Abstract
Background
Allospecific CD154+T-cytotoxic memory cells (CD154+TcM) predict acute cellular rejection (ACR) after liver or intestine transplantation (LTx, ITx) in small cohorts of children and can enhance immunosuppression management, but await validation and clinical implementation.
Methods
To establish safety and probable benefit, CD154+TcM were measured in cryopreserved samples from 214 children <21 years (NCT#1163578). Training set samples, n=158, were tested with research-grade reagents and 122 independent validation set samples were tested with cGMP-manufactured reagents after assay standardization and reproducibility testing. Recipient CD154+TcM induced by stimulation with donor cells were expressed as a fraction of those induced by HLA-non-identical cells in parallel cultures. The resulting immunoreactivity index (IR) if > 1 implies increased rejection-risk.
Results
Training and validation set subjects were demographically similar. Mean coefficient of test variation was <10% under several conditions. Logistic regression incorporating several confounding variables identified separate pre-transplant and post-transplant IR thresholds for prediction of rejection in respective training set samples. An IR ≥ 1.1 in post-transplant training samples, and IR ≥1.23 in pre-transplant training samples predicted LTx or ITx rejection in corresponding validation set samples in the 60-day post-sampling period with sensitivity, specificity, positive and negative predictive values of 84%, 80%, 64%, and 92%, respectively (AUC 0.792), and 57%, 89%, 78%, and 74%, respectively (AUC 0.848). No adverse events were encountered due to phlebotomy.
Conclusions
Allospecific CD154+T-cytotoxic memory cells predict acute cellular rejection after liver or intestine transplantation in children. Adjunctive use can enhance clinical outcomes.
Introduction
Predicting acute cellular rejection (ACR) accurately can enhance safe use of immunosuppression in the rare population of children with liver or intestine transplantation (LTx, ITx). Inadequate immunosuppression can lead to ACR in 30-40% LTx and 30-60% ITx, while over-immunosuppression is a leading cause of late mortality due to life-threatening infections and lymphoma.1-7 Immunosuppression dosing is based on the risk of rejection, which is assessed with a combination of clinical and laboratory findings and biopsy. These parameters lack specificity for rejection-risk. Features of ITx rejection such as fever or diarrhea, or of LTx rejection such as elevated liver function tests are also seen with systemic viral illnesses. The cross-match blood test predicts antibody-mediated rejection, but not ACR. Biopsies detect ongoing rejection, cannot predict a future episode, and are invasive surgical procedures, which can also cause bleeding or perforation.
Non-invasive prediction of rejection can add specificity to clinical rejection-risk assessment, but remains an unmet need and is challenging. Roughly 500 children receive LTx and 50 children receive ITx in the United States each year.8 These low numbers preclude powered organ-specific test evaluation, but qualify such an assay for regulatory consideration as an orphan device, because the disease condition affects ≤4000 patients per year.9 Augmenting analyzable subjects by combining LTx and ITx populations is a potential solution but would require a test system predicated on common mechanisms, for e.g. donor specific alloresponse, a universal mechanism of transplant rejection. The Humanitarian Device exemption regulatory path incentivizes device development for orphan populations by requiring that such a test 1) addresses an unmet need and has no predicate for the intended use, 2) does not pose an unreasonable or significant risk of injury, and 3) demonstrates probable benefit which outweighs the risk of injury or illness related to its intended use.10 Impending regulation of in-vitro diagnostics is likely to foster interest in this mechanism for rare and high-risk diseases.11,12
A prospective immune monitoring protocol at our center (NCT#1163578) shows that allospecific T-cytotoxic memory cells, which express the inflammatory marker, CD154 (CD154+TcM) predict and associate with ACR after several types of transplants with high sensitivity and specificity in training-set validation-set testing of small cohorts.13-16 Described in our previous reports, the innovations in this test system relative to others include co-culture of living responder and stimulator cells pre-labeled with fluorochrome-labeled antibody, inclusion of monensin and detector antibodies to CD154 in the culture medium, and prediction of rejection with CD154+TcM. 13-16 CD154+TcM are measured in recipient peripheral blood leukocytes (PBL) after overnight stimulation with donor and HLA-non-identical PBL in parallel reactions. If donor-induced CD154+TcM exceed those induced by reference PBL, the resulting ratio termed the immunoreactivity index or IR exceeds 1 and implies increased risk of rejection (Figure 1). An index <1 implies decreased risk. This concept was derived from the proliferative mixed lymphocyte culture, in which donor-specific alloreactivity was enhanced among rejection-prone children compared with those who were rejection-free.17,18 The IR is a personalized output because donor-specific CD154+TcM are normalized to those induced by a reference allostimulus for the same recipient. Disease-specificity has been established with regression models, in which CD154+TcM emerged as the best predictor of rejection from among naïve and memory T-helper and T-cytotoxic cells in independent analyses of liver, intestine and renal allograft recipients.13-16 If donor cells are not available for extended testing, PBL from normal human subjects, which match donor at one antigen each at the HLA-A, -B and -DR loci, have been used as “surrogate” donor cells in this test system without compromising rejection-risk assessment.16 Based on these data and unmet clinical need, CD154+TcM received Humanitarian Use Device designation (HUD#08-0206) for the measurement of rejection-risk and the management of immunosuppression in children with LTx or ITx by the FDA's Office of Orphan Products in 2009. Here, we describe pre-clinical performance evaluation of this test system leading to its FDA approval.19 The additional innovations described here include a negative control reaction condition to enhance reliability of the flow cytometry gating strategy, statistical comparison of stimulated and background reaction conditions to enhance reliable detection of true positive CD154+TcM, test standardization with cGMP reagents and extensive reproducibility testing, and validation of test performance in training set samples in independent validation samples. 20, 21
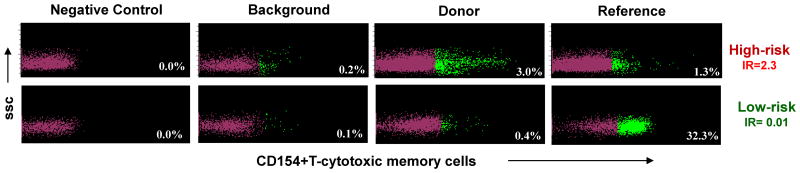
Figure 1.
Upper panel with four scatterplots shows increased risk of rejection, because CD154+TcM induced by stimulation with donor PBL exceed those produced after stimulation with HLA-non-identical PBL in the reference reaction. Lower panel with four scatterplots shows decreased risk of rejection, because donor-induced CD154+TcM are exceeded by those in the reference reaction. The antibody to CD154 is labeled with the fluorochrome, phycoerythrin. T-cytotoxic memory cells which express CD154 (green dots) are separated from those that do not express CD154 (magenta dots) by implementing the gating strategy described in Supplementary Figure 1 in negative control reaction condition. SSC = side scatter.
Methods
Subjects
After informed consent (University of Pittsburgh IRB # 0405628 NCT#1163578), blood samples were obtained prospectively from children <21 years with LTx or ITx to determine immunoreactivity indices of CD154+TcM (IR).
Samples and assay
Samples were obtained before (IR0) or after transplantation during the first 60 days (IR1), days 61-199, and at days 200 onward (IRx) at surveillance visits or “for cause” biopsies. Ficoll-purified PBL from 3-5 ml whole blood were de-identified and cryopreserved in liquid nitrogen for batched analysis of allospecific CD154+TcM with flow cytometry after overnight 16-hour culture with donor cells and HLA-non-identical human cells in parallel reactions, as described previously (Figure 1).13 Because recipients return to referring facilities during days 61-199, sample collection was inconsistent during this period. Therefore, these samples were not analyzed. Samples in which stimulation with donor and HLA-non-identical PBL failed to generate increased CD154+TcM cell counts over background (P≤0.05, Poisson test) were not analyzed.20 Samples with <0.45 million viable PBL after thawing were inadequate for assay setup and were discarded.
Endpoints and terminology
ACR within the 60-day period after sampling or after transplantation was the study endpoint. Biopsy-proven rejection was confirmed by re-review of all biopsies by either one of two senior pathologists (RJ or SR) using established criteria.21 In some LTx recipients who could not be biopsied, elevated liver function tests and absence of bile duct dilatation on ultrasound implied rejection. Subjects with and without ACR in the 60-day post-sampling period were termed rejectors and non-rejectors, respectively.
Study and assay design
The test system was evaluated in three phases between 2006-2012: on training set subject samples, on normal human PBL for assay standardization and precision testing, and on validation set subject samples.
De-identified training set samples were analyzed with research grade fluorochrome-labeled antibodies and the LSRII flow cytometer (BDBiosciences, San Jose, CA) between 2006-2010. Test results were merged with outcomes. Threshold IR values which predicted rejection within 60 days after the sample were established with training set samples. A separate threshold was developed for pre-transplant IR0, when no immunosuppression is used. Post-transplant IR1 and IRx samples were analyzed together because they were obtained from immunosuppressed subjects. Only one sample was used in the pre- or post-transplant periods from any given subject so only independent measurements existed within respective post- and pre-transplant models. To capture as many early rejection events in these rare subjects, the IR1 sample was used preferentially over the IRx sample if both were available from a recipient. The general approach to training-set/validation-set testing is illustrated in Figure 2.
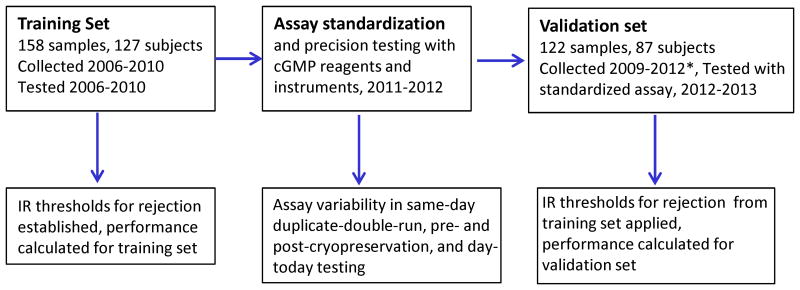
Figure 2.
Flow chart with timelines for testing of training set samples, assay standardization and precision testing, and testing of validation set samples.
Before testing the performance of predictive IR thresholds in validation set samples, a standardized test format was developed between 2011-2012 using assays between HLA-mismatched PBL from normal human subjects. Test reproducibility was established per guidelines of the National Committee of Clinical Laboratory Standards.22 These assays used cGMP-synthesized versions of antibodies used previously, and which were conjugated to brighter fluorochromes (BD Biosciences, San Jose, CA) and the FDA-approved FACS-CANTO flow cytometer (BD Biosciences, San Jose, CA). Stimulator and responder PBL were pre-labeled with an identical clone of anti-Tc antibody conjugated to two different tandem dyes to distinguish responder from stimulator (SDC, Figure 1). The brighter tandem dyes, allophycocyanin-H7 (APCH7, catalog number 641409) for responder Tc and phycoerythrin-cyanin-7 (PECy7, catalog number 335805) for stimulator Tc, prevented loss of cell counts due to dye quenching, and confirmed that the tandems did not dissociate and stain other cells in the culture. Other reagents included the viability dye 7-aminoactinomycin-D, catalog number 559925) and fluorochrome-labeled the T-cell marker CD3 (flourescein isothiacyanate, FITC, catalog number 349201), and the memory marker CD45RO (allophycocyanin, APC, catalog number 340438) (SDC, Figure 1). No change was made to a) the anti-CD154 antibody (catalog number 555700) which is custom conjugated to the fluorochrome phycoerythrin (PE) for our purposes under cGMP conditions by BD Biosciences, San Jose, and b) the cell culture medium consisted of RPMI (Invitrogen, catalog number 22400-089), fetal calf serum (Invitrogen, catalog number 10082-147) and monensin (Golgi stop, BD Biosciences, catalog number 5544724).
In the final assay used for reproducibility studies, recipient PBL pre-labeled with anti-CD8-APCH7 were incubated without (negative control) or with anti-CD154-PE (background) in culture medium. For the variability studies, pre-labeled recipient PBL were also incubated 1: 1 with HLA-non-identical PBL prelabeled with antiCD8-PECy7 (stimulated). The stimulated reaction was replaced with the donor and reference reactions in assays performed in subject samples. The donor and recipient reactions consisted respectively of pre-labeled recipient PBL incubated 1: 1 with pre-labeled donor PBL (donor) and pre-labeled HLA-non-identical PBL (reference). SDC Figure 1 describes the gating strategy for the test system. The preset acceptable upper limit of mean coefficient of variation (%CV) for CD154+TcM induced by stimulation was 20%.
Validation set samples consisted of archived subject samples with ≥ 2 million total cells, which were not tested with or were accrued after testing of training set samples. These samples were obtained between 2009-2012, de-identified by study coordinator (AB), and analyzed with the standardized test format between 2012-2013. Test results were linked to subject identity and outcomes by the statistician (BH), performance determined by applying training set rejection-risk thresholds, and results communicated to senior author (RS).
Overlap in training and validation set time periods
To utilize resources efficiently, testing of some samples obtained during the accrual period for the training set (2006-2010) was deferred pending availability of additional samples from the same subject, or stimulator cells from the appropriate normal human donor. These samples made up the validation set along with those collected after the training set collection period (2009-2012), resulting in overlapping time periods for the two sample sets (Table 1A). There was no contamination of samples between the training and validation data sets for a particular time period, pre- or post-transplant.
Table 1A.
Subject and sample characteristics. Organ type L=liver, LSB=liver-intestine, SB=intestine, LK=liver-kidney. Sample type IR0=pre-transplant sample. IR1=sample obtained on post-transplant days 1-60. IRx=sample obtained on post-days 200-onward. A negative prefix for time between sample and transplant denotes a pre-transplant sample obtained before transplantation. The time between sample and biopsy/event is preceded by a negative symbol or is zero because all samples were obtained before or on the day of the biopsy/event.
| Training set( August 10, 2006 to June 9. 2010) | Validation set( March 19, 2009 to August 16, 2012) | ||||
|---|---|---|---|---|---|
| All | Analyzable | All | Analyzable | p- valuetest vs validation(All subjects) | |
| Parameters | |||||
| Total subjects | 127 | 120 | 87 | 72 | |
| Subject Age (Years) (mean +/-SD, range) | 8.0±6.3,0.2 to 20.9 | 8.0±6.3,0.2 to 20.9 | 8.6±6.7,0.4 to 20.2 | 8.7±6.6,0.4 to 20.1 | NS (0.502) |
| Gender (M:F) | 81:46 | 76:44 | 48:39 | 41:31 | NS (0.255) |
| Race (Caucasian: non-Caucasian) | 92:35 | 86:34 | 68:19 | 55:17 | NS (0.423) |
| Organ (L: LSB: SB: LK: LL) | 83:24:17:3:0 | 79:22:16:3:0 | 71:5:10:0:1 | 61:2:8:0:1 | 0.007 |
| Induction (None: thymo: campath) | 41:73:13 | 38:69:13 | 38:46:3 | 32:37:3 | NS (0.080) |
| FKWB ( ng / ml ) | 9.5 ± 6.3 | 9.3 ± 6.1 | 7.9 ± 5.1 | 7.5 ± 4.6 | NS (0.095) |
| Actual donor: surrogate donor | 24 :103 | 24 : 96 | 6 : 81 | 5 : 67 | 0.015 |
| Total samples | 158 | 147 | 122 | 97 | |
| IR0 samples | 50 | 49 | 43 | 33 | |
| IR1 samples | 54 | 48 | 39 | 30 | |
| IRx samples | 54 | 50 | 40 | 34 | |
| Time between Transplantation and pre-transplant sample (days) IR0 / (range) | -0.4 ±2(-14 to 0) | -0.4 ±2(-14 to 0) | -4.1 ±15.6(-85 to 0) | -5.2 ±17.7(-85 to 0) | |
| Time between Transplantation and post-transplant samples (days) IR1+IRx | 1183 ± 1786(7 to 6226) | 1202 ± 1825(7 to 6226) | 938± 1435(6 to 5360) | 989 ± 1474(6 to 5360) | NS (0.336) |
| Time between sample and biopsy/event (days) / (range) | 14.4±14.5( 0 to 59) | 14.8 ± 14.4(0 to 59) | 15.2±17.2( 0 to 59) | 13.8±17.0(0 to 59) | NS (0.769) |
| Exclusions | |||||
| Failure to generate signal | 11 | 16 | |||
| Technical failure (inadequate cell counts) | 0 | 9 | |||
Statistical analysis
Logistic regression was used to define respective IR thresholds for pre- and post-transplant training set samples at or above which rejection was predicted within the 60-day period after sampling.23-24 To evaluate factors confounding prediction of ACR, covariates in the logistic model included: age, gender, race (Caucasian vs non-Caucasian), type of stimulator cell (actual donor or surrogate donor), organ transplant type (liver, intestine, combined liver-intestine or combined liver-kidney), tacrolimus whole blood concentrations (FKWBC), induction (rabbit antihuman thymocyte globulin (rATG, Genzyme), campath (alemtuzumab, Genzyme), or none), and time between transplantation and outcome. The IR of CD154+TcM, was log10 transformed to reduce the effect of skewness (rejectors: >1 to 46, SDC, Table 1; and non-rejectors: 0 to 7) and achieve normality. Test performance was calculated as sensitivity, specificity, positive and negative predictive values (PPV, NPV) with 95% confidence intervals, as well as area under the receiver-operating-characteristic curve (AUC, ROC). For the ROC analysis, we weighed the sensitivity and specificity equally and selected the cut-point that maximized both of these parameters simultaneously. The pre- and post-transplant logistic regression models both stratified by and including all covariates (described above) were compared to the single CD154+TcM IR variable models for predicting training set samples. All analyses were conducted in the R statistical programming environment.25
Results
Patients
Test performance was evaluated in in 280 total samples from 214 subjects. The training set included 158 samples from 127 subjects (Table 1A). After excluding 11 samples, which failed stimulation, 147 samples from 120 subjects were analyzed. Samples were evenly distributed in pre-transplant or IR0, and the two post-transplant IR1 and IRx periods. The validation set of 122 samples from 87 subjects was similarly reduced to 97 analyzable samples from 72 subjects after excluding 9 samples with inadequate cell counts and 16 samples for failed stimulation. Fewer actual donor cells were used as stimulators in the validation cohort because of fewer living donor LTx in this period. FKWBC were also lower in the validation set. Fewer small-bowel containing allograft recipients were present in the validation set. The groups were similar in all other respects. Sampling occurred at a mean interval of two weeks before a biopsy in either cohort. Differences in donor-recipient HLA-matching between rejectors and non-rejectors did not achieve statistical significance (Table 1B). Three subjects who provided an analyzable pre-transplant (IR0) training set sample also provided an analyzable validation set IRx sample late after transplantation (SDC Figure. 2).
Table 1B. Differences in HLA match at the HLA-A, -B and DR loci between rejectors and non-rejectors for pre- and post-transplant samples in the training and validation sets.
| Rejector | non-rejector | p:value* | ||
|---|---|---|---|---|
| Pre-transplant Training set samples, IR0 (n=49) | N | 25 | 24 | |
| A-match | 0.52 | 0.46 | 0.78 (NS) | |
| B-match | 0.40 | 0.21 | 0.09 (NS) | |
| DR-match | 0.52 | 0.46 | 0.78 (NS) | |
| Post-transplant Training set samples, IR1&IRx (n=98) | N | 24 | 74 | |
| A-match | 0.67 | 0.66 | 0.97 (NS) | |
| B-match | 0.29 | 0.32 | 0.97 (NS) | |
| DR-match | 0.46 | 0.54 | 0.59 (NS) | |
| Pre-transplant Validation set samples, IR0 (n=33) | N | 14 | 19 | |
| A-match | 0.57 | 0.63 | 0.78 (NS) | |
| B-match | 0.29 | 0.42 | 0.53 (NS) | |
| DR-match | 0.50 | 0.42 | 0.87 (NS) | |
| Post-transplant Validation set samples, IR1&IRx (n=64) | N | 19 | 45 | |
| A-match | 0.63 | 0.62 | 0.92 (NS) | |
| B-match | 0.26 | 0.40 | 0.30 (NS) | |
| DR-match | 0.58 | 0.58 | 0.81 (NS) |
p-value: Mann-Whitney test
Immunosuppression
The relative distribution of induction and maintenance immunosuppression among analyzable pre- and post-transplant samples in the training and validation sets are shown in Table 1C. Induction was performed with rabbit anti-human thymocyte globulin (rATG, Genzyme, Cambridge, MA) or alemtuzumab (campath, Genzyme, Cambridge, MA) in all intestine recipients and some liver recipients. A subset of liver recipients did not receive induction therapy. Maintenance immunosuppression was started after transplantation and consisted of Tacrolimus or rapamycin as the primary agent. Steroids and cellcept were used as adjunctive maintenance agents. Three liver recipients, two in the training set and one in the validation set were free of maintenance immunosuppression. Fewer samples were obtained after campath induction in the validation set compared with the training set because of fewer recipients of small bowel allografts in the validation set.
Table 1C. Distribution of induction and maintenance immunosuppressants.
| N (%) | N (%) | N (%) | N (%) | N (%) | ||
|---|---|---|---|---|---|---|
| INDUCTION | TOTAL | Thymo | Campath | None | ||
| Training set | Pre-transplant | 49 (100) | 29 (59.2) | 10 (20.4) | 10 (20.4) | |
| Validation set | Pre-transplant | 33 (100) | 19 (57.5) | 1 (3) | 13 (39.4) | |
| p-value* | 0.999 (NS) | 0.043 | 0.080 (NS) | |||
| Training set | Post-transplant | 98 (100) | 54 (55.1) | 11 (11.2) | 33 (33.7) | |
| Validation set | Post-transplant | 64 (100) | 32 (50) | 3 (4.7) | 29 (45.3) | |
| p-value* | 0.629 (NS) | 0.252 (NS) | 0.142 (NS) | |||
| MAINTENANCE | Tacrolimus | Steroids | Cellcept | Rapamycin | ||
| Training set | Post-transplant | 98 (100) | 91 (92.8) | 36 (36.7) | 4 (4.1) | 5 (5.1) |
| Validation set | Post-transplant | 64 (100) | 61 (95.3) | 29 (45.3) | 0 (0) | 2 (3.1) |
| p-value* | 0.741 (NS) | 0.326 (NS) | 0.154 (NS) | 0.705 (NS) | ||
p-value Fisher exact test
Diagnoses
The diseases leading to end-stage disease and transplantation for liver or intestine-containing allografts are shown in Table 2.
Table 2. Causes of end-organ disease requiring liver or intestine transplantation in 214 study subjects.
| Diagnoses | Liver-containing allografts | Diagnoses | Intestine containing allografts |
|---|---|---|---|
| Biliary Atresia | 48 | Volvulus | 14 |
| Maple syrup urine disease | 22 | Gastroschisis | 12 |
| Hepatoblastoma | 13 | Necrotizing enterocolitis | 9 |
| Fulminant Liver Failure | 9 | Jejunal Atresia | 6 |
| Crigler Najjar Syndrome | 7 | Hirschsprung's | 4 |
| Familial cholestasis | 7 | Pseudoobstruction | 4 |
| Urea cycle defect | 7 | Tufting enteropathy | 2 |
| Cystic Fibrosis | 6 | Trauma | 2 |
| Cryptogenic Cirrhosis | 5 | Microvillous inclusion disease | 2 |
| ARKPD | 4 | SMV thrombosis | 1 |
| Autoimmune hepatitis | 4 | ||
| Primary Scelorisn Cholangitis | 4 | ||
| Alagille's syndrome | 4 | ||
| Caroli's disease | 3 | ||
| Tyrosinemia | 3 | ||
| Wilson's disease | 2 | ||
| Alpha 1 antitrypsin deficiency | 2 | ||
| Neonatal hepatitis | 2 | ||
| Embryonal sarcoma | 1 | ||
| Histiocytosis | 1 | ||
| Neuroendocrine tumor | 1 | ||
| Rhabdomyosarcoma | 1 | ||
| Histiocytosis | 1 | ||
| abernathy | 1 | ||
| TOTAL | 158 | 56 |
Test standardization
Using PBL from normal human subjects, we first confirmed that manufacturer-recommended concentrations of each of the abovementioned fluorochrome-labeled antibodies and 7-AAD were at or exceeded the minimum concentration to detect the highest percentage of positive cells26. Next, we established the specificity of each antibody in the cocktail by measuring the variation in frequencies of CD8+ cells or Tc upon adding each antibody alone and in combination with others. The coefficient of variation (%CV) in the frequency of Tc in PBL from three normal human subjects was 3.5%-12.2% with successive addition of each antibody, except anti-CD154 (Table 3). The acceptable %CV for this and all other phases of reproducibility testing shown below is ≤ 20%. When anti-CD154-PE was added to the remaining fluorochrome-labeled antibodies, the variation in Tc frequency ranged from %CV 1.04-5.9%. Two lots of each antibody were tested for their variability in detecting respective target marker using PBL from three normal human subjects. The %CV ranged from 0.9-15.3%.
Table 3. Effect of multiple antibodies on %CD8+cells labeled with anti-CD8-APCH7.
| Normal control 1 | Normal control 2 | Normal control 3 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lot 1 | Lot 2 | Lot 1 | Lot 2 | Lot 1 | Lot 2 | |||||||
| Tube 1 | CD8-APCH7 | 19.6 | 18.7 | 30.8 | 30.1 | 28.7 | 29.2 | |||||
| Tube 2 | CD8-APCH7 | CD8-PECy7 | 18 | 17.6 | 31 | 29.2 | 39.4 | 32.4 | ||||
| Tube 3 | CD8-APCH7 | CD3-FITC | 18.6 | 19.3 | 32.4 | 30.9 | 30.7 | 31.4 | ||||
| Tube 4 | CD8-APCH7 | CD45RO-APC | 18.2 | 19.6 | 32.7 | 33.4 | 29.6 | 32.7 | ||||
| Tube 5 | CD8-APCH7 | 7-AAD | 18.1 | 19.2 | 31.2 | 31.3 | 30.3 | 31.6 | ||||
| Tube 6 | CD8-APCH7 | CD154 | 16.1 | 16.6 | 25.6 | 25.4 | 28.5 | 27 | ||||
| Tube 7 | CD8-APCH7 | CD3-FITC | CD45RO-APC | 18.4 | 19.3 | 32.8 | 32.9 | 28.3 | 28.5 | |||
| Tube 8 | CD8-APCH7 | CD8-PECy7 | CD3-FITC | CD45RO-APC | 18.4 | 18.4 | 27.4 | 31.4 | 34.6 | 35.8 | ||
| Tube 9 | CD8-APCH7 | CD8-PECy7 | CD3-FITC | CD45RO-APC | 7-AAD | 18.6 | 18 | 27.4 | 29.8 | 35.4 | 32.6 | |
| Tube 10 | CD8-APCH7 | CD8-PECy7 | CD3-FITC | CD45RO-APC | 7-AAD | CD154 | 19.8 | 17.8 | 27.2 | 30.8 | 35 | 33.4 |
| Mean | 18.38 | 18.45 | 29.85 | 30.52 | 32.05 | 31.46 | ||||||
| SD | 1.00 | 0.95 | 2.68 | 2.22 | 3.79 | 2.58 | ||||||
| %CV | 5.5 | 5.2 | 9.0 | 7.3 | 11.8 | 8.2 | ||||||
Reproducibility testing studies were conducted using PBL from normal human subjects, because our clinical subjects many of whom are 6 months in age and weigh 4 kg cannot provide the blood sample volume for multiple replicates. The mean coefficient of variation in allospecific CD154+TcM which were induced by stimulation was evaluated in each study. In addition to the three reproducibility studies described below, reproducibility was also evaluated for samples tested on three different flow cytometers by three different operators (n=21, CV 8.2 ± 4.8%, SDC, Table 2), and for samples tested by two different technicians (n=5, CV 4.8 ± 3%, SDC Table 3).
Effect of cryopreservation
Because test performance was established in cryopreserved archived subject samples, variation due to cryopreservation was established in assays between 20 HLA-mismatched unique pairs of PBL from normal human subjects before and 30-days after cryopreservation. Stimulated CD154+TcM before and after cryopreservation demonstrated an acceptable mean %CV of 8.9%, which was below the pre-specified 20% limit (Tables 4a and 4b).
Table 4a. Mean %CD154+TcM in 20 normal human blood samples tested before and 30 days after cryopreservation.
| Sample type | Reaction | N | Mean | CI-low | CI-up | SD | Median | Min | Max |
|---|---|---|---|---|---|---|---|---|---|
| Before cryopreservation | Background | 20 | 4.2 | 2.4 | 5.9 | 3.8 | 3 | 0.6 | 13.2 |
| Stimulated | 20 | 24.8 | 19.5 | 30 | 11.3 | 25.2 | 9 | 55 | |
| After 30-day cryopreservation | Background | 20 | 6.1 | 4.1 | 8.2 | 4.4 | 4.7 | 1.1 | 17 |
| Stimulated | 20 | 22.8 | 17.7 | 27.9 | 10.9 | 23.2 | 9.1 | 52.6 |
Table 4b. Mean %CV for CD154+TcM in 20 normal human blood samples tested before and after 30-day cryopreservation.
| Sample type | Reaction | N | Mean | CI-low | CI-up | SD | Median | Min | Max |
|---|---|---|---|---|---|---|---|---|---|
| Before vs after 30-day cryopreservation. | Background | 20 | 45.3 | 33.8 | 56.8 | 24.5 | 42.6 | 5 | 87.2 |
| Stimulated | 20 | 8.9 | 5.6 | 12.2 | 7 | 6.5 | 0.8 | 24.9 |
Same-day duplicate testing
Assays between twenty unique pairs of HLA-mismatched PBL from normal human subjects were performed in duplicate (a and b) in each of two runs (run 1 and 2) on the same day to determine within run (a vs. b within runs 1 and 2) and between run (all replicates) variability in CD154+TcM generated in the stimulated reaction. Stimulated CD154+TcM in all replicates of each sample demonstrated an acceptable mean %CV of 6.0%, which was below the pre-specified 20% limit (Tables 5a and 5b).
Table 5a. Mean %CD154+TcM for 20 duplicate assays (a and b) in each of two runs, (1 and 2).
| Run | Reaction | N | Mean | CI-low | CI-up | SD | Median | Min | Max |
|---|---|---|---|---|---|---|---|---|---|
| 1a | Background | 20 | 0.3 | 0.18 | 0.51 | 0.35 | 0.25 | 0 | 1.6 |
| Stimulated | 20 | 24.1 | 19 | 29.2 | 10.8 | 25 | 3.7 | 41.2 | |
| 1b | Background | 20 | 0.3 | 0.19 | 0.42 | 0.25 | 0.3 | 0 | 1.1 |
| Stimulated | 20 | 24.2 | 19 | 29.4 | 11.1 | 24.6 | 3 | 41.3 | |
| 2a | Background | 20 | 0.28 | 0.12 | 0.44 | 0.35 | 0.2 | 0 | 1.5 |
| Stimulated | 20 | 25.1 | 19.6 | 30.5 | 11.6 | 25.2 | 2.9 | 42.5 | |
| 2b | Background | 20 | 0.24 | 0.13 | 0.36 | 0.25 | 0.2 | 0 | 1.1 |
| Stimulated | 20 | 25.2 | 19.9 | 30.4 | 11.3 | 26.4 | 3.2 | 43.1 |
Table 5b. Mean %CV for %CD154+TcM within each of two runs, 1 and 2, and for all replicates performed in both runs.
| Run | Reaction | N | Mean | CI-low | CI-up | SD | Median | Min | Max |
|---|---|---|---|---|---|---|---|---|---|
| 1a & 1b | Background | 20 | 38.9 | 17 | 60.9 | 47 | 27.2 | 0 | 142 |
| Stimulated | 20 | 5.2 | 2.9 | 7.4 | 4.9 | 4 | 0.2 | 16.4 | |
| 2a & 2b | Background | 19 | 43 | 20.1 | 65.9 | 47.5 | 28.4 | 0 | 142 |
| Stimulated | 20 | 5.4 | 3.5 | 7.2 | 4 | 5.9 | 0.6 | 16.2 | |
| 1a-2b | Background | 20 | 61.3 | 42.9 | 79.7 | 39.3 | 54.7 | 18.2 | 200 |
| Stimulated | 20 | 6 | 4.6 | 7.5 | 3.1 | 6 | 1.5 | 11.1 |
Day-to-day variation
Real life patient samples can be tested on the same day (condition 1a), after 24-hour storage at ambient temperature in a reference laboratory if the samples arrive late in the day from a local hospital (condition 1b), or after overnight shipment at ambient temperature (condition 1c). Five unique pairs of HLA-mismatched PBL from normal human subjects were tested under each condition. Stimulated CD154+TcM in all replicates of each sample demonstrated an acceptable mean %CV of 3.2%, which was below the pre-specified 20% limit (Tables 6a and 6b).
Table 6a. Mean %CD154+TcM for each condition of storage/shipment of five samples in day-to-day variation testing.
| Storage condition | Reaction | N | Mean | CI-low | CI-up | SD | Median | Min | Max |
|---|---|---|---|---|---|---|---|---|---|
| 1a (same day) | Background | 5 | 2.1 | -0.7 | 4.9 | 2.3 | 2 | 0.1 | 5.8 |
| Stimulated | 5 | 5.5 | -1.3 | 12.3 | 5.5 | 3.9 | 1.4 | 14.6 | |
| 1b(24h, ambient temp) | Background | 5 | 1.6 | -1.3 | 4.5 | 2.3 | 0.6 | 0.3 | 5.7 |
| Stimulated | 5 | 5.7 | -1.3 | 12.7 | 5.6 | 4.4 | 1.4 | 15.1 | |
| 1c (24 hr, overnight shipment) | Background | 5 | 2.3 | -0.2 | 4.8 | 2 | 2.9 | 0.1 | 3.7 |
| Stimulated | 5 | 5.5 | -1.1 | 12.2 | 5.4 | 4.1 | 1.4 | 14.5 |
Table 6b. %CV for %CD154+TcM between three conditions of storage/shipment for five samples in day to day variation testing.
| Storage condition | Reaction | N | Mean | CI-low | CI-up | SD | Median | Min | Max |
|---|---|---|---|---|---|---|---|---|---|
| 1a, 1b and 1c | Background | 5 | 60.3 | 22.4 | 98.2 | 30.5 | 66.7 | 7.9 | 86.6 |
| Stimulated | 5 | 3.2 | -0.6 | 7.0 | 3.0 | 2.2 | 0 | 6.7 |
Development of multivariate (optimal) and single-variable predictive models in training set
For 98 analyzable post-transplant training set samples, the IR of CD154+TcM (p=0.0008), organ transplant type (p=0.019), and FKWBC (p=0.004) emerged as significant covariates in logistic regression analysis. Stepwise (exhaustive) regression identified the most predictive, yet parsimonious model. The optimal model contained the five variables: time between transplantation and assay (p=0.061), race (p=0.053), organ transplant type (p=0.0028), FKWBC (p=0.0025), and IR of CD154+TcM (p=0.0003). For 49 analyzable pre-transplant training set samples, the IR of CD154+TcM (p=0.0041) emerged as the most significant covariate in logistic regression. In stepwise regression, the optimal model contained the four variables Organ (p=0.16), Gender (p=0.026), Race (p=0.076), and IR of CD154+TcM (p=0.002). For either pre- or post-transplant models, the cut point was identified as the optimal level of both sensitivity and specificity from the ROC curve of this training set predicting training set (i.e., optimal true positive and true negative values). To identify the tradeoff in predictive accuracy between the optimal model with multiple variables and a model with the single most overall predictive variable, the IR of CD154+TcM, performance of these two logistic regression models was compared in the training set (SDC, Tables 4). For the single variable post-transplant or IR1+IRx model, the cut point was determined at a raw IR value of 1.10. The raw IR value for the single variable pre-transplant or IR0 model was 1.23. ROC curves for the single variable model for training and validation set pre- and post-transplant samples are shown in Figure 3.
Figure 3.
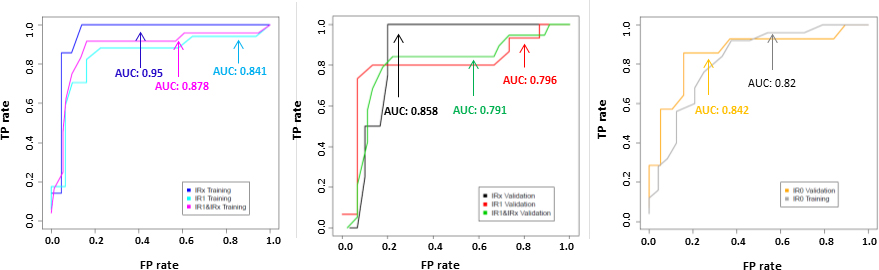
Receiver operator characteristic (ROC) curves for post-transplant (IR1+IRx) training (left panel) and validation (middle panel) and pre-transplant (right panel) data sets using single variable IR value. Each plot also shows ROC curves for corresponding early post-transplant (IR1) and late post-transplant (IRx) samples. TP rate=True positive rate. FP rate=False positive rate. AUC=area under the receiver-operating characteristic curve. IR0=sample obtained before transplantation, IR1=samples obtained between days 1-60 after transplantation, IRx= samples obtained from days 200 onward after transplantation (see Supplementary table SDC 6 for additional details).
Model stability
Given the modest number of rejection events, for e.g. 25 in the post-transplant training set samples, model overfitting is a distinct possibility27. The coefficient of the IR variable in the post-transplant training set samples was 3.41 in the multivariate model and 3.31 in the single variable model based on the IR alone - a difference of ∼3% (SDC Table 5). The error term for this coefficient goes from 0.93 in the multivariate model to 0.77 in the single variable model - a difference of ∼18%. This result and the reproducibility of predictive performance in an independent validation set reassure us that this model is in fact stable and predictive. Additionally, beyond adjusting for potentially confounding variables, we have performed multiple stratified analyses, where the performance of the single variable model is evaluated in subjects subgrouped by the various covariates. The results of stratified subanalyses are shown for the covariates type of organ transplanted, type of induction, whether actual or surrogate donor stimulators were used, and whether rejection or non-rejection were diagnosed by “for-cause” or surveillance biopsy or clinically (SDC Tables S6-S9). These analyses also confirm good stability in model performance.
Replication of test performance in validation samples and final model selection
The optimal models for pre- and post-transplant samples, which incorporated multiple covariates demonstrated inferior performance when applied to corresponding validation set samples (SDC, Tables 4a and 4b). The single variable model demonstrated consistent performance for predicting rejection in the training and validation sets. An IR ≥ 1.1 in post-transplant samples demonstrated sensitivity and specificity of 92% and 84%, respectively in training set and 84% and 80%, respectively in the validation set (Table 7a). An IR ≥ 1.23 in pre-transplant samples predicted lower sensitivity of 57% in the validation set compared with 80% in the training set (Table 7b). However, the respective 95% confidence intervals showed overlap, 30-81% vs 59-92%, and test specificity, PPV and NPV were similar.
Table 7. Performance of single variable post-transplant (7a upper table) and pre-transplant (7b, lower table) models based on IR of CD154+TcM in training and validation sets.
| Cohort | AUC | Cut value | Sensitivity | 95% CI | Specificity | 95% CI | PPV | 95% CI | NPV | 95% CI |
|---|---|---|---|---|---|---|---|---|---|---|
| Training set(n=98) | 0.878 | 0.0420 (1.10 on raw IR scale) | 92% (22/24) | 72%, 99% | 84% (62/74) | 73%, 91% | 65% (22/34) | 46%, 80% | 97% (62/64) | 88%, 99% |
| Validation set(n=64) | 0.791 | 0.042 (1.10 on raw IR scale) | 84% (16/19) | 60%, 96% | 80% (36/45) | 65%, 90% | 64% (16/25) | 43%, 81% | 92% (36/39) | 78%, 98% |
| Cohort | AUC | Cut value | Sensitivity | 95% CI | Specificity | 95% CI | PPV | 95% CI | NPV | 95% CI |
| Training set(n=49) | 0.82 | 0.088 (1.23 on raw IR scale) | 80% (20/25) | 59%, 92% | 71% (17/24) | 49%, 87% | 74% (20/27) | 53%, 88% | 77% (17/22) | 54%, 91% |
| Validation set (n=33) | 0.842 | 0.088 (1.23 on raw IR scale) | 57% (8/14) | 30%, 81% | 89% (17/19) | 65%, 98% | 80% (8/10) | 44%, 96% | 74% (17/23) | 51%, 89% |
Additional analyses to test the effect of confounders
Comparable test performance within the range seen in overall training and validation set samples was also seen in samples sub-grouped by time of sampling after transplantation, the type of stimulator-actual or surrogate donor, organ transplant type, type of induction immunosuppression, and whether rejection or non-rejection were diagnosed by for-cause or surveillance biopsy or clinically (SDC, Tables 6-10). Performance estimates are less likely to be meaningful for those subgroups with small numbers.
Adverse events
No adverse events were encountered due to phlebotomy.
Discussion
Our study shows that a “fine” functional T-cell subset, allospecific CD154+TcM, predicts acute cellular rejection in the rare population of children with liver or intestine transplantation and addresses the unmet need for non-invasive rejection-risk assessment. Developed in samples from 127 children, test performance is replicated in blinded samples from 87 subjects. Test sensitivity, specificity, PPV and NPV of 92%, 84%, 65%, and 97% respectively in post-transplant training set samples, and 84%, 80%, 64% and 92% respectively in blinded independent post-transplant validation set samples, which were tested 18 months later with a standardized assay format with cGMP reagents and instruments represents true replication. Significant attributes of the test system include actionable results after overnight culture, and the potential for indefinite testing with “surrogate” donor stimulators without compromising rejection-risk determination (SDC, Table 8). Other advantages are a personalized test output, the immunoreactivity index, and prediction of early rejection with pre-transplant samples. The lower sensitivity of test predictions with pre-transplant validation set samples of 57% is noteworthy compared with 80% sensitivity in the training set. The smaller numbers of rejectors in the validation set compared with training set, 14 vs 25, and overlap in respective 95% confidence intervals, 30-81% vs 59-91% offer reassurance that actual sensitivity may lie within these estimates. This performance is reasonable given that there is no other non-invasive predictor of cellular rejection for this rare population. The confidence intervals for pre-transplant sensitivity also encompass the performance of the ELISPOT in predicting renal transplant rejection, and suggest that lower predictive sensitivity is a feature of pre-transplant samples.28 Enhanced donor-specific alloreactivity, the mechanism underlying acute cellular rejection in a variety of organ transplants, and its measurement with CD154+TcM, the parameter used to measure rejection-risk makes this test system potentially adaptable to other types of organ transplants. Finally, the test is highly reproducible, with coefficient of variation of 10% or less in simulated daily testing, and after 24-hour storage or overnight shipment.
Several factors may affect test performance. The type of cell stimulator, whether surrogate or actual donor cell was not a significant covariate in the regression analysis, which established predictive thresholds. This is consistent with previously reported stability in rejection-risk assessment in samples tested with both types of stimulators.16 As added evidence, reasonable test performance is also seen in subjects sub-grouped further by surrogate donor or actual donor stimulator cells (SDC, Table 6), and by various other confounders (SDC, Tables 7-10). Further, optimal predictive models, which incorporated the covariate organ type and several other covariates such as type of stimulator, tacrolimus whole blood levels, race, time between transplantation and sample, and type of induction treatment demonstrated inferior performance when applied to validation set samples. In contrast, the single variable model based on the IR of CD154+TcM performed consistently in training and validation sets. Possible reasons include the fact that compared with other T-cell subsets, the alloresponse of CD154+TcM has shown specificity for rejection after three different types of transplants including those evaluated here. Second, by reporting test results as an index which uses a reference alloresponse to normalize donor-induced CD154+TcM from the same patient likely negates the effect of these confounders, which are expected to affect either reaction proportionately.
The effect of opportunistic tissue-invasive infections with cytomegalovirus and Epstein-Barr virus on rejection-risk assessment with CD154+TcM remains unknown. These infections were absent in all but one subject at the time when analyzable blood samples were obtained, likely due to pre-emptive treatment of viremia with evolving surveillance protocols in most centers. This subject experienced Epstein-Barr viral enteritis in the intestine allograft. The post-transplant sample from this subject obtained during this episode failed allostimulation. Therefore no result could be generated. Test formats and thresholds for PCR-based viral load monitoring changed throughout the 6-year study period, precluding reliable assessments of the effect of viremia on test performance during this pre-clinical evaluation. Early performance evaluation (unpublished) during clinical use of this test system in 63 children with liver or intestine transplantation has shown that test predictions have not been confounded by infections. This cohort includes 20 children who were evaluated in the pre-clinical phase and re-tested as a component of clinical care, and 43 new subjects. Among 11 of these 63 children, one experienced biopsy-proven cholangitis, one experienced adenoviral allograft enteritis and nine demonstrated EBV viral replication without tissue-invasive disease with mean (SEM, range) EBV viral load of 10926 copies per ml (4472, range 120-31000) at the time of sampling. No differences were seen between children with infection compared with those without infection in test sensitivity (3/4 or 75%, vs 18/21 or 86%, p=0.527, NS, Fisher's exact test) and specificity (6/7 or 86% vs 31/31 or 100%, p=0.184, NS). CMV viremia was not reported or detected in this clinical cohort on the day of sampling. An expanded clinical evaluation will be the subject of a follow-up report.
Because the determination of rejection-risk is central to the daily management of a transplant recipient, clinical situations most suited for this test system are likely to vary. Our early experience suggests that the adjunctive information provided by non-invasive rejection-risk assessment is likely i) to assist clinical decision-making when minimization of immunosuppression is being considered earlier than indicated by the prevailing clinical protocol, and ii) to better assess the clinical significance of indeterminate, borderline or non-specific inflammatory changes in late surveillance biopsies.29 Additional analysis of data obtained during clinical use will determine whether the test is being used in this way.
In summary, allospecific T-cytotoxic memory cells fulfil an unmet need for personalized prediction of acute cellular rejection in the rare and high-risk population of children with liver or intestine transplantation with clinically acceptable and reproducible performance. The potential benefit of risk-based optimization of immunosuppression with adjunctive information provided by this first-in-class flow cytometric test outweighs the risks of phlebotomy. The additional risks of undetected false positive and false negative results are minimized by using test results as an adjunct with all available clinical and laboratory information, in a manner concurrent with current clinical practice.
Supplementary Material
Acknowledgments
We thank Plexision, Inc., Pittsburgh, PA, for assay standardization and data analyses. We also thank Ms Dale Zecca for manuscript preparation.
Funding: This study was supported in part by the National Institutes of Health under award 5R01AI073895-05 (Dr. Sindhi), a departmental endowment from the Hillman Foundation of Pittsburgh to the Hillman Center for Pediatric Transplantation at the Children's Hospital of Pittsburgh of UPMC, Pittsburgh, PA, the Szalay Foundation, and the Herridge, Giventer-Braff, Snow and Davidson families.
Role of the Funder/Sponsor: The National Institutes of Health, the Hillman Foundation of Pittsburgh, the Szalay Foundation and the families who provided financial support had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
Abbreviations
- ACR
acute cellular rejection
- AUC
area-under-the-receiver-operator-characteristic-curve
- CV
coefficient of variation
- CD154
CD40 ligand
- FDA
Food and Drug Administration
- FKWBC
tacrolimus whole blood concentrations
- HLA
human leukocyte antigen
- IR
immunoreactivity index
- ITx
intestine transplantation
- LTx
liver transplantation
- NCT
National Clinical Trial
- NPV
negative predictive value
- PBL
peripheral blood leukocytes
- PPV
positive predictive value
- rATG
rabbit antihuman thymocyte globulin
- Tc
T-cytotoxic cell
- TcM
T-cytotoxic memory cell
- UPMC
University of Pittsburgh Medical Center
Footnotes
Author Contributions: Dr. Sindhi had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Dr. Higgs and Mr. Qing Sun take responsibility for the accuracy of the data analysis.
Study concept and design: All authors.
Acquisition, analysis, or interpretation of data: All authors.
Drafting of the manuscript: Sindhi, Ashokkumar, Higgs, Sun.
Critical revision of the manuscript for important intellectual content: All authors.
Statistical analysis: Higgs, Sun.
Study supervision: Sindhi.
Data collection during clinical implementation: Boehm, Dirling, Fazzolare, Harris, Hartle, Jativa, Kachmar, Nicely, O'Toole, Remaley, Stanley.
Conflict of Interest Disclosures: Test systems are based on technology described in US Patent 8426146. Inventor: Rakesh Sindhi. Assignee: University of Pittsburgh-of the Commonwealth System of Higher Education, Pittsburgh, PA, and licensed to Plexision, Inc., Pittsburgh 15224, in which the University holds equity. Rakesh Sindhi serves as an unpaid consultant and Chethan Ashokkumar as a paid consultant to licensee without other financial relationships. Disclosed conflicts of interest have been managed in accordance with the University of Pittsburgh's policies and procedures. The rest of the authors have nothing to disclose.
Analyses of pre-clinical evaluation were presented at the plenary session of the annual meeting of the American Association for the Study of Liver Diseases (AASLD) on November 3, 2013, in Washington, DC.
Obtained funding: Sindhi.
Administrative, technical, or material support: Brown, White, Levy, Ashokkumar, Ningappa.
Publisher's Disclaimer: Disclaimer: The content, findings, and conclusions of this report are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health, the Hillman Foundation of Pittsburgh, family foundations or the families who supported this work.
References
- 1.Shepherd RW, Turmelle Y, Nadler M, et al. Risk factors for rejection and infection in pediatric liver transplantation. Am J Transplant. 2008;8:396–403. doi: 10.1111/j.1600-6143.2007.02068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reyes J, Mazariegos GV, Abu-Elmagd K, et al. Intestinal transplantation under tacrolimus monotherapy after perioperative lymphoid depletion with rabbit anti-thymocyte globulin (thymoglobulin) Am J Transplant. 2005;5:1430–36. doi: 10.1111/j.1600-6143.2005.00874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nayyar N, Mazariegos G, Ranganathan S, et al. Pediatric small bowel transplantation. Semin Pediatr Surg. 2010;19:68–77. doi: 10.1053/j.sempedsurg.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Sudan DL, Shaw BW, Jr, Langnas AN. Causes of late mortality in pediatric liver transplant recipients. Ann Surg. 1998;227:289–95. doi: 10.1097/00000658-199802000-00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fridell JA, Jain A, Reyes J, Biederman R, et al. Causes of mortality beyond 1 year after primary pediatric liver transplant under tacrolimus. Transplantation. 2002;74:1721–24. doi: 10.1097/00007890-200212270-00014. [DOI] [PubMed] [Google Scholar]
- 6.Soltys KA, Mazariegos GV, Squires RH, Sindhi RK, Anand R SPLIT Research Group. Late graft loss or death in pediatric liver transplantation: an analysis of the SPLIT database. Am J Transplant. 2007;7:2165–71. doi: 10.1111/j.1600-6143.2007.01893.x. [DOI] [PubMed] [Google Scholar]
- 7.Abu-Elmagd KM, Costa G, Bond GJ, et al. Five hundred intestinal and multivisceral transplantations at a single center: major advances with new challenges. Ann Surg. 2009;250:567–81. doi: 10.1097/SLA.0b013e3181b67725. [DOI] [PubMed] [Google Scholar]
- 8.UNOS OPTN Annual Report. Annual Report of the U.S Organ Procurement and Transplantation Network and the Scientific Registry of Transplant Recipients: Transplant Data 1998-2007. Department of Health and Human Services, Health Resources and Services Administration, Healthcare Systems Bureau, Division of Transplantation; Rockville, MD: United Network for Organ Sharing; Richmond, VA: University Renal Research and Education Association; Ann Arbor, MI: 2008. [Google Scholar]
- 9.Designating Humanitarian Use Devices. [Accessed January, 23, 2015]; http://www.fda.gov/ForIndustry/DevelopingProductsforRareDiseasesConditions/DesignatingHumanitarianUseDevicesHUDS/default.htm.
- 10.Eydelman MB, Chen EA. The FDA's humanitarian device exemption program. Health Aff (Millwood) 2011 Jun 30;:1210–12. doi: 10.1377/hlthaff.2011.0550. author reply 1212. [DOI] [PubMed] [Google Scholar]
- 11.Sharfstein J. FDA Regulation of laboratory-developed diagnostic tests: Protect the public, advance the science. JAMA. 2015 Jan 5; doi: 10.1001/jama.2014.18135. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Evans JP, Watson MS. Genetic testing and FDA regulation: Overregulation threatens the emergence of genomic medicine. JAMA. 2015 Jan 5; doi: 10.1001/jama.2014.18145. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 13.Ashokkumar C, Talukdar A, Sun Q, et al. Allospecific CD154+ T cells associate with rejection risk after pediatric liver transplantation. Am J Transplant. 2009;9:179–91. doi: 10.1111/j.1600-6143.2008.02459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashokkumar C, Gupta A, Sun Q, et al. Allospecific CD154+ T cells identify rejection-prone recipients after pediatric small-bowel transplantation. Surgery. 2009;146:166–73. doi: 10.1016/j.surg.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Sindhi R, Ashokkumar C, Higgs BW, et al. Allospecific CD154 + T-cytotoxic memory cells as potential surrogate for rejection risk in pediatric intestine transplantation. Pediatr Transplant. 2012;16:83–91. doi: 10.1111/j.1399-3046.2011.01617.x. Erratum in: Pediatr Transplant. 2012;16:913. [DOI] [PubMed] [Google Scholar]
- 16.Ashokkumar C, Shapiro R, Tan H, et al. Allospecific CD154+ T-cytotoxic memory cells identify recipients experiencing acute cellular rejection after renal transplantation. Transplantation. 2011;92:433–38. doi: 10.1097/TP.0b013e318225276d. [DOI] [PubMed] [Google Scholar]
- 17.Sindhi R, Magill A, Bentlejewski C, et al. Enhanced donor-specific alloreactivity occurs independently of immunosuppression in children with early liver rejection. Am J Transplant. 2005;5:96–102. doi: 10.1111/j.1600-6143.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- 18.Khera N, Janosky J, Zeevi A, Mazariegos G, Marcos A, Sindhi R. Persistent donor-specific alloreactivity may portend delayed liver rejection during drug minimization in children. Front Biosci. 2007;12:660–63. doi: 10.2741/2090. [DOI] [PubMed] [Google Scholar]
- 19.FDA U.S. Food and Drug Administration, Recently-Approved Devices. http://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfTopic/pma/pma.cfm?num=h130004.
- 20.Roederer M. How many events is enough? Are you positive? Cytometry Part A. 2008;73A:384–85. doi: 10.1002/cyto.a.20549. [DOI] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards (NCCLS) Evaluation of Precision Performance of Quantitative Measurement Methods; Approved Guideline—Second Edition. NCCLS document EP5-A2. 2004 [Google Scholar]
- 22.International Panel: Demetris AJ, Batts KP, Dhillon AP, et al. Banff schema for grading liver allograft rejection: An international consensus document. Hepatology. 1997;25:658–63. doi: 10.1002/hep.510250328. [DOI] [PubMed] [Google Scholar]
- 23.Hosmer DW, Lemeshow S. Applied Logistic Regression. 2. New York, NY: John Wiley & Sons Inc; 2000. [Google Scholar]
- 24.Fawcett T. ROC Graphs: Notes and Practical Considerations for Researchers. [Accessed December 29, 2014];2004 Mar 16;:1–38. http://binf.gmu.edu/mmasso/ROC101.pdf.
- 25.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. URL http://www.R-project.org. [Google Scholar]
- 26.Hulspas R. Titration of Fluorochrome-Conjugated UNIT 6.29 Antibodies for Labeling Cell Surface Markers on Live Cells. In: Paul Robinson J, editor. Current Protocols in Flow Cytometry. 6.29. Vol. 54. 2010. pp. 6.29.1–6. 29.9. [DOI] [PubMed] [Google Scholar]
- 27.van der Ploeg T, Austin PC, Steyerberg EW. Modern modelling techniques are data hungry: a simulation study for predicting dichotomous endpoints. BMC Med Res Methodol. 2014 Dec 22;14:137. doi: 10.1186/1471-2288-14-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Augustine JJ, Siu DS, Clemente MJ, Schulak JA, Heeger PS, Hricik DE. Pre-Transplant IFN-gamma ELISPOTs are associated with post-Transplant renal function in African American renal transplant recipients. Am J Transplant. 2005;5:1971–1975. doi: 10.1111/j.1600-6143.2005.00958.x. [DOI] [PubMed] [Google Scholar]
- 29.Ranganathan S, Celik N, Mazariegos G, Sindhi R. Liver allograft fibrosis and minimization of immunosuppression. Pediatr Transplant. 2015 Sep 3; doi: 10.1111/petr.12590. [Epub ahead of print] No abstract available. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.