Abstract
Background
A newborn screening (NBS) program has been utilized to detect asymptomatic newborns with inherited metabolic diseases (IMDs). There have been some bottlenecks such as false-positives and imprecision in the current NBS tests. To overcome these issues, we developed a multigene panel for IMD testing and investigated the utility of our integrated screening model in a routine NBS environment. We also evaluated the genetic epidemiologic characteristics of IMDs in a Korean population.
Methods
In total, 269 dried blood spots with positive results from current NBS tests were collected from 120,700 consecutive newborns. We screened 97 genes related to NBS in Korea and detected IMDs, using an integrated screening model based on biochemical tests and next-generation sequencing (NGS) called NewbornSeq. Haplotype analysis was conducted to detect founder effects.
Results
The overall positive rate of IMDs was 20%. We identified 10 additional newborns with preventable IMDs that would not have been detected prior to the implementation of our NGS-based platform NewbornSeq. The incidence of IMDs was approximately 1 in 2,235 births. Haplotype analysis demonstrated founder effects in p.Y138X in DUOXA2, p.R885Q in DUOX2, p.Y439C in PCCB, p.R285Pfs*2 in SLC25A13, and p.R224Q in GALT.
Conclusions
Through a population-based study in the NBS environment, we highlight the screening and epidemiological implications of NGS. The integrated screening model will effectively contribute to public health by enabling faster and more accurate IMD detection through NBS. This study suggested founder mutations as an explanation for recurrent IMD-causing mutations in the Korean population.
Keywords: Epidemiology, Founder mutation, Incidence, Inherited metabolic disease, Newborn screening, Next-generation sequencing
INTRODUCTION
Inherited metabolic diseases (IMDs) are a heterogeneous group of rare diseases with a collective incidence of 1 in 500 to 4,000 live births, representing a substantial public health burden [1,2,3,4,5]. Therefore, a newborn screening (NBS) program was introduced to detect presymptomatic newborns with IMDs. Over the past decade, tandem mass spectrometry (MS/MS) has been a major technological breakthrough for the NBS program by providing a way to detect multiple metabolites simultaneously [3,4,5,6,7]. Although the use of MS/MS has enabled cost-effective, rapid IMD identification, there have been some bottlenecks such as false-positives and imprecision [4,5,6,8]. As a second-tier method, enzymatic assays are laborious, time-consuming, and semiquantitative. Sequential Sanger sequencing is hampered by the genetic heterogeneity of IMDs, which results in delayed diagnosis [8,9]. These limitations of the current NBS tests have raised a necessity of rapidly diagnosing IMDs by next-generation sequencing (NGS) [8,9,10,11,12].
Recent studies have demonstrated that NGS is useful for the molecular diagnosis of some IMDs, including hyperphenylalaninemia, lysosomal storage diseases, and mitochondrial diseases [13,14,15]. Furthermore, several previous studies revealed the analytical validity and clinical utility of NGS for newborns from the United States [10,12]. For example, a NGS panel called NBDx, targeting 126 genes for NBS, was developed, and it demonstrated acceptable analytical performance [10]. Additionally, a previous study revealed that NGS leads to improved outcomes in the neonatal intensive care unit, confirming its clinical utility [12].
Currently, NGS is on the verge of being adopted for NBS. To introduce a multigene panel into an NBS program, some important factors should be considered. First, the analytical validity and clinical utility of NGS should be evaluated in a routine NBS environment as previously noted [10,12]. Second, the current biochemical NBS tests cannot be replaced by the exclusive use of NGS. Third, the multigene panel for NBS should be designed with specific considerations of genetic epidemiologic characteristics of the target diseases and population. Until recently, epidemiologic studies of IMDs with NGS in a NBS setting have not been reported.
The epidemiology of IMDs screened by NBS programs varies widely among different ethnic populations [1]. There are considerable differences in the incidence and spectrum of IMDs between Asians and other ethnicities [1,2,3,4,6,16,17]. The collective incidence of IMDs has been reported to be 1 in 2,800 in Korea, 1 in 6,219 in Taiwan, 1 in 6,300 (except hyperphenylalaninemia) in Australia, 1 in 2,000 in Italy, 1 in 4,100 in Germany, and 1 in 500 in a pan-ethnic population [1,2,4,5,18].
In addition to ethnic backgrounds, the application of methods like MS/MS have a strong impact on the results of IMD epidemiologic studies. One study reported that there was increase in the incidence of IMDs after the introduction of MS/MS into NBS [2]. Another study revealed that the incidence of medium-chain-acyl-coenzyme A dehydrogenase (MCAD) deficiency was specifically increased after the implementation of MS/MS [4]. Previous genetic epidemiologic studies of IMDs have focused on evaluating a limited number of diseases using conventional molecular methods [19,20,21,22,23,24,25,26]. Little is known about the incidence and spectrum of IMDs as estimated by the application of NGS as a screening method.
The aim of this study was to develop a multigene panel for detecting IMDs during NBS in Korea, and to evaluate the utility of an integrated screening model based on traditional biochemical tests and a multigene panel in a routine NBS system. In addition, we aimed to investigate genetic epidemiologic characteristics of IMDs in the Korean newborn population using NGS. We determined the overall incidence and mutation spectrum of IMDs based on the integrated screening model and tested for founder effects of recurrent mutations.
METHODS
1. Study participants and study design
We developed a multigene panel called NewbornSeq that integrates DNA isolation, targeted sequencing, variant annotation, and data interpretation. We evaluated the sensitivity and specificity of NewbornSeq, using 37 controls (27 positive controls and 10 negative controls). The positive controls were from confirmed IMD patients, as determined by biochemical tests and Sanger sequencing from the Samsung Medical Center, Seoul, Korea. Negative controls consisted of one healthy volunteer sample and nine samples of patients with other diseases which were not screened for in the current NBS program. Between-run reproducibility was measured by detecting mutations in technical duplicates. Turnaround time (TAT) was compared between NewbornSeq and the current NBS tests.
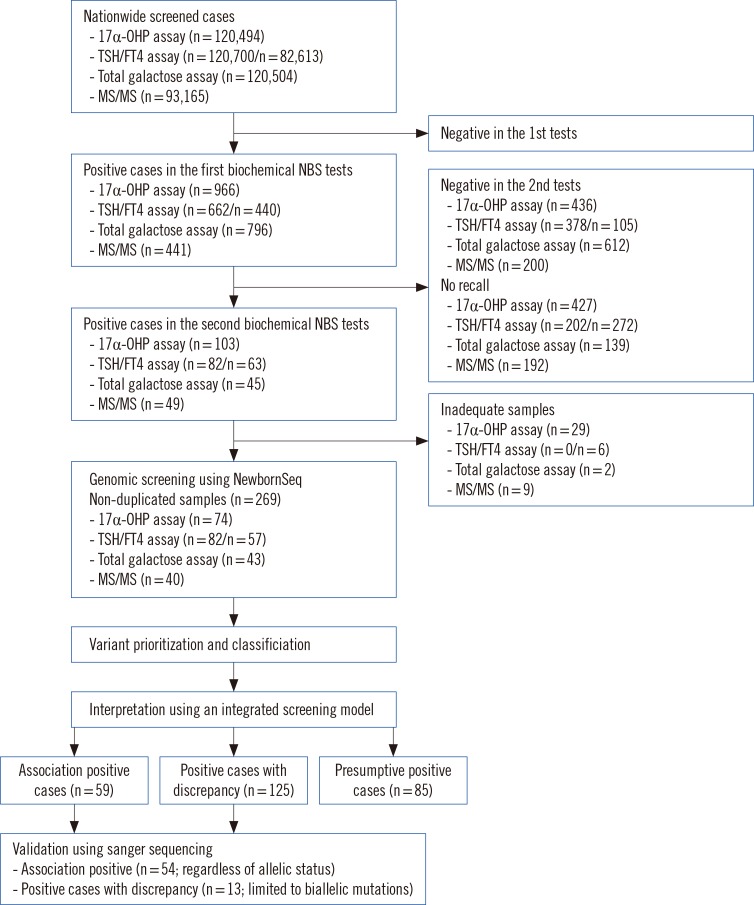
A population study was conducted in 120,700 newborns during routine NBS performed at Green Cross Laboratories in Korea from May 2013 to July 2014. The population in this study represented about 22% of total births (120,700/540,200) in Korea during that period [18]. A total of 269 cases with positive results from current NBS tests were also screened by NewbornSeq. The population samples were applied to an integrated screening model based on biochemical NBS tests and NewbornSeq (Fig. 1). We investigated whether there was any association between abnormal metabolite levels and gene mutations. According to the association results, patients were divided into three groups: (1) the APC group (Association-Positive Cases, with mutations in genes relevant to abnormal metabolite levels), (2) the PCD group (Positive Cases with Discrepancy, with mutations in genes irrelevant to abnormal metabolites), and (3) the PPC group (Presumptive Positive Cases, with metabolite abnormalities only) (Fig. 1). We investigated the mutation incidence and spectrum of the IMDs in the APC group. Researchers were blinded to all information regarding the identification of the newborns and controls. This study was approved by the Institutional Review Board of the Samsung Medical Center; informed consent was exempt because this study was performed by using stored biospecimens.
Fig. 1. Workflow for diagnosing inherited metabolic diseases. The study population represented about 22% of births (120,700/540,200) in Korea during the designated period. Using the integrated screening model, results were interpreted and divided into three groups: Association-Positive Cases (Cases with mutations in genes relevant to metabolites), Positive Cases with Discrepancy (Cases with mutations in genes irrelevant to metabolites), and Presumptive Positive Cases (Cases with only metabolite abnormalities). The numbers in brackets indicate the number of samples.
Abbreviations: 17α-OHP, 17α-hydroxyprogesterone; TSH, thyroid-stimulating hormone; FT4, free thyroxine; MS/MS, tandem mass spectrometry.
2. Current newborn screening pipeline
Dried blood spots (DBS) were collected from a heel stick on day 3-5 after birth. All NBS tests were performed as a part of the routine NBS program in Korea (Supplemental Method 1). The cases with metabolite levels higher than the cutoff were retested. "Presumptive positive" cases were defined as the individuals with abnormal levels of a metabolite detected from two separate samples.
3. NewbornSeq pipeline
1) DNA preparation and targeted sequencing
Genomic DNA from 27 positive controls and 269 newborns was extracted from EDTA-anticoagulated whole blood (WB) and DBS, respectively (Supplemental Method 1). Additional genomic DNA from 10 negative controls was extracted from DBS. Among them, genomic DNA from one healthy control was extracted from WB and DBS to validate the procedure of isolating DNA from DBS. A customized multiplex PCR amplification strategy was applied to analyze the 97 genes in the current Korean NBS panel, by using Ion AmpliSeq Designer software (Life Technologies, Carlsbad, CA, USA; Supplemental Table S1). All exons and intron sequences of 20 bp around each exon were targeted. The genomic regions with known mutations in the regulatory sequences were included, ultimately resulting in a total of 287 kb for analysis. Ninety-seven percent of targeted bases were covered under this protocol. Targeted sequencing was performed by using the Ion PGM platform (Life Technologies) following the manufacturer's instructions (Supplemental Method 1).
2) Bioinformatic analysis, mutation prioritization, and Sanger sequencing
Data were analyzed by using Torrent Suite software (version 4.0.3; Life Technologies). Variant calling was performed by using the "Germ Line-PGM-High Stringency" setting (Supplemental Table S2). The variants were functionally annotated by using the ANNOVAR tool [27,28].
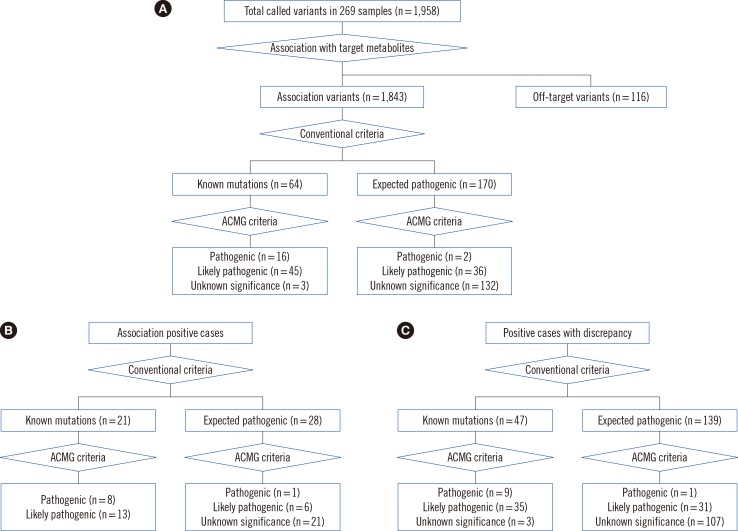
To prioritize pathogenic variants, we sequentially applied the following criteria: selection of allele frequency <0.01 in the 1000 Genome Project (1000GP, http://browser.1000genomes.org/index.html), the Exome Sequencing Project (ESP6500, http://evs.gs.washington.edu/EVS/), and the Exome Aggregation Consortium (ExAC, http://exac.broadinstitute.org/); selection of variants with multiple lines of evidence supporting "deleterious" or "damaging" effects, using Sorting Intolerant From Tolerant (SIFT), Polymorphism Phenotyping v2 (Polyphen-2), likelihood ratio test (LRT), MutationTaster, MutationAssesor, or FATHMM; selection of variants with a Genomic Evolutionary Rate Profiling (GERP) score higher than 2; removal of common polymorphisms reported in dbSNP v.138; selection of protein-impacting mutations such as nonsense mutations, mutations in GT-AG dinucleotides of the canonical splice sites, and frameshift mutations (Fig. 2). To avoid the false exclusion of pathogenic mutations, we manually reviewed the variants. Finally, the prioritized variants were classified as known pathogenic (KP) mutations and expected pathogenic (EP) mutations. "Disease-causing mutations" (DM) in the Human Genome Mutation Database (HGMD) or "pathogenic" mutations in ClinVar were categorized as KP mutations, while other variants were considered EP mutations [29,30]. Novel EP variants were compared to the reference sequence of whole-exome sequencing data (Korean Reference Genome DB, KRGDB, http://152.99.75.168/KRGDB/menuPages/firstInfo.jsp) from 622 healthy Korean individuals. In parallel, we applied the criteria for pathogenicity classification according to the American College of Medical Genetics and Genomics (ACMG) guideline [31], and the prioritized variants were classified as pathogenic variants, likely pathogenic variants, and variants of unknown significance (VUS) (Fig. 2). All prioritized mutations from the APC group and compound heterozygous or homozygous mutations from the PCD group were validated with independent Sanger sequencing (Supplemental Method 1).
Fig. 2. Putative variant prioritization and pathogenicity classification. Pathogenic variants were prioritized based on conventional methods and the American College of Medical Genetics and Genomics criteria in (A) total samples (n=269), (B) association-positive cases (n=125), and (C) in positive cases with discrepancy (n=85). The numbers in brackets indicate the number of different types of variants.
Abbreviation: ACMG, American College of Medical Genetics and Genomics.
4. Haplotype analysis
Haplotype analysis was performed to determine if there were founder effects in recurrent mutations identified from both the APC and PCD groups. The selection criteria of samples and SNPs for genotyping are shown in Supplemental Method 2. A total of 123 SNPs and seven candidate mutations were genotyped on the Sequenom MassARRAY SNP genotyping platform (Sequenom Inc., San Diego, CA, USA) and by Sanger sequencing, respectively (Supplemental Table S3). Haplotypes were constructed by using the software PHASE v2.1.1 (http://stephenslab.uchicago.edu/phase/download.html). Haplotype frequencies in mutation-positive cases were compared with those of 90 control individuals from the Korean HapMap [32].
5. Statistical analyses
Kruskal-Wallis test was used to compare metabolite levels among APC, PCD, and PPC. The statistical analyses were performed with MedCalc version 11.5.1.0 (Mariakerke, Belgium). P values less than 0.05 were considered statistically significant.
RESULTS
1. Performance of the NewbornSeq pipeline
Sequencing quality and coverage statistics using control samples are summarized in Supplemental Table S4. Taking the median, a total of 99% and 93% of bases were covered by at least 1-fold and 20-fold of coverage, respectively. The median percentage of on-target reads was 93% across the samples. There was no difference between the use of DBS and WB as sample type.
The conventional prioritization method reduced the number of variants per sample from a median of 247 to 4 in the 27 positive control samples (reduction rate of 98%). When the ACMG criteria were applied, the number of variants was reduced to a median of three per sample (reduction rate of 99%). NewbornSeq showed 100% sensitivity and specificity for 97 pathogenic variant alleles (54 causative alleles and 43 incidental alleles) in 27 positive control samples. However, only 96% (93/97) of pathogenic variants were reproducible; four pathogenic variants were not replicated in technical duplicates owing to low coverage less than 20 folds (Supplemental Table S5). The TAT was a median of 17 days by Sanger sequencing-based second-tier tests, which was reduced to within five days by the application of NewbornSeq (Supplemental Table S5).
A total of 1,958 variants were called in 269 newborns. We further reduced the number of variants to 244 (0-3 variants/sample) using the conventional criteria. According to the association between the metabolite abnormalities and mutated genes, 59 cases (22%), 125 cases (46%), and 85 cases (32%) were included in the APC, PCD, and PPC groups, respectively (Fig. 1). Sixty-six alleles among 70 mutant alleles from the APC group were confirmed by Sanger sequencing (validation rate of 94%, Supplemental Table S6).
When comparing metabolite levels among the groups, both thyroid-stimulating hormone (TSH) and free thyroxine (FT4) levels among the APC, PCD, and PPC groups were significantly different (P values for TSH and FT4 were 0.0044 and 0.0299, respectively; Supplemental Table S7).
2. Diagnosis of inherited metabolic diseases through the integrated screening model
In the APC group, 54 cases were validated among 59 cases with mutations in genes relevant to metabolite abnormalities, including congenital hypothyroidism (CH, n=34), galactosemia (n=11), type II citrullinemia (CTLN2, n=3), phenylketonuria (PKU, n=1), methylmalonic aciduria (MMA, n=2), and 3-methylcrotonyl-CoA carboxylase deficiency (3-MCC deficiency, n=3) (Table 1). Three cases (IMD_144, IMD_152, and IMD_ 153) had concurrent heterozygous mutations in different genes within the same metabolic pathway (Table 2). The overall positive rate of IMDs was estimated to be 20% (54/269) (Supplemental Table S8).
Table 1. Mutation incidence and frequency of inherited metabolic diseases detected using an integrated screening model.
| Disease/Gene | Mode of inheritance | N of validated cases | Birth prevalence | |||
|---|---|---|---|---|---|---|
| Biallelic mutations | Any mutations | Biallelic mutations | Any mutations | Compatible to mode of inheritance | ||
| Congenital hypothyroidism | ||||||
| TSHR | AD/AR | 1 | 10 | 1in 120,700 | 1in 12,070 | 1in 12,070 |
| PAX8 | AD | 0 | 3 | NA | 1in 40,233 | 1in 40,233 |
| DUOX2 | AD/AR | 2 | 12 (14)* | 1in 60,350 | 1in 8,621 | 1in 8,621 |
| DUOXA2 | AR | 2 | 7 (8)† | 1in 60,350 | 1in 15,088 | 1in 60,350 |
| TPO | AR | 0 | 1 | NA | 1in 120,504 | NA |
| SLC5A5 | AR | 1 | 1 | 1in 120,700 | 1in 120,700 | 1in 120,700 |
| Subtotal | 6 | 34 (37) | 1in 20,117 | 1in 3,550 | 1in 4,023 | |
| Galactosemia | ||||||
| GALE | AR | 1 | 6 | 1in 120,504 | 1in 20,084 | 1in 120,504 |
| GALT | AR | 0 | 3 | NA | 1in 40,168 | NA |
| GALK1 | AR | 0 | 2 | NA | 1in 60,252 | NA |
| Subtotal | 1 | 11 | 1in 120,504 | 1in 10,955 | 1in 120,504 | |
| Citrullinemia type II | ||||||
| SLC25A13 | AR | 1 | 3 | 1in 93,165 | 1in 31,055 | 1in 93,165 |
| Phenylketonuria | ||||||
| PAH | AR | 0 | 1 | NA | 1in 93,165 | NA |
| Methylmalonic aciduria | ||||||
| MUT | AR | 1 | 2 | 1in 93,165 | 1in 46,583 | 1in 93,165 |
| 3-methylcrotonyl-CoA carboxylase deficiency | ||||||
| MCCC1 | AR | 0 | 3 | NA | 1in 31,055 | NA |
| Total | AD/AR | 9 | 54 (57) | 1in 13,411 | 1in 2,235 | 1in 4,828 |
*One case with concurrent TSHR and DUOX2 mutations; †One case with concurrent DUOX2 and DUOXA2 mutations, and the other case with concurrent DUOXA2 and PAX8 mutations.
Abbreviations: AD, autosomal dominant; AR, autosomal recessive; NA, not applicable.
Table 2. Diagnosis of inherited metabolic diseases using an integrated screening model.
| Sample ID | Metabolite | cut-off* | NBS tests* | Gene | NT alteration | AA alteration | Conventional criteria | ACMG category | Zygosity | Disease | Frequency in APC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IMD_26 | C3 | 5 | 10 | MUT | c.2179C > T | p.R727X | KP | P | ComHet | MMA | 1/54 |
| MUT | c.322C > T | p.R108C | KP | LP | MMA | 1/54 | |||||
| IMD_30 | C5OH | 0.6 | 1.102 | MCCC1 | c.475T > C | p.C159R | KP | LP | Het | 3-MCC deficiency | 1/54 |
| IMD_31 | Cit | 55 | 348 | SLC25A13 | c.851delGTAT | p.M285Pfs*2 | KP | P | Het | CTLN2 | 3/54 |
| IMD_32 | Cit | 55 | 430 | SLC25A13 | c.851delGTAT | p.M285Pfs*2 | KP | P | Het | CTLN2 | 3/54 |
| IMD_39 | FT4 | 0.8 | 0.4 | DUOX2 | c.1232G > A | p.R411K | EP | VUS | NA | CH | 1/54 |
| IMD_42 | FT4 | 0.8 | 0.6 | TSHR | c.1454C > A | p.A485D | EP | VUS | NA | CH | 2/54 |
| IMD_44 | TSH | 12 | 23.3 | DUOX2 | c.1588A > T | p.K530X | KP | P | Het | CH | 2/54 |
| IMD_47 | TSH | 12 | 13.1 | DUOX2 | c.2654G > A | p.R885Q | KP | LP | Het | CH | 3/54 |
| IMD_48 | TSH | 12 | 25.5 | DUOXA2 | c.413dupA | p.Y138X | KP | LP | Het | CH | 4/54 |
| IMD_50 | TSH | 12 | 34.6 | TSHR | c.403A > T | p.N135Y | EP | VUS | NA | CH | 1/54 |
| TSHR | c.1349G > A | p.R450H | KP | LP | Het | CH | 4/54 | ||||
| IMD_52 | TSH | 12 | 16.5 | PAX8 | c.300dupTACC | p.M102fs | EP | LP | Het | CH | 1/54 |
| IMD_54 | TSH | 12 | 13.2 | TSHR | c.611C > T | p.A204V | KP | LP | Het | CH | 2/54 |
| IMD_56 | TSH | 12 | 12.1 | DUOXA2 | c.535T > C | p.Y179H | EP | VUS | NA | CH | 1/54 |
| IMD_57 | TSH | 12 | 12.1 | PAX8 | c.192G > C | p.R64S | EP | VUS | NA | CH | 1/54 |
| IMD_66 | TSH | 12 | 21.9 | DUOX2 | c.4010G > T | p.G1337V | EP | VUS | Het | CH | 1/54 |
| DUOX2 | c.1588A > T | p.K530X | KP | P | Het | CH | 2/54 | ||||
| IMD_68 | TSH | 12 | 17 | DUOX2 | c.1462G > A | p.G488R | KP | LP | Het | CH | 4/54 |
| IMD_79 | Gal | 13 | 21.7 | GALE | c.1002G > A | p.W334X | EP | P | Het | Galactosemia | 1/54 |
| IMD_80 | Gal | 13 | 19 | GALT | c.50+1G > A | NA | KP | P | Het | Galactosemia | 1/54 |
| IMD_81 | Gal | 13 | 32.2 | GALE | c.47G > A | p.S16N | EP | VUS | NA | Galactosemia | 1/54 |
| IMD_83 | Gal | 13 | 19.8 | GALE | c.905G > A | p.G302D | KP | LP | Het | Galactosemia | 2/54 |
| IMD_87 | Gal | 13 | 16.5 | GALT | c.998G > A | p.R333Q | KP | LP | Het | Galactosemia | 1/54 |
| IMD_89 | Gal | 13 | 40.4 | GALT | c.1034C > A | p.A345D | KP | LP | Het | Galactosemia | 1/54 |
| IMD_92 | TSH | 12 | 28.5 | TSHR | c.1349G > A | p.R450H | KP | LP | Het | CH | 4/54 |
| IMD_100 | C5OH | 0.6 | 2.571 | MCCC1 | c.288+2T > A | NA | KP | P | Het | 3-MCC deficiency | 2/54 |
| IMD_101 | TSH | 12 | 14.6 | DUOX2 | c.2635G>A | p.E879K | KP | LP | Het | CH | 1/54 |
| IMD_106 | C5OH | 0.6 | 1.016 | MCCC1 | c.288+2T > A | NA | KP | P | Het | 3-MCC deficiency | 2/54 |
| IMD_112 | Gal | 13 | 17 | GALE | c.264delT | p.F88fs | EP | LP | Het | Galactosemia | 1/54 |
| IMD_113 | FT4 | 0.8 | 0.3 | DUOXA2 | c.413dupA | p.Y138X | KP | LP | Het | CH | 4/54 |
| IMD_124 | Gal | 13 | 17.6 | GALE | c.905G > A | p.G302D | KP | LP | Het | Galactosemia | 2/54 |
| IMD_125 | TSH | 12 | 12.9 | TSHR | c.1349G > A | p.R450H | KP | LP | Het | CH | 4/54 |
| IMD_139 | Gal | 13 | 27 | GALE | c.38A > G | p.Y13C | EP | VUS | NA | Galactosemia | 1/54 |
| GALE | c.10A > G | p.K4E | EP | VUS | NA | Galactosemia | 1/54 | ||||
| IMD_142 | Phe | 130 | 142.216 | PAH | c.1065+1G > A | NA | KP | P | Het | PKU | 1/54 |
| IMD_144 | TSH | 12 | 23.1 | TSHR | c.611C > T | p.A204V | KP | LP | Het | CH | 2/54 |
| DUOX2 | c.2654G > A | p.R885Q | KP | LP | Het | CH | 3/54 | ||||
| IMD_149 | TSH | 12 | 14.2 | DUOX2 | c.3616G > A | p.A1206T | EP | VUS | NA | CH | 1/54 |
| DUOX2 | c.1462G > A | p.G488R | KP | LP | Het | CH | 4/54 | ||||
| IMD_152 | TSH | 12 | 12.3 | DUOX2 | c.3329G > A | p.R1110Q | KP | LP | Het | CH | 1/54 |
| DUOXA2 | c.738C > G | p.Y246X | KP | P | Het | CH | 3/54 | ||||
| IMD_153 | TSH | 12 | 13.6 | DUOXA2 | c.738C > G | p.Y246X | KP | P | Het | CH | 3/54 |
| PAX8 | c.739G > A | p.E247K | EP | VUS | NA | CH | 1/54 | ||||
| IMD_159 | Cit | 55 | 128.9 | SLC25A13 | c.1180+1G > A | NA | KP | P | ComHet | CTLN2 | 1/54 |
| SLC25A13 | c.851delGTAT | p.M285Pfs*2 | KP | P | CTLN2 | 3/54 | |||||
| IMD_164 | C3 | 5 | 9.126 | MUT | c.1228A > G | p.I410V | EP | VUS | NA | MMA | 1/54 |
| IMD_186 | TSH/GAL | 12.0/13.0 | 31.3/13.5 | DUOXA2 | c.413dupA | p.Y138X | KP | LP | Hom | CH | 4/54 |
| IMD_189 | TSH/FT4 | 12.0/0.8 | 55.7/0.4 | DUOX2 | c.1462G > A | p.G488R | KP | LP | Het | CH | 4/54 |
| IMD_191 | TSH/17α-OHP/FT4 | 12/12/0.8 | 54.9/18.1/0.2 | SLC5A5 | c.1060A > C | p.T354P | KP | LP | ComHet | CH | 1/54 |
| SLC5A5 | c.1605del | p.G535fs | EP | LP | CH | 1/54 | |||||
| IMD_196 | TSH | 12 | 94.1 | DUOX2 | c.1462G > A | p.G488R | KP | LP | Het | CH | 4/54 |
| IMD_197 | TSH | 12 | 14.8 | TSHR | c.1349G > A | p.R450H | KP | LP | Het | CH | 4/54 |
| IMD_203 | TSH | 12 | 12.4 | TSHR | c.1556G > A | p.R519H | EP | VUS | NA | CH | 1/54 |
| IMD_206 | TSH | 12 | 54.8 | DUOXA2 | c.280C > T | p.R94C | EP | VUS | NA | CH | 1/54 |
| DUOXA2 | c.413dupA | p.Y138X | KP | LP | Het | CH | 4/54 | ||||
| IMD_209 | TSH | 12 | 15.1 | DUOX2 | c.1319G > A | p.S440N | EP | VUS | NA | CH | 1/54 |
| IMD_210 | TSH | 0012 | 0054.8 | TSHR | c.1449C > A | p.N483K | EP | VUS | NA | CH | 1/54 |
| IMD_211 | TSH | 12 | 13 | DUOX2 | c.227C > T | p.P76L | EP | VUS | NA | CH | 1/54 |
| IMD_221 | TSH | 12 | 25.7 | TSHR | c.1454C > A | p.A485D | EP | VUS | NA | CH | 2/54 |
| IMD_234 | Gal | 13 | 27.7 | GALK1 | c.1159G > A | p.A387T | EP | VUS | NA | Galactosemia | 2/54 |
| IMD_235 | Gal | 13 | 19.6 | GALK1 | c.1159G > A | p.A387T | EP | VUS | NA | Galactosemia | 2/54 |
| IMD_237 | FT4 | 0.8 | 0.2 | DUOX2 | c.2654G > A | p.R885Q | KP | LP | Het | CH | 3/54 |
| IMD_238 | FT4 | 0.8 | 0.4 | TPO | c.1061G > T | p.W354L | EP | VUS | NA | CH | 1/54 |
| IMD_264 | 17α-OHP/FT4 | 12.0/0.7 | 17.9/0.4 | DUOXA2 | c.738C > G | p.Y246X | KP | P | Het | CH | 3/54 |
Reference sequences of MUT, MCCC1, SLC25A13, DUOX2, TSHR, DUOXA2, PAX8, GALE, GALT, GALT, PAH, SLC5A5, GALK1, and TPO were NM_000255, NM_001293273, NM_001160210, NM_014080, NM_000369, NM_207581, NM_003466, NM_001127621, NM_001258332, NM_000155, NM_000277, NM_000453, NM_000154, and NM_175722, respectively.
*The metabolite units of C3, C5OH, Phe, Cit, Gal, TSH, FT4 and 17α-OHP were µmol/L, µmol/L, µmol/L, µmol/L, µmol/L, mU/L, ng/dL, ng/mL, respectively. Recurrent mutations are in bold.
Abbreviations: KP, known pathogenic mutation based on the Human Genome Mutation Database (DM) or ClinVar (pathogenic) databases; EP, expected pathogenic mutation based on population frequency, in silico prediction, and mutation type (loss of function mutations); P, pathogenic; LP, likely pathogenic; VUS, variant of unknown significance; NA, not applicable; Het, heterozygous; ComHet, compound heterozygous; Hom, homozygous; Cit, citrulline; GAL, galactose; TSH, thyroid stimulating hormone; FT4, free T4; MMA, Methylmalonic aciduria; 3-MCC deficiency, 3-methylcrotonyl-CoA carboxylase deficiency; PKU, Phenylketonuria; CTLN2, Type II citrullinemia; CH, Congenital hypothyroidism.
We validated 13 cases with biallelic mutations for IMDs in the PCD group. Among them, there were 10 cases with treatable diseases, including ornithine carbamoyltransferase deficiency (OTC deficiency, n=2), type II glutaric aciduria (n=1), lysinuric protein intolerance (n=3), PKU (n=1), CH (n=2), and propionic aciduria (n=1) (Table 3). Multiple lines of evidence supporting deleterious effects of the 18 different mutant alleles are summarized in Supplemental Table S9. Details of the mutations and metabolite abnormalities identified in the PCD group are described in Supplemental Table S10.
Table 3. Unexpected detection of cases with biallelic mutations in genes irrelevant to metabolite abnormalities.
| Sample ID | Metabolites (Level; RR or cut-off) | Gene | NT alteration | AA alteration | Conventional criteria | ACMG category | Status* | Zygosity | Disease name | |
|---|---|---|---|---|---|---|---|---|---|---|
| Abnormal | Relevant | |||||||||
| IMD_35 | C0 (4.06; cut-off 7) | Cit (10.9; RR 2-55), Arg (16.8; RR 0–67) Gln (103; RR 0–300) | OTC | c.298+5G>C | NA | KP | LP | Known | Hom | OTC deficiency |
| IMD_36 | C0 (6.599, cut-off 7) | Glu (258; RR 0–805), C4 (0.23; RR 0–1.2), C6 (0.024; RR 0–0.5), C8 (0.007, RR 0–0.35), C10 (0.023 RR 0–0.5), C12 (0.033; RR 0–0.6), C18 (0.416; RR 0–2.13) | ETFB | c.155insT | p.P52fs | EP | LP | Novel | Hom | GA Type II |
| IMD_132 | Gal (14.8; RR: less than 13) | Arg (1.162; RR 0–67.3), Orn (47.565; RR 0–175) | SLC7A7 | c.498T>G | p.I166M | EP | VUS | Novel | Hom | LPI |
| IMD_162 | C5 (1.909; RR: less than 0.81) | Arg (3.353; RR 0–67.31), Orn (34.551; RR 0–175) | SLC7A7 | c.498T>G | p.I166M | EP | VUS | Novel | Hom | LPI |
| IMD_205 | TSH (12.7; RR: less than 12) | Phe (29.4; RR 0–130), Tyr (34.268; RR 0–299), Phe/Tyr (0.858; RR 0–2.5) | PAH | c.721C>T | p.R241C | KP | LP | Known | ComHet | PKU |
| PAH | c.442-1G>A | NA | KP | P | Known | |||||
| IMD_214 | Gal (21.4; RR: less than 13) | TSH (3.2; RR; less than 12), FT4 (1.8; RR; less than 0.8) | DUOX2 | c.3239T>C | p.I1080T | KP | LP | Known | ComHet | CH |
| DUOX2 | c.2678A>G | p.N893S | EP | VUS | Novel | |||||
| IMD_216 | Gal (13.5; RR: less than 13) | TSH (2.5; RR; less than 12), FT4 (2.3; RR; less than 0.8) | DUOX2 | c.617G>T | p.G206V | KP | VUS | Known | ComHet | CH |
| c.4232G>A | p.C1411Y | KP | VUS | Known | ||||||
| IMD_234 | Gal (27.7; RR: less than 13) | Cit (11.4; RR 2-55), Arg (14.7; RR 0–67), Gln (32; RR 0–300) | OTC | c.298+5G>C | NA | KP | LP | Known | Hom | OTC deficiency |
| IMD_237 | FT4 (0.2; RR less than 0.8) | C3 (0.4; RR 0.2-5) | PCCB | c.1283C>T | p.T428I | KP | LP | Known | ComHet | PA |
| PCCB | c.1316A>G | p.Y439C | KP | LP | Known | |||||
| IMD_243 | FT4 (0.6; RR less than 0.8) | Arg (9.5; RR 0-67.3), Orn (36; RR 0-175) | SLC7A7 | c.498T>G | p.I166M | EP | VUS | Novel | Hom | LPI |
Reference sequences of OTC, ETFB, HAL, SLC7A7, PAH, DUOX2, and PCCB were NM_000531.5, NM_001014763, NM_001258333, NM_001126105, NM_000277, NM_014080, and NM_000532, respectively. The metabolites units of TSH and FT4 were mU/L, ng/dL, ng/mL, respectively. The unit of the other metabolites was µmol/L.
*The mutation status was assessed based on the Human Genome Mutation Database (DM) or ClinVar (pathogenic) databases.
Abbreviations: AA, amino acid; NT, nucleotide; KP, known pathogenic; EP, expected pathogenic based on population frequency, in silico prediction, and mutation type (loss of function mutations); P, pathogenic; LP, likely pathogenic; VUS, variant of unknown significance; RR, reference range; Gal, galactose; TSH, thyroid-stimulating hormone; FT4, free thyroxine; Cit, citrulline; Arg, arginine; Gln, glutamine; Glu, glutamate; Orn, ornithine; Phe, phenylalanine; Tyr, tyrosine; NA, not applicable; Het, heterozygous; ComHet, compound heterozygous; Hom, homozygous; OTC deficiency, Ornithine carbamoyltransferase deficiency; LPI, Lysinuric protein intolerance; CH, Congenital hypothyroidism; PA, Propionic academia, GA type II, Glutaric acidemia type II; PKU, Phenylketonuria.
3. Mutation incidence of inherited metabolic diseases
We estimated the overall incidence of IMDs based on the current NBS tests to be 1 in 449 in the Korean population. The overall mutation incidence of IMDs calculated through an integrated screening model in the APC group was estimated to be one in 2,235 in the Korean population (Table 1). The highest incidences seen for CH and galactosemia were due to DUOX2 mutations and GALE mutations, respectively (Table 1).
4. Frequency and spectrum of pathogenic mutations
A total of 45 different mutations, including 21 known mutations and 24 expected pathogenic variants, were identified in 54 validated APCs (Table 2). In silico analyses results of validated variants are summarized in Supplemental Table S11. Recurrent mutations from the APC group were found in CTLN2 [p.R285 Pfs*2 in SLC25A13 (n=3)], CH [p.R885Q in DUOX2 (n=3), p.K530X in DUOX2 (n=2), p.G488R in DUOX2 (n=2), p.Y138X in DUOXA2 (n=4), p.R450H in TSHR (n=2), p.Y246X in DUOXA2 (n=2), p.A485D in TSHR (n=2), p.A204V in TSHR (n=2)], galactosemia [p.G302D in GALE (n=2), and 3-MCC deficiency [c.288+2T>A in MCCC1 (n=2)] (Table 2).
5. Founder effects
Seven different recurrent mutations, including SLC25A13 (p.R285Pfs*2), DUOXA2 (p.Y138X), GALE (p.G302D), SLC7A7 (p.I166M), PCCB (p.Y439C), and GALT (p.R224Q) were selected to construct haplotypes. Two samples with DUOXA2 mutations and three samples with DUOX2 mutations were added. Haplotype analysis yielded 392.2 kb, 392.6 kb, 775.3 kb, 399.2 kb, and 88.9 kb segments across the following mutations in DUOXA2, DUOX2, PCCB, SLC25A13, and GALT, respectively. All haplotypes were exclusively observed in mutation-containing cases (Table 4).
Table 4. Comparison of mutation-containing haplotypes between cases and controls.
| Genes | Haplotype * | SNPs in haplotype | Physical distance (bp) | % in cases | % in controls |
|---|---|---|---|---|---|
| DUOXA2 | TCCCGCCCCTATMAGTTTATCCTCC | rs397358, rs1473003, rs12913288, rs11635836, rs4775709, rs2467844, rs28662287, rs8024922, rs199138, rs269866, rs269862, rs269856, p.Y138X, rs16977681, rs175088, rs2271435, rs1648314, rs1648306, rs1648298, rs1706828, rs12439643, rs10519018, rs1706767, rs17533116, rs11636114 | 392,600 | 66.7% (8/12) | 0.00% (0/90) |
| DUOX2 | CCTTCTCCCTMATAGTTTATTCTCG | rs397358, rs1473003, rs12913288, rs11635836, rs4775709, rs2467844, rs28662287, rs8024922, rs199138, rs269866, p.R885Q, rs269862, rs269856, rs16977681, rs175088, rs2271435, rs1648314, rs1648306, rs1648298, rs1706828, rs12439643, rs10519018, rs1706767, rs17533116, rs11636114 | 392,600 | 50.0% (3/6) | 0.00% (0/90) |
| PCCB | AATAATGTCGTMGATTC | rs16843560, rs4678435, rs3772390, rs9845457, rs561307, rs16843829, rs2290131, rs576771, rs1279840, rs9856769, rs518972, p.Y439C, rs696520, rs7620314, rs900048, rs4521165, rs7616204 | 775,000 | 100.0% (3/3) | 0.00% (0/90) |
| SLC25A13 | TGGCAMCCCAC | rs184381, rs10267710, rs6465486, rs3779486, rs2301629, p.R285Pfs*2, rs12666465, rs6465496, rs35974282, rs4729249, rs12669236 | 399,200 | 100.0% (4/4) | 0.00% (0/90) |
| GALT | GCCMCCT | rs10972175, rs11791806, rs10814130, p.R224Q, rs3808868, rs1104748, rs2812365 | 37,700 | 100.0% (4/4) | 0.00% (0/90) |
The haplotype frequencies in mutation-positive cases were compared with those in 90 control individuals from the Korean HapMap.
*M represents recurrent mutations (p.Y138X in DUOXA2, p.R885Q in DUOX2, p.Y439C in PCCB, p.R285Pfs*2 in SLC25A13, p.R224Q in GALT).
Abbreviation: SNP, single nucleotide polymorphism.
DISCUSSION
The introduction of NGS is likely to change NBS practices. However, current NBS tests will not be replaced by genomic screening because some diseases, including CH, are not usually genetic conditions despite the fact that some related mutations have been reported. In this study, we noted that TSH levels were higher in PPC group cases than in APC group cases. This indicated the possibility of the presence of true CH in the PPC group. On the other hand, current NBS tests have some shortcomings. Disease risk can be modified by the environment over time, so current biochemical tests could yield false negatives or false positives. To complement the current NBS tests without replacing them, we designed NewbornSeq. NewbornSeq showed superior performance in characteristics important for its application in the NBS program: rapid TAT, small amounts of DNA required, minimally invasive sample type, sensitivity, and specificity.
In this study, we detected IMDs by applying an integrated screening model based on biochemical tests and NewbornSeq. The integrated screening model provided causative mutations in 20% of newborns with positive results from the biochemical tests in a NBS environment. In addition, it is noteworthy that the shortcomings of the current NBS tests, such as overdiagnosis and overtreatment, can be reduced by using the integrated screening model. For instance, galactosemia and a benign variant (known as Duarte galactosemia) cannot be differentiated by using current biochemical NBS tests. Under the current NBS system, a lactose-free diet might be provided to newborns with benign variants. A differential diagnosis between the pathogenic diseases and their benign variants using the integrated screening model could help avoid unnecessary treatment. In this study, we successfully excluded five Duarte galactosemia cases among 43 cases with increased galactose levels, by applying the integrated screening model.
We also identified ten cases with biallelic mutations for preventable IMDs from the PCD group (i.e., secondary findings). These cases would not be detected prior to the implementation of NewbornSeq, suggesting the presence of false negatives in the current NBS pipeline. This might be because some modifiers including prematurity, total parenteral nutrition, or maternal disease may influence the level of metabolites and the age of onset. Although these additional cases were not the primary target of the integrated screening model, they represent an important public health issue. Future studies will determine whether these cases benefited from the early treatment they received.
It should be noted that monoallelic mutations were frequently accompanied by metabolite abnormalities in the APC group. Frequent heterozygote mutations might be attributable to false-negatives in the current NewbornSeq pipeline because: i) missing variants due to low depth of coverage, ii) unidentified mutations in regulatory regions, iii) unidentified mutations in amplification-resistant gene regions, iv) allele dropout due to SNPs in PCR primer-binding sites, or v) structural variations (SV). Actually, we showed the possibility of false-negative results in four pathogenic alleles in control samples due to low depth of coverage. False-negatives can also be attributed to monoallelic mutations described in some of IMDs such as Wilson's disease or non-Mendelian mechanisms, such as synergistic heterozygosity [33,34,35,36].
To the best of our knowledge, this is the first genetic IMD epidemiologic study in the NBS setting using NGS. The representativeness of the population in this study prompted us to investigate the genetic epidemiology of IMDs in Korea. The incidence of IMDs based on the integrated screening model was 1 in 2,235 newborns using the APC group (Table 1). Using the reported data on the false positive rate (5-10 false positives/1 true positive) of MS/MS, the incidence of IMDs was calculated to be 1 in 2,245 from the biochemical incidence (1:449) [6]. The mutation incidence (1 in 2,235) based on the integrated screening model is quite similar to the disease incidence calculated from the biochemical incidence. This indicates that the integrated screening model provides a reliable and robust estimation of the incidence rate of IMDs, although data regarding clinical phenotypes were not used.
We identified founder effects in p.Y138X in DUOXA2, p.R885Q in DUOX2, p.Y439C in PCCB, and p.R224Q in GALT, except the mutation of p.R285Pfs*2 in SLC25A13, which was already reported as a founder mutation in Asians [24]. This study suggested that founder mutations could explain most of the recurrent IMD-related mutations in Koreans. Considering the history of the migration of the Mongoloids, further studies are needed to determine the time of origin and distribution pattern of these founder mutations in East Asian populations, including Chinese and Japanese populations.
This study is the first proof-of-concept study for introducing an integrated screening model into the actual NBS system. Importantly, this study raised some issues that should be considered regarding the introduction of NGS in a routine NBS system. First, there is the need for the clinical interpretation of unintended mutations, which would be frequently identified in genetic screening. This study suggested that the long-term follow-up of newborns with secondary findings or monoallelic mutations is necessary. Furthermore, future studies are recommended to determine whether the cases would benefit from the early treatment they received and to investigate whether the pathogenic variants were significantly associated with disease. Second, there is the need for a system that integrates biological knowledge with clinical information. Functional studies on novel mutations are time-consuming and impractical in the NBS setting. In future studies, a more sophisticated system should be introduced to integrate functional data, variant penetrance, and clinical data. Third, there is the need for another analytical platform to detect unidentified mutations, such as SV and regulatory mutations. In this study, although we found one instance of congenital adrenal hyperplasia (CAH) by the use of the integrated screening model, we could not validate the mutations of CYP21A2. The absence of CAH might be due to false-positives from the biochemical tests, false-negatives from NewbornSeq, or lack of proper validation methods; it is difficult to detect mutations of the CYP21A2 gene owing to its high pseudogene homology and frequently observed SV. Future studies are required to develop an additional platform to analyze SVs and genes with pseudogenes for highly suspicious cases.
In summary, we highlighted the epidemiologic and screening implications of NGS through the first population-based study in an NBS environment. This study has led to concerns about the opportunities and challenges for the implementation of NGS in NBS because it detected additional IMD cases that were not detected with the current NBS tests. The integrated screening model will be an effective public health strategy because it will enable faster and more accurate IMD detection. The future use of the integrated screening model as a first-tier approach will likely be more beneficial than the current NBS tests.
Acknowledgments
This study was supported by a grant from the Korea Health Technology R&D Project, Ministry of Health and Welfare, Republic of Korea (A120030).
Footnotes
Authors' Disclosures of Potential Conflicts of Interest: No potential conflicts of interest relevant to this article were reported.
SUPPLEMENTARY MATERIALS
Disorders related to current newborn screening in Korea
Variant calling criteria
SNPs genotyped for haplotype analysis
Sequencing quality and coverage summary of NewbornSeq
Mutation classification, diagnosis, and turnaround time in 37 control samples
Validation rate of NewbornSeq using Sanger sequencing
Comparison of metabolite level among groups
Detection rate of inherited metabolic diseases using the integrated screening model according to disease category
In silico analyses of variants identified in ten cases with genetic alterations irrelevant to metabolite abnormalities
Cases with genetic alterations irrelevant to metabolite abnormalities
In silico analyses of mutations identified in association positive cases
References
- 1.Feuchtbaum L, Carter J, Dowray S, Currier RJ, Lorey F. Birth prevalence of disorders detectable through newborn screening by race/ethnicity. Genet Med. 2012;14:937–945. doi: 10.1038/gim.2012.76. [DOI] [PubMed] [Google Scholar]
- 2.Yoon HR, Lee KR, Kang S, Lee DH, Yoo HW, Min WK, et al. Screening of newborns and high-risk group of children for inborn metabolic disorders using tandem mass spectrometry in South Korea: a three-year report. Clin Chim Acta. 2005;354:167–180. doi: 10.1016/j.cccn.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 3.Zytkovicz TH, Fitzgerald EF, Marsden D, Larson CA, Shih VE, Johnson DM, et al. Tandem mass spectrometric analysis for amino, organic, and fatty acid disorders in newborn dried blood spots: a two-year summary from the New England Newborn Screening Program. Clin Chem. 2001;47:1945–1955. [PubMed] [Google Scholar]
- 4.Wilcken B, Wiley V, Hammond J, Carpenter K. Screening newborns for inborn errors of metabolism by tandem mass spectrometry. N Engl J Med. 2003;348:2304–2312. doi: 10.1056/NEJMoa025225. [DOI] [PubMed] [Google Scholar]
- 5.Schulze A, Lindner M, Kohlmüller D, Olgemöller K, Mayatepek E, Hoffmann GF. Expanded newborn screening for inborn errors of metabolism by electrospray ionization-tandem mass spectrometry: results, outcome, and implications. Pediatrics. 2003;111:1399–1406. doi: 10.1542/peds.111.6.1399. [DOI] [PubMed] [Google Scholar]
- 6.Mak CM, Lee HC, Chan AY, Lam CW. Inborn errors of metabolism and expanded newborn screening: review and update. Crit Rev Clin Lab Sci. 2013;50:142–162. doi: 10.3109/10408363.2013.847896. [DOI] [PubMed] [Google Scholar]
- 7.Sahai I, Marsden D. Newborn screening. Crit Rev Clin Lab Sci. 2009;46:55–82. doi: 10.1080/10408360802485305. [DOI] [PubMed] [Google Scholar]
- 8.Park KJ, Kim JW. Perspectives on next-generation newborn screening. Lab Med Online. 2015;5:169–175. [Google Scholar]
- 9.Knoppers BM, Sénécal K, Borry P, Avard D. Whole-genome sequencing in newborn screening programs. Sci Transl Med. 2014;6:229cm2. doi: 10.1126/scitranslmed.3008494. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharjee A, Sokolsky T, Wyman SK, Reese MG, Puffenberger E, Strauss K, et al. Development of DNA confirmatory and high-risk diagnostic testing for newborns using targeted next-generation DNA sequencing. Genet Med. 2015;17:337–347. doi: 10.1038/gim.2014.117. [DOI] [PubMed] [Google Scholar]
- 11.Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012;4:154ra135. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willig LK, Petrikin JE, Smith LD, Saunders CJ, Thiffault I, Miller NA, et al. Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: a retrospective analysis of diagnostic and clinical findings. Lancet Respir Med. 2015;3:377–387. doi: 10.1016/S2213-2600(15)00139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernández-Marmiesse A, Morey M, Pineda M, Eiris J, Couce ML, Castro-Gago M, et al. Assessment of a targeted resequencing assay as a support tool in the diagnosis of lysosomal storage disorders. Orphanet J Rare Dis. 2014;9:59. doi: 10.1186/1750-1172-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao YY, Qu YJ, Song F, Zhang T, Bai JL, Jin YW, et al. Fast clinical molecular diagnosis of hyperphenylalaninemia using next-generation sequencing-based on a custom AmpliSeq panel and Ion Torrent PGM sequencing. Mol Genet Metab. 2014;113:261–266. doi: 10.1016/j.ymgme.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Calvo SE, Compton AG, Hershman SG, Lim SC, Lieber DS, Tucker EJ, et al. Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci Transl Med. 2012;4:118ra10. doi: 10.1126/scitranslmed.3003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Applegarth DA, Toone JR, Lowry RB. Incidence of inborn errors of metabolism in British Columbia, 1969-1996. Pediatrics. 2000;105:e10. doi: 10.1542/peds.105.1.e10. [DOI] [PubMed] [Google Scholar]
- 17.Dionisi-Vici C, Rizzo C, Burlina AB, Caruso U, Sabetta G, Uziel G, et al. Inborn errors of metabolism in the Italian pediatric population: a national retrospective survey. J Pediatr. 2002;140:321–327. doi: 10.1067/mpd.2002.122394. [DOI] [PubMed] [Google Scholar]
- 18.Niu DM, Chien YH, Chiang CC, Ho HC, Hwu WL, Kao SM, et al. Nationwide survey of extended newborn screening by tandem mass spectrometry in Taiwan. J Inherit Metab Dis. 2010;33:S295–S305. doi: 10.1007/s10545-010-9129-z. [DOI] [PubMed] [Google Scholar]
- 19.Narumi S, Muroya K, Asakura Y, Aachi M, Hasegawa T. Molecular basis of thyroid dyshormonogenesis: genetic screening in population-based Japanese patients. J Clin Endocrinol Metab. 2011;96:E1838–E1842. doi: 10.1210/jc.2011-1573. [DOI] [PubMed] [Google Scholar]
- 20.Narumi S, Muroya K, Asakura Y, Adachi M, Hasegawa T. Transcription factor mutations and congenital hypothyroidism: systematic genetic screening of a population-based cohort of Japanese patients. J Clin Endocrinol Metab. 2010;95:1981–1985. doi: 10.1210/jc.2009-2373. [DOI] [PubMed] [Google Scholar]
- 21.Narumi S, Muroya K, Abe Y, Yasui M, Asakura Y, Adachi M, et al. TSHR mutations as a cause of congenital hypothyroidism in Japan: a population-based genetic epidemiology study. J Clin Endocrinol Metab. 2009;94:1317–1323. doi: 10.1210/jc.2008-1767. [DOI] [PubMed] [Google Scholar]
- 22.Kalaydjieva L, Perez-Lezaun A, Angelicheva D, Onengut S, Dye D, Bosshard NU, et al. A founder mutation in the GK1 gene is responsible for galactokinase deficiency in Roma (Gypsies) Am J Hum Genet. 1999;65:1299–1307. doi: 10.1086/302611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki M, West C, Beutler E. Large-scale molecular screening for galactosemia alleles in a pan-ethnic population. Hum Genet. 2001;109:210–215. doi: 10.1007/s004390100552. [DOI] [PubMed] [Google Scholar]
- 24.Lu YB, Kobayashi K, Ushikai M, Tabata A, Iijima M, Li MX, et al. Frequency and distribution in East Asia of 12 mutations identified in the SLC25A13 gene of Japanese patients with citrin deficiency. J Hum Genet. 2005;50:338–346. doi: 10.1007/s10038-005-0262-8. [DOI] [PubMed] [Google Scholar]
- 25.Chien YH, Chiang SC, Huang A, Chou SP, Tseng SS, Huang YT, et al. Mutation spectrum in Taiwanese patients with phenylalanine hydroxylase deficiency and a founder effect for the R241C mutation. Hum Mutat. 2004;23:206. doi: 10.1002/humu.9215. [DOI] [PubMed] [Google Scholar]
- 26.Wang T, Okano Y, Eisensmith RC, Harvey ML, Lo WH, Huang SZ, et al. Founder effect of a prevalent phenylketonuria mutation in the Oriental population. Proc Natl Acad Sci U S A. 1991;88:2146–2150. doi: 10.1073/pnas.88.6.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34:E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim YU, Kim SH, Jin H, Park YK, Ji M, Kim YJ. The Korean HapMap Project website. Genomics Inform. 2008;6:91–94. [Google Scholar]
- 33.Satoh M, Aso K, Ogikubo S, Ogasawara A, Saji T. Genetic analysis in children with transient thyroid dysfunction or subclinical hypothyroidism detected on neonatal screening. Clin Pediatr Endocrinol. 2009;18:95–100. doi: 10.1297/cpe.18.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Marco G, Agretti P, Montanelli L, Di Cosmo C, Bagattini B, De Servi M, et al. Identification and functional analysis of novel dual oxidase 2 (DUOX2) mutations in children with congenital or subclinical hypothyroidism. J Clin Endocrinol Metab. 2011;96:E1335–E1339. doi: 10.1210/jc.2010-2467. [DOI] [PubMed] [Google Scholar]
- 35.Coffey AJ, Durkie M, Hague S, McLay K, Emmerson J, Lo C, et al. A genetic study of Wilson's disease in the United Kingdom. Brain. 2013;136:1476–1487. doi: 10.1093/brain/awt035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vockley J. Metabolism as a complex genetic trait, a systems biology approach: implications for inborn errors of metabolism and clinical diseases. J Inherit Metab Dis. 2008;31:619–629. doi: 10.1007/s10545-008-1005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Disorders related to current newborn screening in Korea
Variant calling criteria
SNPs genotyped for haplotype analysis
Sequencing quality and coverage summary of NewbornSeq
Mutation classification, diagnosis, and turnaround time in 37 control samples
Validation rate of NewbornSeq using Sanger sequencing
Comparison of metabolite level among groups
Detection rate of inherited metabolic diseases using the integrated screening model according to disease category
In silico analyses of variants identified in ten cases with genetic alterations irrelevant to metabolite abnormalities
Cases with genetic alterations irrelevant to metabolite abnormalities
In silico analyses of mutations identified in association positive cases