Abstract
Neuroimaging genetics has attracted growing attention and interest, which is thought to be a powerful strategy to examine the influence of genetic variants (i.e., single nucleotide polymorphisms (SNPs)) on structures or functions of human brain. In recent studies, univariate or multivariate regression analysis methods are typically used to capture the effective associations between genetic variants and quantitative traits (QTs) such as brain imaging phenotypes. The identified imaging QTs, although associated with certain genetic markers, may not be all disease specific. A useful, but underexplored, scenario could be to discover only those QTs associated with both genetic markers and disease status for revealing the chain from genotype to phenotype to symptom. In addition, multimodal brain imaging phenotypes are extracted from different perspectives and imaging markers consistently showing up in multimodalities may provide more insights for mechanistic understanding of diseases (i.e., Alzheimer’s disease (AD)). In this work, we propose a general framework to exploit multi-modal brain imaging phenotypes as intermediate traits that bridge genetic risk factors and multi-class disease status. We applied our proposed method to explore the relation between the well-known AD risk SNP APOE rs429358 and three baseline brain imaging modalities (i.e., structural magnetic resonance imaging (MRI), fluorodeoxyglucose positron emission tomography (FDG-PET) and F-18 florbetapir PET scans amyloid imaging (AV45)) from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database. The empirical results demonstrate that our proposed method not only helps improve the performances of imaging genetic associations, but also discovers robust and consistent regions of interests (ROIs) across multi-modalities to guide the disease-induced interpretation.
Keywords: Multimodal Intermediate Phenotypes, Diagnosis-guided, Single Nucleotide Polymorphisms (SNPs), Alzheimer’s Disease
Introduction
Alzheimer’s disease (AD) is the most common type of neurodegenerative disorder characterized by progressive impairment of memory and other cognitive functions in elderly people worldwide (Brookmeyer et al., 2007). Effective prevention and treatment of AD is a major challenge, given that no disease-modifying medicine in AD is available. To address this challenge, many AD studies focus on systems biology of the brain to better understand complex neurobiological systems, from genetic factors, protein products, cellular components, and to the complex interplay of brain structure, function, behavior and cognition (Pasinetti and Hiller-Sturmhofel, 2008).
High throughput genotyping technology, coupled with multimodal brain imaging, holds great promise to investigate the role of genetic variation in brain structure and function. An emerging research field, imaging genetics, focuses on study genetics using imaging measures as intermediate phenotypes, which is different from case-control studies (Glahn et al., 2007; Gottesman and Gould, 2003), and may yield interesting results for us to understand the complex biological mechanism of the disease (i.e., AD).
In prior imaging genetics research, genome-wide association studies (GWAS) have been performed to identify the associations between single nucleotide polymorphisms (SNPs) and imaging quantitative traits (QTs). To address the high dimensionality of the imaging genetics data, some hypothesis-driven approaches have focused on a small number of genetic variables and searched for their QT associations in the whole brain (Brun et al., 2009; Filippini et al., 2009). In contrast, some other studies have focused on a limited number of imaging QTs and searched for their SNP associations in the entire genome (Baranzini et al., 2009; Potkin et al., 2009). In recent studies, taking into account the inherent structure among genotype or phenotype data (e.g., spatial information in images or combining the effect of multiple genetic variants), some researchers have developed several generalized multivariate linear regression analysis or least square kernel machine methods to boost the detection power (Ge et al., 2012; Hibar et al., 2011; Kohannim et al., 2012; Kohannim et al., 2011; Wang et al., 2012b). Although those methods may have potentials to help discover structured phenotypic imaging markers related to some candidate risk SNPs, the problem of existing methods in imaging genetics is that phenotypes could be related to many genetic markers on different pathways that are not all disease specific. A valuable scenario would be to discover only those QTs associated with both genetic markers and disease status to better reveal the biological pathways specific to the disease. Thus, it is an important research topic to incorporate the subjects’ diagnosis information (e.g., class labels), and to discover disease-specific imaging genetic associations on the chain from genetic data to brain to symptom.
More recently, some diagnosis information guided methods have been proposed in the field of imaging genetics. The method proposed in (Vounou et al., 2012; Vounou et al., 2010) employed a two-step procedure for detecting genetic factors associated with imaging biomarkers: 1) firstly, they pre-selected the disease relevant voxel level imaging phenotypes with high classification performance between AD and healthy control (HC) groups using penalized linear discriminant analysis; 2) secondly, they identified the SNPs associated with the multivariate imaging biomarkers identified from the first step. Different from general linear regression models, another framework employed Bayesian theory for detecting genetic variants associated with disease related imaging QTs (Batmanghelich et al., 2013). The Bayesian model performed genetic identification and imaging feature selection simultaneously, and could identify interesting associations along the pathway from gene to imaging and then to symptom.
In addition, most of existing imaging genetic studies (Batmanghelich et al., 2013; Vounou et al., 2012; Vounou et al., 2010; Wang et al., 2012a; Wang et al., 2012b) have focused on the associations between only single imaging modality (e.g., magnetic resonance imaging (MRI)-voxel based morphometry (VBM) or FreeSurfer measures) and SNPs. These methods can identify interesting patterns within a certain modality, but are limited in discovery of consistent regional patterns across multiple modalities.
To address the above challenges, this work aims to identify consistent brain regions whose multimodal imaging measures can serve as intermediate traits between genetic risk factor and disease status. Our goal is to design a simple and powerful model to extract disease-relevant imaging genetic associations. Accordingly, we develop a novel diagnosis-guided multi-modality (DGMM) framework that can discover common regions of interests (ROIs) that are associated with both risk genetic factors and disease status (Hao et al., 2016). In this study, to evaluate the effectiveness and efficiency of our DGMM method, we perform extensive experiments on three modalities of phenotypes, voxel-based measures extracted from structural MRI, fluorodeoxyglucose positron emission tomography (FDG-PET)) scans, and 18-F florbetapir (AV-45) PET scans (i.e., amyloid imaging data). We examine their associations with apolipoprotein E (APOE) SNP rs429358 (the best known AD genetic risk factor, see those in the AlzGene database (www.alzgene.org)) (Filippini et al., 2009; Liu et al., 2015). All the data are downloaded from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort. The empirical results on ADNI show that our method not only yields improved performances under the metrics of correlation coefficient and root mean squared error, but also detects a compact set of consistent and robust ROIs across three imaging modalities which are relevant to the genetic risk marker.
Subjects
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). For up-to-date information, see www.adni-info.org.
In the present study, a total of 913 non-Hispanic Caucasian participants with both imaging and genotyping data available were studied. Table 1 lists the demographics of all these subjects. Diagnosis was made using the standard criteria described in the ADNI-2 procedures manual (http://www.adniinfo.org). Briefly, HC participants had no subjective or informant-based complaint of memory decline and normal cognitive performance. SMC participants had subjective memory concerns as assessed using the Cognitive Change Index (CCI; total score from first 12 items >16), no informant-based complaint of memory impairment or decline, and normal cognitive performance on the Wechsler Logical Memory Delayed Recall (LM-delayed) and the Mini-Mental State Examination (MMSE) (Risacher et al., 2015); EMCI participants had a memory concern reported by the subject, informant, clinician, abnormal memory function approximately 1 standard deviation below normative performance adjusted for education level on the LM-delayed, an MMSE total score greater than 24; Besides a subjective memory concern as reported by subject, study partner or clinician, Clinical Dementia Rating (CDR) on LMCI subjects was 0.5 and Memory Box (MB) score must be at least 0.5 ; MMSE score on AD should be between 20 and 26 and CDR should be 0.5 or 1.0.
Table 1.
Characteristics of the subjects
| Subjects | HC | SMC | EMCI | LMCI | AD |
|---|---|---|---|---|---|
| Number | 211 | 82 | 273 | 187 | 160 |
| Gender(M/F) | 109/102 | 33/49 | 153/120 | 108/79 | 95/65 |
| Age(mean±std) | 76.14±6.53 | 72.45±5.67 | 71.48±7.12 | 73.86±8.44 | 75.18±7.88 |
| Education (mean±std) | 16.45±2.62 | 16.78±2.67 | 16.08±2.62 | 16.38±2.81 | 15.86±2.75 |
Note: HC=Healthy Control, SMC=Significant Memory Concern, ECMI=Early Mild Cognitive Impairment, LCMI=Late Mild Cognitive Impairment, AD=Alzheimer’s disease.
Risk SNP Genotype Data
Genetic risk factors can help scientists focus on relevant biological pathways and networks and form effective hypothesis for drug design. Given a genetic risk SNP, it is also important to identify its quantitative traits at brain structure and functional level to help understand the underlying biological mechanism.
Some researchers have identified a number of genes in addition to APOE ε4 that may increase a person's risk for Alzheimer’s disease (AD), including BIN1, CLU, PICALM, and CR1, see those in Lambert et al. (Lambert et al., 2013) and the AlzGene database (www.alzgene.org). APOE ε4 is called a risk-factor gene because it increases a person's risk of developing the disease. To our knowledge, APOE (located on chromosome 19) has a key role in coordinating the mobilization and redistribution of cholesterol, phospholipids, and fatty acids, and it is implicated in mechanisms such as neuronal development, brain plasticity, and repair functions (Mahley and Rall, 2000). In imaging genetics research experiments, several whole-brain studies focused on mapping this genetic risk factor (Filippini et al., 2009; Liu et al., 2015). Accordingly, in our experiments, we focus on the susceptibility SNP rs429358, which is determined using APOE ε2/ε3/ε4 status information (www.snpedia.com/index.php/APOE) from the ADNI clinical database for each participant. As shown in Figure 1, there are three subtypes according to the relationship between APOE ε2/ε3/ε4 and the two SNPs (i.e., rs429358 and rs7412). And the allelic variant on rs429358 corresponds to the absence or presence of APOE ε4: (1) allele C at rs429358 indicates an ε4 allele, and (2) allele T at rs429358 indicates a none- ε4 allele (i.e., ε2 or ε3).
Figure 1.
Illustration of allelic variants of APOE ε2/ε3/ε4
In our experiments, rs429358 value was coded in an additive fashion as 0, 1 or 2, indicating the number of minor alleles (i.e., C alleles or APOE ε4 copies). If we considered APOE ε4 as our target genetic risk of AD, the value was also coded as 0, 1 or 2, but indicating the number of APOE ε4 copies. Thus, the association study results on rs429358 should be consistent with the results on APOE ε4.
Imaging phenotype data
The MRI, FDG-PET, and AV45-PET data used in this paper were also obtained from the ADNI database (adni.loni.usc.edu). We aligned the preprocessed multi-modality imaging data (VBM, FDG, AV45) to each participant’s same visit scan, and then created normalized gray matter density maps from MRI data in the standard Montreal Neurological Institute (MNI) space as 2×2×2 mm3 voxels, registered the FDG-PET and AV45-PET scans into the same space by SPM software package (Ashburner and Friston, 2007). 116 ROI level measurements of mean gray matter densities, FDG-PET glucose utilization, and AV45 amyloid values were further extracted based on the MarsBaR AAL atlas (Tzourio-Mazoyer et al., 2002). After removal of cerebellum, the imaging measures on each modality (VBM, FDG or AV45) of 90 ROIs were used as QTs in our experiments. All the measures were pre-adjusted for age, gender, and education.
Methods
Associations between Genotype and Phenotype
In this section, we systematically develop our computational models to explore the association between risk candidate SNPs and imaging phenotypes. Our proposed method mainly addresses the problem based on the general linear (least square) regression approach. Given the imaging phenotypes X = [x1, …, xn, …, xN]T ∊ RN×d as input and a pre-selected candidate SNP y= [y1, …, yn, …, yN]T ∊ RN as output in the regression model, where N is the number of participants (sample size) and d is the number of imaging QTs (feature dimensionality). The association model is designed to solve:
| (1) |
where R(w) is a regularization term and λ is the corresponding regularization parameter. The weight vector w measures the relative importance of the imaging QTs in predicting the SNP genotype. To encourage the ‘sparsity’ among features, in the Lasso method a l1-norm regularization is imposed on the coefficients as follows:
| (2) |
where λ is a regularization parameter that controls the sparsity in the solution. The non-zero elements of indicate that the corresponding input features are relevant to the regression outputs. This penalized regression method imposes l1-norm sparisty on the individual variables for feature selection (Kohannim et al., 2012; Tibshirani, 2011).
Following the existing work (Wang et al., 2012a), it’s worth noting that the above mathematical formulations are also used for association studies between genotypes and phenotypes. In this prior work, the goal of the learned regression model was not only on the genotype prediction accuracy, but also on identifying biologically meaningful SNP and imaging markers and discovering the underlying complex biological mechanisms of the diseases, when the linear regression as the formulation of Eq (1) was applied to exploring the imaging genetic associations.
Diagnosis-guided Single-modality Phenotype Associations
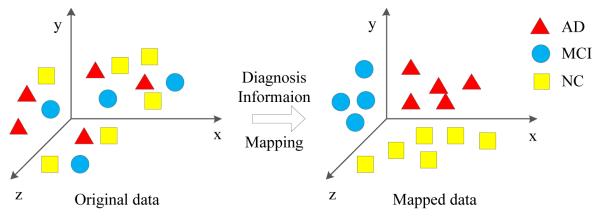
A risk genetic factor may affect multiple imaging QTs that are not all disease specific. We aim to discover only those imaging QTs associated with both the genetic factor and disease status, in order to have a better understanding of the biological pathway specific to the disease. In this study, we consider the relationship between imaging phenotypes and the diagnosis information which are not fully used in conventional imaging genetics methods. More specifically, we will utilize the subjects’ diagnostic information, i.e., HC, SMC, EMCI, LMCI, or AD. If subjects are similar to each other in the original feature space, their respective response values (i.e., predicted genotype values) should be also similar. Figure 2 illustrates an example of embedding the diagnosis information (i.e., clinical status) from original data to the mapped data space. To address this issue, we induce a new regularization term that can preserve the class level diagnosis information:
| (3) |
where S = [Sij] ∊ Rn×n denotes a similarity matrix that measures the diagnostic similarity between every pair of samples. L = D − S is the Laplacian matrix of S, where D is the diagonal matrix with element defined as . Then, the similarity matrix can be defined as:
| (4) |
Figure 2.
Schematic illustration of the diagnosis information (i.e., clinical status) embedded from original data to mapped data.
The penalized term Eq (3) enforces that, after being mapped into the label space, the distance between the within-class data will be small. The similarity between subjects within the same class can be defined as 1 if connected or 0 otherwise, which avoids the necessity of choosing hyper-parameter compared to using heat kernel (Belkin and Niyogi, 2003). Eq (3) can be applied to an existing regression model (e.g., Eq (2)) so that the mapping of the data into the label space will not only be determined by the regression model but also be regularized by Eq (3). The goal of the Eq (3) term is to encourage subjects from the same class to be close to each other in the label space.
With these observations, we induce the diagnosis labels constraint into the single modality phenotypic solution and then formulate the diagnosis-guided single modality (DGSM) phenotype association model as follows:
| (5) |
The strength of DGSM method is that it explicitly models the priori diagnosis information among subjects in the objective function that minimizes distance within each diagnostic class for selecting the disease-relevant QTs associated with the SNP. Especially, the DGSM model can generalize and handle the progressive disease with multi-diagnosis status, comparing to the binary diagnosis analysis methods that were adopted in (Batmanghelich et al., 2013; Vounou et al., 2012).
Concatenating-modality Phenotype Associations
A common practice in data fusion is the concatenation of all features from different modalities into a longer feature vector, which may provide essential complementary information for this association study. Given N training subjects or samples with M modalities of phenotypes, we denote as the data matrix of the m-th modality, as the concatenating matrix of the M modalities and y= [y1, …, yn, …, yN]T ∊ RN be the corresponding response value (i.e. APOE SNP rs429358). Let wc = [w1, …, wm, …, wM]T ∊ RM·d be the linear discriminant function corresponding to the M modalities. Then the concatenating-modality phenotype association model that based on Lasso for sparsity solution can be formulated as follows:
| (6) |
Diagnosis-guided Concatenating-modality Phenotype Associations
Following the diagnosis-guided single modality phenotype associations, we also embed the diagnosis information into the concatenating-modalities to discover the phenotypic associations with an AD genetic risk factor. Thus, we induce the diagnosis label constraint based on Eq (6), and then formulate a diagnosis-guided concatenating-modality (DGCM) phenotype association model as follows:
| (7) |
where denotes a similarity matrix that measures the similarity between every pair of samples on the concatenating M modalities across different subjects. Here, Lc = Dc − Sc represents a Laplacian matrix for the concatenating M modalities, where Dc is the diagonal matrix with element defined as . λ1 and λ2 denote control parameters of the regularization terms, respectively.
Multi-modality Phenotype Associations
Intuitively, since the pathological changes from the same ROIs can be examined through structural and functional radiologic imaging, simultaneously performing ROI feature selections across multimodalities is very helpful to suppress noises in the individual modality features. We assume that there are N training subjects or samples, with each represented by M modalities of phenotypes. Denote as the data matrix of the m-th modality, and y= [y1, …, yn, …, yN]T ∊ RN be the corresponding response value (i.e. APOE SNP rs429358). Let wm ∊ Rd be the linear discriminant function corresponding to the m-th modality. Then the multi-modality phenotype association model can be formulated as follows:
| (8) |
where W = [w1,w2, …, wM] ∊ Rd×M is the weight matrix whose row wj is the vector of coefficients assigned to the j-th feature across different modalities, and is to penalize all coefficients in the same row of matrix W for joint feature selection. It is worth noting that the l2,1-norm regularization term is a “group-sparsity” regularizer, which forces only a small number of features to be selected from different modalities (Yuan and Lin, 2006). Figure 3 shows schematic illustration of diagnosis-guided multi-modality phenotype associations. The parameter λ is a regularization parameter that is used to balance the relative contributions of those two terms in Eq (8).
Figure 3.
Schematic illustration of multi-modality phenotype associations
Diagnosis-guided Multi-modality Phenotype Associations
In this study, we propose to develop a novel diagnosis-guided multi-modality (DGMM) framework to discover the multi-modality phenotypic associations with an AD genetic risk factor, where the framework explicitly models the priori diagnosis information among subjects in the objective function for selecting the disease-relevant and ROI-consistent multi-modality QTs associated with the SNP. Sample label relations and multi-modalities have recently been successfully investigated and applied to design more powerful models on AD classification and clinical scores regression (Jie et al., 2015; Yu et al., 2014; Zhu et al., 2014a; Zhu et al., 2015; Zhu et al., 2013; Zhu et al., 2014b), which are inspired by using multi-task learning framework and taking into account the priori relationship between sample data and the corresponding labels in machine learning community (Belkin et al., 2006). Thus, we induce the diagnosis label constraint into the multi-modality phenotypic solution, and then formulate a diagnosis-guided multi-modality (DGMM) phenotype association model as follows:
| (9) |
where denotes a similarity matrix that measures the similarity between every pair of samples on the m-th modality across different subjects. Here, Lm = Dm − Sm represents a combinational Laplacian matrix for the m-th modality, where Dm is the diagonal matrix with element defined as . λ1 and λ2 denote control parameters of the regularization terms, respectively. Their values can be determined via inner cross-validation on training data. In short, the above model is designed to find the better solution that is robust to noises or outliers via considering both multimodalities and the rich information inherent in the observations.
Optimization Algorithm
A similar model has been used in (Jie et al., 2015; Zhu et al., 2013) for multimodality disease classification. The objective function can be efficiently solved using the Nesterov’s accelerated proximal gradient optimization algorithm (Chen et al., 2009), via solving the optimization problem on the Eq (9). Algorithm 1 shows such an efficient solution.
Firstly, we separate the objective function into the smooth part Eq (10) and non-smooth part Eq (11) as following:
| (10) |
| (11) |
We define the approximation function Eq (12) as following, which is composited by the above smooth part and non-smooth one:
| (12) |
where denotes the Frobenius norm, ∇f(Wi) denotes the gradient of f(W) on point W1 at the i-th iteration, and l is the step size. Finally, the update step of Nesterov’s APG is defined as:
| (13) |
where wj and vj denote the j-th row of the matrix W and V, respectively. NAGP performs a simple step of gradient descent to go from Wi to V, and then it slides a little bit further than
| (14) |
Therefore, through Eq (12), this problem can be decomposed into d separate sub-problems. The key of APG algorithm is how to solve the update step efficiently. The analytical solutions of those sub-problems can be easily obtained:
| (15) |
Instead of performing gradient descent based on Wi, we compute the search point as (Beck and Teboulle, 2009):
| (16) |
where and .
Algorithm 1.
to minimize J in Equation (9)
|
Input: risk
genetics (i.e. APOE) y= [y1,
…, yn, …,
yN]T ∊
RN, multi-modalities , subjects with diagnosis labels (i.e., HC, SMC, EMCI, LMCI or AD) |
| Output: Wi, J* |
| Initialization: l0 = 1, σ = 2, W0 = Z1 = 0, ρ0 = 1, I=1000 |
| For i=1 to max_iteration I |
| End |
| Calculate J* |
Experimental Results
Experimental Settings
In our experiment, 5-fold cross-validation strategy was adopted to evaluate the effectiveness of our proposed method. As for parameters of regularization, we determined their values by nested 5-fold cross-validation on the training set. It was to fine tune the parameters (λ1 and λ2 in Eq(9)) in the range of {10−5, 3×10−5, 10−4, 3×10−4…, 3, 10}. In current studies, we compared SM (denoted as single modality based method with Lasso (Tibshirani, 2011) to detect a sparse significant subset from imaging phenotypic features (i.e., ROI measures)), CM (denoted as concatenating modalities with Lasso to detect a sparse subset from imaging phenotypes), MM (denoted as multi-modality method to detect imaging phenotypes from a sparse subset of common ROIs), and DGSM, DGCM and DGMM (denoting DG as diagnosis-guided added methods corresponding to the standard SM, CM and MM, respectively).
Improved Association between Risk SNP and Multi-modal Phenotypic Imaging Markers
We compare our proposed diagnosis-guided based methods (including DGSM, DGCM and DGMM) with conventional methods (including SM, CM and MM), respectively. The performance on each dataset is assessed with root mean squared error (RMSE) and correlation coefficient (CC) between actual and predicted response values, which are widely used in measuring performances of regression and association analysis. The average results of RMSE and CC among the 5-fold training and testing data on MRI-VBM, FDG-PET and AV45 modalities are calculated respectively as shown in Table 2.
Table 2.
Comparison of regression performances on top risk SNP APOE rs429358 of the competing methods in terms of Root Mean Square Error (RMSE) and Correlation Coefficient (CC)
| Method | RMSE(mean±std) | CC(mean±std) | ||
|---|---|---|---|---|
|
|
||||
| train | test | train | test | |
| SM(VBM) | 0.8628±0.0408 | 0.8723±0.0489 | 0.0658±0.0456 | 0.0184±0.0666 |
| SM(FDG) | 0.8817±0.1061 | 0.9062±0.1668 | 0.0253±0.0679 | 0.0316±0.0941 |
| SM(AV45) | 0.8940±0.0095 | 0.8963±0.0589 | 0.0075±0.0171 | 0.0264±0.0835 |
| DGSM(VBM) | 0.8231±0.0063 | 0.8234±0.0261 | 0.2145±0.0821 | 0.1565±0.0846 |
| DGSM(FDG) | 0.8238±0.0064 | 0.8237±0.0259 | 0.2206±0.0641 | 0.1624±0.0560 |
| DGSM(AV45) | 0.8203±0.0087 | 0.8254±0.0259 | 0.2535±0.0749 | 0.1725±0.0596 |
| CM | 0.9276±0.1485 | 0.9361±0.1560 | 0.0242±0.0556 | 0.0436±0.0727 |
| DGCM | 0.8235±0.0062 | 0.8236±0.0261 | 0.2383±0.0633 | 0.1918±0.1056 |
| MM(VBM) | 0.7644±0.0071 | 0.8752±0.0257 | 0.4370±0.0190 | 0.2107±0.0698 |
| MM(FDG) | 0.7679±0.0090 | 0.8836±0.0265 | 0.4238±0.0172 | 0.1695±0.0527 |
| MM(AV45) | 0.7606±0.0071 | 0.8730±0.0271 | 0.4518±0.0161 | 0.2286±0.0677 |
| DGMM(VBM) | 0.7886±0.0079 | 0.8214±0.0314 | 0.3688±0.0207 | 0.2484±0.0570 |
| DGMM(FDG) | 0.7901±0.0087 | 0.8229±0.0262 | 0.3605±0.0195 | 0.2345±0.0676 |
| DGMM(AV45) | 0.7877±0.0085 | 0.8201±0.0322 | 0.3805±0.0176 | 0.2545±0.0572 |
As shown in Table 2, DGSM yields the RMSE values of 0.8234, 0.8237, 0.8254 and CC values of 0.1565, 0.1624 and 0.1725 on three different modalities, respectively, which are better than those of SM. In addition, DGCM yields the RMSE values of 0.8236 and CC values of 0.1918, which are better than those of CM. Moreover, DGMM achieves the best RMSE values of 0.8214, 0.8229 and 0.8201 and the best CC values of 0.2484, 0.2345 and 0.2545 on three different modalities, respectively, which are better than those of MM. These results indicate that the proposed DG based methods consistently outperform their non-DG based methods in both RMSE and CC performance measures. It’s worth noting that although the concatenation of all features can provide essential complementary information in ideal condition, CM-type models may be not enough for effective combination in this work as they brought more noises in widespread feature space. However, the multi-task strategy (i.e., l2,1-norm constrain) can enhance the robustness of ROI detection, which demonstrates that both diagnosis-guided priori knowledge and multi-modality information make it possible to improve the performances of regression and association from imaging phenotypes to genotype.
Identification of Consistent and Robust ROIs as Intermediate Phenotypes
Besides the improved performances, one major goal of this study is to identify some significant and robust phenotypes that are highly correlated to both risk SNP marker and disease status to capture imaging genetics associations in AD research. Figure 4 shows all comparisons of weight maps for the multi-modalities on 90 ROI associations with APOE SNP rs429358 respect to different methods. As expected, DGMM method can select sparse and significant ROIs associated with APOE rs429358. Although the SNP may affect different sets of ROIs while using different modalities as phenotype, the ROIs selected by our model tend to have all their modalities associated with the SNP and show great potential for further investigation. It is well known that the selected ROIs such as left hippocampus, right precuneus, left superior occipital gyrus and left calcarine gyrus are related to the structure atrophy, pathological amyloid depositions, and metabolic alteration in the brain (Camus et al., 2012; Liu et al., 2015; Reiman et al., 1996; Wishart et al., 2006), showing effectiveness of the proposed method.
Figure 4.
Weight maps for the multi-modalities on 90 ROI associations with APOE SNP rs429358 respect to different methods.
The top 10 selected MRI-VBM imaging features, as well as their average regression coefficients across five cross-validation trials, are visualized in Figure 5 by mapping them onto the human brain. The colors of the selected brain regions indicate the regression coefficients of the corresponding MRI-VBM markers. As expected, left hippocampus and left amygdala have been detected on top 10 ROIs associated with the risk genotype biomarker by the proposed DGMM method. It’s worth noting that these stable markers are in accordance with the previous studies. For example, the reduction of hippocampal gray matter has been correlated with APOE SNP rs429358 (Wishart et al., 2006).
Figure 5.
Visualization of the top 10 VBM ROIs selected by the proposed method. The color represents the regression coefficients of the corresponding VBM markers.
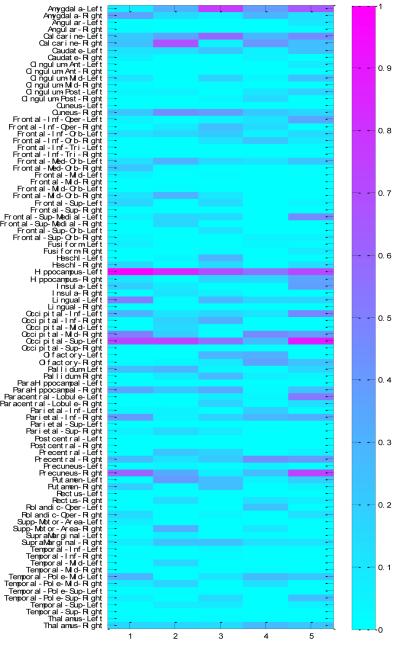
The overall regression coefficients which are combinations of VBM, FDG and AV45 for the 90 ROIs by DGMM test are plotted in Figure 6. The association weight map shows that the selected imaging markers by our proposed method have clear patterns that span across all the five cross-validation trials, and these identified phenotypic markers are from extremely stable ROIs such as left hippocampus, left superior occipital gyrus and left calcarine gyrus. In summary, the identified stable markers strongly agree with the existing findings. For example, the reductions of hippocampal gray matter and glucose metabolism for pathological changes have been shown to be associated with the best established genetic risk factor APOE polymorphism (Camus et al., 2012; Liu et al., 2015; Reiman et al., 1996; Wishart et al., 2006). Hence, these consistent ROIs detected by our proposed DGMM are closer to the underlying pathogenic location of the disease and the drug targeting for treatment in the future.
Figure 6.
Weight maps on the associations between diagnosis-guided consistent 90 ROI imaging markers and APOE rs429358 across five cross-validation trials by proposed DGMM methods.
Discussion
In this paper, we have proposed a novel diagnosis-guided multi-modality (DGMM) framework to detect brain imaging phenotypes as intermediate QTs that are associated with both a certain risk genetic factor and disease status. The experimental results on 913 subjects from ADNI show that our DGMM model can substantially improve the performance of the associations. Specifically, for prediction and association measurements, our proposed method can achieve high average correlation coefficient values of 0.2484, 0.2345 and 0.2545 on VBM, FDG, AV45 modalities, respectively. Besides the improved regression performances, our model can also identify some significant and robust phenotypic ROIs that reveal disease-specific imaging genetic associations on the chain from gene to brain to symptom.
Predictability from Phenotypes to Genotype
Similar to that used in our study for elucidating the associations between genotype and phenotype, the predicting formulations have been used in other imaging genetics association studies. In our work, the goal of the learned regression model is to select more biologically meaningful imaging phenotypes and discover the underlying complex biological mechanisms of the diseases. Here, we apply standard multivariate linear regression and R-square measures to evaluate the imaging genetic associations. In statistics, R-square (explained variation / total variation) is a measure of how close the data are to the fitted regression line. It is also known as the coefficient of determination, which provides a measure of how well observed outcomes are replicated by the model, based on the proportion of total variation of outcomes explained by the model (Draper, 2002). Table 3 provides how successful the fit is in explaining the variation of the data (Putcha and Raton, 2008), which indicate how much of the SNP genotypic variation can be explained by the different phenotypes (VBM, FDG and AV45, respectively). In this case, VBM measurement yields 3.5% (3.4% adjusted) explained capacity from left hippocampus (denoted HippL for short) to APOE, while both FDG and AV45 show weaker predictabilities. This is in accordance with the fact that in the pathology pathway of AD (Wishart et al., 2006). The proportion explained by top 10 consistent ROIs on each modality can achieve half adjusted R-square value of the predictabilities with all independent variables (90 ROIs). These results also demonstrate the promise of the proposed method in terms of its capability to identify the more significant brain ROIs associated with the top risk genotype.
Table 3.
Predictability from Phenotypes to Genotype APOE rs429358 via R-square Statistical measures
| R-square | Adjusted R-square | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| Modality | HippL | Top10ROIs | 90ROIs | HippL | Top10ROIs | 90ROIs |
| VBM | 0.035 | 0.093 | 0.189 | 0.034 | 0.083 | 0.1 |
| FDG | 0.005 | 0.051 | 0.174 | 0.004 | 0.04 | 0.083 |
| AV45 | 0.01 | 0.09 | 0.203 | 0.009 | 0.08 | 0.115 |
Top Risk SNP vs Non-disease related SNP
The APOE polymorphism is the best established genetic risk factor for pathological changes that is also associated with anatomical brain changes (Liu et al., 2015). In our experiment, we test the performance on the top risk SNP APOE rs429358 in the DGMM framework, as reported in Table 2. We also selected a non-AD related SNP rs12410166 as the comparison to evaluate the performance of the proposed model. As shown in Table 4, all methods (including our DGMM framework) yielded very low average correlation coefficients measures, compared to that of top risk SNP APOE reported in Table 2. The model with irrelevant priori information embedding has leaded over-fitting on the badly noisy train data, meanwhile, has lost the power of generalization on test data. The originality of the work is to make full use of the risk genotype and corresponding disease samples to find the intermediate phenotype between an AD genetic marker and the disease status. Therefore, this set of contrast experiment has demonstrated that the pattern of consistent multimodal intermediate phenotypes we learned from the model only by using AD related SNP can capture the potential of interpreting the biological pathway from gene to brain to diagnosis.
Table 4.
Comparison of regression performances on random selected SNP rs12410166 of the competing methods in terms of Correlation Coefficient (CC)
| Method | CC(mean±std) |
|
|---|---|---|
| train | test | |
| SM(VBM) | −0.0394±0.0670 | 0.0224±0.0977 |
| SM(FDG) | 0.0633±0.0311 | −0.0221±0.0650 |
| SM(AV45) | 0.0542±0.0349 | 0.0092±0.0643 |
| DGSM(VBM) | 0.0617±0.0442 | −0.0113±0.0521 |
| DGSM(FDG) | 0.0821±0.0213 | −0.0344±0.0370 |
| DGSM(AV45) | 0.0797±0.0883 | −0.0126±0.0945 |
| CM | 0.0100±0.0205 | 0.0032±0.0918 |
| DGCM | 0.0698±0.0295 | −0.0289±0.0881 |
| MM(VBM) | 0.3470±0.0192 | 0.0372±0.0328 |
| MM(FDG) | 0.3594±0.0104 | 0.0521±0.0585 |
| MM(AV45) | 0.3479±0.0190 | 0.0352±0.0417 |
| DGMM(VBM) | 0.1845±0.1161 | 0.0320±0.0652 |
| DGMM(FDG) | 0.2324±0.0917 | −0.0071±0.0589 |
| DGMM(AV45) | 0.1808±0.1190 | 0.0338±0.0551 |
Multi-locus Genetic Association Models vs Top Risk SNP Associations
In the existing studies, multi-locus genetic associations are used to discover reliable genetic influences with small effect size by adopted multivariate approaches such as statistical methods and machine learning techniques (Hibar et al., 2011; Shen et al., 2014). Here, we will review some multi-locus genetic marker detections in existing literatures which include diagnosis based methods (Batmanghelich et al., 2013; Vounou et al., 2012). In Batmanghelich et al’s Bayesian model, APOE ε4 and APOE ε3 were selected to be strongly correlated with AD under the highest posterior probability. And also, variants on APOC1, TOMM40 and PVRL were among high probability regions (hippocampus and temporal lobe) (Batmanghelich et al., 2013). In Vounou et al.’s two-step procedure identification, the experimental results showed PIK3R3/PIK3CG and PRKCA/PRKCB were important in driving selection of many pathways in the top 30 ranks. In addition, TOMM40, CR1 and APOE were in the top 10 ranking pathways, including ADCY2, ACTN1, ACACA and GNAI1, all of which were associated with AD related changes in hippocampal gene expression (Vounou et al., 2012). However, the focus of our work is not identifying several multi-locus genetic patterns associated with phenotypes. However, it supplies a simple and efficient framework to identify intermediate imaging QTs that can bridge the gap between one top risk gene and a disease, comparing to the case control study.
Limitation
While aiming to develop an intermediate trait identification framework, the current study is limited by two factors. First, we associate only the top risk SNP (i.e., APOE rs429358), while AD is 50-70% heritable with complex genetic underpinnings, and an individual marker explains limited heritability of AD. Therefore, polygenic scores (Dudbridge, 2013; Sabuncu et al., 2012) that is considered as multi-locus genetic effect should be used in our DGMM model.
Second, for fair comparisons among SM-type models, we have reported the prediction accuracy of three imaging modalities separately. Actually, we haven’t designed a least squared error for joint prediction in the objective function. However, motivated by ensemble learning, we have averaged individual outputs learned by our DGMM for joint prediction denoted as DGMM (A-All). As shown in Table 5, the joint prediction results are superior to the separate ones on both training and test data. Furthermore, in order to investigate the relative contribution of each modality, we have extended weighted ensemble predictions denoted as DGMM (W-All). In our empirical study, we constrained the summation of individual weights was 1 and the optimal values were learned based on the training through a grid search using the range from 0 to 1 at a step of 0.1. The optimal weights have demonstrated that VBM and AV45 have larger contributions to the joint predictions. However, the contributed weight for each modality was not optimized from the objective function. Thus, it is an interesting future topic to apply our DGMM model to extend a one-step joint association framework considering the relative contribution of each modality.
Table 5.
performance comparisons of separate and joint association study by our proposed DGMM in terms of Root Mean Square Error (RMSE) and Correlation Coefficient (CC)
| Method | RMSE(mean±std) | CC(mean±std) | ||
|---|---|---|---|---|
|
|
||||
| train | test | train | test | |
| DGMM(VBM) | 0.7886±0.0079 | 0.8214±0.0314 | 0.3688±0.0207 | 0.2484±0.0570 |
| DGMM(FDG) | 0.7901±0.0087 | 0.8229±0.0262 | 0.3605±0.0195 | 0.2345±0.0676 |
| DGMM(AV45) | 0.7877±0.0085 | 0.8201±0.0322 | 0.3805±0.0176 | 0.2545±0.0572 |
| DGMM(A-ALL) | 0.7841±0.0083 | 0.8164±0.0293 | 0.4309±0.0116 | 0.2895±0.0269 |
| DGMM(W-All) | 0.7838±0.0087 | 0.8160±0.0285 | 0.4393±0.0133 | 0.2938±0.0149 |
Third, as different imaging modalities can provide essential complementary information that can improve performances of accuracy, a lot of multi-modality based methods including concatenation and integrating among the feature level or other ensemble methods (Liu et al., 2012) such as kernel combination (Jie et al., 2015; Zhang et al., 2011) and random forest (Gray et al., 2013) have been widely address the classification and prediction problem. However, our goal is to identify major ROIs whose multimodal measures can serve as intermediate traits, ignoring the diversity of the multi-modality phenotypes. We happen to induce existing multi-task learning aspect (Jie et al., 2015; Yuan and Lin, 2006; Zhang and Shen, 2012) for joint selecting the robust and consistent ROIs among different quantitative brain phenotypes for underlying the mechanisms of the disease. In order to balance the aspects of multi-modality phenotypic diversities and consistent ROIs selection in the association analysis above, we expect to use a more powerful model that can examine both individual (one ROI with certain modality phenotype) and shared features (the same ROIs among all modality phenotypes) of the different quantitative brain phenotypes to further improve our multimodal intermediate phenotypes identification. In the future work, we will address the above limitations for further improvement.
Conclusions
In summary, this study developed a diagnosis-guided multi-modality (DGMM) framework for identifying neuroimaging quantitative phenotypes which can server as intermediate traits between a certain risk genetic factor and disease status. This approach explicitly utilized the priori diagnosis information among subjects in the objective function for selecting the most relevant multi-modality QTs associated with top risk SNP (i.e., APOE rs429358) in one step. The empirical experiments on the ADNI database showed that our method improved performances under the metrics of both correlation coefficient and root mean squared error compared with other competing methods. Specifically, the main contribution of this work was to identify a compact set of robust and consistent ROIs across the multimodal phenotypes (i.e., MRI-VBM, FDG-PET and AV45) to have a mechanistic understanding of AD biology. This general DGMM framework can be extended and applied to identify potential intermediate traits for genetic studies of other disorders.
Acknowledgments
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Bio-gen; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
This research is supported by the National Natural Science Foundation of China (Nos. 61422204, 61473149), the Jiangsu Natural Science Foundation for Distinguished Young Scholar (No. BK20130034), the Specialized Research Fund for the Doctoral Program of Higher Education (No. 20123218110009), the NUAA Fundamental Research Funds (No. NE2013105), the Jiangsu Qinglan Project of China and Nanjing University of Aeronautics and Astronautics Ph.D student short-term visiting scholar project.
At Indiana University, this work was supported by NIH R01 LM011360, U01 AG024904, RC2 AG036535, R01 AG19771, P30 AG10133, UL1 TR001108, R01 AG 042437, and R01 AG046171; NSF IIS-1117335; DOD W81XWH-14-2-0151, W81XWH-13-1-0259, and W81XWH-12-2-0012; NCAA 14132004; and CTSI SPARC Program.
Footnotes
Information Sharing Statement
Both the source code and documentation are available on request.
References
- Ashburner J, Friston K. Voxel-Based Morphometry. Statistical Parametric Mapping: The Analysis of Functional Brain Images. 2007:92–98. [Google Scholar]
- Baranzini SE, Wang J, Gibson RA, Galwey N, Naegelin Y, Barkhof F, Radue EW, Lindberg RL, Uitdehaag BM, Johnson MR, Angelakopoulou A, Hall L, Richardson JC, Prinjha RK, Gass A, Geurts JJ, Kragt J, Sombekke M, Vrenken H, Qualley P, Lincoln RR, Gomez R, Caillier SJ, George MF, Mousavi H, Guerrero R, Okuda DT, Cree BA, Green AJ, Waubant E, Goodin DS, Pelletier D, Matthews PM, Hauser SL, Kappos L, Polman CH, Oksenberg JR. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet. 2009;18:767–778. doi: 10.1093/hmg/ddn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batmanghelich NK, Dalca AV, Sabuncu MR, Polina G. Joint modeling of imaging and genetics. Inf Process Med Imaging. 2013;23:766–777. doi: 10.1007/978-3-642-38868-2_64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck A, Teboulle M. A Fast Iterative Shrinkage-Thresholding Algorithm for Linear Inverse Problems. Siam Journal on Imaging Sciences. 2009;2:183–202. [Google Scholar]
- Belkin M, Niyogi P. Laplacian eigenmaps for dimensionality reduction and data representation. Neural Computation. 2003;15:1373–1396. [Google Scholar]
- Belkin M, Niyogi P, Sindhwani V. Manifold regularization: A geometric framework for learning from labeled and unlabeled examples. Journal of Machine Learning Research. 2006;7:2399–2434. [Google Scholar]
- Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer's disease. Alzheimers & Dementia. 2007;3:186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- Brun CC, Lepore N, Pennec X, Lee AD, Barysheva M, Madsen SK, Avedissian C, Chou YY, de Zubicaray GI, McMahon KL, Wright MJ, Toga AW, Thompson PM. Mapping the regional influence of genetics on brain structure variability--a tensor-based morphometry study. Neuroimage. 2009;48:37–49. doi: 10.1016/j.neuroimage.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus V, Payoux P, Barre L, Desgranges B, Voisin T, Tauber C, La Joie R, Tafani M, Hommet C, Chetelat G, Mondon K, de La Sayette V, Cottier JP, Beaufils E, Ribeiro MJ, Gissot V, Vierron E, Vercouillie J, Vellas B, Eustache F, Guilloteau D. Using PET with 18F-AV-45 (florbetapir) to quantify brain amyloid load in a clinical environment. Eur J Nucl Med Mol Imaging. 2012;39:621–631. doi: 10.1007/s00259-011-2021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Pan WK, Kwok JT, Carbonell JG. Accelerated Gradient Method for Multi-Task Sparse Learning Problem. 2009 9th Ieee International Conference on Data Mining.2009. pp. 746–751. [Google Scholar]
- Draper NR. Applied regression analysis. Bibliography update 2000-2001. Communications in Statistics-Theory and Methods. 2002;31:2051–2075. [Google Scholar]
- Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini N, Rao A, Wetten S, Gibson RA, Borrie M, Guzman D, Kertesz A, Loy-English I, Williams J, Nichols T, Whitcher B, Matthews PM. Anatomically-distinct genetic associations of APOE epsilon4 allele load with regional cortical atrophy in Alzheimer's disease. Neuroimage. 2009;44:724–728. doi: 10.1016/j.neuroimage.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Ge T, Feng J, Hibar DP, Thompson PM, Nichols TE. Increasing power for voxel-wise genome-wide association studies: the random field theory, least square kernel machines and fast permutation procedures. Neuroimage. 2012;63:858–873. doi: 10.1016/j.neuroimage.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Thompson PM, Blangero J. Neuroimaging endophenotypes: Strategies for finding genes influencing brain structure and function. Human Brain Mapping. 2007;28:488–501. doi: 10.1002/hbm.20401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: Etymology and strategic intentions. American Journal of Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Gray KR, Aljabar P, Heckemann RA, Hammers A, Rueckert D. Random forest-based similarity measures for multi-modal classification of Alzheimer's disease. Neuroimage. 2013;65:167–175. doi: 10.1016/j.neuroimage.2012.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X, Yan J, Yao X, Risacher SL, Saykin AJ, Zhang D, Shen LI. Diagnosis-Guided Method for Identifying Multi-Modality Neuroimaging Biomarkers Associated with Genetic Risk Factors in Alzheimer's Disease. Pac Symp Biocomput. 2016;21:108–119. [PMC free article] [PubMed] [Google Scholar]
- Hibar DP, Kohannim O, Stein JL, Chiang MC, Thompson PM. Multilocus genetic analysis of brain images. Front Genet. 2011;2:73. doi: 10.3389/fgene.2011.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jie B, Zhang D, Cheng B, Shen D. Manifold regularized multitask feature learning for multimodality disease classification. Human Brain Mapping. 2015;36:489–507. doi: 10.1002/hbm.22642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohannim O, Hibar DP, Stein JL, Jahanshad N, Hua X, Rajagopalan P, Toga AW, Jack CR, Weiner MW, de Zubicaray GI, McMahon KL, Hansell NK, Martin NG, Wright MJ, Thompson PM, Initia, A.D.N. Discovery and replication of gene influences on brain structure using LASSO regression. Frontiers in Neuroscience. 2012:6. doi: 10.3389/fnins.2012.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohannim O, Hibar DP, Stein JL, Jahanshad N, Jack CR, Weiner MW, Toga AW, Thompson PM, Initi, A.s.D.N. Boosting Power to Detect Genetic Associations in Imaging Using Multi-Locus, Genome-Wide Scans and Ridge Regression. 2011 8th Ieee International Symposium on Biomedical Imaging: From Nano to Macro, 1855-1859.2011. [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, European Alzheimer's Disease, I. Genetic, Environmental Risk in Alzheimer's, D. Alzheimer's Disease Genetic, C. Cohorts for, H. Aging Research in Genomic, E. Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O'Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Jr., Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nothen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang D, Shen D. Ensemble sparse classification of Alzheimer's disease. Neuroimage. 2012;60:1106–1116. doi: 10.1016/j.neuroimage.2012.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yu JT, Wang HF, Han PR, Tan CC, Wang C, Meng XF, Risacher SL, Saykin AJ, Tan L. APOE genotype and neuroimaging markers of Alzheimer's disease: systematic review and meta-analysis. J Neurol Neurosurg Psychiatry. 2015;86:127–134. doi: 10.1136/jnnp-2014-307719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW, Rall SC., Jr. Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- Pasinetti GM, Hiller-Sturmhofel S. Systems biology in the study of neurological disorders: focus on Alzheimer's disease. Alcohol Res Health. 2008;31:60–65. [PMC free article] [PubMed] [Google Scholar]
- Potkin SG, Turner JA, Guffanti G, Lakatos A, Torri F, Keator DB, Macciardi F. Genome-wide strategies for discovering genetic influences on cognition and cognitive disorders: methodological considerations. Cogn Neuropsychiatry. 2009;14:391–418. doi: 10.1080/13546800903059829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putcha V, Raton B. Handbook of univariate and multivariate data analysis and interpretation with SPSS. Journal of the Royal Statistical Society Series a-Statistics in Society. 2008;171:317–317. [Google Scholar]
- Reiman EM, Caselli RJ, Yun LS, Chen K, Bandy D, Minoshima S, Thibodeau SN, Osborne D. Preclinical evidence of Alzheimer's disease in persons homozygous for the epsilon 4 allele for apolipoprotein E. N Engl J Med. 1996;334:752–758. doi: 10.1056/NEJM199603213341202. [DOI] [PubMed] [Google Scholar]
- Risacher SL, Kim S, Nho K, Foroud T, Shen L, Petersen RC, Jack CR, Jr., Beckett LA, Aisen PS, Koeppe RA, Jagust WJ, Shaw LM, Trojanowski JQ, Weiner MW, Saykin AJ. APOE effect on Alzheimer's disease biomarkers in older adults with significant memory concern. Alzheimers Dement. 2015;11:1417–1429. doi: 10.1016/j.jalz.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabuncu MR, Buckner RL, Smoller JW, Lee PH, Fischl B, Sperling RA, Neuroimaging, A.s.D. The Association between a Polygenic Alzheimer Score and Cortical Thickness in Clinically Normal Subjects. Cerebral Cortex. 2012;22:2653–2661. doi: 10.1093/cercor/bhr348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Thompson PM, Potkin SG, Bertram L, Farrer LA, Foroud TM, Green RC, Hu X, Huentelman MJ, Kim S, Kauwe JS, Li Q, Liu E, Macciardi F, Moore JH, Munsie L, Nho K, Ramanan VK, Risacher SL, Stone DJ, Swaminathan S, Toga AW, Weiner MW, Saykin AJ. Genetic analysis of quantitative phenotypes in AD and MCI: imaging, cognition and biomarkers. Brain Imaging Behav. 2014;8:183–207. doi: 10.1007/s11682-013-9262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso: a retrospective. Journal of the Royal Statistical Society Series B-Statistical Methodology. 2011;73:273–282. [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Vounou M, Janousova E, Wolz R, Stein JL, Thompson PM, Rueckert D, Montana G, Initia, A.D.N. Sparse reduced-rank regression detects genetic associations with voxel-wise longitudinal phenotypes in Alzheimer's disease. Neuroimage. 2012;60:700–716. doi: 10.1016/j.neuroimage.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vounou M, Nichols TE, Montana G. Discovering genetic associations with high-dimensional neuroimaging phenotypes: A sparse reduced-rank regression approach. Neuroimage. 2010;53:1147–1159. doi: 10.1016/j.neuroimage.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Nie F, Huang H, Yan J, Kim S, Nho K, Risacher SL, Saykin AJ, Shen L. From phenotype to genotype: an association study of longitudinal phenotypic markers to Alzheimer's disease relevant SNPs. Bioinformatics. 2012a;28:i619–i625. doi: 10.1093/bioinformatics/bts411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Nie FP, Huang H, Kim S, Nho K, Risacher SL, Saykin AJ, Shen L, Initi, A.s.D.N. Identifying quantitative trait loci via group-sparse multitask regression and feature selection: an imaging genetics study of the ADNI cohort. Bioinformatics. 2012b;28:229–237. doi: 10.1093/bioinformatics/btr649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart HA, Saykin AJ, McAllister TW, Rabin LA, McDonald BC, Flashman LA, Roth RM, Mamourian AC, Tsongalis GJ, Rhodes CH. Regional brain atrophy in cognitively intact adults with a single APOE epsilon4 allele. Neurology. 2006;67:1221–1224. doi: 10.1212/01.wnl.0000238079.00472.3a. [DOI] [PubMed] [Google Scholar]
- Yu G, Liu Y, Thung KH, Shen D. Multi-Task Linear Programming Discriminant Analysis for the Identification of Progressive MCI Individuals. PLoS One. 2014:9. doi: 10.1371/journal.pone.0096458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. Journal of the Royal Statistical Society Series B-Statistical Methodology. 2006;68:49–67. [Google Scholar]
- Zhang D, Shen D. Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer's disease. Neuroimage. 2012;59:895–907. doi: 10.1016/j.neuroimage.2011.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wang Y, Zhou L, Yuan H, Shen D. Multimodal classification of Alzheimer's disease and mild cognitive impairment. Neuroimage. 2011;55:856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Suk HI, Shen D. A novel multi-relation regularization method for regression and classification in AD diagnosis. Med Image Comput Comput Assist Interv. 2014a;17:401–408. doi: 10.1007/978-3-319-10443-0_51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Suk HI, Wang L, Lee SW, Shen D. A novel relational regularization feature selection method for joint regression and classification in AD diagnosis. Med Image Anal. 2015 doi: 10.1016/j.media.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XF, Huang Z, Yang Y, Shen HT, Xu CS, Luo JB. Self-taught dimensionality reduction on the high-dimensional small-sized data. Pattern Recognition. 2013;46:215–229. [Google Scholar]
- Zhu XF, Suk HI, Shen D. A novel matrix-similarity based loss function for joint regression and classification in AD diagnosis. Neuroimage. 2014b;100:91–105. doi: 10.1016/j.neuroimage.2014.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]