Abstract
Rare mutations, including copy-number variants (CNVs), contribute significantly to autism spectrum disorder (ASD) risk. Although their importance has been established in families with only one affected child (simplex families), the contribution of both de novo and inherited CNVs to ASD in families with multiple affected individuals (multiplex families) is less well understood. We analyzed 1,532 families from the Autism Genetic Resource Exchange (AGRE) to assess the impact of de novo and rare CNVs on ASD risk in multiplex families. We observed a higher burden of large, rare CNVs, including inherited events, in individuals with ASD than in their unaffected siblings (odds ratio [OR] = 1.7), but the rate of de novo events was significantly lower than in simplex families. In previously characterized ASD risk loci, we identified 49 CNVs, comprising 24 inherited events, 19 de novo events, and 6 events of unknown inheritance, a significant enrichment in affected versus control individuals (OR = 3.3). In 21 of the 30 families (71%) in whom at least one affected sibling harbored an established ASD major risk CNV, including five families harboring inherited CNVs, the CNV was not shared by all affected siblings, indicating that other risk factors are contributing. We also identified a rare risk locus for ASD and language delay at chromosomal region 2q24 (implicating NR4A2) and another lower-penetrance locus involving inherited deletions and duplications of WWOX. The genetic architecture in multiplex families differs from that in simplex families and is complex, warranting more complete genetic characterization of larger multiplex ASD cohorts.
Introduction
Genetic variation accounts for a major proportion of the liability to autism spectrum disorder (ASD). Twin studies have estimated the heritability of ASD to be at least 50%, given that concordance of ASD in monozygotic twins ranges from 37% to 95% depending on the study design and the diagnostic criteria used.1, 2, 3, 4 The importance of genetics in ASD susceptibility is also reflected by the recent success of microarray and whole-exome sequencing studies, which have established the role of de novo copy-number variants (CNVs) and de novo protein-disrupting single-nucleotide variants (SNVs) in ASD pathogenesis. Similar contributions have been identified in individuals with intellectual disability (ID) without ASD.5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15 Approximately 3.7% of affected individuals carry a large (>500 kb) de novo CNV, whereas only 0.4%–0.8% of their unaffected siblings do,9, 10, 11, 13, 16 implying that the rate of large de novo CNVs is approximately five times higher in individuals with ASD.
Despite their rarity in ASD, several recurrent large CNVs have been identified and include loci at 1q21.1 (MIM: 612475 and 612474), 3q29 (MIM: 609425), 7q11.23 (MIM: 609757), 15q11–13 (MIM: 608636), 15q13.3 (MIM: 612001 and 608636), 16p11.2 (MIM: 611913 and 614671), and 22q11.2 (MIM: 608363),11, 13, 17 as well as several other very rare CNVs, including the 5q35 deletion (Sotos syndrome [MIM: 117550]) and the 17q12 deletion (MIM: 614527). Although the overall signal for statistical enrichment comes from rare, de novo CNVs in ASD, some known recurrent CNVs can be inherited, as seen with 15q11–13 duplications or 1q21.1 CNVs.18, 19 Some studies have suggested an excess of maternally inherited pathogenic CNVs,15, 20 but an excess of maternal transmission appears to vary by locus rather than being generalizable.9, 13, 16, 20
Some studies have included individuals from families with multiple affected children (multiplex families) in a case-control design9, 12, 21, 22, 23 but have not focused specifically on analyzing the role of CNVs in these families. In fact, most major studies have focused primarily on simplex families who have been carefully selected for the absence of observable relevant clinical phenotypes in first-degree relatives—a select cohort that is likely to have less inherited genetic variation than the general ASD population and especially multiplex families. Generalization from studies in simplex or mixed family types provides most of what we know about specific genetic contributions to ASD.10, 11, 24 Moreover, heritable variation is estimated to provide a significant contribution to ASD risk,4, 25 but studies of simplex families most likely underestimate this component. Multiplex families are estimated to constitute about 11% of ASD-affected families and are hypothesized to have a different genetic architecture.12, 26, 27, 28, 29, 30 One prominent model posits that they are enriched with inherited, potentially dominant variation and therefore exhibit a smaller contribution from de novo events than do simplex families.30 Stoppage, the decision to stop having more children after a diagnosis in one, is also an issue in ASD-affected families.31 In some cases, multiplex families are more likely to include children with milder ASD symptoms, therefore reducing the likelihood of stoppage. This provides an additional hypothesis that multiplex families are enriched with inherited variants of lower impact.
Despite the predicted differences in the genetic architecture of ASD between simplex and multiplex families, few large studies of CNVs have been conducted in large multiplex cohorts.12, 32, 33 We undertook such a study by using the Autism Genetic Resource Exchange (AGRE), the largest cohort of predominantly multiplex ASD-affected families who have been genotyped on microarray platforms that permit genome-wide CNV detection, allowing us to focus on the genetic architecture in multiplex families in a well-powered cohort.
Material and Methods
Samples
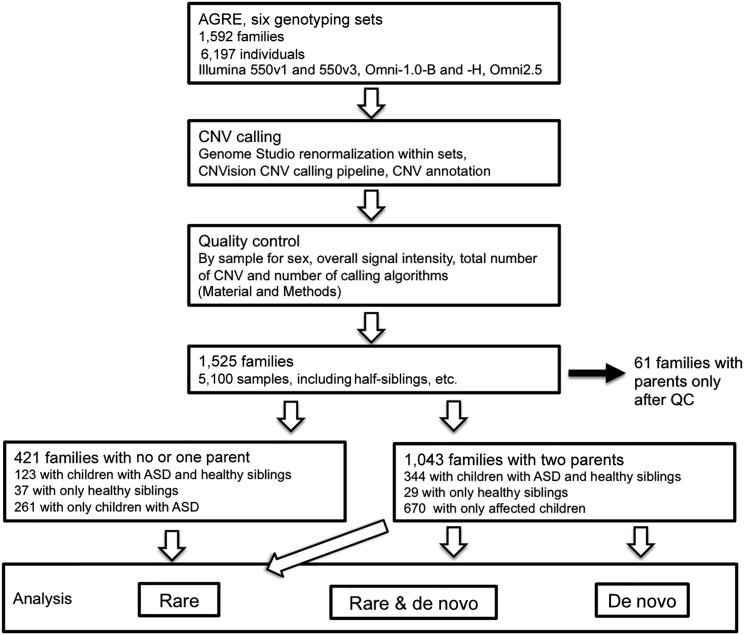
We analyzed an AGRE sample set consisting of 1,532 families, a subset of whom had been previously analyzed with a different, less conservative pipeline or a different focus in earlier studies34 (Table S1). Of the available families, 1,189 (78%) are multiplex, and 343 (22%) are simplex. No restrictions regarding family structure were imposed during sample collection, which included both extended families and half-siblings in addition to nuclear families. The number of individuals with available biomaterials was at minimum one individual per family, which led to a collection of mixed family structures (Figure 1 and Table S1). Pedigrees of families affected by ASD-associated CNVs were drawn with the Madeline 2.0 Pedigree Drawing Engine.35
Figure 1.
Overview of the Analysis Pipeline
We started by renormalizing intensities according to manufacturer recommendations and continued with the CNVision pipeline, followed by quality control (QC) with strict recommended criteria. After QC, we proceeded to annotate CNVs by type and annotate samples by family structure and analyzed the samples in multiplex (at least two affected children) and combined (all samples and family types) analyses for rare (<1% frequency in the DGV) and de novo CNVs. Families in whom the child or children failed QC were excluded from the analyses. Analyses were performed by CNV size (≥500 kb, 100–500 kb, and combined) in the mentioned CNV categories. Inheritance patterns for CNVs could be determined only in families with both parents, and only these families were included in the de novo analyses.
Sample diagnostic criteria are provided on the Internet System of Accessing Autistic Children website (see Web Resources). In summary, the Autism Diagnostic Observation Schedule and Autism Diagnostic Interview-Revised were used for assigning individuals into three categories: (1) a strict autism diagnosis, (2) not quite autism, (3) or broad spectrum. In this study, all individuals in any of these categories were considered affected, in accordance with standard AGRE practice.36 The term “broad spectrum” includes phenotypes that fit former Asperger syndrome and pervasive developmental disorder – not otherwise specified, which are currently considered a part of the ASD spectrum. A more detailed description is available online at the AGRE website (see Web Resources) and in previous publications.36 This study was approved by institutional review boards at the AGRE, University of Washington, and University of California, Los Angeles (UCLA). Written informed consent was obtained from all adult participants, and assent with agreement from their parents was obtained from all underage participants.
Microarrays
Individuals were genotyped on six different platforms according to manufacturer protocols over multiple years. We initially included 4,013 samples from the AGRE Illumina Human 550v1 and 550v3 genotyping set from Wang et al.37 but were only able to retain 2,974 samples after quality control (see below and Table S1). The remainder of the samples (n = 2,284) were genotyped at the UCLA Neuroscience Genomics Core on three different versions of Illumina HumanHap Omni1 arrays (n = 1,265) or the Illumina HumanHap Omni2.5 array (n = 1,019) (Figure 1 and Table S1). DNA for microarrays was extracted from lymphoblastoid cell lines (LCLs) at the Rutgers Repository according to standard protocols. All individual arrays were recalled and calibrated by batch, and sex chromosomes were recalled on GenomeStudio according to manufacturer protocols.
CNV Calling and Annotation
CNVs were called with CNVision11 (a pipeline that uses two “gold-standard” CNV-calling algorithms for microarray data), PennCNV,38 QuantiSNP,39 and GNOSIS, an algorithm based on CNVision integrated outlier detection. All CNVs on each microarray batch were annotated to their respective genome versions with the Bamotate annotation tool11 for overlapping genes, regions with recurrent syndrome-causing CNVs, and select genomic features (such as telomeres, centromeres, and human leukocyte antigen). After annotation to their respective genome versions, all CNVs were lifted from hg18 to hg19 (UCSC Genome Browser) with the UCSC LiftOver tool.
CNV Quality Control
Sample-based quality-metric thresholds were based on the standard recommendations from the developers of PennCNV and QuantiSNP.38, 39 In addition to controlling for basic noise, we excluded all 88 samples whose sex chromosome log R ratios (LRRs) fell over 3 SDs from the mean of the dataset or whose number of CNVs was more or less than 3 SDs from the mean of the respective genotyping set to remove samples with potentially wide-ranging LCL artifacts or poor-quality genotypes. No unexpected sex chromosome syndromes were identified.
Previous studies have shown that CNV calls from microarrays are sensitive to the algorithms used and the size of the CNV. Using overlapping calls from several algorithms increases the reliability of CNV detection.40, 41 CNVs were included initially if they were >50 kb, identified by at least two of the three algorithms, and spanned a minimum number of probes (10 for Illumina 550K arrays and 15 for Illumina HumanHap Omni1 and Omni2.5 arrays). CNVs that overlapped repetitive or rearranged regions and potential LCL artifacts (such as antibody-coding fragments, T cell receptors, telomeres, centromeres, array or genome gap regions, or segmental duplications encompassing >50% of their length) were excluded.
We used two additional validation steps for the CNV calls: visual inspection from microarray intensity data and qPCR validation for all de novo CNVs. We found that although it was conservative and time consuming, visual inspection of intensity data was still an important step in estimating the reliability of CNV calls. We plotted the LRR and B allele frequency by using custom scripts for all available family members of the CNV carrier. False-negative CNVs were added to the data with the same size and location to match the true-positive calls of family members. We excluded ambiguous calls with no clear CNV signal, most of which were located in pericentromeric regions, telomeres, common CNV regions, or regions close to array and genome gap regions, which often represent non-unique regions of the genome. Our visual-inspection step resulted in the inclusion of 53% of all CNV calls, and we found a false-negative rate of 2% with the automatic calling algorithms. For each AGRE genotyping set, 65%–89% of samples remained after quality control (Figure 1 and Table S1). This is comparable to other studies that used similar array types. Our CNV calls were consistent with published data from an earlier analysis in a subset of the AGRE cohorts;12, 34 one exception was a CNV previously called in a telomeric region because it was too noisy to call in our data. Five individuals included in this previous analysis were excluded from our analysis on the basis of general quality metrics. All CNV calls are available in the National Database for Autism Research (see Accession Numbers).
qPCR Validation
All CNVs that were predicted to be de novo events by CNVision and large recurrent events with uncertain inheritance were validated by qPCR. In addition, a randomly chosen set of inherited events of smaller sizes was included for qPCR validation. All primers were designed with Primer3 default settings. Samples were run in quadruplicate with 1 ng/μL DNA for the CNV and RNaseP reference. Each primer was also run in 0.5 and 1 ng/μL of reference DNA mix from ten unaffected individuals, and primers for duplications were also run in 2 ng/μL of reference DNA mix. The accuracy of visual inspection is clear given that all visually confirmed de novo CNVs passed qPCR testing (15/15) (Table S2); however, only 13/42 (31%) of 50–100 kb de novo CNVs and 34/189 (18%) of <50 kb de novo CNVs that were not confirmed by visual inspection were experimentally confirmed, consistent with previous experience.11
In Situ Hybridization
In situ hybridization was performed with radioactive probes for NR4A2 (MIM: 601828; GenBank: BC009288, base pairs 4–2,175]) as previously described.42, 43, 44 Regional annotation of expression was performed with the Atlas of Human Central Nervous System Development.45
Statistical Analyses
All statistical analyses were performed with R (3.0.2). We used a non-linear mixed model and general regression model with affection status as the response variable. The linear mixed model was also used for testing the association between the de novo CNVs and the Raven's nonverbal IQ score, the Social Responsiveness Scale population-normalized Z score,46 the Vineland Adaptive Behavior Scales, the Stanford-Binet Intelligence Scales, and the Peabody Picture Vocabulary Test. For tests of de novo burden, we included all children from all families in whom two parents had array data. For tests of overall burden, we selected one random affected and one random unaffected sample from each family to control for relatedness. Gene- and locus-based analyses were family based, and only one CNV per affected and unaffected sibling was taken into account for each family.
We used the non-parametric Kruskal-Wallis test47 to estimate differences in all tested CNV categories between different arrays for possible detection bias (Table S2 and Figure S1) and discovered a significant effect. Therefore, we included the genotyping set and the first three components of multidimensional-scaling analysis as covariates to account for array effects and population stratification, respectively. For IQ analyses, we also included sex as a covariate. Regression models were fit with the R package lme4, which includes a function to fit general regression models.48
Power analyses were conducted with the DSS Researcher’s Toolkit’s online two-sample estimation tool for percentage values. We used our current sample size, de novo rates from an analysis of the Simons Simplex Collection (SSC),13 and a 5% alpha rate in our estimations.
Samples from the Database of Genomic Variants
The Database of Genomic Variants (DGV) is a public repository of structural chromosomal variation, and its datasets are not necessarily uniformly curated or checked for overlap. We downloaded the DGV data and only included studies with genome-wide detection methods. All samples were inspected for overlap, and after careful pruning, we were left with 27,263 independent human samples.
Results
Using the CNVision pipeline,11 we applied a CNV-calling approach that is based on integrating three algorithms: PennCNV,38 QuantiSNP,39 and GNOSIS11 (Figure 1). This approach has been shown to increase reliability of CNV detection11, 40, 41 and also permits more direct comparisons with studies that use this same pipeline to analyze SSC families.11, 13 Given the resolution of SNP microarrays and our aim of focusing on events more likely to be reliable and pathogenic in neurodevelopmental disorders, we targeted CNVs of ≥100 kb49 and carefully controlled for biases introduced by array type or population structure. After quality control, our dataset consisted of 1,464 families with 1,764 affected and 572 unaffected children (Figure 1). We used all children from the 1,043 families with data from both parents for the de novo CNV analyses.
Large, Rare CNVs in the AGRE
We found 524 rare (population frequency below 1% in the DGV), large (≥500 kb) inherited or de novo CNVs at 214 non-overlapping loci in the non-repetitive unique regions of the genome. We tested all 15 de novo deletions and 10 of 22 duplications in this class by qPCR, and all were confirmed for a 100% validation rate (Table S3).
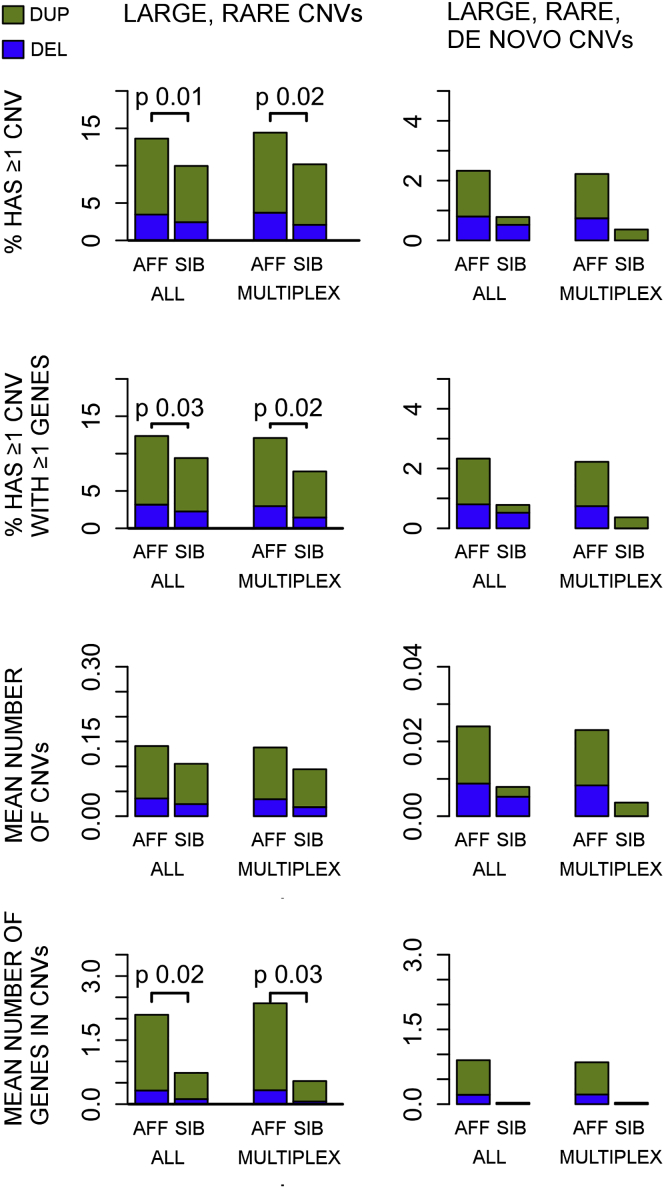
Overall, large, rare CNVs, either inherited or de novo, are more frequent in affected children than in their unaffected siblings (147/1,122 [13.1%] versus 36/402 [8.9%], odds ratio [OR] = 1.7, p = 0.016, logistic regression) (Figure 2 and Tables S4, S5, and S6), as previously observed.11, 13, 22, 23 28% of all large, rare CNVs that we observed in the AGRE are in previously ASD-associated loci (Figure 3A). Consistent with previous studies, we only observed maternal over-transmission of large, rare CNVs at the imprinted 15q11–13 locus, where 7 of 16 such events were maternal, six were de novo, one was paternal, and two were of unknown inheritance because biomaterials were unavailable from the parents.
Figure 2.
Summary of CNV Association Results in the AGRE
Summary of results for all samples combined (simplex and multiplex) and multiplex families only. Detailed sample sizes and summaries by array are available in Table S2. CNVs over 500 kb were considered large, and CNVs with a population frequency below 1% in the DGV were considered rare. CNVs found in a child but not in the parents were considered de novo. Analyses for large, rare CNVs contained inherited or de novo CNVs, whereas de novo analyses contained only de novo CNVs. Only p values smaller than 0.05 are shown in the figure. The fraction of deletions is shown in blue, and the fraction of duplications is shown in green. Abbreviations are as follows: Aff, affected sibling; and Sib, unaffected sibling.
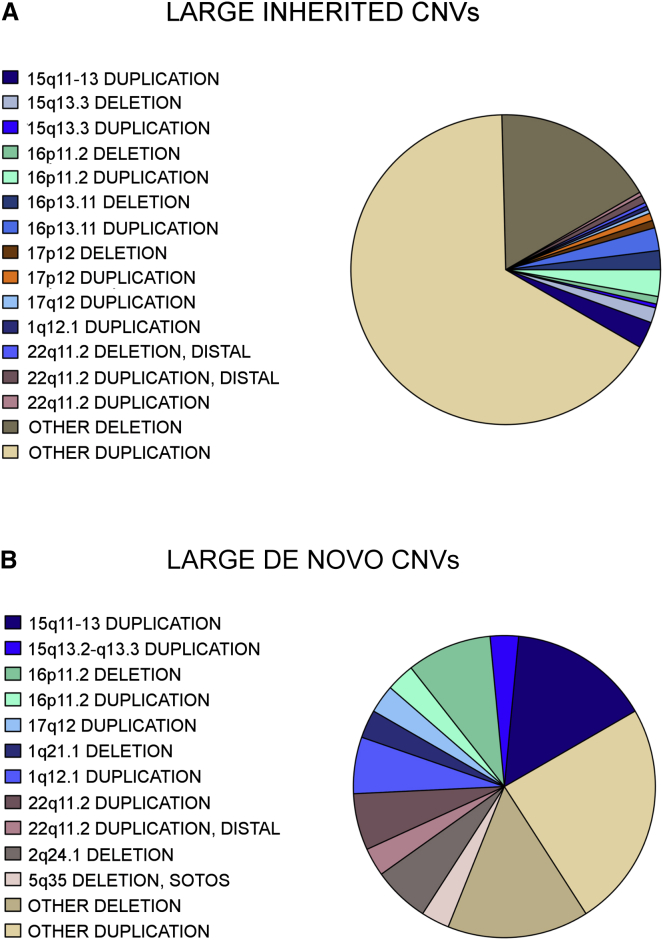
Figure 3.
Distribution of Large, Rare CNVs
The majority of large, rare de novo CNVs in the AGRE are in loci considered to be associated with ASD (55%), and only a minority of the CNVs are loci of unknown pathogenicity (not presently associated with ASD). Inherited CNVs, however, are mostly of unknown pathogenicity, and only 28% are in known ASD-associated loci. Most ASD-associated CNV loci have been identified through the recurrence of de novo events, which would bias our findings toward de novo CNVs. Also, because these loci have large effect sizes on behavior and cognition, we would expect a larger percentage of the CNVs to be de novo at these loci as a result of potential effects on fecundity. Table 1 shows a breakdown of all known ASD-associated loci found in the AGRE, Table S3 shows all large, rare de novo CNVs, and Table S4 shows large, rare inherited CNVs.
Out of a total of 163 non-overlapping large, rare CNV loci identified in either affected or unaffected siblings, 143 were present in affected and 41 were observed in unaffected siblings. 20 of the 143 CNV loci in affected children (13.9%) were not previously associated with ASD and contained one or more additional overlapping CNVs in at least one other affected child, but not in any unaffected sibling in the AGRE. 13 of these 20 were not observed in unaffected siblings in either the SSC or AGRE cohorts and thus are potential candidate ASD risk loci (Table S7). In contrast, none of the loci found in unaffected siblings were restricted to them. Thus, we observed a significant enrichment of these large, rare CNVs as a class in affected siblings (13/143 [13.9%] versus 0/41 [0%], p = 0.041, Fisher’s exact test).
Because the events are rare, present only in a few families, and have a <1% frequency in the DGV, we combined these with published data from the SSC13 to assess their potential association with ASD (Table S7). We found evidence for one locus: CNVs (deletions or duplications) overlapping WWOX (MIM: 605131) were found in affected children in 12 of 3,565 families (0.34%) but in only one unaffected sibling out of 2,633 families (0.04%, p = 0.01, OR = 8.8, Fisher’s exact test) (Figure S2). Published affected individuals from the Autism Genome Project (AGP) did not carry any rare CNVs overlapping WWOX.23 The overall frequency of >100 kb CNVs overlapping WWOX in the DGV is 26/27,263 (0.10%), which is significantly lower than in the ASD individuals. The combined association test for all datasets was nominally significant (p = 0.0148, OR = 2.6, 95% confidence interval [CI] = 1.1–5.4, Fisher’s exact test). Recessive mutations in WWOX have been linked to infantile epileptic encephalopathy, delayed development, and poor eye contact,50 but a history of seizures was reported in only two of the AGRE individuals harboring a CNV that overlaps WWOX (Table S11). Nine of the other large, rare CNVs that were only observed in affected children in the AGRE were also found in affected children from the SSC (and not in unaffected siblings), but none of these loci individually reach statistical significance because of their low frequency. We list these in Table S7 and consider them potential candidate risk loci that require more evidence from future family cohorts. Several of these CNVs contain single genes, including FAM19A, CDH18 (MIM: 603019), CDH13 (MIM: 601364), IGF1R (MIM: 147370), and ARSK (MIM: 610011).
We next assessed the contribution of large, rare de novo CNVs to ASD risk in AGRE multiplex families. We found that siblings with ASD carry at least one large, rare, de novo CNV more frequently (27/1,217 [2.2%]) than their unaffected siblings (1/273 [0.4%]), although this must be considered a trend given that it does not reach our threshold for statistical significance (OR = 7.1, p = 0.06, logistic regression; Figure 2 and Tables S5 and S6). As expected, the enrichment in affected siblings was significantly lower in AGRE than in the SSC families (OR = 6.3; 3.8% in affected siblings versus 0.6% in unaffected SSC siblings),13 consistent with a significantly lower frequency of de novo events in multiplex ASD (p = 0.01, one-sided Fisher’s exact test). We had the power to detect a similar difference for large, rare, de novo CNVs, as seen in the SSC (>95% power to observe 3.7% and 0.8% of ≥500 kb de novo CNVs in affected and unaffected individuals, respectively),11, 13 and therefore this most likely represents a true difference between simplex and multiplex cohorts.
17 of 32 (53%) of all large, de novo events observed in affected children in the AGRE, but not in unaffected siblings, were in loci that have been previously associated with ASD11, 13, 17, 51 (see Table 1, Figure 3B, Table S8, and below). CNVs in these previously identified ASD-associated loci were observed in affected children in 46 of 1,464 (3.1%) families (Table 1 and Table S8). The most frequent recurrent CNV loci in the AGRE were at 15q11–13 (1.1%), 16p13.11 (0.7%), 22q11.2 (0.5%), 16p11.2 (0.4%), and 1q21.1 (0.3%) (Table 1; see also Figure 4).
Table 1.
Large, Rare, Recurrent ASD-Associated CNVs
| Locus | Position (Mb)a | Frequency (n) | Affected | Siblings |
|---|---|---|---|---|
| 1q21.1 | 146.5–147.8 | 0.34% (5) | one de novo deletion; three de novo and two maternal duplications | one paternal duplication |
| 5q35 | 36.2–37.7 | 0.07% (1) | one de novo deletion | not observed |
| 7q11.23 | 72.7–74.1 | 0.07% (1) | one paternal duplication | not observed |
| 15q11–13 | _ | 1.1% (16) | six de novo, seven maternal, one paternal, and two NA2 CNVs | |
| BP2-3b | 23.6–28.9 | 0.68% (10) | four de novo, four maternal, and two NA2 duplications | not observed |
| BP3-5b | 28.9–32.5 | 0.13% (2) | one maternal deletion and one de novo duplication | not observed |
| BP4-5b | 30.9–32.6 | 0.27% (4) | one de novo, two maternal, and one paternal deletion | not observed |
| 16p13.11 | 14.9–16.3 | 0.66% (10) | two maternal and one paternal deletion; one maternal, three paternal, and two NA2 duplications |
one paternal deletion one paternal duplication |
| 16p11.2 | 29.6–30.2 | 0.35% (5) | three de novo deletions; one de novo and one maternal duplication | one maternal duplication |
| 17q12 | 34.8–36.2 | 0.07% (1) | one de novo duplication | not observed |
| 22q11.2 | – | 0.48% (7) | three de novo, one paternal, two maternal, and one NA2 CNVs | |
| Classicb | 18.9–21.5 | 0.27% (4) | two de novo, one paternal, and one NA2 duplication | not observed |
| Distalb | 23.0–25.0 | 0.20% (3) | one de novo and two maternal duplications | not observed |
| Total | – | 3.1% (46) | 19 de novo, 14 maternal, six paternal, and six NA2 CNVs | two maternal and two paternal CNVs |
The table describes the total number (n) of families with known ASD-causing events and how often (frequency) these events occur in affected and unaffected children of these families. Out of a total 1,464 families, 1,398 have affected children, and 539 have unaffected siblings passing quality filters. Fisher’s exact test gave p = 0.02538 for deletions and p = 0.01130 for duplications in affected versus unaffected siblings. The AGRE didn’t contain any of the following known ASD-associated CNVs: 3q29 deletions, 17p11.2 deletions or duplications, 17q11.2 deletions, and 17q21.31 deletions. A significant proportion of the large, ASD-associated events that have a known mode of inheritance are de novo events (37%). This is interesting given that most of the AGRE families have two or more affected children, and the de novo events here are present only in one affected child. The following abbreviation is used: NA, no biomaterials were available from one or more parents.
Position in the human reference sequence (UCSC Genome Browser hg19).
Sub-loci within a larger ASD-associated CNV locus.
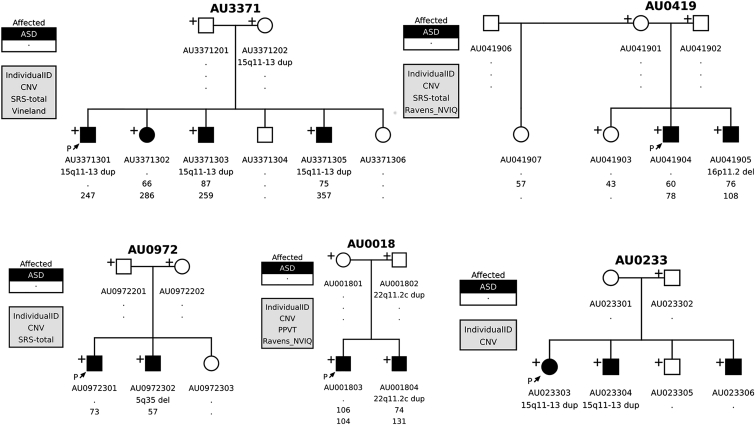
Figure 4.
The Non-segregation of Established ASD Risk CNVs in Multiplex Families
We observed non-segregation of known pathogenic CNVs in multiplex families: all affected children in the same family did not necessarily share the CNV. Most of these (n = 17/21) were de novo CNVs and were present in only one affected child. The cause of ASD in the other affected child remains unknown. Thorough sequencing of all family members will be beneficial for determining the exact phenotypic effects of the pathogenic CNVs and assessing other possible genetic causes of ASD in these families. Plus signs indicate samples with available DNA for testing.
CNVs in ASD-Associated Genes and Candidate Genes
Recent exome sequencing studies in simplex families have identified several genes in which children with ASD carry disruptive de novo point mutations that are associated with the disorder.5, 6, 8, 15, 52, 53 We scrutinized all CNVs in 71 regions and genes with ASD-associated or ASD candidate variants,13 such as NRXN3 (MIM: 600568), down to 100 kb resolution. Because we were interested in gene-disrupting events, we included only CNVs spanning exons, promoter regions, and UTRs in our analyses (Table 2 and Table S8). We identified CNVs, most of which were inherited, in 20 genes that have been observed or suggested to carry most likely disruptive mutations in individuals with ASD. The observed CNVs in affected children in the AGRE included events in ANK2 (MIM: 106410), CNTNAP2 (MIM: 604569), CNTN4 (MIM: 607280), RBFOX1 (MIM: 605104), NRXN1 (MIM: 600565), MACROD2 (MIM: 611567), and AUTS2 (MIM: 607270) (Table 2 and Table S8).
Table 2.
Exon-, UTR-, or Promoter-Overlapping CNVs in ASD-Associated and Candidate Genes
| Genes (MIM) | Families (n) | Families (%) | De Novo |
Inheritance |
Deletions | Duplications | Disrupted Elements | ||
|---|---|---|---|---|---|---|---|---|---|
| Maternal | Paternal | Unknowna | |||||||
| ANK2 (106410)b | 3 | 0.20 | 1 | 0 | 2 | 0 | 1 | 2 | exon, 3′ UTR, promoter |
| ASXL3 (615115) | 1 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | exon |
| AUTS2 (607270)b | 2 | 0.14 | 0 | 2 | 0 | 0 | 1 | 1 | exon, promoter |
| CNTNAP2 (604569)c | 1 | 0.07 | 0 | 0 | 1 | 0 | 1 | 0 | exon |
| CNTN4 (607280) | 5 | 0.34 | 0 | 1 | 3 | 1 | 2 | 3 | exon, promoter |
| CTTNBP2 (609772)b | 1 | 0.07 | 0 | 1 | 0 | 0 | 0 | 1 | exon, 3′ UTR |
| CYFIP1 (606322) | 9 | 0.61 | 2 | 3 | 2 | 2 | 5 | 4 | exon |
| GABRB3 (137192)b | 1 | 0.07 | 0 | 0 | 0 | 1 | 0 | 1 | 5′ UTR |
| GIGYF1 (612064)d | 2 | 0.14 | 0 | 1 | 1 | 0 | 2 | 0 | whole gene, GNB2 (MIM: 139390) |
| KCNMA1 (600150)b,c | 1 | 0.07 | 0 | 1 | 0 | 0 | 0 | 1 | 5′ UTR + |
| MACROD2 (611567)c | 6 | 0.41 | 1 | 2 | 1 | 2 | 6 | 0 | 3′ UTR,4 promoter, 5′ UTR, exon |
| MIB1 (608677)b | 1 | 0.07 | 0 | 0 | 0 | 1 | 0 | 1 | exon, 3′ UTR, miR133 |
| MYO9B (602129)b | 1 | 0.07 | 0 | 0 | 0 | 1 | 1 | 0 | exon, 3′ UTR |
| NCKAP1 (604891) | 1 | 0.07 | 0 | 0 | 0 | 1 | 1 | 0 | whole gene deleted |
| NRXN1 (600565)c | 7 | 0.48 | 0 | 3 | 2 | 2 | 7 | 0 | exon, 5′ UTR, promoter |
| NRXN3 (600568)c | 1 | 0.07 | 0 | 0 | 0 | 1 | 1 | 0 | exon |
| RBFOX1 (605104)c,e | 6 | 0.41 | 1 | 3 | 2 | 0 | 3 | 3 | exon, promoter |
| SLC5A12 (612455) | 1 | 0.07 | 0 | 1 | 0 | 0 | 0 | 1 | exon, 3′ UTR |
| SHANK2 (603290)f | 1 | 0.07 | 0 | 1 | 0 | 0 | 0 | 1 | exon, promoter, DHCR7 (MIM: 602858) |
| Total | 51 | 3.48 | 6 | 19 | 14 | 12 | 32 | 19 | – |
A list of all candidate genes with >50 kb CNVs overlapping a functional region (all events overlap at least one exon, the 5′ UTR, and the promoter or the 3′ UTR). In total, 3.48% of families carry one of these events. Most of the CNVs are inherited.
The parent(s) did not have biomaterials available for genotyping, and the mode of inheritance could not be assessed.
Gene listed as a haploinsufficiency gene in Huang et al.54 No information was available for RBFOX1.
Several families have CNVs in the intronic regions in this gene.
Both parents carry the deletion in a heterozygous state, and the child is a heterozygote. It is not possible to identify the parent of origin.
Families with deletions in the 3′ UTR contain unaffected children with this deletion.
Duplication also encompasses DHCR7. Disruptive mutations in this gene are known to cause ID and Smith-Lemli-Opitz syndrome (MIM: 270400).
We next assessed the frequency of all CNVs affecting these loci in both affected and unaffected siblings in the combined AGRE and SSC collections (Sanders et al.13) and found a higher number of CNVs in this set of ASD-associated genes in affected individuals than in their unaffected siblings (77 in ASD individuals in 3,565 families [2.2%] versus 18 in unaffected siblings in 2,633 families [0.07%], OR = 3.3, 95% CI = 1.9–5.7, p = 9.4 × 10−7) and a signal stronger in deletion carriers (38/3,565 [1.1%] in ASD versus 7/2,633 [0.2%] in unaffected siblings, OR = 4.0, 95% CI = 1.8–10.7, p = 0.0001) than in duplication carriers (Table S8). Most of the CNVs were small to medium sized (range = 57 kb to 4 Mb, median = 270 kb) and restricted to a single gene (Table S8). There was no bias for either maternal or paternal inheritance (21 maternal, 21 paternal, 6 de novo, and 5 with no parental information). Loci containing more than one gene included ANK2 (which was contained within a duplication extending to NEUROG1 [MIM: 601726]), deletions extending to neighboring genes in the CYFIP1 (MIM: 606322) region in several individuals, and a KCNMA1 (MIM: 600150) duplication extending to DLG5. We detected three de novo, non-recurrent CNVs in ASD-associated genes: an exonic 480 kb duplication overlapping CYFIP1 in an affected sibling, a previously identified deletion including the first exon of RBFOX1 in another affected sibling,55 and a 4 Mb duplication including ANK2, a gene harboring recurrent de novo SNV in the SSC, in an unaffected sibling.5, 13, 52
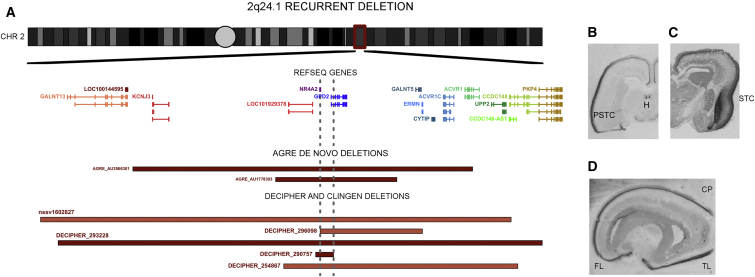
De Novo Deletions at 2q24.1 Are Associated with ASD
Two affected individuals in the AGRE have overlapping large, rare de novo hemizygous deletions at 2q24.1 (Figure 5). CNVs greater than 100 kb are rare at the 2q24.1 locus, given that none have been reported in previous ASD studies or the DGV. We turned to DECIPHER56 and the Clinical Genome Resource (ClinGen), which contain clinically discovered isolated cases, and searched for ≤5 Mb CNVs in 2q24.1; we identified five individuals with 2q24.1 CNVs (Table S9). All were deletions ranging from 170 kb to 4.7 Mb (Figure 5), and all overlapped the deletions in the two ASD individuals in the AGRE. Only two reporting sites had tested the parents for CNVs, and both reported a de novo deletion in the child (Table S9).
Figure 5.
The 2q24.1 Recurrent De Novo Deletion Locus and NR4A2 Expression in the Fetal Brain
(A) AGRE, DECIPHER, and ClinGen CNVs in 2q24.1 all overlap two genes: NR4A2 and GDP2. All samples from DECIPHER or ClinGen were reviewed for additional CNVs, and only samples with no additional ASD risk CNVs were included. The minimal overlap area between gray dashed lines contains all of NR4A2 and the beginning of GPD2. De novo deletions are marked in dark red, and deletions with no information on the mode of inheritance are marked in coral red.
(B–D) Using in situ hybridization, we looked at the localization of NR4A2 expression in fetal brains from 19–20 gestational weeks. Coronal sections (B and C) showed strong expression in the deep layers of the cortical plate (CP; especially in the perisylvian temporal cortex [PSTC]), in the stratified transitional field (STF) (C) that runs through the claustrum and habenula (H) (B) and in the sagittal section (D) of the CP, especially in the frontal (FL) and temporal (TL) lobes.
Both DECIPHER56 and ClinGen, a genomic variant database funded by the National Human Genome Research Institute, are general databases for clinical data and have no restrictions on phenotype. Interestingly, the phenotype information available for four of five samples from the databases and from contact with the primary clinical sites that characterized the individuals revealed that they exhibit both ASD-like symptoms and language delay, similarly to the two affected siblings in the AGRE. More specifically, three of four affected individuals with phenotype data have speech or language delay, and the fourth is reported to have non-specific psychomotor delay. All individuals with the CNV manifest behavioral and/or cognitive symptoms, but only three are described as having either an ASD-like or Sotos-like phenotype (Table S9). Individuals with Sotos syndrome are characterized by pre- and postnatal overgrowth, characteristic facial features, learning disability including delayed language development,57 and sometimes ASD or ASD-like behavior.58 Combining AGRE, SSC (2,591 affected individuals and 2,100 siblings),13 AGP ASD samples (941 non-overlapping affected individuals),23 and filtered DGV data (27,263 population samples) revealed an association between the 2q24.1 deletion and ASD (p = 0.020, Fisher’s exact test). If we combine the ASD individuals with all clinical DECIPHER samples in which one de novo 2q24.1 deletion was observed in 18,819 clinical samples, the signal is stronger (p = 0.0076, Fisher’s exact). Overall, the 2q24.1 deletion is very rare (<0.05% in affected individuals) and thus far appears restricted to de novo events.
The 2q24.1 deletion critical region overlaps two genes (Figure 5A): NR4A2 and GPD2 (MIM: 138430), neither of which has been listed in the list of predicted ASD risk genes by Liu et al.59 However, both mRNAs are bound by FMRP60 and are potentially regulated by CHD8 (MIM: 610528) in human neuronal stem cells, whose regulatory targets are enriched among genes that harbor ASD risk variants.60, 61 Several truncating mutations or deletions have been reported in GPD2 in the Exome Aggregation Consortium (ExAC) Browser and DGV. In contrast, disruptive SNVs are extremely rare in NR4A2 in the ExAC Browser, and no CNVs overlap with NR4A2 in the DGV despite adequate coverage on most microarray platforms. In fact, NR4A2 is among the predicted loss-of-function (LoF)-intolerant (LI) constrained genes in the ExAC Browser (pLI = 0.991)62 and is far less tolerant of truncating and missense variants than most genes (Residual Variation Intolerance Score [RVIS] = −0.63 [17.3rd percentile]),63 whereas GPD2 is predicted to be tolerant of LoF (pLI = 0.000101) and missense and truncating variants (RVIS = 0.22 [68.5th percentile]). These genetic data provide support for mutations in NR4A2 as the more plausible ASD-contributing factor at this two-gene locus.
Orthogonal neurobiological data also highlight the likely role of NR4A2 in human brain development, given that a previous study suggested its involvement in the development of language-related brain regions during human fetal development.44 This is perhaps even more remarkable in combination with the clinical observation that several individuals with the deletion have prominent language delay (Table S9). Therefore, we re-assessed the expression of NR4A2 in the developing human brain, including some new samples along with those from our previous in situ hybridization study.44 We observed strong expression in the claustrum, the habenula, and the deep cortical plate, especially in the frontal and perisylvian temporal regions and in the temporal entorhinal cortex (Figures 5B–5D). The expression pattern reflects the data in the Allen Brain Atlas,64, 65 where NR4A2 is highly expressed in the fetal human posterior, superior temporal gyrus (a region critical for in speech and language),44, 66, 67 and claustrum in the adult human brain.
Intelligence in ASD Individuals Correlates with De Novo CNVs
ID is comorbid with ASD in 31% of cases,68 and recent work has found an association between de novo variants and IQ, especially for protein-disrupting SNVs.5, 7, 30 A similar negative correlation has also been detected between IQ and de novo CNVs, as well as the number of genes contained within de novo CNVs.11, 13
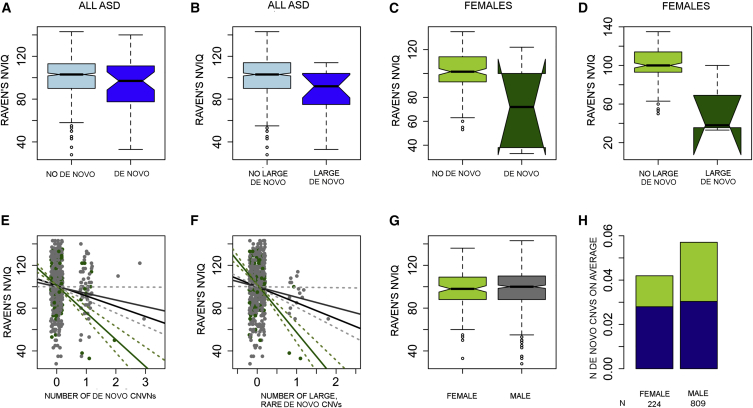
We analyzed the correlation between IQ and de novo CNVs in 142 female and 644 male affected individuals whose Raven's non-verbal IQ (NVIQ) measurements were available. We observed a significant relationship between lower IQ in affected individuals and the number of rare, de novo CNVs (effect size of −10 points per de novo CNV, p = 0.0005, linear regression). However, the signal was stronger for females (effect size of −26 points per de novo CNV, p = 5.8 × 10−5, linear regression) (Figures 6A–6D). No other psychometric tests were associated with CNV status. We then limited the analysis to large, rare, de novo events and observed an even stronger signal in females (effect size of −13.2 for both males and females [p = 0.002] and effect size of −43.1 for females only [p = 0.00004], linear regression) (Figures 6E and 6F). This was not due to a sex difference in IQ distributions because there was no overall difference in the measured Raven's IQ range between affected males and females (Figure 6G). Previous work in simplex families has shown that affected females are enriched with de novo CNVs, but we did not observe this in the AGRE cohort (Figure 6H). This could be due to diminished power, because power was comparable but low in both studies (50.1% [alpha 5%] in AGRE; 52.6% [alpha 5%] in SSC).13
Figure 6.
Raven's NVIQ Is Correlated with the Number of Rare De Novo CNVs in ASD
Overall, the effect of rare de novo CNVs (A) and large, rare de novo CNVs (B) on NVIQ is modest between de novo CNV carriers (light blue) and non-carriers (dark blue), but the effect is stronger in females with de novo CNVs (light green, C) than in females without de novo CNVs (dark green, D). The midline in the boxplots (A–D and E) represents the median, the notches show the 95% confidence interval, the solid box represents the middle 50% of the data points (quantiles two and three), the whiskers extend to show 1.5× the interquartile rage, and the individual dots are outlier values. The number of rare de novo CNVs (E) or large, rare de novo CNVs (F) correlates clearly with NVIQ, more so in females (green) than in males (gray) or the total affected population (black line). SDs are shown with dashed lines. However, there is neither a difference in Raven’s NVIQ between affected females (green) and males (gray) (G) nor an increased number of de novo CNVs in affected females (H) (the portion of large [≥500 kb], rare, de novo CNVs is shown in green, and rare, 100–500 kb de novo CNVs are in blue).
Patterns of Inheritance and Segregation
One expectation that motivates the recruitment of multiplex families for genetic studies is that they will be enriched with inherited factors and subsequently that affected children in these families will share genetic risk factors or a single high-risk event.25, 30, 69, 70, 71 This prediction is supported by a trend toward an excess of inherited rare, large CNVs in affected children in the AGRE (Figure 2 and Tables S5 and S6). In contrast, affected children in simplex families (from the SSC) are enriched with de novo events and do not show a significant signal for the enrichment of rare, inherited CNVs.13 A previous sequencing study in 85 quartet families found that many affected sibling pairs did not share rare, presumed damaging SNVs.33 However, because the overwhelming majority of these variants are of unknown pathogenicity and it is expected that half of any such variants should be different between siblings by chance, the interpretation of these data33 is unclear.
To further investigate assumptions about the genetic architecture of ASD on the basis of family structure, we analyzed the 30 AGRE multiplex families harboring a CNV at an ASD-associated locus, for example, 15q11–13 duplications and 16p11.2 duplications and deletions (Table 1 and Figures S3–S9). We observed that in the majority of AGRE multiplex families affected by these known risk CNVs (21/30 [70%]), a de novo or inherited event was not shared by all affected children. 16 of these de novo CNVs were observed in only one affected individual (76%), whereas the other five were inherited (24%) but did not fully segregate with ASD in the families. To demonstrate this phenomenon, we show multiple examples of CNVs at these loci, including the well-established causal duplication at 15q11–13 (Table 1, Figure 4, Figures S3–S9, and Tables S8 and S10). We observed no consistent effect on severity measures (such as IQ, adaptive functioning, or difficulty in social interactions) to explain the difference between the siblings carrying the known major risk allele and those without it (Table S10).
These results have two potential major interpretations in the case of de novo events: (1) that misfortune has struck twice, and the other affected children harbor undetected de novo events (CNVs or SNVs) or (2) that other inherited factors contribute to ASD in the family. In the latter case, the prediction is that the CNV might also contribute to severity or overall developmental disability but might not be the primary cause of the ASD that is shared among the affected siblings. In other words, we are faced not with the commonly observed phenomenon of reduced penetrance of these major risk alleles but rather with the absence of the risk allele in other affected siblings in the family, even though the families were recruited under the expectation that risk variants would be shared. Such observations would be unlikely under a dominant model, where a single high-risk variant in an affected child is expected to cause ASD in a family.25, 72
To test the likelihood of the dominant single driving genetic factor assumed to be consistent with the structure of AGRE multiplex families,72 we conducted a Bayesian analysis under three priors representing a wide range of distributional assumptions by assuming a single dominant factor with high penetrance. In each case, the current assumption of a single dominant factor is highly unlikely (posterior probability of 1.7 × 10−24). It is evident that in addition to de novo contributions, multiple ASD risk factors could segregate to different affected siblings even within multiplex families with heritable high-penetrance risk variants.
Discussion
Here, we have found that families ascertained for having two or more children with ASD and simplex families from the SSC show distinct patterns of genetic risk: the rate of large, rare de novo CNVs is lower in multiplex families, and there is an increased burden of large, rare inherited CNVs. For the AGRE cohort, we had power to detect a significant difference in the de novo mutation rate, under the assumption that the rates are comparable to those seen in SSC simplex families (>95% power for 3.7% and 0.8% of ≥500 kb de novo CNVs in affected and unaffected siblings, respectively, and >80% power for 2.3% and 0.5% of ≥1 Mb de novo CNVs in affected and unaffected siblings, respectively),11, 13 but neither class of de novo events was significantly increased in affected individuals from AGRE multiplex families. Despite their lower frequency in ASD in multiplex families, de novo CNVs are correlated with lower IQ in both SSC simplex families and AGRE families.55 The correlation is particularly strong in affected females.55 However, unlike in simplex families, the proportion of affected females with de novo CNV is smaller (11/290 [3.8%]) than that of males (58/1,089 [5.3%]) in our data. A similar trend holds for large, rare de novo events (4/290 [1.4%] for females versus 28/1,089 [2.6%] for males) and large, rare, gene-containing CNVs (41/371 [11.1%] for females versus 168/1,393 [12.1%] for males), although these differences do not reach statistical significance.55
Another important observation is that many of the recurrent de novo CNVs identified in previous studies are present as de novo events in affected children in the AGRE as well. This contributes to the remarkable lack of co-inheritance of known ASD-associated CNVs in ASD-affected children in multiplex families. However, this observation is not exclusive to de novo events and also extends to five inherited events, including one duplication at 15q11.2–13.1 and two duplications at 22q11.2. Indeed, non-transmission of a CNV from a parent to an affected child at previously established ASD risk loci was observed in 41% (5/12) of families harboring such loci. These results indicate that affected children in multiplex families harbor a complex burden of risk factors, an important consideration for genetic counseling. The discovery of an ASD-associated de novo CNV in a family does not exclude the possibility that a second affected child in the same family will harbor a different set of genetic and environmental risk factors. This is similar to what has been observed in a smaller sample of 85 families in whom rare exonic variants, mostly of uncertain pathogenicity, showed a similar pattern of discordance in siblings with ASD.33 In the AGRE, the pattern was observed in previously identified ASD risk genes with established pathogenicity, increasing confidence that we are observing discordance for major genetic risk among siblings in multiplex families.
We also identified a risk locus for ASD and language impairment at chromosomal region 2q24.1, whose statistical association we confirmed via combined analysis of multiple cohorts. The clinical manifestations in individuals with a de novo 2q24.1 deletion (Table S9), including ASD or autism-like behavior and language delay, are quite similar, indicating that this CNV causes a distinct clinical syndrome. We also discovered one previous report of a single individual with a hemizygous deletion overlapping NR4A2 and GPD2; this proband displayed intellectual delay and pervasive developmental disorder.73 The authors focused on GPD2 in their follow-up analyses and stated that “NR4A2 is unlikely to cause significant clinical manifestations.”73 On the contrary, we suggest that NR4A2 is a more plausible candidate given its expression in language-related brain regions in both the adult and developing human brains and its intolerance of LoF, missense, and truncating mutations in human populations (e.g., in the ExAC Browser).62, 63 The protein encoded by GPD2 is involved in glycolysis, is broadly localized, and is located at the mitochondrial membrane within cells. It has been suggested to be involved in type II diabetes in rodents, but with the exception of one report of a 2q24.1 deletion overlapping both NR4A2 and GPD2,73 no additional report has suggested a connection between GPD2 and neuropsychiatric or neurodevelopmental disorders. In summary, our observations, when combined with those of previous studies, provide strong statistical support for 2q24.1 deletions as a cause of a highly penetrant form of syndromic ASD consisting of ID, language delay, and ASD-like behavioral and cognitive deficits. As more individuals with this rare mutation are identified and characterized, its penetrance in ASD and the range of its clinical manifestations can be further refined.
Similarly, our observed OR of 8.8 for WWOX suggests that CNVs in this gene are also a potential inherited risk factor for ASD. However, in contrast to NR4A2, WWOX is among the most mutation-tolerant genes according to both Genic Intolerance Score (RVIS = 1.1 [92.1st percentile])63 and the ExAC Browser constraint score (pLI = 1.98 × 10−8),62 indicating that heterozygous mutations in it are unlikely to be strong risk factors. Consistently, affected individuals with a WWOX-overlapping CNV have generally less severe ASD phenotypes and IQs in the mostly normal range (Table S11). Homozygous mutations in WWOX cause an epileptic encephalopathy (MIM: 616211).74, 75 Interestingly, only 2 of 15 (13%) individuals with either a duplication or a deletion in WWOX had a history of seizures: one with febrile seizures and the other with infantile spasms (Table S11). This indicates that the association between WWOX CNVs and ASD is unlikely to be caused by comorbid epilepsy. This locus is reminiscent of the overlap between ASD and other epilepsy risk genes, such SCN2A (MIM: 182390) and GABRB3 (MIM: 137192).52, 76 Further investigation of potential modifier mutations and compound-heterozygous mutations, as well as large replication cohorts, will be needed for better understanding the putative contribution of WWOX to ASD.
Although slightly more than half of the AGRE collection has been included in previous studies (Table S1),17, 19, 20, 22, 23, 24, 32, 34, 77 neither the 2q24.1 de novo deletion nor CNVs in WWOX have been previously associated with ASD. This is not surprising, given that only one sample in the previously published AGRE studies had the 2q24.1 deletion, and neither the SSC nor the AGP contains samples with the 2q24.1 deletion. Although CNVs in WWOX are more common than 2q24.1 deletions, they are still rare, and obtaining adequate power to detect signal will require a combined analysis of two large datasets.
Previous studies in the SSC have observed that females with ASD have more de novo CNVs10, 13 than males, which is consistent with the model whereby a larger genetic “hit” is needed to cause ASD in females.5, 10, 11, 13 Yet, we did not observe this trend in the AGRE. In addition, the CNV burden between mothers and fathers did not reveal a bias toward female carriers of major risk alleles. These observations in the AGRE could reflect sample size or differences in genetic architecture in simplex and multiplex families10 and are consistent with a previously observed higher load of heritable factors in fathers from multiplex ASD-affected families,28 but this needs to be replicated in larger cohorts.
In contrast to some previous studies,22, 24, 78 we did not observe any difference between affected children and control individuals (unaffected siblings or individuals from the general population) for the overall number of duplications and deletions; for gene-containing duplications or deletions; for exonic deletions, exonic duplications, or rare exonic CNVs; or for genes in CNVs in any other size category. Several potential explanations include differences between the genetic architectures in multiplex and simplex families, differences between control siblings and control individuals from the general population, or potential technical confounders introduced in previous studies by differences between platforms used in affected and control individuals (Table S2).21, 22, 78 In the current study, we were careful to control for the array platform used for case-control comparisons, and rather than using unrelated control individuals, we relied on siblings from multiplex families as our comparison set. This comparison set could also have reduced our chances of seeing true-positive signals, especially for inherited variants with small effect sizes and low penetrance. Because the number of families affected by ASD-associated high-risk CNVs is small and microarray platforms have a limited resolution for variant detection, further studies with deep sequencing and extensive phenotyping of multiplex families carrying ASD-associated CNVs are needed for fully assessing the roles of known high-risk ASD-associated factors and assumed inheritance patterns. It is possible that additional shared, strong, and highly penetrant risk factors exist in these families, but this seems unlikely given that a recent whole-genome sequencing study of 85 multiplex families found nearly the same rate of sibling discordance and found few shared risk factors in siblings with ASD.33 Our findings indicate that it will be worthwhile to undertake both larger-scale collection of multiplex families subjected to whole-genome sequencing and larger-scale genome-wide association studies in ASD individuals for low-risk common variation to better define the role of specific heritable factors in ASD susceptibility and how they create risk in individuals.
Acknowledgments
This project received funding from National Institute for Mental Health (NIMH) grants R01 MH081754 and R01 MH100027 (to D.H.G.) and R01 MH074090 (to C.L.M.) and from the Sigrid Jusélius Foundation for Postdoctoral Fellowship (to V.M.L.). We gratefully acknowledge support from the Autism Genetic Resource Exchange (AGRE) and Autism Speaks, the participating AGRE families, and resources provided by the AGRE Consortium, a program of Autism Speaks. The AGRE was supported in part by NIMH grant 1U24MH081810 to Clara M. Lajonchere (principal investigator), as well as previous grant R01 MH064547-01S1 to D.H.G. We gratefully acknowledge the University of California, Los Angeles (UCLA) Department of Statistics and the Institute for Digital Research and Education for statistical and computational support and resources. We thank Brett Abrahams for his work on the in situ hybridizations at UCLA, Jillian Haney for technical assistance, Laura Perez Cano and Neel Parikshak for scripts to ascertain family properties, and Tor Solli-Nowlan for computational support. We acknowledge Erin Kaminsky, Daniel Moreno-De-Luca, Andreas Moreno-De-Luca, and Abby Hare-Harris for technical assistance in the analysis of copy-number-variant pathogenicity in the C.L.M. lab. This study used data generated by the DECIPHER community. A full list of contributing centers is available at http://decipher.sanger.ac.uk and via email at decipher@sanger.ac.uk. Funding for the project was provided by the Wellcome Trust. This study also included data generated by the International Standards for Cytogenomic Arrays Consortium.79 Funding for the project was provided in part by NIH grants HD064525 and MH074090.
Published: August 25, 2016
Footnotes
Supplemental Data include nine figures and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.06.036.
Accession Numbers
The accession number for this study is NDAR: 393 (http://dx.doi.org/10.15154/1238063). The accession numbers for the 2q24.1 deletions reported in this paper are ClinVar: SCV000282106 and SCV000282167. The accession numbers for the ASD-associated CNVs and CNVs in genes with ASD-associated most likely gene-disrupting SNVs reported in this paper are ClinVar: SCV000282087–SCV000282105, SCV000282107–SCV000282151, SCV000282158– SCV000282166, and SCV000282168–SCV000282187.
Web Resources
Autism Genetic Research Exchange (AGRE) clinical information, http://research.agre.org/program/diag.cfm
Clinical Genome Resource (ClinGen) Structural Variation Database Search, https://clinicalgenome.org
Database of Genomic Variants (DGV), http://dgv.tcag.ca/dgv/app/home
DECIPHER, https://decipher.sanger.ac.uk
DSS Research, https://www.dssresearch.com/Home.aspx
DSS Statistical Power calculators, https://www.dssresearch.com/KnowledgeCenter/ToolkitCalculators/StatisticalPowerCalculators
Internet System of Accessing Autistic Children, http://www.autismtools.org
National Database for Autism Research (NDAR), https://ndar.nih.gov/
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
References
- 1.Robinson E.B., Koenen K.C., McCormick M.C., Munir K., Hallett V., Happé F., Plomin R., Ronald A. A multivariate twin study of autistic traits in 12-year-olds: testing the fractionable autism triad hypothesis. Behav. Genet. 2012;42:245–255. doi: 10.1007/s10519-011-9500-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sandin S., Lichtenstein P., Kuja-Halkola R., Larsson H., Hultman C.M., Reichenberg A. The familial risk of autism. JAMA. 2014;311:1770–1777. doi: 10.1001/jama.2014.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colvert E., Tick B., McEwen F., Stewart C., Curran S.R., Woodhouse E., Gillan N., Hallett V., Lietz S., Garnett T. Heritability of Autism Spectrum Disorder in a UK Population-Based Twin Sample. JAMA Psychiatry. 2015;72:415–423. doi: 10.1001/jamapsychiatry.2014.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tick B., Bolton P., Happé F., Rutter M., Rijsdijk F. Heritability of autism spectrum disorders: a meta-analysis of twin studies. J. Child Psychol. Psychiatry. 2016;57:585–595. doi: 10.1111/jcpp.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iossifov I., Ronemus M., Levy D., Wang Z., Hakker I., Rosenbaum J., Yamrom B., Lee Y.H., Narzisi G., Leotta A. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neale B.M., Kou Y., Liu L., Ma’ayan A., Samocha K.E., Sabo A., Lin C.F., Stevens C., Wang L.S., Makarov V. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders S.J., Murtha M.T., Gupta A.R., Murdoch J.D., Raubeson M.J., Willsey A.J., Ercan-Sencicek A.G., DiLullo N.M., Parikshak N.N., Stein J.L. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itsara A., Wu H., Smith J.D., Nickerson D.A., Romieu I., London S.J., Eichler E.E. De novo rates and selection of large copy number variation. Genome Res. 2010;20:1469–1481. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy D., Ronemus M., Yamrom B., Lee Y.H., Leotta A., Kendall J., Marks S., Lakshmi B., Pai D., Ye K. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Sanders S.J., Ercan-Sencicek A.G., Hus V., Luo R., Murtha M.T., Moreno-De-Luca D., Chu S.H., Moreau M.P., Gupta A.R., Thomson S.A. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yoon S., Krasnitz A., Kendall J. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders S.J., He X., Willsey A.J., Ercan-Sencicek A.G., Samocha K.E., Cicek A.E., Murtha M.T., Bal V.H., Bishop S.L., Dong S., Autism Sequencing Consortium Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 15.Krumm N., Turner T.N., Baker C., Vives L., Mohajeri K., Witherspoon K., Raja A., Coe B.P., Stessman H.A., He Z.X. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marshall C.R., Noor A., Vincent J.B., Lionel A.C., Feuk L., Skaug J., Shago M., Moessner R., Pinto D., Ren Y. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreno-De-Luca D., Sanders S.J., Willsey A.J., Mulle J.G., Lowe J.K., Geschwind D.H., State M.W., Martin C.L., Ledbetter D.H. Using large clinical data sets to infer pathogenicity for rare copy number variants in autism cohorts. Mol. Psychiatry. 2013;18:1090–1095. doi: 10.1038/mp.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mefford H.C., Sharp A.J., Baker C., Itsara A., Jiang Z., Buysse K., Huang S., Maloney V.K., Crolla J.A., Baralle D. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N. Engl. J. Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glessner J.T., Wang K., Cai G., Korvatska O., Kim C.E., Wood S., Zhang H., Estes A., Brune C.W., Bradfield J.P. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller D.T., Shen Y., Weiss L.A., Korn J., Anselm I., Bridgemohan C., Cox G.F., Dickinson H., Gentile J., Harris D.J. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J. Med. Genet. 2009;46:242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girirajan S., Johnson R.L., Tassone F., Balciuniene J., Katiyar N., Fox K., Baker C., Srikanth A., Yeoh K.H., Khoo S.J. Global increases in both common and rare copy number load associated with autism. Hum. Mol. Genet. 2013;22:2870–2880. doi: 10.1093/hmg/ddt136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinto D., Delaby E., Merico D., Barbosa M., Merikangas A., Klei L., Thiruvahindrapuram B., Xu X., Ziman R., Wang Z. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 2014;94:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinto D., Pagnamenta A.T., Klei L., Anney R., Merico D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Girirajan S., Dennis M.Y., Baker C., Malig M., Coe B.P., Campbell C.D., Mark K., Vu T.H., Alkan C., Cheng Z. Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder. Am. J. Hum. Genet. 2013;92:221–237. doi: 10.1016/j.ajhg.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaugler T., Klei L., Sanders S.J., Bodea C.A., Goldberg A.P., Lee A.B., Mahajan M., Manaa D., Pawitan Y., Reichert J. Most genetic risk for autism resides with common variation. Nat. Genet. 2014;46:881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Constantino J.N., Zhang Y., Frazier T., Abbacchi A.M., Law P. Sibling recurrence and the genetic epidemiology of autism. Am. J. Psychiatry. 2010;167:1349–1356. doi: 10.1176/appi.ajp.2010.09101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerdts J.A., Bernier R., Dawson G., Estes A. The broader autism phenotype in simplex and multiplex families. J. Autism Dev. Disord. 2013;43:1597–1605. doi: 10.1007/s10803-012-1706-6. [DOI] [PubMed] [Google Scholar]
- 28.Klei L., Sanders S.J., Murtha M.T., Hus V., Lowe J.K., Willsey A.J., Moreno-De-Luca D., Yu T.W., Fombonne E., Geschwind D. Common genetic variants, acting additively, are a major source of risk for autism. Mol. Autism. 2012;3:9. doi: 10.1186/2040-2392-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Virkud Y.V., Todd R.D., Abbacchi A.M., Zhang Y., Constantino J.N. Familial aggregation of quantitative autistic traits in multiplex versus simplex autism. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2009;150B:328–334. doi: 10.1002/ajmg.b.30810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ronemus M., Iossifov I., Levy D., Wigler M. The role of de novo mutations in the genetics of autism spectrum disorders. Nat. Rev. Genet. 2014;15:133–141. doi: 10.1038/nrg3585. [DOI] [PubMed] [Google Scholar]
- 31.Hoffmann T.J., Windham G.C., Anderson M., Croen L.A., Grether J.K., Risch N. Evidence of reproductive stoppage in families with autism spectrum disorder: a large, population-based cohort study. JAMA Psychiatry. 2014;71:943–951. doi: 10.1001/jamapsychiatry.2014.420. [DOI] [PubMed] [Google Scholar]
- 32.Matsunami N., Hadley D., Hensel C.H., Christensen G.B., Kim C., Frackelton E., Thomas K., da Silva R.P., Stevens J., Baird L. Identification of rare recurrent copy number variants in high-risk autism families and their prevalence in a large ASD population. PLoS ONE. 2013;8:e52239. doi: 10.1371/journal.pone.0052239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuen R.K., Thiruvahindrapuram B., Merico D., Walker S., Tammimies K., Hoang N., Chrysler C., Nalpathamkalam T., Pellecchia G., Liu Y. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat. Med. 2015;21:185–191. doi: 10.1038/nm.3792. [DOI] [PubMed] [Google Scholar]
- 34.Bucan M., Abrahams B.S., Wang K., Glessner J.T., Herman E.I., Sonnenblick L.I., Alvarez Retuerto A.I., Imielinski M., Hadley D., Bradfield J.P. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trager E.H., Khanna R., Marrs A., Siden L., Branham K.E., Swaroop A., Richards J.E. Madeline 2.0 PDE: a new program for local and web-based pedigree drawing. Bioinformatics. 2007;23:1854–1856. doi: 10.1093/bioinformatics/btm242. [DOI] [PubMed] [Google Scholar]
- 36.Lajonchere C.M., AGRE Consortium Changing the landscape of autism research: the autism genetic resource exchange. Neuron. 2010;68:187–191. doi: 10.1016/j.neuron.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang K., Zhang H., Ma D., Bucan M., Glessner J.T., Abrahams B.S., Salyakina D., Imielinski M., Bradfield J.P., Sleiman P.M. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K., Li M., Hadley D., Liu R., Glessner J., Grant S.F., Hakonarson H., Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colella S., Yau C., Taylor J.M., Mirza G., Butler H., Clouston P., Bassett A.S., Seller A., Holmes C.C., Ragoussis J. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seiser E.L., Innocenti F. Hidden Markov Model-Based CNV Detection Algorithms for Illumina Genotyping Microarrays. Cancer Inform. 2015;13(Suppl 7):77–83. doi: 10.4137/CIN.S16345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winchester L., Yau C., Ragoussis J. Comparing CNV detection methods for SNP arrays. Brief. Funct. Genomics Proteomics. 2009;8:353–366. doi: 10.1093/bfgp/elp017. [DOI] [PubMed] [Google Scholar]
- 42.Abu-Khalil A., Fu L., Grove E.A., Zecevic N., Geschwind D.H. Wnt genes define distinct boundaries in the developing human brain: implications for human forebrain patterning. J. Comp. Neurol. 2004;474:276–288. doi: 10.1002/cne.20112. [DOI] [PubMed] [Google Scholar]
- 43.Easterday M.C., Dougherty J.D., Jackson R.L., Ou J., Nakano I., Paucar A.A., Roobini B., Dianati M., Irvin D.K., Weissman I.L. Neural progenitor genes. Germinal zone expression and analysis of genetic overlap in stem cell populations. Dev. Biol. 2003;264:309–322. doi: 10.1016/j.ydbio.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 44.Abrahams B.S., Tentler D., Perederiy J.V., Oldham M.C., Coppola G., Geschwind D.H. Genome-wide analyses of human perisylvian cerebral cortical patterning. Proc. Natl. Acad. Sci. USA. 2007;104:17849–17854. doi: 10.1073/pnas.0706128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bayer S.A., Altman J. CRC Press; 2002. Atlas of Human Central Nervous System Development. [Google Scholar]
- 46.Lowe J.K., Werling D.M., Constantino J.N., Cantor R.M., Geschwind D.H. Social responsiveness, an autism endophenotype: genomewide significant linkage to two regions on chromosome 8. Am. J. Psychiatry. 2015;172:266–275. doi: 10.1176/appi.ajp.2014.14050576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kruskal W.H., Wallis W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952;47:583–621. [Google Scholar]
- 48.Bates D., Machler M., Bolker B.M., Walker S.C. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015;67:1–48. [Google Scholar]
- 49.Girirajan S., Brkanac Z., Coe B.P., Baker C., Vives L., Vu T.H., Shafer N., Bernier R., Ferrero G.B., Silengo M. Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet. 2011;7:e1002334. doi: 10.1371/journal.pgen.1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mignot C., Lambert L., Pasquier L., Bienvenu T., Delahaye-Duriez A., Keren B., Lefranc J., Saunier A., Allou L., Roth V. WWOX-related encephalopathies: delineation of the phenotypical spectrum and emerging genotype-phenotype correlation. J. Med. Genet. 2015;52:61–70. doi: 10.1136/jmedgenet-2014-102748. [DOI] [PubMed] [Google Scholar]
- 51.Moreno-De-Luca D., Mulle J.G., Kaminsky E.B., Sanders S.J., Myers S.M., Adam M.P., Pakula A.T., Eisenhauer N.J., Uhas K., Weik L., SGENE Consortium. Simons Simplex Collection Genetics Consortium. GeneSTAR Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am. J. Hum. Genet. 2010;87:618–630. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., Levy R., Ko A., Lee C., Smith J.D. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang N., Lee I., Marcotte E.M., Hurles M.E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin C.L., Duvall J.A., Ilkin Y., Simon J.S., Arreaza M.G., Wilkes K., Alvarez-Retuerto A., Whichello A., Powell C.M., Rao K. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- 56.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ball L.J., Sullivan M.D., Dulany S., Stading K., Schaefer G.B. Speech-language characteristics of children with Sotos syndrome. Am. J. Med. Genet. A. 2005;136A:363–367. doi: 10.1002/ajmg.a.30799. [DOI] [PubMed] [Google Scholar]
- 58.Tatton-Brown K., Douglas J., Coleman K., Baujat G., Cole T.R., Das S., Horn D., Hughes H.E., Temple I.K., Faravelli F., Childhood Overgrowth Collaboration Genotype-phenotype associations in Sotos syndrome: an analysis of 266 individuals with NSD1 aberrations. Am. J. Hum. Genet. 2005;77:193–204. doi: 10.1086/432082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu L., Lei J., Sanders S.J., Willsey A.J., Kou Y., Cicek A.E., Klei L., Lu C., He X., Li M. DAWN: a framework to identify autism genes and subnetworks using gene expression and genetics. Mol. Autism. 2014;5:22. doi: 10.1186/2040-2392-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darnell J.C., Van Driesche S.J., Zhang C., Hung K.Y., Mele A., Fraser C.E., Stone E.F., Chen C., Fak J.J., Chi S.W. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cotney J., Muhle R.A., Sanders S.J., Liu L., Willsey A.J., Niu W., Liu W., Klei L., Lei J., Yin J. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat. Commun. 2015;6:6404. doi: 10.1038/ncomms7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., van de Lagemaat L.N., Smith K.A., Ebbert A., Riley Z.L. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller J.A., Ding S.L., Sunkin S.M., Smith K.A., Ng L., Szafer A., Ebbert A., Riley Z.L., Royall J.J., Aiona K. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dikker S., Silbert L.J., Hasson U., Zevin J.D. On the same wavelength: predictable language enhances speaker-listener brain-to-brain synchrony in posterior superior temporal gyrus. J. Neurosci. 2014;34:6267–6272. doi: 10.1523/JNEUROSCI.3796-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mesgarani N., Cheung C., Johnson K., Chang E.F. Phonetic feature encoding in human superior temporal gyrus. Science. 2014;343:1006–1010. doi: 10.1126/science.1245994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Developmental Disabilities Monitoring Network Surveillance Year 2010 Principal Investigators. Centers for Disease Control and Prevention (CDC) Prevalence of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill. Summ. 2014;63:1–21. [PubMed] [Google Scholar]
- 69.Philippe A., Martinez M., Guilloud-Bataille M., Gillberg C., Råstam M., Sponheim E., Coleman M., Zappella M., Aschauer H., Van Maldergem L. Genome-wide scan for autism susceptibility genes. Paris Autism Research International Sibpair Study. Hum. Mol. Genet. 1999;8:805–812. doi: 10.1093/hmg/8.5.805. [DOI] [PubMed] [Google Scholar]
- 70.Risch N., Spiker D., Lotspeich L., Nouri N., Hinds D., Hallmayer J., Kalaydjieva L., McCague P., Dimiceli S., Pitts T. A genomic screen of autism: evidence for a multilocus etiology. Am. J. Hum. Genet. 1999;65:493–507. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.International Molecular Genetic Study of Autism Consortium A full genome screen for autism with evidence for linkage to a region on chromosome 7q. Hum. Mol. Genet. 1998;7:571–578. doi: 10.1093/hmg/7.3.571. [DOI] [PubMed] [Google Scholar]
- 72.Zhao X., Leotta A., Kustanovich V., Lajonchere C., Geschwind D.H., Law K., Law P., Qiu S., Lord C., Sebat J. A unified genetic theory for sporadic and inherited autism. Proc. Natl. Acad. Sci. USA. 2007;104:12831–12836. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barge-Schaapveld D.Q., Ofman R., Knegt A.C., Alders M., Höhne W., Kemp S., Hennekam R.C. Intellectual disability and hemizygous GPD2 mutation. Am. J. Med. Genet. A. 2013;161A:1044–1050. doi: 10.1002/ajmg.a.35873. [DOI] [PubMed] [Google Scholar]
- 74.Szymańska K., Szczałuba K., Lugowska A., Obersztyn E., Radkowski M., Nowakowska B.A., Kuśmierska K., Tryfon J., Demkow U. The analysis of genetic aberrations in children with inherited neurometabolic and neurodevelopmental disorders. BioMed Res. Int. 2014;2014:424796. doi: 10.1155/2014/424796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Valduga M., Philippe C., Lambert L., Bach-Segura P., Schmitt E., Masutti J.P., François B., Pinaud P., Vibert M., Jonveaux P. WWOX and severe autosomal recessive epileptic encephalopathy: first case in the prenatal period. J. Hum. Genet. 2015;60:267–271. doi: 10.1038/jhg.2015.17. [DOI] [PubMed] [Google Scholar]
- 76.Allen A.S., Berkovic S.F., Cossette P., Delanty N., Dlugos D., Eichler E.E., Epstein M.P., Glauser T., Goldstein D.B., Han Y., Epi4K Consortium. Epilepsy Phenome/Genome Project De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morrow E.M., Yoo S.Y., Flavell S.W., Kim T.K., Lin Y., Hill R.S., Mukaddes N.M., Balkhy S., Gascon G., Hashmi A. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Prasad A., Merico D., Thiruvahindrapuram B., Wei J., Lionel A.C., Sato D., Rickaby J., Lu C., Szatmari P., Roberts W. A discovery resource of rare copy number variations in individuals with autism spectrum disorder. G3 (Bethesda) 2012;2:1665–1685. doi: 10.1534/g3.112.004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kaminsky E.B., Kaul V., Paschall J., Church D.M., Bunke B., Kunig D., Moreno-De-Luca D., Moreno-De-Luca A., Mulle J.G., Warren S.T. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet. Med. 2011;13:777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.