Abstract
The origin of Tibetans remains one of the most contentious puzzles in history, anthropology, and genetics. Analyses of deeply sequenced (30×–60×) genomes of 38 Tibetan highlanders and 39 Han Chinese lowlanders, together with available data on archaic and modern humans, allow us to comprehensively characterize the ancestral makeup of Tibetans and uncover their origins. Non-modern human sequences compose ∼6% of the Tibetan gene pool and form unique haplotypes in some genomic regions, where Denisovan-like, Neanderthal-like, ancient-Siberian-like, and unknown ancestries are entangled and elevated. The shared ancestry of Tibetan-enriched sequences dates back to ∼62,000–38,000 years ago, predating the Last Glacial Maximum (LGM) and representing early colonization of the plateau. Nonetheless, most of the Tibetan gene pool is of modern human origin and diverged from that of Han Chinese ∼15,000 to ∼9,000 years ago, which can be largely attributed to post-LGM arrivals. Analysis of ∼200 contemporary populations showed that Tibetans share ancestry with populations from East Asia (∼82%), Central Asia and Siberia (∼11%), South Asia (∼6%), and western Eurasia and Oceania (∼1%). Our results support that Tibetans arose from a mixture of multiple ancestral gene pools but that their origins are much more complicated and ancient than previously suspected. We provide compelling evidence of the co-existence of Paleolithic and Neolithic ancestries in the Tibetan gene pool, indicating a genetic continuity between pre-historical highland-foragers and present-day Tibetans. In particular, highly differentiated sequences harbored in highlanders’ genomes were most likely inherited from pre-LGM settlers of multiple ancestral origins (SUNDer) and maintained in high frequency by natural selection.
Keywords: Tibetan, Sherpa, genetic history, next-generation sequencing, archaic ancestry, Denisovan, Neanderthal
Introduction
Current knowledge of the origin and population history of Tibetan highlanders is still very much in its infancy and controversial. In particular, two key questions remain unsolved: (1) who did Tibetans descend from, and (2) how long have human beings been living at the Tibetan Plateau? Both archaeological and genetic studies have documented a human presence on the plateau as early as 30,000 years before present (YBP).1, 2 Linguistic studies have suggested that the Tibetan and Chinese people share a common root ancestor and that the Tibetan-Chinese split took place ∼6,000 YBP.3 A recent genetic study utilizing exome sequencing data estimated a divergence time of 2,750 years between Tibetans and Han Chinese.4 Existing archaeological and genetic data are not sufficient to address the discrepancy between times estimated by different studies. Analysis of Y chromosome and mitochondrial DNA (mtDNA) has demonstrated the co-existence of both Paleolithic and Neolithic ancestries in the paternal and maternal lineages of present-day Tibetans5, 6 but has not explicitly determined whether those Paleolithic lineages originated from anatomically modern human (AMH) or non-AMH (archaic hominid) species. Further investigation is required to confirm whether there is a genetic continuity, or merely a continuity of culture, between the pre-historical and present-day populations of Tibetans.
A recent study based on sequencing a single gene in Tibetans suggested that a particular haplotype motif consisting of five SNPs surrounding EPAS1 (MIM: 603349) was introgressed from Denisovans, an extinct non-modern human species.7 This indicated the existence of a non-AMH-derived sequence in Tibetan genomes. Nonetheless, the extent to which the non-AMH sequences contribute to the genetic makeup is unknown. Neither is it clear where the non-AMH sequences in Tibetans originated. Were these sequences introduced indirectly by recent migrations of populations with both AMH and non-AMH ancestries? Or were the archaic sequences inherited directly from early Tibetan Plateau inhabitants and are thus independent of those found in other East Asian populations? Answering these questions is indispensable for understanding the demographic history of Tibetans and elucidating the mechanism of their adaptation to high-altitude environments.
Existing archaeological and genetic data are not sufficient to address the fundamental questions mentioned above. This manuscript describes a whole-genome deep-sequencing effort of Tibetan highlanders and samples of Han Chinese, a lowland-residing group that shares close ancestry with the Tibetan people. A careful analysis of these genomes, together with available data on archaic and modern humans, can provide further insights into the genetic prehistory and adaptations of Tibetans.
Material and Methods
Populations and Samples
Peripheral-blood samples of 33 Tibetans (TBN) and 5 Sherpas (SHP) were collected from six prefectures (Lhasa, Chamdo, Nagqu, Nyingchi, Shannan, and Shigatse) in the Tibet Autonomous Region (Figure S1 and Table S1), and blood samples of 39 Han Chinese (HAN) were collected from diverse regions in China (Figure S1 and Table S1). Each individual was the offspring of a non-consanguineous marriage of members of the same nationality within three generations. All samples were collected with informed consent and approved by the Biomedical Research Ethics Committee of the Shanghai Institutes for Biological Sciences. Prior to sequencing and analysis, all samples were stripped of personal identifiers (if any existed). All procedures were in accordance with the ethical standards of the Responsible Committee on Human Experimentation (approved by the Biomedical Research Ethics Committee of the Shanghai Institutes for Biological Sciences) and the Helsinki Declaration of 1975 (revised in 2000).
Genome Sequencing and Data Processing
Whole-genome sequencing, with high target coverage (30×–60×) for 150 bp paired-end reads, was carried out on an Illumina HiSeq X Ten according to Illumina-provided protocols with standard library preparation at WuXi NextCODE (Shanghai). Each sample was run on a unique lane with at least 90 GB of data that had passed filtering, and read data were quality controlled so that 80% of the bases achieved at least a base quality score of 30. Reads were merged, adaptor trimmed, and mapped to the human reference genome (GRCh37) with the Burrows-Wheeler Aligner.8 Variant calling was carried out with the HaplotypeCaller module in the Genome Analysis Toolkit (GATK).9, 10
We filtered the results of joint variant calling of HAN and TIB (TBN and SHP) samples and retained autosomal single-nucleotide variants (SNVs) that (1) were bi-allelic in HAN and TIB and (2) had a missing rate less than 20% in any one of the HAN, TBN, and SHP populations. Finally, we were left with 11,452,436 SNVs for estimating minor allele frequency, heterozygosity, inbreeding coefficient, and runs of homozygosity, etc. Individual heterozygosity, identity by state, identity by descent, and inbreeding coefficient were estimated with PLINK v.1.07.11
To determine the ancestral and derived allele of each variant obtained from our sequenced genomes and the public data integrated in our analysis, we downloaded the ancestral sequences from the 1000 Genomes Project; these were inferred from six primates in the Enredo-Pecan-Ortheus (EPO) pipeline. We employed BEDTools v.2.24.012, 13 to get the ancestral allele for each locus in the variant call format (VCF) files from the ancestral sequences. In the analyses related to the derived allele states, we only chose the loci with an ancestral allele (A, C, T, and G or a, c, t, and g) matching the “REF” or “ALT” allele in the VCF files. A SNP’s ancestral state was not used if the SNP mapped to more than one genomic region. Finally, 11,021,340 SNVs with an unambiguously derived allele remained for comparison of derived-allele frequency.
Public and Published Data
Ancient genomes used in the analysis included those of an Altai Neanderthal,14 a Denisovan,15 and Ust’-Ishim, a 45,000-year-old modern human from Siberia.16 The Affymetrix Human Origins genotyping dataset17 and variants from phase 3 of the 1000 Genomes Project18 were also included in the data analysis of this study (see Tables S3 and S4).
Determining Y Chromosomal and mtDNA Lineages
Y chromosomal and mtDNA haplogroups were determined on the basis of the key mutations commonly used for nomenclature. We developed an algorithm to search all possible combinations of the key mutations used for nomenclature from our sequence data to determine the fine-scale paternal and maternal haplogroup affiliated with each sample (Tables S5 and S6). To compare the frequency distribution of worldwide populations (because fine-scale sub-lineages of most worldwide populations were not available), we combined sub-lineages under each major paternal or maternal haplogroup.
Estimating Genetic Differences between Populations
Genetic differences between populations were measured with FST according to Weir and Cockerham;19 this metric accounts for differences in the sample size in each population. The SNP-specific FST between Tibetan and Han Chinese for each SNP was also calculated with the same formula. Confidence intervals of the FST over loci were calculated by bootstrap resampling with 1,000 replications. To reduce the influence of large sample-size differences between populations, we discarded HuOrigin populations17 with a sample size smaller than five for pairwise comparison; these included Himba (4), Kikuyu (4), Datog (3), Oromo (4), Italian_South (1), Canary_Islanders (2), Saami_WGA (1), Scottish (4), Dolgan (3), Tlingit (4), Australian (3), Chane (1), Chilote (4), Inga (2), Piapoco (4), Ticuna (1), and Wayuu (1). We also discarded SNPs with a missing rate larger than 20% in any single population. This left us with 563,816 autosomal SNPs, which we analyzed to estimate pairwise FST for 188 populations.
Principal-Component Analysis
Principal-component analysis (PCA) was performed at the individual level with EIGENSOFT v.3.0.20 To investigate fine-scale population structure and individual genetic affinities, we performed a series of PCAs by gradually removing “outliers” on the basis of a plot of the first and second principal components (PCs) and re-analyzing the remaining samples on the basis of the same set of SNV markers.
Using Outgroup f3 Statistics to Test Ancient Ancestry
We assessed the genetic relatedness between ancient and TIB genomes by computing outgroup f3 statistics; this method is insensitive to genetic drift and less affected by sample-size bias when ancient genomes are involved.21
We computed f3(X; ancient, African) according to Reich et al.21 to find the admixture signal from ancient genomes in modern human populations. Ancient genomes used in the analysis included those of an Altai Neanderthal,14 a Denisovan,15 and Ust’-Ishim.16 BAM files of the ancient genomes were downloaded from the respective references and managed with Picard v.1.112, and the genotypes were called with GATK v.3.3.9 Data on DNA polymorphisms were stored in VCF files and managed with VCFtools.22 X represents worldwide non-African modern human populations (23 Native American, 23 Central Asian and Siberian, 25 East Asian, 4 Oceanian, 22 South Asian, and 58 West Eurasian). A significantly negative f3(X; ancient, YRI [Yoruba in Ibadan, Nigeria]) (|Z| > 2) suggests ancient gene introgression. There is no significant admixture signal from any of the ancient genomes (Altai Neanderthal, Denisovan, or Ust’-Ishim), given that all f3 values are significantly positive (|Z| > 2). Therefore, these analyses do not provide clear insight into the ancient admixture in modern humans, possibly because the analysis design of f3(X; ancient, African) is not powerful enough or the data are insufficient, for example, the admixture proportion from ancient genomes is small or admixture occurred too long ago.
Further, we used an “outgroup” f3 statistic, f3(African; ancient, X), according to Raghavan et al.23 to examine the relationship between modern human populations and ancient genomes (Altai Neanderthal, Denisovan, and Ust’-Ishim). Without gene flow from an ancient population, f3(YRI; ancient, X) has the same value for a different X. A larger f3 value suggests more allele sharing with ancient individuals and ancient gene introgression.
Estimation of Ne and Divergence Time
We applied pairwise sequentially Markovian coalescent (PSMC) analysis24 and multiple sequentially Markovian coalescent (MSMC) analysis25 to infer the long-term effective population sizes (Ne) and time of divergence (Tdivergence) from the high-coverage genomes. PSMC analysis was implemented according to the original procedure.25 For MSMC analysis, rather than phasing the data from bamCaller.py directly, we used only those SNVs with allele states identical to those of genotypes called by bamCaller.py from the VCF results from GATK analysis, and we phased the data with SHAPEIT2.26
Our estimation of effective population size over time and that of divergence times and time to the most recent common ancestor (TMRCA) in this study depends on estimates of the germline mutation rate (μ). To overcome uncertain date estimates due to a lack of confidence in the true value of the mutation rate, we scaled time in units of 2μT and effective population size in units of 2μNe × 103 (where μ is the mutation rate, Ne is the effective population size, and T is time in generations). We also calculated absolute estimation by assuming a fast mutation rate of 1.0 × 10−9 per site per year (the commonly used phylogenetic mutation rate, i.e., ∼2.5 × 10−8 per base per human generation for a generation time of 25 years)27 or a slow mutation rate of 0.5 × 10−9 per site per year (∼1.25 × 10−8 per base per human generation for a generation time of 25 years).28, 29, 30 In most occasions, we adopted a slow mutation rate (0.5 × 10−9 per site per year) because results based on the slow mutation rate agree better with the paleoanthropological record and with estimates from mtDNA.31
To study the influence of the non-modern human sequences on the estimation of Ne and divergence time, we removed non-AMH sequences from the genomes used for analysis. To facilitate the analysis and avoid any bias of the fluctuation of sample size (or haplotype number) during removal of non-modern human sequences, we directly masked the genomic regions on the basis of S∗ results with a signature of non-modern human ancestry identified in any of the genomes for analysis. Therefore, we removed genomic regions with non-modern human ancestry rather than partially removing haplotypes.
Ancestry Analysis with ADMIXTURE
The model-based ancestry-estimation method ADMIXTURE32 was applied on the merged HuOrigin, TIB, and HAN dataset consisting of 2,422 individuals from 205 worldwide populations. ADMIXTURE, which infers individual genetic-ancestry coefficients by conditioning on a specific number (K) of “ancestral populations,” is a useful tool for analyzing population structure. We used it mainly to identify the structure of TIB and HAN in the context of worldwide and regional populations. Because the model in ADMIXTURE does not consider linkage disequilibrium (LD), we used PLINK v.1.0711 to prune the original dataset with dense SNPs. After we assigned an r2 threshold of 0.4 in every continuous window of 200 SNPs advanced by 25 SNPs (--indep-pairwise 200 25 0.4), 288,979 SNPs remained for the ADMIXTURE analysis.32 We ran ADMIXTURE with a random seed for the merged dataset from K = 2 to K = 20 with default parameters (--cv = 5) in ten replicates for each K.
Because the individual ancestry proportion varied significantly when K was large, we used CLUMPP33 to examine the convergence of different runs in the ten replicates for each K; the optimal ancestry proportion was defined by the single replicate that showed the closest value to the ancestry proportion that appeared most commonly among the ten replicates. We also assessed the cross-validation error in the ten replicates to find the best K of the ancestral populations.
Estimating Ancient Gene Flow on the Basis of f4 Statistics
We used an f4 test (D test), D(African, X, Neanderthal, Denisovan), according to Patterson et al.34, 35 to determine the main source of gene flow (either Neanderthal or Denisovan) to the target populations (particularly TIB). A significantly negative D value indicates more Neanderthal than Denisovan allele sharing in population X, a non-African modern human population. In contrast, a significantly positive D value (Z > 2) suggests more gene introgression from Denisovans than from Neanderthals. With the exception of Oceanian populations, non-African populations always share significantly more alleles with Neanderthals than with Denisovans.
Using D(African, ancient, TIB, X) according to Durand et al.,17, 34 we further determined whether a certain ancient genome contributes more ancestry to TIB than to any other non-African modern human population. A significantly positive D value indicates that ancient genomes share a higher proportion of alleles with population X than with TIB.
Finally, we based four designs (four equations) on f4 statistics to quantitatively estimate the ancient ancestry in modern human populations to find out whether there is excess archaic ancestry in TIB. We first randomly selected a population as the reference population (ref) and obtained a series of ratio values. The population showing the smallest ratio value, which is always negative, might have the relatively smallest gene introgression from ancient populations. Then, we used these reference populations to estimate ancient gene flow in the other non-African populations:
| (Equation 1) |
As in Equation 1, we used chimpanzee (chimp) as an outgroup and used Africans (YRI) as a reference population, which was assumed to have no ancient gene flow:
| (Equation 2) |
In view of the similarity between Neanderthals and Denisovans, the previous formulas might overestimate the gene flow of each ancient population. So, we also used another ancient population as an outgroup to control such an effect:
| (Equation 3) |
In addition, we can also estimate the non-ancient admixture proportion. Here, Neanderthals and Denisovans are reference populations for each other:
| (Equation 4) |
Detecting Archaic Sequences in Modern Human Genomes
We further locally identified genomic segments of non-modern human origins by using the S∗ statistic38 and ArchaicSeeker, a method developed in this study. The principle of this method is based on the high divergence between modern humans and archaic hominins38, 39, 40 and the finding that archaic hominin introgressions are absent from sub-Saharan Africans.41 We defined “E-allele” as an allele absent from Africans but observed in Eurasians.38, 39, 40 In simulations, we found that the E-allele rate (defined as the number of E-alleles on a segment divided by the number of polymorphic sites in the region spanning the segment) is much higher on archaic introgressive segments than on modern human segments. We further implemented a hidden-Markov-model-based method to partition each Eurasian chromosome into segments with different E-allele rates and labeled each segment with one of the two hidden states. One hidden state represents segments that are of modern human origin and have a lower E-allele rate, and the other one represents segments that are of archaic hominin origin and have a higher E-allele rate. The process of the algorithm was as follows. We first set a penalty for state transitions (λ) of the algorithm and initial E-allele rates for two states. The initial E-allele rates are the observed E-allele rates on modern Eurasian and archaic hominin genomes. Second, we implemented the classic Viterbi algorithm to partition the Eurasian chromosomes into segments and labeled each segment with a hidden state. Third, with segments from step two, we re-estimated the E-allele rate for both hidden states. Fourth, we repeated steps two and three until convergence. In the Viterbi algorithm, we used the same initial probability for each state. We set the transition probability between two states as e−λ and set the emission probability of both states as their E-allele rates.
After we partitioned each chromosome into segments and labeled a hidden state for each segment, we selected segments in a state with a higher E-allele rate as candidates for archaic introgressions and implemented a three-stepped approach to filter false positives.42, 43, 44 For each candidate segment, we first reconstructed a phylogenetic tree for the candidate segment, along with Neanderthal, Denisovan, African, and chimpanzee sequences. To detect Neanderthal-like and Denisovan-like sequences, we required the segment to coalesce first with Neanderthals and Denisovans, respectively. Second, we estimated the divergence time between the candidate and Neanderthal segments, denoted as tNean (or between candidate and Denisovan segments, denoted as tDeni). To detect Neanderthal-like sequences, we required tNean to be less than 550,000 years ago14 and tNean to be less than tDeni. To detect Denisovan-like sequences, we required tDeni to be less than 550,000 years ago and tDeni to be less than tNean. Third, we required the genetic length of the segments to be consistent with an introgression time later than 550,000 years ago to reject the ancestral polymorphism model.
On the basis of E-alleles and the algorithm described above, we further extended the method to the haplotype level and implemented the algorithm in a computer program, ArchaicSeeker, which is a more heuristic method of detecting archaic DNA sequences in present-day human genomes. For those detected archaic genomic segments, we classified them into three groups: Neanderthal-like, Denisovan-like, and unknown archaic. The first two groups were results from the three methods based on comparative analyses of the archaic genomes (Denisovan or Neanderthal). However, the unknown archaic segments, which could be detected only by the S∗ method, were defined as archaic-like signals that cannot be attributed to either modern human genomes or sequenced archaic genomes.
Local-Ancestry Inference
To understand the genetic makeup of some interesting regions on a finer scale, we developed a method on the basis of the results of ChromoPainter (cp)45 to obtain the fine-scale ancestry makeup of a given genomic region. To infer the local ancestries of TIB with cp, we selected Han Chinese genomes and some available archaic genomes, including an Altai Neanderthal genome14 and a Denisovan genome,15 as reference data. Moreover, because we observed that Tibetans shared a considerable proportion of ancestry with Siberian populations, we also included the Ust'-Ishim as a reference.16 In addition, to avoid the false-positive inference of archaic ancestral segments, we included full sequence data of YRI from phase 3 of the 1000 Genomes Project in our reference population panel. With this reference panel, we classified the local genomes into six categories regarding their ancestry: Denisovan-like, Neanderthal-like, Ust’-Ishim-like, Han-Chinese-like, African-like, and uncertain ancestry.
Analyzing the Correlation between Ancestry and Altitude
To investigate whether there could be any relationship between the ancestral composition of individuals and the altitude where they reside, we grouped Tibetan individuals according to geographical regions and calculated the correlation between percentages of non-modern human sequences and altitude. In this analysis, Han Chinese and Sherpa samples were not included, although a much stronger correlation would be expected if they were. The Tibetan samples were grouped into seven regional populations according to the information provided in Table S1; Shigatse samples were grouped into two populations because three individuals were from a rather different altitude (Dingri, 4,300 m). Therefore, seven regional populations—Lhasa (3,650 m), Nyingchi (3,000 m), Chamdo (3,240 m), Shannan (3,573 m), Shigatse (3,853 m), Nagqu (4,522 m), and Dingri (4,300 m)—were used for this analysis. The altitude for each region was averaged over the number of individuals residing in a given region. Accordingly, the proportion of sequences from each category (non-modern human sequences, Neanderthal-like sequences, Denisovan-like sequences, Neanderthal-specific sequences, Denisovan-specific sequences, and unknown archaic sequences) was averaged over the number of individuals in the same region. Pearson’s correlation coefficient (R) and coefficient of determination (R2) were calculated on the basis of the estimated ancestry proportions and altitude data.
Estimating TMRCA
To estimate the TMRCA of a group of sequences (or haplotypes) in a certain region, we first calculated the average pairwise nucleotide differences of these sequences according to the following formula:
where n is the number of sequences analyzed in a given genomic region ab, and πij is the nucleotide difference between two sequences i and j (i ≠ j). The TMRCA can be estimated with the following formula:
where μab is the local mutation rate of a genomic region with length lab from positions a to b. This mutation rate can be estimated with the formula below:
where dHum − AncHumChimp denotes the nucleotide difference between the human reference genome and that of the most recent common ancestor of humans and chimpanzees of region ab. THum − HumChimp is the divergence time of humans and chimpanzees, and 13 million years was used in this study.
Results
Genetic Affinities of TIB in the Context of Global Populations
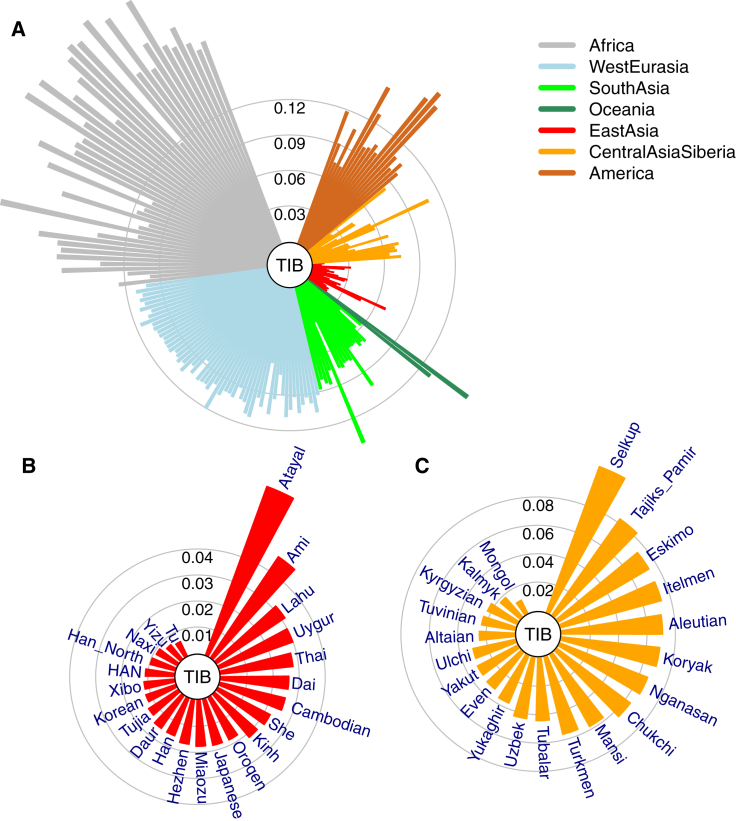
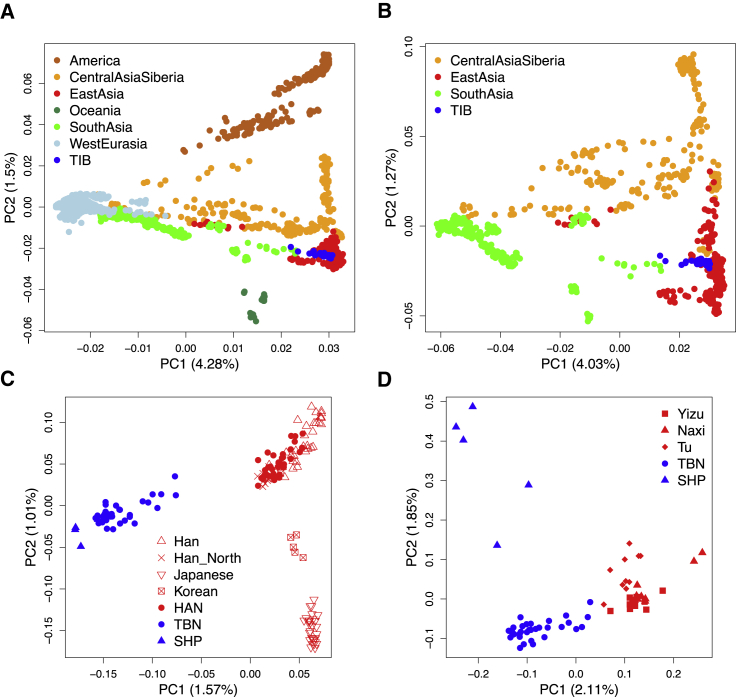
A comparison of genetic differences measured by unbiased FST19 between TIB and 188 worldwide populations17 (Tables S3 and S4) showed that TIB is most closely related to East Asian populations (Figures 1A and 1B), followed by Central Asian and Siberian populations (Figure 1C). These relationships remained consistent in separate analyses of TBN and SHP samples (Figures S2 and S3). In addition, these results were also confirmed by PCA (Figure 2 and Figures S4 and S5) and outgroup f3 statistics21 (Figure S6). Interestingly, TBN and SHP are distinguishable in a two-dimensional PC plot (Figure 2D), indicating that they are two distinct groups with genetic differences (FST = 0.010), slightly smaller than those between TBN and HAN (FST = 0.011). The overall genetic makeup of TIB is closer to surrounding populations living in the plateau, such as Tu (FST = 0.006), Yizu (FST = 0.007), and Naxi (FST = 0.009), than to any other population worldwide (Figure 1B and Tables S7 and S8), most likely as a result of direct ancestry sharing or reciprocal gene flow between these populations.
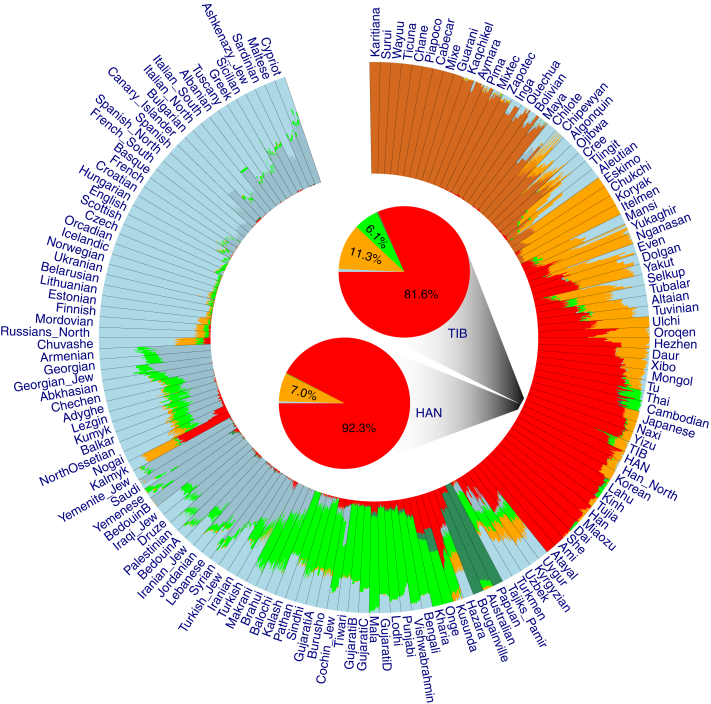
Figure 1.
Genetic Affinities of TIB in the Context of Worldwide Populations
(A) A fan-like chart showing genetic differences (FST) between TIB and worldwide populations. Each branch represents a comparison between TIB and 1 of the 256 populations; lengths are proportional to the FST values, indicated by gray circles. The populations are classified by geographical regions and indicated with the colors shown in the legend. The populations in each region are presented in a clockwise order according to great-circle distance to Tibet.
(B) A fan-like chart showing FST between TIB and East Asian populations.
(C) A fan-like chart showing FST between TIB and Central Asian and Siberian populations.
Figure 2.
Analysis of the First Two PCs of TIB Individuals and Other Population Samples
Geographical regions where the individuals are located are indicated with the colors shown in the legend. Numbers in brackets denote the variance explained by each PC. The HAN samples are classified into East Asian populations and are not highlighted in this illustration.
(A) In this two-dimensional PC plot, TIB samples are located closely in the East Asian cluster and share similar PC coordinates with some populations in South Asia and Central Asia and Siberia.
(B) PCA of TIB individuals and Eurasian samples (western Eurasian samples were excluded).
(C) PCA of TIB individuals and some East Asian samples.
(D) PCA of TBN and SHP individuals and samples of some closely related surrounding populations (Tu, Yizu, and Naixi).
We caution that the relationship revealed by an overall analysis of the genome-wide data only reflects a limited aspect of the origins of the populations studied. The results should not be directly interpreted as the genetic origins and history of the Tibetans; rather, they reflect the population’s present-day genetic makeup, which could be substantially influenced by recent gene flow from surrounding populations. This highlights the need to account for admixture when inferring population history.
Genetic Relationship and Divergence between TIB and HAN
Among the lowland populations, the data indicated that Han Chinese are most closely related to TIB (FST = 0.011) (Figure 1B and Table S7); the two populations had an overall strong correlation in the allele-frequency spectrum (Figure S7), but Y chromosome and mtDNA data showed substantial differences between them (Tables S5 and S6). Y chromosome haplogroup D-M174 is an East-Asian-specific male lineage and is rare in populations from regions bordering East Asia (Central Asia, North Asia, and the Middle East), usually less than 5%.6, 46, 47, 48 In our data, D-M174 lineages were not observed in the 39 HAN samples (frequency was 0) but had a high frequency in TIB (66.6%; Table S5), indicating a very large difference in Y DNA lineages between the two groups. Such a large difference was not observed in mtDNA, but some haplogroups also showed considerable differences between groups. For example, the frequency of B5 in TBN (0%) was quite different from that in HAN (10.26%) (Table S6). Some haplogroups, such as A and M5, also showed substantial differences between SHP and TBN; however, because sample sizes, especially that of SHP, in our data were small, estimation of haplotype frequency is not reliable.
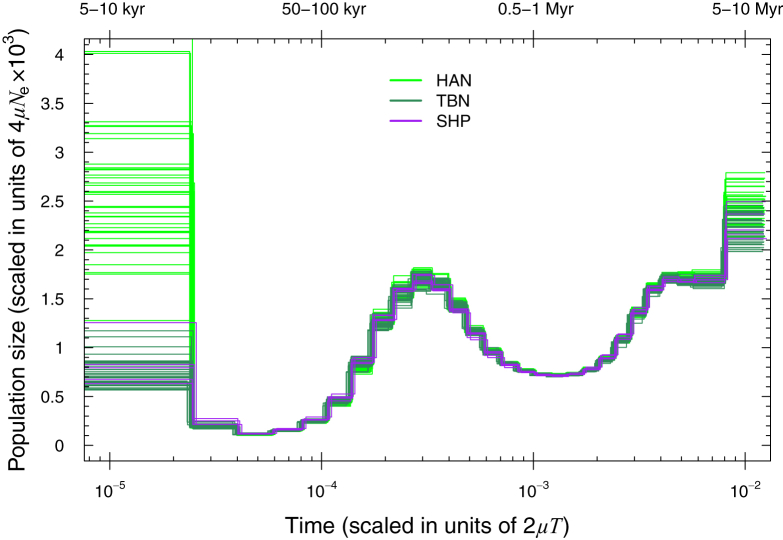
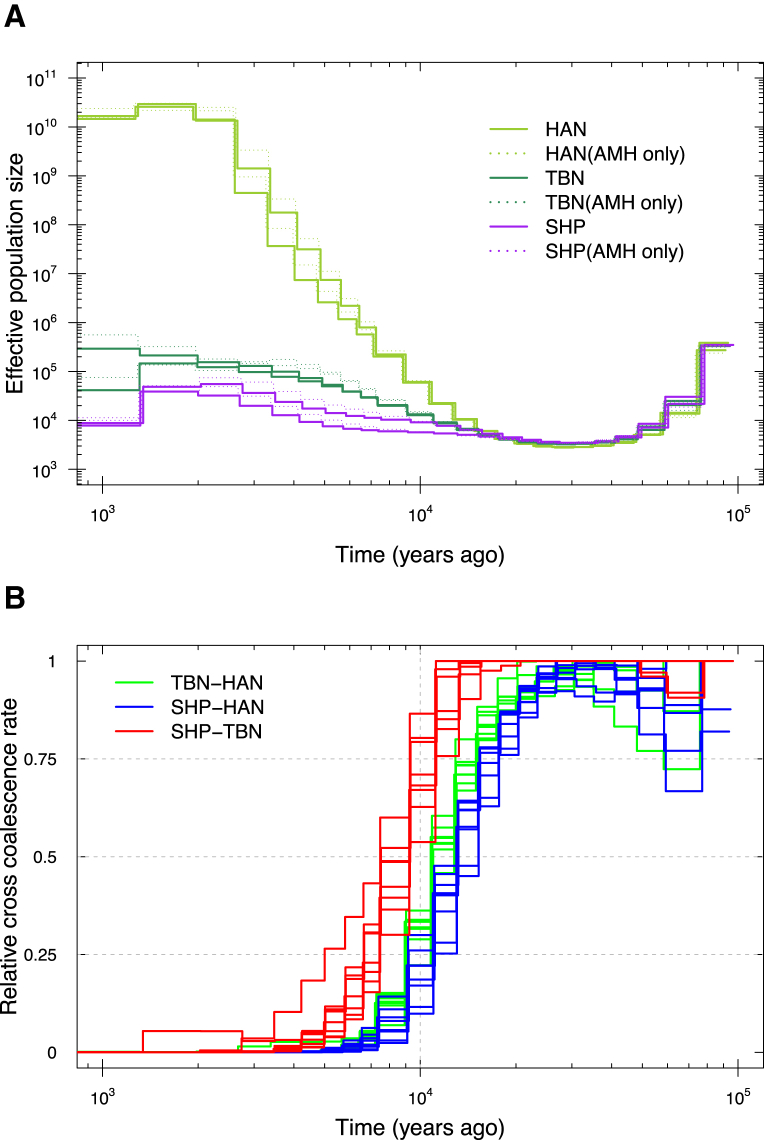
TIB showed an overall lower genetic diversity than HAN, including a lower level of heterozygosity, a higher level of runs of homozygosity (Figure S8), and a consistently smaller effective population size over the last ∼15,000 years (Figures 3 and 4A), most likely because TIB is isolated on the Tibetan Plateau, whereas HAN has experienced recent population expansion. The average time of divergence between TBN and HAN was estimated to be ∼15,000–9,000 YBP on the basis of a sequentially Markovian coalescent analysis24, 25 of high-coverage genomes. This divergence time between Tibetans and Han Chinese is thus much earlier than the estimate of 2,750 years ago by a previous study.4 In contrast, the estimated divergence time between SHP and HAN was ∼16,000–11,000 YBP, and that between TBN and SHP was ∼11,000–7,000 YBP (Figure 4B). The divergence between TIB and HAN most likely resulted from recent migration to the Tibetan Plateau after the Last Glacial Maximum (LGM), a period of intense cold from ∼26,500 to 19,000 YBP.49 Subsequent gene flow between TBN and HAN or continuous migrations to the plateau most likely caused the divergence time between TBN and HAN to be slightly shorter than that between SHP and HAN.
Figure 3.
Estimated Changes in Effective Population Size over Time
The estimation was based on PSMC analysis of single genomes from three modern human groups with deeply sequenced genomes in this study (33 TBN, 5 SHP, and 39 HAN). To overcome uncertain date estimates due to a lack of confidence in the true value of the mutation rate, we scaled time in units of 2 μT (where μ is the mutation rate and T is time in generations). We also provide an absolute estimation of time (top) under the assumption of a fast mutation rate of 1.0 × 10−9 or a slow mutation rate of 0.5 × 10−9 per site per year.
Figure 4.
Estimated Changes in Historical Effective Population Size and Divergence Time between Populations
(A) The estimation was based on MSMC analysis of four genomes (eight haploid genomes) randomly selected from each of the three modern human groups with deeply sequenced genomes in this study (TBN, SHP, and HAN). The analysis was repeated twice with different combinations of different individuals, and two curves are displayed for each group in the plot with different colors. Curves with dashed lines denote the estimation based on the same set of genomes but with non-modern human sequences removed (AMH only).
(B) The estimation was based on the MSMC algorithm. Two individual genomes were randomly selected from each group in the group pairs (TBN versus HAN, SHP versus HAN, or SHP versus TBN), and in total four genomes (eight haploid genomes) were used for each MSMC analysis. The analysis was repeated eight times, and eight curves are displayed for each group in the plot with different colors. Here, we show the results based on absolute estimation of time under the assumption of a slow mutation rate of 0.5 × 10−9 per site per year.
Ancestral Makeup of TIB
Ancestry analysis with ADMIXTURE32 suggested that present-day Tibetans share the majority of their ancestry makeup with populations from East Asia (∼82%), Central Asia and Siberia (∼11%), and South Asia (∼6%) and have minor ancestral relationships with western Eurasian (<1%) and Oceanian (<0.5%) populations (Figure 5 and Figures S9–S11). In contrast, HAN share much less ancestry with Siberian (∼7%), South Asian (<0.5%), and Oceanian (∼0%) populations but higher ancestry with East Asian populations (>90%).
Figure 5.
Summary Plot of Genetic Admixture
The results of individual admixture proportions estimated from 592,799 autosomal SNPs with genotype data available for 38 TIB, 39 HAN, and 2,345 HuOrigin samples (African samples were not included). Each individual is represented by a single line broken into K = 7 colored segments with lengths proportional to the K = 7 inferred clusters. The population IDs are presented outside of the circle of the plot. The results of population-level admixture of TIB and HAN are summarized and displayed in the two pie charts in the center of the circle plot; admixture proportions are denoted as percentages and with different colors.
We applied outgroup f3 statistics21 and f4 statistics17, 34 to assess ancient ancestral contributions to TIB by analyzing available ancient genomes, including those of an Altai Neanderthal,14 a Denisovan,15 and Ust’-Ishim, a 45,000-year-old anatomically modern human from Siberia16 (see Material and Methods). TIB was found to share slightly more alleles with ancient genomes than many worldwide populations, except for Oceanian populations, which showed significantly higher shared archaic (both Denisovan- and Neanderthal-like) ancestry than TIB, and East Asian populations (especially those surrounding TIB: Yizu, Tu, and Naxi), which had similar or even higher levels of shared archaic ancestry than TIB (Tables S9, S10, S11, S12, S13, S14, and S15 and Figures S12 – S26). In particular, no modern human populations shared more alleles with Ust’-Ishim than TIB (Table S13 and Figure S14). Although the exact estimates varied and the absolute values are not comparable, these genome-wide analyses indicate that TIB shares consistently higher ancestry with ancient non-AMH genomes than with HAN, the lowland population that has the closest modern relationship with TIB.
Individual Archaic Ancestry Is Higher in TIB Than in HAN
Genomic segments of non-AMH origins were identified with the S∗ statistic,38 as well as two methods developed in this study (see Material and Methods). The total amount of non-AMH sequences in the TIB gene pool (6.17%) was significantly higher than that in the HAN gene pool (5.86%) (p < 10−5) (Figures 6A and Figure S27A). Restricting the comparison to Neanderthal-like and Denisovan-like sequences, we did not observe any significant differences between TIB and HAN as a percentage of Neanderthal-like sequences per individual (Figure 6B), although we did observe some marginally significant differences when we restricted the comparison to Neanderthal-specific sequences (p = 0.01; Figure S27B). Significant differences were also observed between TIB and HAN on the basis of Denisovan-like sequences per individual (p = 0.001; Figure 6C), and this difference was more pronounced when the comparison was restricted to Denisovan-specific sequences (p < 10−5; Figure S27C). Together, this analysis suggests that the major differences between TIB and HAN resulted from the contribution of Denisovan-like and unknown non-AMH sequences (Figure 6 and Figure S27).
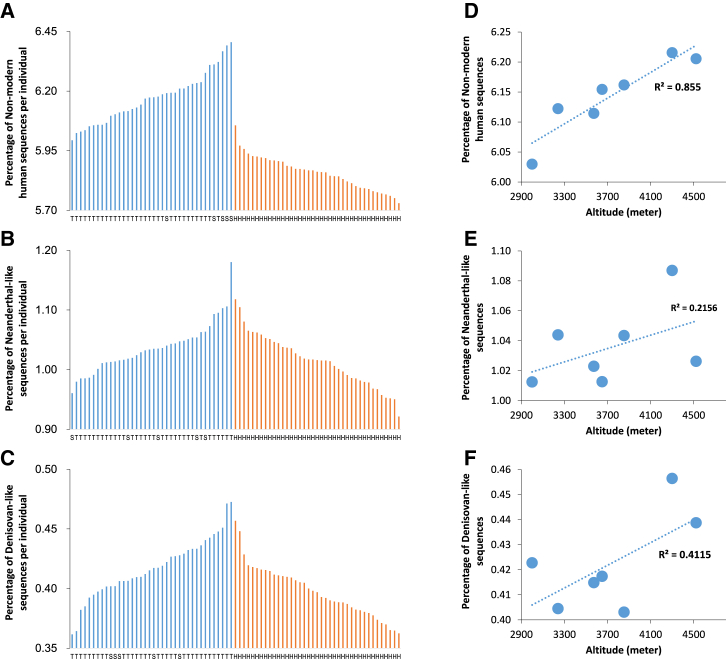
Figure 6.
Distribution of Non-modern Human Sequences among Individuals and Correlations with Altitude
(A) A comparison of the percentage of non-modern human sequences per individual between TIB (6.17 ± 0.10) and HAN (5.86 ± 0.07) demonstrates significant differences (p < 10−5). On the horizontal axis, T, S, and H denote Tibetan, Sherpa, and Han Chinese, respectively.
(B) A comparison of the percentage of Neanderthal-like sequences per individual between TIB (1.04 ± 0.04) and HAN (1.02 ± 0.04) shows significant differences (p = 0.379). On the horizontal axis, T, S, and H denote Tibetan, Sherpa, and Han Chinese, respectively.
(C) A comparison of the percentage of Denisovan-like sequences per individual between TIB (0.42 ± 0.02) and HAN (0.40 ± 0.02) shows slightly significant differences (p = 0.001); the statistical significance (p value) was obtained by permutation tests repeated 100,000 times. On the horizontal axis, T, S, and H denote Tibetan, Sherpa, and Han Chinese, respectively.
(D) Correlation between the average proportion of non-modern human sequences in regional Tibetan populations and altitude (Pearson R2 = 0.855; p = 0.0083).
(E) Correlation between the average proportion of Neanderthal-like sequences per individual and altitude (Pearson R2 = 0.216; p = 0.354).
(F) Correlation between the average proportion of Denisovan-like sequences per individual and altitude (Pearson R2 = 0.412; p = 0.170).
Altitudinal Correlation of Archaic Ancestry
Interestingly, when the Tibetan samples were grouped into seven regional populations according to geographical location (see Material and Methods), a strong correlation was observed between the average proportion of non-AMH sequences present in Tibetan individuals and altitude (Pearson R2 = 0.855; p = 0.0083; Figure 6D), but no significant correlation was observed in relation to Neanderthal-like or Denisovan-like sequences (Figures 6E and 6F). These results indicate that non-AMH sequences have some association with altitude, possibly contributing to the adaptation of Tibetans, but the main contribution seems to originate from unknown non-AMH ancestry (Figure S27D) rather than from Neanderthal-like or Denisovan-like sequences. A similar pattern and conclusion was apparent when the analysis was restricted to Neanderthal-specific and Denisovan-specific sequences (Figures S27E and S27F).
A Highly Differentiated Genomic Region with Elevated Archaic Ancestry in TIB
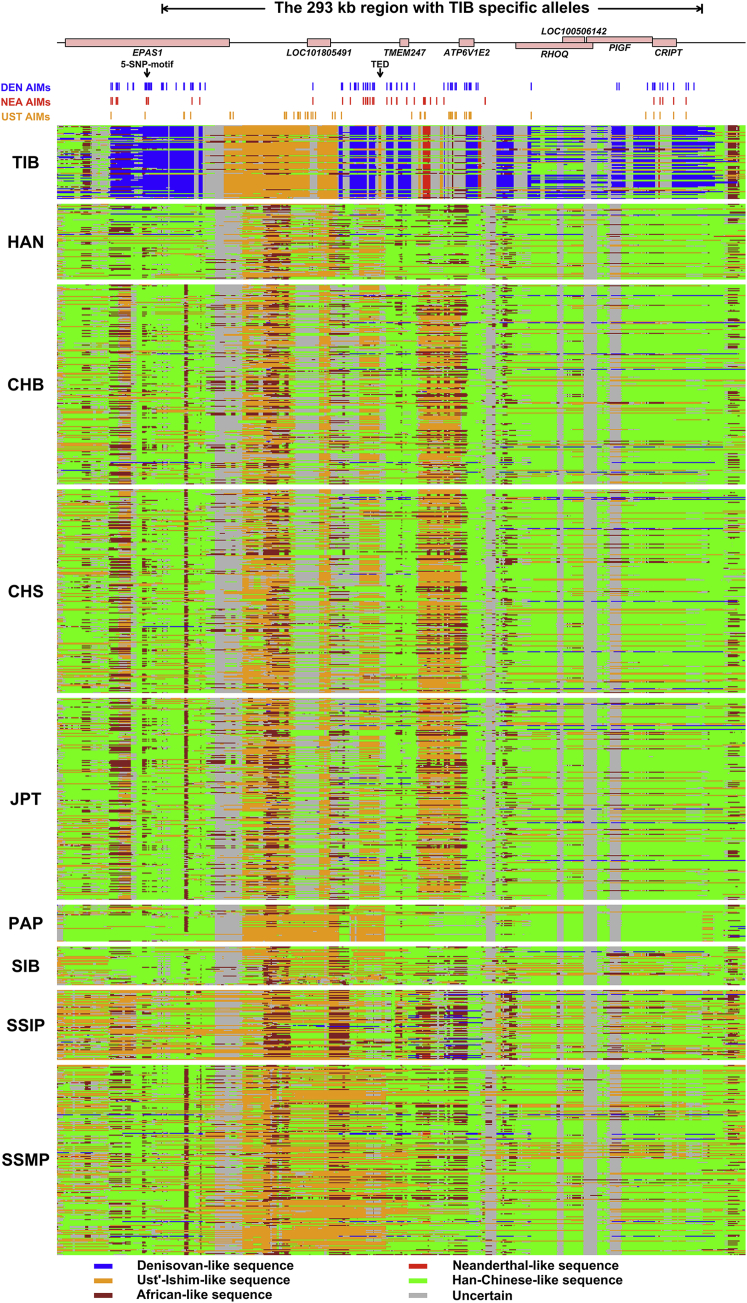
The above results reveal that non-AMH-derived sequences not only exist in the Tibetan genomes but are also very common. However, it is not clear whether these non-AMH sequences derived from recent migrations to Tibet with both AMH and non-AMH ancestries or were inherited from the ancient colonization of the Tibetan Plateau. The overall spatial distributions of non-AMH-derived sequences are similar between TIB and HAN (Figure S28), and the TMRCA for non-AMH sequences in both TIB and HAN was estimated to be >40,000 years (see Material and Methods). These results might be not surprising, given that TIB and HAN share recent genetic drift (Figures 1 and 2) and the majority of their genetic makeup (Figure 5). Nonetheless, when frequency was taken into account, a ∼300 kb region was found to be considerably different between TIB and HAN, such that both Denisovan-like and Neanderthal-like ancestries were significantly elevated in TIB (Figure S29). This ∼300 kb region is located on chromosome 2, encompasses eight genes (EPAS1, LOC101805491, TMEM247, ATP6V1E2, RHOQ [MIM: 605857], LOC100506142, PIGF [MIM: 600153], and CRIPT [MIM: 604594]), and has extreme differences between TIB and HAN (FST > 0.65) for SNVs with a high frequency of derived alleles. These differences are in sharp contrast to the genome-wide average (FST = 0.011).
Entangled Ancestries and Their Ancient Origins in the ∼300 kb Region
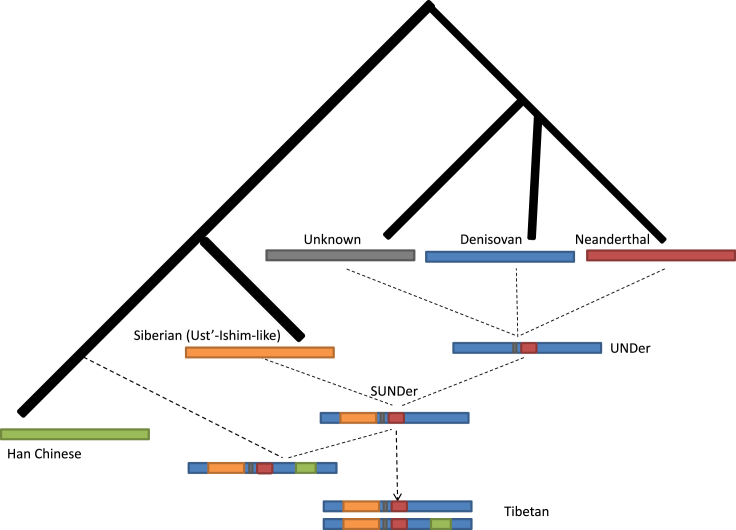
The ancestral pattern in the ∼300 kb region is extremely complicated, such that it contains a mix of Denisovan, Neanderthal, ancient Siberian, and unknown archaic ancestries, which are elevated in TIB (Table S16 and Figure 7). The unusually high frequency of archaic sequences and substantial differences between TIB and HAN—as well as other worldwide populations—cannot be explained by recent gene flows or incomplete linage sorting (Figure 7 and Table S17). These highly differentiated sequences harbored in the genomes of present-day Tibetan highlanders were most likely “inherited” from earlier settlers rather than “introgressed” by later arrivers. The complicated ancestral architectures in these regions have many implications for Tibetan origins and their pre-history. The surviving archaic sequences in the ∼300 kb region trace their ancestries back to ∼62,000–38,000 years ago (Table S17), pre-dating the LGM49 and indicating that the colonization of humans in Tibet is much more ancient than previously thought. The mixed archaic ancestries in this ∼300 kb region and simulation studies indicate that Tibet has been a human melting pot since the Paleolithic age, despite its inhospitable environment and the fact that interbreeding occurred among different hominin groups before the post-LGM arrivals. We suggest that SUNDer, an unknown ancient group of multiple ancestral origins, introduced the ancient haplotypes that are unique to present-day Tibetan highlanders (Figure 8).
Figure 7.
Local-Ancestry Inference of a ∼300 kb Region in TIB and Eight Other Populations
The local ancestry was jointly inferred with ArchaiSeeker and ChromoPainter45 (see Material and Methods). The upper panel shows genes located in the ∼300 kb region (chr2: 46,577,796–46,870,806). The two vertical arrows indicate the two previously identified Tibetan-specific variant tags, the five-SNP motif,7 and the Tibetan-enriched deletion (TED).50 The positions of ancestry-informative markers (AIMs) for the Denisovan (DEN AIM), the Neanderthal (NEA AIM), and Ust’-Ishim (UST AIM) are also displayed in the following rows. For the AIMs of ancient samples identified here (DEN, NEA, and UST), we required the frequency of the ancient samples’ alternative allele (with respect to reference genome GRCh37) to be >0.5 in TIB and <0.1 in other worldwide populations. The lower panels exhibit the inferred ancestry of haplotypes in TIB, HAN, CHB (Han Chinese in Bejing, China; 1000 Genomes), CHS (Southern Han Chinese; 1000 Genomes), JPT (Japanese in Tokyo, Japan; 1000 Genomes), PAP (Papuan),51 SIB (Siberians),51, 52, 53 SSIP (Singapore Indians),54 and SSMP (Singapore Malay).55 Each row represents a haplotype with ancestry derived from Neanderthals14 (red), Ust’-Ishim16 (orange), Denisovans15 (blue), or Han Chinese (green) or uncertain ancestry (including unknown archaic and uncertain modern human; gray).
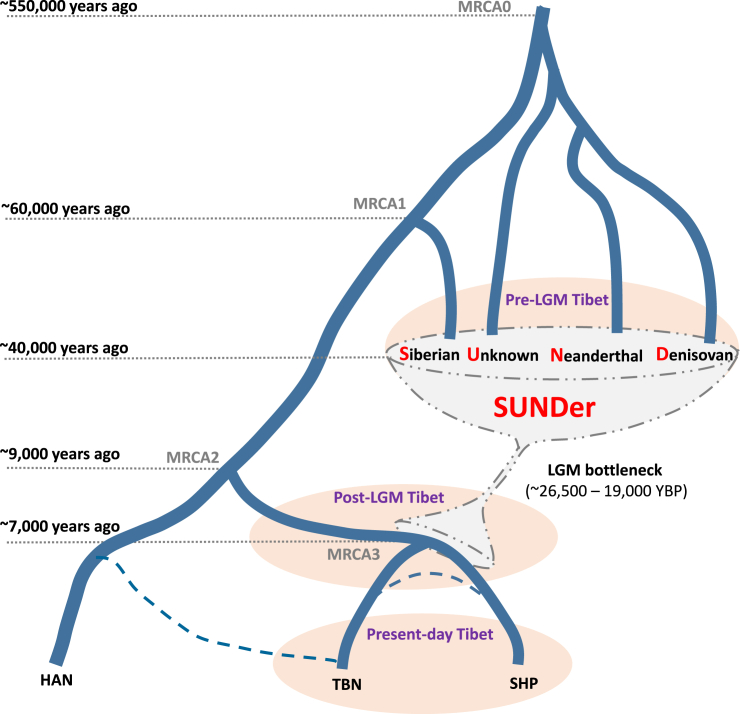
Figure 8.
A Model for Reconstructing the Evolutionary History of SUNDer
This model was constructed on the basis of the observed ancestry information and haplotype pattern in the ∼300 kb region (see Figure 7). The ancient group represented by SUNDer was generated by two ancient admixture events: one occurred among a Denisovan-like group, a Neanderthal-like archaic group, and one or more unknown archaic hominin groups and resulted in an admixed group (UNDer), and the other occurred between UNDer and an ancient Siberian (modern human) group represented by Ust’-Ishim and eventually resulted in SUNDer. We assume that these two admixture events occurred before the LGM and contributed ancient ancestral components, including Neanderthal-like (red), Denisovan-like (blue), unknown archaic (gray), and Ust’-Ishim-like Siberian (orange) ancestry, to the Tibetans’ gene pool. The post-LGM admixture occurred between SUNDer and lowland modern human groups represented by Han Chinese and introduced the majority of the ancestry (green) into the Tibetans’ gene pool. Finally, this evolutionary history is reflected in EPAS1, encompassed in the ∼300 kb region, and its downstream region in the Tibetan genome. Two major Tibetan haplotypes are shown in the present-day Tibetans: the old one is SUNDer-like (frequency: 17/76), and the new one is the SUNDer haplotype with a Han Chinese component (frequency: 23/76).
A Two-Wave “Admixture of Admixture” Model
Our data support that the Tibetan genome appears to have arisen from a mixture of multiple ancestral gene pools, but the ancestral composition is much more complicated, and its history can be traced back considerably earlier than previously suspected. For instance, a recent study has suggested that “Tibetans are a mixture of ancestral populations related to the Sherpa and Han Chinese.”56 We propose a two-wave “admixture of admixture” (AoA) model, despite its simplicity, to help explain the ancestral makeup and pre-history of Tibetans and Sherpas (Figure 9). An ancient wave of admixture occurred in a pre-LGM era >40,000 YBP, which could have resulted in the unique mosaic pattern of Paleolithic ancestries observed in the Tibetan genomes. The ancient gene pool of the Tibetans originated from an ancient admixed population, SUNDer, which was a group of hybrids of ancient Siberians (modern humans) and several archaic populations—including Denisovan-like, Neanderthal-like, and most likely a few unknown non-modern human groups that currently have not been identified by archeological or genetic studies. The admixture events that eventually formed SUNDer could have occurred on the Tibetan Plateau or in lowland areas before the SUNDer arrived at the plateau at least ∼40,000 YBP, before the LGM (∼26,500–19,000 YBP).49 Between ∼40,000 and ∼15,000 YBP, few new migrations occurred between the lowland and the plateau as a result of the LGM. However, from about ∼15,000 to ∼9,000 YBP (Figure 4), many more migrations to the plateau from the lowland included modern human ancestry. Therefore, another more recent wave of admixture occurred between Paleolithic and Neolithic ancestries, which probably resulted from a post-LGM migration to the plateau, most likely a population split from the common ancestor of Tibetans and Han Chinese. The divergence of Tibetans and Sherpas occurred ∼11,000 to ∼7,000 YBP (Figure 4), no earlier than the divergence between Tibetans and Han Chinese, and thus does not support that Tibetans are a mixture of Sherpa and Han Chinese.
Figure 9.
A Sketch Map for the Origins and Demographic History of Sherpas and Tibetans
This simplified model of the origins and evolutionary history of Sherpas and Tibetans is based on the observations and estimations from this study. The two dashed lines connecting HAN and TBN and connecting TBN and SHP represent possible gene flow between populations. Abbreviations are as follows: MRCA0, most recent common ancestor of modern human and archaic hominoids;14 MRCA1, most recent common ancestor of Eurasians; MRCA2, most recent common ancestor of HAN and TIB; MRCA3, most recent common ancestor of TBN and SHP; SUNDer: a tentative label for the early settlers who contributed ancient or archaic ancestry to present-day Tibetan highlanders.
Discussion
We provide compelling evidence for the co-existence of both Paleolithic and Neolithic ancestries on a genome-wide scale in the modern Tibetan gene pool, which supports a genetic continuity between pre-LGM highland-foragers and present-day Tibetans. We have explicitly revealed a prevalent non-AMH ancestry of the Paleolithic lineages, significantly advancing our understanding of the genetic prehistory of human colonization in Tibet as suggested by previous Y chromosomal and mtDNA studies.5, 6
The Paleolithic ancestries in the modern Tibetan gene pool entangle Denisovan-like, Neanderthal-like, ancient-Siberian-like, and unknown archaic sequences, indicating that Tibet remained a human melting pot where interbreeding occurred among different hominine groups before the LGM, although the motivations for prehistoric people to settle at the environmentally inhospitable plateau are still not clear. The results of this study indicate that plateau colonization and the altitudinal adaptation of human beings were considerably earlier and more complicated than had previously been suspected.
Non-AMH ancestries, despite being present in low proportions, composed a substantial part of the Tibetan gene pool and shaped the genetic architecture of present-day Tibetans and Sherpas. However, it is noteworthy that the Neolithic ancestries, which are dominant in contemporary Tibetans, might also contain non-AMH lineages via genetic introgression that occurred in the common ancestors of Tibetans and Han Chinese much earlier than the divergence of the two groups.35, 38, 39 This is why we observed that overall spatial distributions of non-AMH-derived sequences are similar between TIB and HAN. This admixture pattern in Tibetan genomes is very complex but unsurprising given that archaeological data have already suggested an ancient initial occupation of the plateau, followed by multiple migrations at different times and from different places, which have created a complex, mosaic population history.1
Taking advantage of the whole-genome sequence data that we generated simultaneously in both Tibetans and Han Chinese, we estimate that Tibetans diverged from Han Chinese with an average coalescence time of ∼15,000–9,000 years. This estimation is much earlier than 2,750 years ago, estimated by a recent study.4 Our estimation is less likely to be affected by the archaic sequences harbored in the whole-genome data, which could potentially confound the estimation of population divergence, because the non-AMH sequences were excluded from the genomic data of both Tibetans and Han Chinese in this analysis. Therefore, this time estimation largely reflects the divergence of modern human ancestry in Tibetans and Han Chinese since the two populations split from their shared ancestral population. However, subsequent gene flows from other populations and between the two populations are expected to influence the estimation of population divergence, which we were not able to fully evaluate and control here. Further efforts are also needed to elucidate the genetic relationship between Tibetans and Sherpas; for instance, the reported relationship by recent studies between Sherpa and Tibetan groups are controversial.56, 57, 58 Even though we observed some degree of differentiation between the two groups, uncovering their population structure and inferring their demographic history will require larger sample sizes.
Acknowledgments
This work acknowledges financial support from the Strategic Priority Research Program of the Chinese Academy of Sciences (CAS) (XDB13040100 to S.X.), the National Natural Science Foundation of China (91331204 to S.X., 31501011 to Y.L., 31601046 to H.L., 31260252 and 31460286 to L.K., and 31260263 to Y.G.), the National Science Fund for Distinguished Young Scholars (31525014 to S.X.), the Program of Shanghai Academic Research Leader (16XD1404700 to S.X.), the Science and Technology Commission of Shanghai Municipality (14YF1406800 to Y.L. and 16YF1413900 to H.L.), and the National High-Tech Research and Development Program (2012AA021802 to Y.Y.). S.X. is a Max Planck Independent Research Group Leader and a member of the CAS Youth Innovation Promotion Association. S.X. also gratefully acknowledges support from the National Program for Top-Notch Young Innovative Talents of the “Wanren Jihua” Project. We thank Dr. Mark Stoneking for his helpful discussion and Dr. Yundi Chen and his colleagues from WuxiNextCODE for their technical assistance during whole-genome sequencing. The funders had no role in study design, data collection or analysis, decision to publish, or preparation of the manuscript.
Published: August 25, 2016
Footnotes
Supplemental Data include 29 figures and 17 tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.07.002.
Accession Numbers
The whole-genome sequences reported in this paper have been deposited in the Genome Sequence Archive (GSA) and National Omics Data Encyclopedia (NODE) under accession numbers GSA: PRJCA000246 and NODE: ND00000013EP, respectively.
Web Resources
1000 Genomes GRCh37 human reference genome, ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/technical/reference/human_g1k_v37.fasta.gz
1000 Genomes Project phase 3 data, http://www.1000genomes.org/data
ArchaicSeeker, http://www.picb.ac.cn/PGG/resource.php
Data for Siberia (Ust’-Ishim), http://cdna.eva.mpg.de/ust-ishim/VCF/
EPO (Enredo-Pecan-Ortheus) pipeline, ftp://ftp.1000genomes.ebi.ac.uk/
Genome Sequence Archive, http://gsa.big.ac.cn
International Society of Genetic Genealogy Y-DNA Haplotype Tree, http://www.isogg.org/tree/
National Omics Data Encyclopedia, http://www.biosino.org/node/
OMIM, http://www.omim.org
Supplemental Data
References
- 1.Aldenderfer M. Peopling the Tibetan plateau: insights from archaeology. High Alt. Med. Biol. 2011;12:141–147. doi: 10.1089/ham.2010.1094. [DOI] [PubMed] [Google Scholar]
- 2.Qi X., Cui C., Peng Y., Zhang X., Yang Z., Zhong H., Zhang H., Xiang K., Cao X., Wang Y. Genetic evidence of paleolithic colonization and neolithic expansion of modern humans on the tibetan plateau. Mol. Biol. Evol. 2013;30:1761–1778. doi: 10.1093/molbev/mst093. [DOI] [PubMed] [Google Scholar]
- 3.Wang W.S.-Y. Language and the Evolution of Modern Humans. In: Omoto K., Tobias P.V., editors. The Origins and Past of Modern Humans. World Scientific; 1998. pp. 247–262. [Google Scholar]
- 4.Yi X., Liang Y., Huerta-Sanchez E., Jin X., Cuo Z.X., Pool J.E., Xu X., Jiang H., Vinckenbosch N., Korneliussen T.S. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao M., Kong Q.P., Wang H.W., Peng M.S., Xie X.D., Wang W.Z., Jiayang, Duan J.G., Cai M.C., Zhao S.N. Mitochondrial genome evidence reveals successful Late Paleolithic settlement on the Tibetan Plateau. Proc. Natl. Acad. Sci. USA. 2009;106:21230–21235. doi: 10.1073/pnas.0907844106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi H., Zhong H., Peng Y., Dong Y.L., Qi X.B., Zhang F., Liu L.F., Tan S.J., Ma R.Z., Xiao C.J. Y chromosome evidence of earliest modern human settlement in East Asia and multiple origins of Tibetan and Japanese populations. BMC Biol. 2008;6:45. doi: 10.1186/1741-7007-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huerta-Sánchez E., Jin X., Asan, Bianba Z., Peter B.M., Vinckenbosch N., Liang Y., Yi X., He M., Somel M. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 2014;512:194–197. doi: 10.1038/nature13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quinlan A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinformatics. 2014;47:1–34, 34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P.H., de Filippo C. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer M., Kircher M., Gansauge M.T., Li H., Racimo F., Mallick S., Schraiber J.G., Jay F., Prüfer K., de Filippo C. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu Q., Li H., Moorjani P., Jay F., Slepchenko S.M., Bondarev A.A., Johnson P.L., Aximu-Petri A., Prüfer K., de Filippo C. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 2014;514:445–449. doi: 10.1038/nature13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazaridis I., Patterson N., Mittnik A., Renaud G., Mallick S., Kirsanow K., Sudmant P.H., Schraiber J.G., Castellano S., Lipson M. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weir B.S., Cockerham C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 20.Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reich D., Thangaraj K., Patterson N., Price A.L., Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raghavan M., Skoglund P., Graf K.E., Metspalu M., Albrechtsen A., Moltke I., Rasmussen S., Stafford T.W., Jr., Orlando L., Metspalu E. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature. 2014;505:87–91. doi: 10.1038/nature12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiffels S., Durbin R. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 2014;46:919–925. doi: 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Connell J., Gurdasani D., Delaneau O., Pirastu N., Ulivi S., Cocca M., Traglia M., Huang J., Huffman J.E., Rudan I. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 2014;10:e1004234. doi: 10.1371/journal.pgen.1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nachman M.W., Crowell S.L. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roach J.C., Glusman G., Smit A.F., Huff C.D., Hubley R., Shannon P.T., Rowen L., Pant K.P., Goodman N., Bamshad M. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong A., Frigge M.L., Masson G., Besenbacher S., Sulem P., Magnusson G., Gudjonsson S.A., Sigurdsson A., Jonasdottir A., Jonasdottir A. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conrad D.F., Keebler J.E., DePristo M.A., Lindsay S.J., Zhang Y., Casals F., Idaghdour Y., Hartl C.L., Torroja C., Garimella K.V., 1000 Genomes Project Variation in genome-wide mutation rates within and between human families. Nat. Genet. 2011;43:712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scally A., Durbin R. Revising the human mutation rate: implications for understanding human evolution. Nat. Rev. Genet. 2012;13:745–753. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- 32.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakobsson M., Rosenberg N.A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 34.Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin P., Stoneking M. Denisovan Ancestry in East Eurasian and Native American Populations. Mol. Biol. Evol. 2015;32:2665–2674. doi: 10.1093/molbev/msv141. [DOI] [PubMed] [Google Scholar]
- 36.Durand E.Y., Patterson N., Reich D., Slatkin M. Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 2011;28:2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu Q., Hajdinjak M., Moldovan O.T., Constantin S., Mallick S., Skoglund P., Patterson N., Rohland N., Lazaridis I., Nickel B. An early modern human from Romania with a recent Neanderthal ancestor. Nature. 2015;524:216–219. doi: 10.1038/nature14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vernot B., Akey J.M. Resurrecting surviving Neandertal lineages from modern human genomes. Science. 2014;343:1017–1021. doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]
- 39.Sankararaman S., Mallick S., Dannemann M., Prüfer K., Kelso J., Pääbo S., Patterson N., Reich D. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seguin-Orlando A., Korneliussen T.S., Sikora M., Malaspinas A.S., Manica A., Moltke I., Albrechtsen A., Ko A., Margaryan A., Moiseyev V. Paleogenomics. Genomic structure in Europeans dating back at least 36,200 years. Science. 2014;346:1113–1118. doi: 10.1126/science.aaa0114. [DOI] [PubMed] [Google Scholar]
- 41.Green R.E., Krause J., Briggs A.W., Maricic T., Stenzel U., Kircher M., Patterson N., Li H., Zhai W., Fritz M.H. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding Q., Hu Y., Xu S., Wang J., Jin L. Neanderthal introgression at chromosome 3p21.31 was under positive natural selection in East Asians. Mol. Biol. Evol. 2014;31:683–695. doi: 10.1093/molbev/mst260. [DOI] [PubMed] [Google Scholar]
- 43.Hu Y., Ding Q., He Y., Xu S., Jin L. Reintroduction of a Homocysteine Level-Associated Allele into East Asians by Neanderthal Introgression. Mol. Biol. Evol. 2015;32:3108–3113. doi: 10.1093/molbev/msv176. [DOI] [PubMed] [Google Scholar]
- 44.Ding Q., Hu Y., Xu S., Wang C.C., Li H., Zhang R., Yan S., Wang J., Jin L. Neanderthal origin of the haplotypes carrying the functional variant Val92Met in the MC1R in modern humans. Mol. Biol. Evol. 2014;31:1994–2003. doi: 10.1093/molbev/msu180. [DOI] [PubMed] [Google Scholar]
- 45.Lawson D.J., Hellenthal G., Myers S., Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8:e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Underhill P.A., Passarino G., Lin A.A., Shen P., Mirazón Lahr M., Foley R.A., Oefner P.J., Cavalli-Sforza L.L. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann. Hum. Genet. 2001;65:43–62. doi: 10.1046/j.1469-1809.2001.6510043.x. [DOI] [PubMed] [Google Scholar]
- 47.Underhill P.A., Shen P., Lin A.A., Jin L., Passarino G., Yang W.H., Kauffman E., Bonné-Tamir B., Bertranpetit J., Francalacci P. Y chromosome sequence variation and the history of human populations. Nat. Genet. 2000;26:358–361. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- 48.Su B., Xiao C., Deka R., Seielstad M.T., Kangwanpong D., Xiao J., Lu D., Underhill P., Cavalli-Sforza L., Chakraborty R., Jin L. Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum. Genet. 2000;107:582–590. doi: 10.1007/s004390000406. [DOI] [PubMed] [Google Scholar]
- 49.Clark P.U., Dyke A.S., Shakun J.D., Carlson A.E., Clark J., Wohlfarth B., Mitrovica J.X., Hostetler S.W., McCabe A.M. The Last Glacial Maximum. Science. 2009;325:710–714. doi: 10.1126/science.1172873. [DOI] [PubMed] [Google Scholar]
- 50.Lou H., Lu Y., Lu D., Fu R., Wang X., Feng Q., Wu S., Yang Y., Li S., Kang L. A 3.4-kb Copy-Number Deletion near EPAS1 Is Significantly Enriched in High-Altitude Tibetans but Absent from the Denisovan Sequence. Am. J. Hum. Genet. 2015;97:54–66. doi: 10.1016/j.ajhg.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raghavan M., Steinrücken M., Harris K., Schiffels S., Rasmussen S., DeGiorgio M., Albrechtsen A., Valdiosera C., Ávila-Arcos M.C., Malaspinas A.-S. POPULATION GENETICS. Genomic evidence for the Pleistocene and recent population history of Native Americans. Science. 2015;349:aab3884. doi: 10.1126/science.aab3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raghavan M., DeGiorgio M., Albrechtsen A., Moltke I., Skoglund P., Korneliussen T.S., Grønnow B., Appelt M., Gulløv H.C., Friesen T.M. The genetic prehistory of the New World Arctic. Science. 2014;345:1255832. doi: 10.1126/science.1255832. [DOI] [PubMed] [Google Scholar]
- 53.Martin A.R., Costa H.A., Lappalainen T., Henn B.M., Kidd J.M., Yee M.C., Grubert F., Cann H.M., Snyder M., Montgomery S.B., Bustamante C.D. Transcriptome sequencing from diverse human populations reveals differentiated regulatory architecture. PLoS Genet. 2014;10:e1004549. doi: 10.1371/journal.pgen.1004549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong L.P., Lai J.K., Saw W.Y., Ong R.T., Cheng A.Y., Pillai N.E., Liu X., Xu W., Chen P., Foo J.N. Insights into the genetic structure and diversity of 38 South Asian Indians from deep whole-genome sequencing. PLoS Genet. 2014;10:e1004377. doi: 10.1371/journal.pgen.1004377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong L.P., Ong R.T., Poh W.T., Liu X., Chen P., Li R., Lam K.K., Pillai N.E., Sim K.S., Xu H. Deep whole-genome sequencing of 100 southeast Asian Malays. Am. J. Hum. Genet. 2013;92:52–66. doi: 10.1016/j.ajhg.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeong C., Alkorta-Aranburu G., Basnyat B., Neupane M., Witonsky D.B., Pritchard J.K., Beall C.M., Di Rienzo A. Admixture facilitates genetic adaptations to high altitude in Tibet. Nat. Commun. 2014;5:3281. doi: 10.1038/ncomms4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang L., Zheng H.X., Chen F., Yan S., Liu K., Qin Z., Liu L., Zhao Z., Li L., Wang X. mtDNA lineage expansions in Sherpa population suggest adaptive evolution in Tibetan highlands. Mol. Biol. Evol. 2013;30:2579–2587. doi: 10.1093/molbev/mst147. [DOI] [PubMed] [Google Scholar]
- 58.Bhandari S., Zhang X., Cui C., Bianba, Liao S., Peng Y., Zhang H., Xiang K., Shi H., Ouzhuluobu Genetic evidence of a recent Tibetan ancestry to Sherpas in the Himalayan region. Sci. Rep. 2015;5:16249. doi: 10.1038/srep16249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.