Abstract
While ribosomal proteins (RP) are thought to primarily facilitate biogenesis of the ribosome and its ability to synthesize protein, emerging evidence suggests that individual RP can perform critical regulatory functions that control developmental processes. We showed previously that despite the ubiquitous expression of the RP, Rpl22, germline ablation of Rpl22 in mice causes a selective, p53-dependent block in the development of αβ, but not γδ, T cell progenitors. Nevertheless, the basis by which Rpl22 loss selectively induces p53 in αβ T cell progenitors remained unclear. We show here that Rpl22 regulates the development of αβ T cells by restraining endoplasmic reticulum (ER) stress responses. In the absence of Rpl22, ER stress is exacerbated in αβ, but not γδ, T cell progenitors. The exacerbated ER stress in Rpl22-deficient αβ T lineage progenitors is responsible for selective induction of p53 and their arrest, as pharmacological induction of stress is sufficient to induce p53 and replicate the selective block of αβ T cells, and attenuation of ER stress signaling by knockdown of PERK, an ER stress sensor, blunts p53 induction and rescues development of Rpl22-deficient αβ T cell progenitors. Rpl22-deficiency appears to exacerbate ER stress by interfering with the ability of ER stress signals to block new protein synthesis. Our finding that Rpl22-deficiency exacerbates ER stress responses and induces p53 in αβ T cell progenitors provides insight into how a ubiquitously expressed RP can perform regulatory functions that are selectively required by some cell lineages but not others.
INTRODUCTION
Ribosomal proteins (RP) are ubiquitous proteins that play critical roles in facilitating ribosome biogenesis and its core function of synthesizing protein (1). Mutations in RP cause a group of diseases called ribosomopathies that are generally thought to be the consequence of impairment of either assembly or function of the ribosome (2). Ribosomopathies are characterized by disrupted hematopoiesis resulting in bone marrow failure and anemia in early life, increased risk of developing leukemias or solid tumors, and skeletal or craniofacial abnormalities (3-6). These anomalies are thought to result from the loss of the general, supportive functions of RP (7). Nevertheless, it is becoming increasingly understood that RP possess regulatory capabilities whose loss might also contribute to the developmental anomalies observed in ribosomopathies (8, 9). However, loss-of-function approaches to study RP eliminate both the general role of the RP in supporting the biogenesis and function of the ribosome, as well as any regulatory function it might have (10, 11). This makes it particularly difficult to disentangle whether developmental anomalies accompanying ablation of an RP gene result from generalized impairment of ribosome function or loss of the regulatory roles.
We have identified a ribosomal protein, Rpl22, which represents an opportunity to distinguish developmental anomalies resulting from loss of essential, supportive RP functions from those resulting from loss of RP activities that are more regulatory in nature (12). Rpl22 is a widely expressed component of the 60S large ribosome subunit, but it is not essential for the core ribosome function of global protein synthesis (13). Moreover, germline ablation of the Rpl22 gene is not lethal, as Rpl22-deficient mice are of normal size, and are fertile and healthy (13). However, Rpl22-deficient mice display a remarkable reduction in thymic size and cellularity. The reduction in thymic cellularity in Rpl22-deficient mice results from a selective, and highly-penetrant block in the development of αβ, but not γδ, T cell lineage progenitors (13). The block in αβ lineage T cell progenitors results from selective induction of p53 protein in αβ lineage cells, since the developmental arrest is completely alleviated by p53-deficiency (13). Moreover, the function of p53 in arresting development appears to be mediated primarily through induction of apoptosis, as it is alleviated by the elimination of pro-apoptotic p53 targets, but not by those that regulate cell cycle progression (14).
The selective requirement for Rpl22 function in αβ, but not γδ, T cell progenitors is surprising, since both of these lineages arise from a common progenitor in the thymus (15, 16). Early T cell progenitors lack expression of either CD4 or CD8, and are called double negative (DN) thymocytes. DN thymocytes progress through four stages of differentiation characterized by expression of different surface markers (DN1, CD44+CD25−; DN2, CD44+CD25+; DN3, CD44−CD25+; DN4, CD44−CD25−) (17, 18). Concurrent with their commitment to the T lineage, DN1 (CD44+CD25−) cells up-regulate CD25 and begin to rearrange their T cell receptor (TCR) γ, δ, and β genes (Tcrg, Tcrd, and Tcrb) via V(D)J recombination (19, 20). The divergence of the αβ and γδ lineages occurs between the initiation of TCR gene rearrangement at the DN2 stage and arrival at the DN3 stage (16). The separation of these lineages is controlled by different TCR complexes expressed by the progenitors (21). Progenitors that productively rearrange their TCRγ and δ loci and express the γδ TCR adopt the γδ fate in response to the transduction of stronger or more prolonged TCR signals (22). Following γδ lineage commitment, the progenitors remain DN, mature as indicated by the downregulation of CD24, and then exit the thymus (23). Conversely, progenitors that productively rearrange the TCRβ locus and express the pre-TCR complex adopt the αβ fate in response to the transduction of weaker or more transient TCR signals. The weak pre-TCR signals enable these progenitors to traverse the β-selection checkpoint at the DN3 stage and differentiate to the DP stage, which both involves and depends on extensive proliferation (24). Rpl22-deficiency selectively blocks the development of αβ lineage progenitors as they attempt to traverse the β-selection checkpoint (13). The pre-TCR dependence of αβ T cell development distinguishes it from that of γδ progenitors. While no alterations in pre-TCR function were noted in Rpl22-deficient thymocytes, perturbation of the downstream signals linked to the pre-TCR has not been excluded as a cause of the selective arrest observed in Rpl22-deficient αβ progenitors.
The basis for selective arrest of αβ, but not γδ, T lineage progenitors in Rpl22-deficient mice remains unexplained. Since pre-TCR signaling and adoption of the αβ lineage are accompanied by extensive proliferation (24), it is possible that proliferation activates cellular stresses in these cells that are not experienced by other cell types, and this could be responsible for the selectivity of Rpl22-dependence. One stress response of particular interest is the unfolded protein or endoplasmic reticulum (ER) stress response, which represents a homeostatic process that serves to match the cellular protein-folding burden to the capacity of the cellular chaperones responsible for protein folding (25). There are three ER stress signaling pathways; PERK, IRE1α and ATF6 (26). ER stress pathways are of interest as a possible determiner of the tissue specificity of p53-dependent developmental arrest in Rpl22-deficient mice because ER stress signaling is activated as αβ lineage progenitors traverse the β-selection checkpoint, their excessive activation has been linked to p53 induction, and Rpl22-deficiency in yeast has been linked to perturbation of cell growth in a manner dependent upon the yeast ortholog of ATF4 (27). ATF4 is a transcription factor activated by ER stress (25).
In this study, we discovered a novel, causal link between Rpl22-deficiency, exacerbation of ER stress, and tissue restricted induction of p53 in αβ lineage T cell precursors. Specifically, we found that: 1) Rpl22 loss exacerbates ER stress selectively in αβ, but not γδ lineage progenitors; 2) pharmacologic induction of ER stress is capable of replicating the effects on thymocyte development caused by Rpl22 loss; and 3) alleviation of ER stress by attenuating signaling through the ER stress sensor, PERK, blunts p53 induction and restores the development of αβ lineage T cells. These findings suggest that Rpl22 loss exacerbates ER stress in some cell types, but not others, and that exacerbated stress is responsible for the selective induction of p53 that causes the developmental arrest.
MATERIALS AND METHODS
Mice
All mouse strains were housed in the Laboratory Animal Facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care at Fox Chase Cancer Center. Mice were handled in accordance with Institutional Animal Care and Use Committee approved protocols. The following mouse strains were used: Rpl22-deficient (13), KN6 γδ TCR transgenic (Tg) (28); Pre-Tα-deficient (a gift from Dr. H von Boehmer Comment, Dana Farber Cancer Institute) (29); and p53-deficient mice (a gift from Dr. Maureen Murphy, Wistar Institute) (30). AB strain zebrafish were bred and maintained under standard aquaculture conditions at Fox Chase core Zebrafish Facility.
Flow Cytometry
Single-cell suspensions of thymic cells were stained with the following fluorochrome-conjugated antibodies: anti-CD4 (clone GK1.5), anti-CD25 (clone 7D4), anti-CD44 (clone IM7) anti-γδ TCR (clone GL3), anti-CD24 (clone 30-F1) and anti-CD8 (Clone 53-6.7) (Biolegend, San Diego, CA). Multiparametic flow data was collected using an LSRII (BD, Biosciences) and analyzed using FlowJo software (Tree Star, Ashland, OR). Flow cytometric purification of cells was performed using a FACS Aria II (BD Biosciences).
Methionine Labeling
Thymocytes or thymic lymphoma cell lines were metabolically labeled with [35S]-methionine for 30 minutes. Following extraction with NP-40 Lysis Buffer (1% NP-40, 50mM TRIS pH 8.0, 150mM NaCl), the clarified extracts were either subjected to TCA precipitation to determine total counts incorporated or were immunoprecipitated with anti-p53 antibody as described (13).
RNA Immunoprecipitation
Clarified NP40 lysates of thymocytes were immunoprecipitated with rabbit anti-Rpl22 antibody prepared by immunization with the N-terminal 15 amino acids of Rpl22 coupled to chicken serum albumen. Immunoprecipitated RNA was washed four times in lysis buffer containing 1M urea and isolated using Qiagen RNAeasy mini kits with an on-the-column DNase digestion. p53 and actin mRNA was quantified by real time PCR using stock primers from ABI.
Biosensor assay in zebrafish embryos
The murine p53 fragment containing the Rpl22 binding hairpin sequence (nucleotides 455-767) was fused in-frame to EGFP and subcloned into pCS2+. An equivalent biosensor was created in which the Rpl22 hairpin target sequence was mutated and disrupted using the Gene Taylor kit (Invitrogen). The full-length mRpl22 coding sequence was also cloned into pCS2+ and in vitro transcribed, capped mRNAs for microinjection were synthesized using the mMessage mMachine kit (Ambion). 100pg of mRNA each for both Rpl22 and the biosensor constructs were injected into one-cell stage embryos, which were photographed at 6hpf. Images were taken using a Nikon SMZ1500 stereomicroscope equipped with DS-Fi1 digital camera and Nikon AR imaging software.
RNAse protection
DNA templates used for RNAse protection and in vitro transcription reactions were prepared using primers appended to the T7 promoter, and cloned into pCR2.1 vector (Thermofisher). Transcription reactions were performed on EcoRI digested fragments (New England Biolab), using the MAXIscript T7 kit (Ambion), in presence of 10 μCi of 32P-UTP (Perkin Elmer), and purified by G-50 columns (Illustra Probe-Quant, GE Healthcare). Labeled transcripts were re-natured in TE buffer for 2 min at 95 ºC and then placed on ice. Re-natured transcript (1 μl) was pre-incubated on ice for 12 min in HLA/Terasaki plate in 7 μl reaction with 5 μl of 2x RPA buffer (20 mM Tris pH 8.0, 100 mM NaCl, 1.5 mM MgCl2, 1.6 mM DTT, 5% glycerol), 0.5 μl of tRNA (10mg/ml, baker’s yeast; Roche), and 22 μM (+) and 36 μM (++) of GST-purified protein. After 30 min of UV crosslinking, the samples were digested at 37ºC for 20 min with 1 μl of freshly prepared RNase A solution comprising 4 μl RNase A (Qiagen, 7,500U/μl, 100 mg/ml) and 0.8 μl of 5x RNase A buffer (100 mM Tris pH 7.0, 10 mM MgCl2, 1 M KCl, and 1.2 μl of ddH2O). Samples were denatured with 2 μl of loading buffer for 2 min at 95ºC, resolved by SDS-PAGE and visualized using a Fuji BAS-2500 phosphoimager plate reader.
Immunoblot analysis
Single cell suspensions of primary thymocytes or lymphoma cells were lysed in NP-40 or RIPA lysis buffers (20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM beta-glycerophosphate, 1 mM Na3VO4, 1 μg/ml leupeptin) containing complete mini-tab protease and phosphatase inhibitor tablets (Roche, Basel, Switzerland). Samples were resolved on NuPage Novex Bis-Tris gels (Invitrogen) and blotted with the following antibodies: anti-PERK (Cell Signaling Technologies, Beverly, MA) anti-phospho PERK (Cell signaling Technologies, Beverly, MA), total and phospho-serine 51 of eIF2α, anti-p53 (Clone #IMX25, Leica Microsystems, UK), anti-IRE1α and phospho-IRE1α (Abcam), anti-ATF6 (Abcam), anti-BiP (Santa Cruz), anti-CHOP (Santa Cruz Bio.), anti-XBP-1s (Santa Cruz), anti-ATF4 (Abcam, MA), anti-β-actin (Santa Cruz), anti-GAPDH, and anti-calnexin. Blocking was performed with either in 5% milk or 5% BSA prepared in TBS with 0.1% Tween-20 per manufacturer’s guidelines. For LI-COR analysis, manufacturers guidelines were followed and fluoresceinated secondary antibodies conjugated with either IR-Dye 488 or IR-Dye 648 were used to visualize bound primary antibody.
Quantitative Real-Time PCR
Total RNA was isolated from primary cells using the RNA-Easy system (Qiagen). Purified mRNA was converted to cDNA using Superscript II reverse transcription PCR with oligo dT primers (Invitrogen). Quantitative real-time PCR was performed on the Prism 7700 thermocycler (Applied Biosystems) using Taqman Real-time PCR primer/probe sets specific to murine targets. The identity of probe and primer sets will be provided upon request. Analysis for each primer/probe set was performed for each cell type in triplicate. Bar graphs show the fold change (2−ΔΔCt or RQ value + RQ max) over control.
Anti-CD3 treatment of Rag2−/− mice
Rag2−/− mice were injected i.p. with either PBS or with anti-CD3 antibody (145-2C11) at 10μg/g body weight. At 24 hours after treatment, explanted thymi were formalin fixed, sectioned, and stained with antibodies reactive with fibrillarin to mark the position of nucleoli and NPM to assess nucleolar integrity.
Cell viability and drug treatment
Thymic lymphoma cells were generated as described (31). Cell lines or primary thymocytes were treated with the indicated doses of either DMSO vehicle, Thapsigargin (THG), or Tunicamycin (TUN).
Constructs and cloning
miR30 based short hairpin RNA (shRNA) constructs were cloned into the pMLS vector as previously described (13). The primers used will be provided upon request. The shRNA constructs were retrovirally-transduced into either primary fetal liver progenitors or thymic lymphoma cell lines as described (13). Transduced, GFP+ cells were isolated by flow cytometry.
OP9 cultures and retroviral transduction of fetal thymocytes
Viral particles were produced by transient calcium-phosphate transfection of Phoenix cells with shRNAs expressed in the MSCV-based vectors, MLP or MLS, as described (32). Hematopoietic progenitors from the fetal livers of embryonic day 14 Rpl22+/+ and Rpl22−/− mice were expanded in IL7 (5ng/mL) and Flt-3 (5ng/mL) on OP9-DL1 expressing monolayers. After four days, cells were spin-infected with retrovirus containing shRNA targeting PERK (MLS-PERK), IRE1α (MLS-IRE1α), ATF6 (MLS-ATF6), or an irrelevant control (MLS-H2T10/22). For p53 knockdown, precursors were infected with a control vector (MLP) or MLP-p53 (provided by S. Lowe, Cold Spring Harbor Laboratory). Developmental progression was monitored by flow cytometry on days 5, 8, 12, and 16 of culture by electronically gating on GFP+ cells.
Statistics
All experiments were assayed in triplicate (n = 3). Data are expressed as mean ± SEM. All statistical analyses were performed using GraphPad Pro Prism 5.0 (GraphPad, SanDiego, CA). Student’s t-test and two-way ANOVA were employed to analyze the differences between sets of data. p <0.05 was considered statistically significant. The following standard was use for the representation of asterisk: *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
RESULTS
Rpl22-deficiency selectively increases p53 synthesis in αβ T cell progenitors
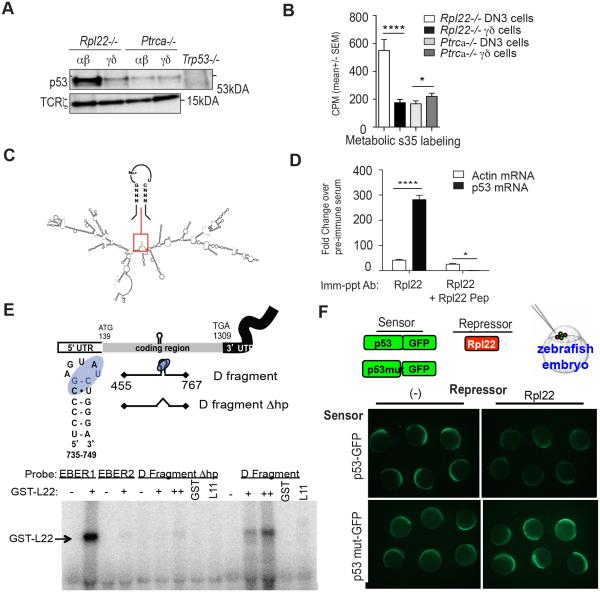
We have previously shown that the p53 induction observed in Rpl22-deficient αβ T lineage progenitors occurs post-transcriptionally, and results from increased protein synthesis (13); however, we had not previously determined whether the lack of p53 induction in Rpl22-deficient γδ cells occurred because p53 synthesis was not upregulated. To address this question, we performed [35S]-methionine metabolic labeling on purified αβ and γδ lineage precursors. We observed that while synthesis of p53 protein was markedly increased in Rpl22-deficient αβ lineage (DN3) thymocytes relative to that in Rpl22-expressing Ptcra−/− DN3, it was not increased in Rpl22-deficient γδ lineage thymocytes (Figure 1A and B). We next asked how Rpl22 loss was influencing p53 synthesis. Rpl22 is an RNA binding protein that has been reported to directly regulate p53 synthesis through direct binding to p53 mRNA (33). Rpl22 binds RNA targets through recognition of a stem loop structure with a G-C-U at the neck of the stem (34). M-fold structure prediction revealed the presence of a consensus Rpl22 binding site in p53 mRNA, suggesting that binding by Rpl22 was possible (Figure 1C) (35). To determine whether Rpl22 could bind p53 mRNA, we performed co-immunoprecipitation analysis using an anti-Rpl22 antibody. Indeed, anti-Rpl22 antibody co-precipitated p53 mRNA, but not control β-actin mRNA, suggesting that Rpl22 associates with p53 mRNA in cells (Figure 1D). Further, we mapped the Rpl22 binding site in p53 mRNA in vitro by conducting RNAse protection analysis (RPA) using recombinant GST-Rpl22 fusion protein. We observed that Rpl22, but not Rpl11, can directly bind p53 mRNA and that the binding was abrogated by mutagenesis of the consensus Rpl22 binding site (Figure 1E). Further, through the use of biosensors containing intact or mutant Rpl22 binding sites, we determined that when Rpl22 mRNA was co-injected into zebrafish embryos with the biosensors, Rpl22 was capable of silencing the expression of the biosensor containing an intact Rpl22 binding site, but not one in which the binding site had been destroyed by mutagenesis (Figure 1F). Collectively, these data indicate that Rpl22 can directly bind p53 mRNA and regulate its expression. Nevertheless, direct regulation of p53 translation by Rpl22 is insufficient to explain why the loss of Rpl22 from all cells results in the selective translational de-repression of p53 in only certain cell types, like αβ lineage T cell progenitors.
Figure 1. Rpl22-deficient αβ thymocytes exhibit a tissue restricted increase in p53 synthesis.
(A) αβ (γδTCR− DN3) and γδ T cell precursors (γδTCR+ DN) isolated from mice of the indicated genotypes were metabolically labeling for 30 minutes with [35S] methionine. Detergent extracts of the metabolically labeled cells were immunoprecipitated with anti-p53 or anti-TCRζ following which the immune complexes were resolved on SDS-PAGE gels. (B) Aliquots of the detergent extracts from metabolically labeled cells were TCA precipitated in triplicate to assess total radioactive incorporation. The mean ± standard deviation from three experiments was depicted graphically. Results are representative of at least 3 independent experiments. *p-value<0.05; ****p-value<0.0001. (C) M-fold computational analysis of p53 mRNAs showing G-C-U Rpl22 binding motif boxed in red. (D) Rpl22 binding of p53 mRNA was assessed by co-immunoprecipitation of p53 mRNA with anti-Rpl22 antibody from detergent extracts of culured thymocytes. Co-immunoprecipitation is specific as it was blocked by the peptide immunogen against which the anti-Rpl22 antiserum was raised. (E) Rpl22 binding to p53 mRNA was assessed by RNAse protection assay by co-incubation of recombinant GST-Rpl22 with the D fragment of p53 mRNA (nucleotides 455-767). Binding was abolished by deletion of Rpl22 binding motif. (F) The fragment of p53 containing the Rpl22 binding site was appended to GFP to create a biosensor. Upon co-injection of mRNA encoding the biosensor into zebrafish embryos with mRNA encoding Rpl22, the GFP signal at 6 hours post fertilization was markedly decreased. The decrease in fluorescence caused by Rpl22 co-expression was abrogated when the Rpl22 binding site in the biosensor was deleted.
Induction of p53 in Rpl22−/− αβ lineage T cell progenitors is not associated with impaired ribosome biogenesis or nucleolar stress
Since mutations in some RP have been shown to activate p53 by impairing ribosome biogenesis, it was possible that Rpl22-loss was impairing ribosome biogenesis. While the tissue restriction of p53 induction in Rpl22-deficient mice suggested that this possibility was unlikely, we nevertheless investigated whether Rpl22-deficiency affected processing of ribosomal RNA (rRNA), which is defective under circumstances where ribosome biogenesis is impaired (36-38). Pulse-chase analysis revealed that Rpl22-deficieny did not delay the processing of rRNA precursors into the mature 28S, 18S, and 5S species (Figure S1A). Defects in ribosome biogenesis also compromise the integrity of the nucleolus and activate p53 by liberating components that impair p53 degradation (36, 37, 39). Specifically, the loss of nucleolar integrity results in the release of proteins such as p19ARF and nucleophosmin (NPM) into the nucleoplasm (40). Consistent with our assessment of ribosome biogenesis above, we found that Rpl22-deficiency did not result in the escape of NPM into the nucleoplasm, as it remained restricted to the nucleolus, as marked by fibrillarin staining (Figure S1B) (41). Finally, to genetically assess whether compromised nucleolar integrity was linked to p53-induction in Rpl22-deficient progenitors, we asked if elimination of p19ARF (Cdkn2a), one of the proteins that is lost from the nucleolus under stress, rescued the developmental arrest caused by Rpl22-deficiency. Upon induction of cellular insults, p19ARF leaves the nucleolus and binds to the p53 ubiquitin ligase, Mouse Double Minute 2 (MDM2), thereby attenuating its ability to promote the degradation of p53 and resulting in increased p53 levels (42, 43). Consistent with the absence of any biochemical or microscopic indication that ribosome biogenesis or nucleolar integrity were compromised, ablation of the Cdkn2a gene failed to rescue thymocyte development in Rpl22-deficient mice (Figure S1C-H). These data suggest that the p53 induction observed in Rpl22-deficient αβ thymocytes occurs via an ARF (Cdkn2a)-independent mechanism and that Rpl22-deficiency is not inducing p53 through impaired ribosome biogenesis or nucleolar stress.
The arrest of αβ T cell development by Rpl22-deficiency is not mediated by effects on the pre-TCR complex
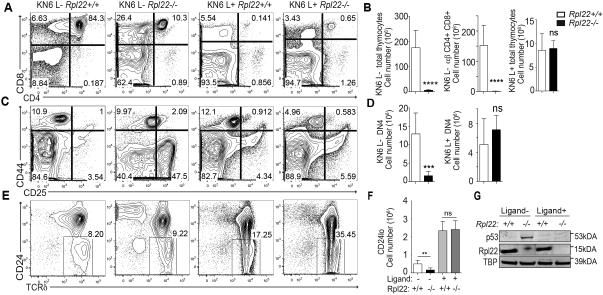
One possible explanation for the selective arrest of αβ but not γδ lineage progenitors in Rpl22-deficient mice would be if Rpl22 ablation interfered with the function of the pre-TCR complex, which is dispensable for γδ development but is critical for αβ lineage T cells to traverse the β-selection checkpoint at the DN3 stage. To address this possibility, we employed the KN6 γδTCR Tg mouse model, in which the γδTCR Tg expressed on a Rag2−/− background is capable of instructing progenitors to adopt either the γδ or αβ fate, depending on the nature of the TCR signals transduced (28). Indeed, in the absence of the selecting ligand for the KN6 γδTCR, H-2T10/22 (KN6 L−), progenitors adopt the αβ fate and develop to the DP stage (Figure 2A,B). Importantly, this differentiation process, which is independent of the function of the pre-TCR complex because RAG2-deficiency blocks rearrangement of endogenous Tcr loci, was blocked by the loss of Rpl22, since Rpl22-deficient progenitors were arrested at the β-selection checkpoint at the DN3 stage, and failed to develop to the DP stage (Figure 2A-D). In contrast, KN6 γδTCR Tg progenitors that encounter ligand (KN6 L+), adopt the γδ fate, remain DN, differentiate to the DN4 stage, and mature, as evidenced by their down-modulation of CD24 (Figure 2A-F). Adoption of the γδ fate by these γδTCR Tg progenitors and differentiation into mature CD24lo cells is not blocked by Rpl22-deficiency (Figure 2A-F). Given that the αβ fate is not specified by the pre-TCR in this model, these results suggest that Rpl22 is not blocking αβ development by interfering with pre-TCR function. Further, the block in αβ development in Rpl22-deficient ligand-deficient KN6 γδTCR Tg mice is associated with p53 induction, which is not observed in γδ precursors, as was true for γδ precursors from non-transgenic Rpl22-deficient mice (Figure 2G). These data indicate that the disruption of αβ T cell development by Rpl22-deficiency does not result from impaired pre-TCR expression or function, but rather is a consequence of interference with the αβ lineage differentiation program.
Figure 2. The arrest of Rpl22-deficient αβ lineage progenitors is not mediated by alterations in pre-TCR function.
(A-D) Single-cell suspensions of thymocytes from KN6 Tg mice of the indicated genotypes that either express (L+) or lack ligand (L−) were stained with antibodies reactive with CD4, CD8, CD44, and CD25 and analyzed by flow cytometry. Gate frequencies are listed on the histograms and used to calculate absolute numbers of the αβ lineage DP thymocytes (A, B) or DN4 thymocytes that have traversed the β-selection checkpoint (C, D). The results are depicted graphically and represent mean± standard deviation of 3-8 mice per group. (E,F) The number of mature CD24loTCRδ+ γδ lineage thymocytes was determined by staining thymocytes from mice with the indicated genotypes with anti-CD24 and anti-TCRδ antibodies. Gate frequencies are listed on the histogram and were used to determine the absolute number of mature γδ lineage thymocytes, which is depicted graphically on the right. These results are representative of three experiments of 3-8 mice per group. (G). Detergent extracts of DN thymocytes from mice of the indicated genotypes were immunoblotted with anti-p53, anti-Rpl22 and anti-GAPDH antibodies. **p-value<0.01, ***p-value<0.001; ****p-value<0.0001.
Rpl22-deficiency selectively exacerbates ER stress in αβ lineage progenitors
To gain insight into how Rpl22 loss selectively induces p53 in αβ progenitors, we explored pathways linked to Rpl22 function. Loss of Rpl22 in yeast alters growth control in a manner dependent on the yeast ortholog of ATF4, a transcriptional effector of ER stress signaling (44). Moreover, ER stress is induced as αβ progenitors traverse the β-selection checkpoint (45), and exacerbation of ER stress has previously been linked to p53 induction in cell line models (25, 26, 46). Thus, we examined whether Rpl22 loss exacerbates ER stress responses in αβ progenitors. There are three ER stress signaling pathways relevant to thymocytes, PERK, ATF6, and IRE-1α, that are activated to alleviate proteotoxic stress (Figure 3A) (25, 46-49). We found that Rpl22-deficiency activates the PERK pathway, as evidenced by the increased phosphorylation of the PERK kinase, enhanced phosphorylation of the PERK substrate eIF2α, and the consequent translational induction of its effector transcription factor, ATF4 (Figure 3B,C). Moreover, ATF4 induction resulted in increased expression of its targets, CHOP, ATF3, Trib3, and the chaperone BiP (Grp78) (Figure 3B-D). We also observed an increase in activation of IRE1α, marked by its phosphorylation in Rpl22-deficient thymocytes, and subsequent induction of its effector transcription factor, XBP-1, which is activated by splicing of its mRNA (Figure 3E,F) (48). XBP-1 induction also resulted in transactivation of its stress alleviating targets including Dnajb9 and Dnajc3 (Figure 3G). In contrast to the PERK and IRE1 pathways, the ATF6 pathway displayed minimal activation as evidenced either by the generation of active, cleaved ATF6 (ATF6f) or the induction of its targets (Figure S2A, B). Importantly, exacerbation of ER stress signaling in Rpl22-deficient progenitors was not a consequence of p53 induction, as it was still observed in Rpl22/p53-double deficient progenitors (Figure S2C). This indicates that activation of ER stress signals in Rpl22-deficient progenitors is upstream of p53, as would be expected if ER stress were responsible for p53 induction. Importantly, the exacerbation of ER signaling observed in Rpl22-deficient αβ T cell progenitors was not observed in Rpl22-deficient KN6 Tg γδ T cell progenitors, whose commitment to the γδ fate in the presence of ligand (KN6 Ligand +) was not impaired by Rpl22-deficiency (Figures 2 and 3H-J). Conversely, in Rpl22-deficient KN6 γδ T cell progenitors whose commitment to the αβ fate in the absence of ligand (KN6 Ligand −) is blocked by Rpl22-deficiency, ER stress signaling was exacerbated (Figures 2 and 3H,J). ER stress signaling in γδ lineage progenitors was assessed using the KN6 model described above, as it enables the isolation of sufficient γδ lineage progenitors to perform biochemical analysis. Taken together, these data suggest that induction of stress appears to be restricted to the population of cells in which Rpl22-deficiency also causes a p53-dependent arrest in development.
Figure 3. Rpl22-deficiency selectively exacerbates ER stress in αβ, but not γδ lineage progenitors.
(A) Schematic of ER stress signaling pathways. (B,C) Activation of the PERK-eIF2α-ATF4 pathway in CD4-CD8- DN thymocytes from Rpl22+/+ or Rpl22−/− mice was assessed by immunoblotting using the indicated antibodies. Band intensity was quantified relative to β-actin loading control using LI-COR and/or ImageJ. (D) mRNA encoding the indicated targets of the PERK ER stress pathway were quantified by performing real time PCR on RNA extracted from DN thymocytes of indicated genotypes. mRNA levels of PERK ER stress signals were normalized to that of GAPDH. The graph indicates fold change of mRNA expression in Rpl22−/− thymocytes (2−ΔΔCt or RQ) relative to that in Rpl22+/+ thymocytes. (E-G) Activation of the IRE1α-XBP-1 pathway in Rpl22−/− thymocytes was assessed using the indicated antibodies as above. (H-J) The activation of ER stress in Rpl22+/+ and Rpl22−/− KN6 γδTCR Tg progenitors adopting the γδ fate in the presence of ligand (KN6 L+) or the αβ fate in the absence of ligand (KN6 L−) was assessed by immunoblotting using the indicated antibodies and quantifying the level of ER stress induced target mRNA by real time PCR. Statistical significance was assessed using the Student’s t test and results are representative of at least three experiments performed. *p-value<0.05; **p-value<0.01, ***p-value<0.001; ****p-value<0.0001.
Pharmacologic induction of ER stress activates p53 and selectively blocks development of Rpl22-sufficient αβ T lineage thymocytes
To determine whether excessive ER stress signaling was responsible for p53 induction and the arrest of Rpl22-deficient αβ T cell development, we pharmacologically induced ER stress in Rpl22-sufficient thymocytes using two pharmacologic agents that induce ER stress, the sarco/endoplasmic reticulum Ca2+-ATPase inhibitor, thapsigargin (THG) (50) and the inhibitor of protein glycosylation, tunicamycin (TUN) (51). Indeed, both stress inducers activated the PERK pathway and induced p53 (Figure 4A and data not shown). Moreover, treatment with these two inducers of ER stress also caused a selective block in development of Rpl22-expressing αβ, but not γδ, lineage progenitors that were cultured on OP9-DL1 monolayers, a culture system capable of supporting the development of T cells in vitro (Figure 4B-E). Importantly, the ER stress-mediated blockade of αβ T cell development appears to be at least partially p53-dependent as the developmental arrest is substantially alleviated by p53-deficiency (Figure 4F-G). The alleviation of the arrest caused by TUN treatment is more profound than for that caused by THG treatment, presumably because THG-mediated release of the ER calcium stores can also induce mitogenic signals that could alter development. These data suggest that pharmacologic induction of ER stress is sufficient to induce p53 expression and selectively block the development of αβ lineage T cells.
Figure 4. Pharmacologic induction of ER stress can activate p53 and block development of Rpl22-sufficient αβ T lineage thymocytes.
(A) DN Rpl22+/+ thymocytes were cultured with either DMSO (vehicle) or 0.1 μM Thapsigargin (THG) for 4 hrs., following which detergent extracts were blotted with the indicated antibodies to assess ER stress and p53 induction. (B-E) Rpl22+/+ DN thymocytes treated with DMSO, 0.01, 0.1 μM THG, or 0.5μg/ml Tunicamycin (TUN) were cultured on OP9-DL1 monolayers for 5 days following which effects on development were assessed by flow cytometry as in Figure 2. Gate frequencies are listed on the histograms. (F-G) Rpl22+/+Trp53+/+ and Rpl22+/+Trp53−/− thymocytes were treated with ER stress activators at the doses indicated above and plated on OP9 monolayers to monitor progression of DN to DP. The results are graphically depicted as above, represent mean ± standard deviation, and are representative of three experiments performed. Students T test was used to calculate the p-values; **p-value<0.01; ***p-value<0.001; ****p-value<0.0001.
Attenuation of PERK signaling rescues the development of Rpl22−/− thymocytes in vitro
If excessive ER stress is responsible for the p53-mediated impairment of αβ T cell development in Rpl22−/− mice, then alleviation of stress should restore the development of αβ T precursors. We therefore asked whether knocking down each ER stress sensor individually using shRNA would rescue development of Rpl22−/− fetal liver precursors on OP9-DL1 monolayers (52). Fetal liver progenitors isolated from day 14 Rpl22+/+ and Rpl22−/− embryos were transduced with shRNA targeting PERK, IRE1α, or ATF6 (Figure S3), and cultured on OP9-DL1 monolayers to assess the effect on development (Figure 5A-D). A p53-targeting shRNA served as a positive control for rescue of development of Rpl22−/− αβ lineage T cells. Attenuation of PERK and IRE1α signaling in Rpl22-expressing progenitors caused some impairment of the development of Rpl22+/+ αβ lineage progenitors to the DP stage, suggesting that these pathways contribute to the development of normal progenitors (Figure 5A). Importantly, shRNA targeting two distinct regions of PERK displayed a clear rescue of development of Rpl22−/− progenitors to the DP stage of αβ T cell development (Figures 5C,D and S3B, C), suggesting that attenuating the PERK arm of ER stress signals was sufficient to alleviate the developmental arrest of Rpl22-deficient progenitors. Consistent with this observation, attenuation of IRE1α and ATF6 signaling failed to rescue development of Rpl22-deficient progenitors (Figures 5C,D and S3B). The ability of PERK knockdown to rescue development of Rpl22−/− αβ lineage T cell progenitors appeared to be mediated by attenuating p53 induction, since the expression of the p53 targets PUMA and NOXA was diminished upon PERK knockdown, but not by knockdown of IRE1α (Figure 5E and S3D). These data suggest that the selective arrest of αβ T lineage progenitors in Rpl22-deficient mice results from the restricted activation of PERK ER stress signaling, which induces p53 in those progenitors.
Figure 5. Attenuation of PERK signaling rescues the development of Rpl22−/− thymocytes in vitro.
(A-D) The role of ER stress signaling in the arrest of Rpl22−/− thymocytes was assessed by knocking down ER stress sensors PERK and Ire1α using shRNA. Rp22+/+ (A, B) and Rpl22−/− (C, D) fetal liver precursors from day 14.5 of gestation were differentiated to the DN3 stage on OP9-DL1 monolayers and then transduced with control shRNA (shCon) or those targeting p53 (Shp53), PERK (ShPERK-1) or IRE1α (ShIRE1α−1). Developmental progression was monitored by flow cytometry on day 16 of culture by electronically gating on GFP+ transduced cells. Gate frequencies of populations are listed on the histograms and were used to calculate the fold increases in absolute number of DP thymocytes, which is depicted graphically on the right as the mean ± standard deviation. Results are representative of at least three experiments performed. (E) The effect of attenuating PERK signaling on p53 activation was assessed by quantitating the expression level of p53 targets by real-time PCR analysis, as described in Figure 3. Analysis for each primer/probe set was performed for each cell type in triplicate and normalized to GAPDH. Bar graphs show the fold change relative to the expression level in control transduced Rpl22+/+ (2−ΔΔCt or RQ value + RQ max). Results are representative of 3 independent experiments. Student’s t test was used to calculate p-values: *p-value < 0.05 and **p-value < 0.01
Rpl22 modulates ER stress pathways by facilitating stress-induced translational repression
Rpl22 loss blocks development of αβ lineage progenitors by exacerbating ER stress; however, the mechanism by which Rpl22 influences ER stress signaling remained unclear. ER stress signaling enables cells to eliminate proteotoxic stress by simultaneously inducing stress-alleviating genes, while attenuating new protein synthesis (25). The attenuation of new protein synthesis is particularly important to enable cells to cope with the existing proteotoxic burden, while not continuing to make additional proteins that would add to the proteotoxic stress. To gain insight into how Rpl22 loss might be preventing the resolution of stress, we performed time course analysis of stress signaling in Rpl22-sufficient and Rpl22-deficient thymic lymphoma cell lines that had been treated with the pharmacologic stress inducer, THG. We found that the ER stress signaling was more pronounced in Rpl22-deficient cells and its resolution was delayed (Figure 6A). Given that the induction of stress alleviating genes was occurring in these cells, we asked whether another manifestation of ER stress signals, attenuation of new protein synthesis, was intact. To address this, we pharmacologically induced ER stress using THG and then measured protein synthesis by performing metabolic labeling analysis. Interestingly, though ER stress induction substantially reduced new protein synthesis in Rpl22-expressing thymic lymphomas, this was not observed in Rpl22-deficient thymic lymphomas (Figure 6B). Importantly, the inability of ER stress signals to repress protein synthesis was due to the absence of Rpl22 in these cells, as its re-introduction into Rpl22−/− thymic lymphoma cells both restored the ability of ER stress to repress new protein synthesis and enabled these cells to alleviate ER stress as indicated by the reduced expression of the targets of ER stress signals (Figure 6B,C). These data suggest that Rpl22-deficiency exacerbates ER stress by disabling the capacity of those stress signals to attenuate new protein synthesis, thereby causing the continuing generation of unfolded proteins that add to the proteotoxic burden (Figure 6D).
Figure 6. Rpl22-deficiency impairs the shutdown of protein synthesis that normally accompanies ER stress signaling.
(A) Thymic lymphomas derived from mice in which an AKT2 oncogene was expressed in T cell precursors from Rpl22+/+ and Rpl22−/− mice were treated with 0.1μM THG, following which detergent extracts were blotted with the indicated antibodies to assess the effect on ER stress signaling through the PERK pathway, as above. (B,C) The effect of Rpl22-deficiency on ER stress mediated suppression of protein synthesis was evaluated by treating the indicated AKT thymic lymphomas with 0.1μM THG for 2-4hrs and then metabolically labeling them with [35S]-methionine for 30 min. Methionine incorporation into new protein was quantified by performing TCA precipitation of triplicate samples of detergent extracts of the cells. Results are representative of 3 independent experiments. Rpl22 was re-expressed in Rpl22−/− lymphoma cells by retroviral transduction with an Rpl22 cDNA (pMiY-L22). The level of Rpl22 expression in the transduced cells was verified by immunoblotting. The effect of Rpl22-deficiency and reconstitution of ER stress signaling was assessed by quantifying the expression of ER stress response genes by real time PCR on the cell lines above. Triplicate measurements were made for each target gene, normalized to GAPDH and expressed graphically as the mean ± standard deviation. **p-value < 0.01; ***p-value < 0.001, ****p-value<0.0001. (D) A model schematizing the way that Rpl22 loss prevents the shutdown of new protein synthesis by ER stress signals and the way that continued protein synthesis contributes to prolongation and exacerbation of ER stress signaling.
DISCUSSION
In this study, we provide insight into the molecular basis by which germline ablation of the widely expressed ribosomal protein, Rpl22, results in tissue-restricted developmental defects. The tissue restriction is related to selective exacerbation of ER stress responses in the affected lineage, αβ T cell progenitors, but not in the closely related yet resistant population of γδ T lineage progenitors. In stark contrast to other RP mutations, which disrupt ribosome biogenesis and cause widespread induction of p53 through altering its stability (37, 40, 42), Rpl22 loss selectively induces p53 by controlling its translation in a manner that does not involve disruptions in ribosome biogenesis, or impairment of general protein synthesis. We show that Rpl22 is able to bind to p53 mRNA through a specific stem-loop motif and repress its translation. However, such a direct mechanism of regulating p53 translation predicts that germline ablation of Rpl22 would cause widespread p53 induction, which was not observed. This led us to elucidate the basis for the tissue restriction of p53 induction in Rpl22-deficient mice. Indeed, we found that the translational induction of p53 is restricted to αβ T cell progenitors in Rpl22-deficient mice by exacerbation of the ER stress responses that are normally linked to their differentiation program. Rpl22-deficiency exacerbates two of the three ER stress-signaling pathways, PERK and IRE1α; however, the PERK pathway is primarily responsible for developmental arrest since the arrest can be alleviated by attenuation of PERK signaling. αβ T lineage progenitors appear to be particularly susceptible to the dysregulation of PERK signaling as they traverse the β-selection checkpoint at the DN3 stage. Interestingly, at this stage, the cells transition from quiescence to robust proliferation, which activates ER stress signaling, and is associated with a marked increase in Rpl22 levels (13, 49). Accordingly, one important role that Rpl22 plays during normal T cell development is to modulate ER stress signaling during this transition by enabling the progenitors to repress new protein synthesis, an issue that γδ lineage progenitors apparently need not confront.
A critical question is why αβ lineage progenitors are susceptible to exacerbation of ER stress signals, while γδ progenitors appear to be resistant. For the αβ lineage, pre-TCR signaling and passage through the β-selection checkpoint is a critical stage in thymocyte development that is accompanied by, and dependent upon, a rapid burst of proliferation (49). Interestingly, γδ T cells, which are not arrested by Rpl22 loss, do not undergo such a burst in proliferation (24). Thus, it may be the abrupt transition from quiescence to rapid proliferation that sensitizes Rpl22-deficient αβ lineage progenitors to arrest. Consistent with this notion, we observe a similar arrest at the equivalent stage of B lymphoid development, where pre-B cell receptor signals induce a comparably abrupt transition to rapid proliferation (53). Nevertheless, we have not yet been able to test the causal relationship between proliferation and arrest, which could entail blunting the capacity of Rpl22-deficient αβ lineage progenitors to proliferate.
While the exacerbation of ER stress signals clearly plays a critical role in tissue-restriction of the developmental defects caused by Rpl22-deficiency, the mechanistic link between ER stress and p53 induction remains unclear. PERK is the critical ER stress sensor responsible for inducing p53 and arresting T cell development, as evidenced by the ability of PERK knockdown to alleviate the developmental arrest. PERK signaling has previously been shown to enhance p53 synthesis; however, the molecular basis for the increase in p53 translation has not been elucidated (54). Nevertheless, p53 induction by PERK is likely to result from one of the following modes of regulation. First, p53 could be activated by PERK through its transcriptional effector ATF4, which may transactivate a factor that controls the translation of p53 mRNA. A number of trans-acting factors have been implicated in regulating p53 translation, including Rpl26 and nucleolin (55); however, whether the expression of these and other factors is regulated by ATF4 is unknown at present. Alternatively, PERK might also activate p53 through translational controls induced by phosphorylation of eIF2α. eIF2α phosphorylation increases expression of ATF4 by initiating translation through an upstream open reading frame, which utilizes non-standard translational start codons and depends upon eIF2A (56). This mode of regulation might also be capable of either controlling p53 translation directly, or may indirectly regulate p53 translation by controlling the expression of a distinct trans-acting factor that modulates p53 translation. Ribosome profiling has revealed that this mode of regulation is quite common (57). Efforts to distinguish these possibilities are in progress.
Our analysis reveals that one of the critical ways that Rpl22 facilitates αβ T cell development is by regulating the homeostatic ER stress responses encountered when those progenitors traverse the β-selection checkpoint (45). We show that it is the loss of this regulatory function of Rpl22 that is responsible for arresting αβ T cell development. Nevertheless, an important unanswered question is how Rpl22 regulates ER stress signaling. It appears to be tightly linked to a failure to arrest new protein synthesis in response to ER stress signaling. Indeed, ER stress signaling is exacerbated upon Rpl22-loss despite the profound induction of stress-alleviating genes, presumably because the unabated synthesis of nascent proteins continues to add to the proteotoxic burden. When Rpl22 is re-expressed in Rpl22-deficient cells, the repression of protein synthesis in response to ER stress stimuli is re-established and stress-signaling processes return to baseline. However, the mechanism by which Rpl22 contributes to the shutdown of new protein synthesis following ER stress signaling remains to be established. Phosphorylation of eIF2α in response to proteotoxic stress represses new protein synthesis through sequestration of the guanine nucleotide exchange factor, eIF2B (58, 59). eIF2B is required to initiate new protein synthesis and so global, cap-dependent protein synthesis is blocked when eIF2B is sequestered through physical association with phospho-eIF2α (60). The ability of eIF2α phosphorylation to sequester eIF2B and shut down protein synthesis can be disabled when eIF2B expression rises to super-stoichiometric levels (59). Accordingly, one way Rpl22-deficiency could block the shutdown of protein synthesis could be to enhance either eIF2B expression or function. The shutdown of protein synthesis also depends on the function of cellular stress granules and so another possibility is that Rpl22 loss might block the shutdown of new protein synthesis by disabling stress granule function (61). Although the role of Rpl22 in stress granules remains unexplored, there is evidence showing Rpl22 is able to associate with argonaute 2 via an RNA bridge (62). Argonaute 2 plays an important role in translational repression by stress granules and efforts are currently underway to determine if Rpl22-loss interferes with stress granule formation or function (63).
Our findings illustrate the critical lineage-restricted regulatory functions of RP and reveal ER stress pathways as a novel set of signaling pathways that control the fate of developing αβ T cells. The basis for the dependence of certain lineages on Rpl22 remains unresolved but could relate to the extent to which proteotoxic stress is encountered during normal development. Consistent with this view, we observe that Rpl22-dependence occurs in both B and T lineage progenitors as they transition from quiescence to rapid proliferation in response to pre-receptor signals. Rpl22 may facilitate the cell’s ability to manage this transition through its ability to contribute to the shutdown of protein synthesis in response to stress. Critical future efforts will focus on determining how Rpl22 does so, which will entail unraveling the basis for the regulatory functions of Rpl22 and whether they are mediated from within specialized ribosomes or, alternatively, in an extraribosomal fashion. This will likely provide a new perspective on how RP function in regulating normal development and progression to disease.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Drs. Balachandran, Sykes and Steel for critical review of manuscript, and Drs. Vaidya and Bouchard for helpful suggestions. We gratefully acknowledge the assistance of the following core facilities of the Fox Chase Cancer Center: Cell Culture, DNA Sequencing, Flow Cytometry, Genomics, and Laboratory Animal.
ABBREVIATIONS
- Ago2
Argonaute RISC Catalytic Component 2
- ATF3
Activating Transcription Factor-3
- ATF4
Activating Transcription Factor-4
- ATF6
Activating Transcription Factor-6
- CHOP/DDIT3
DNA damage-inducible transcript-3
- DN
double negative
- DP
double positive
- EIF2A
Eukaryotic translation initiation factor 2-alpha
- EIF2B
Eukaryotic translation initiation factor 2- beta
- ER
Stress Endoplasmic Reticulum Stress
- HERP
Homocysteine-Inducible ER Stress-Inducible Ubiquitin-Like Domain
- HRD-1
HMG-CoA reductase degradation protein 1
- IRE1a
inositol-requiring enzyme 1-alpha
- Noxa
NADPH oxidase activator 1
- PERK
protein kinase RNA-like endoplasmic reticulum kinase
- pre-TCR
pre-T cell receptor
- PUMA
p53-upregulated modulator of apoptosis
- RAG-1/2
recombination-activated genes 1/2
- RP
Ribosomal protein
- RpL22
ribosomal protein L22
- TCA
Tricarboxylic acid
- TCR
T cell receptor
- Tg
Transgenic
- THG
Thapsigargin
- TUN
Tunicamycin
- XBP-1
X-box binding protein 1
Footnotes
This work was supported by NIH grants R01AI110985 and core grant P30CA006927, Leukemia and Lymphoma Society Grant 6057-14, and an appropriation from the Commonwealth of Pennsylvania.
REFERENCES
- 1.Raiser DM, Narla A, Ebert BL. The emerging importance of ribosomal dysfunction in the pathogenesis of hematologic disorders. Leukemia & lymphoma. 2014;55:491–500. doi: 10.3109/10428194.2013.812786. [DOI] [PubMed] [Google Scholar]
- 2.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig T, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, Tentler D, Mohandas N, Carlsson B, Dahl N. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nature genetics. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 4.Ellis SR, Gleizes PE. Diamond Blackfan anemia: ribosomal proteins going rogue. Seminars in hematology. 2011;48:89–96. doi: 10.1053/j.seminhematol.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Danilova N, Sakamoto K, Lin S. Ribosomal protein L11 mutation in zebrafish leads to haematopoietic and metabolic defects. British journal of haematology. 2011;152:217–228. doi: 10.1111/j.1365-2141.2010.08396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakhoul H, Ke J, Zhou X, Liao W, Zeng SX, Lu H. Ribosomopathies: mechanisms of disease. Clinical medicine insights. Blood disorders. 2014;7:7–16. doi: 10.4137/CMBD.S16952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danilova N, Gazda HT. Ribosomopathies: how a common root can cause a tree of pathologies. Disease models & mechanisms. 2015;8:1013–1026. doi: 10.1242/dmm.020529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warner J, McIntosh K. How common are extraribosomal functions of ribosomal proteins? Molecular cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCann KL, Baserga SJ. Genetics. Mysterious ribosomopathies. Science. 2013;341:849–850. doi: 10.1126/science.1244156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nature reviews. Molecular cell biology. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X, Liao JW, Liao MJ, Liao P, Lu H. Ribosomal proteins: functions beyond the ribosome. Journal of Molecular Cell Biology. 2015;7:92–104. doi: 10.1093/jmcb/mjv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fahl SP, Wang M, Zhang Y, Duc AC, Wiest DL. Regulatory Roles of Rpl22 in Hematopoiesis: An Old Dog with New Tricks. Critical reviews in immunology. 2015;35:379–400. doi: 10.1615/critrevimmunol.v35.i5.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson S, Lauritsen J, Hartman M, Foushee A, Lefebvre J, Shinton S, Gerhardt B, Hardy R, Oravecz T, Wiest D. Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity. 2007;26:759–772. doi: 10.1016/j.immuni.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 14.Stadanlick J, Zhang Z, Lee S-Y, Hemann M, Biery M, Carleton M, Zambetti G, Anderson S, Oravecz T, Wiest D. Developmental arrest of T cells in Rpl22-deficient mice is dependent upon multiple p53 effectors. Journal of immunology (Baltimore, Md. : 1950) 2011;187:664–675. doi: 10.4049/jimmunol.1100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrie H, Scollay R, Shortman K. Commitment to the T cell receptor-alpha beta or -gamma delta lineages can occur just prior to the onset of CD4 and CD8 expression among immature thymocytes. European journal of immunology. 1992;22:2185–2188. doi: 10.1002/eji.1830220836. [DOI] [PubMed] [Google Scholar]
- 16.Ciofani M, Knowles GC, Wiest DL, von Boehmer H, Zuniga-Pflucker JC. Stage-specific and differential notch dependency at the alphabeta and gammadelta T lineage bifurcation. Immunity. 2006;25:105–116. doi: 10.1016/j.immuni.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 17.Wiest D, Berger M, Carleton M. Control of early thymocyte development by the pre-T cell receptor complex: A receptor without a ligand? Seminars in immunology. 1999;11:251–262. doi: 10.1006/smim.1999.0181. [DOI] [PubMed] [Google Scholar]
- 18.Takahama Y. Journey through the thymus: stromal guides for T-cell development and selection. Nature reviews. Immunology. 2006;6:127–135. doi: 10.1038/nri1781. [DOI] [PubMed] [Google Scholar]
- 19.Livak F, Tourigny M, Schatz DG, Petrie HT. Characterization of TCR gene rearrangements during adult murine T cell development. J Immunol. 1999;162:2575–2580. [PubMed] [Google Scholar]
- 20.Taccioli GE, Rathbun G, Shinkai Y, Oltz EM, Cheng H, Whitmore G, Stamato T, Jeggo P, Alt FW. Activities involved in V(D)J recombination. Curr Top Microbiol Immunol. 1992;182:107–114. doi: 10.1007/978-3-642-77633-5_13. [DOI] [PubMed] [Google Scholar]
- 21.Lee SY, Stadanlick J, Kappes DJ, Wiest DL. Towards a molecular understanding of the differential signals regulating alphabeta/gammadelta T lineage choice. Semin Immunol. 2010;22:237–246. doi: 10.1016/j.smim.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong GW, Zuniga-Pflucker JC. gammadelta and alphabeta T cell lineage choice: resolution by a stronger sense of being. Semin Immunol. 2010;22:228–236. doi: 10.1016/j.smim.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Pereira P, Zijlstra M, McMaster J, Loring JM, Jaenisch R, Tonegawa S. Blockade of transgenic gamma delta T cell development in beta 2-microglobulin deficient mice. EMBO Journal. 1992;11:25–31. doi: 10.1002/j.1460-2075.1992.tb05023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 25.Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nature reviews. Molecular cell biology. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 26.Woehlbier U, Hetz C. Modulating stress responses by the UPRosome: a matter of life and death. Trends in biochemical sciences. 2011;36:329–337. doi: 10.1016/j.tibs.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, Pak DN, Fields S, Kennedy BK, Kaeberlein M. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haks MC, Lefebvre JM, Lauritsen JP, Carleton M, Rhodes M, Miyazaki T, Kappes DJ, Wiest DL. Attenuation of gammadeltaTCR signaling efficiently diverts thymocytes to the alphabeta lineage. Immunity. 2005;22:595–606. doi: 10.1016/j.immuni.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Fehling HJ, Krotkova A, Saint-Ruf C, von Boehmer H. Crucial role of the pre-T-cell receptor alpha gene in development of alpha beta but not gamma delta T cells. Nature. 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- 30.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Current biology : CB. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 31.Rao S, Lee S-Y, Gutierrez A, Perrigoue J, Thapa RJ, Tu Z, Jeffers JR, Rhodes M, Anderson S, Oravecz T, Hunger SP, Timakhov RA, Zhang R, Balachandran S, Zambetti GP, Testa JR, Look TA, Wiest DL. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood. 2012;120:3764–3773. doi: 10.1182/blood-2012-03-415349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nature genetics. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 33.Rashkovan M, Vadnais C, Ross J, Gigoux M, Suh W-K, Gu W, Kosan C, Möröy T. Miz-1 regulates translation of Trp53 via ribosomal protein L22 in cells undergoing V (D) J recombination. Proceedings of the National Academy of Sciences. 2014;111 doi: 10.1073/pnas.1412107111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobbelstein M, Shenk T. In vitro selection of RNA ligands for the ribosomal L22 protein associated with Epstein-Barr virus-expressed RNA by using randomized and cDNA-derived RNA libraries. J Virol. 1995;69:8027–8034. doi: 10.1128/jvi.69.12.8027-8034.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic acids research. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol. 2001;21:4246–4255. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubbi C, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. The EMBO journal. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29:4253–4260. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 39.Boulon S, Westman B, Hutten S, Boisvert F-M, Lamond A. The nucleolus under stress. Molecular cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 41.Yuan X, Zhou Y, Casanova E, Chai M, Kiss E, Gröne H-J, Schütz G, Grummt I. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Molecular cell. 2005;19:77–87. doi: 10.1016/j.molcel.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Wolf G, Bhat K, Jin A, Allio T, Burkhart W, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Molecular and cellular biology. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tao W, Levine AJ. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci U S A. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steffen K, MacKay V, Kerr E, Tsuchiya M, Hu D, Fox L, Dang N, Johnston E, Oakes J, Tchao B, Pak D, Fields S, Kennedy B, Kaeberlein M. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brunsing R, Omori S, Weber F, Bicknell A, Friend L, Rickert R, Niwa M. B- and T-cell development both involve activity of the unfolded protein response pathway. The Journal of biological chemistry. 2008;283:17954–17961. doi: 10.1074/jbc.M801395200. [DOI] [PubMed] [Google Scholar]
- 46.Rutkowski D, Kaufman R. A trip to the ER: coping with stress. Trends in cell biology. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 47.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Developmental cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 48.Yani C, Federica B. IRE1: ER stress sensor and cell fate executor. Trends in Cell Biology. 2013;23 doi: 10.1016/j.tcb.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kreslavsky T, Gleimer M, Miyazaki M, Choi Y, Gagnon E, Murre C, Sicinski P, von Boehmer H. beta-Selection-induced proliferation is required for alphabeta T cell differentiation. Immunity. 2012;37:840–853. doi: 10.1016/j.immuni.2012.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thastrup O, Cullen PJ, Drøbak BK, Hanley MR, Dawson AP. Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2 (+)-ATPase. Proceedings of the National Academy of Sciences. 1990;87:2466–2470. doi: 10.1073/pnas.87.7.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heifetz A, Keenan RW, Elbein AD. Mechanism of action of tunicamycin on the UDP-GlcNAc:dolichyl-phosphate Glc-NAc-1-phosphate transferase. Biochemistry. 1979;18:2186–2192. doi: 10.1021/bi00578a008. [DOI] [PubMed] [Google Scholar]
- 52.Zúñiga-Pflücker J. T-cell development made simple. Nature reviews. Immunology. 2004;4:67–72. doi: 10.1038/nri1257. [DOI] [PubMed] [Google Scholar]
- 53.Fahl SP, Harris B, Coffey F, Wiest DL. Rpl22 Loss Impairs the Development of B Lymphocytes by Activating a p53-Dependent Checkpoint. Journal of immunology (Baltimore, Md. : 1950) 2015;194:200–209. doi: 10.4049/jimmunol.1402242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bourougaa K, Naski N, Boularan C, Mlynarczyk C, Candeias M, Marullo S, Fåhraeus R. Endoplasmic reticulum stress induces G2 cell-cycle arrest via mRNA translation of the p53 isoform p53/47. Molecular cell. 2010;38:78–88. doi: 10.1016/j.molcel.2010.01.041. [DOI] [PubMed] [Google Scholar]
- 55.Chen J, Guo K, Kastan M. Interactions of nucleolin and ribosomal protein L26 (RPL26) in translational control of human p53 mRNA. The Journal of biological chemistry. 2012;287:16467–16476. doi: 10.1074/jbc.M112.349274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, Martins-Green M, Shastri N, Walter P. Translation from the 5' untranslated region shapes the integrated stress response. Science (New York, N.Y.) 2016;351 doi: 10.1126/science.aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kimball SR, Fabian JR, Pavitt GD, Hinnebusch AG, Jefferson LS. Regulation of guanine nucleotide exchange through phosphorylation of eukaryotic initiation factor eIF2alpha. Role of the alpha- and delta-subunits of eiF2b. J Biol Chem. 1998;273:12841–12845. doi: 10.1074/jbc.273.21.12841. [DOI] [PubMed] [Google Scholar]
- 59.Balachandran S, Barber GN. Defective translational control facilitates vesicular stomatitis virus oncolysis. Cancer Cell. 2004;5 doi: 10.1016/s1535-6108(03)00330-1. [DOI] [PubMed] [Google Scholar]
- 60.Pavitt GD, Ramaiah KVA, Kimball SR. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine–nucleotide exchange. Genes & …. 1998 doi: 10.1101/gad.12.4.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 62.Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Molecular cell. 2008 doi: 10.1016/j.molcel.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson P, Kedersha N, Ivanov P. Stress granules, P-bodies and cancer. Biochim Biophys Acta. 2015;1849:861–870. doi: 10.1016/j.bbagrm.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.