Abstract
Objective
The current study identified novel combinations of genetic and psychological factors that predicted 12-month post-operative pain and disability outcomes following arthroscopic shoulder surgery.
Methods
A prospective pre-surgical cohort (n = 150) was recruited to complete validated psychological questionnaires and have their DNA collected from saliva. DNA was genotyped for a priori selected genes involved with pain modulation (ADRB2, OPRM1, AVPR1A, GCH1, and KCNS1) and inflammation (IL1B, TNF/LTA, and IL6). The outcome measures of interest were the Brief Pain Inventory (BPI) and Disabilities of the Arm, Shoulder, and Hand (DASH) questionnaire. Follow up for the cohort was at 3, 6, and 12 months post-operatively. After controlling for age, sex, race, and pre-operative status, genetic and psychological factors were entered as main effects and interaction terms in separate general linear models for predicting post-operative pain and disability outcomes.
Results
Seven interactions involving pain modulatory genes were identified. Three provided strong statistical evidence for different outcomes including; 1) KCNS1 and kinesiophobia for pre-operative pain intensity, 2) ADRB2 and depressive symptoms for post-operative course, and 3) GCH1 and anxiety symptoms for 12 month pain intensity outcome. Ten interactions involving inflammatory genes were identified. Three provided strong statistical evidence for the 12 month post-operative course outcome including; 1) 2 different IL6 SNP’s and pain catastrophizing, and 2) IL6 and depressive symptoms.
Conclusion
The current study identified novel genetic and psychological interactions that can be used in future studies to further understand the development of persistent post-operative pain and investigate the effectiveness of tailored treatment.
The etiology of chronic pain conditions is complex, influenced by many different biopsychosocial factors (i.e. genetic, psychological, social, and environmental) interacting to determine an individual’s risk.1, 2 Specific to the purposes of this study, persistent post-operative pain is a subset of chronic pain that results in undesirable surgical outcomes.3 Persistent post-operative shoulder pain occurs following orthopedic surgery, even when arthroscopy is used.4 Prediction of those at risk for persistent post-operative pain is difficult and improvement in this area has been highlighted as a research priority.3
We have investigated genetic and psychological factors in pre-clinical5, 6 and clinical cohorts7 with the overarching goal of improving prediction of post-operative outcomes. In our line of research we selected a priori candidate genes known to be involved in pain modulation (e.g. COMT and AVPR1A) or pro-inflammatory responses (e.g. TNF/LTA and IL1B), as well as psychological constructs (e.g. fear of pain and pain catastrophizing) that are established precursors to chronic musculoskeletal pain conditions. In our pre-clinical study involving 190 healthy adults we induced pain by eccentrically exercising the rotator cuff muscles to fatigue and then monitoring shoulder pain and upper-extremity disability ratings over several consecutive days. In this pre-clinical study we observed multiple interactions between genes and psychological factors that improved prediction of pain related phenotypes.5, 6 From these pre-clinical analyses we identified risk subgroups, and one comprised of COMT SNP rs6269 and pain castastrophizing not only predicted heightened shoulder pain in the pre-clinical cohort but also predicted 12-month post-operative outcomes among patients undergoing shoulder surgery.7
The identification of a robust high-risk COMT and pain catastrophizing subgroup was encouraging, but there are many other genetic and psychological interactions that could improve prediction of post-operative outcomes. Our prior approach was translational in nature, with the goal being to first identify candidate interactions in a pre-clinical cohort and then carry those forward for validation in a clinical cohort.7 This past approach, while rigorous, did not identify genetic and psychological interactions directly in a clinical cohort, which is traditionally how prognostic factors are identified. Therefore, further investigation of these interactions is warranted given that our pre-clinical pain model was not a perfect approximation of post-operative pain. Indeed, there is ample biological plausibility for additional genetic and psychological interactions influencing post-operative outcomes. Therefore, the purpose of the current analysis was to investigate whether combinations of a priori selected genetic and psychological factors predicted 12-month post-operative outcomes for patients receiving arthroscopic shoulder surgery. These additional analyses are warranted because they will identify novel and potentially clinically relevant interactions that advance our understanding of shoulder pain conditions.
PATIENTS AND METHODS
Overview
This study was reported following STREGA extension 8 of STROBE guidelines9 and registered prospectively at http://clinicaltrials.gov/ct2/show/NCT00187863. The University of Florida’s Human Subject Institutional Review Board (IRB-01) approved this study and all participants provided informed consent prior to enrollment. The current paper is the primary analysis for identifying genetic and psychological predictors in the post-operative shoulder pain cohort.
Subjects
Consecutive individuals with shoulder pain were evaluated by orthopedic surgeons at the University of Florida’s Orthopaedic and Sports Medicine Institute between 2009 and 2012. Surgical candidacy was determined by poor response to conservative treatment, diagnostic imaging, and physician examination. Individuals that were surgical candidates were then further screened by one of the authors (WHG) for study eligibility. Eligible participants were then consented, and scheduled for a baseline study session within 1 week of surgery. Demographic data were captured by self-report questionnaires and included sex, age, and race. Participants underwent shoulder arthroscopy, and returned for study sessions at 3 months, 6 months and 1 year post-operatively.
Inclusion criteria were: 1) between 18 and 85 years of age, 2) complaints of pain limited to anterior, lateral, or posterior shoulder, and 3) scheduled for arthroscopic surgery. Additionally participants met one of the following criteria: 4) documented or suspected rotator cuff tendinopathy (evidence from clinical examination or imaging studies) including small (<1 cm), medium (1-3 cm), and large (3-5 cm) tears, or 5) documented or suspected adhesive capsulitis (evidence from clinical examination or imaging studies), or 6) documented or suspected superior labrum from anterior to posterior lesion or isolated anterior/posterior labral tears (evidence from clinical examination or imaging studies).
Exclusion criteria were: 1) current complaints of pain lasting longer than the past 3 months involving neck, elbow, hand, low back, hip, knee, or ankle, or 2) massive rotator cuff tear (>5 cm), or 3) documented shoulder OA or RA, or 4) prior shoulder surgery within the past year or currently complaining of pain from prior shoulder surgery, or 5) current shoulder fracture, tumor, or infection, or 6) previously diagnosed chronic pain disorder (including, but not limited to irritable bowel syndrome, fibromyalgia, temporomandibular disorder, chronic low back pain etc.), or 7) current psychiatric management, or 8) current gastrointestinal or renal illness.
Predictive Measures
Genetic and psychological factors that were used as predictors in this study were a priori selected as briefly described below and in more detail in prior pre-clinical studies.5, 6
Genetic Measures
Originally 19 single nucleotide polymorphisms (SNPs) from 10 pain candidate genes (OPRM1, COMT, ADRB2, AVPR1A, GCH1, KCNS1, TNF/LTA, TNF-308, IL1B, and IL6) were considered in prior pre-clinical analyses.5, 6 These genes were selected because of their established links with pain sensitivity, musculoskeletal pain, or the potential to develop chronic pain and there was an a priori plan to investigate them in pre-clinical and clinical cohorts. The genes reported in the current paper include 15 SNPs from 9 candidate genes not previously considered as predictors in the post-operative cohort. The COMT gene was not further considered because it has already been included in a validated risk subgroup, and this would be redundant with our prior study.7 Each of the remaining genes is associated with biological process that could plausibly interact with psychological constructs to impact the reporting of shoulder pain. Therefore, these genes were considered appropriate for further investigation in the post-operative cohort. Conceptually, OPRM1, ADRB2, AVPR1A, GCH1, and KCNS1 were investigated as pain modulatory genes and TNF/LTA, TNF-308, IL1B, and IL6 were investigated as pro-inflammatory genes just as they were in our pre-clinical studies.5, 6 Genotyping of the selected single nucleotide polymorphisms (SNPs) of these genes was performed as described in our previous reports using subject DNA extracted from buccal epithelial cells.5, 6 Hardy-Weinberg equilibrium was calculated and found acceptable for each SNP.
Psychological Measures
Psychological measures were selected to represent constructs commonly identified as predictors of poorer outcome for musculoskeletal pain conditions. Depressive symptoms and anxiety were assessed with the Patient Health Questionnaire (PHQ)10, 11 and the State-Trait Anxiety Inventory (STAI).12 Fear of pain was assessed with the Fear of Pain Questionnaire (FPQ-III)13-15 and for sake of brevity we used a shortened 9-item version that has performed similarly to the full scale.16 Fear of movement was assessed with the shortened version of the Tampa Scale of Kinesiophobia (TSK-11).17, 18 Finally, pain catastrophizing was assessed with the Pain Catastrophizing Scale (PCS).19
Outcome Measures
Pain and disability measures were collected pre-operatively and at 3, 6, and 12 months post-operatively. The Brief Pain Inventory (BPI) is a widely accepted measure of pain intensity that has good test-rest reliability over short intervals.20 The BPI consists of rating pain intensity on an 11-point numerical rating scale ranging from 0 (no pain) to 10 (worst pain imaginable). In addition, the Disabilities of the Arm, Shoulder, and Hand Questionnaire (DASH) was used to assess upper-extremity disability.21 We used a validated abridged version of the DASH (the QuickDASH) which consists of 11 functional items with total scores ranging from 0 (not disability) to 100 (complete disability).22
Sample Size
Sample size was determined a priori based on an estimation of effect parameters for the genetic and psychological factors as well as their interactions on outcomes, which were specified in terms of R-square of the full model and R-square difference between the full and the reduced models. The SAS Power procedure was adopted to evaluate the required sample sizes to achieve a target power of 80% to test each effect at a type-I error level of 0.005. We found that the proposed sample size of 180 would enable us to detect all but two anticipated effect sizes based on estimates from our pilot data. However, for pragmatic reasons we recruited 150 into the post-operative cohort. This alteration in sample size did not adversely affect power because the number of predictor variables included in analyses was reduced. Specifically, based on our previous findings, we reduced the number of covariates included in the regression models to exclude medication status and rotator cuff size as they did not impact findings in the clinical cohort.7
Data Analysis
All statistical analyses were conducted using SAS version 9.2 (SAS Institute, Cary, North Carolina). Summary statistics were calculated for all demographic, genetic, psychological, and outcome measures. For every gene, a general linear model was fitted to assess its main effect (genotype level) and a series of expanded models were fitted to study its interaction with the psychological factors for each shoulder pain outcome of interest. The predictors in the linear models included demographic variables (age, gender, and race), pain candidate genes as individual SNPs, psychological factors in their original continuous metric, and the gene × psychology interaction. Therefore, each linear model had the same structure with 4 increments including 1) demographic data, 2) genotype, 3) psychological factor, and 4) the gene by psychological factor interaction. The BPI and DASH were used as dependent variables in the linear models as a) pre-operative status; b) post-operative course by area under the curve (AUC, computed with 3, 6, and 12 month outcome scores); and c) 12 month outcome score. Three different versions of the outcome measures were used in the analysis to distinguish between factors predictive of pre-operative status, post-operative course, and final post-operative status respectively. Pre-operative values were used as covariates in linear models that included post-operative measures as dependent variables.
With this analytical approach the gene by psychological interaction effect was determined individually after accounting for other relevant predictor variables, thereby identifying unique prediction of variability for the dependent variable of interest. In our linear modeling, we conducted a total of 18 independent tests to determine if interactions (9 genetic factors by 2 psychological factors) improved prediction for each shoulder pain outcome. Bonferroni correction would yield a threshold alpha level of 0.0027 for each outcome. While this might be a conservative correction for genetic studies, the value of 0.0027 provides a convenient benchmark in assessing the outcome of the analyses. Specifically, interaction terms with p values <0.0027 were considered “strong” statistical evidence for predicting the pain phenotype of interest, while those with p values ≥0.0027 but <0.05 as showing “moderate” statistical evidence for predicting the pain phenotype of interest. Interaction terms with p values ≥0.05 were not reported in this paper. This same approach for p-value adjustment and interpretation was used in our pre-clinical studies.5, 6
Results
A total of 150 patients were consented, with 120 (80.0%), 123 (82.0%), and 120 (80.0%) providing follow up at 3, 6, and 12 months respectively. Descriptive data for this cohort are reported in Tables 1 and 2.
Table 1.
Description of Pain Modulatory and Inflammatory Genes
| SNP | Frequency | Percent | SNP | Frequency | Percent | ||
|---|---|---|---|---|---|---|---|
| Pain Modulatory | Inflammatory | ||||||
|
ADRB2
rs1042713 |
AA | 18 | 12.2% |
IL1B
rs16944 |
AA | 20 | 13.5% |
| AG | 79 | 53.7% | AG | 64 | 43.2% | ||
| GG | 50 | 34.0% | GG | 64 | 43.2% | ||
|
ADRB2
rs1042714 |
CC | 52 | 35.1% |
IL1B
rs1143627 |
AA | 63 | 42.6% |
| CG | 76 | 51.4% | AG | 65 | 43.9% | ||
| GG | 20 | 13.5% | GG | 20 | 13.5% | ||
|
OPRM1
rs1799971 |
AA | 109 | 73.7% |
IL1B
rs1143634 |
AA | 10 | 6.8% |
| AG | 36 | 24.3% | AG | 46 | 31.1% | ||
| GG | 3 | 2.0% | GG | 92 | 62.2% | ||
|
AVPR1
rs1042615 |
AA | 22 | 15.0% |
IL6
rs1800797 |
AA | 16 | 10.8% |
| AG | 58 | 39.5% | AG | 68 | 46.0% | ||
| GG | 67 | 45.6% | GG | 64 | 43.2% | ||
|
AVPR1
rs10877969 |
CC | 5 | 3.4% |
IL6
rs2069840 |
CC | 56 | 38.6% |
| CT | 44 | 29.7% | CG | 77 | 53.1% | ||
| TT | 99 | 66.9% | GG | 12 | 8.3% | ||
|
GCH1
rs3783641 |
AA | 9 | 6.1% |
TNF-308
rs1800629 |
AA | 3 | 2.1% |
| AT | 44 | 29.9% | AG | 38 | 26.0% | ||
| TT | 94 | 64.0% | GG | 105 | 71.9% | ||
|
KCNS1
rs734784 |
8 | 27 | 18.4% |
TNF/LTA
rs2229094 |
CC | 12 | 8.2% |
| CT | 64 | 43.5% | CT | 69 | 46.9% | ||
| TT | 56 | 38.1% | TT | 66 | 44.9% | ||
|
TNF/LTA
rs1800683 |
AA | 17 | 11.6% | ||||
| AG | 70 | 47.6% | |||||
| GG | 60 | 40.8% | |||||
Table 2.
Descriptive Summary of Operative Shoulder Pain Cohort
| Pre-Operative | 3 months | 6 months | 12 months | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Variable | Mean±SD or Frequency |
Median [Min, Max] or Percent |
Mean±SD or Frequency |
Median [Min, Max] or Percent |
Mean±SD or Frequency |
Median [Min, Max] or Percent |
Mean±SD or Frequency |
Median [Min, Max] or Percent |
|
| Age | 42.7±17.4 | 42 [18,81] | |||||||
| Gender | Female | 51 | 34% | ||||||
| Male | 99 | 66% | |||||||
| Race | White | 127 | 85% | ||||||
| Black or African American |
11 | 7% | |||||||
| Other | 12 | 8% | |||||||
| Dominant Hand |
Right | 132 | 88% | ||||||
| Left | 18 | 12% | |||||||
| Surgical Side |
Right | 75 | 50% | ||||||
| Left | 75 | 50% | |||||||
| FPQ | 20.9±6.0 | 21 [9, 36] | |||||||
| PCS | 10.8±8.6 | 9 [0, 42] | |||||||
| PHQ | 3.6±4.0 | 2 [0, 25] | |||||||
| STAI | 45.9±3.4 | 46 [35, 56] | |||||||
| TSK-11 | 23.7±6.1 | 24 [12, 40] | |||||||
| Average BPI | 3.3±2.4 | 3 [0, 10] | 1.5±1.5 | 1 [0, 8.7] | 1.3±1.5 | 0.7 [0, 7.3] | 1.1±1.6 | 0.5 [0, 8] | |
| Worst BPI | 5.4±2.6 | 6 [0, 10] | 3.0±2.2 | 3 [0, 0] | 2.6±2.4 | 2 [0, 10] | 2.0±2.3 | 1 [0, 10] | |
| Average DASH | 34.3±17.9 | 31.8 [2.3, 79.5] |
24.4±14.5 | 21.5 [0, 75] |
14.6±11.6 | 11.4 [ 0, 50] |
12.0±14.1 | 6.8 [0, 81.8] |
|
| Peak DASH | 37.6±12.4 | 36.5 [13, 74] |
30.9±12.1 | 28.0 [13, 68] |
25.1±8.8 | 24.0 [13, 49] |
23.7±10.2 | 20.0 [11, 68] |
|
Table 2 Key: FPQ = Fear of Pain Questionnaire; PCS = Pain Catastrophizing Scale; PHQ = Patient Health Questionnaire; STAI = State Trait Anxiety Inventory; TSK-11 = Tampa Scale of Kinesiophobia; BPI = Brief Pain Inventory; DASH = Disability of Arm, Shoulder, and Hand questionnaire
Pain Modulatory Genes
Regression analyses for the pain modulatory genes are reported in Table 3. The final models that contained interactions explained between 25% to 44% variance (p’s < 0.005). Interactions that met our criterion for “strong” statistical evidence are depicted graphically in Figure 1.
Table 3.
Regression analyses for prediction of clinical outcomes by pain modulatory genes and psychological factors
| Dependent Variable |
Genetic and Psychological Predictors |
Demographics Only | Add Baseline Variable |
Add Genotype | Add Psychological Factor |
Add Interaction | P- Value of Full Model |
R2 of Full Model |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R2 | P- value |
R2 | P- value |
R2
Increment |
P- value |
R2
Increment |
P- value |
R2
Increment |
P- value |
||||
| Pre-Operative Status | |||||||||||||
| Average Pain Intensity |
KCNS1(rs734784) + TSK-11 |
0.246 | <.0001 | 0.000 | 0.957 | 0.039 | 0.007 | 0.055 | 0.004 | <.0001 | 0.341 | ||
| Worst Pain Intensity |
KCNS1(rs734784) +
TSK-11 |
0.163 | <.0001 | 0.009 | 0.480 | 0.081 | 0.000 | 0.071 | 0.001 | <.0001 | 0.324 | ||
| 12 Month Post-Operative Course (AUC) | |||||||||||||
| Average Pain Intensity |
ADRB2(rs1042713) + PHQ |
0.188 | <.0001 | 0.160 | <.0001 | 0.001 | 0.863 | 0.044 | 0.002 | 0.048 | 0.0036 | <.0001 | 0.442 |
|
AVPR1A(rs1042615) + TSK-11 |
0.188 | <.0001 | 0.160 | <.0001 | 0.010 | 0.338 | 0.005 | 0.304 | 0.036 | 0.0197 | <.0001 | 0.399 | |
| Quick-DASH Average |
ADRB2(rs1042713)+
PHQ |
0.102 | 0.004 | 0.110 | <.0001 | 0.018 | 0.193 | 0.028 | 0.024 | 0.091 | 0.0001 | <.0001 | 0.349 |
| Quick-DASH Peak |
ADRB2(rs1042713) + PHQ |
0.136 | 0.001 | 0.021 | 0.077 | 0.029 | 0.109 | 0.002 | 0.543 | 0.058 | 0.0106 | <.0001 | 0.246 |
| 12 Month Post-Operative Status | |||||||||||||
| Average Pain Intensity |
GCH1(rs3783641) +
STAI |
0.097 | 0.020 | 0.039 | 0.026 | 0.032 | 0.122 | 0.012 | 0.199 | 0.086 | 0.0025 | 0.000 | 0.267 |
Table 3 Key: *Bolded variables met our criterion for strong statistical evidence for an interaction (p < 0.0027 for addition of interaction term); AUC = area under curve; DASH = Disability of Arm, Shoulder, and Hand questionnaire; FPQ = Fear of Pain Questionnaire; PCS = Pain Catastrophizing Scale; PHQ = Patient Health Questionnaire; STAI = State Trait Anxiety Inventory; TSK-11 = Tampa Scale of Kinesiophobia
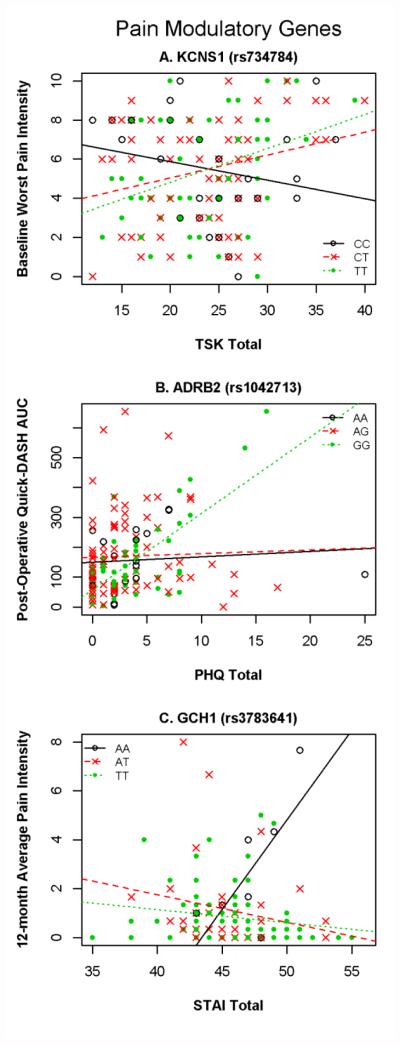
Figure 1. Pain Modulatory Gene × Psychology Interactions that Strongly Predicted Pain and Disability Outcomes.
Figure 1 Key: TSK-11 = Tampa Scale of Kinesiophobia; DASH = Disability of Arm, Shoulder, and Hand; AUC = Area Under the Curve; PHQ = Patient Health Questionnaire; STAI = State Trait Anxiety Inventory; BPI = Brief Pain Inventory
In predicting pre-operative status one genetic and psychological interaction emerged with strong statistical evidence and one with moderate statistical evidence. These interactions both involved the KCNS1 SNP interacting with the TSK-11 to predict worst and average pain intensity ratings. These interaction terms contributed an additional 7.1% (p = 0.001) and 5.5% (p = 0.004) variance to the full models for the strong and moderate evidence respectively. Increasing TSK-11 scores were more strongly associated with increasing pain intensity for the CT and TT genotypes in comparison with those in the CC genotype (Fig. 1A).
In predicting post-operative outcome course one genetic and psychological interaction showed strong statistical evidence, and three showed moderate statistical evidence. The strong statistical evidence was for an interaction between ADRB2 and PHQ predicting post-operative upper-extremity disability AUC by adding 9.1% (p = 0.0001) variance to the final model. Increasing PHQ scores were more strongly associated with increasing upper-extremity disability for the AA genotype in comparison with those in the AG and GG genotypes (Fig. 1B). The ADRB2 and PHQ interaction also added 5.8% variance (p = 0.0106) to peak post-operative upper-extremity disability scores. Pain intensity AUC was predicted by two different interactions that added significant variance to the final model. The first interaction was ADRB2 and PHQ (4.8% and p = 0.0036) and the second interaction was AVPR1A and TSK-11 (3.6% and p = 0.0197).
In predicting 12-month post-operative status only one genetic and psychological interaction was detected and it met our criterion for strong statistical evidence. This interaction involved GCH1 and the STAI and the term added 8.6% (p = 0.0025) variance to the full model. Increasing STAI scores were more strongly associated with increasing average pain intensity for the AA genotype in comparison with those in the AT and TT genotypes (Fig. 1C).
Inflammatory Genes
Regression analyses for the inflammatory genes are reported in Table 4. The final models that contained interactions explained between 21% to 45% variance (p’s < 0.005). There were no genetic and psychological interactions that met our criterion for predicting pre-operative status. Interactions that met our criterion for “strong” statistical evidence are depicted graphically in Figure 2.
Table 4.
Regression analyses for prediction of clinical outcomes by inflammatory genes and psychological factors
| Dependent Variable |
Genetic and Psychological Predictors |
Demographics Only |
Add Baseline Variable |
Add Genotype | Add Psychological Factor |
Add Interaction | P- Value of Full Model |
R2 of Full Model |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R2 | P- value |
R2 | P- value |
R2
Increment |
P- value |
R2
Increment |
P- value |
R2
Increment |
P- value |
||||
| Pre-Operative Status (none) | |||||||||||||
| 12 Month Post-Operative Course (AUC) | |||||||||||||
| Average Pain Intensity |
TNF/LTA(rs2229094) + FPQ | 0.197 | <.0001 | 0.158 | <.0001 | 0.009 | 0.368 | 0.002 | 0.496 | 0.032 | 0.0312 | <.0001 | 0.399 |
| IL6(rs1800797) + PCS | 0.197 | <.0001 | 0.158 | <.0001 | 0.002 | 0.793 | 0.015 | 0.076 | 0.036 | 0.0190 | <.0001 | 0.408 | |
| IL6(rs2069840) + FPQ | 0.197 | <.0001 | 0.158 | <.0001 | 0.001 | 0.867 | 0.003 | 0.467 | 0.036 | 0.0215 | <.0001 | 0.395 | |
| IL6(rs2069840) + PCS | 0.197 | <.0001 | 0.158 | <.0001 | 0.001 | 0.867 | 0.014 | 0.081 | 0.077 | 0.0002 | <.0001 | 0.448 | |
| IL6(rs2069840) + PHQ | 0.197 | <.0001 | 0.158 | <.0001 | 0.001 | 0.867 | 0.036 | 0.005 | 0.051 | 0.0027 | <.0001 | 0.444 | |
| Quick-DASH Average |
IL1B(rs1143627) + PHQ | 0.109 | 0.003 | 0.105 | <.0001 | 0.008 | 0.516 | 0.023 | 0.045 | 0.058 | 0.0048 | <.0001 | 0.303 |
| IL6(rs1800797) + PCS | 0.109 | 0.003 | 0.105 | <.0001 | 0.001 | 0.931 | 0.043 | 0.006 | 0.045 | 0.0162 | <.0001 | 0.303 | |
| IL6(rs2069840) + FPQ | 0.109 | 0.003 | 0.105 | <.0001 | 0.012 | 0.354 | 0.006 | 0.291 | 0.054 | 0.0075 | <.0001 | 0.287 | |
| IL6(rs2069840) + PCS | 0.109 | 0.003 | 0.105 | <.0001 | 0.012 | 0.354 | 0.040 | 0.008 | 0.081 | 0.0004 | <.0001 | 0.347 | |
| IL6(rs2069840) + PHQ | 0.109 | 0.003 | 0.105 | <.0001 | 0.012 | 0.354 | 0.024 | 0.041 | 0.067 | 0.0019 | <.0001 | 0.317 | |
| Quick DASH Peak |
IL6(rs2069840) + FPQ | 0.132 | 0.001 | 0.023 | 0.066 | 0.001 | 0.945 | 0.003 | 0.498 | 0.059 | 0.0117 | 0.001 | 0.218 |
| 12 Month Post-Operative Status | |||||||||||||
| Quick-DASH Average |
TNF/LTA(rs2229094) + STAI | 0.104 | 0.014 | 0.019 | 0.122 | 0.031 | 0.141 | 0.022 | 0.089 | 0.054 | 0.027 | 0.001 | 0.229 |
| IL6(rs2069840) + FPQ | 0.104 | 0.014 | 0.019 | 0.122 | 0.003 | 0.839 | 0.009 | 0.302 | 0.073 | 0.009 | 0.004 | 0.207 | |
Table 4 Key: *Bolded variables met our criterion for strong statistical evidence for an interaction (p < 0.0027 for addition of interaction term); AUC = area under curve; DASH = Disability of Arm, Shoulder, and Hand questionnaire; FPQ = Fear of Pain Questionnaire; PCS = Pain Catastrophizing Scale; PHQ = Patient Health Questionnaire; STAI = State Trait Anxiety Inventory; TSK-11 = Tampa Scale of Kinesiophobia
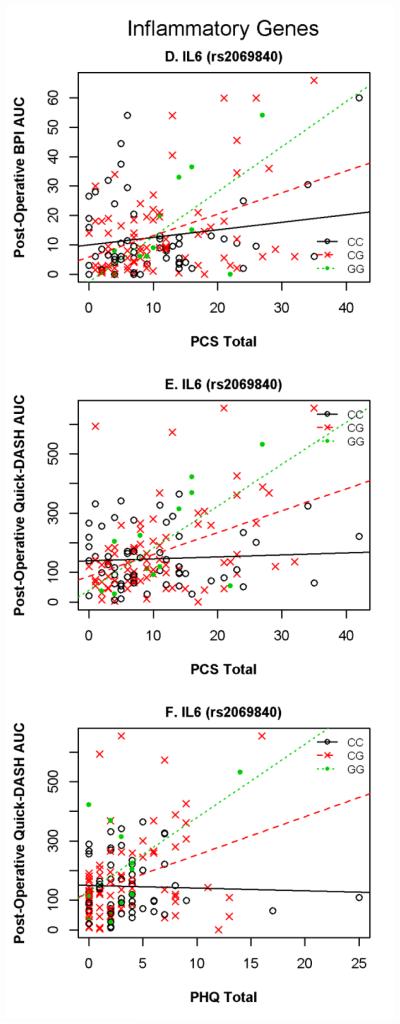
Figure 2. Inflammatory Gene × Psychology Interactions that Strongly Predicted Pain and Disability Outcomes.
Figure 2 Key: PCS = Pain Catastrophizing Scale; DASH = Disability of Arm, Shoulder, and Hand; AUC = Area Under the Curve; PHQ = Patient Health Questionnaire; BPI = Brief Pain Inventory
In predicting post-operative outcome course three genetic and psychological interactions showed strong statistical evidence, and 10 with moderate evidence. The interactions with strong statistical evidence were between IL6 and the PCS and between IL6 and the PHQ for two different outcomes. The interaction between IL6 and PCS added 7.7% variance (p = 0.0002) to the final model for pain intensity AUC and 8.1% variance (p = 0.0004) to the final model for upper-extremity disability AUC. The interaction between IL6 and PCS added 8.1% variance (p = 0.0004) and interaction between IL6 and PHQ added 6.7% variance (p = 0.0019) to the final model for upper-extremity disability AUC. For all three interactions, increasing PCS (or PHQ) scores were associated with increasing in pain outcomes intensity as follows: strongly positive for GG genotype, less but positive for CG, non-significant for CC. (Fig. 1D-F). A summary of the 10 other interactions for post-operative outcomes includes 7 interactions involving IL6 SNPs with other psychological measures for predicting pain and disability, 2 interactions involving IL1B and the PHQ for predicting disability, and 1 interaction for TNF/LTA and the FPQ for predicting pain. These interactions are reported in detail in Table 3.
In predicting 12-month post-operative status two genetic and psychological interactions were detected and both met our criterion for moderate statistical evidence. Both of these interactions involved prediction of 12-month disability scores, with the TNF/LTA and STAI interaction adding 5.4% variance (p = 0.027) and the IL6 and FPQ interaction adding 7.3% variance (p = 0.009) to the final models respectively.
Discussion
In this study we identified novel interactions between genetic and psychological factors that improved prediction of 12 month post-operative clinical outcomes following arthroscopic shoulder surgery. These findings advanced our previous translational work by identifying interactions directly in a clinical cohort. Our results indicated strong statistical evidence for 3 different interactions between pain modulatory genes and psychological factors for predicting pre-operative, post-operative course, and 12 month outcomes (KCNS1 × TSK-11, ADRB2 × PHQ, and GCH1 × STAI, respectively). Our results also indicated strong statistical evidence for 3 interactions involving an inflammatory gene for predicting post-operative course (2 for IL6 × PCS and IL6 × PHQ). These interactions added between 6-9% additional variance to full regression models, magnitude which suggests these findings may have clinical relevance for improving post-operative outcome prediction. Strong statements regarding the clinical implications of these findings must await future replication of the results; however, we believe this study has high potential clinical relevance in two areas. First, we expect these results to provide guidance on incorporating genetic and psychological factors to improve prediction of post-operative outcomes. Second, we expect the subgroups derived from these results will allow for the development and testing of the effectiveness of tailored treatment approaches based on genetic and psychological risk factors.
Interactions with Pain Modulatory Genes
The current analysis revealed additional interactions that improved prediction of post-operative outcomes beyond the previously identified COMT × PCS risk subgroup.7 Our results suggest these interactions may have phenotypic specificity, as each interaction with strong statistical evidence predicted a different clinical outcome. In our parallel analysis for predicting pre-clinical shoulder pain phenotypes in healthy subjects following exercise-induced shoulder injury, KCNS1 and ADRB2 interacted with catastrophizing and depression, respectively, to predict disability and pain duration, respectively.6 While the exact psychological factors involved with these interactions differed from those observed in the clinical cohort, the replication of these particular genes interacting with related psychological factors in separate cohorts suggests they are robust.
ADRB2 was the most frequent pain modulatory gene to interact with psychological factors and it was commonly coupled with a measure of depressive symptoms. This finding converges with previous research in which ADRB2 haplotype was associated with psychological traits, and predictive of those developing temporomandibular disorder.23 Furthermore, there is converging evidence that ADRB2 genotype is associated with chronic widespread pain24 and disabling neck and back pain.25 The current study adds to the existing literature by indicating ADRB2 has a role in the development of persistent post-operative disability via its interactions with psychological factors. The other pain modulatory genes identified in these interactions do not have as many comparison studies available. KCNS1 was identified in peripheral neuropathic pain models that were further investigated and determined to increase risk of low back pain with the “C” risk allele.26 Interestingly, our findings compliment this previous report by indicating increased risk with the “T” allele when combined with fear of movement. Previous research has identified pain protective genotypes/haplotypes of GCH1 in preclinical and clinical models27-29 and this study demonstrated that GCH1 homogenous for the “A” allele interacted with anxiety to predict 12 month pain outcomes. The novelty and specificity of these findings for KCNS1 and GCH1 need further confirmation in future studies that predict post-operative outcomes.
Interactions with Inflammatory Genes
In contrast to the pain modulatory genes, there was notable consistency for inflammatory gene interaction findings. IL6 was the only gene with strong statistical evidence for an interaction predicting post-operative course for pain and disability measures and the “G” allele was consistently predictive of higher risk. Furthermore IL6 was included in 10/12 of gene × psychological interactions indicating robust findings with potential clinical relevance for improving outcome prediction by collecting this information pre-operatively.
Scant literature is available in patient populations to aid the interpretation of the inflammatory gene findings, but there are precedents in pre-clinical studies. For example, in healthy subjects experiencing experimental pain higher pain catastrophizing predicted increased IL6 serum levels, providing biological plausibility for interactions between pain-related psychological constructs and inflammatory responses.30 In our pre-clinical analyses predicting shoulder pain phenotypes we identified TNF/LTA and IL1B as interacting with psychological factors.5 IL6 was not predictive of any of the pre-clinical phenotypes, which was a stark contrast to the clinical findings. This lack of translation could be an indication that exercise-induced injury may be a better choice for general musculoskeletal pain processes, as evidenced by examples of translation for pain modulatory genes. A pre-clinical model of exercise-induced injury may not be a good choice for mimicking inflammatory processes that lead to the development of persistent post-operative pain and disability. Collectively these findings indicate that IL6 polymorphisms indicating a pro-inflammatory predisposition should be considered in concert with psychological functioning as strong candidates for having clinical relevance in future predictive models of post-operative pain and disability.
Future Research
These findings provide direction for future study. Models for predicting post-operative outcomes could start with the established COMT × PCS interaction, and then add other interactions identified in these analyses to determine if prediction improved. It is conceivable that consideration of both inflammatory and pain modulatory factors in prediction models would allow for more accurate prediction, but this approach has yet to be tested. Future study could also determine how prediction of specific outcomes changes when including the outcome specific gene by psychology interactions identified in this study. For example, a future study could determine if the addition of the GCH1 × TSK-11 interaction improves the precision of 12 month pain intensity outcome prediction after considering the COMT × PCS interaction. Investigation of these questions in an independent cohort would provide valuable information on what are the parameters for optimal outcome prediction when using such interactions. Finally, future research could add biological phenotypes so that a better understanding of the mechanisms underlying these interactions could be gained.
Limitations
The strengths of this study include the investigation of a priori selected genetic and psychological factors, the intentional link to a pre-clinical model, and 12 month follow up. There are, of course, limitations to consider. First, in our predictive models we did not control for pain medication or rotator cuff tear size. These factors were included in an earlier analysis7 and did not impact outcomes so it was decided to not include them in the current analysis for sake of parsimony. Furthermore we did not consider any psychological × psychological interactions in the regression models even though there is likely interaction amongst the constructs included in this study. Second, we do not have associated biological measures so can’t discuss physiological implications of these findings. This concern is mitigated by using genetic factors with known functional consequences but still limits what can be determined mechanistically from this study. Third, we can’t interpret the identified interactions as being indicative of chronic pain per se because subjects had pre-operative pain that varied greatly in duration. Therefore, the interactions identified in this study are best interpreted as being predictive of persistent post-operative pain.
Conclusions
Persistent post-operative pain is a common and undesirable outcome of surgery that remains difficult to predict,3 even when arthroscopy is used.4 Given the complex nature of pain, multiple factors must be considered when determining risk for developing chronic or persistent pain conditions.1, 2 The current study identified 6 interactions genetic and psychological interactions that improved prediction of pain and disability outcomes. Although clinical relevance of these interactions will ultimately be determined in future studies our findings indicate they can be used to improve the identification of persistent post-operative pain and to design tailored pain management strategies.
Significance and Innovation.
In a planned follow up to our pre-clinical studies we identified interactions between genetic and psychological factors that improved prediction of 12 month pain and disability outcomes.
Interactions with the pain modulatory genes were similar to those in our pre-clinical model, and each interaction was predictive of a different outcome measure.
Interactions with the inflammatory genes differed from our pre-clinical model and consistently predicted the same outcome (12 month post-operative course).
These results provide important direction for future prognostic studies involving post-operative outcomes and/or development of tailored pain management strategies.
Acknowledgments
Financial Support
This study was completed with funding from the National Institutes of Health; NIAMS (AR055899). All authors were independent from this funding source and the funding source played no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review or approval of the manuscript.
Footnotes
Conflict of Interest
The authors have no conflict of interest, financial or otherwise, to declare with submission of this manuscript.
Reference List
- (1).Mogil JS. Pain genetics: past, present and future. Trends Genet. 2012 Jun;28(6):258–266. doi: 10.1016/j.tig.2012.02.004. [DOI] [PubMed] [Google Scholar]
- (2).Diatchenko L, Fillingim RB, Smith SB, Maixner W. The phenotypic and genetic signatures of common musculoskeletal pain conditions. Nat Rev Rheumatol. 2013 Jun;9(6):340–350. doi: 10.1038/nrrheum.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Kehlet H, Jensen TS, Woolf CJ. Persistent postsurgical pain: risk factors and prevention. Lancet. 2006 May 13;367(9522):1618–1625. doi: 10.1016/S0140-6736(06)68700-X. [DOI] [PubMed] [Google Scholar]
- (4).Simanski CJ, Althaus A, Hoederath S, et al. Incidence of Chronic Postsurgical Pain (CPSP) after General Surgery. Pain Med. 2014 Jul;15(7):1222–1229. doi: 10.1111/pme.12434. [DOI] [PubMed] [Google Scholar]
- (5).George SZ, Parr JJ, Wallace MR, et al. Inflammatory Genes and Psychological Factors Predict Induced Shoulder Pain Phenotype. Med Sci Sports Exerc. 2014 Oct;46(10):1871–1881. doi: 10.1249/MSS.0000000000000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).George SZ, Parr JJ, Wallace MR, et al. Biopsychosocial influence on exercise-induced injury: genetic and psychological combinations are predictive of shoulder pain phenotypes. J Pain. 2014 Jan;15(1):68–80. doi: 10.1016/j.jpain.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).George SZ, Wallace MR, Wu SS, et al. Biopsychosocial influence on shoulder pain: risk subgroups translated across preclinical and clinical prospective cohorts. Pain. 2015 Jan;156(1):148–156. doi: 10.1016/j.pain.0000000000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Little J, Higgins JP, Ioannidis JP, et al. Strengthening the reporting of genetic association studies (STREGA): an extension of the STROBE Statement. Hum Genet. 2009 Mar;125(2):131–151. doi: 10.1007/s00439-008-0592-7. [DOI] [PubMed] [Google Scholar]
- (9).von EE, Altman DG, Egger M, Pocock SJ, Gotzsche PC, Vandenbroucke JP. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet. 2007 Oct 20;370(9596):1453–1457. doi: 10.1016/S0140-6736(07)61602-X. [DOI] [PubMed] [Google Scholar]
- (10).Huang FY, Chung H, Kroenke K, Delucchi KL, Spitzer RL. Using the Patient Health Questionnaire-9 to measure depression among racially and ethnically diverse primary care patients. J Gen Intern Med. 2006 Jun;21(6):547–552. doi: 10.1111/j.1525-1497.2006.00409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kroenke K, Spitzer RL. The PHQ-9: a new depression diagnostic and severity measure. Psychiatr Annals. 2002;32:509–521. [Google Scholar]
- (12).Spielberger CD, Gorsuch RL, Lushene RE, Vagg PR, Jacobs GA. Manual for the state and trait anxiety inventory (form Y) Consulting Psychologists Press; Palo Alto, CA: 1983. [Google Scholar]
- (13).Albaret MC, Munoz Sastre MT, Cottencin A, Mullet E. The Fear of Pain questionnaire: factor structure in samples of young, middle-aged and elderly European people. Eur J Pain. 2004 Jun;8(3):273–281. doi: 10.1016/j.ejpain.2003.09.005. [DOI] [PubMed] [Google Scholar]
- (14).McNeil DW, Rainwater AJ. Development of the Fear of Pain Questionnaire--III. J Behav Med. 1998 Aug;21(4):389–410. doi: 10.1023/a:1018782831217. [DOI] [PubMed] [Google Scholar]
- (15).Osman A, Breitenstein JL, Barrios FX, Gutierrez PM, Kopper BA. The Fear of Pain Questionnaire-III: further reliability and validity with nonclinical samples. J Behav Med. 2002 Apr;25(2):155–173. doi: 10.1023/a:1014884704974. [DOI] [PubMed] [Google Scholar]
- (16).Parr JJ, Borsa PA, Fillingim RB, et al. Pain-related fear and catastrophizing predict pain intensity and disability independently using an induced muscle injury model. J Pain. 2012 Apr;13(4):370–378. doi: 10.1016/j.jpain.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Goubert L, Crombez G, Van Damme S, Vlaeyen JW, Bijttebier P, Roelofs J. Confirmatory factor analysis of the Tampa Scale for Kinesiophobia: invariant two-factor model across low back pain patients and fibromyalgia patients. Clin J Pain. 2004 Mar;20(2):103–110. doi: 10.1097/00002508-200403000-00007. [DOI] [PubMed] [Google Scholar]
- (18).Woby SR, Roach NK, Urmston M, Watson PJ. Psychometric properties of the TSK-11-11: a shortened version of the Tampa Scale for Kinesiophobia. Pain. 2005 Sep;117(1-2):137–144. doi: 10.1016/j.pain.2005.05.029. [DOI] [PubMed] [Google Scholar]
- (19).Van Damme S, Crombez G, Bijttebier P, Goubert L, Van Houdenhove B. A confirmatory factor analysis of the Pain Catastrophizing Scale: invariant factor structure across clinical and non-clinical populations. Pain. 2002 Apr;96(3):319–324. doi: 10.1016/S0304-3959(01)00463-8. [DOI] [PubMed] [Google Scholar]
- (20).Keller S, Bann CM, Dodd SL, Schein J, Mendoza TR, Cleeland CS. Validity of the brief pain inventory for use in documenting the outcomes of patients with noncancer pain. Clin J Pain. 2004 Sep;20(5):309–318. doi: 10.1097/00002508-200409000-00005. [DOI] [PubMed] [Google Scholar]
- (21).Hudak PL, Amadio PC, Bombardier C. Development of an upper extremity outcome measure: the DASH (disabilities of the arm, shoulder and hand) [corrected]. The Upper Extremity Collaborative Group (UECG) Am J Ind Med. 1996 Jun;29(6):602–608. doi: 10.1002/(SICI)1097-0274(199606)29:6<602::AID-AJIM4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- (22).Gummesson C, Ward MM, Atroshi I. The shortened disabilities of the arm, shoulder and hand questionnaire (QuickDASH): validity and reliability based on responses within the full-length DASH. BMC Musculoskelet Disord. 2006;7:44. doi: 10.1186/1471-2474-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Diatchenko L, Anderson AD, Slade GD, et al. Three major haplotypes of the beta2 adrenergic receptor define psychological profile, blood pressure, and the risk for development of a common musculoskeletal pain disorder. Am J Med Genet B Neuropsychiatr Genet. 2006 Jul 5;141B(5):449–462. doi: 10.1002/ajmg.b.30324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hocking LJ, Smith BH, Jones GT, Reid DM, Strachan DP, Macfarlane GJ. Genetic variation in the beta2-adrenergic receptor but not catecholamine-O-methyltransferase predisposes to chronic pain: results from the 1958 British Birth Cohort Study. Pain. 2010 Apr;149(1):143–151. doi: 10.1016/j.pain.2010.01.023. [DOI] [PubMed] [Google Scholar]
- (25).Skouen JS, Smith AJ, Warrington NM, et al. Genetic variation in the beta-2 adrenergic receptor is associated with chronic musculoskeletal complaints in adolescents. Eur J Pain. 2012 Oct;16(9):1232–1242. doi: 10.1002/j.1532-2149.2012.00131.x. [DOI] [PubMed] [Google Scholar]
- (26).Costigan M, Belfer I, Griffin RS, et al. Multiple chronic pain states are associated with a common amino acid-changing allele in KCNS1. Brain. 2010 Sep;133(9):2519–2527. doi: 10.1093/brain/awq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Tegeder I, Costigan M, Griffin RS, et al. GTP cyclohydrolase and tetrahydrobiopterin regulate pain sensitivity and persistence. Nat Med. 2006 Nov;12(11):1269–1277. doi: 10.1038/nm1490. [DOI] [PubMed] [Google Scholar]
- (28).Belfer I, Dai F, Kehlet H, et al. Association of functional variations in COMT and GCH1 genes with postherniotomy pain and related impairment. Pain. 2015 Feb;156(2):273–279. doi: 10.1097/01.j.pain.0000460307.48701.b0. [DOI] [PubMed] [Google Scholar]
- (29).Campbell CM, Edwards RR, Carmona C, et al. Polymorphisms in the GTP cyclohydrolase gene (GCH1) are associated with ratings of capsaicin pain. Pain. 2009 Jan;141(1-2):114–118. doi: 10.1016/j.pain.2008.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Edwards RR, Kronfli T, Haythornthwaite JA, Smith MT, McGuire L, Page GG. Association of catastrophizing with interleukin-6 responses to acute pain. Pain. 2008 Nov 15;140(1):135–144. doi: 10.1016/j.pain.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]