Abstract
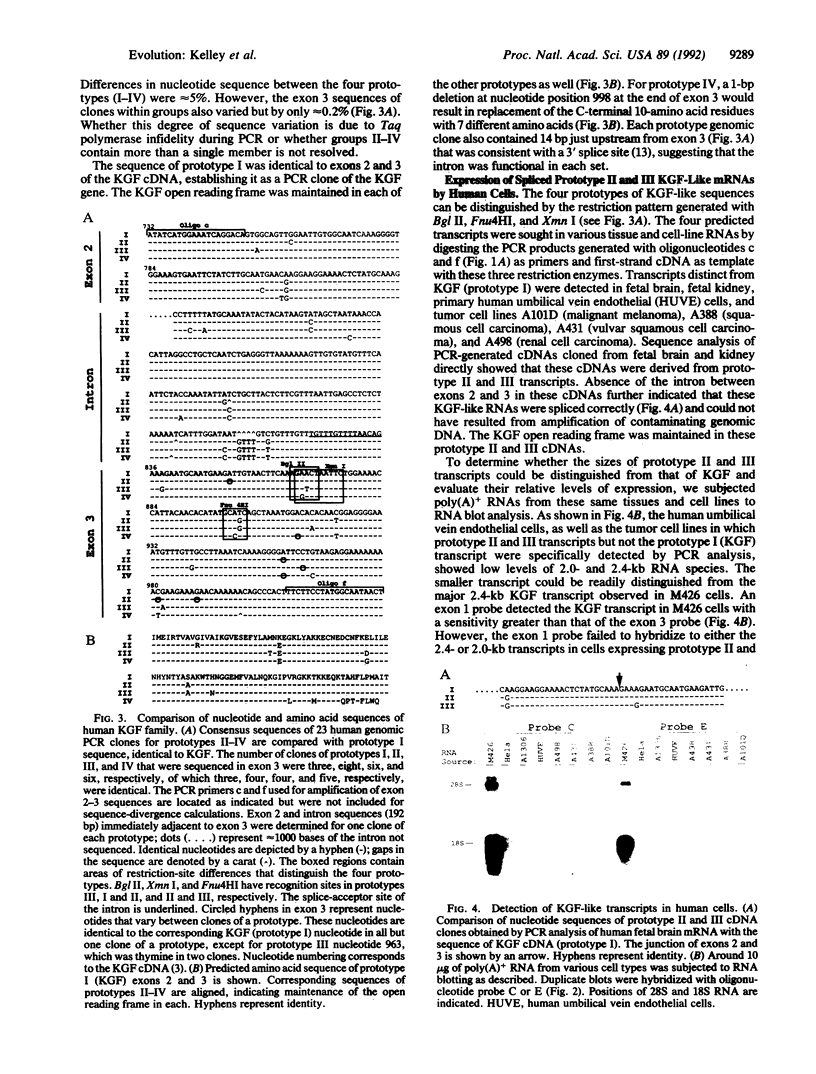
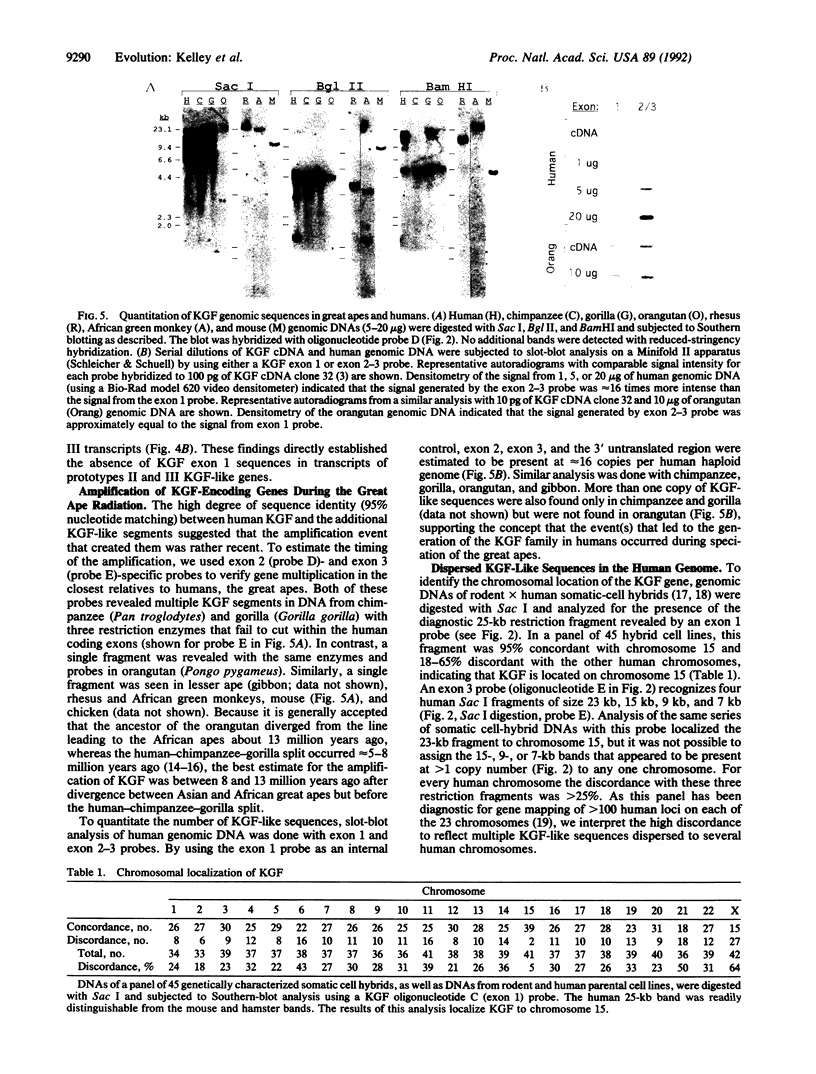
The structural gene for human keratinocyte growth factor (KGF), a member of the fibroblast growth factor family, consists of three coding exons and two introns typical of other fibroblast growth factor loci. A portion of the KGF gene, located on chromosome 15, is amplified to approximately 16 copies in the human genome, and these highly related copies (which consist of exon 2, exon 3, the intron between them, and a 3' noncoding segment of the KGF transcript) are dispersed to multiple human chromosomes. The KGF-like sequences are transcriptionally active, differentially regulated in various tissues, and composed of three distinct classes of coding sequences that are 5% divergent from each other and from the authentic KGF sequence. Multiple copies of KGF-like genes were also discovered in the genomic DNAs of chimpanzee and gorilla but were not found in lesser apes (gibbon), Old World monkeys (African green monkey and macaques), mice, or chickens. The pattern of evolutionary occurrence suggests that a primordial KGF gene was amplified and chromosomally dispersed subsequent to the divergence of orangutan from African apes but before the trichotomous divergence of human, chimpanzee, and gorilla 5-8 million years ago. The appearance of a transcriptionally active and chromosomally dispersed multigene KGF family may have implications in the evolution of the great apes and humans.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham J. A., Whang J. L., Tumolo A., Mergia A., Friedman J., Gospodarowicz D., Fiddes J. C. Human basic fibroblast growth factor: nucleotide sequence and genomic organization. EMBO J. 1986 Oct;5(10):2523–2528. doi: 10.1002/j.1460-2075.1986.tb04530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelaide J., Mattei M. G., Marics I., Raybaud F., Planche J., De Lapeyriere O., Birnbaum D. Chromosomal localization of the hst oncogene and its co-amplification with the int.2 oncogene in a human melanoma. Oncogene. 1988 Apr;2(4):413–416. [PubMed] [Google Scholar]
- Anagnou N. P., O'Brien S. J., Shimada T., Nash W. G., Chen M. J., Nienhuis A. W. Chromosomal organization of the human dihydrofolate reductase genes: dispersion, selective amplification, and a novel form of polymorphism. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5170–5174. doi: 10.1073/pnas.81.16.5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews P. Fossil evidence on human origins and dispersal. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):419–428. doi: 10.1101/sqb.1986.051.01.050. [DOI] [PubMed] [Google Scholar]
- Benharroch D., Birnbaum D. Biology of the fibroblast growth factor gene family. Isr J Med Sci. 1990 Apr;26(4):212–219. [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden P., Jaenichen R., Zachau H. G. Structural features of transposed human VK genes and implications for the mechanism of their transpositions. Nucleic Acids Res. 1990 Apr 25;18(8):2101–2107. doi: 10.1093/nar/18.8.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes S., Smith R., Casey G., Dickson C., Peters G. Sequence organization of the human int-2 gene and its expression in teratocarcinoma cells. Oncogene. 1989 Apr;4(4):429–436. [PubMed] [Google Scholar]
- Burgess W. H., Maciag T. The heparin-binding (fibroblast) growth factor family of proteins. Annu Rev Biochem. 1989;58:575–606. doi: 10.1146/annurev.bi.58.070189.003043. [DOI] [PubMed] [Google Scholar]
- Childs G., Maxson R., Cohn R. H., Kedes L. Orphons: dispersed genetic elements derived from tandem repetitive genes of eucaryotes. Cell. 1981 Mar;23(3):651–663. doi: 10.1016/0092-8674(81)90428-1. [DOI] [PubMed] [Google Scholar]
- Finch P. W., Rubin J. S., Miki T., Ron D., Aaronson S. A. Human KGF is FGF-related with properties of a paracrine effector of epithelial cell growth. Science. 1989 Aug 18;245(4919):752–755. doi: 10.1126/science.2475908. [DOI] [PubMed] [Google Scholar]
- Giard D. J., Aaronson S. A., Todaro G. J., Arnstein P., Kersey J. H., Dosik H., Parks W. P. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst. 1973 Nov;51(5):1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- Glisin V., Crkvenjakov R., Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974 Jun 4;13(12):2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Honjo T., Habu S. Origin of immune diversity: genetic variation and selection. Annu Rev Biochem. 1985;54:803–830. doi: 10.1146/annurev.bi.54.070185.004103. [DOI] [PubMed] [Google Scholar]
- Janczewski D. N., Goldman D., O'Brien S. J. Molecular genetic divergence of orang utan (Pongo pygmaeus) subspecies based on isozyme and two-dimensional gel electrophoresis. J Hered. 1990 Sep-Oct;81(5):375–387. [PubMed] [Google Scholar]
- Kimelman D., Kirschner M. Synergistic induction of mesoderm by FGF and TGF-beta and the identification of an mRNA coding for FGF in the early Xenopus embryo. Cell. 1987 Dec 4;51(5):869–877. doi: 10.1016/0092-8674(87)90110-3. [DOI] [PubMed] [Google Scholar]
- Lötscher E., Zimmer F. J., Klopstock T., Grzeschik K. H., Jaenichen R., Straubinger B., Zachau H. G. Localization, analysis and evolution of transposed human immunoglobulin V kappa genes. Gene. 1988 Sep 30;69(2):215–223. doi: 10.1016/0378-1119(88)90432-5. [DOI] [PubMed] [Google Scholar]
- Marics I., Adelaide J., Raybaud F., Mattei M. G., Coulier F., Planche J., de Lapeyriere O., Birnbaum D. Characterization of the HST-related FGF.6 gene, a new member of the fibroblast growth factor gene family. Oncogene. 1989 Mar;4(3):335–340. [PubMed] [Google Scholar]
- Mergia A., Tischer E., Graves D., Tumolo A., Miller J., Gospodarowicz D., Abraham J. A., Shipley G. D., Fiddes J. C. Structural analysis of the gene for human acidic fibroblast growth factor. Biochem Biophys Res Commun. 1989 Nov 15;164(3):1121–1129. doi: 10.1016/0006-291x(89)91785-3. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C., Roux D., Mattei M. G., de Lapeyriere O., Goldfarb M., Birnbaum D., Jordan B. R. The FGF-related oncogenes hst and int.2, and the bcl.1 locus are contained within one megabase in band q13 of chromosome 11, while the fgf.5 oncogene maps to 4q21. Oncogene. 1988 Dec;3(6):703–708. [PubMed] [Google Scholar]
- O'Brien S. J., Bonner T. I., Cohen M., O'Connell C., Nash W. G. Mapping of an endogenous retroviral sequence to human chromosome 18. Nature. 1983 May 5;303(5912):74–77. doi: 10.1038/303074a0. [DOI] [PubMed] [Google Scholar]
- O'Brien S. J., Nash W. G., Goodwin J. L., Lowy D. R., Chang E. H. Dispersion of the ras family of transforming genes to four different chromosomes in man. Nature. 1983 Apr 28;302(5911):839–842. doi: 10.1038/302839a0. [DOI] [PubMed] [Google Scholar]
- Pech M., Jaenichen H. R., Pohlenz H. D., Neumaier P. S., Klobeck H. G., Zachau H. G. Organization and evolution of a gene cluster for human immunoglobulin variable regions of the kappa type. J Mol Biol. 1984 Jun 25;176(2):189–204. doi: 10.1016/0022-2836(84)90420-0. [DOI] [PubMed] [Google Scholar]
- Rubin J. S., Osada H., Finch P. W., Taylor W. G., Rudikoff S., Aaronson S. A. Purification and characterization of a newly identified growth factor specific for epithelial cells. Proc Natl Acad Sci U S A. 1989 Feb;86(3):802–806. doi: 10.1073/pnas.86.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley C. G., Ahlquist J. E. DNA hybridization evidence of hominoid phylogeny: results from an expanded data set. J Mol Evol. 1987;26(1-2):99–121. doi: 10.1007/BF02111285. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stark G. R., Debatisse M., Giulotto E., Wahl G. M. Recent progress in understanding mechanisms of mammalian DNA amplification. Cell. 1989 Jun 16;57(6):901–908. doi: 10.1016/0092-8674(89)90328-0. [DOI] [PubMed] [Google Scholar]
- Studencki A. B., Wallace R. B. Allele-specific hybridization using oligonucleotide probes of very high specific activity: discrimination of the human beta A- and beta S-globin genes. DNA. 1984;3(1):7–15. doi: 10.1089/dna.1.1984.3.7. [DOI] [PubMed] [Google Scholar]
- Yoshida T., Miyagawa K., Odagiri H., Sakamoto H., Little P. F., Terada M., Sugimura T. Genomic sequence of hst, a transforming gene encoding a protein homologous to fibroblast growth factors and the int-2-encoded protein. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7305–7309. doi: 10.1073/pnas.84.20.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunis J. J., Prakash O. The origin of man: a chromosomal pictorial legacy. Science. 1982 Mar 19;215(4539):1525–1530. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]
- Zhan X., Bates B., Hu X. G., Goldfarb M. The human FGF-5 oncogene encodes a novel protein related to fibroblast growth factors. Mol Cell Biol. 1988 Aug;8(8):3487–3495. doi: 10.1128/mcb.8.8.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]






