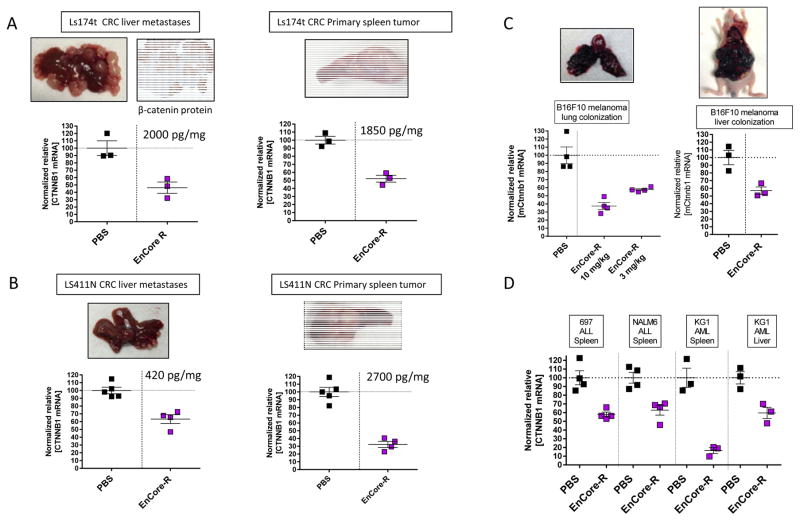
While subcutaneously implanted tumors are useful for evaluating novel antitumor agents, the complexity of nanoparticle-mediated delivery necessitates investigation of delivery to tumors in different organ sites. We sought to explore whether the EnCore LNP can provide functional delivery of DsiRNA payloads to tumors in different organ sites, and to those of epithelial or hematopoietic origin. To this end, LS411N and LS174t tumors were surgically implanted into the spleens of nude mice (representative images of tumors from untreated animals are included in Figure 3A–B), and evaluated for the ability of EnCore-R/CTNNB1 to silence mRNA in both the primary splenic tumor as well as in liver metastases which develop over time (Figure 3A–B). Next, to determine the performance of EnCore-R in disseminated solid tumors in immunocompetent animals, we employed a murine syngeneic model. Applying a commonly used model of experimental metastasis, mouse B16F10 melanoma cells were implanted intravenously into wild type mice to generate lung tumors, or injected hydrodynamically (42) to generate large liver tumors (Figure 3C). For both the lung and liver B16F10 tumors, a single dose of EnCore-R/CTNNB1 DsiRNA effectively reduced CTNNB1 mRNA levels. Finally, experimental models of ALL and AML are also useful for evaluating the range of tumors permissive for LNP delivery, as they can also colonize the spleen and/or liver of immunocompromised mice after IV injection of tumor cells (Figure 3D). Again, EnCoreR/CTNNB1 DsiRNA treatment caused significant CTNNB1 mRNA KD in these leukemia-derived lung and liver tumors, showing that activity of this formulation is not limited to the subcutaneous compartment or to tumors of epithelial origin. It should be noted that, for this series of experiments, we elected to administer EnCore-R/CTNNB1 at a comparatively higher dose level (3–10mg/kg) to obtain a preliminary read in tumors with increased heterogeneity than conventional subcutaneous xenografts. Taken together, these data demonstrate the ability of EnCore-R to deliver pharmacological levels of DsiRNA to tumors of diverse tissue origin.
Figure 3. EnCore-R delivers CTNNB1 DsiRNA to metastatic and disseminated cell line-derived tumors.
Colorectal cancer (CRC) liver metastases were generated by surgically implanting Ls174t (A) and LS411N (B) cells in spleens of nude mice. The splenic primary tumor and liver metastases were independently evaluated for CTNNB1 mRNA 24 hours after dosing EnCoreR/CTNNB1 or PBS. Representative tissue images to show tumor morphology are included, as well as a representative 4× immunohistochemistry image for human β-catenin protein to confirm the identity of the LS174t tumor in (A). Also indicated on the plots are the mean [DsiRNA] levels measured in the primary spleen and metastatic liver tumors at the time of harvest. In (C), murine B16F10 melanoma cells were injected intravenously into wild-type mice and allowed to colonize the lung (left panel) or injected hydrodynamically to form large tumors in the liver (right panel). Mouse Ctnnb1 (mCtnnb1) was measured in the tumor 24 hours after administering EnCore-R/CTNNB1 or PBS. In (D), human acute lymphoblastic leukemia (ALL) or acute myelogenous leukemia (AML) cells were injected intravenously into immunocompromised mice. After allowing for the tumor cells to colonize the spleen or liver, as indicated, EnCoreR/CTNNB1 DsiRNA was administered and mRNA knockdown was evaluated after 24 hours. n=3 or 4 animals per cohort in all panels. All cohorts received a single dose of 10 mg/kg except where indicated; p<0.05 vs. PBS for all experiments shown in this figure.
EnCore LNP/CTNNB1 treatment yields tumor growth inhibition
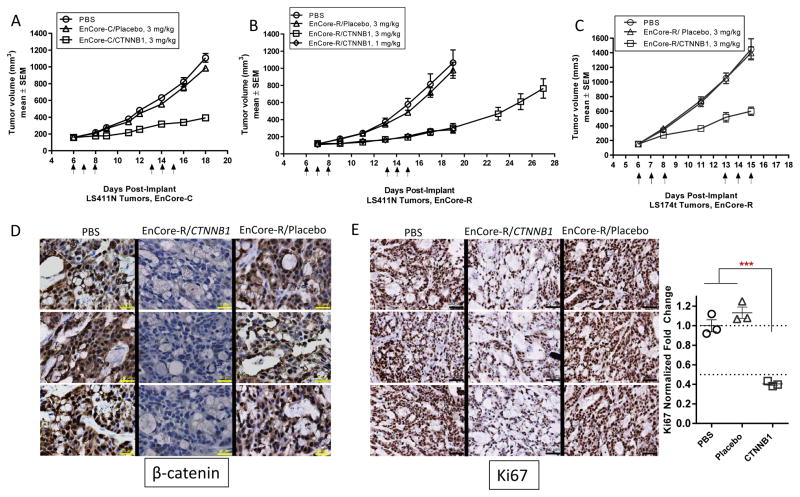
To determine if CTNNB1 silencing can result in anti-tumor efficacy in Wnt/β-catenin activated tumors, LS411N tumor-bearing mice (n=8/cohort) were given either an irrelevant control DsiRNA (“Placebo”) or CTNNB1 DsiRNA formulated in EnCore-LNPs (3 daily doses per weekly cycle). After two weekly dosing cycles, EnCore-C (3 mg/kg, Figure 4A) caused tumor growth inhibition of 75% relative to vehicle-treated animals, whereas EnCore-R (1 mg/kg, Figure 4B) caused tumor growth inhibition of 82%. In a cohort where therapy was discontinued after the second cycle, tumor growth resumed to a level roughly comparable to untreated subjects (Figure 4B), suggesting that long-term suppression of β-catenin is required for maintenance of efficacy in this model. Comparable tumor growth inhibition was observed in a second Wnt-activated colorectal tumor model, Ls174t, which harbors direct CTNNB1 mutations (43) (Figure 4C). Efficacy in Ls174t tumors correlated to a sharp reduction of tumor β-catenin protein levels by immunohistochemistry after a single dosing cycle. Importantly, the reduction in tumor β-catenin occurs in all subcellular compartments, and most strongly in the nucleus where the activated oncoprotein resides (Figure 4D). Finally, the decrease in β-catenin protein is associated with a significant decrease in Ki67 protein, indicating an effect on the proliferative capacity of the tumor cells (Figure 4E).
Figure 4. CTNNB1 DsiRNA inhibits tumor growth in colorectal cancer xenografts.
LS411N (A, B) or Ls174t (C) tumor-bearing mice were treated with EnCore-C and EnCore-R containing CTNNB1 or an irrelevant Placebo DsiRNA, or a phosphate-buffered saline (PBS) control (n=6–8 per cohort). Intravenous doses were given at times indicated by the arrows at 1 or 3 mg/kg. Dosing was initiated at a starting volume of 150–200 mm3. Tumor volume was determined at regular intervals throughout the studies. (D), 72 hours after the final dose of a qdx3 cycle (3 mg/kg), Ls174t tumors from animals treated with EnCore-R containing CTNNB1 or Placebo DsiRNA were subjected to immunohistochemistry for β-catenin protein (n=3/cohort). Images were acquired at 40× magnification (yellow scale bars = 30 μm). (E), 72 hours after the final dose of a qdx3 cycle (3 mg/kg), Ls174t tumors from animals treated with EnCore-R containing CTNNB1 or Placebo DsiRNA were subjected to immunohistochemistry for Ki67 protein (n=3/cohort). Quantitation of the Ki67 stain intensity is plotted.
To determine if EnCore-R/CTNNB1 also yields in vivo efficacy in other subcutaneous models that harbor defects in Wnt signaling or downstream effectors, we tested similar dosing regimens in SW403 (APC mutant) colorectal tumors (35) and in Hep3B hepatocellular tumors (Figure S4A &B). Interestingly, Hep3B tumors are not known to harbor classical mutations in Wnt/β-catenin signaling nodes; however, they are highly sensitive to inhibition of MYC, a well-characterized effector and a direct transcriptional target of β-catenin in tumorigenesis (13, 30, 44). Robust tumor growth inhibition was observed in both models. Efficacy in SW403 tumors also correlated to a near complete depletion of tumor β-catenin protein levels by immunohistochemistry after 2 dosing cycles (Figure S4C). Overall, these studies suggest broad applicability for Wnt/β-catenin-dependent tumors regardless of their tissue origin.
As a control for target specificity, we also tested EnCore-R/CTNNB1 in RKO tumors (35). RKO tumors are wild type for APC and CTNNB1, and therefore do not contain activated Wnt signaling, unlike >90% of human colorectal tumors. Importantly, no antitumor efficacy was observed in RKO tumors (Figure S5), suggesting that, as predicted, only Wnt-dependent tumors will respond to β-catenin inhibition.
Apolipoprotein E-independent delivery of DsiRNA to tumors
The B16F10 syngeneic melanoma model (Figure 3C) offers the opportunity to investigate LNP-mediated DsiRNA delivery in germline knockout and transgenic animals, as it eliminates the requirement to establish tumors in immunocompromised mice. To explore the mechanism of EnCore-mediated delivery to tumors, we established B16F10 tumors in both wild-type mice and in apolipoprotein E (ApoE)-deficient mice. LNP-mediated delivery to normal hepatocytes has been previously shown to be highly dependent on opsonization of ApoE onto the particle surface, which facilitates endocytosis of the nanoparticle via the low-density lipoprotein receptor (LDLR) (45). While silencing of the mouse Ctnnb1 gene in the liver was completely abrogated in the ApoE −/− mice, loss of ApoE did not affect the DsiRNA activity in B16F10 subcutaneous tumors (Figure S6A), nor in B16F10 experimental lung metastases (Figure S6B). To our knowledge this is the first demonstration of ApoE-independent delivery for an LNP, and demonstrates that the cell internalization mechanism of LNPs are context-dependent.
CTNNB1 DsiRNA causes a dampening of Wnt/β-catenin signaling in patient-derived xenograft (PDX) tumors
From a drug delivery perspective, PDX tumors are widely accepted to better recapitulate the heterogeneous architecture of human cancers than cell-line-derived tumors (46). To determine how EnCore-mediated delivery to a PDX tumor compares in efficiency to CLDX tumors, tumor fragments from a patient with HCC patient (model LI0050) were orthotopically implanted into the livers of immunocompromised mice. This particular tumor is a Grade 4 HCC derived from a 49-year-old female and was previously determined to be unresponsive to the cytotoxic agents doxorubicin and cisplatin (data not shown).
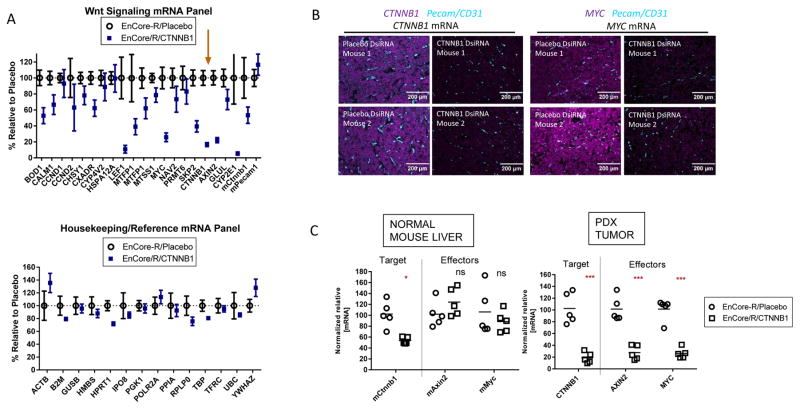
After a single 5 mg/kg dose of EnCore-R/CTNNB1 DsiRNA, multiplexed gene expression analysis showed a dramatic effect on known Wnt/β-catenin-responsive mRNAs (2, 47) relative to treatment with a Placebo DsiRNA (Figure 5A, upper panel). Overall, 12 out of 20 transcripts included in the panel were reduced by greater than 30%, including the well-characterized effectors LEF1, MYC, AXIN2 and GLUL (glutamine synthase). Additionally, we included the murine Ctnnb1 gene on the panel (Figure 5A, “mCtnnb1”). While the mean KD of human CTNNB1 in the treated cohort was 85%, knockdown of mCtnnb1 was only 46%, suggesting that EnCore-R can deliver the DsiRNA payload to host nonparenchymal cells within the tumor (e.g., stromal cells, macrophages, T-cells), albeit with reduced efficiency. A mouse vascular endothelial marker, Pecam (CD31), was unaffected (Figure 5A, “mPecam”). As an important control, CTNNB1 DsiRNA did not significantly affect a panel of unrelated ubiquitously expressed mRNAs (Figure 5A, bottom). In situ hybridization for both CTNNB1 mRNA and downstream effector MYC mRNA in fixed tumor sections suggested homogeneous silencing and tumor penetration (Figure 5B). Importantly, the RNA interference-based mechanism of action was confirmed by the 5′RACE method (20, 32) followed by next-generation sequencing (NGS), which detected significant levels of the predicted Ago2-mediated cleavage product of CTNNB1 mRNA at single-nucleotide resolution (Figure S7).
Figure 5. CTNNB1 DsiRNA activity in an orthotopic patient-derived xenograft (PDX) model of hepatocellular carcinoma.
Patient-derived hepatocellular carcinoma (HCC) tumor fragments were implanted orthotopically in the livers of immunocompromised mice. The implanted tumors were monitored for growth using a satellite cohort and human alpha-fetoprotein levels, and allowed to grow to a mean of ~300mm3. A single 5 mg/kg dose was administered to animals with established tumors. (A), RNA isolated from the orthotopic tumors (24 hours post-dose) was subjected to low density array (LDA) mRNA profiling for transcripts associated with Wnt/β-catenin signaling (upper panel) or ubiquitously expressed reference/housekeeping genes (lower panel). The arrow highlights CTNNB1 mRNA, which was included on the panel. The mean values +/− SEM (n=5) are shown as normalized to the Placebo DsiRNA cohort (B), PDX tumor samples were subjected to in situ hybridization for CTNNB1 or MYC transcripts (magenta) as indicated. The slides were co-stained for murine Pecam/CD31 mRNA (cyan) and images were acquired at 20× magnification. (C), Normalized relative RNA levels for murine Ctnnb1, Axin2, and Myc in the normal adjacent liver (left), or their human counterparts in the orthotopic tumor (ns=not significant).
To investigate the ability of EnCore-R/CTNNB1 DsiRNA to selectively target the PDX tumor over normal adjacent liver, we measured mRNA levels for CTNNB1 and downstream effectors AXIN2 and MYC in the normal adjacent mouse liver as well as in the orthotopic tumor (Figure 5C). While mCtnnb1 KD was observed in the normal liver, mAxin2 and mMyc expression were unaffected. These data suggest that the DsiRNA potency was insufficient to have a functional consequence in normal tissue, thereby providing a potential tolerability advantage over LNPs that were developed originally for liver indications (29).
CTNNB1 DsiRNA is efficacious in CTNNB1/KRAS-driven hepatocellular carcinoma (HCC)
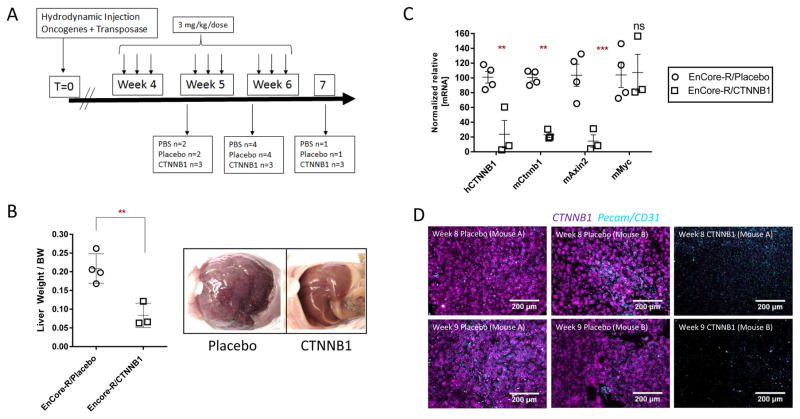
Based on the PDX data (Figure 5), we decided to further model the potential of EnCoreR/CTNNB1 DsiRNA for HCC. Plasmids encoding constitutively activated CTNNB1 (S45Y mutation) and KRAS (G12D mutation) were hydrodynamically injected into the livers of wild-type mice, yielding aggressive hepatocellular tumors (Figure 6). In this model, random genomic integration of the transgenes is induced by the Sleeping Beauty transposon system, which has been employed to study both hepatoblastoma and HCC (33, 48). Dosing was initiated 5 weeks after hydrodynamic injection (HDI), corresponding to the time that tumor nodes become clearly visible (data not shown). The dosing regimen is shown on Figure 6A. At the time of study termination, extensive tumor burden was observed in the Placebo-treated mice, as indicated by grossly enlarged livers and hundreds of individual tumor nodes (Figure 6B). In contrast, EnCore-R/CTNNB1 DsiRNA-treated animals displayed significantly reduced tumor burden (Figure 6B). This phenotype coincided with robust mRNA KD of the mutant human CTNNB1 mRNA transgene (hCTNNB1), the endogenous murine Ctnnb1 mRNA, and downstream effector mAxin2 (Figure 6C). Curiously, cMyc expression was unaffected in this setting, consistent with previous observations that MYC-driven HCC and Wnt-driven HCC may represent two distinct molecular subclasses (49). In situ hybridization for CTNNB1 mRNA in fixed tumor sections also confirmed the homogeneous silencing and tumor penetration in these tumors (Figure 6D). Comparably efficient delivery was also observed in a second Sleeping Beauty transposon-derived model driven by activated Ctnnnb1 (ΔN90/β-catenin) and Yap (YapS127A), which histologically resembles human hepatoblastoma (Figure S8) (33). In addition to demonstrating potency and efficacy, preliminary studies show that EnCore-R LNP demonstrated an acceptable tolerability profile in non-human primates upon repeat administration (Figure S9), suggesting a path forward for clinical development.
Figure 6. CTNNB1 DsiRNA activity in a spontaneous genetic model of hepatocellular carcinoma (HCC).
HCC tumors were generated by hydrodynamically injecting plasmids containing human oncogenes CTNNB1-S45Y and KRAS-G12D, and the components of the Sleeping Beauty transposon system for genome integration into wild-type mice. (A), Timeline of the study showing the dosing scheme and harvest/necropsy times for mice treated with EnCoreR/CTNNB1 DsiRNA or EnCore-R/Placebo DsiRNA. The Placebo DsiRNA is known by the identifier “CK-placebo” as indicated. (B), Tumor growth inhibition as measured by the Liver Weight/Body Weight ratio at time of harvest (left) and representative images of the liver in the peritoneal cavity at time of harvest (Week 9). (C), Normalized relative mRNA levels of mouse Ctnnb1, the mouse Axin2, and the human CTNNB1 transgene. (D), hybridization for human CTNNB1 (magenta) and mouse Pecam/CD31 (cyan) mRNAs in livers from Placebo DsiRNA or CTNNB1 DsiRNA-treated animals.
Discussion
The β-catenin/Wnt pathway is among the most functionally validated targets for hepatocellular (HCC) and colorectal (CRC) cancers. Using an elegant genetic model, Dow et. al. (50) recently demonstrated that inhibition of activated Wnt signaling (through restoration of APC tumor suppressor gene expression) causes tumor regression and terminal differentiation in well-established colorectal tumors. Additionally, inducible CTNNB1 shRNA experiments demonstrated that a normal crypt progenitor phenotype is restored upon suppression of CTNNB1 mRNA in established CRC (35). In HCC, even incomplete suppression of CTNNB1 through use of a research-grade antisense oligonucleotide yielded a complete anti-tumor response in Wnt-driven tumors (51). It is also worth noting that CTNNB1 inhibition may have applications in other tumor types, including immune checkpoint therapy-resistant melanoma, where activated Wnt signaling appears to suppress T-cell infiltration to promote resistance (52). While numerous attempts have been made to target the Wnt pathway and even β-catenin directly (53), most have encountered potency or tolerability concerns, due in part to their lack of tumor selectivity. Transcriptional modulators are historically difficult to target pharmacologically, and their pleotropic effects sometimes make targeting upstream signals or downstream effectors insufficient. Therefore, targeting CTNNB1 at the mRNA level with a tumor-selective RNAi agent is an appealing option for this significant unmet medical need.
LNPs represent the most advanced class of RNAi delivery vehicles in clinical oncology (29), but their tendency to be pharmacologically active in normal liver tissue could be problematic for certain targets. For example, in healthy liver, β-catenin regulates the expression of genes involved in many metabolic pathways, zonation and homeostasis (33). To our knowledge, the optimized EnCore formulations described in this report represents the first LNP for which functional delivery is more efficient in the tumor than in normal adjacent tissue. While the molecular basis for this tumor selectivity is still being elucidated, we have determined that, unlike SNALP-class LNPs (45), functional delivery to tumors does not require apolipoprotein E optimization. This suggests that the mechanism of cell internalization for EnCore LNPs is different in tumor versus normal liver, and that an exogenous targeting ligand is not necessary to drive homogeneous delivery in the difficult tumor microenvironment (19). Continued progress in understanding the basis of delivery, along with our systematic medicinal chemistry approach, will yield LNPs of higher potency and selectivity. Other non-lipid-based RNAi delivery vehicles, including cyclodextrin polymers (54), have also shown promise in preclinical tumor models, although early clinical development appears to have stalled (20).
From a pharmaceutical perspective, the modular nature of RNAi and LNP technology makes it appealing for personalized medicine applications in oncology. DsiRNAs targeting combinations of historically undruggable oncogenes including MYC, KRAS and CTNNB1 in a tumor-selective manner are currently being investigated in additional preclinical models, including genetically defined PDX and advanced metastasis models.
Supplementary Material
Acknowledgments
Financial support: This work was funded in part by Center for Strategic Scientific Initiatives, National Cancer Institute (CSSI, NCI; 1R43CA186410 - 01A1) to B. D. Brown and colleagues at Dicerna Pharmaceuticals, Inc.
The authors would like to thank Hannah Bogich for additional technical assistance. The authors would also like to thank David Miller and James Weissman for helpful suggestions during the review process.
Abbreviations List
- RNAi
RNA interference
- DsiRNA
Dicer-substrate siRNA
- LNP
Lipid Nanoparticles
- PDX
Patient derived xenograft
- CRC
Colorectal Cancer
- TCF/LEF
T-cell factor/lymphoid enhancing factor
- GMP
Good manufacturing practices
- SAR
Structure-activity relationship
- CLDX
Cell line derived xenograft
- FBS
Fetal bovine serum
- PK/PD
Pharmacokinetic/pharmacodynamic
- AFP
Alpha-fetoprotein
- IACUC
Institutional Animal care and use committee
- AAALAC
Association for Assessment & Accreditation of Laboratory Animal care
- APC
Adenomatous polyposis coli
- KD
Knockdown
- IV
Intravenous
- LDLR
Low-density lipoprotein receptor
- NGS
Next generation sequencing
- HCC
Hepatocellular carcinoma
- HDI
Hydrodynamic injection
Footnotes
Disclosure of Potential Conflicts of Interest:
All authors except B.Y., J.T. and S.M. are full-time employees and shareholders of Dicerna Pharmaceuticals. S.M. participates on a Scientific Advisory Board of Dicerna Pharmaceuticals.
References
- 1.Taketo MM. Shutting down Wnt signal-activated cancer. Nat Genet. 2004;36:320–2. doi: 10.1038/ng0404-320. [DOI] [PubMed] [Google Scholar]
- 2.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 3.Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17:45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25:7531–7. doi: 10.1038/sj.onc.1210059. [DOI] [PubMed] [Google Scholar]
- 5.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–90. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 6.Polakis P. Drugging Wnt signalling in cancer. EMBO J. 2012;31:2737–46. doi: 10.1038/emboj.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dihlmann S, von Knebel Doeberitz M. Wnt/beta-catenin-pathway as a molecular target for future anti-cancer therapeutics. Int J Cancer. 2005;113:515–24. doi: 10.1002/ijc.20609. [DOI] [PubMed] [Google Scholar]
- 8.Luu HH, Zhang R, Haydon RC, Rayburn E, Kang Q, Si W, et al. Wnt/beta-catenin signaling pathway as a novel cancer drug target. Curr Cancer Drug Targets. 2004;4:653–71. doi: 10.2174/1568009043332709. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe K, Dai X. Winning WNT: race to Wnt signaling inhibitors. Proc Natl Acad Sci U S A. 2011;108:5929–30. doi: 10.1073/pnas.1103102108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borna H, Imani S, Iman M, Azimzadeh Jamalkandi S. Therapeutic face of RNAi: in vivo challenges. Expert Opin Biol Ther. 2015;15:269–85. doi: 10.1517/14712598.2015.983070. [DOI] [PubMed] [Google Scholar]
- 11.Bramsen JB, Kjems J. Engineering small interfering RNAs by strategic chemical modification. Methods Mol Biol. 2013;942:87–109. doi: 10.1007/978-1-62703-119-6_5. [DOI] [PubMed] [Google Scholar]
- 12.Rose SD, Kim DH, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, et al. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–56. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudek H, Wong DH, Arvan R, Shah A, Wortham K, Ying B, et al. Knockdown of beta-catenin with dicer-substrate siRNAs reduces liver tumor burden in vivo. Mol Ther. 2014;22:92–101. doi: 10.1038/mt.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snead NM, Wu X, Li A, Cui Q, Sakurai K, Burnett JC, et al. Molecular basis for improved gene silencing by Dicer substrate interfering RNA compared with other siRNA variants. Nucleic Acids Res. 2013;41:6209–21. doi: 10.1093/nar/gkt200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster DJ, Barros S, Duncan R, Shaikh S, Cantley W, Dell A, et al. Comprehensive evaluation of canonical versus Dicer-substrate siRNA in vitro and in vivo. RNA. 2012;18:557–68. doi: 10.1261/rna.031120.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cairns R, Papandreou I, Denko N. Overcoming physiologic barriers to cancer treatment by molecularly targeting the tumor microenvironment. Mol Cancer Res. 2006;4:61–70. doi: 10.1158/1541-7786.MCR-06-0002. [DOI] [PubMed] [Google Scholar]
- 17.Tredan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst. 2007;99:1441–54. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- 18.Choi IK, Strauss R, Richter M, Yun CO, Lieber A. Strategies to increase drug penetration in solid tumors. Front Oncol. 2013;3:193. doi: 10.3389/fonc.2013.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain RK. Normalizing tumor microenvironment to treat cancer: bench to bedside to biomarkers. J Clin Oncol. 2013;31:2205–18. doi: 10.1200/JCO.2012.46.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–70. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdul Ghafoor Raja M, Katas H, Jing Wen T. Stability, Intracellular Delivery, and Release of siRNA from Chitosan Nanoparticles Using Different Cross-Linkers. PLoS One. 2015;10:e0128963. doi: 10.1371/journal.pone.0128963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuellar TL, Barnes D, Nelson C, Tanguay J, Yu SF, Wen X, et al. Systematic evaluation of antibody-mediated siRNA delivery using an industrial platform of THIOMAB-siRNA conjugates. Nucleic Acids Res. 2015;43:1189–203. doi: 10.1093/nar/gku1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Podesta JE, Al-Jamal KT, Herrero MA, Tian B, Ali-Boucetta H, Hegde V, et al. Antitumor activity and prolonged survival by carbon-nanotube-mediated therapeutic siRNA silencing in a human lung xenograft model. Small. 2009;5:1176–85. doi: 10.1002/smll.200801572. [DOI] [PubMed] [Google Scholar]
- 24.Doan C, Le L, Hoang S, Do S, Le D. Simultaneous silencing of VEGF and KSP by siRNA cocktail inhibits proliferation and induces apoptosis of hepatocellular carcinoma Hep3B cells. Biol Res. 2014;47:70. doi: 10.1186/0717-6287-47-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Xu Z, Guo S, Zhang L, Sharma A, Robertson GP, et al. Intravenous delivery of siRNA targeting CD47 effectively inhibits melanoma tumor growth and lung metastasis. Mol Ther. 2013;21:1919–29. doi: 10.1038/mt.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue W, Dahlman JE, Tammela T, Khan OF, Sood S, Dave A, et al. Small RNA combination therapy for lung cancer. Proc Natl Acad Sci U S A. 2014;111:E3553–61. doi: 10.1073/pnas.1412686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ait-Oudhia S, Mager DE, Straubinger RM. Application of pharmacokinetic and pharmacodynamic analysis to the development of liposomal formulations for oncology. Pharmaceutics. 2014;6:137–74. doi: 10.3390/pharmaceutics6010137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol. 2013;31:638–46. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 29.Tabernero J, Shapiro GI, LoRusso PM, Cervantes A, Schwartz GK, Weiss GJ, et al. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013;3:406–17. doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]
- 30.Tolcher AW, Papadopoulos KP, Patnaik A, Rasco DW, Martinez D, Wood DL, et al. Safety and activity of DCR-MYC, a first-in-class Dicer-substrate small interfering RNA (DsiRNA) targeting MYC, in a phase I study in patients with advanced solid tumors. J Clin Oncol. 2015;33:15s. (suppl; abstr TPS11006) [Google Scholar]
- 31.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abrams MT, Koser ML, Seitzer J, Williams SC, DiPietro MA, Wang W, et al. Evaluation of efficacy, biodistribution, and inflammation for a potent siRNA nanoparticle: effect of dexamethasone co-treatment. Mol Ther. 2010;18:171–80. doi: 10.1038/mt.2009.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao J, Calvisi DF, Ranganathan S, Cigliano A, Zhou L, Singh S, et al. Activation of beta-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology. 2014;147:690–701. doi: 10.1053/j.gastro.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson SC, Baryza JL, Reynolds AJ, Bowman K, Keegan ME, Standley SM, et al. Real time measurement of PEG shedding from lipid nanoparticles in serum via NMR spectroscopy. Mol Pharm. 2015;12:386–92. doi: 10.1021/mp500400k. [DOI] [PubMed] [Google Scholar]
- 35.Scholer-Dahirel A, Schlabach MR, Loo A, Bagdasarian L, Meyer R, Guo R, et al. Maintenance of adenomatous polyposis coli (APC)-mutant colorectal cancer is dependent on Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A. 2011;108:17135–40. doi: 10.1073/pnas.1104182108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjaer N, et al. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867–81. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan C, Allen TM, Cullis PR. Lipid nanoparticle delivery systems for siRNA-based therapeutics. Drug Deliv Transl Res. 2014;4:74–83. doi: 10.1007/s13346-013-0161-z. [DOI] [PubMed] [Google Scholar]
- 38.Gindy ME, Feuston B, Glass A, Arrington L, Haas RM, Schariter J, et al. Stabilization of Ostwald ripening in low molecular weight amino lipid nanoparticles for systemic delivery of siRNA therapeutics. Mol Pharm. 2014;11:4143–53. doi: 10.1021/mp500367k. [DOI] [PubMed] [Google Scholar]
- 39.Budzik BW, Colletti SL, Davis JR, Hills ID, Seifried DD, Stanton MG. Google Patents. 2014. Low molecular weight cationic lipids for oligonucleotide delivery. [Google Scholar]
- 40.Zhang Y, Arrington L, Boardman D, Davis J, Xu Y, DiFelice K, et al. The development of an in vitro assay to screen lipid based nanoparticles for siRNA delivery. J Control Release. 2014;174:7–14. doi: 10.1016/j.jconrel.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Garbuzenko O, Zalipsky S, Qazen M, Barenholz Y. Electrostatics of PEGylated micelles and liposomes containing charged and neutral lipopolymers. Langmuir. 2005;21:2560–8. doi: 10.1021/la0479105. [DOI] [PubMed] [Google Scholar]
- 42.Li J, Yao Q, Liu D. Hydrodynamic cell delivery for simultaneous establishment of tumor growth in mouse lung, liver and kidney. Cancer Biol Ther. 2011;12:737–41. doi: 10.4161/cbt.12.8.16442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mologni L, Dekhil H, Ceccon M, Purgante S, Lan C, Cleris L, et al. Colorectal tumors are effectively eradicated by combined inhibition of {beta}-catenin, KRAS, and the oncogenic transcription factor ITF2. Cancer Res. 2010;70:7253–63. doi: 10.1158/0008-5472.CAN-10-1108. [DOI] [PubMed] [Google Scholar]
- 44.Lin CP, Liu JD, Chow JM, Liu CR, Liu HE. Small-molecule c-Myc inhibitor, 10058-F4, inhibits proliferation, downregulates human telomerase reverse transcriptase and enhances chemosensitivity in human hepatocellular carcinoma cells. Anticancer Drugs. 2007;18:161–70. doi: 10.1097/CAD.0b013e3280109424. [DOI] [PubMed] [Google Scholar]
- 45.Akinc A, Querbes W, De S, Qin J, Frank-Kamenetsky M, Jayaprakash KN, et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol Ther. 2010;18:1357–64. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cassidy JW, Caldas C, Bruna A. Maintaining Tumor Heterogeneity in Patient-Derived Tumor Xenografts. Cancer Res. 2015 doi: 10.1158/0008-5472.CAN-15-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herbst A, Jurinovic V, Krebs S, Thieme SE, Blum H, Goke B, et al. Comprehensive analysis of beta-catenin target genes in colorectal carcinoma cell lines with deregulated Wnt/beta-catenin signaling. BMC Genomics. 2014;15:74. doi: 10.1186/1471-2164-15-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X, Calvisi DF. Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am J Pathol. 2014;184:912–23. doi: 10.1016/j.ajpath.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–92. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dow LE, O’Rourke KP, Simon J, Tschaharganeh DF, van Es JH, Clevers H, et al. Apc Restoration Promotes Cellular Differentiation and Reestablishes Crypt Homeostasis in Colorectal Cancer. Cell. 2015;161:1539–52. doi: 10.1016/j.cell.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delgado E, Okabe H, Preziosi M, Russell JO, Alvarado TF, Oertel M, et al. Complete response of Ctnnb1-mutated tumours to beta-catenin suppression by locked nucleic acid antisense in a mouse hepatocarcinogenesis model. J Hepatol. 2015;62:380–7. doi: 10.1016/j.jhep.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–5. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 53.Kahn M. Can we safely target the WNT pathway? Nat Rev Drug Discov. 2014;13:513–32. doi: 10.1038/nrd4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan TL, Fellmann C, Lee CS, Ritchie CD, Thapar V, Lee LC, et al. Development of siRNA payloads to target KRAS-mutant cancer. Cancer Discov. 2014;4:1182–97. doi: 10.1158/2159-8290.CD-13-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.