Abstract
Via whole-exome sequencing, we identified six females from independent families with a common neurodevelopmental phenotype including developmental delay, intellectual disability, autism, hypotonia, and seizures, all with de novo predicted deleterious variants in the nuclear localization signal of Heterogeneous Nuclear Ribonucleoprotein H2, encoded by HNRNPH2, a gene located on the X chromosome. Many of the females also have seizures, psychiatric co-morbidities, and orthopedic, gastrointestinal, and growth problems as well as common dysmorphic facial features. HNRNPs are a large group of ubiquitous proteins that associate with pre-mRNAs in eukaryotic cells to produce a multitude of alternatively spliced mRNA products during development and play an important role in controlling gene expression. The failure to identify affected males, the severity of the neurodevelopmental phenotype in females, and the essential role of this gene suggests that male conceptuses with these variants may not be viable.
Keywords: HNRNPH2, pre-mRNA, alternative splicing, autism, developmental delay, microcephaly, neurodevelopment, X chromosome
Main Text
A number of X-linked disorders have been extensively associated to intellectual disability as well as neurodevelopmental disabilities including autism.1, 2, 3, 4, 5, 6 Here we describe six unrelated females who have de novo missense variants in two specific amino acids predicted to be deleterious in the protein product of Heterogeneous Nuclear Ribonucleoprotein H2 (HNRNPH2 [MIM: 300610]). HNRNPH2 (GenBank: NM_019597.4) is located at Xq22.1 and encodes a member of a family of ubiquitous heterogeneous nuclear ribonucleoproteins (HNRNP). The HNRNPs are a large group of RNA binding proteins that have distinct nucleic acid binding properties that act as a shuttle between the nucleus and the cytoplasm and act on pre-mRNA to positively or negatively affect spliceosome assembly at nearby splice sites, thereby controlling pre-mRNA splicing.7, 8 These proteins are designated HNRNPA1 through HNRNPU, and each contains one or more RNA binding domains, such as RNA recognition motifs (RRM) or ribonucleoprotein (RNP) consensus sequences (Figure 1).9, 10 Alternative RNA splicing allows for the production of multiple mRNAs from a single gene, allowing for control of gene expression by cell type and developmental stage. Aberrant splicing causes many neurological diseases, including various hematolymphoid neoplasias, autosomal-dominant retinitis pigmentosa, microcephalic osteodysplastic primordial dwarfism type 1 (MOPD1 [MIM: 210710]), amyotrophic lateral sclerosis (ALS [MIM: 105400]), spinal muscular atrophy (SMA [MIM: 600354]), and X-linked syndromic mental retardation (XLMR [MIM: 309530]).11, 12, 13 We report an association of de novo predicted pathogenic missense variants in HNRNPH2 with a range of neurodevelopmental features including developmental delay (DD), intellectual disability (ID), autism, hypotonia, and seizures.
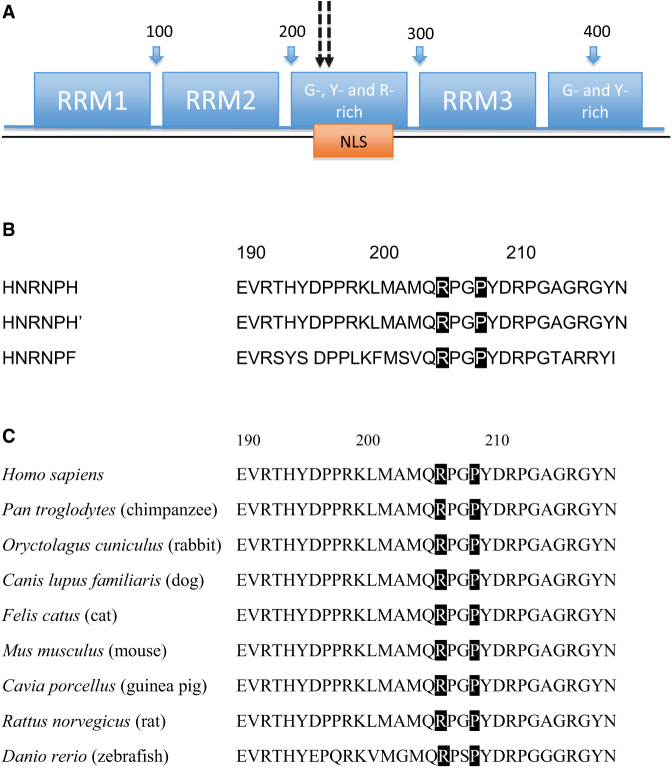
Figure 1.
HNRNPH2 Domains and Conservation among Various Family Members and across Species
(A) HNRNPH2 domains. The numbers above (100, 200, 300, and 400) represent the amino acid sequence. Dotted arrows indicate the location of de novo variants at amino acid positions 206 and 209. Abbreviations are as follows: NLS, nuclear localization sequence; RRM, RNA recognition motifs; G, glycine-rich domain; Y, tyrosine-rich domain; R, arginine-rich domain.
(B) Highly conserved sequences among members of the HNRNPH family, including HNRNPs H1 (H), H2 (H’), and F proteins. Highlighted boxes indicate amino acid residues 206 and 209 reported.
(C) Cross species conservation of amino acids 206 and 209. Highlighted boxes indicate amino acid residues 206 and 209 reported.
Informed consent was obtained from all participants included in the study and the study was approved by the Institutional Review Board of Columbia University. Magnetic resonance imaging (MRI) and photographs of affected individuals were obtained with consent from each individual’s guardians. For the five females tested at GeneDx, a commercial clinical genetic testing laboratory, genomic DNA was extracted from whole blood from the affected children and their parents. Exome sequencing was performed on exon targets captured using the Agilent SureSelect Human All Exon V4 (50 Mb) kit or the Clinical Research Exome kit (Agilent Technologies) as previously described.14 After automated filtering of variants with a MAF of >10%, manual curation was performed to filter less common variants with MAF of 1%–10%, single variants in genes inherited from unaffected parents, evaluate predicted effects of rare variants and known function of the genes and associated human conditions, and examine overlapping phenotypes of individuals with de novo variants in the same gene as previously described.14 All de novo variants were confirmed by Sanger sequencing. The general assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission page.
In the GeneDx cohort, five female individuals were identified out of 2,030 females and 2,486 males with DD and/or ID analyzed by WES with de novo predicted pathogenic missense variants in HNRNPH2 (Table 1). WES on these five samples produced an average of ∼10 GB of sequence per sample. Mean coverage of captured regions was ∼138× per sample, with >98% covered with at least 10× coverage, an average of 91% of base call quality of Q30 or greater, and an overall average mean quality score of >Q36. Filtering of common SNPs (>10% frequency present in 1000 Genomes database) resulted in ∼5,050 variants per proband sample. Among these five case subjects, four had de novo missense variants at amino acid 206: three with c.616C>T (p. Arg206Trp) (GenBank: NM_019597.4) and one with c.617G>A (p.Arg206Gln) (Figure 1). Individual 4 harbored a de novo missense variant located three amino acids away (c.626C>T [p.Pro209Leu]). According to a model that predicts gene-specific background mutation rate based on sequence context,15 the expected number of de novo missense variants in HNRNPH2 among 2,030 unrelated females is about 0.051. Using a previously established mutation rate and Poisson test,15 the observed five de novo missense variants in these individuals is highly significant (p value = 2.3 × 10−9). Putative de novo variants were filtered for variant call quality and then for presence in unaffected internal control WES samples. In two case subjects, HNRNPH2 was the only de novo change whereas in the other three case subjects, there were a small number of other variants. These other de novo variants were manually evaluated at the gene level in the context of the individuals’ phenotypes and at the variant level for cross-species conservation and in silico predicted pathogenicity. These other variants were ruled out as likely contributing to the individuals’ phenotypes. A table of the filtering and remaining variants is provided in Table S1. One additional individual (individual 6) was identified with the same c.616C>T (p.Arg206Trp) variant by WES at another laboratory, and sequencing methods for that individual have been previously described.19 It is notable that five of the six missense variants alter the same amino acid. The missense variants are not present in ExAC, and there are no truncating variants in HNRNPH2 in the NHLBI Exome Variant Server or ExAC (accessed December 2015) or in a local database of more than 23,000 exomes.
Table 1.
Predicted Pathogenicity and Allele Frequencies of HNRNPH2 Variants
| Nucleic Acid Variant | cDNA Change | Amino Acid Change | Position | SIFT | PolyPhen (HDIV Score) | Mutation Taster | CADD Phred | ExAC Allele Frequency |
|---|---|---|---|---|---|---|---|---|
| CGG>TGG | c.616C>T | p.Arg206Trp | chrX: 100,667,592 | damaging (.97) | tolerated (0.025) | disease causing | 20.6 | 0 |
| CGG>CAG | c.617G>A | p.Arg206Gln | chrX: 100,667,593 | damaging (.99) | tolerated (0.025) | disease causing | 22.1 | 0 |
| CCC>CTC | c.626C>T | p.Pro209Leu | chrX: 100,667,602 | damaging (1) | damaging (0.938) | disease causing | 22.3 | 0 |
These six individuals range in age from 2 years to 34 years and all have DD or ID (Table 2). Three individuals have histories of developmental regression. Three individuals were diagnosed with autism spectrum disorders and three have behavioral disturbances, ranging from aggressive or self-injurious behavior to psychiatric diagnoses such as anxiety, attention deficit hyperactivity disorder (ADHD), obsessive-compulsive disorder (OCD), or stereotyped behaviors. Neurological exams were notable for tone abnormalities, including all six with hypotonia and one also with hypertonia (spasticity), as well as ataxia and gait disturbance. Three of the individuals were diagnosed with seizures and two with acquired microcephaly. Two had cerebellar anomalies on brain imaging whereas the MRIs of two others were unremarkable (Figure 2). All individuals have dysmorphic facial features, and Figure 3 illustrates three individuals (#1, #2, and #5) who share many similar facial features including almond-shaped eyes, short palpebral fissures, a short philtrum, full lower lip, long columella, hypoplastic alae nali, and micrognathia. Orthopedic problems were common and included scoliosis, lordosis, pectus carinatum, pes planus, and joint laxity and likely are at least partially secondary to the neurological problems. Feeding difficulties, gastresophageal reflux disease, and constipation were noted in several individuals, as was short stature.
Table 2.
Clinical Characteristics of Individuals with De Novo HNRNPH2 Variants
| Individual | Variant | Age (Years) | Microcephaly? (Congenital or acquired?) | DD/ID | Regressions | ASD | ADHD | Other Psychiatric Co-morbidities | Seizures | Tone Abnormalities (Hypo or Hyper) | Other Neurological Findings | MRI Findings | Ophthalmologic Findings | Dysmorphic Features | Skeletal Anomalies | GI Symptoms and Growth Parameters | Other Clinical Findings |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | c.616C>T (p.Arg206Trp) | 34 | Y (A) | Y | Y | Y | Y | anxiety, OCD, aggressive behavior, self-injurious | Y | hypo, hyper | abnormal gait, ankle clonus | normal | Y, exotropia | hypotelorism, wide mouth, full lips, high narrow nasal bridge, curly hair | lordosis, short stature, bilateral femoral osteotomies, arachnodactyly | FTT, short stature | N |
| 2 | c.616C>T (p.Arg206Trp) | 8 | N | Y | N | Y | N | repetitive stereotypic behavior | Y | hypo | ataxia, muscle weakness | aplasia /hypoplasia cerebellar vermis | ? | epicanthal folds, midface hypoplasia, almond shaped eyes | N | constipation | mild hearing loss, anemia, epistaxis |
| 3 | c.617G>A (p.Arg206Gln) | 4 | N | Y | Y | N | N | N | N | hypo | N | NA | N | small palpebral fissures | pes planus | GERD as an infant | happy demeanor, sensitive to noise, hand flapping |
| 4 | c.626C>T (p.Pro209Leu) | 6 | Y (A) | Y | Y | N | N | anxiety | Y | hypo | incoordination | normal | N | hypertelorism, fetal finger pads | short stature, talus valgus | FTT, feeding issues | cardiac (atrial septal defect and MVP), sensory disorder |
| 5 | c.616C>T (p.Arg206Trp) | 21 | N | Y | N | Y | Y | N | N | hypo | N | normal | N | highly arched palate, mild micrognathia, elongated fingers | pectus carinatum, pes planus, scoliosis, stretchable skin, joint laxity | GERD as a child; underweight | cardiac (MVP) |
| 6 | c.616C>T (p.Arg206Trp) | 2 | N | Y | N | N | N | N | N | hypo | torticollis, dystonic posturing left hand, dyspraxia | possible distorted cerebellar vermis | N | symmetrically concave eyebrow, pseudo-fissure in upper lip | N | feeding difficulties, drooling | N |
Abbreviations are as follows: GERD, gastroesophageal reflux disease; FTT, failure to thrive; MVP, mitral valve prolapse; ?, unknown; A, acquired; NA, not applicable.
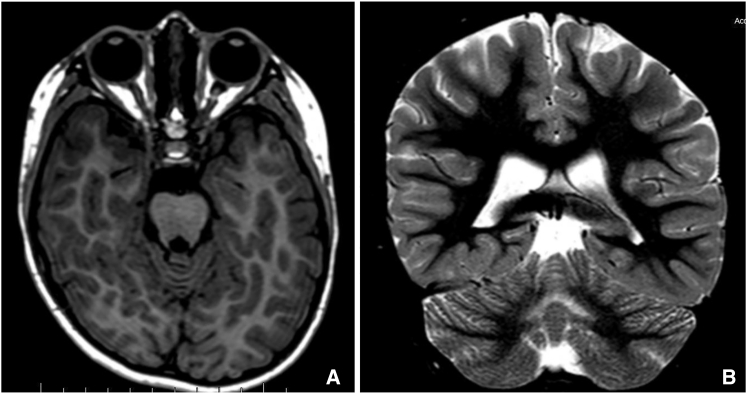
Figure 2.
Magnetic Resonance Images of Affected Individuals
Axial T1-weighted (A) and coronal (B) STIR images of individual 2 demonstrating hypoplasia of cerebellar vermis at age 7 years old. Images obtained on a 1.5 Tesla MRI.
Figure 3.
Dysmorphic Features in Affected Individuals
Photographs of individual 1 (A–C) and individual 5 (D–F) with dysmorphic features including almond shaped eyes, short palpebral fissures, short philtrum, full lower lip, long columella, and hypoplastic alae nasi. Individual 1 is shown at 5 years of age (A) and 34 years of age (B, C). Individual 5 is shown at 21 years of age. Individual 2 (G) is shown at 8 years of age.
HNRNPH2 is located on chromosome Xq22.1, contains 449 amino acids, and is a member of the HNRNP family. The protein product has several different domains, including three quasi-RRMs that bind RNA and two glycine-rich domains (Figure 1A).16 Of note, there are highly conserved sequences between HNRNPH, H2 (also referred to as H’), and F (Figure 1B) and therefore are often referred to as HNRNP H/F. HNRNP H and F proteins are highly conserved in the RRM and have an extensive glycine-rich region near the carboxyl terminus, which further allows members of the H/F family to homo- or heterodimerize.9 Furthermore, these glycine-rich domains in this gene are critical for nuclear localization of the protein.16 A nuclear localization sequence (NLS) located within a glycine-rich domain between amino acids 194 and 220 has been identified to interact with the import receptor transportin 1 (Trn1). This NLS encompasses our two missense variants, a region highly conserved across species (Figure 1C).16 Proteins in the HNRNP family localize to the nucleus, bind to heterogeneous nuclear RNA, and shuttle between the nucleus and cytoplasm for pre-mRNA processing and transport. This protein is ubiquitous and highly co-localized in the nuclear compartments in the brain, gastrointestinal tract, lung, skin, spleen, and testes, which may be the foundation for the multisystem abnormalities noted in this study. Most studies for HNRNPH2 have been in vitro functional assays, and there have been no knockout mouse models to further study gene function.
Five do novo variants identified in our series are located at amino acid 206, and the last variant involves nearby amino acid 209. Both of these amino acids are located within the NLS, suggesting that this is a critical region for proper gene function. Furthermore, given that all six individuals are female, it is possible that these variants are lethal in males and that only heterozygous females survive, similar to our recently reported findings with DDX3X (MIM: 300160).17
HNRNPs mediate pre-mRNA alternative splicing, and aberrations in splicing are increasingly recognized as a cause of developmental disorders.18 HNRNPs A1B2, H, and F have each been identified in neurons and are important in cellular proliferation, differentiation, and apoptosis. Developmentally, HNRNP H/F splicing factors have been proposed as “master orchestrators” of differentiation, integral to the developmental programming in nervous system differentiation.19, 20 There have not been any studies specifically demonstrating HNRNPH2 expression throughout development. However, other HNRNPs have been more studied. For example, HNRNP A2 and HNRNP E1 co-localize in oligodendrocytes and neuronal dendrites. Overexpression of or microinjection of exogenous HNRNPE1 in rat neural cells inhibited translation of cis-acting A2 response element (A2RE) RNAs, which forms a complex with HNRNPA1 called a RNA granule.20 These granules are then transported along microtubules to distal dendrites to regulate myelin basic protein production.20
As in the adult neurodegeneration literature, it is possible that mutant HNRNPH2 leads to trafficking problems that keep HNRNPH2 in the cytoplasm.21, 22, 23 Recently, several RNA binding proteins including FUS, ataxin 2, HNRNPA1, and HNRNPB2 have been implicated in the frontotemporal dementia/amyotrophic lateral sclerosis (FTD/ALS) phenotype pathology. The proposed pathogenesis includes deposition of poorly soluble assemblies on the mutant RNA-binding protein in the nucleus and cytoplasm of neurons in the brain and spinal cord of ALS/FTD-affected individuals with mis-localization of the protein due to incorrect nuclear localization signaling.22, 23 Specifically, sequestration of HNRNPH by potentially toxic GGGGCC intronic repeat expansions in ALS/FTD disrupts appropriate RNA processing and contributes to neurodegeneration.24 Similarly, a family was identified with a missense variant in HNRNPA2B1 with dominantly inherited degeneration called multisystem proteinopathy, also known as inclusion body myopathy with frontotemporal dementia (IBMPFD [MIM: 615424]) which affects muscle, bone, brain, and motor neurons.21 Wild-type HNRNP A1 and A2 have an intrinsic tendency to assemble into self-seeding fibrils as it contains a distinctive prion-like domain (PrLD) enriched in uncharged polar amino acids and glycine, but mutant proteins greatly accelerate this process.21 Pathogenic variants in PrLDs of HNRNPs A2B1 and A1 have been described in families with this inherited neurodegeneration.21 The reported disease variants promote excess incorporation of HNRNPA2 and HNRNPA1 into stress granules and drive the formation of cytoplasmic inclusions in Drosophila and mouse models that may initiate the degenerative disease. In another related study, HNRNPA2B1 has been previously implicated in fragile-X-associated tremor ataxia syndrome (FXTAS [MIM: 300623]), and the protein sequesters with RNA foci in postmortem human tissue and then is recapitulated in Drosophila models.25, 26, 27 Three individuals in our series have developmental regression suggesting an underlying neurodegenerative process, and the lack of consistent neurodegenerative phenotype across individuals could be due to the random X inactivation pattern.
Here we describe six females with de novo missense variants in HNRNPH2, a gene on the X chromosome that is critical for pre-mRNA processing and trafficking between the nucleus and cytoplasm, allowing for heterogeneity of protein products in different tissues and at different developmental checkpoints. All of the individuals share a common neurological phenotype of developmental delay, some with regression, autism, and tone abnormalities. Many have additional neurological problems such as seizures or psychiatric co-morbidities such as ADHD, anxiety, and OCD. Most of the individuals also have systemic abnormalities, including orthopedic, gastrointestinal, and growth problems. Five of the individuals share the same altered amino acid, 206, and the sixth individual carries a variant at the nearby amino acid position 209, both of which are located within a glycine-rich domain containing a very particular nuclear localization sequence. Recent studies show that similar HNRNP proteins that are dysfunctional can accumulate in the cytoplasm, leading to neurodegenerative conditions in adults. We propose a similar gain-of-function variant in these six individuals that produces a distinct neurodevelopmental phenotype. Critical checkpoints in the development of the brain as well as other organ systems are managed by the HNRNP family members, and aberrant gene function can lead to aberrant development of such systems. In the case of this gene located on the X chromosome, variants in males may be embryonic lethal, and the corresponding phenotype in females leads to one with a range of neurodevelopmental disability, including developmental delay, intellectual disability, autism, psychiatric disease, tone abnormalities, seizures, and other systemic abnormalities. More functional assays including animal models of this critical mRNA regulator is imperative to better understand the implications of alterations in this developmental pathway and potential neurodegenerative implications.
Conflicts of Interest
M.T.C., A.T., K.R., and K.G.M. are employees of GeneDx. W.K.C. is a former employee of GeneDx.
Acknowledgments
We gratefully acknowledge all the individuals and their families. Funding support provided by the Simons Foundation.
Published: August 18, 2016
Footnotes
Supplemental Data include case reports and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.06.028.
Accession Numbers
The ClinVar accession numbers for the reported sequences in this paper are SCV000267837, SCV000267838, and SCV000267839.
Web Resources
ClinVar submission page (GeneDx), http://www.ncbi.nlm.nih.gov/clinvar/submitters/26957/
ExAC Browser, http://exac.broadinstitute.org/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Lombardi L.M., Baker S.A., Zoghbi H.Y. MECP2 disorders: from the clinic to mice and back. J. Clin. Invest. 2015;125:2914–2923. doi: 10.1172/JCI78167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassani S., Zapata J., Gerosa L., Moretto E., Murru L., Passafaro M. The neurobiology of X-linked intellectual disability. Neuroscientist. 2013;19:541–552. doi: 10.1177/1073858413493972. [DOI] [PubMed] [Google Scholar]
- 3.des Portes V. X-linked mental deficiency. Handb. Clin. Neurol. 2013;111:297–306. doi: 10.1016/B978-0-444-52891-9.00035-X. [DOI] [PubMed] [Google Scholar]
- 4.Santoro M.R., Bray S.M., Warren S.T. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu. Rev. Pathol. 2012;7:219–245. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- 5.Ropers H.H. Genetics of intellectual disability. Curr. Opin. Genet. Dev. 2008;18:241–250. doi: 10.1016/j.gde.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Ropers H.H. Genetics of early onset cognitive impairment. Annu. Rev. Genomics Hum. Genet. 2010;11:161–187. doi: 10.1146/annurev-genom-082509-141640. [DOI] [PubMed] [Google Scholar]
- 7.Cáceres J.F., Stamm S., Helfman D.M., Krainer A.R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 8.Sohail M., Xie J. Evolutionary emergence of a novel splice variant with an opposite effect on the cell cycle. Mol. Cell. Biol. 2015;35:2203–2214. doi: 10.1128/MCB.00190-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi Y.D., Grabowski P.J., Sharp P.A., Dreyfuss G. Heterogeneous nuclear ribonucleoproteins: role in RNA splicing. Science. 1986;231:1534–1539. doi: 10.1126/science.3952495. [DOI] [PubMed] [Google Scholar]
- 10.Chou M.Y., Rooke N., Turck C.W., Black D.L. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng D., Xie J. Aberrant splicing in neurological diseases. Wiley Interdiscip. Rev. RNA. 2013;4:631–649. doi: 10.1002/wrna.1184. [DOI] [PubMed] [Google Scholar]
- 12.Linder B., Fischer U., Gehring N.H. mRNA metabolism and neuronal disease. FEBS Lett. 2015;589:1598–1606. doi: 10.1016/j.febslet.2015.04.052. [DOI] [PubMed] [Google Scholar]
- 13.Singh R.K., Cooper T.A. Pre-mRNA splicing in disease and therapeutics. Trends Mol. Med. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka A.J., Cho M.T., Millan F., Juusola J., Retterer K., Joshi C., Niyazov D., Garnica A., Gratz E., Deardorff M. Mutations in SPATA5 are associated with microcephaly, intellectual disability, seizures, and hearing loss. Am. J. Hum. Genet. 2015;97:457–464. doi: 10.1016/j.ajhg.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Dusen C.M., Yee L., McNally L.M., McNally M.T. A glycine-rich domain of hnRNP H/F promotes nucleocytoplasmic shuttling and nuclear import through an interaction with transportin 1. Mol. Cell. Biol. 2010;30:2552–2562. doi: 10.1128/MCB.00230-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snijders Blok L., Madsen E., Juusola J., Gilissen C., Baralle D., Reijnders M.R., Venselaar H., Helsmoortel C., Cho M.T., Hoischen A., DDD Study Mutations in DDX3X are a common cause of unexplained intellectual disability with gender-specific effects on Wnt signaling. Am. J. Hum. Genet. 2015;97:343–352. doi: 10.1016/j.ajhg.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Homsy J., Zaidi S., Shen Y., Ware J.S., Samocha K.E., Karczewski K.J., DePalma S.R., McKean D., Wakimoto H., Gorham J. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dardenne E., Polay Espinoza M., Fattet L., Germann S., Lambert M.P., Neil H., Zonta E., Mortada H., Gratadou L., Deygas M. RNA helicases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation. Cell Rep. 2014;7:1900–1913. doi: 10.1016/j.celrep.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Kosturko L.D., Maggipinto M.J., Korza G., Lee J.W., Carson J.H., Barbarese E. Heterogeneous nuclear ribonucleoprotein (hnRNP) E1 binds to hnRNP A2 and inhibits translation of A2 response element mRNAs. Mol. Biol. Cell. 2006;17:3521–3533. doi: 10.1091/mbc.E05-10-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H.J., Kim N.C., Wang Y.D., Scarborough E.A., Moore J., Diaz Z., MacLea K.S., Freibaum B., Li S., Molliex A. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami T., Qamar S., Lin J.Q., Schierle G.S., Rees E., Miyashita A., Costa A.R., Dodd R.B., Chan F.T., Michel C.H. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu C., Zhang J., Gao F., Yang L., Jia M., Zhu H., Gong W. FUS-NLS/Transportin 1 complex structure provides insights into the nuclear targeting mechanism of FUS and the implications in ALS. PLoS ONE. 2012;7:e47056. doi: 10.1371/journal.pone.0047056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee Y.B., Chen H.J., Peres J.N., Gomez-Deza J., Attig J., Stalekar M., Troakes C., Nishimura A.L., Scotter E.L., Vance C. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–1186. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwahashi C.K., Yasui D.H., An H.J., Greco C.M., Tassone F., Nannen K., Babineau B., Lebrilla C.B., Hagerman R.J., Hagerman P.J. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- 26.Jin P., Duan R., Qurashi A., Qin Y., Tian D., Rosser T.C., Liu H., Feng Y., Warren S.T. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sofola O.A., Jin P., Qin Y., Duan R., Liu H., de Haro M., Nelson D.L., Botas J. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.