Abstract
Acquired tamoxifen (TAM) resistance is a significant clinical problem in treating patients with estrogen receptor α (ERα) + breast cancer. We reported that ERα increases nuclear respiratory factor-1 (NRF-1), which regulates nuclear-encoded mitochondrial gene transcription, in MCF-7 breast cancer cells and NRF-1 knockdown stimulates apoptosis. Whether NRF-1 and target gene expression is altered in endocrine resistant breast cancer cells is unknown. We measured NRF-1and metabolic features in a cell model of progressive TAM-resistance. NRF-1 and its target mitochondrial transcription factor A (TFAM) were higher in TAM-resistant LCC2 and LCC9 cells than TAM-sensitive MCF-7 cells. Using extracellular flux assays we observed that LCC1, LCC2, and LCC9 cells showed similar oxygen consumption rate (OCR), but lower mitochondrial reserve capacity which was correlated with lower Succinate Dehydrogenase Complex, Subunit B in LCC1 and LCC2 cells. Complex III activity was lower in LCC9 than MCF-7 cells. LCC1, LCC2, and LCC9 cells had higher basal extracellular acidification (ECAR), indicating higher aerobic glycolysis, relative to MCF-7 cells. Mitochondrial bioenergetic responses to estradiol and 4-hydroxytamoxifen were reduced in the endocrine-resistant cells compared to MCF-7 cells. These results suggest the acquisition of altered metabolic phenotypes in response to long term antiestrogen treatment may increase vulnerability to metabolic stress.
Keywords: breast cancer, NRF-1, mitochondrial oxidative phosphorylation, tamoxifen-resistance
1. Introduction
Approximately 40% of breast cancer patients whose primary tumors express estrogen receptor α (ERα) and are initially responsive to endocrine therapies, i.e., tamoxifen (TAM) or aromatase inhibitors (AI), relapse with acquired endocrine resistant disease progression(1). Defining the mechanisms of TAM-resistance remains an important clinical issue for breast cancer patients. Although AIs have replaced TAM as the first-line treatment for postmenopausal women with ERα+ breast tumors, ten years of TAM is recommended as adjuvant therapy for ERα+ premenopausal breast cancer patients and for postmenopausal women who have relapsed on or cannot tolerate AI therapy (2). There are thousands of breast cancer survivors who have received TAM as a mono-adjuvant therapy and are at unknown risk for developing TAM-resistant metastatic disease as a late recurrence, an emergence from dormancy (3). TAM is a selective ER modulator (SERM) with agonist and antagonist activities mediated by ERα, ERβ, and G-protein coupled ER (GPER) (4). Multiple mechanisms contribute to the evolution of cells resistant to the growth inhibiting, anti-estrogenic effects of TAM and AIs (reviewed in (4,5)).
Mitochondrial–nuclear crosstalk is critical for the maintenance of cellular homeostasis and is dysregulated in cancer (6). Epithelial tumor cell metabolism is supported by fuel sources from cancer-associated fibroblasts and adipocytes (7). Epithelial breast cancer cells have been suggested to have increases in mitochondrial number, anabolic function, oxidative phosphorylation (OXPHOS) (8), and nuclear respirator factor 1 (NRF-1) (9). NRF-1 is a master regulator of nuclear-encoded mitochondrial gene transcription, including genes for mitochondrial bioenergetic function (10). We reported that estradiol (E2) stimulates NRF-1 transcription, which in turn, increases the transcription of its targets including the mitochondrial transcription factor TFAM (transcription factor, mitochondrial) in MCF-7 and T47D (both are ERα+/PR+,HER2−, luminal A (11)) breast cancer cells and mouse mammary gland (12,13).
MCF-7 cells chronically exposed to 25 μM H2O2 (simulating oxidative stress in a tumor) showed increased NRF-1 and TFAM expression, decreased ERα expression and increased colony-forming potential (14). E2 was reported to increase ROS thus activating AKT which phosphorylated and activated NRF-1 (p-NRF-1) and increased transcription of its cell cycle targets: CDC2, PRC1, PCNA, cyclin B1, and CDC25C in MCF-7 cells (15). No one has evaluated NRF-1 expression or that of its targets involved in mitochondrial bioenergetics in TAM-resistant breast cancer cells.
A recent publication cataloged changes in the RNA levels of key genes in glycolysis, gluconeogenesis, glycogen synthesis and degradation, and the pentose phosphate pathway (PPP) in MCF-7 and BT-474 (ERα+,PR+,HER2+) breast cancer cell lines that had undergone EMT (16). However, this study did not examine functional consequences of these changes in bioenergetic parameters. Studies in transgenic mouse mammary tumor virus (MMTV)–polyoma middle T (PyMT) mice with different mtDNA but identical nuclear DNA revealed that the mtDNA background directly affected primary tumorigenicity and metastatic efficiency, although the precise mechanism(s) are still unknown (17). Overall, the contribution of metabolic reprogramming in the development of endocrine resistance in breast cancer is poorly understood.
The goals of this study were to evaluate the expression of NRF-1 and its target TFAM in TAM-resistant cells derived from MCF-7 cells and to identify and characterize bioenergetic differences of intact endocrine-sensitive versus TAM-resistant cells. We compared MCF-7 ERα+/PR+,HER2−, luminal A breast cancer cells with LCC1 (ERα+/PR+,HER2−; E2-independent, TAM- and fulvestrant-sensitive), LCC2 (ERα+/PR+,HER2−; E2-independent, TAM-resistant, fulvestrant-sensitive), and LCC9 (ERα+/PR+,HER2-; E2-independent, TAM- and fulvestrant-resistant) breast cancer cell lines, which are derived from MCF-7 cells, as a cellular models of progression to endocrine-resistance (18). Our results reveal increased NRF-1 and TFAM in endocrine-resistant cells as well differences in bioenergetic phenotypes in TAM-resistant breast cancer cells.
2. Materials and Methods
2.1. Reagents and antibodies
17β-estradiol (E2), 4-hydroxytamoxifen (4-OHT), Oligomycin A, Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP), Rotenone, Antimycin A were purchased from Sigma-Aldrich (St. Louis, MO., USA). Antibodies were purchased as follows: NRF-1, Rockland Immunochemicals, Inc (Pottstown, PA, USA); TFAM (DO1P), Abnova; total OXPHOS WB antibody cocktail (Abcam, Cambridge, MA, USA);α-tubulin, Neomarkers; β-actin, Sigma-Aldrich; GAPDH (Santa Cruz Biotechnology, Dallas, TX, USA); UQCRC2 (PA530204, Thermo Fisher, Waltham, MA, USA).
2.2 Cell culture and treatments
MCF-7 and T47D cells were purchased from ATCC (Manassas, VA, USA), LCC2, and LCC9 cell lines were derived from MCF-7 cells by cultivation with the antiestrogens 4-hydroxytamoxifen, ICI 182,780 (Fulvestrant) and LY 117018 respectively, and were graciously provided as a gift by Dr. Robert Clarke, Georgetown University (18). LCC1 cells are derived from MCF-7 as E2-independent, TAM-sensitive cells and were also a gift from Dr. Robert Clarke (19). MCF-7, LCC1, LCC2, and LCC9 cells were maintained in IMEM (Cellgro, Manassas, VA, USA) containing 5% fetal bovine serum (FBS, Atlanta Biologicals, Norcross, GA, USA) and 1% Penicillin/Streptomycin (Cellgro). T47D were grown in RPMI (Cellgro) containing 5% FBS and 25 nM insulin (Sigma). Where indicated, cells were treated 24 and 48 h with vehicle control (EtOH .001%), 10 nM E2, or 100 nM 4-OHT. For other experiments, cells were grown in phenol red-free medium containing 5% dextran coated charcoal (DCC)-stripped FBS for 48 h prior to treatment as indicated.
2.3. Metabolic analysis with Seahorse XF24 Extracellular Flux Analyzer
To measure mitochondrial bioenergetics profile in breast cancer cells, a Seahorse Bioscience XF24 Extracellular Flux Analyzer was used (Seahorse Bioscience, North Billerica, MA, USA). Cells were plated on XF24 plates for 24 h prior to assay. One hour prior to the assay, the cells were switched to DMEM assay media containing 2 mM Glutamax (ThermoFisher), 1 mM sodium pyruvate, 25 mM glucose , and 1.85 g/L NaCl (all from Sigma), pH 7.4, and maintained at 37°C in an non-CO2 incubator. Sensor cartridges were pre-incubated overnight in XF24 Calibrant solution (Seahorse Bioscience). After measurement of basal ECAR (extracellular acidification rate, mpH/min) and OCR (oxygen consumption rates, pMoles O2/min), mitochondrial function was interrogated by the sequential injection of Oligomycin A (1.5 μM), FCCP (0.5 μM), and Antimycin A (10 μM) in combination with Rotenone (2 μM), as described previously (20). This allowed for the calculation of ATP-linked O2 consumption, proton leak, maximal respiratory capacity, reserve capacity and non-mitochondrial respiration (20). Appropriate concentrations for oligomycin and FCCP were determined for each cell type ((21) and Supplementary Figure 1). Antimycin A was injected at 10 μM to ensure complete inhibition of complex III. After each experiment, the protein concentrations in each well were measured by BioRad DC™ Protein Assay (BioRad, Hercules, CA, USA). All OCR and ECAR values were normalized to protein concentration. ATP-linked OCR, reserve capacity, proton leak, non-mitochondrial OCR, maximum mitochondrial capacity, glycolytic reserve, state apparent, respiratory control ratio (RCR) basal and RCRmax were calculated as described (21,22).
2.4. Mitochondrial:Nuclear DNA Ratios
Total DNA was isolated from untreated MCF-7 and T47D, cells. Quantitative, real time polymerase chain reaction (QRT-PCR) for mitochondrial DNA content was determined using SYBR Green ROX qPCR Mastermix (Qiagen, Valencia, CA, USA) for measuring the mitochondrial-encoded nicotinamide adenine dinucleotide dehydrogenases: MTND1 and MTND2, neither of which show deletions, duplications, or mutations in studies in human diseases including breast cancer (23), and mitochondrial-encoded cytochrome c oxidase I MTCO1, which shows a low mutation rate in cancers, including breast tumors (24), relative to nuclear-encoded gene 18S rRNA (12,21). Each sample was analyzed in triplicate in the Viia7 Real-Time PCR system (ThermoFisher). Values represent mean fold change ± SEM calculated from the equation 2−ΔΔCT and normalized to EtOH values.
2.5. ADP/ATP ratio: The ADP/ATP ratio was measured using the ADP/ATP Ratio Assay Kit
(catalog no. MAK135) from Sigma-Aldrich following the manufacturer’s instructions. Briefly, cells were seeded (5,000/well) in a 96-well, flat-bottom, black plate with clear bottoms (Corning-Costar, Tewksbury, MA, USA) and grown for 48 h in non-serum starve (NS) or ‘serum starve’ medium, as above. All analyses were measured in quadruplicate (21).
2.6. Protein Isolation and Western blot
Whole cell lysates were prepared in radioimmunoprecipitation (RIPA) buffer with added protease inhibitors (Roche). Protein concentrations were determined using the Bio-Rad DC Protein Assay (Bio-Rad). 30-40 μg of protein lysates were separated on 10 % (lab-made) or 14 % (ThermoFisher) SDS PAGE gels and electroblotted to PVDF membranes. Data were captured and analyzed by Carestream Image Station 4000 R Pro with Carestream Molecular Imaging Software, version 5.0, (Carestream Health, Inc). The values from regions of interest (ROI) normalized to the loading control and the normalized value of MCF-7 cells was set to 1 for comparison of cell lines between separate experiments.
2.7. Complex III activity assay
Complex III activity was determined as previously described (25), with some modification. Briefly, 250 μg cell lysate were added to reaction mixture (25 mM potassium phosphate pH 7.4, 100 μM decyclubiquinol, 75 μM cytochrome c, 500 μM KCN, and 100 μM Na2EDTA) in 1.5 mL cuvette and change in absorbance was read at 550 nm for 4 min using a Synergy 2 (BioTek) spectrophotometer. Myxothiozol (13 μM) was added to the reaction mixture to determine myxothiozol insensitive activity and subtracted from total activity to determine myxothiozol specific complex III activity.
2.8. Transcript levels
RNA was isolated from MCF-7 and LCC9 breast cancer cells using the Exiqon miRCURY™RNA Isolation kit (Woburn, MA, USA). RNA concentration was assessed using a NanoDrop spectrophotometer. The Truseq Stranded mRNA kit (Illumina, San Diego, CA, USA) was used to prepare mRNA libraries from 2 μg total RNA. Libraries were confirmed on the Agilent 2100 Bioanalyzer (Santa Clara, CA, USA) and quantitated using the Illumina Library Quantification Kit, ABI Prism qPCR Mix from Kapa Biosystems and the ABI7900HT real-time PCR instrument. 75-76 cycle single read sequencing was performed with the 500 High-output v2 (75cycle) sequencing kit on the Illumina NextSeq500 instrument. The sequence reads were mapped to the human reference genome, version GRCh37.1 using the mapping algorithm tophat (26) version 2.0.2. The expression levels were quantified at loci specified by the annotation found at ENSEMBL, Homo_sapiens.GRCh37.73.gtf using cufflinks version 2.2.1. Contributions to the annotation file from both ribosomal RNA (rRNA) and mitochondrial RNA (mtRNA) were removed from the gtf file prior to use. Differential analyses between the specified conditions was performed using cuffdiff version 2.2.1. The raw data of our RNA-seq are available at Gene Expression Omnibus (GEO) database: accession number GSE81620. These RNA seq data were examined for the expression of 68 (of 79) nuclear-encoded genes that are not regulated by miR-29 (excluded: NDUFC2, NDUFS6. IQCR1−. IQCR11. IQCRQ. ATP5C1, ATP5F1, ATP5G1, ATP5G3, ATP5L, ATPIF1) in the mitochondrial respiratory chain complexes I-V using the list located at http://www.genenames.org/genefamilies/mitocomplex.
2.9. Mining of publicly available breast cancer microarray data
Analysis of breast cancer data sets used BreastMark (27) to generate Kaplan-Meier survival curves at http://glados.ucd.ie/BreastMark/index.html.
2.10. Statistical analysis
Data are represented as mean ± standard error of the mean (SEM) of at least three independent experiments. Statistical analyses were performed using GraphPad Prism 5 (Graph Pad Software, Inc., LaJolla, CA, USA). One-way analysis of variance (ANOVA) was followed by Tukey post hoc test.
3. Results
3.1. Comparison of NRF-1 and TFAM protein expression in TAM-sensitive versus TAM-resistant cell lines
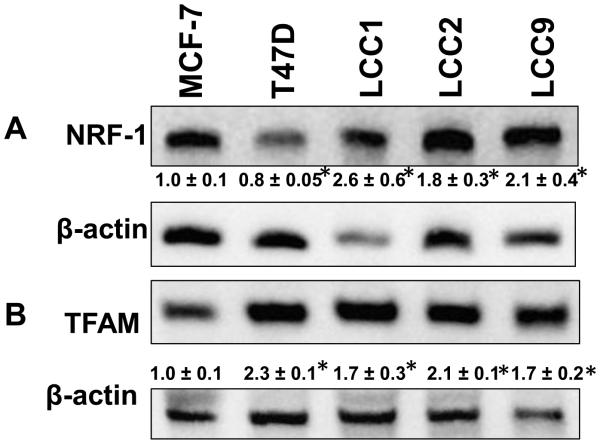
NRF-1 is a nuclear-encoded transcription factor that regulates the transcription of nuclear-encoded mitochondrial transcription factors, e.g., TFAM, and components of the electron transport chain (ETC) (10). We reported that E2 stimulated NRF-1 transcription in MCF-7 and T47D luminal A breast cancer cells (12), but no one has examined the expression of NRF-1 or TFAM in endocrine-resistant breast cancer cells. Western blot showed that NRF-1 expression was higher in LCC1, LCC2, and LCC9 than MCF-7 cells (Fig. 1A). As reported previously, T47D cells have lower NRF-1 than MCF-7 cells (12). TFAM, an NRF-1-regulated, nuclear-encoded mitochondrial transcription factor, was, like NRF-1, higher in LCC1, LCC2, and LCC9 cells relative to MCF-7 cells (Fig. 1B). The higher TFAM in T47D compared to MCF-7, despite lower NRF-1, was suggested to result from the low ERα/ERβ ratio in T47D cells (28), although nuclear NRF-1 protein was not evaluated.
Figure 1.
Comparison of protein levels of nuclear-encoded proteins regulating transcription of genes for mitochondrial function in TAM-sensitive and TAM-resistant breast cancer cell lines cells. Cells were maintained in normal growth medium and 30 μg protein from whole cell lysates were separated on 10% SDS gels. Representative western blots of NRF-1 and TFAM expression are shown. The membranes were stripped and re-probed for β-actin or α-tubulin for normalization. Values below the blots are the ratio of target protein/ β-actin in that gel normalized to MCF-7 NS (set to one). Values are the average of 3 separate experiments ± SEM. * p < 0.05 versus MCF-7.
3.2. Mitochondrial bioenergetics profiles in tamoxifen-sensitive and resistant breast cancer cell lines
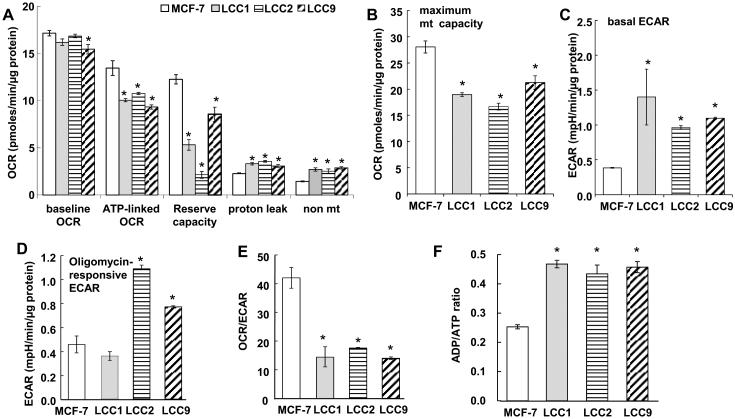
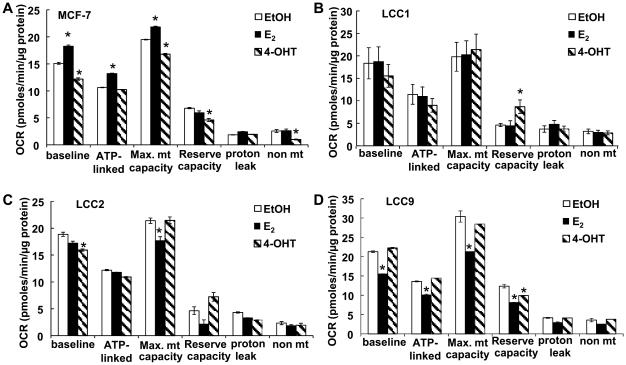
To determine relative mitochondrial and glycolytic activity in TAM-sensitive MCF-7 versus estrogen-independent/TAM-sensitive LCC1, and TAM-resistant LCC2, and LCC9 cells, we measured oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) by extracellular flux analysis (Fig. 2, Supplementary Fig. 1 and 2). A limitation of this study is that cellular bioenergetics were measured in IMEM which contains non-physiological levels of glucose and glutamine (2000 and 292 mg/L, respectively) and is not supplemented with lactate, but contains 110 mg/L sodium pyruvate. OCR and ECAR were measured under basal conditions and after sequential addition of the ATP synthesis inhibitor oligomycin, the uncoupler FCCP, and rotenone (Rot) + antimycin A (A.A.), inhibitors of complexes I and III, respectively. The number of cells/well and concentrations of oligomycin, FCCP, antimycin A, and rotenone were optimized for each cell line (21) (Supplementary Fig. 1).
Figure 2.
Cellular and mitochondrial bioenergetic paramenters in TAM-sensitive MCF-7 and TAM-resistant breast cancer cells. MCF-7, LCC1, LCC2, and LCC9 human breast cancer cells were plated in XF-24 plates and equilibrated for 24 h prior to running the extracellular flux assay in the Seahorse XF-24 bioanalyzer. A and B) OCR parameters. C and D) ECAR parameters. A-E) Each bar is the average of 3-8 separate experiments ± SEM. F) ADP/ATP ratio was measured in quadruplicate. *p < 0.05 versus MCF-7 for the indicated parameter. Analysis used one-way ANOVA followed by Tukey test.
Although basal OCR was statistically lower in LCC9 versus MCF-7 cells, the difference was only 5% (Fig. 2A). Larger differences were detected in ATP-linked OCR and mitochondrial reserve capacity, also called spare or reserve respiratory capacity (29), which were lower in the three endocrine-resistant cell lines than the parental MCF-7 cell line (Fig. 2A). Proton leak was higher in LCC1, LCC2, and LCC9 than MCF-7 cells (Fig. 2A). Basal proton leak is cell type specific and corresponds to the levels of ANT (adenine nucleotide translocase) and UCPs (uncoupling proteins) (30). Non-mitochondrial OCR, likely attributable to cytoplasmic oxidases, was higher in the endocrine-resistant cells as well. The coupling efficiency, also called coupling ratio, is defined as (oligomycin-sensitive OCR)/(basal OCR) (31) was lower in LCC1 and the TAM-R LCC2 and LCC9 cells versus MCF-7, suggesting lower ATP demand (Table 1) and was inversely correlated with proton leak, NRF-1, and TFAM (correlation coefficients = −0.854, −0.890, −0.834, respectively). These data suggest differences in mitochondrial energetics in estrogen- and TAM- sensitive and -resistant cell lines.
Table 1.
Coupling efficiency, State apparent, intact cell basal respiratory control ratio (RCRbasal), and maximum RXR (RCRmax) were calculated as described (21,22).. Values are the mean ± SEM of 3-4 replicate experiments. * P < 0.05 versus MCF-7 by one way ANOVA followed by Tukey test.
| Cell line | Coupling Efficiency |
State apparent | RCR basal | RCR max |
|---|---|---|---|---|
| MCF-7 | 0.78 ± 0.01 | 3.49 ± 0.06 | 5.15 ± 0.09 | 9.18 ± 0.15 |
| LCC1 | 0.62 ± 0.02 | 3.35 ± 0.07 | 3.87 ± 0.22 * | 5.40 ± 0.12 * |
| LCC2 | 0.64 ± 0.01 | 3.17 ± 0.04 | 3.90 ± 0.04 * | 4.48 ± 0.05 * |
| LCC9 | 0.60 ± 0.02 | 3.48 ± 0.11 | 3.82 ± 0.12 * | 6.42 ± 0.21 * |
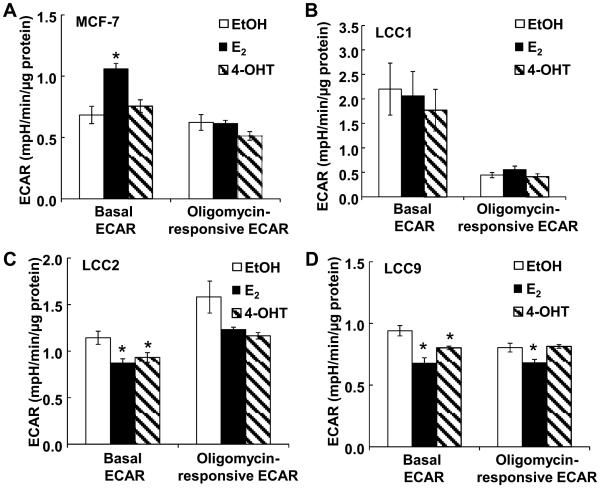
We previously reported that ECAR measurements agree with metabolic measurements of the conversion of [5-3H]-glucose to [3H]2O in MCF-7 and T47D cells, demonstrating that ECAR is a reliable measurement of glycolysis in these cells (21). Basal ECAR was lower in MCF-7 than any of the other cell lines (Fig. 2C). These data suggest an increase in glycolysis in E2-resistant LCC1 and TAM-resistant LCC2 and LCC9 cells derived from MCF-7 cells. Following oligomycin addition, OCR decreased and ECAR increased in all cells, reflecting coupling of mitochondrial function and glycolysis in the cells (Supplementary Fig.2). The glycolytic response to oligomycin, formerly considered as the glycolytic reserve (32), was higher in the TAM-resistant LCC2 and LCC9 cells compared with either TAM-sensitive MCF-7 or LCC1 cells (Fig. 2D). MCF-7 cells showed the highest OCR/ECAR ratio, corresponding to their low basal ECAR (Fig. 2E), suggesting relatively less reliance on glycolysis for energy compared with the other cell types. To evaluate if these bioenergetic differences equate to differences in phosphorylated adenylates, we measured ATP and ADP The ADP/ATP ratio was higher in LCC1, LCC2, and LCC9 compared to MCF-7 cells (Fig. 2F), corresponding with lower coupling efficiency in these cells (Table 1). Together, these data suggest that loss of E2-dependence (LCC1) and gain of TAM-resistance (LCC2 and LCC9) increases ECAR and apparent glycolytic reserve while producing less or consuming more ATP.
3.3. Differences in mitochondrial DNA content in TAM-resistant cells
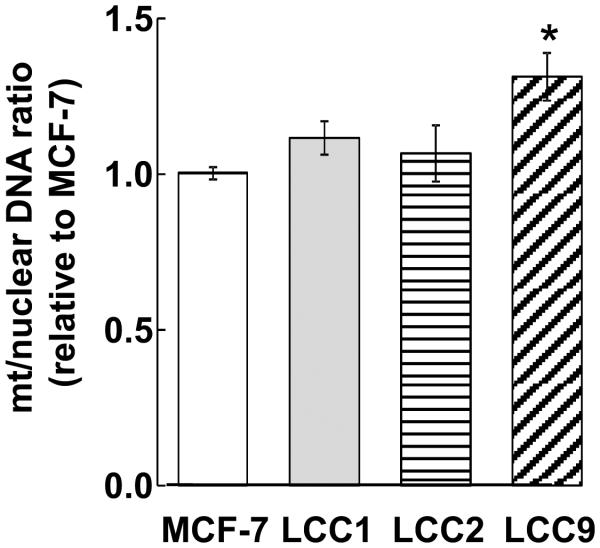
Higher OCR/ECAR in MCF-7 cells may reflect higher mitochondrial content relative to the other cells. Therefore, we examined the mitochondrial/nuclear DNA as an index of mitochondrial abundance (33). The mitochondrial DNA content of the TAM-resistant LCC9 cells was significantly higher than MCF-7 cells (Fig. 3).
Figure 3.
Mitochondrial DNA in breast cancer cells. Genomic DNA was extracted and mtDNA content as estimated as the ratio of the mt-encoded genes MTND1, MTND2, and 16S rRNA normalized to nuclear-encoded 18S as determined by qPCR. Bars are the average of 3 ± SEM separate determinations. Bars are the mean ± SEM of 3 separate experiments. *p < 0.05 versus MCF-7 cells. Analysis used one-way ANOVA followed by Tukey Test.
3.4. Expression of oxidative phosphorylation (OXPHOS) subunits in TAM-sensitive versus TAM-resistant cell lines
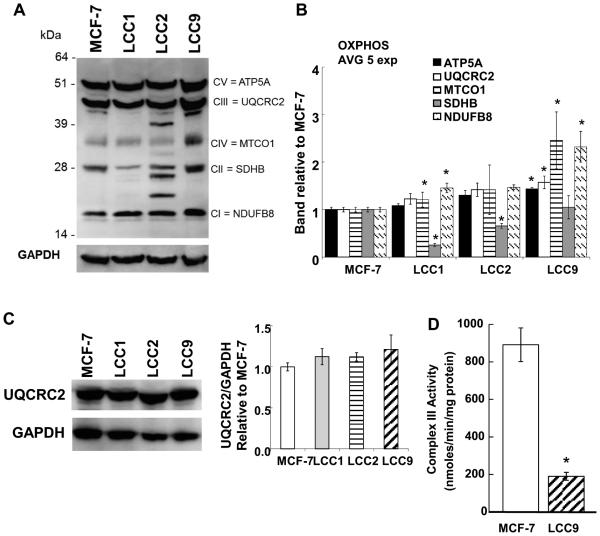
The protein expression of OXPHOS subunits, including NDUF8 (Complex I), SDHB (Complex II), UQCRC2 (Complex III), MTCOI (Complex IV), and ATP5A (Complex V) was examined by Western blot analysis using an antibody cocktail (Fig. 4A). LCC2 cells displayed additional bands with lower MW than UQCRC2 and SDHB. SDHB protein levels were significantly lower in LCC1 and LCC2 than MCF-7 and while SDHB was lower in LCC9, the difference was not significant. Complex II has been suggested to influence mitochondrial reserve capacity (34). Indeed, SDHB protein was correlated with reserve capacity in the 4 cell lines (correlation coefficient 0.642). An additional band was seen below the UQCRC2 band in LCC9 cells. UQCRC2 has 7 alternative splicing variants (www.genecards.org). Higher levels of MTCO1 were detected in the TAM-resistant LCC9 cells compared to MCF-7, LCC1, or LCC2 cells (Fig 4B). This observation at the protein level agrees with MTCO1 transcript levels (Fig. 3). Examination of UQCRC2 protein using a different antibody showed no statistical difference in UQCRC2 protein between the cell lines (Fig. 4C). It is possible that the lower MW band seen below UQCRC2’s expected migration, and the 2 bands below SDHB in LCC2 (Fig. 4A) are non-specific, or the result of protein degradation.
Figure 4.
OXPHOS protein expression in breast cancer cell lines. A) Cells were maintained in normal growth medium and 40 μg of whole cell extract proteins were separated on NuPAGE 4-12% Bis-Tris gels. A representative western blot is shown. For A and C, the membranes were stripped and re-probed for GAPDH for normalization. B) Values are the ratio of target protein/ GAPDH normalized to each value for MCF-7, set to one. Values are the average of 5 separate experiments ± SEM. C) Western blot for UQCRC2. D) Complex III activity was measured in triplicate in three biological replicate samples for each cell line. Thus values are the avg. of 9 separate determinations ± SEM. For B, C, and D: * p < 0.05 versus MCF-7.
3.5. Lower Complex III activity in TAM-resistant versus TAM-sensitive cell lines
Since LCC9 cells showed the most changes in OXPHOS proteins relative to MCF-7 (Fig. 4B), complex III activity was measured in MCF-7 and LCC9 cell lysates (Fig. 4D). LCC9 cells had significantly lower complex III activity than MCF-7 cells
3.6. RNA seq of mitochondrial complex genes in MCF-7 versus TAM-resistant LCC9 cells
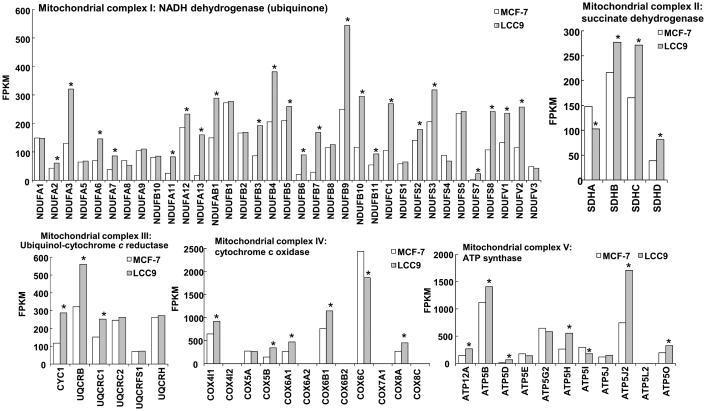
We have examined the transcriptomes of MCF-7 and LCC9 cells by RNA sequencing (RNA seq, whole transcriptome shotgun sequencing, GSE81620). Among the top ten GO (gene ontology) terms identified when comparing transcriptomes from MCF-7 versus LCC9 are respiratory electron transport chain, ATP synthesis coupled electron transport, oxidative phosphorylation, cellular respiration, and mitochondrial translation (Table 2). To identify gene expression changes that may be involved in the differences seen in OCR between MCF-7 and LCC9 cells, we examined the expression of 68 nuclear-encoded genes in the mitochondrial respiratory chain complexes I-V from RNA sequencing experiments in these cell lines (GSE81620) (Fig. 5). Overall, LCC9 cells showed higher expression of 46 of the 68 of the expressed genes than MCF-7 cells. Further studies will be required to identify if these changes play a role (if any) in endocrine-resistance.
Table 2.
Gene Ontology (GO) Biological Processes in the transcriptomes of MCF-7 and LCC9 cells were identified using categoryCompare (45)
| Energy derived by oxidation of organic compounds |
|---|
| Respiratory electron transport chain |
| Electron transport chain |
| ATP synthesis coupled electron transport |
| Oxidative phosphorylation |
| Mitochondrial ATP synthesis coupled electron transport |
| Cellular respiration |
| Mitochondrial translation: Mitochondrial translational initiation, elongation and termination, |
Figure 5.
Comparison of mitochondrial OXPHOS complex gene transcript levels in MCF-7 versus LCC9 cells. Values are FPKM (Fragments Per Kilobase of transcript per Million mapped reads) from RNA-seq analysis in GSE81620. Values are the average of 9 separate experiments. *p value < 0.05 Fisher’s exact test.
3.7. Dysregulated E2 and 4-OHT mitochondrial bioenergetics profiles in TAM-resistant breast cancer cell lines.
To examine the bioenergetics responses of TAM-sensitive versus resistant breast cancer cells to E2 and 4-OHT, the cells were grown in phenol red-free medium supplemented with 5% DCC-FBS for 48 h prior to ligand treatment to minimize the effects of steroid hormones in the FBS or xenoestrogens in the medium prior to hormone treatment (35). E2 increased whereas 4-OHT inhibited basal OCRand maximal mitochondrial capacity in MCF-7 cells (Fig. 6A). E2 increased ATP-linked OCR in MCF-7 cells. In estrogen-independent LCC1 cells, neither E2 nor 4-OHT affected basal or ATP-linked OCR, maximal mitochondrial capacity, proton leak, or non-mt OCR (Fig. 6B). 4-OHT increased reserve capacity in LCC1 cells. 4-OHT reduced basal OCR, but not ATP-linked OCR in TAM-resistant LCC2 cells (Fig. 6C). E2 reduced the maximal mitochondrial capacity in LCC2 and LCC9 TAM-resistant cells (Fig. 6C and 6D). E2 also reduced basal and ATP-linked OCR and reserve capacity in LCC9 cells (Fig. 6D). 4-OHT reduced reserve capacity in LCC9 cells (Fig. 6D).
Figure 6.
Effects of E2 and 4-OHT on mitochondrial bioengergetic functions in TAM-sensitive versus TAM-resistant breast cancer cells. TAM-sensitive MCF-7 (A), estrogen-independent/TAM-sensitive LCC1 (B), and TAM-resistant LCC2 (C) and LCC9 (D) breast cancer cells were grown in phenol-red-free IMEM containing 5% DCC-FBS (‘serum-starved’) for 48 h prior to 24 h treatment with EtOH (vehicle control), 10 nM E2, or 100 nM 4-OHT followed by extracellular flux analysis. After baseline OCR measurements were collected, oligomycin (1.5 μM), FCCP (0.5 μM) and rotenone (2 μM) were injected sequentially as in Supplementary Figure 2. Measurements of mitochondrial function were calculated. All values were normalized to protein/well at the conclusion of the assay. Each point the avg. of 5 separate wells ± SEM within one experiment. * p < 0.05 versus EtOH. Analysis used one-way ANOVA followed by Newman-Keuls Multiple Comparison Test.
E2 increased basal ECAR in MCF-7 cells (Fig. 7A). In contrast, E2 did not affect basal ECAR in LCC1 cells (Fig. 7B). LCC1 cells have a lower oligomycin-responsive ECAR compared with the other cells. Although statistically significant, the magnitude of decrease in basal ECAR in LCC2 and LCC9 cells with E2 and 4-OHT are small compared to the E2-stimulation of ECAR in MCF-7 cells (Fig. 7A, 7C, 7D).
Figure 7.
Effects of E2 and 4-OHT on ECAR in TAM-sensitive versus TAM -resistant breast cancer cells. TAM-sensitive MCF-7 (A), estrogen-independent/tamoxifen-sensitive LCC1 (B), and TAM-resistant LCC2 (C) and LCC9 (D) breast cancer cells were grown in phenol-red-free IMEM containing 5% DCC-FBS (‘serum-starved’) for 48 h prior to 24 h treatment with EtOH (vehicle control), 10 nM E2, or 100 nM 4-OHT followed by extracellular flux analysis. After baseline ECAR measurements were collected, oligomycin (1.5 μM), FCCP (0.5 μM) and rotenone (2 μM) were injected sequentially as in Supplementary Figure 2. All values were normalized to protein/well at the conclusion of the assay. Baseline ECAR and glycolytic reserve capacity were calculated. Each point the avg. of 5 separate wells ± SEM within one experiment. * p < 0.05 versus EtOH. Analysis used one-way ANOVA followed by Newman-Keuls Multiple Comparison Test.
4. Discussion
Acquired resistance to endocrine therapies in breast cancer occurs primarily without the loss of ERα expression but arises from cellular adaptation to the stress experienced by the cells from TAM’s antiestrogen activity or estrogen deprivation due to aromatase inhibition (36). Although cancer cells have unique metabolic properties to support the malignant phenotype, whether metabolic changes are indicative or causative of acquired endocrine-resistance is unknown.
Here we observed that NRF-1 expression was higher in LCC1, LCC2, and LCC9 than MCF-7 cells, a result corresponding with protein levels of NRF-1-regulated, nuclear-encoded TFAM. NRF-1 is a master regulator of nuclear-encoded mitochondrial gene transcription (10) and is involved in regulating the mitochondrial unfolded protein response that is cytoprotective in breast cancer cells (37). TFAM regulates the transcription of mitochondrial-encoded genes including MTCO1 which correlated with TFAM (correlation coefficient 0.97).
Despite higher NRF-1 and TFAM, basal OCR was not higher in LCC1, LCC2, or LCC9 cells relative to MCF-7, suggesting that the increase in NRF-1 and TFAM does not correspond to increased mitochondrial bioenergetics. In fact, ATP-linked OCR was lower in LCC1, LCC2, and LCC9 cells and these cells showed lower mitochondrial reserve capacity compared to MCF-7 cells, suggesting that these cells could be vulnerable to cellular/metabolic stress (29). In mouse neonatal cardiac myocytes, mitochondrial reserve capacity was shown to depend on complex II activity, which is regulated by glucose uptake and by carnitine palmitoyltransferase I (CPT1B) for fatty acid uptake and by SIRT3, the mitochondrial sirtuin (34). Complex II is encoded by 4 genes (Figure 5) with only SDHA showing lower expression in LCC9 than MCF-7. CPT1B transcript levels were lower in LCC9 than MCF-7, but SIRT3 expression was higher in LCC9 (Supplementary Fig. 3A). Although Complex III activity was low in LCC9 compared with MCF-7 cells, the transcript levels of 7 of the 9 nuclear-encoded subunits of ubiquinol-cytochrome c reductase complex were higher in LCC9 than MCF-7 and UQCRC2 protein was similar in all cells. In the heart, defects in MTCYB (cytochrome b) are thought to be responsible for decreased complex III activity seen with aging (38), but there appear to be no studies of mitochondrial complex III defects in breast cancer.
Proton leak was higher in the LCC1, LCC2, and LCC9 versus MCF-7. Basal proton leak corresponds to the levels of ANT (adenine nucleotide translocase) and UCPs (uncoupling proteins) (30). ANT transcript levels were higher in LCC9 than MCF-7 cells (Supplementary Fig. 3B). We did not detect UCP1 or UCP3 transcripts in our RNA seq analysis of MCF-7 and LCC9 cells, but UCP2 was expressed in both cell lines (Supplementary Fig. 3B). Interestingly, UCP2 overexpression in ERα+ breast tumors is a poor prognostic indicator and UCP2 knockdown enhanced TAM-stimulated cytotoxicity in MCF-7 cells (39). Future studies will be required to examine these proteins in endocrine-resistant breast tumors and cells.
We observed that MTCO1 protein was higher in LCC9 than in MCF-7, LCC1, and LCC2 cells. We also detected higher transcript levels of 5 of the 12 nuclear encoded subunits of cytochrome c oxidase in LCC9 versus MCF-7 cells. A strong negative selection was reported for MTCO1 in human cancers suggesting that tumor cells are dependent on MTCO1 function (24). Our results in LCC9 cells are in agreement with a report showing higher staining of MTCO1 and SDHB in human breast tumor epithelial cells than adjacent normal tissue (40). These proteins/genes have not been evaluated in de novo or acquired endocrine-resistant tumors.
TAM-resistant LCC2 and LCC9 cells showed a higher ECAR response to oligomycin than TAM-sensitive LCC1 or MCF-7 cells, suggesting an increased ability to switch from mitochondrial respiration to glycolysis in endocrine-resistance. These data are in agreement with a recent report showing increased lactate production and glucose consumption in LCC2 and LCC9 cells (41) and with a report showing significant increases in proteins involved in ‘energy metabolism’ in ERα+/PR+ human breast tumors, relative to HER2+ and TNBC, including multiple components of the ETC, e.g., NDUF, UQCR, SDH, and COX subunits and ATP synthase (ATP5 and ATP6), while proteins linked to glycolysis, serine synthesis (PHGDH), and glutamine consumption (GLS) were lower (42).
Little is known about the bioenergetic responses of endocrine-resistant breast cancer cells to E2 or 4-OHT. We observed that the mitochondrial bioenergetic responses of MCF-7 and LCC1 cells to E2 and 4-OHT generally reflect the effect of these ERα ligands on cell proliferation and proliferative gene transcription (43), e.g., E2 increasing and 4-OHT repressing OCR and ECAR. Similarly, 4-OHT reduced basal OCR in LCC1 cells, but both E2 and 4-OHT increased maximal mitochondrial capacity and reserve capacity, suggesting an adaptive response in LCC1 cells that may favor survival under cellular stress conditions, i.e., the absence of E2 used to select these cells. Mitochondrial bioenergetic responses of the TAM-resistant LCC2 cells to E2 and 4-OHT differ from those in MCF-7 cells, in agreement with the altered transcriptional and proliferative responses of these cells to these ligands. The mechanism by which E2 decreased the maximal mitochondrial capacity in LCC2 and LCC9 cells is currently unknown. In serum-deprived MCF-7 cells, E2 directly regulates mitochondrial RNA metabolism via increasing mitochondrial ERα and its interaction with mitochondrial HSD17B10 (17β-hydroxysteroid dehydrogenase) resulting in increased mitochondrial RNA processing and OCR (44). It will be interesting to examine if LCC2 and LCC9 have reduced mitochondrial ERα. Given the role for complex II in mitochondrial respiratory reserve (34), it will be important to examine if E2 inhibits complex II activity in LCC2 and LCC9 cells. To our knowledge, no one has examined complex II activity in endocrine-resistant breast cancer cells or tumors.
In summary, we observed higher expression of NRF-1 and TFAM in LCC1, LCC2, and LCC9 than parental MCF-7 cells, suggesting a role for increased NRF-1 in endocrine-resistance. Specific targets regulated by NRF-1 and their role in endocrine resistance remain to be identified, but do not appear to stimulate mitochondrial respiration. Our results demonstrate that LCC1, LCC2, and LCC9 cells showed no or marginal change in basal OCR relative to MCF-7, but had significantly lower ATP-linked OCR and lower reserve capacity than the parental MCF-7 cell line, suggesting that these cells may be susceptible to stress, which could be a point of therapeutic intervention. The reduced mitochondrial reserve capacity was correlated with lower SDHB complex II protein expression. We also detected increased glycolysis in E2-resistant LCC1 and TAM-resistant LCC2 and LCC9 cells. As expected based on previously published studies on the proliferative responses of these cells, E2 and 4-OHT differentially regulate OCR and ECAR in TAM-sensitive MCF-7, E2-independent LCC1, and TAM-resistant LCC2, and LCC9 cells. We conclude that different metabolic phenotypes occur in TAM resistant cells which could provide unique targets for breast cancer treatment.
Supplementary Material
Highlights.
NRF-1 and TFAM expression are higher in endocrine-resistant breast cancer cells
Oxygen consumption rate is similar in endocrine-sensitive and resistant cells
Mitochondrial reserve capacity is lower in endocrine-resistant cells
Endocrine-resistant breast cancer cells have increased glycolysis
Bioenergetic responses to E2 and tamoxifen are lower in endocrine-resistant cells
Acknowledgements
We thank Kristen H. Luken for performing some of the initial optimization studies in these cell lines and DuPont-Manual High School student Vandad Sharbaiani for his assistance to B.N.R. in some experiments. We thank Kendra Swope and Stephany Vittitow for assisting with searching for selected genes in GSE81620.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
The authors declare that there is no conflict of interest.
Funding
This research was supported by the National Institutes of Health grant R01 DK053220 to C.M.K., a grant from the University of Louisville School of Medicine to C.M.K., and an Internal Research Grant from the Office of the Executive Vice President for Research and Innovation of the University of Louisville to C.M.K. Huy Xuan Mai was supported by a summer research fellowship from the National Institutes of Health T35 DK072923 to C.M.K.
Author contributions
B.N.R., M.M.I., H.X.M., and N.A-R. performed experiments; M.M.I., B.G.H., and C.M.K. designed experiments; N.A-R. and C.M.K. performed statistical analyses; P.M. performed the experiments for the data in GSE81620 that were analyzed by T.S.K. and E.R.C. C.M.K. wrote the manuscript with editing by B.G.H.
References
- 1.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 2.Burstein HJ, Temin S, Anderson H, Buchholz TA, Davidson NE, Gelmon KE, Giordano SH, Hudis CA, Rowden D, Solky AJ, Stearns V, Winer EP, Griggs JJ. Adjuvant Endocrine Therapy for Women With Hormone Receptor–Positive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline Focused Update. J Clin Oncol. 2014 doi: 10.1200/JCO.2013.54.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang XH, Giuliano M, Trivedi MV, Schiff R, Osborne CK. Metastasis dormancy in estrogen receptor-positive breast cancer. Clin Cancer Res. 2013;19:6389–6397. doi: 10.1158/1078-0432.CCR-13-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan P, Maximov PY, Curpan RF, Abderrahman B, Jordan VC. The molecular, cellular and clinical consequences of targeting the estrogen receptor following estrogen deprivation therapy. Mol Cell Endocrinol. 2015;408:245–263. doi: 10.1016/j.mce.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke R, Tyson JJ, Dixon JM. Endocrine resistance in breast cancer – An overview and update. Mol Cell Endocrinol. 2015;418:220–234. doi: 10.1016/j.mce.2015.09.035. Part 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horan MP, Cooper DN. The emergence of the mitochondrial genome as a partial regulator of nuclear function is providing new insights into the genetic mechanisms underlying age-related complex disease. Hum Genet. 2014;133:435–458. doi: 10.1007/s00439-013-1402-4. [DOI] [PubMed] [Google Scholar]
- 7.Romero IL, Mukherjee A, Kenny HA, Litchfield LM, Lengyel E. Molecular Pathways: Trafficking of Metabolic Resources in the Tumor Microenvironment. Clin Cancer Res. 2015;21:680–686. doi: 10.1158/1078-0432.CCR-14-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sotgia F, Martinez-Outschoorn UE, Pavlides S, Howell A, Pestell RG, Lisanti MP. Understanding the Warburg effect and the prognostic value of stromal caveolin-1 as a marker of a lethal tumor microenvironment. Breast Cancer Res. 2011;13:213. doi: 10.1186/bcr2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ertel A, Tsirigos A, Whitaker-Menezes D, Birbe RC, Pavlides S, Martinez-Outschoorn UE, Pestell RG, Howell A, Sotgia F, Lisanti MP. Is cancer a metabolic rebellion against host aging? In the quest for immortality, tumor cells try to save themselves by boosting mitochondrial metabolism. Cell Cycle. 2012;11:253–263. doi: 10.4161/cc.11.2.19006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scarpulla RC, Vega RB, Kelly DP. Transcriptional integration of mitochondrial biogenesis. Trends in Endocrinology & Metabolism. 2012;23:459–466. doi: 10.1016/j.tem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, Gray JW, Bissell MJ. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1:84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattingly KA, Ivanova MM, Riggs KA, Wickramasinghe NS, Barch MJ, Klinge CM. Estradiol stimulates transcription of Nuclear Respiratory Factor-1 and increases mitochondrial biogenesis. Mol Endocrinol. 2008;22:609–622. doi: 10.1210/me.2007-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanova MM, Radde BN, Son J, Mehta FF, Chung S-H, Klinge CM. Estradiol and tamoxifen regulate NRF-1 and mitochondrial function in mouse mammary gland and uterus. J Mol Endocrinol. 2013;51:233–246. doi: 10.1530/JME-13-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahalingaiah PK, Ponnusamy L, Singh KP. Chronic oxidative stress causes estrogen-independent aggressive phenotype, and epigenetic inactivation of estrogen receptor alpha in MCF-7 breast cancer cells. Breast Cancer Res Treat. 2015;153:41–56. doi: 10.1007/s10549-015-3514-0. [DOI] [PubMed] [Google Scholar]
- 15.Okoh VO, Garba NA, Penney RB, Das J, Deoraj A, Singh KP, Sarkar S, Felty Q, Yoo C, Jackson RM, Roy D. Redox signalling to nuclear regulatory proteins by reactive oxygen species contributes to oestrogen-induced growth of breast cancer cells. Br J Cancer. 2015;112:1687–1702. doi: 10.1038/bjc.2014.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondaveeti Y, Guttilla Reed IK, White BA. Epithelial–mesenchymal transition induces similar metabolic alterations in two independent breast cancer cell lines. Cancer Lett. 2015;364:44–58. doi: 10.1016/j.canlet.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 17.Feeley KP, Bray AW, Westbrook DG, Johnson LW, Kesterson RA, Ballinger SW, Welch DR. Mitochondrial Genetics Regulate Breast Cancer Tumorigenicity and Metastatic Potential. Cancer Res. 2015;75:4429–4436. doi: 10.1158/0008-5472.CAN-15-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunner N, Boysen B, Jirus S, Skaar TC, Holst-Hansen C, Lippman J, Frandsen T, Spang-Thomsen M, Fuqua SA, Clarke R. MCF7/LCC9: an antiestrogen-resistant MCF-7 variant in which acquired resistance to the steroidal antiestrogen ICI 182,780 confers an early cross-resistance to the nonsteroidal antiestrogen tamoxifen. Cancer Res. 1997;57:3486–3493. [PubMed] [Google Scholar]
- 19.Brunner N, Boulay V, Fojo A, Freter CE, Lippman ME, Clarke R. Acquisition of hormone-independent growth in MCF-7 cells is accompanied by increased expression of estrogen-regulated genes but without detectable DNA amplifications. Cancer Res. 1993;53:283–290. [PubMed] [Google Scholar]
- 20.Hill BG, Dranka BP, Zou L, Chatham JC, Darley-Usmar VM. Importance of the bioenergetic reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem J. 2009;424:99–107. doi: 10.1042/BJ20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radde BN, Ivanova MM, Mai HX, Salabei JK, Hill BG, Klinge CM. Bioenergetic differences between MCF-7 and T47D breast cancer cells and their regulation by estradiol and tamoxifen. Biochem J. 2015;465:49–61. doi: 10.1042/BJ20131608. [DOI] [PubMed] [Google Scholar]
- 22.Dranka BP, Benavides GA, Diers AR, Giordano S, Zelickson BR, Reily C, Zou L, Chatham JC, Hill BG, Zhang J, Landar A, Darley-Usmar VM. Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic Biol Med. 2011;51:1621–1635. doi: 10.1016/j.freeradbiomed.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Damas J, Samuels DC, Carneiro J, Amorim A, Pereira F. Mitochondrial DNA rearrangements in health and disease--a comprehensive study. Hum Mutat. 2014;35:1–14. doi: 10.1002/humu.22452. [DOI] [PubMed] [Google Scholar]
- 24.Stafford P, Chen-Quin EB. The pattern of natural selection in somatic cancer mutations of human mtDNA. J Hum Genet. 2010;55:605–612. doi: 10.1038/jhg.2010.76. [DOI] [PubMed] [Google Scholar]
- 25.Spinazzi M, Casarin A, Pertegato V, Salviati L, Angelini C. Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat Protocols. 2012;7:1235–1246. doi: 10.1038/nprot.2012.058. [DOI] [PubMed] [Google Scholar]
- 26.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madden SF, Clarke C, Gaule P, Aherne ST, O'Donovan N, Clynes M, Crown J, Gallagher WM. BreastMark: an integrated approach to mining publicly available transcriptomic datasets relating to breast cancer outcome. Breast Cancer Res. 2013;15:R52. doi: 10.1186/bcr3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sastre-Serra J, Nadal-Serrano M, Pons DG, Valle A, Oliver J, Roca P. The Effects of 17beta-estradiol on Mitochondrial Biogenesis and Function in Breast Cancer Cell Lines are Dependent on the ERalpha/ERbeta Ratio. Cell Physiol Biochem. 2012;29:261–268. doi: 10.1159/000337607. [DOI] [PubMed] [Google Scholar]
- 29.Hill BG, Benavides GA, Lancaster JR, Jr., Ballinger S, Dell'Italia L, Jianhua Z, Darley-Usmar VM. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol Chem. 2012;393:1485–1512. doi: 10.1515/hsz-2012-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jastroch M, Divakaruni AS, Mookerjee S, Treberg JR, Brand MD. Mitochondrial proton and electron leaks. Essays Biochem. 2010;47:53–67. doi: 10.1042/bse0470053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brand MD, Nicholls DG. Assessing mitochondrial dysfunction in cells. Biochem J. 2011;435:297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mookerjee SA, Nicholls DG, Brand MD. Determining Maximum Glycolytic Capacity Using Extracellular Flux Measurements. PLoS One. 2016;11:e0152016. doi: 10.1371/journal.pone.0152016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kemper MF, Zhao Y, Duckles SP, Krause DN. Endogenous ovarian hormones affect mitochondrial efficiency in cerebral endothelium via distinct regulation of PGC-1 isoforms. J Cereb Blood Flow Metab. 2013;33:122–128. doi: 10.1038/jcbfm.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfleger J, He M, Abdellatif M. Mitochondrial complex II is a source of the reserve respiratory capacity that is regulated by metabolic sensors and promotes cell survival. Cell Death Dis. 2015;6:e1835. doi: 10.1038/cddis.2015.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biswas R, Vonderhaar BK. Role of serum in the prolactin responsiveness of MCF-7 human breast cancer cells in long-term tissue culture. Cancer Res. 1987;47:3509–3514. [PubMed] [Google Scholar]
- 36.Ring A, Dowsett M. Mechanisms of tamoxifen resistance. Endocr Relat Cancer. 2004;11:643–658. doi: 10.1677/erc.1.00776. [DOI] [PubMed] [Google Scholar]
- 37.Papa L, Germain D. Estrogen receptor mediates a distinct mitochondrial unfolded protein response. J Cell Sci. 2011;124:1396–1402. doi: 10.1242/jcs.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lesnefsky EJ, Chen Q, Hoppel CL. Mitochondrial Metabolism in Aging Heart. Circ Res. 2016;118:1593–1611. doi: 10.1161/CIRCRESAHA.116.307505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pons DG, Nadal-Serrano M, Torrens-Mas M, Valle A, Oliver J, Roca P. UCP2 inhibition sensitizes breast cancer cells to therapeutic agents by increasing oxidative stress. Free Radic Biol Med. 2015;86:67–77. doi: 10.1016/j.freeradbiomed.2015.04.032. [DOI] [PubMed] [Google Scholar]
- 40.Whitaker-Menezes D, Martinez-Outschoorn UE, Flomenberg N, Birbe RC, Witkiewicz AK, Howell A, Pavlides S, Tsirigos A, Ertel A, Pestell RG, Broda P, Minetti C, Lisanti MP, Sotgia F. Hyperactivation of oxidative mitochondrial metabolism in epithelial cancer cells in situ: Visualizing the therapeutic effects of metformin in tumor tissue. Cell Cycle. 2011;10:4047–4064. doi: 10.4161/cc.10.23.18151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woo YM, Shin Y, Lee EJ, Lee S, Jeong SH, Kong HK, Park EY, Kim HK, Han J, Chang M, Park JH. Inhibition of Aerobic Glycolysis Represses Akt/mTOR/HIF-1alpha Axis and Restores Tamoxifen Sensitivity in Antiestrogen-Resistant Breast Cancer Cells. PLoS One. 2015;10:e0132285. doi: 10.1371/journal.pone.0132285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyanova S, Albrechtsen R, Kronqvist P, Cox J, Mann M, Geiger T. Proteomic maps of breast cancer subtypes. Nat Commun. 2016;7 doi: 10.1038/ncomms10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riggs KA, Wickramasinghe NS, Cochrum RK, Watts MB, Klinge CM. Decreased Chicken Ovalbumin Upstream Promoter Transcription Factor II Expression in Tamoxifen-Resistant Breast Cancer Cells. Cancer Res. 2006;66:10188–10198. doi: 10.1158/0008-5472.CAN-05-3937. [DOI] [PubMed] [Google Scholar]
- 44.Sanchez MI, Shearwood AM, Chia T, Davies SM, Rackham O, Filipovska A. Estrogen-mediated regulation of mitochondrial gene expression. Mol Endocrinol. 2015;29:14–27. doi: 10.1210/me.2014-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flight RM, Harrison BJ, Mohammad F, Bunge MB, Moon LD, Petruska JC, Rouchka EC. categoryCompare, an analytical tool based on feature annotations. Front Genet. 2014;5:98. doi: 10.3389/fgene.2014.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.