Abstract
Background
Anti-hepatitis C virus antibody (anti-HCV) assays are recommended for screening HCV-infected persons. The VIDAS Anti-HCV Assay (bioMérieux, France), based on the enzyme-linked fluorescence test principle, was recently introduced in Korea. We evaluated the clinical performance of the VIDAS assay.
Methods
One hundred HCV-positive and 1,002 HCV-negative blood samples confirmed by Architect anti-HCV (Abbott Laboratories, USA) and COBAS TaqMan HCV real-time PCR (Roche Diagnostics, USA) or the Procleix Ultrio Plus Assay (Gen-Probe Incorporated, USA) were obtained from the Human Serum Bank (HSB) and tested by VIDAS. In case of discrepant results, we conducted a recombinant immunoblot assay (RIBA).
Results
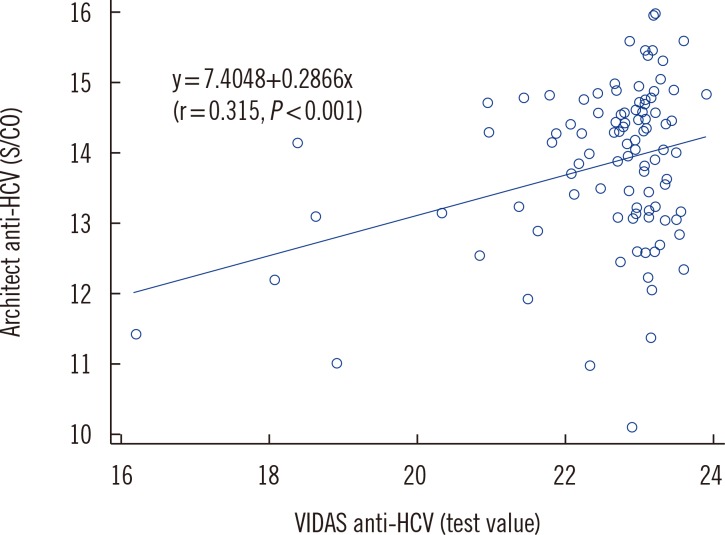
The agreement rates for known HCV-positive and HCV-negative samples between the VIDAS assay and the HSB testing were 100% (95% confidence interval [CI]: 96.4-100%) and 99.5% (95% CI: 98.8-99.8%), respectively. One of the five discrepant samples was positive for Core 2+ and NS3-2 2+ reactivity, two samples were negative, and the other two were indeterminate regarding NS4 2+ reactivity in RIBA. We observed a significant but weak positive correlation between the titers of VIDAS and Architect assays (r=0.315, P<0.001).
Conclusions
The VIDAS anti-HCV assay, developed on the VIDAS automated immunoassay platform based on the ready-to-use, single-sample test concept may be useful in small-to-medium-sized laboratories. It showed good agreement with Architect anti-HCV and COBAS PCR assays and is therefore useful for detection of HCV infection. Weakly test-positive (ambiguous) samples require additional testing by another anti-HCV, RIBA, or HCV RNA assay.
Keywords: Hepatitis C, Hepatitis C antibodies, Immunoassay, Immunoenzyme techniques, Immunoblotting, Real-time polymerase chain reaction
INTRODUCTION
Hepatitis C is a liver disease caused by the hepatitis C virus (HCV) and can progress to chronic liver disease. HCV poses a significant public health problem worldwide: approximately 150 million people have chronic HCV infection, and 500,000 people die every year from the chronic complications of hepatitis C [1]
Initial HCV infection is commonly asymptomatic; thus, laboratory assays are necessary for early diagnosis. Anti-HCV antibody assays are recommended for screening to identify persons with HCV infection [2,3]. Numerous anti-HCV antibody immunoassays have been developed, and sensitivity and specificity have been markedly improved across three generations. Recently, rapid anti-HCV antibody assays have also been developed and are used in the clinic for HCV screening [4].
The VIDAS Anti-HCV Assay (bioMérieux, Marcy-L'Etoile, France) was recently developed as a third-generation immunoassay using antigens corresponding to the HCV Core, NS3, and NS4 proteins for qualitative detection of anti-HCV antibodies in human serum or plasma. The VIDAS test uses the enzyme-linked fluorescence assay (ELFA) principle, combining the enzyme-linked immunosorbent assay with final blue fluorescence detection, leading to excellent sensitivity and specificity [5]. Since it is a fully automated system and uses a single-dose, ready-to-use reagent, it may be useful in small- to medium-sized laboratories. In this study, our aim was to evaluate clinical performance of the VIDAS Anti-HCV assay. We compared the results of VIDAS Anti-HCV Assay with the results of other anti-HCV assay and HCV PCR, and analyzed the correlation between the titers of two anti-HCV assays.
METHODS
1. Clinical samples for performance analysis
One hundred HCV-positive and 1,002 HCV-negative blood samples were obtained from the Human Serum Bank (HSB: NRF-2015M3A9B8030780) of Chung-Ang University Hospital (http://hsb.knrrc.or.kr), which were collected between September 2010 and August 2015. The test-positive samples were confirmed by Architect Anti-HCV (Abbott Laboratories, Abbott Park, IL, USA) and COBAS TaqMan HCV Real-time PCR (Roche Diagnostics, Indianapolis, IN, USA) assays. The test-negative samples were confirmed by Architect Anti-HCV and Procleix Ultrio Plus Assay (Gen-Probe Incorporated, San Diego, CA, USA), which is a qualitative nucleic-acid assay for detection of HIV-1 RNA, HCV RNA, and HBV DNA in blood samples. All our samples were tested and interpreted according to the manufacturer's instructions. The results of the 100 HCV-positive samples detected by Architect Anti-HCV and COBAS PCR assays were 13.87±1.13 signal-to-cut-off (S/CO) ratio and 5.68±0.84 log10 IU/mL, respectively. This study protocol was approved by the Institutional Review Board of Dongtan Sacred Heart Hospital (decision no. 2014-310).
2. The VIDAS anti-HCV assay
The VIDAS anti-HCV assay uses the ELFA principle, combining a two-step enzyme immunoassay sandwich method with final fluorescence detection. The solid-phase receptacle (SPR) serves as the solid phase for the reaction and is coated with antigens representing the HCV Core, NS3, and NS4 proteins. Additionally, the SPR acts as a pipetting device. Assay reagents are ready to use and are predispensed in the sealed reagent strips. All of the assay steps are performed automatically by the instrument. At each stage of the reaction, the reaction medium is cycled in and out of the SPR several times.
After the sample is diluted, it is processed on the SPR. The anti-HCV antibodies present in the sample bind to the antigens coating the interior of the SPR. The antibodies are detected by means of mouse monoclonal anti-human IgG antibodies in Fab form and the substrate 4-methyl-umbelliferyl phosphate. The fluorescence of the product is measured at 450 nm. The results are analyzed and calculated automatically; a test value (TV) <1.00 indicates a negative result and a TV ≥1.00 a positive result.
3. Confirmatory test for discrepancy resolution
In case of discrepancy between the VIDAS assay and HSB (Architect anti-HCV and COBAS TaqMan HCV PCR or Procleix Ultrio Plus assays) results, we carried out a MP Diagnostics HCV Blot (hereafter, "MP blot"; MP Biomedicals, Santa Ana, CA, USA) assay. MP blot is a qualitative enzyme immunoblot assay, based on the immobilization of HCV recombinant antigens and synthetic peptides from Core, NS3 (NS3-1, NS3-2 subunits), NS4, and NS5 proteins as individual bands onto a nitrocellulose strip. The samples were tested and interpreted according to the manufacturer's instructions. The absence of bands of 1+ or greater reactivity indicates a negative result; 1+ or greater reactivity of two or more HCV antigens or 2+ or greater reactivity of the Core band only indicates a positive result; and any single band having 1+ or greater reactivity but not meeting the positive-result criteria is interpreted as indeterminate.
4. Statistical analyses
The agreement between HSB and VIDAS assay results was assessed on the basis of the positive agreement rate, the negative agreement rate, total agreement rate, and kappa coefficient (κ). The correlation between titers of VIDAS (TVs) and Architect (S/CO ratios) assays was evaluated by using Pearson's correlation test and linear regression. Data with P values <0.05 were considered statistically significant. Statistical analyses were performed in the IBM SPSS Statistics software, version 23 (IBM Corporation, Armonk, NY, USA) or in MedCalc (MedCalc Software, Ostend, Belgium).
RESULTS
1. Comparison of the VIDAS anti-HCV assay with the results of Human Serum Bank
The 100 HCV-positive samples were all test-positive (TV 22.56±1.24) and 997 of 1,002 HCV-negative samples were test-negative according to the VIDAS assay. Five samples showed discrepant results between the VIDAS assay and HSB data. These five all tested negative in the Architect assay and COBAS PCR, but were test-positive according to the VIDAS assay. Therefore, the positive and negative agreement rates of the VIDAS assay were 100% (95% confidence interval [CI]: 96.4-100%) and 99.5% (95% CI: 98.8-99.8%), respectively (Table 1). Total agreement rate of the results between the VIDAS assay and HSB was 99.5%, and the κ coefficient was 0.973 (95% CI: 0.950-0.997).
Table 1. Comparison of the results of the VIDAS anti-HCV assay with those of the Human Serum Bank.
| Results of HSB | VIDAS Anti-HCV | N of specimen | Results (TVs measured by VIDAS anti-HCV: mean ± SD) |
|---|---|---|---|
| Positive | Positive | 100 | 22.56 ± 1.24 |
| Positive | Negative | 0 | |
| Negative | Positive | 5 | 2.58 ± 1.89 |
| Negative | Negative | 997 | |
| Total | 1,102 |
Sensitivity of VIDAS Anti-HCV: 100% (95% confidence interval: 96.4-100%); Specificity of VIDAS Anti-HCV: 99.5% (95% confidence interval: 98.8-99.8%).
Abbreviations: HCV, hepatitis C virus; HSB, Human Serum Bank of Chung-Ang University Hospital; TV, test value.
2. Analysis of samples showing discrepant results
The results on the five samples showing discrepancy among tests are shown in Table 2. All five samples were weakly reactive in the VIDAS assay (TV 1.05-6.23). One sample showed Core 2+ and NS3-2 2+ reactivity on MP blot and was considered a true positive result. Two samples tested negative on MP blot and were considered false positive results of the VIDAS assay. The other two samples showed indeterminate results on the MP blot; both of them showed NS4 2+ reactivity.
Table 2. Comparison between the results of VIDAS anti-HCV, Architect anti-HCV, Roche COBAS TaqMan HCV PCR, and MP Diagnostics HCV Blot for five discrepant samples.
| Architect Anti-HCV | Roche COBAS TaqMan HCV PCR | VIDAS Anti-HCV (TVs) | MP Diagnostics HCV Blot |
|---|---|---|---|
| Negative | Negative | Positive (1.05) | Positive (Core 2+, NS3-2 2+) |
| Negative | Negative | Positive (6.23) | Negative |
| Negative | Negative | Positive (1.30) | Negative |
| Negative | Negative | Positive (2.42) | Indeterminate (NS4 2+) |
| Negative | Negative | Positive (1.91) | Indeterminate (NS4 2+) |
Abbreviations: HCV, hepatitis C virus; TV, test value.
3. The correlation between titers of VIDAS and Architect anti-HCV assays
The titers (TVs) of the 100 samples test-positive according to the VIDAS assay were consistently higher than those of the Architect assay (S/CO ratios; Fig. 1). We found a significant but weak positive correlation between the titers of the VIDAS and Architect assays (r=0.315, P<0.001). The line in Fig. 1 represents the linear regression of data.
Fig. 1. Correlation between the titers of test-positive samples (n=100) detected by the VIDAS and Architect anti-HCV assays.
Abbreviations: HCV, hepatitis C virus; S/CO, signal-to-cut-off.
DISCUSSION
In the present study, the positive and negative agreement rates of the VIDAS anti-HCV assay were 100% (95% CI: 96.4-100%) and 99.5% (95% CI: 98.8-99.8%), respectively. We compared these positive and negative agreement rates with the sensitivity and specificity of other studies, respectively. Several studies have evaluated a currently available automated chemiluminescence immunoassay analyzer, with sensitivity and specificity values ranging from 99.5% to 100% and from 96.5% to 99.9%, respectively [6,7,8]. A recent study of the VIDAS system showed 99.7% sensitivity and 99.0-99.67% specificity [5]. In our study, the sensitivity and specificity are comparable to or higher than those reported in other studies [5,6,7,8].
There were five samples (0.5%) with discrepancy between the VIDAS assay and HSB results in our study. These five samples tested negative in both Architect and Procleix PCR assays, but were weakly test-positive in the VIDAS assay (TV 1.05-6.23). Such discrepant results between immunoassays have been reported elsewhere, especially for samples with low S/CO ratios, and may be due to random errors, differences in antigens or epitopes, or differences in labeling substances for signal detection in the assays [7,8,9,10]. The Architect test is a two-step immunoassay based on the chemiluminescent microparticle immunoassay and employs the Hcr43 recombinant fusion protein (Core, NS3; expressed in E. coli) and the c100-3 recombinant protein (NS4; expressed in yeast) as antigens. On the other hand, VIDAS is a two-step enzyme immunoassay sandwich method with final fluorescence detection and uses the NS3 recombinant protein expressed in E. coli and Core, NS4A, and NS4B peptides as antigens. One of the five discrepant samples in our study showed very weak reactivity in the VIDAS assay (TV 1.05) and Core 2+ and NS3-2 2+ reactivity on MP blot, indicating a true positive result. We retested the Architect Anti-HCV Assay on this sample and obtained a positive result (S/CO 1.3), which was different from the original Architect result. We analyzed the sample using additional methods: Elecsys Anti-HCV II (Roche Diagnostics, Mannheim, Germany) and ADVIA Centaur HCV (Siemens Healthcare Diagnostics, Marburg, Germany) assays, both of which yielded a negative result. Other two discrepant samples (VIDAS TVs 1.30 and 6.23) showed negative results on the MP blot. The three discrepant results described above may be caused by the differences in antigens or epitopes between the assays rather than the random error.
The other two samples (VIDAS TVs 1.91 and 2.42) showed only NS4 2+ reactivity and were interpreted as indeterminate on the MP blot. Indeterminate results of the immunoblot assay are defined as any single band having 1+ or greater reactivity (except 2+ or greater reactivity for the Core band only, which indicates a positive result in the MP Diagnostics HCV Blot assay) and can be seen in a cleared HCV infection or during early seroconversion and may be due to nonspecific reactivity [11]. Isolated Core or NS3 reactivity means a higher probability of true presence of anti-HCV antibodies and may be the sign of acute or chronic HCV infection in case of a high S/CO ratio and strong reactivity on the immunoblot assay; in contrast, isolated NS4 reactivity is considered a false positive result (does not indicate HCV infection) [11,12,13]. Because the two samples that showed indeterminate results in our study were weakly test-positive according to VIDAS and showed NS4 2+ reactivity on the MP blot, these results likely indicate nonspecific (false) reactions.
According to the United States Centers for Disease Control and Prevention (CDC) guidelines for laboratory testing and result reporting on antibodies to hepatitis C virus [4,14,15], an anti-HCV antibody assay with high sensitivity should be used for screening tests, and test-positive results should be verified by a more specific serologic test or PCR test, particularly in populations with low HCV prevalence. On the other hand, these recommendations have not been followed by the majority of clinical laboratories for multiple reasons. Therefore, the US CDC has recommended the use of S/CO ratios for anti-HCV antibody screening assays to minimize the number of samples that require supplemental testing, and specific S/CO ratios for the US Food and Drug Administration-licensed or approved anti-HCV immunoassays were provided. In case of the VIDAS assay, however, the TVs have not been evaluated yet and weakly test-positive samples would need to be confirmed or followed up with another anti-HCV, immunoblot, or HCV RNA assay.
Correlation analysis of the titers of VIDAS (TVs) and Architect (S/CO ratios) anti-HCV antibody assays for 100 HCV-positive samples revealed a weak positive correlation (r = 0.315). Several studies have analyzed the correlation between Architect, Elecsys, and Vitros (Ortho-Clinical Diagnostics, Raritan, NJ, USA) anti-HCV antibody assays and showed a strong correlation between Architect and Vitros assays because of the use of the same recombinant antigens under contract agreement between the two manufacturers [7,8,10,16]. The weak correlation between VIDAS and Architect assays was probably due to the differences in antigens or epitopes or labeling substances for signal detection, as mentioned above. To our knowledge, this is the first study on the correlation between VIDAS and Architect assays.
In conclusion, the VIDAS Anti-HCV assay showed excellent agreement with the Architect Anti-HCV Assay and Roche COBAS TaqMan HCV PCR Assay. These findings indicate that the VIDAS test is a reliable tool for screening of the population for HCV infection. Nonetheless, weakly test-positive samples need to be confirmed or followed up with another anti-HCV, immunoblot, or HCV RNA assay.
Acknowledgments
The authors are grateful to bioMérieux for providing the reagents and to Ji Sun Noh for the excellent technical assistance. We also thank Human Serum Bank of Chung-Ang University Hospital for distributing the blood samples.
Footnotes
Authors' Disclosures of Potential Conflicts of Interest: No potential conflicts of interest relevant to this study were reported.
References
- 1.WHO. Hepatitis C. Fact sheet No. 164. [Updated on Jul 2015]. http://www.who.int/mediacentre/factsheets/fs164/en.
- 2.WHO. Guidelines for the screening care and treatment of persons with hepatitis c infection. WHO Press; 2014. pp. 1–222. http://www.who.int/hiv/pub/hepatitis/hepatitis-c-guidelines/en/ [PubMed] [Google Scholar]
- 3.European Association for Study of L. EASL Clinical Practice Guidelines: management of hepatitis C virus infection. J Hepatol. 2014;60:392–420. doi: 10.1016/j.jhep.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Kamili S, Drobeniuc J, Araujo AC, Hayden TM. Laboratory diagnostics for hepatitis C virus infection. Clin Infect Dis. 2012;55(Sl):S43–S48. doi: 10.1093/cid/cis368. [DOI] [PubMed] [Google Scholar]
- 5.Seignères B, Descamps F, Croise R, Barlet V, Bouvier-Alias M, Chevaliez S, et al. Multicenter clinical evaluation of the new 3rd generation assay for detection of antibodies against hepatitis C virus on the VIDAS system. J Clin Virol. 2016;78:20–26. doi: 10.1016/j.jcv.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Alborino F, Burighel A, Tiller FW, van Helden J, Gabriel C, Raineri A, et al. Multicenter evaluation of a fully automated third-generation anti-HCV antibody screening test with excellent sensitivity and specificity. Med Microbiol Immunol. 2011;200:77–83. doi: 10.1007/s00430-010-0171-0. [DOI] [PubMed] [Google Scholar]
- 7.Kim S, Kim JH, Yoon S, Park YH, Kim HS. Clinical performance evaluation of four automated chemiluminescence immunoassays for hepatitis C virus antibody detection. J Clin Microbiol. 2008;46:3919–3923. doi: 10.1128/JCM.01603-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park Y, Seok Y, Choi J, Kim HS. Performance evaluation of the Vitros anti-hepatitis C virus antibody assay for use in clinical laboratories. Clin Biochem. 2012;45:175–177. doi: 10.1016/j.clinbiochem.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 9.Watterson JM, Stallcup P, Escamilla D, Chernay P, Reyes A, Trevino SC. Evaluation of the Ortho-Clinical Diagnostics Vitros ECi Anti-HCV test: comparison with three other methods. J Clin Lab Anal. 2007;21:162–166. doi: 10.1002/jcla.20119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger A, Rabenau H, Allwinn R, Doerr HW. Evaluation of the new ARCHITECT anti-HCV screening test under routine laboratory conditions. J Clin Virol. 2008;43:158–161. doi: 10.1016/j.jcv.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Kiely P, Kay D, Parker S, Piscitelli L. The significance of third-generation HCV RIBA-indeterminate, RNA-negative results in voluntary blood donors screened with sequential third-generation immunoassays. Transfusion. 2004;44:349–358. doi: 10.1111/j.1537-2995.2003.00671.x. [DOI] [PubMed] [Google Scholar]
- 12.Lemaire JM, Courouce AM, Defer C, Bouchardeau F, Coste J, Agulles O, et al. HCV RNA in blood donors with isolated reactivities by third-generation RIBA. Transfusion. 2000;40:867–870. doi: 10.1046/j.1537-2995.2000.40070867.x. [DOI] [PubMed] [Google Scholar]
- 13.Pawlotsky JM, Bastie A, Pellet C, Remire J, Darthuy F, Wolfe L, et al. Significance of indeterminate third-generation hepatitis C virus recombinant immunoblot assay. J Clin Microbiol. 1996;34:80–83. doi: 10.1128/jcm.34.1.80-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alter MJ, Kuhnert WL, Finelli L Centers for Disease Control and Prevention. Guidelines for laboratory testing and result reporting of antibody to hepatitis C virus. Centers for Disease Control and Prevention. MMWR Recomm Rep. 2003;52:1–13. 15. quiz CE1-4. [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. Testing for HCV infection: an update of guidance for clinicians and laboratorians. MMWR Morb Mortal Wkly Rep. 2013;62:362–365. [PMC free article] [PubMed] [Google Scholar]
- 16.Yang R, Guan W, Wang Q, Liu Y, Wei L. Performance evaluation and comparison of the newly developed Elecsys anti-HCV II assay with other widely used assays. Clin Chim Acta. 2013;426:95–101. doi: 10.1016/j.cca.2013.09.010. [DOI] [PubMed] [Google Scholar]