Abstract
β-adrenergic signaling can regulate macrophage involvement in several diseases and often produces anti-inflammatory properties in macrophages, which are similar to M2 properties in a dichotomous M1 vs. M2 macrophage taxonomy. However, it is not clear that β-adrenergic-stimulated macrophages may be classified strictly as M2. In this in vitro study, we utilized recently published criteria and transcriptome-wide bioinformatics methods to map the relative polarity of murine β-adrenergic-stimulated macrophages within a wider M1–M2 spectrum. Results show that β-adrenergic-stimulated macrophages did not fit entirely into any one predefined category of the M1–M2 spectrum but did express genes that are representative of some M2 side categories. Moreover, transcript origin analysis of genome-wide transcriptional profiles located β-adrenergic-stimulated macrophages firmly on the M2 side of the M1–M2 spectrum and found active suppression of M1 side gene transcripts. The signal transduction pathways involved were mapped through blocking experiments and bioinformatics analysis of transcription factor binding motifs. M2-promoting effects were mediated specifically through β2-adrenergic receptors and were associated with CREB, C/EBPβ, and ATF transcription factor pathways but not with established M1–M2 STAT pathways. Thus, β-adrenergic-signaling induces a macrophage transcriptome that locates on the M2 side of the M1–M2 spectrum but likely accomplishes this effect through a signaling pathway that is atypical for M2-spectrum macrophages.
Keywords: macrophage, M1, M2 spectrum, β-adrenergic, transcriptome, bioinformatics, CREB
1. Introduction
Previous studies have found that the sympathetic nervous system (SNS) and β-adrenergic receptor signaling can regulate macrophage involvement in diseases that include cancer,1,2 heart disease,3 and AIDS.4 The earliest investigations came from a line of research that found β-adrenergic signaling can suppress the ability of macrophages to control Mycobacterium avium infection, a significant cause of morbidity and mortality in AIDS patients.4,5 More recently, a mechanism by which macrophage infiltration contributes to increased risk of atherosclerotic cardiovascular disease (i.e., monocyte expression of beta2-integrins for vascular endothelium adhesion) was up-regulated by experimentally-induced anger with higher plasma norepinephrine levels in healthy participants.3 Accordingly, a mouse model of atherosclerosis showed that SNS-induced β-adrenergic signaling promotes macrophage accumulation in atherosclerotic plaques.6 In a murine model of breast cancer, we found that chronic stress can promote accumulation of tumor-associated macrophages (TAMs) in the primary tumor of mice through β-adrenergic signaling.1 Similarly, others have also found that the same chronic stress paradigm in mice will increase TAM prevalence in ovarian tumors through increases in monocyte chemotactic protein 1, a chemokine that is up-regulated in ovarian cancer cells by β-adrenergic signaling.2
TAMs often exhibit properties of an M2 macrophage rather than the classical M1 phenotype of microbicidal macrophages.7 In a non-malignant environment, M2 macrophages facilitate wound healing by tapering immune cell attack, clearing tissue debris, and building new vasculature.7 However, in malignancy, those M2 properties may forestall immune cell attack, break down normal tissue for malignant expansion, and build new vasculature to support cancer cell metabolism and distant metastasis.7,8 To this point, we found that β-adrenergic signaling increased gene expression of the prototypical M2 marker, Arg1, in primary tumors with elevated accumulation of TAMs and confirmed that norepinephrine increases Arg1 expression in macrophages outside the tumor microenvironment (i.e., in bone marrow-derived macrophages).1 Similarly, both norepinephrine and epinephine were found to alter the phenotype of LPS-activated M1-like macrophages by increasing the expression of Arg1.9
However, it is not clear that β-adrenergic-stimulated macrophages may accurately be called M2 macrophages, because (1) M1 and M2 macrophages are not defined by any single emblematic marker,10 (2) rather than being just two separate phenotypes, M1 and M2 macrophages are thought to constitute a spectrum of several related phenotypes,11 and (3) canonical M1 and M2 macrophage activation (i.e., stimulated by IFNγ and IL-4, respectively) is driven primarily by transcription factors that are different from β-adrenergic-related transcription factors recently found to mediate expression of Arg1 and other anti-inflammatory genes.12,13,14 In this latter regard, whereas M1 macrophages exhibit high levels of STAT1 transcription factor and M2 macrophages exhibit high levels of STAT6,10 the well-defined downstream transcription factor of β-adrenergic signaling, CREB,15 in conjunction with the transcription factor C/EBPβ, has been found to regulate expression of genes in macrophages that are associated with an anti-inflammatory phenotype, including Arg1 and Il10.12,13 ATF1, a transcription factor closely-related to CREB, has also been linked to Il10 transcription.14
In order to facilitate accurate description of macrophage polarity phenotypes across diverse experimental scenarios, macrophage biologists have proposed standards for defining multiple distinct categories of M2-like and M1-like macrophages in a linear spectrum (see Figure 1 and Murray et al., 2014),10 which may be used to approximate the spectrum location of β-adrenergic-stimulated macrophages. These categories are based on the extracellular macrophage activator and subsequent transcriptional regulators. For example, canonical M2 macrophages activated by IL-4 are classified as M(IL-4) macrophages, but macrophages activated by other established activators are also considered M2-like. These include immune complex-activated macrophages, M(Ic), IL-10-activated macrophages, M(IL-10), glucocorticoid and transforming growth factor β-activated macrophages, M(GC+TGF-β), and glucocorticoid alone-activated macrophages, M(GC). On the M1 side of the spectrum, canonical M1 macrophages activated by IFNγ are classified as M(IFNγ), and other closely-related M1-like macrophages include lipopolysaccharide and IFNγ-activated macrophages, M(LPS+IFNγ), and lipopolysaccharide alone-activated macrophages, M(LPS). Each of the categories are further defined by the activator’s consensus set of gene expression markers to facilitate comparison of the categories to gene expression from any novel macrophage stimulation scenario.
Figure 1.
(A) M1 - M2 spectrum categories of macrophage activation, based on the activator and subsequent known transcription factors, as proposed by Peter J. Murray and colleagues following the International Congress of Immunology in Milan in 2013. At the far end of the M2 side of the spectrum sits IL-4-activated macrophages, M(IL-4), followed by immune complex-activated macrophages, M(Ic), IL-10-activated macrophages, M(IL-10), glucocorticoid and transforming growth factor β-activated macrophages, M(GC+TGF-β), and glucocorticoid alone-activated macrophages, M(GC). At the far end of the M1 side of the spectrum sits IFNγ-activated macrophages, M(IFNγ), followed by lipopolysaccharide and IFNγ-activated macrophages, M(LPS+IFNγ), and lipopolysaccharide alone-activated macrophages, M(LPS). M(-) indicative of non-activated macrophages. (B) Example markers from each category’s current consensus gene expression set. + signs indicative of relative expression, -ve sign indicative of no expression. Adapted from Immunity, Vol 41, Macrophage Activation and Polarization: Nomenclature and Experimental Guidelines, pp.14–20, Copyright (2014), with permission from Elsevier.
Given these newly proposed standards for defining the relative polarity of a specifically-stimulated macrophage, we selected gene transcripts a priori from each category in the spectrum, as defined for mouse macrophages, to test for differential induction by β-adrenergic signaling in mouse bone marrow-derived macrophages (BMDMs) using quantitative real-time RT-PCR. Furthermore, because no study has examined the effect of β-adrenergic signaling on the total array of transcriptome dynamics involved in M1–M2 polarity, we followed this targeted a priori approach with genome-wide transcriptional profiling in order to more comprehensively assess the spectrum location of β-adrenergic-stimulated macrophages relative to canonical IL-4-stimulated M2 macrophages vs. canonical IFNγ-stimulated M1 macrophages. Finally, given that STAT family transcription factors are established as primary drivers of gene expression in the M1–M2 spectrum,10 we utilized bioinformatic inferences of activity for these transcription factors, as well as for transcription factors related to β-adrenergic-signaling, to assess their role in the macrophage polarization response to β-adrenergic-stimulation. Given the previous findings noted above, we hypothesized that β-adrenergic-stimulated macrophages exhibit a transcriptome that locates on the M2 side of the M1–M2 spectrum and that this M2-spectrum transcriptome is regulated in part by CREB, C/EBPβ, ATF, and STAT family transcription factors.
2. Methods
2.1. Bone marrow derived macrophages (BMDMs)
Flushed bone marrow from female Balb/c mice (Charles River, 8–10 weeks) in RPMI-1640 with L-glutamine (Cellgro-Corning, Inc., #10-040-CV) was passed through a 30-μm cell strainer (Miltenyi, #130-041-407) and subjected to red blood cell lysis buffer (BD Biosciences, #555899). White blood cells were counted by hemocytometry and seeded at 0.25 × 106 cells/mL in a total of 4 mL/well of RPMI-1640 with L-glutamine supplemented with 10% FBS (Atlanta Biologicals, #S11550H), 100 IU penicillin/mL, 100 μg streptomycin/mL (Cellgro-Corning, #30-002-CI), at 37°C, 5% CO2, in a 6-well polystyrene low attachment plate (Costar, #3471) with 10 ng/mL of recombinant mouse M-CSF (Gibco, #PMC2044) for 7 days (media replenished after 2, 5, and 7 days). Flow cytometry was used to confirm macrophage phenotype of resultant BMDMs with fluorescence-conjugated antibodies against murine F4/80 (BD Biosciences, #565411) after mouse Fc blocking (BD Bioscience, #553141) using a FACSAria II High-Speed Cell Sorter with FACSDiva software (BD Biosciences) for analysis of total live cells from gating based on forward- versus side-scatter profiles.
2.2. Macrophage stimulation
To examine the effect of β-adrenergic agonism on gene transcripts indicative of macrophages in the M1–M2 spectrum, BMDMs were incubated with the non-selective β-adrenergic agonist isoproterenol (Sigma, #I2760) for 24 hours. Control BMDMs were incubated in media only during the same 24 hour period. To comprehensively compare the global gene expression profile of β-adrenergic-stimulated macrophages to M1 and M2 transcriptomes, separate BMDMs were incubated with either 20 ng/mL of recombinant mouse IFN-γ (Gibco, #PMC4034) or 20 ng/mL of recombinant mouse IL-4 (Gibco, #PMC0045) for 24 hours. In each of three independent experiments, duplicate wells were prepared for each condition (control, isoproterenol, IL-4, IFN-γ).
To determine the specific receptor subtype necessary for isoproterenol to induce up-regulation of select gene transcripts differentially regulated in a priori experiments and in the global gene expression profile, selective β1-, β2-, and β3-adrenergic antagonists, atenolol (Sigma, #A7655), ICI 118,551 (Sigma, #I127), and L-748,337 (Santa Cruz, #sc-204044), respectively, were used. Antagonists were added to wells 15 minutes before isoproterenol. In each of three independent experiments, duplicate wells were prepared for each condition (control, isoproterenol, control+atenolol, isoproterenol+atenolol, control+ICI 118,551, isoproterenol+ICI 118,551, control+L-748,337, isoproterenol+L-748,337). To determine whether β2-adrenergic receptor signaling was sufficient to induce up-regulation of select gene transcripts, the selective β2-adrenergic agonist, formoterol (Sigma, #F9552), was used. In each of three independent experiments, duplicate wells were prepared for each condition (control, formoterol).
2.3. qRT-PCR
To quantify gene expression, total RNA from BMDMs was extracted (Qiagen RNeasy Mini Kit, #74104), cleared of contaminating DNA with on-column DNase digestion (Qiagen RNase-Free DNase Set, #79254), and quantified by spectrophotometry (NanoDrop ND-1000, Thermo Scientific). Gene transcripts indicative of recently suggested key marker systems for activated macrophages in the M1–M2 spectrum (Murray et al., 2014; Figure 1) were selected a priori for examination in β-adrenergic-stimulated macrophages by qRT-PCR using one-step assay reagents (Qiagen Quantitect Probe RT-PCR, #204443) and TaqMan Gene Expression Assay primer-probes for mouse Arg1, Retnla, Il10, Il4ra, Nos2, and Ido1 (Life Technologies/Applied Biosystems, Mm00475988_m1, Mm00445109_m1, Mm00440502_m1, Mm00439614_m1, Mm01275139_m1, Mm00492586_m1, respectively). Five gene transcripts with previously reported importance for M1–M2 macrophage biology were selected for independent verification of differential expression by qRT-PCR after exhibiting differential expression in the global gene expression profile. These included Cxcl4/Pf4, Ccl24, Dusp1, Il1rn, and Cd74 (Mm00451315_g1, Mm00444701_m1, Mm00457274_g1, Mm00446186_m1, Mm00658576_m1, respectively). Following reverse transcription of RNA template, resulting product underwent 50 PCR amplification cycles of 15 seconds of strand separation at 94°C and 60 seconds of annealing and extension at 60°C. Triplicate determinations were quantified by threshold cycle analysis of FAM fluorescence intensity using iCycler software (Bio-Rad), normalized to values of beta-actin mRNA amplified in parallel (Actb, #Mm00607939_s1). Student’s t test was used to analyze the effects of β-adrenergic-activation on gene expression. Univariate analysis of variance was used to analyze the effects of selective β1-, β2-, and β3-adrenergic antagonists on isoproterenol-induced gene expression with Tukey’s adjustment for multiple comparisons.
2.4. Transcript origin analysis (TOA)
Total RNA (~1 μg ) was assayed using Illumina MouseRef-8 v2.0 Expression Beadchips in the University of California, Los Angeles Neuroscience Genomics Core (UNGC). Quantile normalization16 was applied to values of the 18,138 assayed transcripts, and differentially expressed genes were identified by ≥ 25% difference in mean (log2) expression levels in macrophages treated with isoproterenol vs. controls. Genes that were differentially expressed by isoproterenol were identified based on biological effect size (i.e., difference = mean isoproterenol – mean control) rather than statistical effect size (e.g., t statistic or P value), because previous research has shown that biological effect size-based criteria yield more replicable results than do statistical effect size criteria (e.g., t statistics, P values, or false discovery rate q values).17,18,19,20 Effect size point estimates for individual gene transcripts serve only as input into higher-order bioinformatics analyses testing gene set hypotheses regarding M1- and M2-diagnostic groups in TOA and transcription factor binding motif analysis with TELiS as described below.21 Gene expression data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO; accession no. GSE80185).
For TOA, we defined a cell type diagnosticity score, as previously described,22 for each gene (indexed g = 1 to G, g ∈ 18,134 mouse gene transcripts assayed in the Illumina MouseRef-8 v2.0 Expression Beadchip). In the current experiment, a cell type diagnosticity score indicates the extent to which a given gene transcript is predominately expressed by an M2-polarized macrophage relative to an M1-polarized macrophage in the reference wells of our study (i.e., BMDMs stimulated only by IL-4 or IFNγ, respectively). Positive values indicate relative over-expression of the gene in M2 macrophages and negative values indicate relative over-expression of the gene in M1 macrophages. The mean diagnosticity score for all genes that were up-regulated by β-adrenergic-stimulation and the mean diagnosticity score for all genes down-regulated by β-adrenergic-activation were then tested for statistically significant deviation from the mean population diagnosticity score for all mouse genes using a single-sample t test.23
2.5. Transcription factor analysis
We used a two-sample variant of the Transcription Element Listening System (TELiS; www.telis.ucla.edu) as previously described24 to compare the prevalence of transcription factor-binding motifs (TFBMs) for CREB, C/EBPβ, ATF, and STAT transcription factors in the promoters of genes that were up-regulated and down-regulated in macrophages by isoproterenol stimulation relative to control treatment. Binding motif definitions were retrieved from the TRANSFAC database. Analyses averaged results derived from nine parametric variations of promoter length (−300 bp relative to RefSeq transcription start site, −600 bp, and −1000 bp to +200) and target TFBM match stringency (MatSim = 0.80, 0.90, 0.95). Differential TFBM prevalence ratios of up-regulated to down-regulated genes were then tested for significant deviation from a null population mean ratio of 1 by two-tailed P values in a single-sample t test.
3. Results
3.1. M1–M2 spectrum gene transcripts in the β-adrenergic-stimulated macrophage
To determine whether β-adrenergic-stimulated macrophages fall squarely into one category of the M1–M2 spectrum, we derived F4/80+ macrophages from murine bone marrow cells (Suppl. Figure 1) and exposed them to 1 μM isoproterenol. We then used qRT-PCR to assay expression for two genes from each category’s consensus set of gene markers for mouse macrophages (Figure 1B; see Murray et al., 2014 for full gene sets). We chose two genes because one gene can appear in more than one category’s consensus gene set. For categories on the M2 side of the spectrum, Arg1 and Retnla were measured for M(IL-4); Il10 and Nos2 for M(Ic); Il10 and Il4ra for M(IL-10) (note: M(GC+TGFβ) and M(GC) categories on the M2 side are currently undefined for mouse macrophages). For M(IL-4), isoproterenol induced a 10.4-fold increase in Arg1 gene expression (P < 0.001) but had no significant effect on Retnla expression (P = 0.316; Figure 2). For M(Ic), isoproterenol induced a 2.2-fold increase in Il10 gene expression (P = 0.005) but decreased Nos2 expression by 2.1-fold (P = 0.022; Figure 2).
Figure 2.
Effect of β-adrenergic signaling on select gene transcripts that constitute macrophage activation categories along the M1–M2 spectrum in Murray et al., 2014. Isoproterenol at 1 μM. Data represent mean ± SE of three independent experiments. ***P < 0.001, **P < 0.01.
For M(IL-10), isoproterenol had no significant effect on Il4ra expression (P = 0.411; Figure 2). For categories on the M1 side of the spectrum, Ido1 and Nos2 were measured for M(IFNγ), and Nos2 and Arg1 were measured for both M(LPS+IFNγ) and M(LPS). For M(IFNγ), isoproterenol had no significant effect on Ido1 expression (P = 0.407) and, as already noted above, had a suppressive effect on Nos2 expression (Figure 2). M(LPS+IFNγ) and M(LPS) are both also defined in part by increases in Nos2 expression, which was decreased by isoproterenol treatment, and relatively small increases in Arg1 expression, which was increased significantly by isoproterenol treatment. Collectively, the results of this targeted approach of M1–M2 spectrum genes suggest that β-adrenergic-stimulated macrophages do not fit discretely into any one pre-defined category in the proposed spectrum. Given the up-regulation of both Arg1 and Il10, it may be that β-adrenergic-stimulated macrophages express a profile that shares elements of both M(IL-4) and M(IL-10) categories on the M2 side. Also, given that β-adrenergic signaling down-regulates Nos2 but this gene is up-regulated in all three categories of the M1 side, it may be that β-adrenergic signaling suppresses some elements of M1-spectrum macrophages. To address these questions, we followed this targeted a priori approach with genome-wide transcriptional profiling in order to more comprehensively define the M1–M2 spectrum location of β-adrenergic-stimulated macrophages.
3.2. Comprehensive localization of the β-adrenergic-stimulated macrophage transcriptome in the M1–M2 spectrum
To provide a more systematic and unbiased approach to localization of β-adrenergic-stimulated macrophages within the M1–M2 spectrum, we used transcript origin analysis (TOA) to examine global gene expression profiles derived from Illumina bead arrays. Diagnosticity scores were calculated for each gene in the array, i.e., scores that quantify the extent to which each and every gene in the array is expressed predominately by M2 macrophages relative to M1 macrophages. These diagnosticity scores are listed in Dataset S1, where positive values indicate predominant expression of that gene in M2 macrophages and negative values indicate predominant expression of that gene in M1 macrophages.
We found that isoproterenol up-regulated the expression of 128 genes by ≥ 25% compared to controls (Dataset S2) and that these genes were significantly representative of M2 macrophages (mean diagnosticity score = 2.44, P < 0.0001; Figure 3). Conversely, isoproterenol down-regulated the expression of 113 genes by ≥ 25% difference (Dataset S2), and these genes were significantly representative of M1 macrophages, although as can be seen in Figure 3, this M1 suppressive effect was smaller in comparison to the M2 promoting effect (mean diagnosticity score = −0.86, P < 0.001). Confirmatory qRT-PCR results verified ≥ 25% differential expression for 5 target genes assayed (P values < 0.05; Figure 4). Thus, this transcriptome-wide approach confirms results from the targeted a priori analyses above in locating β-adrenergic-stimulated macrophages on the M2 side of the M1–M2 spectrum and demonstrates that β-adrenergic signaling accomplishes its M2 effect not only by promoting M(IL-4) gene expression on the M2 side but also by actively suppressing M(IFNγ) gene expression on the M1 side.
Figure 3.
Mean diagnosticity score for genes that were up-regulated or down-regulated by β-adrenergic signaling. Diagnosticity scores for each gene transcript quantified the extent to which that transcript was predominately expressed by an M2-polarized macrophage relative to an M1-polarized macrophage.
Figure 4.
Independent verification of differential expression by genes in the global gene expression profile with qRT-PCR. Isoproterenol at 1 μM. Data represent mean ± SE of three independent experiments. ***P < 0.001, *P < 0.05.
3.3. Transcriptional regulation in the β-adrenergic-stimulated macrophage
To assess the role of established M1–M2 spectrum STAT family transcription factors, as well as hypothesized β-adrenergic-related transcription factors, in the effect of β-adrenergic signaling on macrophage polarization, we tested for over-representation of transcription factor binding motifs for CREB, C/EBPβ, ATF, and STAT transcription factors in the promoters of genes that were up- and down-regulated by isoproterenol. We found that CREB binding motifs (TRANSFAC V$CREB_01) were significantly more prevalent among genes up-regulated by isoproterenol vs. down-regulated genes (mean fold difference [MFD] = 1.39, P = 0.026; Figure 5). Similarly, C/EBPβ and ATF binding motifs (TRANSFAC V$CEBPB_02 and ATF_01) were significantly more prevalent among up-regulated genes (MFD = 1.36, P = 0.008 and MFD = 1.34, P = 0.039, respectively; Figure 5). These results are in accord with previous research implicating CREB, C/EBPβ, and ATF1 in the up-regulation of M2-spectrum genes.12–14
Figure 5.
Promoter-based bioinformatic analysis of transcription factors in genes differentially-regulated in β-adrenergic-stimulated vs. control macrophages. Data represent mean fold difference (mean ratio of isoproterenol/control) ±SE of transcription factor binding motifs, averaged over nine parametric combinations of promoter length and motif detection stringency. P values, two-tailed difference from null difference of 1.
In contrast, STAT3 binding motifs (TRANSFAC V$STAT3_01) were significantly more prevalent among genes down-regulated by isoproterenol (MFD = 0.41, P = 0.024; Figure 5), which may be considered aberrant for an M2-spectrum macrophage, given the established role of STAT3 in promoting M2-like gene expression in M(IL-10) macrophages. STAT1 binding motifs (TRANSFAC V$STAT1_01) were also more prevalent among down-regulated genes, as might be expected, given its established role in mediating gene expression in M1-spectrum macrophages, but this result was not significant (MFD = 0.84, P = 0.421; Figure 5). Binding motifs that are receptive to the whole family of STAT transcription factors, i.e., that don’t discriminate STAT3 from STAT1 (TRANSFAC V$STAT_01), were also more prevalent among down-regulated genes (MFD = 0.58, P = 0.003). Considered together, these results suggest that established M1–M2 STAT transcription factors do not play a role in the effect of β-adrenergic signaling on M2-spectrum gene expression in macrophages.
3.4. Receptor specificity in the β-adrenergic signaling pathway for M2 macrophage gene expression
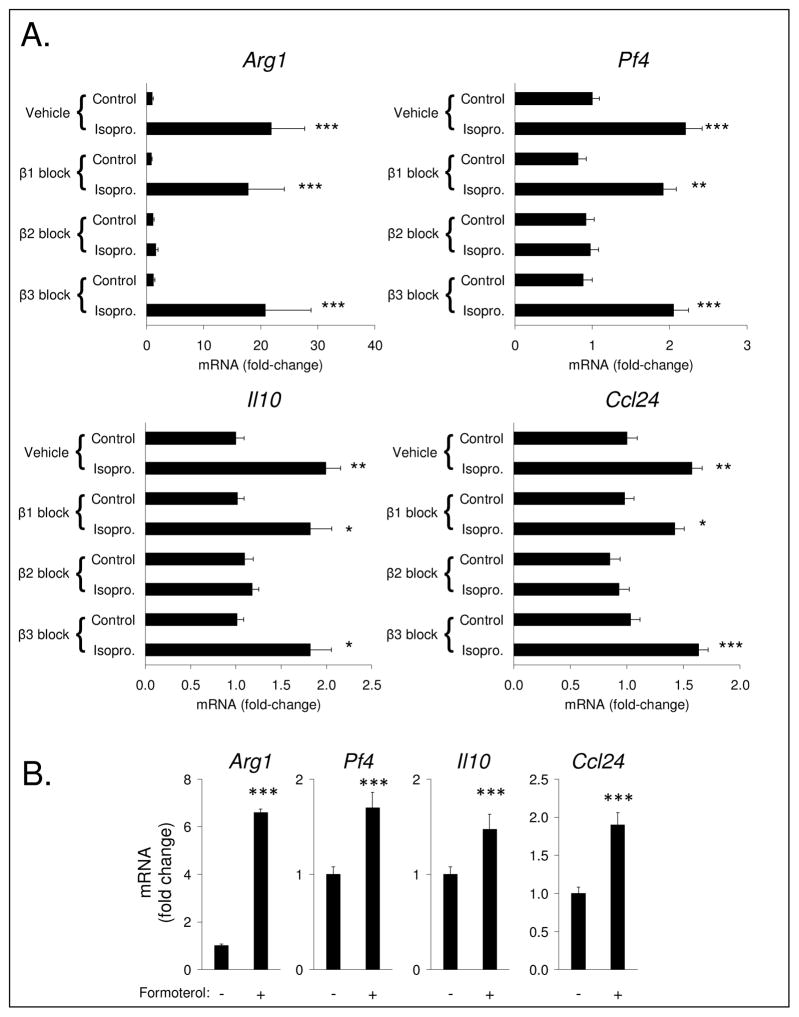
Given that β-adrenergic ligands may signal through any of three receptor subtypes,25 we used selective β1-, β2-, and β3-adrenergic antagonists to determine which subtype mediated isoproterenol effects on expression of specific gene transcripts that were previously found to be up-regulated by isoproterenol in the a priori qRT-PCR assays and in the genome-wide transcriptional profile. As shown in Figure 6A, β2-adrenergic antagonist, ICI 118,551, abrogated isoproterenol-induced M2-spectrum gene expression (average suppression: Arg1, 98 ± 2%, P = 0.0005; Il10, 92 ± 8%, P = 0.032; Pf4, 95 ± 9%, P = 0.0001; Ccl24, 86 ± 9%, P = 0.001). In contrast, neither β1- nor β3-adrenergic antagonism significantly inhibited the effect of isoproterenol on the same gene transcripts (P values > 0.97; Figure 6A). Given the evident necessity of β2-adrenergic signaling in the effect of isoproterenol on select gene transcripts, we further determined the sufficiency of β2-adrenergic signaling by examining the effect of selective β2-adrenergic agonist, formoterol, on the same gene transcripts. As shown in Figure 6B, formoterol significantly increased expression of all transcripts (P values < 0.001).
Figure 6.
(A) Effects of selective β-adrenergic antagonists on isoproterenol-induced M2 gene expression. (B) Effects of selective β2-adrenergic agonist, formoterol, on M2 gene expression. All adrenergic reagents at 100 nM. Vehicle DMSO at 0.01%. Data represent mean ± SE of three independent experiments. ***P < 0.001, **P < 0.01, *P < 0.05 vs. vehicle control.
4. Discussion
β-adrenergic-stimulated macrophages did not fit cleanly into any one pre-defined category of the M1–M2 spectrum as previously defined (Murray et al., 2014; Figure 1), but did express genes that are representative of M(IL-4) and M(IL-10) categories on the M2 side of that spectrum. Comprehensive transcript origin analysis of genome-wide transcriptional profiles confirmed that β-adrenergic-stimulated macrophages primarily express transcripts indicative of M2 macrophages while suppressing transcripts indicative of M1 macrophages. The M2-promoting effects were mediated specifically through β2-adrenergic receptors and were associated with bioinformatic indications of increased CREB, C/EBPβ, and ATF transcription factor pathways, which have previously been shown to regulate M2-associated genes in other contexts.12–14 However, these effects did not appear to be associated with the more established M1–M2 spectrum transcription factors of the STAT family. Together, these results firmly locate β-adrenergic-stimulated macrophages on the M2 side of the M1–M2 spectrum and suggest a selective transcriptional pathway for their polarity that is atypical for M2-spectrum macrophages.
These results are consistent with previous studies that found β-adrenergic signaling regulates macrophage activity in cancer and AIDS via mechanisms that could be interpreted as M2-promoting and/or M1-suppressing. For example, in the context of Mycobacterium avium infection, which disproportionately affects AIDS patients, it was found that epinephrine decreases macrophage expression of MHC class II proteins,5 which feature prominently in classical macrophage activation scenarios by presenting microbial peptides to T-cells.26 Further investigation in this line of research found that β-adrenergic signaling also suppresses nitric oxide (NO) production in M(IFNγ) macrophages, which is used to combat infectious organisms in a classical macrophage response.4 Thus, β-adrenergic mechanisms in these instances are decidedly M1-suppressing. One possible application of the current findings then would be to investigate whether psychological and/or pharmacological interventions aimed at reducing β-adrenergic signaling in AIDS patients could reverse M1 suppression and reduce Mycobacterium avium comorbidity in such patients. Although a review of psychological interventions for persons with HIV has found that salutary effects on neuroendocrine regulation tend to associate with improved immune status, specific examination of macrophage function in these studies was not reported.27
In the context of cancer, the M2-promoting mechanisms of β-adrenergic signaling are salient. β-adrenergic-stimulated macrophages have been found to up-regulate TGF-β,1 a well-established immunosuppressive mechanism that is being investigated for potential therapeutic blockade in cancer patients.28 Consistent with that immunosuppressive result, the current study found that β-adrenergic signaling increased gene expression of platelet factor 4 (PF4) in macrophages (alias, Chemokine C-X-C ligand 4 [CXCL4]), which facilitates the re-growth of colon cancer after chemotherapy by suppressing anti-tumor immunity.29 Thus, given the evidence that TAMs are necessary partners for cancer cell invasion and metastasis,8 these findings underscore the notion that β-adrenergic signaling exerts its facilitative effects on cancer progression, at least in part, by promoting a phenotype in TAMs that is on the immunosuppressive M2 side of the M1–M2 spectrum. Although several observational studies of β-blocker usage in cancer patients exemplify possible application of these findings, i.e., where it was found that β-blocker usage associated with increased survival,30 it is not known to what extent these links may be mediated by inhibition of M2 spectrum properties in tumor associated macrophages.
In the context of heart disease, the effects of β-adrenergic signaling on macrophage polarity are less clear. Although compelling research shows that SNS-induced β-adrenergic signaling increases both myelopoiesis31 and subsequent macrophage accumulation in the aortas of atherosclerosis-prone mice,6 how direct β-adrenergic stimulation of such macrophages affects their polarity and subsequent contribution to progression of atherosclerosis is uncertain. Review of macrophage polarity in atherosclerosis suggests that both M1-like and M2-like macrophages are present in atherosclerotic lesions but also suggests that phenotype plasticity in such lesions is dependent on predominating local factors in the micro-environment.32 This latter finding could mean that β-adrenergic stimulation continues to pull such macrophages toward the M2 side of the spectrum. However, that even being the case, possible application of the β-adrenergic/M2 finding in this context is still ambiguous because the evidence is mixed as to which macrophage phenotype may be harmful vs. protective in atherosclerosis.32
The present finding that β2-adrenergic receptors mediate the effect of isoproterenol on select gene transcripts from the M2 spectrum (Arg1, Il10, Pf4, Ccl24) is consistent with previous studies that show β2-adrenergic receptors are the predominant functional β-adrenergic receptors in macrophages and induce several anti-inflammatory molecules.9,33,34 Additional research has shown that pharmacologic inhibition of mediators downstream of β2-adrenergic receptor signaling, i.e., cAMP and PKA, can block the M1-suppressing effect of β2-adrenergic signaling on NO production in M(IFNγ) macrophages.4 Given that CREB transcription factor is a primary target of the cAMP-PKA signaling pathway,15 those previous results are consistent with the present study’s indication that a β2-adrenergic-induced M2-spectrum transcriptome associates with CREB activation. Also, given the findings by others that CREB can induce C/EBPβ, which in turn can drive expression of genes associated with the M2 spectrum (i.e., Arg1, Il10, Il13ra, Msr1, and Tgm2),12,13 the association found between the C/EBPβ transcription factor and the M2-spectrum transcriptome induced by β-adrenergic signaling in the current study is coherent. However, none of the STAT family transcription factors assessed here showed an association with the up-regulation of M2-spectrum genes by β-adrenergic signaling. In fact, the opposite was true for STAT3, as its binding motif prevalence was over-represented among down-regulated genes. Given the established role of STAT3 in mediating M2-like gene expression in M(IL-10) macrophages (see Figure 1), the failure of β-adrenergic signaling to activate this pathway may explain why selected genes from the consensus gene set for M(IL-10) macrophage (Il10 and Il4ra) were not both increased by isoproterenol in our targeted a priori approach (i.e., Il4ra was not up-regulated; see Figure 2). Similarly, a binding motif that is receptive to the whole family of STAT transcription factors was significantly over-represented among down-regulated genes. Given that result, it is tempting to speculate that the suppressive effect of β-adrenergic signaling on M1-spectrum gene expression may be mediated through suppression of the prototypical M1 transcription factor, STAT1. However, prevalence of the binding motif specific to STAT1, though slightly greater among down-regulated genes, did not reach statistical significance in this study. Thus, the current data on STAT family transcription factors in this study suggests that these factors likely do not play a role in β-adrenergic-induced up-regulation of M2-spectrum gene expression. As for their role in β-adrenergic-induced down-regulation of M1-spectrum genes, future research will be required to address their potential involvement and identify other transcription factors that may be actively inhibited by β-adrenergic signaling.
It should be noted that extrapolation from in vitro models, like the one in the present study, is necessarily limited. Whereas SNS expression of catecholamines in vivo includes both norepinephrine and epinephrine, which can stimulate both α- and β-adrenergic receptors at varying doses, we utilized only isoproterenol at a dose strong enough to induce reliable effects across experiments in this study. Given the in vivo evidence that suggests β-adrenergic blockade is sufficient to inhibit catecholamine effects on macrophage activity in animal models of cancer,1 atherosclerosis,6 and sepsis,9 we limited our focus to β-adrenergic signaling in this study. However, we note the possibility that α-adrenergic signaling could play a role in those in vivo effects. Also, we only looked at changes in gene expression at one time point (24 hours) to analyze the first order (direct) effects of β-adrenergic signaling on the basal macrophage transcriptome. However, it is possible that ongoing episodes of stimulation lasting days or weeks, such as in a chronic stress paradigm, could have effects on macrophages that are not accounted for in the current study.
In summary, the present study demonstrates that β2-adrenergic-stimulation induces a macrophage transcriptome that locates on the M2 side of the M1–M2 spectrum and implicates an atypical signaling pathway for this macrophage that involves CREB, ATF, and C/EBPβ activation in the absence of STAT family regulators. These findings may help illuminate the pathways by which SNS-induced β-adrenergic signaling affects macrophage-related disease processes and may suggest pharmacologic strategies for redirecting the actions of an “M(ADRB2)” macrophage toward more health-promoting trajectories.
Supplementary Material
F4/80+ cells were quantified by flow cytometry after bone marrow cells were cultured with M-CSF for 7 days.
Highlights.
Genome-wide transcriptional profiles of β-adrenergic-stimulated macrophages were analyzed
β-adrenergic-stimulated macrophages located on the M2-side of the M1–M2 macrophage spectrum
β-adrenergic signaling effects were mediated specifically through the β2-adrenergic receptor
Effects were associated with CREB, C/EBPβ, and ATF transcription factor pathways
Established M1–M2 spectrum STAT transcription factors were not associated with these effects
Acknowledgments
Flow cytometry was performed within the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility that is supported by NIH awards CA16042 and AI28697, and by the JCCC, the UCLA AIDS Institute, and the David Geffen School of Medicine at UCLA. Microarray assays were performed by the UCLA Neuroscience Genomics Core. This research was supported by National Institutes of Health grants K07CA188237-01A1, R01CA160890, and R01AG043404-02; Australian National Health and Medical Research Council grants 1008865 and 1053535; Australian Research Council grant LE110100125; and funding from the UCLA Cousins Center for Psychoneuroimmunology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sloan EK, Priceman SJ, Cox BF, Yu S, Pimentel MA, Tangkanangnukul V, Arevalo JM, Morizono K, Karanikolas BD, Wu L, Sood AK, Cole SW. The sympathetic nervous system induces a metastatic switch in primary breast cancer. Cancer Research. 2010;70:7042–52. doi: 10.1158/0008-5472.CAN-10-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armaiz-Pena GN, Gonzalez-Villasana V, Nagaraja AS, Rodriguez-Aguayo C, Sadaoui NC, Stone RL, Matsuo K, Dalton HJ, Previs RA, Jennings NB, Dorniak P, Hansen JM, Arevalo JM, Cole SW, Lutgendorf SK, Sood AK, Lopez-Berestein G. Adrenergic regulation of monocyte chemotactic protein 1 leads to enhanced macrophage recruitment and ovarian carcinoma growth. Oncotarget. 2015;6:4266–73. doi: 10.18632/oncotarget.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greeson JM, Lewis JG, Achanzar K, Zimmerman E, Young KH, Suarez EC. Stress-induced changes in the expression of monocytic beta2-integrins: the impact of arousal of negative affect and adrenergic responses to the Anger Recall Interview. Brain Behav Immun. 2009;23:251–6. doi: 10.1016/j.bbi.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boomershine CS, Lafuse WP, Zwilling BS. Beta2-adrenergic receptor stimulation inhibits nitric oxide generation by Mycobacterium avium infected macrophages. J Neuroimmunol. 1999;101:68–75. doi: 10.1016/s0165-5728(99)00134-4. [DOI] [PubMed] [Google Scholar]
- 5.Zwilling BS, Brown D, Feng N, Sheridan J, Pearl D. The effect of adrenalectomy on the restraint stressed induced suppression of MHC class II expression by murine peritoneal macrophages. Brain Behav Immun. 1993;7:29–35. doi: 10.1006/brbi.1993.1003. [DOI] [PubMed] [Google Scholar]
- 6.Heidt T, Sager HB, Courties G, Dutta P, Iwamoto Y, Zaltsman A, von Zur Muhlen C, Bode C, Fricchione GL, Denninger J, Lin CP, Vinegoni C, Libby P, Swirski FK, Weissleder R, Nahrendorf M. Chronic variable stress activates hematopoietic stem cells. Nat Med. 2014;20:754–8. doi: 10.1038/nm.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–55. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 8.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–6. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Grailer JJ, Haggadone MD, Sarma JV, Zetoune FS, Ward PA. Induction of M2 regulatory macrophages through the β2-adrenergic receptor with protection during endotoxemia and acute lung injury. J Innate Immun. 2014;6:607–18. doi: 10.1159/000358524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, Locati M, Mantovani A, Martinez FO, Mege JL, Mosser DM, Natoli G, Saeij JP, Schultze JL, Shirey KA, Sica A, Suttles J, Udalova I, van Ginderachter JA, Vogel SN, Wynn TA. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–69. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruffell D, Mourkioti F, Gambardella A, Kirstetter P, Lopez RG, Rosenthal N, Nerlov C. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci U S A. 2009;106:17475–80. doi: 10.1073/pnas.0908641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayakawa K, Wang X, Lo EH. CD200 increases alternatively activated macrophages through CREB - C/EBP-beta signaling. J Neurochem. 2016 Mar;136:900–6. doi: 10.1111/jnc.13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ananieva O, Darragh J, Johansen C, Carr JM, McIlrath J, Park JM, Wingate A, Monk CE, Toth R, Santos SG, Iversen L, Arthur JS. The kinases MSK1 and MSK2 act as negative regulators of Toll-like receptor signaling. Nat Immunol. 2008;9:1028–36. doi: 10.1038/ni.1644. [DOI] [PubMed] [Google Scholar]
- 15.Cole SW, Sood AK. Molecular pathways: beta-adrenergic signaling in cancer. Clin Cancer Res. 2012;18:1201–6. doi: 10.1158/1078-0432.CCR-11-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallón S, Loubes JM, Maza E. Statistical properties of the quantile normalization method for density curve alignment. Math Biosci. 2013;242:129–42. doi: 10.1016/j.mbs.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Cole SW, Galic Z, Zack JA. Controlling false-negative errors in microarray differential expression analysis: A PRIM approach. Bioinformatics. 2003;19:1808–16. doi: 10.1093/bioinformatics/btg242. [DOI] [PubMed] [Google Scholar]
- 18.Witten DM, Tibshirani R. A Comparison of Fold-Change and the t-Statistic for Microarray Data Analysis. Stanford University; Stanford, CA: 2007. [Accessed November 9, 2011]. Available at www.stat.stanford.edu/~tibs/ftp/daniela-fold.pdf. [Google Scholar]
- 19.Shi L, et al. MAQC Consortium. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol. 2010;28:827–38. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi L, et al. The balance of reproducibility, sensitivity, and specificity of lists of differentially expressed genes in microarray studies. BMC Bioinformatics. 2008;9:S10. doi: 10.1186/1471-2105-9-S9-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole SW. Elevating the perspective on human stress genomics. Psychoneuroendocrinology. 2010;35:955–62. doi: 10.1016/j.psyneuen.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cole SW, Hawkley LC, Arevalo JM, Cacioppo JT. Transcript origin analysis identifies antigen presenting cells as primary targets of socially regulated gene expression in leukocytes. Proc Natl Acad Sci U S A. 2011;108:3080–5. doi: 10.1073/pnas.1014218108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller RG. Beyond ANOVA: Basics of Applied Statistics. Wiley; New York: 1986. [Google Scholar]
- 24.Cole SW, Yan W, Galic Z, Arevalo J, Zack JA. Expression-based monitoring of transcription factor activity: The TELiS database. Bioinformatics. 2005;21:803–10. doi: 10.1093/bioinformatics/bti038. [DOI] [PubMed] [Google Scholar]
- 25.Cernecka H, Sand C, Michel MC. The odd sibling: features of β3-adrenoceptor pharmacology. Mol Pharmacol. 2014;86:479–84. doi: 10.1124/mol.114.092817. [DOI] [PubMed] [Google Scholar]
- 26.Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109:S21–33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 27.Carrico AW, Antoni MH. Effects of psychological interventions on neuroendocrine hormone regulation and immune status in HIV-positive persons: a review of randomized controlled trials. Psychosom Med. 2008;70:575–84. doi: 10.1097/PSY.0b013e31817a5d30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wrzesinski SH, Wan YY, Flavell RA. Transforming growth factor-beta and the immune response: implications for anticancer therapy. Clin Cancer Res. 2007;13:5262–70. doi: 10.1158/1078-0432.CCR-07-1157. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Gao J, Wang X, Deng S, Ye H, Guan W, Wu M, Zhu S, Yu Y, Han W. CXCL4 mediates tumor regrowth after chemotherapy by suppression of antitumor immunity. Cancer Biol Ther. 2015;16:1775–83. doi: 10.1080/15384047.2015.1095404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cole SW, Nagaraja AS, Lutgendorf SK, Green PA, Sood AK. Sympathetic nervous system regulation of the tumour microenvironment. Nat Rev Cancer. 2015;15:563–72. doi: 10.1038/nrc3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powell ND, Sloan EK, Bailey MT, Arevalo JM, Miller GE, Chen E, Kobor MS, Reader BF, Sheridan JF, Cole SW. Social stress up-regulates inflammatory gene expression in the leukocyte transcriptome via β-adrenergic induction of myelopoiesis. Proc Natl Acad Sci U S A. 2013;110:16574–9. doi: 10.1073/pnas.1310655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolfs IM, Donners MM, de Winther MP. Differentiation factors and cytokines in the atherosclerotic plaque micro-environment as a trigger for macrophage polarisation. Thromb Haemost. 2011;106:763–71. doi: 10.1160/TH11-05-0320. [DOI] [PubMed] [Google Scholar]
- 33.Izeboud CA, Mocking JA, Monshouwer M, van Miert AS, Witkamp RF. Participation of beta-adrenergic receptors on macrophages in modulation of LPS-induced cytokine release. J Recept Signal Transduct Res. 1999;19:191–202. doi: 10.3109/10799899909036645. [DOI] [PubMed] [Google Scholar]
- 34.Verhoeckx KC, Gaspari M, Bijlsma S, van der Greef J, Witkamp RF, Doornbos RP, Rodenburg RJ. In search of secreted protein biomarkers for the anti-inflammatory effect of beta2-adrenergic receptor agonists: application of DIGE technology in combination with multivariate and univariate data analysis tools. J Proteome Res. 2005;4:2015–23. doi: 10.1021/pr050183u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
F4/80+ cells were quantified by flow cytometry after bone marrow cells were cultured with M-CSF for 7 days.