Abstract
The interaction between splicing factors and the transcriptional machinery provides an intriguing link between the coupled processes of transcription and splicing. Here, we show that two components of the SF3B complex that forms part of the U2 small nuclear ribonucleoprotein particle (snRNP), SF3B3 and SF3B5, are also subunits of the Spt-Ada-Gcn5 acetyltransferase (SAGA) transcriptional coactivator complex in Drosophila melanogaster. Whereas SF3B3 had previously been identified as a human SAGA subunit, SF3B5 had not been identified as a component of SAGA in any species. We show that SF3B3 and SF3B5 bind to SAGA independent of RNA, and interact with multiple SAGA subunits including Sgf29 and Spt7 in a yeast two-hybrid assay. Through analysis of sf3b5 mutant flies, we show that SF3B5 is necessary for proper development and cell viability, but not for histone acetylation. Although SF3B5 does not appear to function in SAGA’s histone modifying activities, SF3B5 is still required for expression of a subset of SAGA-regulated genes independent of splicing. Thus, our data support an independent function of SF3B5 in SAGA’s transcription coactivator activity that is separate from its role in splicing.
Keywords: Splicing factors, SF3B3, SF3B5, SAGA, chromatin
Graphical Abstract
Introduction
Splicing occurs co-transcriptionally and is affected by transcription rate, chromatin modifications, and nucleosome occupancy [1–8]. Emerging evidence suggests that components involved in splicing can also modulate transcription [9–11]. Moreover, several splicing factors have been shown to interact with chromatin remodelers [12], histone marks [13–16], or RNA polymerase II itself [17], indicating that there are multiple mechanisms that couple transcription to splicing.
One intriguing link between splicing and transcription is provided by components that are present both in the spliceosome machinery and in transcriptional regulators. For example, a major component of the spliceosome, the U1 snRNA, interacts with the general transcription initiation factor TFIIH to stimulate transcription initiation [10]. In addition, the U2 small nuclear ribonucleoprotein particle (snRNP) interacts with the transcription elongation factor TAT-SF1, and extracts depleted for U2 snRNP show a decrease in transcriptional activity [11]. Further, the SF3B3 (Splicing Factor 3b, subunit 3) subunit of the SF3B complex within the U2 snRNP associates with the transcription factors ERG and TFIIS [18, 19]. Strikingly, SF3B3 is also a subunit of the mammalian Spt-Ada-Gcn5-acetyltransferase (SAGA) transcriptional co-activator complex [20, 21]. Thus, SF3B3 functions as a shared subunit of the U2 snRNP and SAGA.
SF3B3 is highly conserved from yeast to humans, and its homolog in Saccharomyces cerevisiae, Rse1, is an essential gene that is necessary for proper splicing [22–25]. SF3B3 functions as part of the SF3B complex that contributes to recognition of the intron branch site in the pre-mRNA by the U2 snRNP during the first step of splicing [26–29]. Whereas the SF3B complex itself plays a well-defined role in the first step of splicing, the function of its SF3B3 subunit that is shared with SAGA, either in splicing or in transcription, has not been well defined.
Here, we report the identification of a second SF3B component, SF3B5 (Splicing Factor 3b, subunit 5), as a subunit of SAGA in Drosophila melanogaster. Since our study shows that two independent SF3B subunits are components of metazoan SAGA, we sought to determine the function of these shared SF3B subunits in SAGA. SAGA is highly conserved from yeast to humans and possesses several distinct activities that regulate different aspects of transcription activation [30]. First, SAGA contains the histone acetyltransferase Gcn5 that acetylates histone H3 [31]. Gcn5 is also found in a second transcriptional coactivator complex in flies and humans, the Ada2a-containing complex (ATAC) that is distinct from SAGA [32]. Second, SAGA contains a histone deubiquitinase, Ubp8 (Nonstop in Drosophila), that deubiquitinates monoubiquitinated histone H2B (ubH2B) [33]. Independent of these histone modifying activities, SAGA is also a direct coactivator that recruits RNA polymerase II to promoters [34, 35]. Although SAGA is best characterized with regard to its roles at promoters, SAGA also co-localizes with RNA polymerase II on transcribed regions [36–38]. Thus, SAGA and the SF3B complex share a common spatial and temporal distribution during the coupled processes of transcription and splicing.
In this study, we examine the function of the shared SAGA/U2 snRNP subunit, SF3B5, in SAGA-regulated histone modification and gene expression. We show that SF3B5 is required for expression of a subset of SAGA-regulated genes, including a gene that contains no introns in its coding region, but is not required for SAGA-mediated histone acetylation. Thus, our findings are consistent with a function for SF3B5 in SAGA-dependent transcription but not histone modification, independent of its function in the U2 snRNP.
Results and Discussion
Identification of two SF3B proteins within the Drosophila SAGA complex
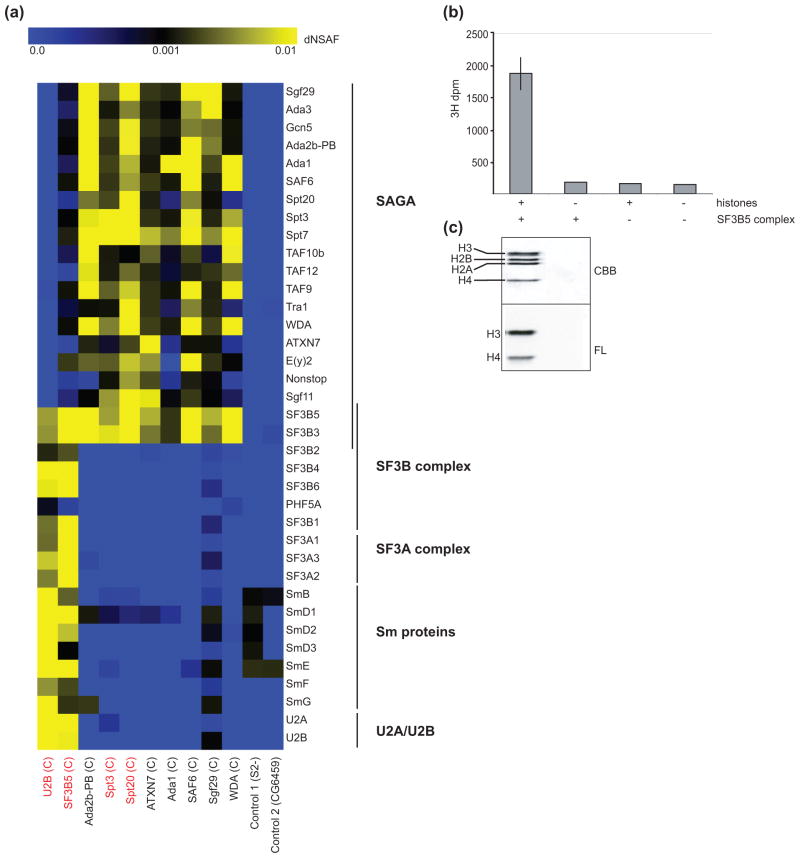
To identify novel Drosophila SAGA subunits, we isolated SAGA using tandem FLAG-HA affinity purification from S2 cell nuclear extracts with the SAGA-specific subunits Spt3 and Spt20 as bait proteins and examined the composition of affinity purified SAGA by Multidimensional Protein Identification Technology (MudPIT) [39]. Purifications using the SAGA-specific subunits Ada2b (isoform PB), ATXN7, Ada1, SAF6, WDA and the shared ATAC/SAGA subunit Sgf29 as bait proteins were previously described and are shown for comparison [40, 41]. Peptides from two proteins were consistently identified in affinity-purified SAGA: CG11985 and CG13900. These two proteins correspond to components of the U2 snRNP spliceosomal complex: SF3B3 (Splicing Factor 3b subunit 3, GeneID: CG13900; FlyBase ID: FBgn0035162) and SF3B5 (Splicing Factor 3b subunit 5, GeneID: CG11985; FlyBase ID: FBgn0040534) (Fig. 1a, Table 1) [42, 43]. SF3B3 and SF3B5 were present at similar dNSAF (distributive normalized spectral abundance factor) levels to those of the core SAGA subunits Spt7 and TAF9, and were not identified in control purifications from cells expressing a non-specific tagged bait protein, or in samples from cells lacking tagged protein (Fig. 1a, Supplemental Table S1).
Fig 1. SF3B3 and SF3B5 are novel components of Drosophila SAGA.
(a) Heat map showing the relative spectral abundance of SAGA and spliceosomal subunits expressed as dNSAF (distributive normalized spectral abundance factor) in tandem FLAG-HA purifications from S2 cells using U2B, SF3B5, Ada2B-PB, Spt3, Spt20, ATXN7, Ada1, SAF6, Sgf29 and WDA as bait proteins, relative to control purifications from untagged S2 cells (S2 -) or S2 cells expressing non-specific tagged protein CG6459. Bait proteins were C-terminally tagged as indicated (C). Bait proteins new to this study are highlighted in red. The dNSAF scale is shown at the top of panel (a) with the highest abundance subunits represented in yellow, and absent or under-represented subunits in blue. dNSAF values used to generate the heat map are provided in Supplemental Table S1. (b, c) The HAT activity of FLAG-purified SF3B5-complexes was assayed in vitro by incorporation of 3H-acetyl CoA into core histones. Core histones and/or FLAG-purified SF3B5-complex were included in each HAT assay as indicated by +/− below the graph in panel b, and 3H-acetyl CoA incorporation assayed for each reaction using both scintillation counting (b) and fluorography (c). Lanes in panel (c) correspond to reactions from above (panel b). Reactions containing complex and histones were performed in triplicate and compared to background levels of single control reactions lacking histones or complex as part of the set of HAT assays previously described for WDA- and SAF6-purified SAGA [40]. Error bars in panel (b) for + SF3B5-complex + histones represent standard deviation of the mean for three technical replicates. (c) Histones were separated by SDS-PAGE, stained with Coomassie Brilliant Blue (CBB) to determine the migration of each histone (upper panel), and 3H-acetyl CoA incorporation for each histone examined using fluorography (FL).
Table 1. Sequence coverage (%) and number of peptides (spectral count) for each polypeptide identified in MudPIT analysis of affinity purifications using U2B, SF3B5, Spt3 and Spt20 as bait proteins.
X, protein not identified.
| Bait: % (spectral count)
|
|||||||
|---|---|---|---|---|---|---|---|
| FBgn ID | CG number | Protein | U2B | SF3B5 | Spt3 | Spt20 | Length (aa) |
| FBgn0050390 | CG30390 | Sgf29 | X | 25.95%(10) | 59.52%(74) | 57.44%(97) | 289 |
| FBgn0030891 | CG7098 | Ada3 | X | 10.97%(13) | 34.53%(59) | 32.91%(101) | 556 |
| FBgn0020388 | CG4107 | Gcn5 | X | 17.34%(31) | 39.98%(162) | 51.91%(253) | 813 |
| FBgn0037555 | CG9638 | Ada2b-PB | X | 16.94%(27) | 42.88%(91) | 44.14%(174) | 555 |
| FBgn0051866 | CG31866 | Ada1 | X | 12.66%(8) | 39.94%(60) | 40.26%(73) | 308 |
| FBgn0031281 | CG3883 | SAF6 | X | 17.85%(45) | 33.33%(123) | 42.4%(153) | 717 |
| FBgn0036374 | CG17689 | Spt20 | X | 8.6%(25) | 25.2%(385) | 36.04%(1423) | 1873 |
| FBgn0037981 | CG3169 | Spt3 | X | 15.1%(17) | 42.19%(693) | 26.56%(149) | 384 |
| FBgn0030874 | CG6506 | Spt7 | X | 24.23%(32) | 33.43%(210) | 36.77%(186) | 359 |
| FBgn0026324 | CG3069 | TAF10b | X | 23.29%(4) | 23.29%(16) | 18.49%(5) | 146 |
| FBgn0011290 | CG17358 | TAF12 | X | 5.63%(1) | 36.25%(18) | 36.25%(25) | 160 |
| FBgn0000617 | CG6474 | TAF9 | 3.96%(1) | 32.73%(18) | 33.09%(72) | 36.69%(74) | 278 |
| FBgn0053554 | CG33554 | Tra1 (Nipped-A) | X | 16.15%(117) | 28.02%(344) | 45.33%(1364) | 3790 |
| FBgn0039067 | CG4448 | WDA | X | 24.5%(43) | 47.51%(240) | 48.86%(373) | 743 |
| FBgn0031420 | CG9866 | ATXN7 | X | 2.47%(5) | 18.02%(46) | 33.88%(108) | 971 |
| FBgn0000618 | CG15191 | E(y)2 | X | 34.65%(13) | 42.57%(25) | 51.49%(26) | 101 |
| FBgn0013717 | CG4166 | Nonstop | X | 4.84%(7) | 23.76%(64) | 28.59%(148) | 703 |
| FBgn0036804 | CG13379 | Sgf11 | X | 8.67%(4) | 44.9%(73) | 44.9%(139) | 196 |
| FBgn0040534 | CG11985 | SF3B5 | 83.53%(46) | 67.06%(89) | 67.06%(33) | 83.53%(32) | 85 |
| FBgn0035162 | CG13900 | SF3B3 | 54.12%(629) | 58.92%(2997) | 51.83%(639) | 51.02%(715) | 1227 |
| FBgn0031493 | CG3605 | SF3B2 (SF3b145) SF3B4 | 37.12%(146) | 47.4%(123) | X | X | 749 |
| FBgn0015818 | CG3780 | (SF3b149/Spx) | 28.24%(353) | 23.63%(410) | X | X | 347 |
| FBgn0035692 | CG13298 | SF3B6 (SF3b14a) | 49.59%(92) | 55.37%(201) | X | X | 121 |
| FBgn0031822 | CG9548 | PHF5A (SF3b14b) | 33.33%(8) | 7.21%(1) | X | X | 111 |
| FBgn0031266 | CG2807 | SF3B1 (SF3b155) | 52.76%(557) | 54.93%(1200) | X | X | 1340 |
| FBgn0266917 | CG16941 | SF3A1 (SF3a120) | 52.42%(313) | 49.74%(340) | X | X | 784 |
| FBgn0014366 | CG2925 | SF3A3 (SF3a60/noi) | 54.27%(337) | 56.26%(281) | X | X | 503 |
| FBgn0036314 | CG10754 | SF3A2 (SF3a66) | 46.21%(120) | 34.47%(222) | X | X | 264 |
| FBgn0262601 | CG5352 | SmB | 49.25%(354) | 29.65%(37) | 7.04%(2) | 10.55%(1) | 199 |
| FBgn0261933 | CG10753 | SmD1 (snRNP69D) | 52.42%(576) | 35.48%(56) | 16.13%(5) | 16.13%(2) | 124 |
| FBgn0261789 | CG1249 | SmD2 | 56.3%(323) | 47.9%(39) | X | X | 119 |
| FBgn0023167 | CG8427 | SmD3 | 35.76%(773) | 6.62%(7) | X | X | 151 |
| FBgn0261790 | CG18591 | SmE | 71.28%(524) | 67.02%(51) | 15.96%(1) | X | 94 |
| FBgn0000426 | CG16792 | SmF (DebB) | 48.86%(45) | 39.77%(13) | X | X | 88 |
| FBgn0261791 | CG9742 | SmG | 57.89%(196) | 28.95%(9) | X | X | 76 |
| FBgn0033210 | CG1406 | U2A | 61.89%(320) | 57.74%(114) | 23.02%(5) | X | 265 |
| FBgn0003449 | CG4528 | U2B (snf) | 43.06%(2520) | 29.17%(87) | X | X | 216 |
To determine whether these splicing proteins were indeed SAGA subunits, we purified tagged SF3B5 from S2 cell nuclear extract and analyzed the resulting complexes by MudPIT. FLAG-HA tandem affinity chromatography of SF3B5 co-purifies all of the known subunits of both the SAGA and the U2 snRNP complexes (Fig. 1a, Table 1, SF3B5 bait column). Although MudPIT analysis of purified SF3B5 identifies all known SAGA subunits, TAF12 is significantly under-represented and was identified in only one of the two technical replicate MudPIT analyses. It is unclear whether this is due to interference of the FLAG-HA epitope tag on SF3B5 with the binding of TAF12 to SAGA, or if SF3B5-SAGA complexes indeed contain reduced levels of TAF12. Despite this, our observations indicate that SF3B3 and SF3B5 are bona fide subunits of both SAGA and the U2 snRNP.
SF3B3 had previously been identified as a component of the mammalian SAGA complex [20, 21], but SF3B5 represents a novel subunit of SAGA. Additionally, both SF3B3 and SF3B5 were identified as interacting with mammalian Sgf29 (CCDC101) [44], which is a shared subunit of the Gcn5-containing SAGA and ATAC complexes [30]. Thus, these data collectively support that both SF3B3 and SF3B5 are subunits of metazoan SAGA. Notably, neither SF3B3 (Rse1) nor SF3B5 (Ysf3) associate at detectable levels with TAP-purified SAGA or SLIK in S. cerevisiae [45]. Thus, the presence of spliceosomal proteins within SAGA appears to be unique to higher eukaryotes.
SF3B3 and SF3B5 are independent subunits of SAGA and the U2 snRNP
Since SF3B3 and SF3B5 are known components of the SF3B complex within the larger U2 snRNP spliceosomal machinery [46, 47], we next asked whether SAGA and other U2 snRNP subunits were physically associated. To do this, we examined the SAGA-specific purifications for the presence of other SF3B proteins, SF3A complex subunits, Sm proteins, or the U2B protein itself. Additional components of the U2 snRNP were not identified reproducibly in purifications using the SAGA-specific subunits Ada2B (isoform PB), Spt3, Spt20, ATXN7, Ada1, SAF6 or WDA (Fig. 1a, Supplemental Table S1). This indicates that the majority of SAGA does not stably associate with other subunits of the SF3B complex or the larger U2 snRNP under the conditions used for our purifications. However, affinity purifications of the shared ATAC and SAGA subunit Sgf29 contained low numbers of peptides for many of the components of the U2 snRNP (Fig. 1a, Supplemental Table S1). This suggests that there is potential cross-talk between the U2 snRNP and a subset of SAGA complexes involving Sgf29. It is unlikely that this represents an interaction between the U2 snRNP and the alternative Gcn5-containing complex, ATAC, because SF3B3 and SF3B5 are not detected in purifications using ATAC-specific subunits as bait [48]. Further, when we purify the U2 snRNP using U2B as bait protein, we only identify proteins from the U2 snRNP spliceosomal complex including SF3B and SF3A complex subunits (Fig. 1a, Table 1). Although one peptide for TAF9 is identified in the U2B purification, we do not identify peptides from any other SAGA subunits including Sgf29 (Table 1). Thus, SF3B3 and SF3B5 are independently associated with SAGA and the U2 snRNP, and do not mediate a stable interaction between these two complexes under the conditions used for our purifications.
SF3B5-containing SAGA complexes acetylate histones
We next asked whether SF3B5-purified SAGA complexes had histone acetyltransferase (HAT) activity. To do this, we performed HAT assays using SF3B5-purified SAGA complex on HeLa core histones as substrate. SF3B5-purified SAGA demonstrated HAT activity on core histones (Fig. 1b), predominantly on histone H3 and to a lesser extent on histone H4 (Fig. 1c). This HAT activity shows a similar histone preference to SAGA purified through SAGA-specific subunits such as SAF6 or WDA [40, 49]. Since the SF3B5-complex HAT assays were performed as part of the same set of HAT assays described in Weake et al. (2009) for WDA and SAF6-purified SAGA, we can compare the HAT activity of these SAGA complexes on histones [40]. Notably, the level of HAT activity of SF3B5-purified SAGA is 2 – 3 fold lower than that of SAGA purified using the core SAGA subunits WDA or SAF6 as bait proteins. However, the SF3B5-purified complex also contains much lower levels of Gcn5 relative to WDA or SAF6-purified SAGA because SF3B5 also co-purifies components of the U2 snRNP in addition to SAGA (compare dNSAF values for Gcn5 in each purification in Supplemental Table S1). Our data therefore indicate that SF3B5-containing SAGA complexes contain the full complement of SAGA subunits and are capable of acetylating histones in a SAGA-specific pattern, suggesting that these complexes purified through SF3B5 represent functional SAGA complexes.
The association of SF3B3 and SF3B5 with SAGA does not require RNA
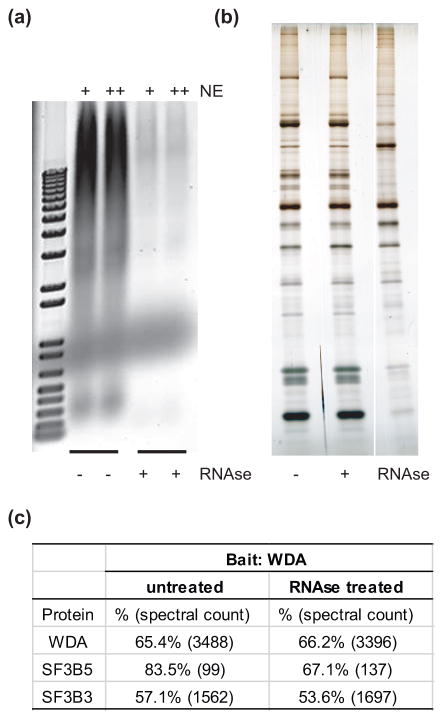
Next, we sought to determine if the association of SF3B3 and SF3B5 with SAGA requires the presence of RNA since the U2 snRNA is a core component of the U2 snRNP [50]. To do this, we isolated SAGA from S2 cell nuclear extracts in the presence and absence of ribonuclease (RNAse) using tandem FLAG-HA affinity chromatography against the bait protein WDA. RNAse treatment reduces nucleic acids in the soluble nuclear extract to levels that are not detectable by ethidium bromide staining following agarose gel electrophoresis (Fig. 2a). However, the composition of SAGA purified via WDA from nuclear extract treated with RNAse appears identical to SAGA purified in the absence of RNAse by SDS-PAGE and silver staining (Fig. 2b). To examine whether SF3B3 and SF3B5 remained present in SAGA following RNAse treatment, we examined the composition of SAGA purified in the presence of RNAse by MudPIT analysis. Notably, similar levels of peptides as determined by spectral counts for SF3B3 and SF3B5 are observed in the SAGA purifications from nuclear extract treated with RNAse relative to the untreated nuclear extract (Fig. 2c). Thus, the association of SF3B3 and SF3B5 with SAGA does not require RNA. This finding is consistent with the lack of annotated RNA-interacting domains in SF3B3 and SF3B5, and with observations that suggest that SF3B3 and SF3B5 are not directly involved in pre-mRNA branch-point recognition [28].
Fig 2. SF3B3 and SF3B5 bind SAGA independent of RNA.
(a, b) SAGA was FLAG-HA purified from S2 cells using WDA as bait protein following treatment of the soluble nuclear extract with RNAse A. An ethidium bromide stained agarose gel of the soluble nuclear extract (NE, 10 μL, + and 20 μL, ++) used for immunoprecipitation with and without RNAse treatment (+/− respectively) is shown in panel (a), and a silver stained SDS-PAGE gel of the purified WDA-complexes +/− RNAse treatment is shown in panel (b). (c) Peptides from SF3B3 and SF3B5 are identified at similar levels in SAGA purifications from S2 cells using WDA as bait in the presence and absence of RNAse treatment. Sequence coverage (%) and number of peptides (spectral count) are shown for each polypeptide, relative to the bait protein WDA.
SF3B3 and SF3B5 interact with Sgf29 and Spt7 in SAGA
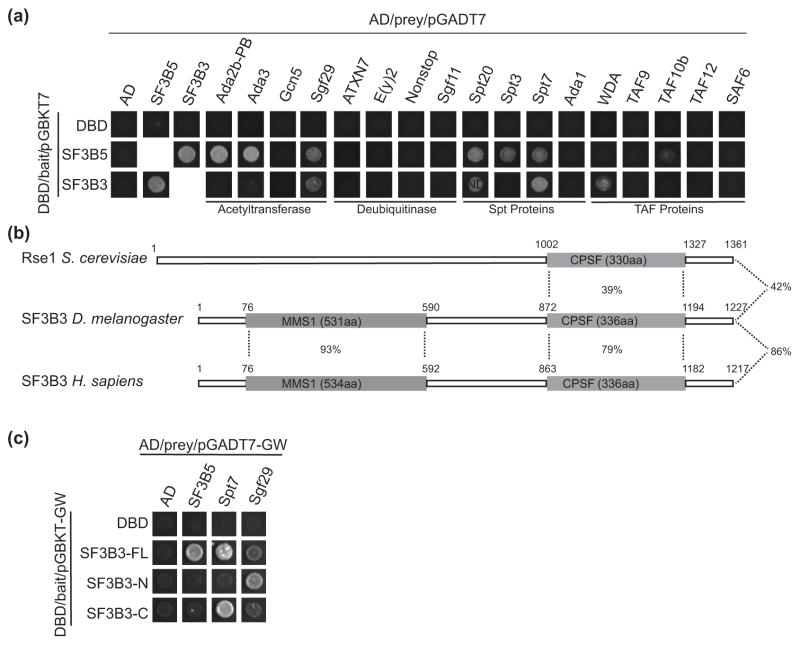
Since the incorporation of SF3B3 and SF3B5 within SAGA is independent of RNA, we next sought to identify the protein subunits in SAGA that interacted with these spliceosomal proteins. We hypothesized that SF3B3 and SF3B5 would interact with SAGA-specific subunits, since these proteins are not found in the related ATAC complex [48]. To test the pair-wise interaction between SF3B3, SF3B5 and each SAGA subunit, we performed a yeast two-hybrid assay with 17 of the characterized Drosophila SAGA subunits as prey, and either SF3B3 or SF3B5 as bait. We did not analyze the SAGA subunit Tra1 (NippedA), which is also a component of the Drosophila Tip60 complex [51], in this assay due to the large size of its coding sequence and high probability of auto-activation. When we examined the pair-wise interaction of SF3B3 and SF3B5 by yeast two-hybrid analysis, we observed a strong reciprocal interaction between SF3B3 and SF3B5 (Fig. 3a). Importantly, neither SF3B3 nor SF3B5 auto-activate transcription of the reporter genes since co-expression of either SF3B3 or SF3B5 fused to the Gal4 DNA-binding domain (DBD) with the plasmid encoding the Gal4 Activating Domain (AD) alone does not result in growth on selective media (Fig. 3a, left column). We next examined the interaction of SF3B3 and SF3B5 with the 17 SAGA subunits. The SAGA subunits assayed also do not auto-activate reporter gene transcription because co-expression of SAGA subunits fused to the AD with the plasmid encoding the DBD alone does not result in growth on selective media (Fig. 3a, top row). Interestingly, we observed interactions between SF3B5 and several proteins within SAGA; Ada2b, Ada3, Sgf29, Spt20, Spt3 and Spt7 (Fig. 3a). We observed fewer interactions between SF3B3 and SAGA subunits, with only Sgf29, Spt7, and WDA showing growth on selective media (Fig. 3a). The binding of Spt20 to SF3B3 was unable to be determined because we did not observe a consistent growth phenotype. Since Sgf29 and Spt7 were identified as interacting with both SF3B3 and SF3B5, these proteins provide the most likely candidates for SAGA subunits that mediate the incorporation of these two spliceosomal subunits into the SAGA complex. This finding is not consistent with our hypothesis since Sgf29 is also a subunit of the Gcn5-containing ATAC complex. Because the yeast two-hybrid assay tests binding of proteins in vivo, we cannot exclude the possibility that the interaction between Drosophila SAGA subunits and SF3B3 or SF3B5 is mediated by endogenous yeast proteins. Despite this caveat, the results from this yeast two-hybrid assay indicate that Sgf29 and Spt7, potentially in conjunction with some of the HAT module subunits and core components Spt3, Spt20 and WDA, provide a binding surface for SF3B3 and SF3B5 within Drosophila SAGA.
Fig. 3. SF3B3 and SF3B5 interact with Sgf29 and Spt7 by yeast two-hybrid analysis.
(a) Yeast two-hybrid assay was performed to test the interaction of SAGA subunits fused to the Gal4 activating domain (AD) with SF3B3 or SF3B5 fused to the Gal4 DNA binding domain (DBD). Empty plasmids containing only the activating domain (AD, left column) or DNA binding domain (DBD, top row) were used to test for auto-activation of each protein. Approximately 30,000 cells were spotted on media lacking leucine, tryptophan, adenine and histidine for each tested interaction between AD- and DBD-fusion proteins (boxes). Images are shown for representative spots for each tested interaction (black boxes) indicating growth or no growth. ND, not determined. (b) Protein alignment of SF3B3 in S. cerevisiae (Rse1), D. melanogaster and H. sapiens. Motifs were identified using Pfam and are shown in grey boxes with the length of each domain indicated in parenthesis and percentage similarity for domains between species shown flanked by dotted lines. The numbers above the proteins denote the amino acids in the sequence showing placement of the domains. Overall percent sequence similarity for each full-length protein pair is shown to the right of the schematic. (c) Yeast two-hybrid assay was performed as described in panel a. Plasmids used in this panel are gateway compatible vectors denoted “GW”. SF3B3-FL contains the full length SF3B3 construct, SF3B3-N contains amino acids 1 - 746 and SF3B3 contains amino acids 747 - 1227 in the pGBKT7-GW plasmid.
The interaction of SF3B3 with Sgf29 and Spt7 is mediated by different domains
Since the association of SF3B3 and SF3B5 with SAGA is observed in Drosophila and humans, but not in S. cerevisiae, we wondered whether there were differences in the yeast and metazoan versions of these proteins that might account for this differential interaction. To examine this possibility, we first compared yeast, Drosophila and human SF3B5 using BLAST [52]. Yeast Ysf3 shares 53% sequence similarity with Drosophila SF3B5 and 50% sequence similarity with human SF3B5, while Drosophila and human SF3B5 share 91% sequence similarity. Additionally, there are no identifiable domains in any of the SF3B5 orthologs. Next, we compared yeast, Drosophila and human SF3B3. While Drosophila and human SF3B3 share 86% sequence similarity, yeast Rse1 shares 42% sequence similarity with Drosophila SF3B3. However, the N-terminal region of Drosophila and human SF3B3 contains a domain that is absent in yeast: the Mono-functional DNA-alkylating methyl methanesulfonate (MMS1) domain (Fig. 3b). The MMS1 domain is found in proteins that protect against replication-dependent DNA damage [53]. A second domain, the cleavage and polyadenylation specificity factor (CPSF) domain, whose namesake is necessary for proper 3’ end processing and pre-mRNA splicing [54], is conserved in all three species (Fig. 3b). Since the MMS1 domain of Drosophila and human SF3B3 is not present in yeast SF3B3, we hypothesized that this domain is required for binding of SF3B3 to SAGA in Drosophila.
To test if the N-terminal region of Drosophila SF3B3 that contains the MMS1 domain was necessary for its binding within SAGA, we repeated our yeast two-hybrid analysis with the N- and C-terminal domains of SF3B3 as bait proteins, and SF3B5, Spt7 and Sgf29 as prey proteins. The bait proteins used in this assay consist of full length SF3B3 (SF3B3-FL), the N-terminal domain of SF3B3 (SF3B3-N, aa1 - 746) and the C-terminal domain of SF3B3 (SF3B3-C, aa747 – 1227). Consistent with our previous yeast two-hybrid analysis, we observe growth on selective media when SF3B5, Sgf29 or Spt7 are co-expressed with full-length SF3B3. However, surprisingly we did not observe an interaction between SF3B5 and either the SF3B3 N- or C-terminal regions (Fig. 3c), indicating that neither of these domains are sufficient for this interaction. This lack of interaction is unlikely to be due to expression problems, since we observe interactions between the SF3B3 C-terminal domain (SF3B3-C) and Spt7, and the SF3B3 N-terminal domain (SF3B3-N) and Sgf29 respectively. Thus, in the yeast two-hybrid assay, the C-terminal region of SF3B3 that contains the conserved CPSF domain is sufficient to interact with Spt7, whereas the N-terminal region of SF3B3 that contains the metazoan-specific MMS1 domain is sufficient to interact with Sgf29. This unexpected result indicates that the presence of the MMS1 domain in metazoan SF3B3 is not sufficient to account for the presence of SF3B3 in Drosophila SAGA but not yeast SAGA. Thus, both the N- and C-terminal regions of SF3B3 contribute to its association with SAGA through independent binding to Spt7 and Sgf29.
SF3B5 is necessary for proper development and cell viability
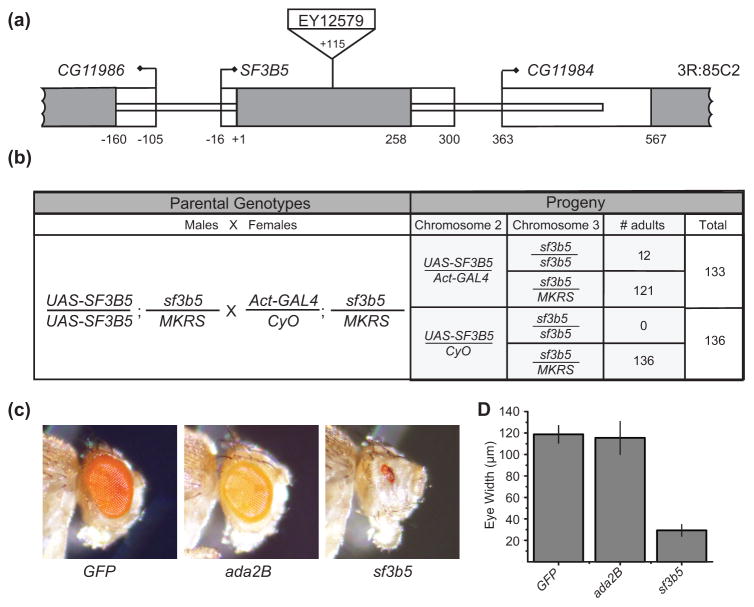
Whereas SAGA is not essential for viability in S. cerevisiae, mutations that disrupt SAGA in Drosophila are lethal during the larval or early pupal stages of development [40, 41, 49, 55–59]. To determine if SF3B5 was also required for development, we sought to identify a loss of function mutation in the SF3B5 gene. We identified a P-element insertion in the coding region of the intronless SF3B5 gene, EY12579 [60] (Fig. 4a). We were only able to identify flies carrying the balancer chromosome in this stock, suggesting that this insertion is homozygous lethal. We will hereafter refer to flies carrying this EY12579 transposon insertion as sf3b5EY12579 mutant flies (Supplemental Table S2).
Fig 4. SF3B5 is necessary for organismal and cell viability.
(a) Schematic representation of the SF3B5 (CG11985) locus on chromosome 3R showing the position of the P-transposon EY12579. The single exon of the SF3B5 gene is represented by the grey box. Translated sequences are filled with grey, and 5' and 3' untranslated regions are shown as open boxes. The +1 position corresponds to the ATG of the translation start site. (b) Genetic crosses were conducted with flies carrying the UAS-SF3B5 rescue construct or the actin5C-GAL4 driver on chromosome 2, and the sf3b5EY12579 allele on chromosome 3. Surviving adult progeny were scored for the presence of the balancer chromosomes using the curly wing phenotype (CyO) and the bristle marker stubble (MKRS). The number of surviving adult progeny and the total number of flies scored are shown for each genotype. (c) Mutant fly eyes were generated using the GMR-hid technique with the following genotypes, Ubi-nlsGFP (wild type), ada2b and sf3b5EY12579. A representative image from a single male fly of each indicated genotype is shown. (d) Mean eye widths of mutant fly eyes generated as described in panel (c) were determined for each indicated genotype. The widths of four separate fly eyes from four independent animals (one eye per animal) were measured, and standard deviation is indicated by error bars. Full genotypes of flies are shown in Supplemental Table S2.
To determine if the lethality in the sf3b5EY12579 flies resulted from loss of SF3B5 function, we generated transgenic flies that express wild-type SF3B5 under GAL4/UAS regulatory control (UAS-SF3B5) [61]. We then crossed flies carrying the sf3b5EY12579 allele and the UAS-SF3B5 transgene with sf3b5EY12579 flies that also ubiquitously express GAL4 under control of the Actin5C promoter as outlined in Figure 4b. Since the UAS-SF3B5 transgene and actin5C-GAL4 driver are on chromosome 2, and the sf3b5EY12579 allele is on chromosome 3, there are four different potential phenotypes in the resulting progeny from this cross: Half of the progeny will have the actin5C-Gal4 driver on chromosome 2 and will therefore express UAS-SF3B5 ubiquitously, while the other half of the progeny will have the CyO balancer and will not express UAS-SF3B5. In each of these halves of the resulting progeny, flies will also either be homozygous or heterozygous for the sf3b5EY12579 allele on chromosome 3, which can be distinguished by the presence of the stubble marker on the MKRS balancer chromosome. As expected since the sf3b5EY12579 allele is homozygous lethal, 100% of CyO progeny from this cross contained the stubble marker, indicating presence of the balancer chromosome (Fig. 4b). If expression of the UAS-SF3B5 transgene is sufficient to rescue viability of the sf3b5EY12579 mutant, then we would expect to see adult progeny that lack the stubble marker only in those flies that also lack the CyO balancer chromosome, as determined by the curly wing marker. When we examined the flies that lacked the CyO balancer chromosome, we found that 9% of flies without the CyO balancer also lacked the stubble marker, indicating that expression of SF3B5 rescues lethality of the sf3b5EY12579 allele (Fig. 4b). Thus we conclude that the lethality associated with the sf3b5EY12579 allele is due to loss of function of SF3B5.
Mutations in other SAGA subunits result in lethality in different stages of larval development, most probably due to residual maternal load of mRNAs for these subunits. For example, ada2b and nonstop homozygotes die as pupae, gcn5 homozygotes die as third instar larvae, and saf6 and wda homozygotes die as second instar larvae [40, 49, 57, 62, 63]. To compare sf3b5EY12579 flies with other SAGA mutants, we sought to determine the developmental stage at which homozygous sf3b5EY12579 flies die. To do this, we generated flies that carried the sf3b5EY12579 allele over a balancer chromosome marked with the green fluorescent protein (GFP). We then identified homozygous sf3b5EY12579 embryos by lack of GFP expression. We observed growth of homozygous sf3b5EY12579 embryos until the first instar larval stage, but we did not observe any further growth, indicating that loss of SF3B5 results in lethality at the first instar larval stage of development.
Since SF3B5 is necessary for viability on an organismal level, we next wanted to determine if SF3B5 is also necessary for cell viability. In yeast, YSF3 is an essential gene for growth, suggesting that the function of SF3B5 in splicing plays a critical role for cell survival [24, 25]. To test whether SF3B5 is necessary for cell viability in Drosophila, we generated mosaic flies that are heterozygous for sf3b5EY12579 in all tissues except the eyes, in which the cells are homozygous for the sf3b5EY12579 allele [64]. Using this approach, we would expect to observe full or partial eye ablation in flies carrying homozygous sf3b5EY12579 cells in the eye if SF3B5 is necessary for cell viability or cell division. As a control, we generated eyes carrying two copies of a non-essential transgene, GFP, on an otherwise wild-type chromosome. The eyes of these control flies were similar to those of wild-type flies (Fig. 4c). However, flies homozygous for sf3b5EY12579 showed dramatic eye ablation (Fig. 4c). To quantify this eye ablation, we measured the width of these eyes in each genotype (n = 4) and found that sf3b5EY12579 eyes were approximately four-fold smaller than those of the wild-type (GFP) control (Fig. 4d). Since these results indicate that SF3B5 is likely to be required for cell viability, we next asked if SAGA subunits were also required for cell viability. To do this, we generated mosaic flies using the same approach that contain eyes homozygous for a mutation in Ada2B. Ada2b is a SAGA-specific subunit that interacts with Gcn5 and is necessary for H3 acetyltransferase activity by SAGA [62, 65]. In contrast to sf3b5EY12579, ada2b eyes are similar in size to the GFP control (Fig. 4c, d). Similar results were observed for nonstop mutations that disrupt the deubiquitinase activity of SAGA (data not shown). Thus, we conclude that SF3B5, but not other SAGA subunits, is required for cell viability. This finding is consistent with the essential role of SF3B3 (Rse1) and SF3B5 (Ysf3) in yeast, and suggests that the requirement of SF3B5 for cell viability in Drosophila results from its function in splicing rather than in SAGA.
SF3B5 is not necessary for H3 acetylation
Although SF3B5’s function in splicing is likely to be more critical for cell function, we wondered whether SF3B5 is also required for any of the known activities of SAGA. SAGA has well characterized histone modifying activities including its HAT activity toward predominantly histone H3 Lysine 9 (H3K9ac) and Lysine 14, and deubiquitinase activity against ubH2B. To determine if SF3B5 is required for SAGA’s HAT or deubiquitinase activities, we compared levels of H3K9ac and ubH2B in sf3b5EY12579 mutant larvae with those of wild-type (OregonR or w1118) larvae. As controls, we also examined ubH2B and H3K9ac levels in nonstop and wda mutants, which disrupt SAGA deubiquitinase activity and HAT activity resulting in elevated ubH2B levels in late third instar larvae, and decreased levels in H3K9ac in embryos respectively [49, 58].
To do this, we first acid extracted histones from wild-type, sf3b5EY12579 and nonstop first instar larvae and performed western blotting analysis using antibodies against H3K9ac and histone H2B (Fig. 5a). Based on this analysis, we find that sf3b5EY12579 larvae show no change in global H3K9ac levels as compared to wild-type first instar larvae (H3K9ac/H2B ratio is 113% of wild-type levels) (Fig. 5a). In contrast, mutations in wda that disrupt SAGA HAT activity [49] result in a clear decrease in H3K9ac to 51% of wild-type levels (H3K9ac/H2B ratio) by the end of embryogenesis (Fig. 5a). Thus, we conclude that SF3B5 is not required for SAGA’s HAT activity.
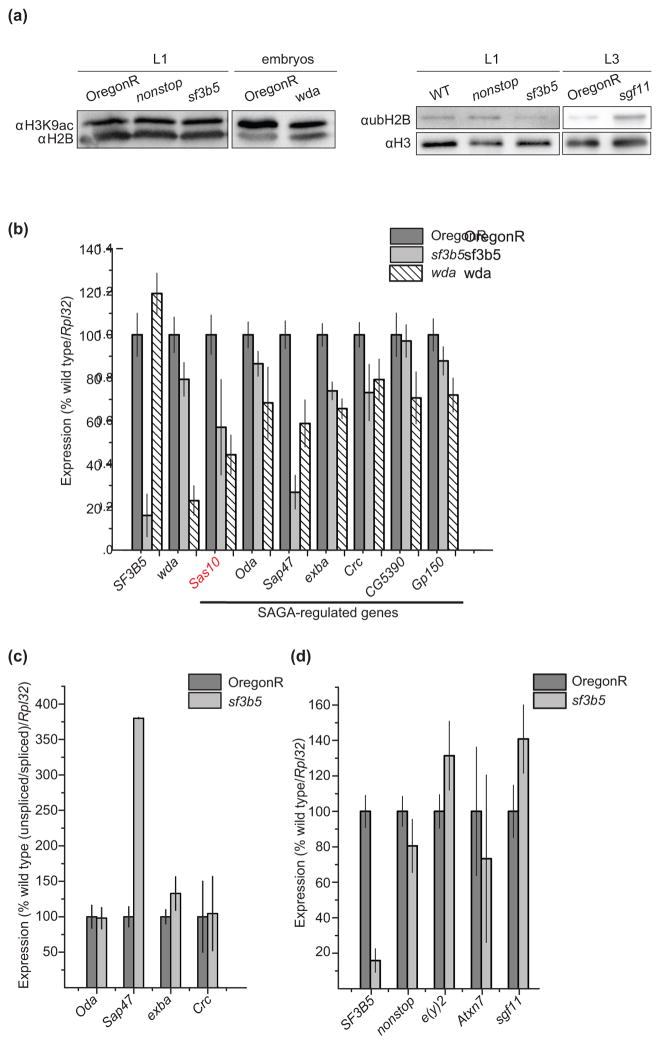
Fig 5. SF3B5 is necessary for expression of a subset of SAGA-regulated genes independent of histone acetylation and splicing.
(a) Acid-extracted histones from wild-type (OregonR or w1118, WT), nonstop and sf3b5EY12579 first instar larvae (L1), OregonR and wda embryos, and OregonR and sgf11 third instar larvae (L3) were analyzed by SDS-PAGE and western blotting using antibodies against H3K9ac and H2B, or ubH2B and H3. (b) RNA was isolated from OregonR (wild-type), sf3b5EY12579 and wda 18 - 24 h embryos and qRT-PCR was performed on oligodT-reverse transcribed cDNA. Mean expression levels are normalized to RpL32 and shown relative to OregonR, which is set as 100%. Error bars denote standard error of the quotient for four biological experiments, and p-values for each comparison determined using ANOVA and Tukey’s honest significant difference (HSD) test are shown in Supplemental Table S3. (c) qRT-PCR was performed as described in panel (b) with random hexamer-reverse transcribed cDNA and primers designed to amplify exon/intron junctions to detect unspliced transcripts. Mean expression levels of the ratio of unspliced to spliced transcripts are normalized to RpL32, and shown relative to OregonR, which is set as 100%. Error bars denote standard error of the quotient for four biological experiments, and p-values for each comparison are shown in Supplemental Table S3. (d) qRT-PCR was performed on OregonR and sf3b5 first instar larvae as described for panel (b). Error bars denote standard error of the quotient for three biological experiments, and p-values for each comparison are shown in Supplemental Table S3.
Next, we examined ubH2B levels in sf3b5EY12579 larvae using antibodies specific for ubH2B relative to histone H3. As a control for the specificity of the ubH2B antibody, we examined ubH2B levels in acid-extracted histones from sgf11 third instar larvae. Histones from sgf11 third instar larvae have ~370% of ubH2B relative to the wild type, indicating that we can detect an increase in ubH2B in SAGA deubiquitinase mutants [58]. However, neither sf3b5EY12579 nor nonstop first instar larvae show strong increases in ubH2B levels relative to wild-type larvae (ubH2B/H3 ratios of 99% and 130% of wild-type levels in sf3b5EY12579 and nonstop respectively) (Fig. 5a). Previously, we were also unable to detect an increase in ubH2B levels in sgf11 embryos [36]. These data suggest that it is not possible to detect strong global changes in the accumulation of ubH2B at first instar larvae when SAGA deubiquitinase activity is defective, potentially due to the lag in accumulation of this modification. Thus, based on this analysis, we cannot conclude definitively whether SF3B5 is required for SAGA deubiquitinase activity.
SF3B5 is necessary for SAGA-activated expression of some SAGA-regulated genes
Although SF3B5 is not required for SAGA’s HAT activity, it is possible that SF3B5 could function in transcription coactivation by SAGA independent of histone modification. Several studies in yeast have shown that SAGA is required for recruitment of the general transcription factors such as TBP to promoters independent of its HAT activity [66, 67]. In addition, the Drosophila SAGA subunit SAF6 is required for SAGA-regulated gene expression independent of either HAT or deubiquitinase activity [40]. Therefore, we sought to determine if SF3B5 is necessary for SAGA’s function in activating gene expression. To test this, we examined transcript levels of SAGA-regulated genes in sf3b5EY12579 embryos using quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis. We had previously identified several SAGA-regulated genes that were co-regulated by the core SAGA subunits SAF6 and WDA in late stage embryos [40]. Thus, we compared transcript levels of a subset of these SAGA-regulated genes in the sf3b5EY12579 embryos with those in wda and wild-type (OregonR) embryos.
First, we examined transcript levels of the SF3B5 and wda genes in each genotype. We observe lower transcript levels of SF3B5 and wda genes in the sf3b5EY12579 and wda mutants respectively, as compared to the wild type (OregonR). However, levels of RpL32, which has previously been shown not to be regulated by SAGA [36, 40, 68], were similar in all three genotypes (99% and 127% of wild type in wda and sf3b5EY12579 respectively). Notably, sf3b5EY12579 embryos have about 80% of wild-type levels of transcript encoding the core SAGA subunit WDA (Fig. 5b). However, it is unlikely that this decrease in transcript results in a strong decrease in WDA protein levels since in contrast to wda mutants, H3K9ac levels are not reduced in sf3b5EY12579 larvae (Fig. 5a).
Next, we examined transcript levels of six SAGA-regulated genes that were previously shown to require WDA for full expression in late stage embryos [40]. Notably, four out of the six genes, Oda, Sap47, exba and Crc, were downregulated in both sf3b5EY12579 and wda embryos relative to the wild type (Fig. 5b, Supplemental Table S3). While most of these genes were downregulated to similar levels in both mutants relative to the wild type, Sap47 showed significantly stronger downregulation in sf3b5 relative to wda embryos (Supplemental Table S3). Interestingly, two of the seven genes examined, CG5390 and Gp150, were significantly downregulated (p-value < 0.05) in wda embryos but not in sf3b5EY12579 embryos (Fig. 5b, Supplemental Table S3). This suggests that SF3B5 is required for full expression of a subset of SAGA-regulated genes.
Since splicing affects transcript levels, SF3B5 could be required for full expression of these SAGA-regulated genes either through its role in SAGA or in the U2 snRNP. Thus, to test whether loss of SF3B5 affected splicing at these SAGA-regulated genes, we examined levels of unspliced transcripts relative to spliced transcripts at Oda, Sap47, exba and Crc (Fig. 5c). To do this, we generated cDNA using random hexamer primers and performed qRT-PCR with primers that anneal within an exon and its adjacent intron to amplify an intron/exon boundary (unspliced transcript). As a control, we used primers that anneal within two adjacent exons to amplify the spliced transcript. Notably, we observe a large increase in unspliced Sap47 transcript in sf3b5EY12579 embryos relative to the wild-type control (Fig. 5c). This is consistent with the stronger reduction in Sap47 expression in sf3b5EY12579 embryos relative to wda embryos, and suggests that SF3B5 is required for proper splicing of this SAGA-regulated gene. However, in contrast to the results observed for Sap47, we do not detect an increase in unspliced transcript levels for three of the SAGA-regulated genes tested, Oda, exba and Crc (Fig. 5c). This result indicates that SF3B5 is required for full expression of these three genes through its role in SAGA rather than in the U2 snRNP.
Next, we asked if SF3B5 would only function to regulate gene expression in the context of active splicing. To test this, we examined transcript levels of Sas10, which does not contain any introns, in sf3b5EY12579 embryos. Levels of Sas10 transcripts were significantly lower (p-value < 0.05, Supplemental Table S3) in both sf3b5EY12579 and wda embryos relative to the wild type (Fig. 5b). This result indicates that SF3B5 can regulate gene expression at genes that lack introns. However, since the SF3B complex is known to play a role in pre-mRNA processing events of intronless genes [69], and since splicing factors can be required for nuclear export of intronless mRNAs [70], it is possible that SF3B5 regulates Sas10 expression via U2 snRNP-mediated processing rather than SAGA-regulated transcription. Further studies would be required to distinguish between these possibilities.
Together, our observations indicate that SF3B5 is required for proper transcriptional activation of a subset of SAGA-regulated genes, independent of active splicing. It is not clear however, why genes respond differently to SF3B5 relative to other SAGA subunits. Future global analysis of the transcriptome of sf3b5EY12579 embryos relative to other SAGA mutants may provide insight into the role that SF3B5 plays in SAGA-activated gene expression.
One potential indirect explanation for the decrease in SAGA-regulated gene expression in sf3b5EY12579 mutants is if SF3B5 is required for splicing of SAGA subunits, thereby affecting levels of these proteins. However, our data argue against an indirect role for SF3B5 in affecting SAGA coactivation activity through regulating levels of SAGA subunits such as WDA. Whereas mutations in wda reduce global levels of H3K9ac in late stage embryos (Fig. 5a), we do not observe a decrease in global levels of histone acetylation in sf3b5EY12579 embryos, suggesting that SAGA remains intact and functional with regards to HAT activity and recruitment to gene promoters. In addition, transcript levels of the genes encoding the deubiquitinase module of SAGA, e(y)2, nonstop, sgf11 and Atxn7, are not reduced in sf3b5EY12579 first instar larvae (Fig. 5d). Thus, we conclude that SF3B5 is likely to be required directly for expression of a subset of SAGA-regulated genes.
SF3B5 is required for SAGA-regulated gene expression independent of histone acetylation and splicing
In this study we identify the spliceosomal components SF3B3 and SF3B5 as subunits of Drosophila SAGA. A previous study had identified a potential role for SF3B3 in the recruitment of SAGA to UV-damaged DNA [20] while this finding was not supported in a second study [21]. However, a second component of the SF3B complex, SF3B1, interacts with BRCA1 following DNA damage to enhance splicing of BRCA1-target genes [71], also supporting crosstalk between the DNA damage and spliceosomal machinery. Here, we show that SF3B5 is required for proper development and cell viability in Drosophila. Notably, our findings indicate that SF3B5 is required for SAGA-mediated transcriptional activation at a subset of SAGA-regulated genes, independent of SAGA’s HAT activity. These observations therefore place SF3B5 in a similar functional role in SAGA as SAF6, which is required for transcription activation independent of both of SAGA’s histone modifying activities [40]. We cannot exclude the possibility that SF3B5 is also required for SAGA deubiquitinase activity, since ubH2B levels do not accumulate to sufficient levels by the larval stage examined. Future studies to examine the requirement of SF3B5 in ubH2B-deubiquitination will be of interest because ubH2B has been shown to be important for co-transcriptional splicing. In humans, the ubH2B histone deubiquitinase USP49 is required for proper splicing of a large number of genes [72].
Based on our findings, we conclude that SF3B3 and SF3B5 play dual roles within the cell in splicing and in transcription activation by SAGA. There are several precedents for SAGA subunits that function in other complexes. For example, the HAT component of SAGA, Gcn5, is shared with the transcription coactivator complex, ATAC [32]. In addition, Sus1 (Drosophila E(y)2), which is required for SAGA deubiquitinase activity, also functions in RNA export as part of the TRanscription-Export (TREX) complex [73, 74]. Our findings indicate that similarly to these other SAGA subunits that are shared between multiple complexes, SF3B3 and SF3B5 have independent roles in the spliceosome and in SAGA. Despite this independent role, we cannot formally exclude the possibility that these subunits mediate transient interactions between SAGA and the U2 snRNP during co-transcriptional splicing. Further studies to examine the SAGA-specific role of SF3B3 and SF3B5 by generating mutations that disrupt the interaction of these components with SAGA but not the U2 snRNP will be necessary to fully define the role of these spliceosomal proteins in metazoan SAGA.
Materials and Methods
Generation of stable cell lines
Drosophila S2 cells were maintained in Hyclone SFX media at 25°C. Stable S2 cell lines expressing Spt3 (CG3169, NP_650146), Spt20 (CG17689, NP_648659), SF3B5 (CG11985, NP_652189.1) and U2B (sans fille; CG4528, NP_511045.1) in the pRmHa3-CHA2FL2 vector were generated by co-transfection with pCoBlast (1:10 ratio) using FuGENE HD transfection reagent (Promega). Selection was carried out in SFX media supplemented with 10% Fetal Bovine Serum in the presence of 25 - 30 μg/mL blastidicin for 2 – 4 weeks.
Affinity purification and MudPIT analysis
Tandem FLAG-HA affinity purification and MudPIT analysis was conducted as described previously [40]. Stable S2 cell lines expressing FLAG-HA tagged bait proteins in the pRmHa3-CHA2FL2 vector were grown in SFX media with low/no copper induction, and soluble nuclear extracts were prepared from 4 L of cells grown to a density of 1 x 107 cells/mL. Cells were harvested by centrifugation, washed in 10 mM HEPEs [Na+], pH 7.5; 140 mM NaCl, and resuspended in 40 mL of Buffer I (15 mM HEPEs [Na+] pH 7.5; 10 mM KCl, 5 mM MgCl2; 0.1 mM EDTA; 0.5 mM EGTA; 350 mM sucrose; supplemented with 20 μg/mL leupeptin, 20 μg/mL pepstatin and 100 μM PMSF). Nuclei were released by Dounce homogenization (40 strokes with loose pestle) and pelleted by centrifugation at 10,400 x g for 15 min at 4ºC. Nuclei were washed once with Buffer I and then resuspended in 20 mL of Extraction Buffer (20 mM HEPEs [Na+], pH 7.5; 10% glycerol; 350 mM NaCl; 1 mM MgCl2; 0.1% TritonX-100; supplemented with 20 μg/mL leupeptin, 20 μg/mL pepstatin and 100 μM PMSF). Nuclei were incubated in Extraction Buffer for 1 h at 4ºC with rotation, and the insoluble chromatin fraction was pelleted by sequential centrifugation steps at 18,000 x g 10 min 4ºC and 40,000 rpm 1.5 h 4ºC (50.2Ti rotor, Beckman). For affinity purification, soluble nuclear extracts were diluted to a final salt concentration of 150 mM NaCl. Where indicated, 250 μg/mL RNAse A was added to soluble nuclear extract prior to immunoprecipitation. Nuclear extracts were incubated with 200 μL (packed bead volume) of anti-FLAG M2 agarose (Sigma) for 4 h – 16 h with rotation, then washed 3 times in Extraction Buffer containing 150 mM NaCl. FLAG-bound proteins were eluted 4 x with 200 μL each of Extraction Buffer (150 mM NaCl) containing 0.5 mg/mL FLAG3 peptide (3XFLAG: NH2-DYKDDDDKGDYKDDDDKGDYKDDDDK-COOH, synthesized by Macromolecular Core Facility, Penn State College of Medicine) for 10 min at 25°C. Pooled FLAG-elutions were incubated with 60 μL (packed bead volume) of EZview anti-HA affinity gel (Sigma) for 4 h – 16 h with rotation, then washed 3 times in Extraction Buffer containing 150 mM NaCl. HA-bound proteins were eluted 6 x with 150 μL each of Extraction Buffer (150 mM NaCl) containing 0.2 mg/mL HA3 peptide (3XHA: NH2-YPYDVPDYAGYPYDVPDYAGYPYDVPDYA-COOH, synthesized by Macromolecular Core Facility, Penn State College of Medicine) for 10 min at 25°C. HA-elutions were pooled, and 5 – 10% o f the pooled elutions (~200 μL) were treated with 0.1 U benzonase for 30 min at 37ºC, and then precipitated with 200 μL of ice-cold 100 mM Tris-HCl, pH 8.5 and 100 μL ice-cold trichloroacetic acid for 16 – 24 h at 4ºC. Precipitated proteins were collected by centrifugation at > 20,000 x g for 30 min at 4ºC and washed twice in 1 mL of ice-cold acetone, followed by centrifugation at 20,000 x g for 10 min at 4ºC. The identity and relative abundance of proteins present in the tandem FLAG-HA affinity purifications was determined using MudPIT [39]. Relative protein levels were estimated using dNSAFs calculated for each protein as described in [75, 76]. Merged data are shown representing two technical replicates of the MudPIT analysis. Heat maps were generated using MultiExperiment Viewer (MeV) software.
HAT assays
HAT assays were performed using FLAG-purified SF3B5-complexes and 500 ng HeLa core histones as substrate as previously described [77]. Each 30 μL HAT reaction contains 50 mM Tris-HCl, pH 8.0, 5% glycerol, 0.1 mM EDTA, pH 8.0, 1 mM DTT, 1 mM PMSF, 0.25 μCi 3H Acetyl Coenzyme A (NET290250UC, PerkinElmer), +/− 500 ng HeLa core histones +/− FLAG-purified SF3B5-complex. Reactions were incubated at 30°C for 30 min and 15 μL was spotted onto P81 phosphocellulose filter paper, washed three times for 5 min each in 50 mM NaHC03-NaCO3 buffer, pH 9.2, and rinsed in acetone. Dried P81 filter papers were subjected to scintillation counting in 4 mL of ScintiSafe EconoF (FisherChemical). The remaining 15 μL of the HAT reaction was separated by SDS-PAGE (18% gel), stained with Coomassie Brilliant Blue, incubated with EN3HANCE autoradiography enhancer (PerkinElmer), dried and exposed to X-ray film for gel fluorography.
Yeast Two-Hybrid
A yeast two-hybrid assay was performed with the Matchmaker Gold Yeast two-hybrid system (Clontech). cDNAs were cloned into pGADT7 and pGBKT7 and were validated by sequencing. For the SF3B3 domain analysis, the Gateway-compatible yeast two-hybrid vectors pGADT7-GW and pGBKT7-GW were used [78]. Plasmids were transformed into S. cerevisiae Y2Hgold and selected for by growth on media lacking leucine and tryptophan. Interaction in the yeast two-hybrid assay was determined by growth on selective media lacking leucine, tryptophan, adenine and histidine according to the manufacturer’s instructions.
Genetics
The sf3b5EY12579 fly stock, y1 w67c23; P59CG11985EY12579/TM3, Sb1 Ser1, was obtained from the Bloomington Drosophila Stock Center at Indiana University (BL21381). The sf3b5EY12579 mutant was crossed to w1118; Drmio/TM3, P{w+mC=GAL4-twi.G}2.3, P{UAS-2xEGFP}AH2.3, Sb1Ser1 (BL6663) to generate an EGFP balanced stock, which was used to identify homozygous mutant embryos and larvae as previously described [49]. The SF3B5 cDNA was cloned into the pUAST-attB vector and transgenic flies were generated using the phiC31 site-specific integration system in the attP40 site on chromosome 2L [79]. Flies carrying the sf3b5EY12579 allele on chromosome 3 and either UAS-SF3B5 (w; P{w+mC=UAS-SF3B5}attP40; P{w+mC y+mDint2=EPgy2}CG11985EY12579/MKRS) or actin5C-GAL4 (w; P{w+mC=Act5C-GAL4}25FO1, P{w+mC=UAS-GFP.nls}14/CyO; P{w+mC y+mDint2=EPgy2}CG11985EY12579/MKRS) on chromosome 2 were generated using standard genetic techniques. Recombinant flies carrying the sf3b5EY12579 allele with FRT82B were generated using standard genetic techniques. Mosaic eyes consisting of sf3b5EY12579, ada2b1 or UAS-GFPnls cells were generated by crossing y1 w*; P{w+m*=GAL4-ey.H}3-8, P{w+mC=UAS-FLP1.D}JD1; P{ry+t7.2=neoFRT}82B P{w+mC=GMR-hid}SS4, l(3)CL-R1/TM2 flies with the following genotypes: (1) yd2 w1118;; P{ry+t7.2=neoFRT}82B, P{w+mC y+mDint2=EPgy2}CG11985EY12579/TM6b Tb1, (2) ada2B1, P{ry+t7.2=neoFRT}82B /TM3 Sb1 Ser1 or (3) w1118;; P{ry+t7.2=neoFRT}82B P{w+mC=Ubi-GFP(S65T)nls}3R/TM6B, Tb1. A complete description of fly genotypes used in this study is provided in Supplemental Table S2.
Histone Western Blot
Histones were acid-extracted from chromatin prepared from larvae or embryos using a modified version of the soluble nuclear extraction protocol as described previously [40]. Briefly, nuclei were isolated as described for affinity purification and MudPIT analysis with two minor modifications: miracloth was used to filter extracts prior to centrifugation, and buffers were supplemented with 10 mM sodium butyrate. Acid-soluble proteins were extracted from the insoluble chromatin pellet by incubation with 0.4 M HCl for 45 min at 25ºC, concentrated using trichloroacetic acid precipitation, and analyzed by SDS-PAGE and western blotting using the following antibodies: anti-histone H2B (Rabbit, 1:1000, Active Motif #39125), anti-acetylated H3 Lys-9 (Rabbit, 1:2000, Millipore 07-352), anti-Ubiquityl-Histone H2B antibody (1:3000, Millipore 17-650) and anti-Histone H3 (1:3000. Active Motif 61277). Relative levels of ubH2B/H3 and H3K9ac/H2B were quantified using Image Lab Software 5.0 (BioRad) within a single blot or cut membrane.
qRT-PCR analysis
RNA was isolated using the Zymoprep Direct-zol RNA MicroPrep kit (Zymo Research) and treated with DNAse I as per the kit protocol. cDNA was generated from 250 ng of RNA using Episcript Reverse Transcriptase (Epicentre) using either oligo dTs or random hexamer primers as indicated. qPCR was conducted using Evagreen 2X Mix (Biotium) and the CFX Connect Real-time system (Biorad). Quantities were determined relative to a 4-fold dilution series of wild-type (OregonR) cDNA. Primers against SAGA-regulated genes were taken from previous studies [40]. New primers used in this study are as follows: SF3B5 5′-GCAAAATGGGTGAACGCTAC-3′ and 5′-AGCCACTCGAACTTTGTGGT-3′, Sas10 5′-ACCGGTGCTCAACTACGTTC-3′ and 5′-GCTCCTCGATCAGATCCTTG-3′, Oda (unspliced) 5’-CCGTGCAAAAAGTGAATGTG-3’ and 5’-GCCAACCTGGAGAACGTCTA -3’, Sap47 (unspliced) 5’-ATCGATATTCCGCTTGTTGC-3’ and 5’-GCGCAAGTTTGATATTGTCG-3’, exba (unspliced) 5’-GAGCCCAAGGACAGGATTG-3’ and 5’-TGCTTGAACGTCTGGAACAG-3’, Crc (unspliced) 5’-′CGGACGAGTTGTCAACAGAA-3’ and 5’-′TCTGAAGATGCACCGAATTG-3’.
Supplementary Material
Highlights.
Spliceosomal components SF3B3 and SF3B5 are components of Drosophila SAGA
SF3B components bind to SAGA independent of RNA
SF3B5 is necessary for organismal and cell viability
SF3B5 is necessary for transcription of a subset of SAGA-regulated genes
Acknowledgments
Fly stocks from the Bloomington Drosophila Stock Center (NIH P40OD018537), and information from FlyBase and FlyMine were used in this study. Support from the American Cancer Society Institutional Research Grant (IRG #58-006-53) and NIH P30 CA023168 to the Purdue University Center for Cancer Research are gratefully acknowledged. PJS was supported by Purdue University Center for Cancer Research Summer Undergraduate Research Program funded by the Carroll County Cancer Association. This work was initiated with support from the National Institutes of Health GM99945-01 to Susan M. Abmayr and Jerry L. Workman. Support from the National Institutes of Health R01EY024905 to VMW is gratefully acknowledged.
Abbreviations
- H3K9ac
acetylated histone H3 Lysine 9
- GFP
green fluorescent protein
- HAT
histone acetyltransferase
- ubH2B
monoubiquitinated histone H2B
- MudPIT
Multidimensional Protein Identification Technology
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
- RNAse
ribonuclease
- SAGA
Spt-Ada-Gcn5 acetyltransferase
- snRNP
small nuclear ribonucleoprotein particle
Footnotes
Conflict of Interest: The authors declare no competing financial interests.
Accession Numbers
The complete MudPIT dataset (raw files, peak files, search files, as well as DTASelect result files) can be obtained from the MassIVE database via ftp://massive.ucsd.edu/ using the accession number MSV000079597 as username with password VMW70974.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–65. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 2.Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT, 2nd, Rosbash M. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes & development. 2011;25:2502–12. doi: 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, et al. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome research. 2012;22:1616–25. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, et al. A slow RNA polymerase II affects alternative splicing in vivo. Molecular cell. 2003;12:525–32. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Howe KJ, Kane CM, Ares M., Jr Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. Rna. 2003;9:993–1006. doi: 10.1261/rna.5390803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, et al. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome research. 2011;21:390–401. doi: 10.1101/gr.111070.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kornblihtt AR. Coupling transcription and alternative splicing. Advances in experimental medicine and biology. 2007;623:175–89. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nature structural & molecular biology. 2009;16:990–5. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 9.de Almeida SF, Carmo-Fonseca M. Design principles of interconnections between chromatin and pre-mRNA splicing. Trends Biochem Sci. 2012;37:248–53. doi: 10.1016/j.tibs.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Kwek KY, Murphy S, Furger A, Thomas B, O'Gorman W, Kimura H, et al. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat Struct Biol. 2002;9:800–5. doi: 10.1038/nsb862. [DOI] [PubMed] [Google Scholar]
- 11.Fong YW, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–33. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- 12.Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nature structural & molecular biology. 2006;13:22–9. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 13.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nature genetics. 2009;41:376–81. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS genetics. 2012;8:e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Molecular cell. 2007;28:665–76. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.David CJ, Boyne AR, Millhouse SR, Manley JL. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes & development. 2011;25:972–83. doi: 10.1101/gad.2038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S, Kollipara RK, Srivastava N, Li R, Ravindranathan P, Hernandez E, et al. Ablation of the oncogenic transcription factor ERG by deubiquitinase inhibition in prostate cancer. Proc Natl Acad Sci U S A. 2014;111:4251–6. doi: 10.1073/pnas.1322198111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27:262–74. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brand M, Moggs JG, Oulad-Abdelghani M, Lejeune F, Dilworth FJ, Stevenin J, et al. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 2001;20:3187–96. doi: 10.1093/emboj/20.12.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, et al. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–95. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen EJ, Frand AR, Chitouras E, Kaiser CA. A link between secretion and pre-mRNA processing defects in Saccharomyces cerevisiae and the identification of a novel splicing gene, RSE1. Mol Cell Biol. 1998;18:7139–46. doi: 10.1128/mcb.18.12.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caspary F, Shevchenko A, Wilm M, Séraphin B. Partial purification of the yeast U2 snRNP reveals a novel yeast pre-mRNA splicing factor required for pre-spliceosome assembly. EMBO J. 1999;18:3463–74. doi: 10.1093/emboj/18.12.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dowell RD, Ryan O, Jansen A, Cheung D, Agarwala S, Danford T, et al. Genotype to phenotype: a complex problem. Science. 2010;328:469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–91. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 26.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harbor perspectives in biology. 2011:3. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, et al. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Molecular cell. 2009;36:593–608. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 28.Golas MM, Sander B, Will CL, Lührmann R, Stark H. Molecular architecture of the multiprotein splicing factor SF3b. Science. 2003;300:980–4. doi: 10.1126/science.1084155. [DOI] [PubMed] [Google Scholar]
- 29.Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–43. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- 30.Spedale G, Timmers HT, Pijnappel WW. ATAC-king the complexity of SAGA during evolution. Genes Dev. 2012;26:527–41. doi: 10.1101/gad.184705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–50. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 32.Guelman S, Suganuma T, Florens L, Swanson SK, Kiesecker CL, Kusch T, et al. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol Cell Biol. 2006;26:871–82. doi: 10.1128/MCB.26.3.871-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–63. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laprade L, Rose D, Winston F. Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3 TBP allele-specific interactions and bypass of Spt8. Genetics. 2007;177:2007–17. doi: 10.1534/genetics.107.081976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15:1946–56. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weake VM, Dyer JO, Seidel C, Box A, Swanson SK, Peak A, et al. Post-transcription initiation function of the ubiquitous SAGA complex in tissue-specific gene activation. Genes Dev. 2011;25:1499–509. doi: 10.1101/gad.2046211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, et al. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell. 2007;27:275–88. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 38.Bonnet J, Wang CY, Baptista T, Vincent SD, Hsiao WC, Stierle M, et al. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev. 2014;28:1999–2012. doi: 10.1101/gad.250225.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Florens L, Washburn MP. Proteomic analysis by multidimensional protein identification technology. Methods Mol Biol. 2006;328:159–75. doi: 10.1385/1-59745-026-X:159. [DOI] [PubMed] [Google Scholar]
- 40.Weake VM, Swanson SK, Mushegian A, Florens L, Washburn MP, Abmayr SM, et al. A novel histone fold domain-containing protein that replaces TAF6 in Drosophila SAGA is required for SAGA-dependent gene expression. Genes Dev. 2009;23:2818–23. doi: 10.1101/gad.1846409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohan RD, Dialynas G, Weake VM, Liu J, Martin-Brown S, Florens L, et al. Loss of Drosophila Ataxin-7, a SAGA subunit, reduces H2B ubiquitination and leads to neural and retinal degeneration. Genes Dev. 2014;28:259–72. doi: 10.1101/gad.225151.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mount SM, Salz HK. Pre-messenger RNA processing factors in the Drosophila genome. J Cell Biol. 2000;150:F37–44. doi: 10.1083/jcb.150.2.f37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herold N, Will CL, Wolf E, Kastner B, Urlaub H, Lührmann R. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol Cell Biol. 2009;29:281–301. doi: 10.1128/MCB.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–80. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 45.Lee KK, Sardiu ME, Swanson SK, Gilmore JM, Torok M, Grant PA, et al. Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol Syst Biol. 2011;7:503. doi: 10.1038/msb.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das BK, Xia L, Palandjian L, Gozani O, Chyung Y, Reed R. Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Mol Cell Biol. 1999;19:6796–802. doi: 10.1128/mcb.19.10.6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Will CL, Urlaub H, Achsel T, Gentzel M, Wilm M, Lührmann R. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002;21:4978–88. doi: 10.1093/emboj/cdf480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suganuma T, Gutiérrez JL, Li B, Florens L, Swanson SK, Washburn MP, et al. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat Struct Mol Biol. 2008;15:364–72. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- 49.Guelman S, Suganuma T, Florens L, Weake V, Swanson SK, Washburn MP, et al. The essential gene wda encodes a WD40 repeat subunit of Drosophila SAGA required for histone H3 acetylation. Mol Cell Biol. 2006;26:7178–89. doi: 10.1128/MCB.00130-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hadjiolov AA, Venkov PV, Tsanev RG. Ribonucleic acids fractionation by density-gradient centrifugation and by agar gel electrophoresis: a comparison. Anal Biochem. 1966;17:263–7. doi: 10.1016/0003-2697(66)90204-1. [DOI] [PubMed] [Google Scholar]
- 51.Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, et al. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–7. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 52.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 53.Hryciw T, Tang M, Fontanie T, Xiao W. MMS1 protects against replication-dependent DNA damage in Saccharomyces cerevisiae. Mol Genet Genomics. 2002;266:848–57. doi: 10.1007/s00438-001-0605-x. [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Chen ZY, Wang W, Baker CC, Krug RM. The 3'-end-processing factor CPSF is required for the splicing of single-intron pre-mRNAs in vivo. RNA. 2001;7:920–31. doi: 10.1017/s1355838201010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Georgieva S, Nabirochkina E, Dilworth FJ, Eickhoff H, Becker P, Tora L, et al. The novel transcription factor e(y)2 interacts with TAF(II)40 and potentiates transcription activation on chromatin templates. Mol Cell Biol. 2001;21:5223–31. doi: 10.1128/MCB.21.15.5223-5231.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ciurciu A, Komonyi O, Pankotai T, Boros IM. The Drosophila histone acetyltransferase Gcn5 and transcriptional adaptor Ada2a are involved in nucleosomal histone H4 acetylation. Mol Cell Biol. 2006;26:9413–23. doi: 10.1128/MCB.01401-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carré C, Szymczak D, Pidoux J, Antoniewski C. The histone H3 acetylase dGcn5 is a key player in Drosophila melanogaster metamorphosis. Mol Cell Biol. 2005;25:8228–38. doi: 10.1128/MCB.25.18.8228-8238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weake VM, Lee KK, Guelman S, Lin CH, Seidel C, Abmayr SM, et al. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J. 2008;27:394–405. doi: 10.1038/sj.emboj.7601966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gause M, Eissenberg JC, Macrae AF, Dorsett M, Misulovin Z, Dorsett D. Nipped-A, the Tra1/TRRAP subunit of the Drosophila SAGA and Tip60 complexes, has multiple roles in Notch signaling during wing development. Mol Cell Biol. 2006;26:2347–59. doi: 10.1128/MCB.26.6.2347-2359.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–81. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 62.Pankotai T, Komonyi O, Bodai L, Ujfaludi Z, Muratoglu S, Ciurciu A, et al. The homologous Drosophila transcriptional adaptors ADA2a and ADA2b are both required for normal development but have different functions. Mol Cell Biol. 2005;25:8215–27. doi: 10.1128/MCB.25.18.8215-8227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin KA, Poeck B, Roth H, Ebens AJ, Ballard LC, Zipursky SL. Mutations disrupting neuronal connectivity in the Drosophila visual system. Neuron. 1995;14:229–40. doi: 10.1016/0896-6273(95)90281-3. [DOI] [PubMed] [Google Scholar]
- 64.Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–9. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qi D, Larsson J, Mannervik M. Drosophila Ada2b is required for viability and normal histone H3 acetylation. Mol Cell Biol. 2004;24:8080–9. doi: 10.1128/MCB.24.18.8080-8089.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dudley AM, Rougeulle C, Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 1999;13:2940–5. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhaumik SR, Green MR. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 2001;15:1935–45. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zsindely N, Pankotai T, Ujfaludi Z, Lakatos D, Komonyi O, Bodai L, et al. The loss of histone H3 lysine 9 acetylation due to dSAGA-specific dAda2b mutation influences the expression of only a small subset of genes. Nucleic Acids Res. 2009;37:6665–80. doi: 10.1093/nar/gkp722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Friend K, Lovejoy AF, Steitz JA. U2 snRNP binds intronless histone pre-mRNAs to facilitate U7-snRNP-dependent 3' end formation. Mol Cell. 2007;28:240–52. doi: 10.1016/j.molcel.2007.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blanchette M, Labourier E, Green RE, Brenner SE, Rio DC. Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF50 in the nuclear export of intronless mRNAs. Mol Cell. 2004;14:775–86. doi: 10.1016/j.molcel.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 71.Savage KI, Gorski JJ, Barros EM, Irwin GW, Manti L, Powell AJ, et al. Identification of a BRCA1-mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Mol Cell. 2014;54:445–59. doi: 10.1016/j.molcel.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Z, Jones A, Joo HY, Zhou D, Cao Y, Chen S, et al. USP49 deubiquitinates histone H2B and regulates cotranscriptional pre-mRNA splicing. Genes & development. 2013;27:1581–95. doi: 10.1101/gad.211037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Köhler A, Pascual-García P, Llopis A, Zapater M, Posas F, Hurt E, et al. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol Biol Cell. 2006;17:4228–36. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rodríguez-Navarro S, Fischer T, Luo MJ, Antúnez O, Brettschneider S, Lechner J, et al. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. doi: 10.1016/s0092-8674(03)01025-0. [DOI] [PubMed] [Google Scholar]
- 75.Swanson SK, Florens L, Washburn MP. Generation and analysis of multidimensional protein identification technology datasets. Methods Mol Biol. 2009;492:1–20. doi: 10.1007/978-1-59745-493-3_1. [DOI] [PubMed] [Google Scholar]
- 76.Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal Chem. 2010;82:2272–81. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- 77.Eberharter A, John S, Grant PA, Utley RT, Workman JL. Identification and analysis of yeast nucleosomal histone acetyltransferase complexes. Methods. 1998;15:315–21. doi: 10.1006/meth.1998.0635. [DOI] [PubMed] [Google Scholar]
- 78.Lu Q, Tang X, Tian G, Wang F, Liu K, Nguyen V, et al. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 2010;61:259–70. doi: 10.1111/j.1365-313X.2009.04048.x. [DOI] [PubMed] [Google Scholar]
- 79.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–83. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.