Abstract
Homozygous loss of SMN1 causes spinal muscular atrophy (SMA), the most common and devastating childhood genetic motor-neuron disease. The copy gene SMN2 produces only ∼10% functional SMN protein, insufficient to counteract development of SMA. In contrast, the human genetic modifier plastin 3 (PLS3), an actin-binding and -bundling protein, fully protects against SMA in SMN1-deleted individuals carrying 3–4 SMN2 copies. Here, we demonstrate that the combinatorial effect of suboptimal SMN antisense oligonucleotide treatment and PLS3 overexpression—a situation resembling the human condition in asymptomatic SMN1-deleted individuals—rescues survival (from 14 to >250 days) and motoric abilities in a severe SMA mouse model. Because PLS3 knockout in yeast impairs endocytosis, we hypothesized that disturbed endocytosis might be a key cellular mechanism underlying impaired neurotransmission and neuromuscular junction maintenance in SMA. Indeed, SMN deficit dramatically reduced endocytosis, which was restored to normal levels by PLS3 overexpression. Upon low-frequency electro-stimulation, endocytotic FM1-43 (SynaptoGreen) uptake in the presynaptic terminal of neuromuscular junctions was restored to control levels in SMA-PLS3 mice. Moreover, proteomics and biochemical analysis revealed CORO1C, another F-actin binding protein, whose direct binding to PLS3 is dependent on calcium. Similar to PLS3 overexpression, CORO1C overexpression restored fluid-phase endocytosis in SMN-knockdown cells by elevating F-actin amounts and rescued the axonal truncation and branching phenotype in Smn-depleted zebrafish. Our findings emphasize the power of genetic modifiers to unravel the cellular pathomechanisms underlying SMA and the power of combinatorial therapy based on splice correction of SMN2 and endocytosis improvement to efficiently treat SMA.
Introduction
Mutations in some ubiquitously expressed housekeeping genes have the seemingly paradoxical capability to impair mainly one specific tissue or cell type. A particularly remarkable example of this phenomenon is autosomal-recessive spinal muscular atrophy (SMA), a common and devastating motor-neuron (MN) disorder caused by a deficit of the ubiquitously expressed housekeeping protein “survival motor neuron” (SMN).
SMN is a highly conserved essential protein involved in snRNP biogenesis and splicing; when homozygously knocked out in mice, it causes early embryonic lethality.1, 2, 3 In contrast to mice, humans have two SMN genes, SMN1 [MIM: 600354] and an almost identical copy gene, SMN2 [MIM: 601627]. Because of a silent variant affecting an exonic splicing enhancer, SMN2 produces only about 10% of correctly spliced full-length transcript and protein.4 Importantly, full-length transcripts of both SMN genes produce an identical, functional SMN protein. Most individuals with SMA harbor homozygous deletions or gene conversions of SMN1 or, rarely, other SMN1 mutations, and disease severity is mainly determined by the number of the additional SMN2 copies.5, 6
Individuals with SMA present with a large phenotypic variability categorized into four types: type I (SMA1 [MIM: 253300]) severe form, which has an onset of <6 months of age and in which affected individuals are never able to sit or walk, have a life expectancy of <2 years of age, and are largely dependent on nutritional and respiratory support; type II (SMA2 [MIM: 253550]) intermediate form, which has an onset of >6 months of age and in which affected individuals are able to sit but never to walk; type III (SMA3 [MIM: 253400]) mild form, which has an onset >18 months of age and in which affected individuals are able to sit and walk; and type IV (SMA4 [MIM: 271150]) adult form, which has an onset of >20 years of age and in which affected individuals are able to sit and walk.7 Most individuals with type I SMA carry two SMN2 copies, those with type II SMA three SMN2 copies, those with type III SMA four SMN2 copies, and those with type IV SMA four to six SMN2 copies.8, 9
SMA is considered a MN disorder because structural and functional disturbances in MNs and MN circuitry, particularly at the presynaptic site of neuromuscular junctions (NMJs), lead to common SMA-related features such as the development of muscle weakness and atrophy of proximal voluntary muscles.10, 11, 12, 13, 14, 15 However, extensive studies have shown that in severe forms of SMA, affected humans and mouse models both display impairment in non-neuronal organs such as heart, lung, intestine, pancreas, and bones.16 In contrast, in milder forms of SMA, the effect of SMN deficiency is limited to MN function. Therefore, only a mild form of SMA can be considered “pure” MN disease.17 Despite this knowledge, and the 20 years that have passed since SMN1 was discovered to be the SMA-determining gene, it remains unclear why MNs are predominantly affected and which signaling pathways and cellular functions are responsible for SMA.
Here we make use of a striking finding related to SMA protective modifiers to unravel the cellular mechanism disturbed in SMA. In 2008, we identified the first SMA-protective modifier, plastin 3 (PLS3 [MIM: 300131]), by using differential expression analysis in SMA-discordant families in whom asymptomatic and SMA-II- or –III-affected siblings carry identical homozygous SMN1 deletions and the same number of SMN2 copies.18 PLS3, located on chromosome Xq23, was highly upregulated in lymphoblastoid cell lines derived from asymptomatic siblings—who were all women—but not symptomatic ones, whereas no difference was observed in fibroblasts, suggesting a tissue-specific regulation.18 The generation of induced pluripotent stem cell lines from fibroblasts of two discordant families has shown that the PLS3 expression is also highly elevated in differentiated MNs generated from asymptomatic but not symptomatic siblings. The latter finding further strengthened the role of PLS3 as a modifier of SMA in MNs.19
PLS3 is a Ca2+-dependent F-actin-binding and -bundling protein that influences the G/F-actin ratio.20 F-actin dynamics are important in many cellular processes, including axon development, cell polarity, migration, vesicle trafficking, and endocytosis.21, 22 Overexpression of PLS3, either in primary MN culture from SMA mice or in zebrafish smn morphants, significantly restored the impaired axonal growth and motor-axon truncation.18, 23 We analyzed the protective effect of PLS3 in the severe Taiwanese SMA mouse model carrying two human SMN2 copies on one allele in a murine Smn-knockout background. Heterozygous overexpression of a PLS3 transgenic allele in this mouse model showed significant improvement in all F-actin-dependent processes related to neurotransmitter release and vesicle recycling at the presynaptic site.24 In addition, PLS3 delays axon pruning, thus counteracting the poor presynaptic connectivity at the NMJ in SMA mice.24 Despite the improved motor abilities observed in the severe SMA mice overexpressing PLS3, survival could not be rescued on a congenic C57BL/6N background and was only moderately elevated from 14 to 18 days on a mixed C57BL/6N (50%):FVB/N (50%) background. Furthermore, these mice had severe multi-organ dysfunction that was not rescued by increased PLS3 expression.24 In contrast, a recent report of a randomly integrated PLS3 allele expressed in the severly affected Δ7-SMA mouse model failed to show motoric improvement or extended survival.25
Hence, reduction of SMN under a certain threshold in severe SMA mice leads to multi-organ impairment, which is predictable given the essential role of SMN in snRNP biogenesis and splicing.1, 2 Moreover, an increasing number of pathways are impaired in type I SMA cells, including transcription, RNA transport, microRNA expression, translation, Ca2+ homeostasis, and survival.26, 27, 28, 29, 30, 31, 32 Instead, in mildly affected SMA individuals, three to four copies of SMN2 appear to be sufficient to maintain proper function of all cell types but MNs; consequently, unraveling the main cellular pathway causing sole MN dysfunction would seem better achieved with mildly affected SMA animal models. In this work, we followed three main lines: first, we hypothesized that the generation of a milder SMA model that reflects the human situation in asymptomatic discordant families would finally prove that PLS3 overexpression has beneficial effects, including prolonged survival. For this, we used a combinatorial therapy of low-dose SMN antisense oligonucleotide (ASO) and PLS3 overexpression. Second, we made use of the finding of a second modifier in SMA (Wirth lab, unpublished data) in combination with a finding in yeast, where the knockout of the plastin ortholog Sac6p causes impaired endocytosis.33 Because synaptic vesicle recycling at the NMJ involves various endocytic processes and is essential for neurotransmission,34 we assumed that reduced SMN amount might impair endocytosis, which might be rescued by increased PLS3 expression. Third, we postulated that by understanding the interactome of PLS3 and identifying new PLS3 interacting partners, we would be able to identify additional disease modifiers that might further support our hypothesis that endocytosis is crucial in SMA.
Material and Methods
Animal Models and Genotyping
The Taiwanese SMA mouse model FVB.Cg-Tg (SMN2)2Hung Smn1tm1Hung/J, stock number 005058 (here named SmnKO/KO;SMN2tg/tg) was purchased from Jackson Laboratory. We backcrossed these mice for more than seven generations to obtain a congenic C57BL/6N background. We maintained the breeding colony by crossing SmnKO/KO;SMN2tg/tg mice and SmnKO/WT mice with WT mice.35 PLS3-overexpressing transgenic animals were used for generating SmnKO/KO;SMN2tg/0;PLS3tg/0 (SMA-PLS3het) and SmnKO/KO;SMN2tg/0;PLS3tg/tg (SMA-PLS3hom) mice as well as HET-PLS3het and HET-PLS3hom mice as shown in Figure S1B, and animals were genotyped as described.24 All zebrafish experiments were performed with the transgenic line tg (mnx1-GFP)ml2TG.36 Animal care and all surgical procedures were performed according to the institutional animal care committee guidelines and the German animal welfare laws and approved under the reference numbers 84-02.04.2014.A006, 84-02.05.20.12.120, 84-02.04.2015.A378, and 84-02.04.2012.A251 of the LANUV (Landesamt für Natur, Umwelt und Verbraucherschutz NRW) state agency of North Rhine-Westphalia.
ASO Injection of Mice
The SMN-ASO and Ctrl-ASO (IONIS Pharmaceuticals,37) were diluted in sterile PBS, and the concentration of 10 μg/ml (working solution) was calculated with photometric density (AD260). The subcutaneous injections (10–50 μg) were performed on postnatal days 2 and 3 (P2 and P3) with a MICROLITER Syringe (Hamilton) as reported.37 All pups of each litter were double blindly injected.
Histology
Organ collection, hematoxilin and eosin (H&E) staining of paraffin sections of lung, intestine, and heart, imaging, and analysis were performed as previously described.24
Immunohistochemistry of MNs and NMJs
For NMJ staining, TVA muscle was fixed in 4% PFA for 20 min. The tissue was rinsed three times in PBS for 10 min so that excess PFA would be removed. From this step on, the staining protocol followed the procedure described below for MN staining. Primary antibody dilutions were mouse anti-SV2 (1:100, Hybridoma Bank) together with mouse anti-neurofilament (1:100, Hybridoma Bank). The secondary antibody was donkey anti-mouse Alexa fluor 488 (1:500, Invitrogen). To outline endplates in muscle tissue, we used BTX conjugated with Rhodamine (1.5 ng/μL, Invitrogen). For proprioceptive input staining, the spinal cord was dissected from euthanized mice and fixed in 4% paraformaldehyde (PFA)/PBS for 1 day. The lumbar L4–L5 region was quickly rinsed in PBS, embedded in tissue-freezing medium (Jung) after cryoprotection (first day—20% sucrose; second day—30% sucrose) and sliced into 40-μm-thick sections (cryostat, Leica). Samples were permeabilized and blocked in PBS containing 4% BSA, 1% Triton X-100, and PBS for 1 hr. Finally, samples were incubated with goat anti-CHAT (1:300, Millipore) and rabbit anti-VGLUT1 (1:300, Synaptic Systems) antibodies overnight. Samples were washed and incubated with secondary antibodies (donkey anti-rabbit Alexa fluor 488 [1:750, Invitrogen] and donkey anti-goat Alexa fluor 568 [1:750, Invitrogen]). Samples were mounted in Mowiol (Kuraray) for further analysis.
Image Acquisition and Analysis
We performed imaging of NMJs with a Zeiss microscope (Axio Imager.M2) with the Apotome.2 system to mimic confocality together with a 40× and 63× oil immersion objective lens with 1.4 NA.
Images of MN soma and proprioceptive inputs were taken with the META 510 confocal microscope (Zeiss). Z-stacks of 50–60 slices were created with ZEN software (Zeiss). Proprioceptive input numbers on MNs and MN soma size were quantified with ImageJ (Fiji) software. All experiments were double blinded.
Motoric-Ability Testing and Weight Measurement
To analyze the motoric ability of animals, we performed tube and grip-strength tests.38 Via the tube test, the proximal hind limb muscle strength, weakness, and fatigue in neonates was assessed. The test is performed in two consecutive trials, in which the animal is placed head down into a 50 ml Falcon tube so that the animal hangs by its hind limbs. Then, the hind limb score (HLS) is evaluated on the basis of the positioning of the hind limbs toward each other.
The Grip Strength Meter (TSE Systems) is a system for determining the gripping strength of small laboratory animals (such as rats and mice). The animal pulls a special height-adjustable grip (with two paws) that is mounted on a high-precision force sensor, and muscle force is recorded in pounds.39 The grip strength test was performed at P36 and P108.
Generation of PLS3 Stable Cell Line, Co-immunoprecipitation, and Pull-Down Experiments
To produce cell lines stably overexpressing PLS3, we transfected HEK293T cells with Flag/His-PLS3 pcDNA6 vector. To select cells in which PLS3 was integrated into the genome, we treated cells with 20 μg/ml of Blasticidin for three weeks. After each passage (every 5 days), we analyzed the amounts of PLS3 by immunoblotting with Flag antibodies.
For co-IP experiments, control and Flag/His-PLS3-overexpressing HEK293T cells were harvested. The cells were lysed, incubated on ice for 30 min, and centrifuged for 30 min at 4°C. After 2 hr of incubation with FLAG M2 affinity beads (Sigma Aldrich) on a shaker at 4°C, the beads were washed, and bound fractions of protein in both control and test co-IPs were eluted in an excess of Flag peptide (100 μg/mL of Flag peptide in TBS buffer).
For pull-down experiments, we cloned PLS3 (NCBI Gene ID: 5358) and tropomodulin 3 (TMOD3, [MIM: 605112], [NCBI gene ID: 29766]) inserts into either PGEX4T3 or PET30BZ vectors by using Mlu1 and Not1 restriction sites. Inserts were confirmed by sequencing. To produce recombinant PLS3 and TMOD3, we induced bacteria containing the PGEX4T3-PLS3 or PET30BZ-TMOD3 with 1 mM isopropyl-β-D-thiogalactosid (IPTG) for 4 hr at 25°C. The tagged proteins were purified as described.40, 41 Finally, proteins bound to the GST or Ni-NTA resins were eluted in excess of reduced glutathione or imidazole, respectively. The amount of protein in the eluted fractions was quantified via the Bradford protein assay. The different deletion constructs containing different regions of coronin 1C (CORO1C, [MIM: 605269], [NCBI gene ID: 23603]) were cloned into the pEGFP-C1 vector. HEK293T cells were transfected with control EGFP, EGFP-CORO1C (ΔC), EGFP-CORO1C (ΔN), or EGFP-CORO1C vectors. For the pull-down assay, recombinant EGFP, EGFP-CORO1C (ΔC), EGFP-CORO1C (ΔN) or EGFP-CORO1C-FL proteins were bound to GFP microbeads. The recombinant GST-PLS3 protein was circulated over the μ columns for 20 min. Afterward, μ columns were washed, and the bound protein fraction was eluted with 95°C Laemmli buffer. Samples were separated by SDS-PAGE, and the membrane was probed with anti-GFP and anti-GST antibodies (1:1000). To investigate PLS3-TMOD3 interaction, we carried out a similar pull-down assay with purified recombinant proteins.
Mass Spectrometry and Analysis
The co-purified proteins with anti-Flag immunoprecipitation from both control and Flag/HisPLS3-overexpressing HEK293T cells were collected (<10 μg). Protein digestion was performed with Lys-C followed by trypsin in combination with filter-aided sample preparation technology (FASP, 10 kDa42). Prior to nano-LC ESI-MS/MS (nanoscale liquid chromatographic electrospray ionization tandem mass spectrometry), peptides were desalted by stage tipping as described.43 Eluted peptides were concentrated by vacuum-centrifugation and diluted to a volume of 15.0 μl with 0.5% acetic acid. Sample analysis was performed on an LTQ Orbitrap Discovery mass spectrometer (Thermo Scientific) coupled to an EASY-nLC II nano-LC system (Proxeon, part of Thermo Scientific). Raw files were processed with the Sequest search algorithm implemented in Proteome Discoverer software (Thermo Scientific) and the database for Homo sapiens. The following search parameters were applied: trypsin as proteolytic enzyme; up to two missed cleavages; carbamidomethylation at cysteine residues as fixed modification; and oxidation at methionine residues and phosphorylation at serine, threonine, and tyrosine residues as variable modifications. Peptide mass tolerance was 10 ppm for intact peptide masses detected in the Orbitrap and 0.8 Da for fragment ions detected in the linear ion trap. We filtered the lists of identified peptides so that they contained only high-confidence (1% false-positive rate) rank-one peptides with matching score-versus-charge-state criteria (charge state +2: ≥ 2.0, +3: ≥ 2.25, +4: ≥ 2.5), a mass deviation of ≤5 ppm, and sequence length of at least six amino acid residues.44
In Vivo Assay of G/F-Actin Ratio
5 × 105 HEK293T or 3 × 105 NSC34 cells were seeded in a six-well plate and transfected with either control or SMN siRNAs. After 48 hr cells were collected and lysed. Quantification of F-actin was performed according to the manufacturer’s instructions (BKo37 kit, Cytoskeleton). Immunoblots were carried out with similar amounts of supernatant and pellet lysate from each experiment. Changes in the amount of F-actin and G-actin were quantified by ImageJ software.
siRNA and Vector Transfection
5 × 105 HEK293T or NSC34 cells were seeded into six-well plates. 2 μg of plasmid DNA were transfected with DharmaFECT according to the manufacturer’s instructions. Cells were harvested 72 hr after transfection for further analysis. For the siRNA experiment, the same transfection strategy was followed, and 50 pmol of either control or other siRNAs, including PLS3, CORO1C, TMOD3, and SMN siRNAs, were added. The siRNAs were purchased from QIAGEN: control 5′-AATTCTCCGAACGTGTCAACGT-3′, Smn1 5′-AAGAAGGAAAGTGCTCACATA-3′ (mouse), SMN1 5′-TGGGATGATACAGCACTGATA-3′ (human), PLS3 5′-CAGGACTAGCTTATCATGAGA-3′ (human), CORO1C 5′-CCCGTACGTCCACTACCTCAA-3′ (human), and TMOD3 5′-ATGCGTTAAGAGATAATGAAA-3′ (human).
Primary Cell Cultures
Primary MNs18 and murine embryonic fibroblasts (MEFs) were isolated and cultured as described.24
Immunoblot and Immunostaining
Fluorescence-based immunostaining and immunoblots were performed in primary cells (MEFs or MNs), cell lines (NSC34), zebrafish (znp1 staining allowed visualization of motor axons), and several mouse tissues via standard protocols. Primary and secondary antibodies were as follows: monoclonal mouse α-β-actin, (60008, Proteintech), monoclonal mouse α-CORO1C (Hybridoma supernatant, gift from C. Clemen, Biochemistry I, University of Cologne), mouse α-GST (SC-459, Santa Cruz), mouse α- GFP (gift, hybridoma supernatant, Biochemistry II, University of Cologne), monoclonal mouse α-SMN (S55920, BD Transduction Lab), α-PLS3, polyclonal rabbit (Eurogentec, custom made18), monoclonal mouse α-FLAG, (1804, Sigma), mouse α-TMOD3, (Sab2102483, Sigma), goat α-CHAT (Ab144p, Milipore), rabbit α-zebrafish actin, (Novus Biologicals), monoclonal mouse α-zebrafish Znp1, (Hybridoma Bank), HRP-conjugated goat α-mouse IgG (115035000, Dianova), HRP-conjugated goat α-rabbit IgG (31460, Pierce), and HRP-conjugated goat α-rabbit IgG (7074, Cell Signaling).
Endocytosis Assays
To unravel differences in endocytosis performance, FITC-dextran uptake after respective incubation times was quantified by FACS. In brief NSC34 or HEK293T cells were transfected with control or SMN siRNAs for 48 hr. The cells were starved by serum depletion for 2 hr and incubated with 5 mg/mL of FITC-dextran (46945, Sigma) for 10 or 20 min at 37°C. Cells were washed with PBS and trypsinized (T3924, Sigma) on ice. After two washes with 1% BSA solution in PBS, uptake of FITC-dextran was measured with the BD FACS Calibur machine. Dead cells were excluded by propidium iodide staining (10 μg/mL, Sigma). To study the effect of low temperature and latrunculin A as endocytosis destabilizers, we performed the FITC-dextran uptake at 4°C or in the presence of 200 ng/ml latrunculin A. FACS data were analyzed with Cyflogic software (see Web Resources).
FM1-43 Endocytosis Experiments
TVA was dissected in HBSS solution (14025, GIBCO) and then incubated for 20 min in external physiological solution containing the following (mM): 145 NaCl, 5 KCl, 10 HEPES, 2 CaCl2, 1 MgCl2, and 10 glucose. The N. intercostalis innervating the TVA muscle was stimulated via a suction electrode pulled from borosilicate glass tubes (GB150T-8P, Science Products GmbH) and mounted in a bipolar electrode holder (model MEW-F15B, Warner Instruments) connected to a universal stimulus isolator (model MI401) and a universal digital stimulator (model MS501). Both MI401 and MS501 were custom-made by the electronics workshop of the Zoological Institute of the University of Cologne. Contraction of the muscles upon nerve stimulation was checked visually under the miscroscope. After incubation with 3 μM FM1-43 (F35355, Molecular Probes), we applied a current pulse train either at 20 Hz or 5 Hz for 1 s (1 mA amplitude, 0.5 ms pulse duration) to load the endocyted synaptic vesicles. Subsequently, we washed muscles with external solution that did not contain CaCl2 or 3 mM MgCl2 but did contain ADVASEP-7 (A3723, Sigma) to efficiently remove the non-internalized FM1-43, and we fixed the muscles with 4% PFA. We also incubated TVA muscles with BTX-647 (1 ng/μL; B-35450, Life Technologies) to stain postsynaptic terminals and easily localize the NMJs, then mounted them on slides. We used three animals per genotype and stimulation set. Imaging was performed as described above. All imaging processes and analyses were blinded. We analyzed images with Fiji on a macro setting and applied the Li threshold method to the postsynaptic terminals to delineate the area of interest in the presynaptic site. Because at P10 the distribution of vesicles is quite homogenous along the NMJ, we quantified the mean intensity of the FM1-43 dye at the presynaptic terminal, which excluded possible variability due to the size.
Overexpression and Knockdown Experiment in Zebrafish
CORO1C, TMOD3, and PLS3 cDNAs were cloned into a pCS2+ vector. Inserts were confirmed by Sanger sequencing. To synthesize mRNAs, vectors were linearized by Not1 digestion and cleaned with a PCR purification kit (QIAGEN). The SP6 mMessage kit (Ambion) was used for transcribing mRNAs of PLS3, CORO1C, and TMOD3 in vitro. Capped mRNAs were further purified via the RNeasy kit (QIAGEN). For knockdown experiments, control or smn morpholino (2 ng; GeneTools) was injected into the yolk sac.45 For overexpression experiments, the capped mRNA (300 pg) was injected into the yolk sac. To track the efficiency of injection in zebrafish eggs, we mixed the mRNA or morpholino with phenol red and rhodamine dyes. Adding rhodamine served as a check of whether the injected solutions were homogenously dispersed in the egg and allowed exclusion of uninjected eggs. Adding 1-phenyl 2-thiourea (PTU) at a final concentration of 200 μM to the medium prevented pigmentation, and eggs were incubated in a 28°C incubator for 34 hr.
Motor-Neuron Staining and Quantification in Zebrafish
Fish were de-chorionated after 34 hr under the binocular microscope. De-chorionated fish larvae were fixed in 4% PFA on a rotary shaker at 4°C overnight. On the next day, the fish were washed in PBS-T (PBS+0.1% Tween20) and dehydrated with methanol at −20°C overnight. On the third day, samples were rehydrated with gradually decreasing concentrations of methanol in BST-T and partially digested with 10 μg/ml of proteinase K solution for 20 min. In the next step, samples were blocked (blocking solution: PBS-T, 1% DMSO, 2% BSA, and 5% FCS). For the staining of MNs, fish were incubated with Znp1 antibody in the blocking solution in a rotary shaker at 4°C overnight (antibody concentration 1:150). After being washed, they were incubated with the secondary antibody in the blocking solution overnight and then washed again. For the imaging, 3–4 fish were mounted on a slide, and the first ten motor neurons after the yolk sac were considered for quantification. On the basis of MN axon appearance, they were categorized into normal, branched, truncated, and severely truncated forms.
Statistics
If not mentioned otherwise, statistical analyses were performed in Excel 2013 (Microsoft), GraphPad Prism (GraphPad Software), and Sigma Plot 11 (Systat Software). ANOVA, the Mann-Whitney U-test, Fisher's exact test, and unpaired Student’s t tests were used. All data are represented as means ± SEM.
Results
PLS3 Overexpression Rescues Survival on a SMN-ASO-Induced Intermediate SMA Mouse Model
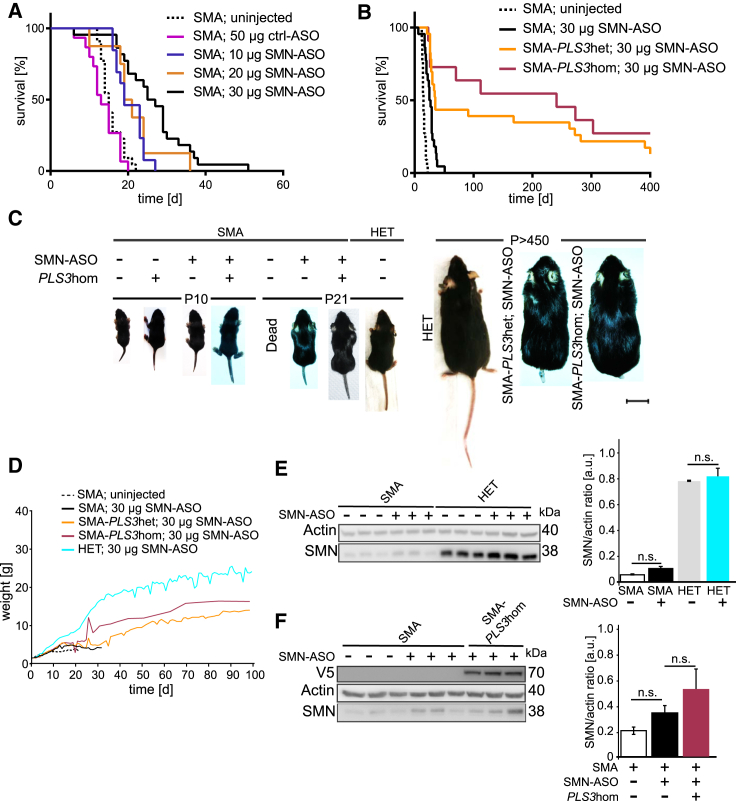
Our previous data have shown that ubiquitous overexpression of one PLS3 transgenic allele in the severe Taiwanese SMA mouse model restores MN and NMJ function as well as motoric abilities but fails to rescue survival, most likely because of a dramatic multi-organ dysfunction that could not be rescued by PLS3 overexpression.24 Therefore, we generated a SMN-ASO-induced milder SMA mouse model—mimicking the human situation of asymptomatic SMN1-deleted siblings—to confirm the beneficial impact of PLS3 observed in humans. We made use of SMN ASOs, which dose dependently elevate the amount of full-length, functional SMN from the human SMN2 transgene in the severe Taiwanese SMA mouse model. This approach corrects SMN2 splicing, includes exon 7, and fully rescues the SMA phenotype when the ASOs are intracerebroventricularly and subcutaneously injected at high doses into pre-symptomatic pups.37 Accordingly, we subcutaneously injected suboptimal doses of 10–50 μg of SMN-ASO on P2 and P3 in SMA mice on a congenic C57BL/6N background in order to produce an intermediate SMA mouse model. Because 40 and 50 μg were shown to prolong survival too much (data not shown), we restricted our extended analysis to SMA mice injected with 10, 20, and 30 μg SMN-ASO and compared survival to that of uninjected and control (ctrl)-ASO-injected mice (Figure 1A). We found that 30 μg SMN-ASO injection on P2 and P3 is an adequate dosage for generating an intermediate SMA mouse model surviving approximately four weeks (26 ± 9.48 days). Using the same injection scheme, we observed a much larger increase in survival at each dose in SMA mice on a congenic FVB/N background, emphazising the relevance of the genetic background in influencing SMA disease severity (Figure S1A). We therefore performed all experiments with SMA mice on a C57BL/6N background to reliably dissect the modifying effect of PLS3. Next, the PLS3 transgenic allele24 was crossed into the Taiwanese SMA mouse strain.35 We generated a SMA mouse (SmnKO/KO;SMN2tg/0) overexpressing PLS3 (here named SMA-PLS3het for SmnKO/KO;SMN2tg/0;PLS3tg/0 and SMA-PLS3hom for SmnKO/KO;SMN2tg/0;PLS3tg/tg), as well as Smn heterozygous mice (here named HET for SmnKO/WT;SMN2tg/0) overexpressing PLS3 (HET-PLS3het for SmnKO/WT;SMN2tg/0;PLS3tg/0 and HET-PLS3hom for SmnKO/WT;SMN2tg/0;PLS3tg/tg). HET mice were used as controls. The breeding scheme is shown in Figure S1B. All pups were injected subcutaneously with 30 μg SMN-ASO at P2 and P3. Strikingly, more than 60% of SMA-PLS3hom mice survived >250 days, and 30% were still alive at >400 days (mean survival: 219 ± 176.78 days). SMA-PLS3het mice showed a less prolonged survival (169 ± 176.11 days), reflecting a PLS3-dosage effect (Figure 1B). SMA-PLS3het and SMA-PLS3hom mice continuously gained weight within the observed period of time (Figure 1D). Although SMA-PLS3hom mice were slightly heavier than SMA-PLS3het mice, they did not reach the body weight of HET mice. SMA-PLS3het and SMA-PLS3hom mice developed ear and tail necrosis over time, starting from one month of age (Figure 1C, right part), in similarity to SmnKO/KO;SMN2tg/tg mice.46 Additionally, these animals showed normal behavior in feeding and grooming. SMN expression upon SMN-ASO injection was verified by immunoblot analysis at P10 so that variability among animals could be excluded. Interestingly, SMN-ASO injection produced very little increase in SMN amount in the brain (data not shown) or spinal cord (Figure 1E), whereas the liver of the same animals clearly showed increased SMN expression (Figure S1C). Furthermore, upon SMN-ASO injection, no significant difference in SMN amount in the spinal cord was found between SMA and SMA-PLS3hom (Figure 1F). This demonstrates an adequate systemic distribution but relatively low penetration into the central nervous system at the doses tested. SMN-ASO injection ameliorated the multi-organ dysfunction of the lung, heart, and intestine in all SMA mice compared to ctrl-ASO injected mice (Figure 2). Furthermore, SMA-PLS3het and SMA-PLS3hom mice showed a tendency toward increased heart size (normalized to body weight) (Figures 2A and 2B).
Figure 1.
PLS3 Rescues Survival in an Intermediate SMA Mouse Model
(A) Kaplan-Meier curves show the survival rate of SMA mice injected at P2 and P3 with different SMN-ASO doses. Uninjected: 16 ± 2.85 days, n = 22. 50 μg ctrl-ASO: 14 ± 3.89 days, n = 15. 10 μg SMN-ASO: 21 ± 3.95 days, n = 11. 20 μg SMN-ASO. 21 ± 7.37 days, n = 8. 30 μg SMN-ASO: 26 ± 9.48 days, n = 22.
(B) Kaplan-Meier curves show the survival rate of PLS3-overexpressing or –non-overexpressing SMA mice treated with 30 μg of SMN-ASO at P2 and P3. PLS3 overexpression drastically increased the survival to 169 ± 176.11 days (n = 23) for SMA-PLS3het and to 219 ± 176.78 days (n = 11) for SMA-PLS3hom in comparison to SMA mice without PLS3 overexpression. A log-rank (Mantel-Cox) test was used.
(C) PLS3 overexpression improved phenotypical development in the intermediate SMA mouse model, similar to results for HET mice. The scale bar represents 2 cm.
(D) Weight progression of SMA mice in comparison to HET mice (n ≥ 10).
(E) Immunoblot analysis of spinal-cord lysates from uninjected and SMN-ASO-treated SMA and HET mice. Note SMN-ASO only very slightly increased the SMN amount (n = 3).
(F) Immunoblot of spinal-cord lysates shows that PLS3 overexpression does not induce SMN elevation (n = 3). n.s., non-significant by a two-tailed Student’s t test.
Error bars represent SEM.
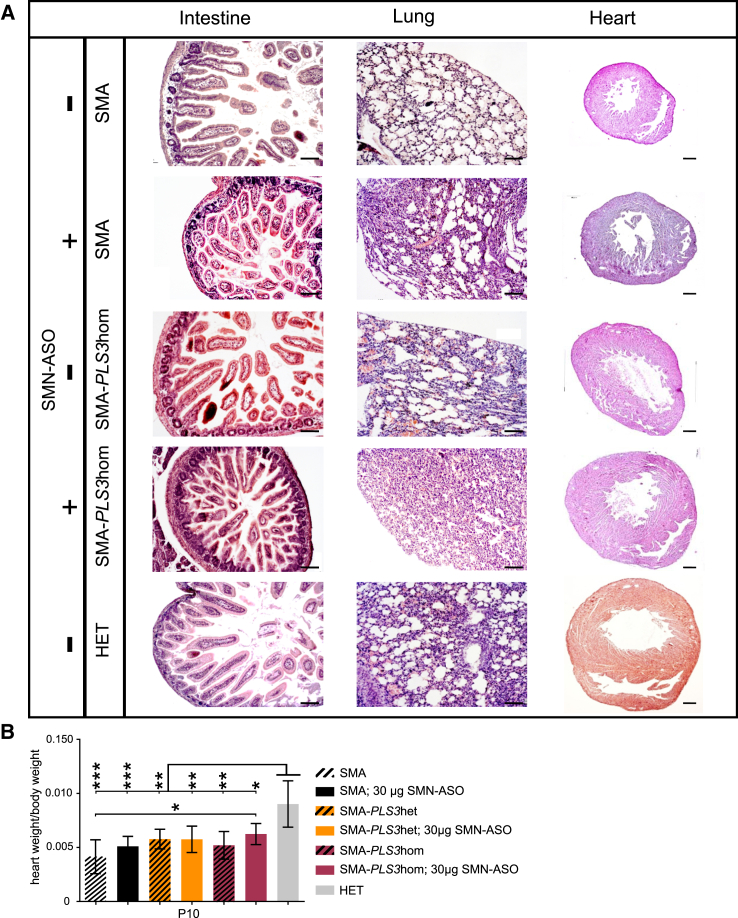
Figure 2.
Low-Dose SMN-ASO Together with PLS3 Overexpression Improves Multi-Organ Dysfunction in the SMA Mouse Model
(A) Representative pictures of histological sections from intestine, lung, and heart (P10). Injection of SMN-ASO and PLS3 overexpression improved intestine, lung, and heart phenotypes in SMA mice. An increased number of intact intestinal villi as well as a better organization of the secretory cells, less emphysema with ruptured alveolar septa, enlarged alveolar spaces in the lung, and an increased heart size can be observed. The scale bar represents 100 μm.
(B) An accumulative effect on the heart size was observed in SMN-ASO-injected SMA mice. The size further increased when PLS3 was overexpressed (n ≥ 3 per genotype). ∗p < 0.05; two-tailed Student’s t test.
Error bars represent SEM.
Our data show that a combinatorial therapy involving SMN-ASO and PLS3 overexpression has a highly beneficial ability to counteract SMA, as already demonstrated by natural protection in human asymptomatic SMN1-deleted siblings.
PLS3 Overexpression Increased NMJ Size, Number of Proprioceptive Inputs, and Motoric Ability in the Intermediate SMA Mouse Model
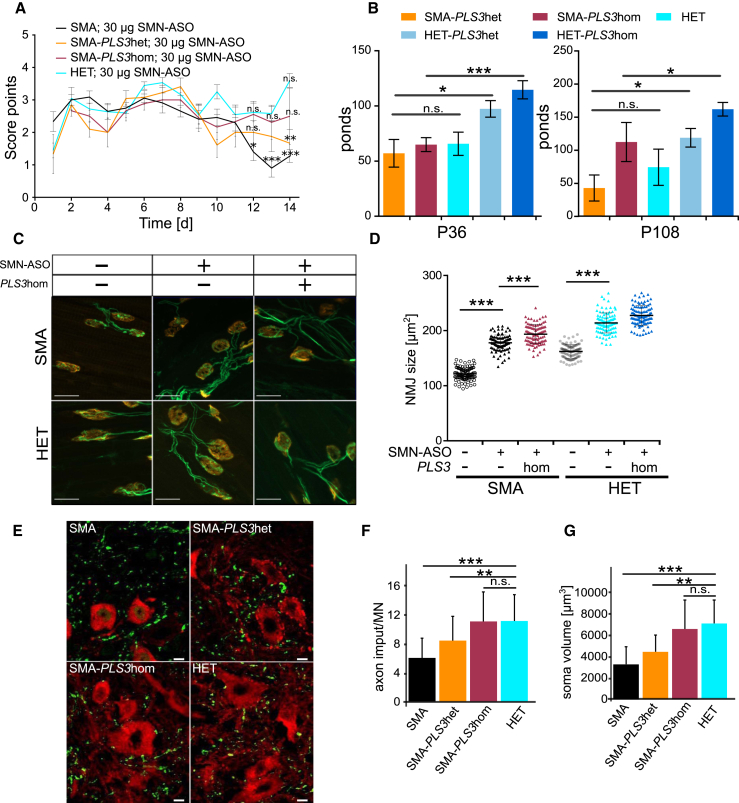
To better understand the impact of PLS3 overexpression in the intermediate SMA mouse model, we characterized motoric performance by using the tube test and grip-strength test. Moreover, we carried out morphological studies comprising NMJ size measurements, as well as quantification of proprioceptive inputs per MN.
In tube test performance there was no significant difference between SMN-ASO-injected SMA, SMA-PLS3het, SMA-PLS3hom, and HET mice at very early postnatal time-points. In contrast, at P13 a significant difference between SMN-ASO-injected SMA and HET mice was observed and was partially rescued by heterozygous and homozygous PLS3 overexpression (Figure 3A). In addition, we measured the grip strength at P36 and P108 and found that although P36 SMA-PLS3het and SMA-PLS3hom mice performed the test with diminished grip strength in comparison to the corresponding control group (HET, HET-PLS3het and HET-PLS3hom, this parameter significantly improved in SMA-PLS3hom mice at P108 (Figure 3B). SMN-ASO-injected SMA-PLS3het or SMA-PLS3hom mice that survived >250 days did not show any abnormal motoric movement or paralysis.
Figure 3.
Tube Test, Grip-Strength Test, NMJ Size, and Proprioceptive-Input Measurements Confirm Improvement in the Intermediate SMA Mouse Model upon PLS3 Overexpression
(A) Tube test of neonatal SMN-ASO-injected mice (P1–P14). SMA-PLS3het and SMA-PLS3hom mice, but not SMA mice, show an improvement in performance (P12–P14) (n ≥ 10).
(B) Grip strength test at P36 and P108 was fully restored in SMA-PLS3hom mice in comparison to HET and HET-PLS3het mice (n ≥ 5).
(C) Representative pictures of NMJ stained with SV2 and NF (green) for the neuronal part and bungarotoxin (red) for the postsynaptic part. The scale bar represents 20 μm.
(D) Quantification shows that injection of SMN-ASO significantly increased the NMJ size in comparison to that of NMJs of untreated SMA mice at P10. Upon PLS3 overexpression, an accumulated effect of SMN-ASO and PLS3 overexpression was observed at the NMJ level (n = 5 per genotype, 100 NMJs measured per animal).
(E) Representative pictures of MN soma (CHAT, red) and proprioceptive input (VGLUT1, green) derived from SMN-ASO-injected mice (P21). The scale bar represents 10 μm.
(F and G) Quantification shows that SMN-ASO injection and PLS3 overexpression significantly improved the number of proprioceptive inputs on the MN soma (n ≥ 70). n.s., non-significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001, two-tailed Student’s t test.
Error bars represent SEM.
Analysis of the NMJ structures revealed that injection of SMN-ASO significantly increased the NMJ size in both SMA and HET mice (Figures 3C and 3D). An accumulative effect of SMN-ASO and PLS3 overexpression on NMJ size was observed at P10 (Figure 3D), and this effect was consistent with our previous findings regarding the role of PLS3 in NMJ structure and function.24 Furthermore, quantification of the vesicular glutamate transporter 1 (VGLUT1) puncta on the MN soma revealed an increased number of proprioceptive inputs and MN soma volume upon SMN-ASO injection. Overexpression of PLS3 together with SMN-ASO has an additive effect and is able to restore the reduced number of proprioceptive inputs and MN soma volume size observed in SMA animals (Figures 3E, 3F, and 3G).
SMA Impairs Endocytosis, which Is Rescued by PLS3 Overexpression
PLS3 is an F-actin-binding and -bundling protein that is involved in many cellular processes.20, 21 Knockdown of Sac6p, the ortholog of PLS3 in yeast, leads to disturbed endocytosis.33 Moreover, F-actin, which is important for all types of endocytosis,22, 47 has been shown to be disturbed in SMA.24, 48, 49 Therefore, we hypothesized that reduced SMN amount might impair endocytosis and can be rescued by PLS3 overexpression.
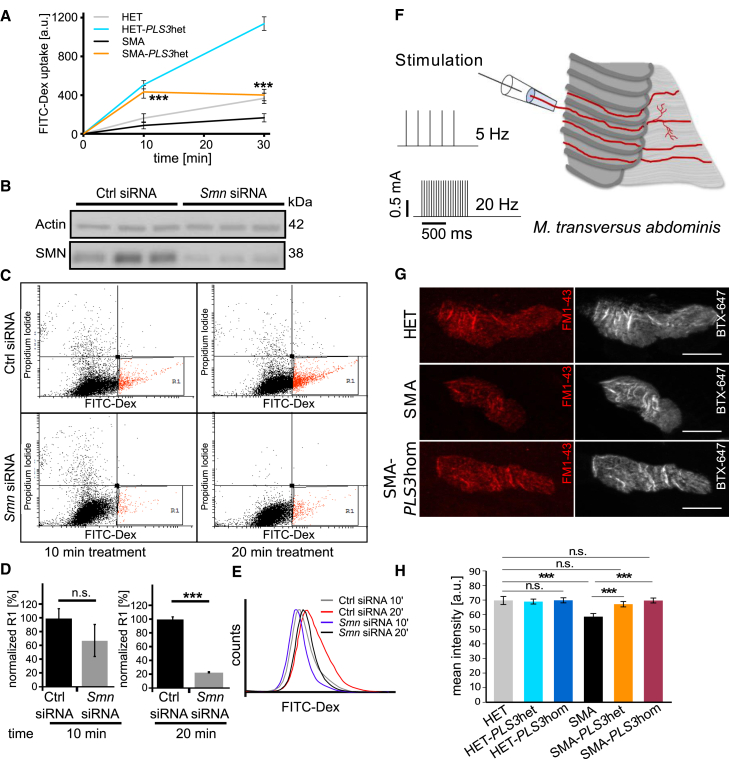
To verify this theory, we first analyzed the uptake of fluorescently labeled dextran (FITC-Dex) by fluid-phase endocytosis47 in murine embryonic fibroblast cell lines (MEFs) derived from SMA, SMA-PLS3het, HET, and HET-PLS3het mice. We used fluorescence microscopy imaging to quantify FITC-Dex uptake in cells fixed at different time points. A strong reduction in endocytic uptake of FITC-Dex was observed in SMA compared to HET MEFs, but uptake was significantly restored by PLS3 overexpression (Figure 4A). These results were further complemented by a fluorescence-activated cell sorting (FACS)-based analysis of FITC-Dex uptake by endocytosis in MN-like NSC34 cells. The method was first optimized in NSC34 cells with two known endocytosis-disrupting conditions as controls: low temperature (4°C) and latrunculin A, an F-actin-depolymerizing reagent.50, 51, 52 Cells were treated with FITC-Dex, and its uptake was quantified at different time points within the highly endocytic cells, which are shown in the region 1 (R1) gate (Figure S2A). Under these unfavorable conditions, a marked decrease in endocytic uptake was found in these cells in comparison to untreated cells or cells grown at 37°C (Figures S2A and S2B).
Figure 4.
PLS3 Overexpression Rescues Impaired Endocytosis in SMN-Depleted NSC34 Cells and at Presynaptic Sites of NMJs in SMA Mice
(A) Quantification of FITC-Dex uptake in MEFs was performed via fluorescence-intensity analysis (n = 3 per genotype and time point, 100 cells measured per cell line). The amount of uptake in SMA versus SMA-PLS3het mice was significantly different at 10 and 30 min.
(B) Immunoblot analysis shows efficient Smn siRNA-mediated knockdown in NSC34 cells.
(C) Representative dot plots showing FITC-Dex uptake in SMN-depleted NSC34 cells at 10 and 20 min (n = 3, 104 cells measured per FACS experiment).
(D and E) Quantification of R1 population and histogram plots show a significant reduction in uptake upon SMN downregulation.
(F) Experimental setup for stimulation of the N. intercostalis innervating the TVA muscle.
(G) Representative pictures of NMJs from HET, SMA, and SMA-PLS3hom muscles. Staining of postsynaptic receptors (BTX-Alexa647, gray) helped to define the area in which FM1-43 uptake (red) at the presynaptic terminals was analyzed (n = 3 per genotype, ∼100 NMJs measured per genotype). The scale bar represents 10 μm.
(H) Quantification of the FM1-43 mean intensity at the presynaptic terminals at P10 in TVA muscles without ASO injection under low-frequency stimulation (5 Hz, 1 s). n.s., non-significant; ∗∗∗p < 0.001, two-tailed Student’s t test.
Error bars represent SEM.
Next, we analyzed the impact of a reduced amount of SMN on endocytic FITC-Dex uptake at different time points in NSC34 cells treated with mouse SMN siRNAs as well as in HEK293T cells (non-neuronal cells), treated with human SMN siRNAs in comparison to control siRNA. The efficiency of SMN downregulation was confirmed by immunoblot analysis (Figure 4B). Analysis of R1-gated cells showed a decrease in the rate and amount of FITC-Dex uptake upon SMN knockdown at 10 and 20 min (Figures 4C, 4D, and 4E). A similar reduction with SMN downregulation was observed in HEK293T cells, which illustrates that impaired endocytosis is a general phenomenon caused by SMN deficiency (Figures S2C, S2D, S2E, and S2F).
To confirm that impaired endocytosis in SMN-depleted cells also occurs in MNs—very specialized cells where synaptic vesicle recycling is highly regulated—we analyzed endocytosis at the NMJ by measuring FM1-43 dye uptake at the presynaptic terminal upon electrical stimulation.53 We performed the experiments in the Transversus abdominis (TVA) muscle at P10. We performed ex vivo preparations in physiological solution to keep the muscles alive for the subsequent stimulation. Electrical stimulation of the N. intercostalis innervating the TVA muscle (see sketch of experimental procedure, Figure 4F) was being executed in two sets: a low-frequency set at 5 Hz and a high-frequency set at 20 Hz for analysis of FM1-43 uptake. In hippocampal neurons low-frequency stimulation mainly induces clathrin-mediated endocytosis (CME), whereas high-frequency stimulation triggers clathrin-independent endocytosis (CIE) or activity-dependent bulk endocytosis (ADBE).54 The mean FM1-43 intensity in the presynaptic terminal was determined by delimitation of the corresponding area from the postsynaptic site, stained with bungarotoxin conjugated with Alexa647 (BTX-647) (Figure 4G). Presynaptic uptake of FM1-43 dye without electrical stimulation was excluded (Figure S3A). Because NMJs in TVA are reported to be smaller in SMA mice than in HET mice,13, 55 we correlated FM1-43 intensity to NMJ size and confirmed that there is no relation between these two parameters (Figure S3C). At low-frequency stimulation, FM1-43 intensity was significantly reduced in SMA compared to HET mice (Figures 4G and 4H). Most importantly, in SMA-PLS3het and SMA-PLS3hom NMJs, endocytosis was restored to HET levels upon 5 Hz stimulation (Figure 4H). At a high-frequency stimulation of 20 Hz, endocytosis was also reduced in SMA compared to HET mice, and PLS3 overexpression (in both homo- and heterozygous mice) rescued the impaired phenotype (Figure S3B). These results demonstrate that endocytosis is disturbed in the NMJ of SMA mice and that this disturbance is counteracted by PLS3 overexpression.
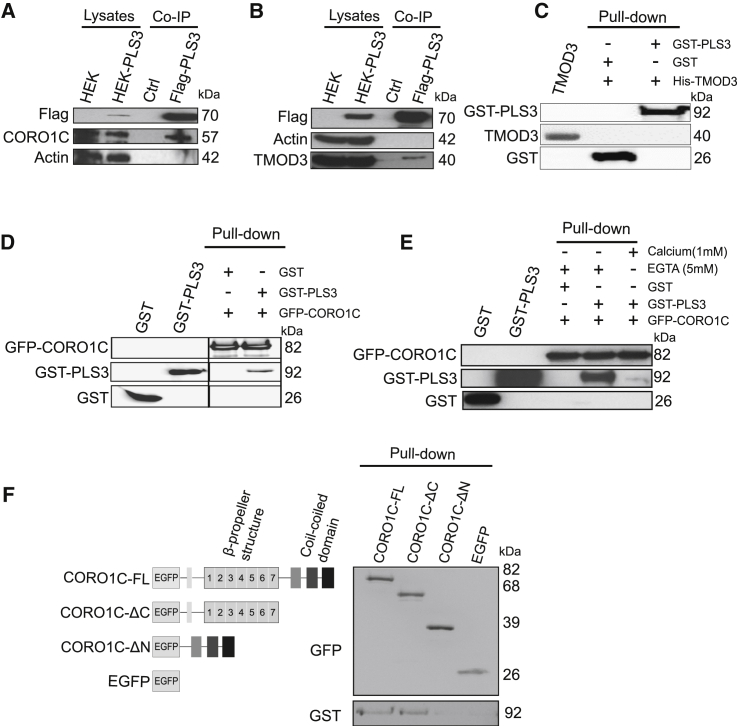
PLS3 Co-precipitates CORO1C and TMOD3 but Directly Binds Only CORO1C
Because SMN deficit impairs endocytosis and the impairment is restored by PLS3 overexpression, we hypothesized that unravelling the interactome of PLS3 would help us to identify further modifiers involved in endocytosis.
Proteome analyses were carried out with a HEK293T cell line stably expressing Flag- and His-tagged PLS3 (Figures S4A and S4B) in conjunction with co-immunoprecipitation (co-IP) and mass spectrometry. We purified the potential PLS3 binding partners by using RSB-100 buffer with high salt concentration. The efficiency of anti-Flag co-IP and purification of PLS3 was verified by immunoblot and silver staining (Figure S4C). Comparison between co-immunoprecipitated proteins from PLS3-overexpressing cells and control cells revealed the presence of 14 proteins, which were only present in the PLS3 co-IP (Table S1).
We selected two actin-related binding partners of PLS3, CORO1C, and TMOD3; these binding partners were also detected by mass spectrometry experiments using a label-free quantification approach (data not shown). Recent data have demonstrated that CORO1C is also involved in endocytosis and has a second actin-binding site that confers co-operative binding to F-actin. The presence of more than one F-actin binding site enables CORO1C to act as an F-actin-binding and -bundling protein similar to PLS3.56, 57, 58 TMOD3 is known to cap pointed ends of actin filaments and to be enriched in leading-edge ruffles and lamellipodia.59, 60 We selected these two PLS3 binding partners as candidates to gain better insight into the role of endocytosis in rescuing SMA.
We performed an additional co-IP and demonstrated that both CORO1C and TMOD3 co-precipitated with PLS3 (Figures 5A and 5B). To validate these interactions, pull-down assays were carried out and showed no direct interaction between His-TMOD3 and GST-PLS3 (Figure 5C), but they did show a direct interaction between EGFP-CORO1C and GST-PLS3 (Figure 5D and Figure S4D). Because the EF-hand domains of PLS3 have Ca2+-binding ability and modulate the function of PLS3 in a Ca2+-dependent manner,61 we analyzed whether Ca2+ affects PLS3-CORO1C interaction. Performing a pull-down assay in the presence (1 mM Ca2+) or absence (5 mM EGTA) of Ca2+, we found that the CORO1C-PLS3 interaction was disrupted in the presence of Ca2+ (Figure 5E). CORO1C contains a C-terminal coiled-coil domain and seven WD repeats, which form a β-propeller structure.62, 63 The C-terminal coiled-coil domain has a self-oligomerization function, and the β-propeller structure forms a docking platform for protein-protein interactions. To elucidate which region of CORO1C is responsible for its interaction with PLS3, we generated full-length CORO1C (CORO1C-FL) and GFP-tagged deletion constructs of C- and N-terminal regions of CORO1C (CORO1C-ΔC and CORO1C-ΔN, respectively). Pull-down assays showed that the N-terminal part of CORO1C, containing the β-propeller structure, directly interacts with GST-PLS3 (Figure 5F).
Figure 5.
PLS3 Interacts with CORO1C and TMOD3 in HEK293T Cells, but only CORO1C Directly Interacts with PLS3
(A and B) Immunoblots of co-IP experiments show the interaction of PLS3 with CORO1C (A) and TMOD3 (B).
(C) Pull-down experiments with His-TMOD3 and GST-PLS3. Immunoblots probed with GST and TMOD3 antibodies show that PLS3 does not interact directly with TMOD3.
(D) Pull-down experiments with EGFP-CORO1C and GST-PLS3 show direct interaction of PLS3 with CORO1C.
(E) PLS3-CORO1C interaction is disrupted in the presence of 1 mM Ca+2.
(F) Pull-down assay of purified protein domains of CORO1C with GST-PLS3 shows that the interaction of PLS3 and CORO1C is mediated through the N-terminal part of CORO1C.
Immunostainings in MEFs derived from PLS3het mice revealed colocalization of PLS3 with both CORO1C and TMOD3 along F-actin filaments. In addition, PLS3 and CORO1C are strongly enriched in lamellipodia structures at growing edges under the plasma membrane (Figures S5A, S5B, and S5C). Because PLS3 was shown to localize in growth cones and axons,18 we further analyzed the expression level of CORO1C in primary MN culture. PLS3 and CORO1C were highly elevated in MNs, and both were detected in the cell body, axon, and growth cone (Figures S6A and S6B). To further investigate whether PLS3 overexpression had an effect on the expression level of its binding partners, we analyzed spinal-cord samples from P10 HET mice with and without PLS3 overexpression by immunoblot. Spinal-cord lysates from HET-PLS3het and HET-PLS3hom mice indicated a tendency toward increased expression of CORO1C and TMOD3 when these mice were compared to HET mice (Figure S7).
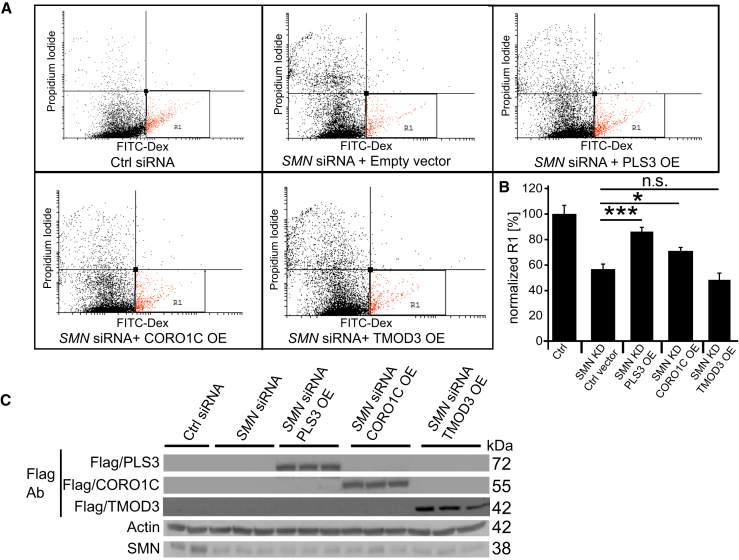
CORO1C and PLS3 but Not TMOD3 Rescue Impaired Endocytosis as Well as Actin Dynamics in SMN-Deficient Cells
Direct interaction of PLS3 with CORO1C suggested a similar mode of action for both proteins and a likely beneficial effect of CORO1C in SMA cells. To further investigate the possible endocytosis-rescuing role of CORO1C in SMN-deficient cells, we again performed fluid-phase endocytosis assays in HEK293T cells, which proved to be a suitable system and, in contrast to NSC34 cells or primary MNs, allows for efficient transfection with plasmid DNA. Quantification of flow-cytometry data indicated that overexpression of CORO1C as well as PLS3 but not TMOD3 was able to rescue endocytosis in SMN-deficient cells (Figures 6A and 6B). Efficiency of SMN siRNA-mediated downregulation and overexpression of CORO1C, TMOD3, and PLS3 was confirmed by immunoblot analysis (Figure 6C).
Figure 6.
Overexpression of PLS3 and CORO1C but Not of TMOD3 Improve Endocytosis in SMN-Deficient Cells
(A and B) Quantification of the R1-gated population shows that PLS3 and CORO1C significantly increase endocytosis in SMN-deficient HEK293T cells after 20 min treatment.
(C) Immunoblots show siRNA-mediated knockdown of SMN and overexpression of PLS3, CORO1C, and TMOD3 in HEK293T cells (n = 5, 104 cells measured per FACS experiment). n.s., non-significant; ∗p < 0.05; ∗∗∗p < 0.001, two-tailed Student’s t test.
Error bars represent SEM.
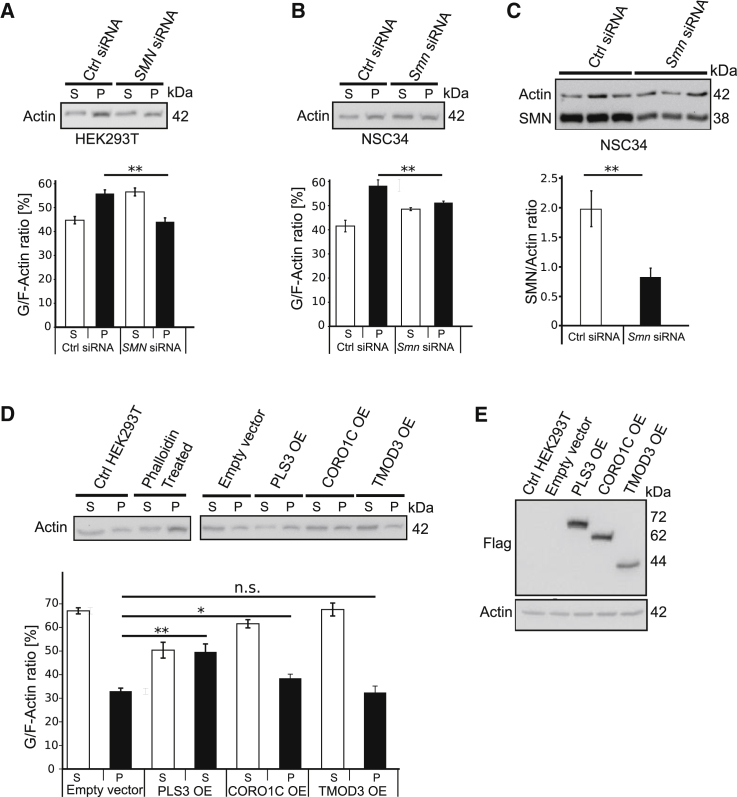
We analyzed the impact of siRNA-mediated knockdown of PLS3, CORO1C, or TMOD3 on endocytosis in HEK293T cells. Consistently, knockdown of PLS3 and CORO1C but not of TMOD3 decreased endocytic FITC-Dex uptake (Figures S8A, S8B, and S8C). Reduced PLS3, CORO1C, or TMOD3 were confirmed by immunoblot analysis (Figures S8D, S8E, and S8F). Although the involvement of F-actin on endocytosis is well documented,22 as is the impact of reduced amounts of SMN on F-actin dynamics and localization,24, 48 we addressed the question about the role of PLS3, CORO1C, and TMOD3 on F-actin dynamics and their ability to restore impaired F-actin amounts caused by reduced amounts of SMN. Our in vivo assay of the G/F-actin ratio revealed that upon SMN knockdown in NSC34 and HEK293T, the amount of F-actin is significantly reduced (Figures 7A–7C). To investigate the effect of PLS3 and its binding partners on F-actin dynamics, we transiently transfected PLS3, CORO1C, TMOD3, or control plasmids into HEK293T cells. Measurement of F-actin amounts showed that overexpression of PLS3 and CORO1C but not TMOD3 significantly increased F-actin amounts (Figures 7D and 7E). Furthermore, our previous studies in SMA mice showed that the amount of F-actin is reduced at the presynaptic site at the NMJ structure and that overexpression of PLS3 compensates for it.24 Taken together, these findings support the idea that PLS3 and CORO1C but not TMOD3 play an important role in endocytosis by restoring F-actin-dependent processes.
Figure 7.
SMN Knockdown Decreases F-actin Amount, which Is Increased by PLS3 and CORO1C but Not TMOD3
(A) Representative immunoblots of in vivo G/F-actin ratio assay in SMN-knockdown HEK293T cells. Blot quantification indicates a reduction (12%) in F-actin amount upon SMN knockdown in HEK293T cells.
(B) Knockdown of SMN in murine MN-like NSC34 cells also decreases the amount of F-actin (7%) in comparison to that in control siRNA-treated cells.
(C) Immunoblot analysis shows a significant reduction in SMN amount upon Smn siRNA-mediated knockdown in NSC34 cells.
(D) An in vivo G/F-actin assay shows that overexpression of PLS3 and CORO1C but not TMOD3 significantly increased the amount of F-actin in comparison to control vector.
(E) Immunoblot analysis shows the overexpression (OE) of PLS3, CORO1C, and TMOD3 in HEK293T cells (n = 5). n.s., non-significant; ∗p < 0.05; ∗∗p < 0.01; two-tailed Student’s t test.
Error bars represent SEM.
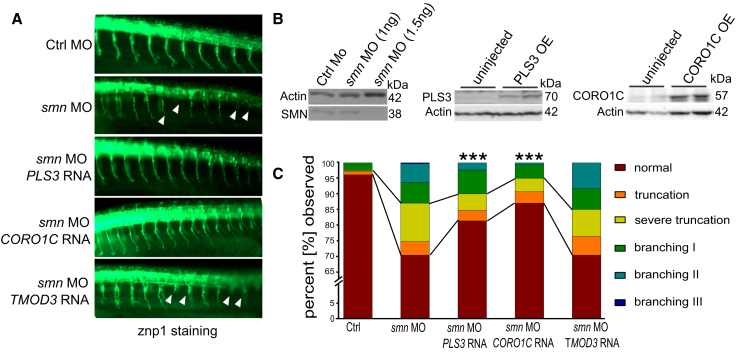
CORO1C but Not TMOD3 Ameliorates SMA Phenotype in Zebrafish Smn Morphants
Zebrafish serve as an excellent alternative vertebrate model to help us understand the genetics and molecular mechanisms of MN disorders.64 To functionally characterize the role of PLS3 interacting partners on the SMA phenotype, we investigated the modifying effect of CORO1C or TMOD3 on the axonal defects caused by loss of Smn. We co-injected CORO1C, TMOD3 or, as a positive control, PLS3 mRNA together with smn antisense morpholino oligonucleotide (MO) into mnx1:eGFP transgenic zebrafish embryos. Importantly, endocytosis-driven internalization of membrane and proteins at the leading edge of the growth cone is crucial for axonal outgrowth and branching.65 As previously shown, injection of smn-MO induced truncations and increased branching of MN axons,18, 23, 61 and both effects were rescued by concomitant PLS3 overexpression. Strikingly, a rescue comparable to that with PLS3 mRNA was obtained upon co-injection of smn-MO with CORO1C mRNA, whereas TMOD3 mRNA had no effect (Figures 8A and 8C). The effect of smn-MO knockdown and PLS3 and CORO1C overexpression was confirmed by immunoblot (Figure 8B).
Figure 8.
CORO1C Rescues the Motor-Neuron Phenotype in SMN-Depleted Fish
(A) Lateral view at 10–12 somites directly posterior to the yolk sac of 34 hpf zebrafish embryos injected with control-MO, smn-MO, smn-MO + PLS3 mRNA, smn-MO + TMOD3 mRNA, and smn-MO + CORO1C mRNA. MN axons in Smn-depleted fish evidence truncation and branching phenotypes when these fish are compared to control-MO fish.
(B) Immunoblots show, from left to right, the dose-dependent effect of smn-MO knockdown, overexpression of PLS3, and overexpression of CORO1C.
(C) Quantitative analysis of MN axons shows that PLS3 and CORO1C significantly improved the axonal truncation and branching phenotypes in Smn-depleted fish (branching types I, II, and III correspond to mild, intermediate, and severe axonal branching, respectively. Evaluated axons: n ≥ 300). ∗∗∗p < 0.001, Fisher’s exact test.
Discussion
The main findings of our work are the following: (1) low amounts of subcutaneously injected SMN-ASO in the severe Taiwanese SMA mouse model enable the generation of an intermediate SMA mouse model with a prolonged lifespan due to an ameliorated systemic organ impairment; (2) PLS3 rescues survival and motoric abilities in this intermediate SMA model, proving that PLS3 is a protective modifier if elevated in the appropriate context; (3) reduced SMN amounts impair endocytosis in MN-like cells and synaptic-vesicle recycling at NMJ in SMA mice, and both effects are rescued by PLS3 overexpression; (4) mass-spectrometry analyses and protein-interaction studies revealed that PLS3 directly binds to CORO1C in a calcium-dependent manner, whereas TMOD3 only associates with PLS3; (5) CORO1C, but not TMOD3, rescues endocytosis in cells; (6) reduced F-actin amounts due to SMN deficiency are restored by overexpression of PLS3 and CORO1C; and (7) similar to what was previously shown for PLS3, CORO1C, but not TMOD3, rescues the SMA phenotype in Smn-depleted zebrafish.
This work demonstrates the power of genetic modifiers and their ability to unravel key cellular mechanisms and protein networks that counteract disease-causing processes (Figure 9). Most importantly, this knowledge might open new therapeutic avenues in the treatment of individuals with SMA, by allowing the use of genetic modifiers involved in endocytosis in addition to small molecules or pharmacological compounds that induce SMN expression or stability.
Figure 9.
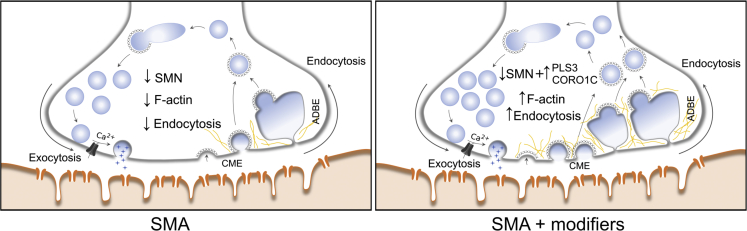
A Model Presenting the Compensatory Role of PLS3 and CORO1C in the Process of Endocytosis in SMA
CME and ADBE stand for clathrin-mediated endocytosis and activity-dependent bulk endocytosis, respectively.
PLS3 Rescues Survival in an SMN-ASO-Induced Intermediate SMA Mouse Model
PLS3 is the first and only SMA protective modifier to have been reported in humans to date.18 Asymptomatic in comparison to symptomatic siblings show elevated PLS3 expression in lymphoblastoid cell lines but not in fibroblasts.18 However, iPSCs generated from these fibroblasts and differentiated into MNs present a high upregulation of PLS3, clearly supporting a protective role of PLS3 in MNs and particularly in growth cones.19 Meanwhile, additional asymptomatic SMN1-deleted individuals in whom PLS3 upregulation has been excluded have been described, suggesting that other SMA modifiers do exist within the human population (Bernal et al.66 and Wirth, unpublished data). The presence of at least 3–4 SMN2 copies in all asymptomatic SMN1-deleted individuals implies that a modifier exerts its protective function only if a certain amount of SMN protein, indispensable in every cell, is available. If the SMN level falls below a certain threshold, a plethora of pathways are disturbed, affecting not only MN but also a number of other cell types and organs and resulting in severe organ dysfunction that cannot be rescued by protective modifiers.16, 67 We therefore considered it unsuitable to search for the cellular mechanism causing NMJ vulnerability and MN dysfunction by using cells derived from individuals with type I SMA or severely affected SMA mouse models. This view is supported by the finding that PLS3 overexpression in both the severe Taiwanese SMA and the Δ7SMA mouse models failed to increase survival,24, 25 despite ameliorating NMJ function and motoric abilities in the Taiwanese SMA model.24
In individuals with milder SMA, the SMN amount appears to be sufficient to exert its housekeeping function in all cells except MNs.47 Most remarkable are individuals with type IV SMA; in these individuals, the disease onset is after the second decade of life,7 which suggests a problem in MN and/or NMJ maintenance but not in development. Because the requirement of SMN protein in humans and mice differs, it turned out to be rather complicated to produce an appropriate intermediate or mild SMA mouse model by varying the SMN2 copy number. Two human SMN2 copies on a murine Smn-null background cause a very severe SMA phenotype, whereas mice with three SMN2 copies are already asymptomatic.35, 46, 68 Therefore, production of an intermediate or mild SMA mouse model was challenging.
It has been reported that pre-symptomatic injection of higher doses of SMN-ASOs or morpholino in different SMA models carrying two SMN2 copies fully rescues the phenotype via a correction of SMN2 splicing, except that mouse size remains reduced.69, 70 The influence of SMN-ASO on exon 7 inclusion steadily decreases until completely vanishing six months after injection.37 Contrary to humans, who can develop SMA later in life, SMN ASO- or morpholino-treated mice showed normal life expectancy, suggesting that low amounts of SMN are sufficient to maintain functional integrity of cells in older mice. Most importantly, turning on SMN expression in an inducible SMA model at various time points has shown an absolute requirement for SMN before P17, which matches with the maturation of NMJs.71 In line with this, later administration of drugs or small molecules that elevate SMN amounts have only moderate beneficial effect, and this only in a low-dose-pre-treated Δ7-SMA mouse or intermediate mouse model.72, 73, 74
As reported in our previous studies, PLS3 seems to compensate for the SMN deficit: NMJs of SMA-PLS3het mice have shown substantial increases in connectivity at the presynaptic level and delayed axonal pruning, allowing the otherwise poorly developed SMA NMJs to become properly innervated and mature adequately.24 Using SMN-ASO at suboptimal doses allowed us to generate an intermediate mouse model surviving for about 1 month. Most importantly, PLS3 overexpression prolonged survival to more than 400 days in 30% and 250 days in 60% of mice. Because the PLS3 transgene has been introduced into the ROSA26 locus on chromosome 6 and is equally expressed in both genders, we did not find any gender-specific effect in mice (data not shown). Despite PLS3 overexpression, the suboptimal amount of SMN caused premature death indistinguishable from that in SMA mice in about 25% of animals. We assume that the distribution of the suboptimal SMN-ASO failed to uniformly disperse in every single organ and that this non-uniform dispersal have caused the earlier death. However, in the majority of the animals, PLS3 overexpression led to a marked increase in survival, suggesting that PLS3 is able to counteract the cellular defects caused by SMN deficit. Recently, a genetically engeneered intermediate mouse model, which could be an interesting model for future studies of modifiers and drugs, has been generated.74 SMN-ASO or chemical compounds restoring SMN2 splicing are currently being used in several ongoing large clinical trials in individuals with type I to III SMA (Web Resources).
Most importantly, our study suggests that a combinatorial therapy, SMN-ASO plus PLS3 overexpression, or a drug or small molecule that restores endocytosis might be a more successful strategy for fully protecting individuals from SMA rather than only amelioriating it. In type I SMA—half of all individuals with SMA—only two SMN2 copies do not allow a sufficient elevation of SMN amounts to provide full MN function. These individuals will mostly benefit from a combined therapy. Because in SMA mice SMN upregulation showed only moderate beneficial effect after disease onset,72, 73, 74 individuals with type II and III SMA might also benefit from a therapy that acts independently of SMN to counteract impaired endocytosis and/or restore F-actin amounts; one such therapy might involve ROCK inhibitors.75
Endocytosis and Synaptic-Vesicle Recycling at the NMJ Are Impaired by Reduced SMN Level and Rescued by PLS3 and CORO1C Overexpression
PLS3 and CORO1C are both F-actin-binding and -bundling proteins.57, 76 F-actin dynamics is pivotal for cellular integrity and is involved in cellular shape, migration, vesicular trafficking, RNA translation, endocytosis, and other processes.21, 22, 47 In the highly polarized MNs all these processes are particularly relevant. We confirmed that SMN-depleted cells show reduced F-actin amounts that might be due to a disturbed transport of β-actin mRNA along the axons.24, 48, 49 Moreover, SMN interacts with profilin, and reduced SMN amounts activate the RhoA pathway in a direct or indirect manner, further hinting toward an involvement of actin dynamics in the pathology of SMA.28, 49, 77 In severely affected SMA mice, zebrafish, worm, and fly models, many F-actin-dependent processes, including axonal growth, axonal connectivity at the NMJ, neurotransmission, F-actin caging, synaptic-vesicle recycling, and proprioceptive input at MN soma are impaired.11, 18, 24, 45, 78, 79 All these processes were either restored or ameliorated by PLS3 overexpression, as shown across species.18, 23, 24, 79
The most prominent phenotype in pls3 KO yeast is impaired endocytosis.33 Here, we demonstrate that reduced SMN levels cause a reduction of endocytosis in various cell types, including NSC34 cells and at the NMJs, as shown by FM1-43 endocytic uptake under low- and high-frequency stimulation. Instead, overexpression of PLS3 and CORO1C restores endocytosis and the SMA-related phenotype in mouse and/or zebrafish. Moreover, siRNA-mediated knockdown of both PLS3 and CORO1C reduces endocytic uptake in various cells. In humans, PLS3 mutations cause osteoporosis and osteoporotic fractures [MIM: 300910], implying a specific important role of PLS3 in bone development and remodeling.80 The cellular mechanism by which PLS3 loss causes osteoporosis remains elusive. In this respect, fibroblasts derived from a male individual harboring a PLS3 nonsense mutation showed reduced endocytotic uptake (Wirth lab, unpublished data).
In neurons, synaptic vesicles are organized in different pools in the presynaptic terminal. The process of endocytosis is crucial for replenishing the recycling pool (RP), which supplies vesicles to the readily releasable pool (RRP) for neurotransmission.81, 82 In SMA mice, the organization and amount of docked vesicles are significantly reduced in the presynaptic site, causing reduced neurotransmitter release at NMJ.12, 24, 83 Moreover, the RRP size is significantly reduced in SMA, and the depletion and refilling time constants of this pool tend to be slower.84 Therefore, we hypothesized that endocytosis might be the key cellular process disturbed in SMA and the cause of the reduced synaptic vesicle number in the terminals. F-actin is essential in all types of endocytosis,47, 85 and its inhibition reduces the endocytosis in neurons under high-rate stimulation.86 Accordingly, PLS3 overexpression is able to rescue the impaired endocytosis in SMA through the crucial role of F-actin in endocytosis.
Because PLS3 and CORO1C interact Ca2+ dependently and because disturbed calcium homeostasis has been shown in SMA MNs,15 decreased endocytosis might be a result of the combination of reduced calcium influx and reduced F-actin dynamics.31 This hypothesis is supported by the fact that overexpression of just the EF-hand domain of PLS3 can be still protective and ameliorate the SMA phenotype in the Smn-depleted fish.61 On the other hand, overexpression of PLS3 without calcium-binding ability does not compensate for SMN loss, implying that calcium is indispensable for the PLS3-rescuing function.61
In an unbiased screen for modifiers of SMA in C. elegans, several other modifiers with direct roles in endocytosis have been identified.79 We therefore cannot exclude the possibility that the reduced endocytosis is solely caused by reduced F-actin in the presynaptic site. Further studies need to be done if we are to better understand the major impaired type of endocytosis and endocytotic traffic in SMA.
Disturbance in actin dynamics and endocytic pathways is not restricted to SMA, as evidenced by the fact that mutations in profilin 1 (PFN1 [MIM: 176610]) cause amyotrophic lateral sclerosis 18 (ALS18 [MIM: 614808]), mutations in alsin 2 (ALS2 [MIM: 606352]) cause ALS2 [MIM: 205100], and mutations in bicaudal 2 Drosophila-related (BICD2 [MIM: 609797]) lead to autosomal-dominant lower-extremity-predominant spinal muscular atrophy-2 (SMALED2 [MIM: 615290]).87, 88, 89
The Power of Genetic Modifiers
Genetic protective modifiers act either directly (upstream) on the expression or function of a disease-causing gene, transcript, or protein or downstream to compensate for disturbances in a pathway affected by the main disease-causing protein. They can work in the same protein complex with the disease-causing protein or independently to compensate or improve the affected pathway.90 Because next-generation technologies have made exome or genome sequencing feasible, numerous pathogenic mutations have been unexpectedly identified in healthy individuals (often centenarians), opening a fully new era and allowing us to better understand diverse counteracting protein networks and their regulation, which ultimately will result in development of novel therapeutic strategies.
Our data strongly support this concept and should be followed by future studies in healthy individuals carrying pathogenic mutations; such studies will eventually help to understand disease-relevant pathways in humans.
Acknowledgments
We thank CECAD for help with mass-spectrometry-based proteomics and imaging. This work was supported by grants from the Deutsche Forschungsgemeinschaft Wi-945/13-1, Wi-945/14-1, RTG 1970 (B.W.) and KY96/1-1 (M.J.K.), SMA Europe (M.R.), EU FP7 NEUROMICS (B.W.), CMMC (B.W.), IGSDHD (A.K.), and AFM-Telethon (L.T.B.). C.F.B. and F.R. are employees of IONIS Pharmaceuticals.
Published: August 4, 2016
Footnotes
Supplemental Data includes eight figures and one table and are available with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.07.014.
Web Resources
Clinical Trials, http://www.clinicaltrials.gov
Cyflogic, www.cyflogic.com
ImageJ, https://imagej.nih.gov/ij/
OMIM, http://www.omim.org
UCSC Genome Browser, http://genome.uscs.edu
Supplemental Data
References
- 1.Pellizzoni L., Kataoka N., Charroux B., Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- 2.Liu Q., Fischer U., Wang F., Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 3.Schrank B., Götz R., Gunnersen J.M., Ure J.M., Toyka K.V., Smith A.G., Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. USA. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 6.Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA) Hum. Mutat. 2000;15:228–237. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 7.Lunn M.R., Wang C.H. Spinal muscular atrophy. Lancet. 2008;371:2120–2133. doi: 10.1016/S0140-6736(08)60921-6. [DOI] [PubMed] [Google Scholar]
- 8.Feldkötter M., Schwarzer V., Wirth R., Wienker T.F., Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wirth B., Brichta L., Schrank B., Lochmüller H., Blick S., Baasner A., Heller R. Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum. Genet. 2006;119:422–428. doi: 10.1007/s00439-006-0156-7. [DOI] [PubMed] [Google Scholar]
- 10.Lotti F., Imlach W.L., Saieva L., Beck E.S., Hao le T., Li D.K., Jiao W., Mentis G.Z., Beattie C.E., McCabe B.D., Pellizzoni L. An SMN-dependent U12 splicing event essential for motor circuit function. Cell. 2012;151:440–454. doi: 10.1016/j.cell.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mentis G.Z., Blivis D., Liu W., Drobac E., Crowder M.E., Kong L., Alvarez F.J., Sumner C.J., O’Donovan M.J. Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy. Neuron. 2011;69:453–467. doi: 10.1016/j.neuron.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong L., Wang X., Choe D.W., Polley M., Burnett B.G., Bosch-Marcé M., Griffin J.W., Rich M.M., Sumner C.J. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J. Neurosci. 2009;29:842–851. doi: 10.1523/JNEUROSCI.4434-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray L.M., Comley L.H., Thomson D., Parkinson N., Talbot K., Gillingwater T.H. Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2008;17:949–962. doi: 10.1093/hmg/ddm367. [DOI] [PubMed] [Google Scholar]
- 14.Kariya S., Park G.H., Maeno-Hikichi Y., Leykekhman O., Lutz C., Arkovitz M.S., Landmesser L.T., Monani U.R. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2008;17:2552–2569. doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruiz R., Casañas J.J., Torres-Benito L., Cano R., Tabares L. Altered intracellular Ca2+ homeostasis in nerve terminals of severe spinal muscular atrophy mice. J. Neurosci. 2010;30:849–857. doi: 10.1523/JNEUROSCI.4496-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamilton G., Gillingwater T.H. Spinal muscular atrophy: going beyond the motor neuron. Trends Mol. Med. 2013;19:40–50. doi: 10.1016/j.molmed.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Wirth B., Barkats M., Martinat C., Sendtner M., Gillingwater T.H. Moving towards treatments for spinal muscular atrophy: hopes and limits. Expert Opin. Emerg. Drugs. 2015;20:353–356. doi: 10.1517/14728214.2015.1041375. [DOI] [PubMed] [Google Scholar]
- 18.Oprea G.E., Kröber S., McWhorter M.L., Rossoll W., Müller S., Krawczak M., Bassell G.J., Beattie C.E., Wirth B. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320:524–527. doi: 10.1126/science.1155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heesen L., Peitz M., Torres-Benito L., Holker I., Hupperich K., Dobrindt K., Jungverdorben J., Ritzenhofen S., Weykopf B., Eckert D. Plastin 3 is upregulated in iPSC-derived motoneurons from asymptomatic SMN1-deleted individuals. Cell. Mol. Life Sci. 2015;73:2089–2104. doi: 10.1007/s00018-015-2084-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delanote V., Vandekerckhove J., Gettemans J. Plastins: versatile modulators of actin organization in (patho)physiological cellular processes. Acta Pharmacol. Sin. 2005;26:769–779. doi: 10.1111/j.1745-7254.2005.00145.x. [DOI] [PubMed] [Google Scholar]
- 21.Pollard T.D., Borisy G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 22.Engqvist-Goldstein A.E., Drubin D.G. Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 23.Hao le T., Wolman M., Granato M., Beattie C.E. Survival motor neuron affects plastin 3 protein levels leading to motor defects. J. Neurosci. 2012;32:5074–5084. doi: 10.1523/JNEUROSCI.5808-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ackermann B., Kröber S., Torres-Benito L., Borgmann A., Peters M., Hosseini Barkooie S.M., Tejero R., Jakubik M., Schreml J., Milbradt J. Plastin 3 ameliorates spinal muscular atrophy via delayed axon pruning and improves neuromuscular junction functionality. Hum. Mol. Genet. 2013;22:1328–1347. doi: 10.1093/hmg/dds540. [DOI] [PubMed] [Google Scholar]
- 25.McGovern V.L., Massoni-Laporte A., Wang X., Le T.T., Le H.T., Beattie C.E., Rich M.M., Burghes A.H. Plastin 3 Expression Does Not Modify Spinal Muscular Atrophy Severity in the Δ7 SMA Mouse. PLoS ONE. 2015;10:e0132364. doi: 10.1371/journal.pone.0132364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bricceno K.V., Martinez T., Leikina E., Duguez S., Partridge T.A., Chernomordik L.V., Fischbeck K.H., Sumner C.J., Burnett B.G. Survival motor neuron protein deficiency impairs myotube formation by altering myogenic gene expression and focal adhesion dynamics. Hum. Mol. Genet. 2014;23:4745–4757. doi: 10.1093/hmg/ddu189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou J., Barahmand-pour F., Blackburn M.L., Matsui Y., Chansky H.A., Yang L. Survival motor neuron (SMN) protein interacts with transcription corepressor mSin3A. J. Biol. Chem. 2004;279:14922–14928. doi: 10.1074/jbc.M309218200. [DOI] [PubMed] [Google Scholar]
- 28.Rossoll W., Jablonka S., Andreassi C., Kröning A.K., Karle K., Monani U.R., Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J. Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akten B., Kye M.J., Hao le T., Wertz M.H., Singh S., Nie D., Huang J., Merianda T.T., Twiss J.L., Beattie C.E. Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc. Natl. Acad. Sci. USA. 2011;108:10337–10342. doi: 10.1073/pnas.1104928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kye M.J., Niederst E.D., Wertz M.H., Gonçalves Ido.C., Akten B., Dover K.Z., Peters M., Riessland M., Neveu P., Wirth B. SMN regulates axonal local translation via miR-183/mTOR pathway. Hum. Mol. Genet. 2014;23:6318–6331. doi: 10.1093/hmg/ddu350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.See K., Yadav P., Giegerich M., Cheong P.S., Graf M., Vyas H., Lee S.G., Mathavan S., Fischer U., Sendtner M., Winkler C. SMN deficiency alters Nrxn2 expression and splicing in zebrafish and mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2014;23:1754–1770. doi: 10.1093/hmg/ddt567. [DOI] [PubMed] [Google Scholar]
- 32.Vyas S., Béchade C., Riveau B., Downward J., Triller A. Involvement of survival motor neuron (SMN) protein in cell death. Hum. Mol. Genet. 2002;11:2751–2764. doi: 10.1093/hmg/11.22.2751. [DOI] [PubMed] [Google Scholar]
- 33.Kübler E., Riezman H. Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J. 1993;12:2855–2862. doi: 10.1002/j.1460-2075.1993.tb05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haucke V., Neher E., Sigrist S.J. Protein scaffolds in the coupling of synaptic exocytosis and endocytosis. Nat. Rev. Neurosci. 2011;12:127–138. doi: 10.1038/nrn2948. [DOI] [PubMed] [Google Scholar]
- 35.Riessland M., Ackermann B., Förster A., Jakubik M., Hauke J., Garbes L., Fritzsche I., Mende Y., Blumcke I., Hahnen E., Wirth B. SAHA ameliorates the SMA phenotype in two mouse models for spinal muscular atrophy. Hum. Mol. Genet. 2010;19:1492–1506. doi: 10.1093/hmg/ddq023. [DOI] [PubMed] [Google Scholar]
- 36.Flanagan-Steet H., Fox M.A., Meyer D., Sanes J.R. Neuromuscular synapses can form in vivo by incorporation of initially aneural postsynaptic specializations. Development. 2005;132:4471–4481. doi: 10.1242/dev.02044. [DOI] [PubMed] [Google Scholar]
- 37.Hua Y., Sahashi K., Rigo F., Hung G., Horev G., Bennett C.F., Krainer A.R. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El-Khodor B.F., Edgar N., Chen A., Winberg M.L., Joyce C., Brunner D., Suarez-Farinas M., Heyes M.P. Identification of a battery of tests for drug candidate evaluation in the SMNDelta7 neonate model of spinal muscular atrophy. Exp Neurol. 2008;212:29–43. doi: 10.1016/j.expneurol.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 39.Passini M.A., Bu J., Roskelley E.M., Richards A.M., Sardi S.P., O’Riordan C.R., Klinger K.W., Shihabuddin L.S., Cheng S.H. CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2010;120:1253–1264. doi: 10.1172/JCI41615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith D.B., Johnson K.S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 41.Crowe J., Masone B.S., Ribbe J. One-step purification of recombinant proteins with the 6xHis tag and Ni-NTA resin. Mol. Biotechnol. 1995;4:247–258. doi: 10.1007/BF02779018. [DOI] [PubMed] [Google Scholar]
- 42.Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 43.Rappsilber J., Mann M., Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 44.Colaert N., Barsnes H., Vaudel M., Helsens K., Timmerman E., Sickmann A., Gevaert K., Martens L. Thermo-msf-parser: an open source Java library to parse and visualize Thermo Proteome Discoverer msf files. J. Proteome Res. 2011;10:3840–3843. doi: 10.1021/pr2005154. [DOI] [PubMed] [Google Scholar]
- 45.McWhorter M.L., Monani U.R., Burghes A.H., Beattie C.E. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J. Cell Biol. 2003;162:919–931. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., Li H. A mouse model for spinal muscular atrophy. Nat. Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 47.Doherty G.J., McMahon H.T. Mechanisms of endocytosis. Annu. Rev. Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 48.Gabanella F., Pisani C., Borreca A., Farioli-Vecchioli S., Ciotti M.T., Ingegnere T., Onori A., Ammassari-Teule M., Corbi N., Canu N. SMN affects membrane remodelling and anchoring of the protein synthesis machinery. J. Cell Sci. 2016;129:804–816. doi: 10.1242/jcs.176750. [DOI] [PubMed] [Google Scholar]
- 49.Bowerman M., Shafey D., Kothary R. Smn depletion alters profilin II expression and leads to upregulation of the RhoA/ROCK pathway and defects in neuronal integrity. J. Mol. Neurosci. 2007;32:120–131. doi: 10.1007/s12031-007-0024-5. [DOI] [PubMed] [Google Scholar]
- 50.Gachet Y., Hyams J.S. Endocytosis in fission yeast is spatially associated with the actin cytoskeleton during polarised cell growth and cytokinesis. J. Cell Sci. 2005;118:4231–4242. doi: 10.1242/jcs.02530. [DOI] [PubMed] [Google Scholar]
- 51.Shurety W., Stewart N.L., Stow J.L. Fluid-phase markers in the basolateral endocytic pathway accumulate in response to the actin assembly-promoting drug Jasplakinolide. Mol. Biol. Cell. 1998;9:957–975. doi: 10.1091/mbc.9.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ayscough K.R., Stryker J., Pokala N., Sanders M., Crews P., Drubin D.G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaffield M.A., Betz W.J. Imaging synaptic vesicle exocytosis and endocytosis with FM dyes. Nat. Protoc. 2006;1:2916–2921. doi: 10.1038/nprot.2006.476. [DOI] [PubMed] [Google Scholar]
- 54.Kononenko N.L., Puchkov D., Classen G.A., Walter A.M., Pechstein A., Sawade L., Kaempf N., Trimbuch T., Lorenz D., Rosenmund C. Clathrin/AP-2 mediate synaptic vesicle reformation from endosome-like vacuoles but are not essential for membrane retrieval at central synapses. Neuron. 2014;82:981–988. doi: 10.1016/j.neuron.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 55.Torres-Benito L., Neher M.F., Cano R., Ruiz R., Tabares L. SMN requirement for synaptic vesicle, active zone and microtubule postnatal organization in motor nerve terminals. PLoS ONE. 2011;6:e26164. doi: 10.1371/journal.pone.0026164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chan K.T., Roadcap D.W., Holoweckyj N., Bear J.E. Coronin 1C harbours a second actin-binding site that confers co-operative binding to F-actin. Biochem. J. 2012;444:89–96. doi: 10.1042/BJ20120209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tilley F.C., Williamson R.C., Race P.R., Rendall T.C., Bass M.D. Integration of the Rac1- and actin-binding properties of Coronin-1C. Small GTPases. 2015;6:36–42. doi: 10.4161/21541248.2014.992259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xavier C.P., Rastetter R.H., Blömacher M., Stumpf M., Himmel M., Morgan R.O., Fernandez M.P., Wang C., Osman A., Miyata Y. Phosphorylation of CRN2 by CK2 regulates F-actin and Arp2/3 interaction and inhibits cell migration. Sci. Rep. 2012;2:241. doi: 10.1038/srep00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Littlefield R., Almenar-Queralt A., Fowler V.M. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat. Cell Biol. 2001;3:544–551. doi: 10.1038/35078517. [DOI] [PubMed] [Google Scholar]
- 60.Fischer R.S., Fritz-Six K.L., Fowler V.M. Pointed-end capping by tropomodulin3 negatively regulates endothelial cell motility. J. Cell Biol. 2003;161:371–380. doi: 10.1083/jcb.200209057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lyon A.N., Pineda R.H., Hao le T., Kudryashova E., Kudryashov D.S., Beattie C.E. Calcium binding is essential for plastin 3 function in Smn-deficient motoneurons. Hum. Mol. Genet. 2014;23:1990–2004. doi: 10.1093/hmg/ddt595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gandhi M., Goode B.L. Coronin: The double-edged sword of actin dynamics. Subcell. Biochem. 2008;48:72–87. doi: 10.1007/978-0-387-09595-0_7. [DOI] [PubMed] [Google Scholar]
- 63.McArdle B., Hofmann A. Coronin structure and implications. Subcell. Biochem. 2008;48:56–71. doi: 10.1007/978-0-387-09595-0_6. [DOI] [PubMed] [Google Scholar]
- 64.Babin P.J., Goizet C., Raldúa D. Zebrafish models of human motor neuron diseases: advantages and limitations. Prog. Neurobiol. 2014;118:36–58. doi: 10.1016/j.pneurobio.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 65.Hall A., Lalli G. Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb. Perspect. Biol. 2010;2:a001818. doi: 10.1101/cshperspect.a001818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bernal S., Also-Rallo E., Martínez-Hernández R., Alías L., Rodríguez-Alvarez F.J., Millán J.M., Hernández-Chico C., Baiget M., Tizzano E.F. Plastin 3 expression in discordant spinal muscular atrophy (SMA) siblings. Neuromuscul. Disord. 2011;21:413–419. doi: 10.1016/j.nmd.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 67.Sleigh J.N., Gillingwater T.H., Talbot K. The contribution of mouse models to understanding the pathogenesis of spinal muscular atrophy. Dis. Model. Mech. 2011;4:457–467. doi: 10.1242/dmm.007245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Monani U.R., Sendtner M., Coovert D.D., Parsons D.W., Andreassi C., Le T.T., Jablonka S., Schrank B., Rossoll W., Prior T.W. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(-/-) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 69.Hua Y., Sahashi K., Hung G., Rigo F., Passini M.A., Bennett C.F., Krainer A.R. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Porensky P.N., Mitrpant C., McGovern V.L., Bevan A.K., Foust K.D., Kaspar B.K., Wilton S.D., Burghes A.H. A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum. Mol. Genet. 2012;21:1625–1638. doi: 10.1093/hmg/ddr600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kariya S., Obis T., Garone C., Akay T., Sera F., Iwata S., Homma S., Monani U.R. Requirement of enhanced Survival Motoneuron protein imposed during neuromuscular junction maturation. J. Clin. Invest. 2014;124:785–800. doi: 10.1172/JCI72017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Feng Z., Ling K.K., Zhao X., Zhou C., Karp G., Welch E.M., Naryshkin N., Ratni H., Chen K.S., Metzger F. Pharmacologically induced mouse model of adult spinal muscular atrophy to evaluate effectiveness of therapeutics after disease onset. Hum. Mol. Genet. 2016;25:964–975. doi: 10.1093/hmg/ddv629. [DOI] [PubMed] [Google Scholar]
- 73.Zhou H., Meng J., Marrosu E., Janghra N., Morgan J., Muntoni F. Repeated low doses of morpholino antisense oligomer: an intermediate mouse model of spinal muscular atrophy to explore the window of therapeutic response. Hum. Mol. Genet. 2015;24:6265–6277. doi: 10.1093/hmg/ddv329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bogdanik L.P., Osborne M.A., Davis C., Martin W.P., Austin A., Rigo F., Bennett C.F., Lutz C.M. Systemic, postsymptomatic antisense oligonucleotide rescues motor unit maturation delay in a new mouse model for type II/III spinal muscular atrophy. Proc. Natl. Acad. Sci. USA. 2015;112:E5863–E5872. doi: 10.1073/pnas.1509758112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coque E., Raoul C., Bowerman M. ROCK inhibition as a therapy for spinal muscular atrophy: understanding the repercussions on multiple cellular targets. Front. Neurosci. 2014;8:271. doi: 10.3389/fnins.2014.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Delanote V., Van Impe K., De Corte V., Bruyneel E., Vetter G., Boucherie C., Mareel M., Vandekerckhove J., Friederich E., Gettemans J. Molecular basis for dissimilar nuclear trafficking of the actin-bundling protein isoforms T- and L-plastin. Traffic. 2005;6:335–345. doi: 10.1111/j.1600-0854.2005.00276.x. [DOI] [PubMed] [Google Scholar]
- 77.Sharma A., Lambrechts A., Hao le T., Le T.T., Sewry C.A., Ampe C., Burghes A.H., Morris G.E. A role for complexes of survival of motor neurons (SMN) protein with gemins and profilin in neurite-like cytoplasmic extensions of cultured nerve cells. Exp. Cell Res. 2005;309:185–197. doi: 10.1016/j.yexcr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 78.Praveen K., Wen Y., Matera A.G. A Drosophila model of spinal muscular atrophy uncouples snRNP biogenesis functions of survival motor neuron from locomotion and viability defects. Cell Rep. 2012;1:624–631. doi: 10.1016/j.celrep.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dimitriadi M., Sleigh J.N., Walker A., Chang H.C., Sen A., Kalloo G., Harris J., Barsby T., Walsh M.B., Satterlee J.S. Conserved genes act as modifiers of invertebrate SMN loss of function defects. PLoS Genet. 2010;6:e1001172. doi: 10.1371/journal.pgen.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van Dijk F.S., Zillikens M.C., Micha D., Riessland M., Marcelis C.L., de Die-Smulders C.E., Milbradt J., Franken A.A., Harsevoort A.J., Lichtenbelt K.D. PLS3 mutations in X-linked osteoporosis with fractures. N. Engl. J. Med. 2013;369:1529–1536. doi: 10.1056/NEJMoa1308223. [DOI] [PubMed] [Google Scholar]
- 81.Südhof T.C. The synaptic vesicle cycle revisited. Neuron. 2000;28:317–320. doi: 10.1016/s0896-6273(00)00109-4. [DOI] [PubMed] [Google Scholar]
- 82.Rizzoli S.O., Betz W.J. Synaptic vesicle pools. Nat. Rev. Neurosci. 2005;6:57–69. doi: 10.1038/nrn1583. [DOI] [PubMed] [Google Scholar]
- 83.Ruiz R., Tabares L. Neurotransmitter release in motor nerve terminals of a mouse model of mild spinal muscular atrophy. J. Anat. 2014;224:74–84. doi: 10.1111/joa.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Torres-Benito L., Ruiz R., Tabares L. Synaptic defects in SMA animal models. Dev. Neurobiol. 2012;72:126–133. doi: 10.1002/dneu.20912. [DOI] [PubMed] [Google Scholar]
- 85.Mooren O.L., Galletta B.J., Cooper J.A. Roles for actin assembly in endocytosis. Annu. Rev. Biochem. 2012;81:661–686. doi: 10.1146/annurev-biochem-060910-094416. [DOI] [PubMed] [Google Scholar]
- 86.Watanabe S., Rost B.R., Camacho-Pérez M., Davis M.W., Söhl-Kielczynski B., Rosenmund C., Jorgensen E.M. Ultrafast endocytosis at mouse hippocampal synapses. Nature. 2013;504:242–247. doi: 10.1038/nature12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hadano S., Hand C.K., Osuga H., Yanagisawa Y., Otomo A., Devon R.S., Miyamoto N., Showguchi-Miyata J., Okada Y., Singaraja R. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat. Genet. 2001;29:166–173. doi: 10.1038/ng1001-166. [DOI] [PubMed] [Google Scholar]
- 88.Wu C.H., Fallini C., Ticozzi N., Keagle P.J., Sapp P.C., Piotrowska K., Lowe P., Koppers M., McKenna-Yasek D., Baron D.M. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488:499–503. doi: 10.1038/nature11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martinez-Carrera L.A., Wirth B. Dominant spinal muscular atrophy is caused by mutations in BICD2, an important golgin protein. Front. Neurosci. 2015;9:401. doi: 10.3389/fnins.2015.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wirth B., Garbes L., Riessland M. How genetic modifiers influence the phenotype of spinal muscular atrophy and suggest future therapeutic approaches. Curr. Opin. Genet. Dev. 2013;23:330–338. doi: 10.1016/j.gde.2013.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.