Abstract
Purpose
Regulatory T (Treg) cells are key modulators in the immune system. Recent studies have shown that atopic dermatitis (AD) patients have higher numbers of Treg cells; however, little is known about the specific phenotype and function of Treg cells in AD.
Materials and Methods
To identify differentially expressed proteins in peripheral induced Treg cells in AD and naturally derived Treg cells in normal controls, CD4+CD25+ Treg cells were isolated from thymus tissue of normal mice and the spleens of AD mice. Membrane proteins were extracted, and quantitative proteomics labeling with Tandem Mass Tags (TMT) was performed, followed by one-dimensional liquid chromatography/tandem mass spectrometry analysis.
Results
Using TMT labeling, we identified 510 proteins, including 63 membrane proteins and 16 plasma membrane proteins. CD47 was one of the upregulated proteins in Treg cells in AD spleens. Although CD47 was expressed in all CD4+ and CD8+ T cells, a significantly higher expression of CD47 was observed in the Treg cells of AD mice and AD patients than in those of normal mice and healthy controls. Furthermore, Treg cells from the spleen showed a significantly higher expression of CD47 than those from the thymus.
Conclusion
We found that CD47 is highly expressed in the Treg cells of AD mice, particularly in the spleen. Based on our results, we propose that CD47high Treg cells are likely induced Treg cells and that upregulated CD47 in the Treg cells of AD patients may play a role in the increased population of Treg cells in AD.
Keywords: Atopic dermatitis, regulatory T cells, CD47, Tandem Mass Tags (TMT)
INTRODUCTION
Atopic dermatitis (AD) is a highly pruritic, chronic relapsing inflammatory skin disease characterized by dry itchy skin.1 An imbalance of Th1 and Th2 immune responses plays a critical role in the pathogenesis of AD.2,3,4 CD25+CD4+ regulatory T (Treg) cells are key modulators of self-tolerance and immune homeostasis via the suppression of excessive immune responses.5,6 Treg cells have been reported to play an important role in many autoimmune and allergic diseases.7 In AD patients, Treg cells are increased compared to healthy controls.8,9 In addition, an elevation of Treg cells correlates significantly with AD severity.10,11 While Treg cells in AD patients have an immunosuppressive function similar to those in healthy controls, CCR6+ Treg cells in AD promote a Th2 immune response.12,13 The underlying reason for the increase of Treg cells in AD and the function of these cells in AD pathogenesis, however, remain unclear.
In this study, we sought to identify proteins that are differentially expressed in peripheral induced Treg cells in AD and in naturally derived Treg cells in the thymus. We employed the recently developed gel-free proteomic approach, which uses isobaric labeling reagents, such as Tandem Mass Tags (TMT), to quantitatively analyze the proteome.14 TMT labeling enables genome-wide quantification of protein expression levels as well as the identification and analysis of a small fraction of the proteins, such as membrane proteins, from the whole proteome.
MATERIALS AND METHODS
Induction of AD skin lesions in NC/Nga mice using Dermatophagoides farinae ointment
Six-week-old female NC/Nga mice were purchased from Central Lab Animal Incorporation (Seoul, Korea) and housed under specific pathogen-free conditions with a stable temperature (22±3℃) and humidity (55±15%). After a week of stabilization, the hair on the back was removed using electric clippers and hair removal cream. One day after hair removal, 150 µL of 4% sodium dodecyl sulfate (SDS) was topically applied to disrupt the skin barrier. Two hours later, Biostir AD cream [Dermatophagoides farinae (D. farinae) body extracts, Biostir, Kobe, Japan] was applied to the dorsal surface. The removal of hair, application of SDS, and treatment using Biostir AD cream were repeated twice a week for 8 weeks. NC/Nga mice without dust mite application were used as a control group.
Evaluation of AD severity
The severity of AD-like skin lesions was measured using the SCORAD index for mice. This index ranges from 0 to 12. In brief, the SCORAD index includes scores based on the presence of erythema or hemorrhage, scarring or dryness, excoriation or erosion, and edema. Each symptom was graded on a scale of 0 to 3 (0, none; 1, mild; 2, moderate; and 3, severe). The score was the sum of individual item scores. The severity of dermatitis was assessed once weekly by two independent researchers.
Enzyme-linked immunosorbent assay (ELISA) for total serum IgE
Total immunoglobulin E (IgE) levels in serum were measured with an enzyme-linked immunosorbent assay (ELISA) MAX™ Deluxe Set (BioLegend, San Diego, CA, USA) in accordance with the manufacturer's instructions. In brief, wells of a 96-well plate were coated with an IgE-specific monoclonal antibody and then incubated overnight at 4℃. Standards and serum samples were added to the plate, which was then incubated at room temperature for 2 h. Captured IgE molecules were detected using biotinylated anti-mouse IgE detection antibody. Avidin-horseradish peroxidase was subsequently added, followed by TMB substrate solution. Absorbance (as optical density) of each well was measured at 450 nm with a microplate reader.
Preparation of splenocytes and thymocytes using high-gradient magnetic cell sorting (MACS)
To identify differentially expressed proteins in peripheral induced Treg cells in AD and in naturally derived Treg cells in normal controls, CD4+CD25+ Treg cells were isolated from thymus tissue of normal mice and the spleens of AD mice. The spleen and thymus were removed from each mouse and disrupted using a syringe plunger to prepare a cell suspension. Blood cells were lysed using blood cell lysis buffer (Sigma-Aldrich, St. Louis, MO, USA) containing 8.3 g/L ammonium chloride and 0.01 M Tris-HCl buffer (pH 7.5±0.2). Murine CD4+CD25+ T cells were isolated from splenocytes and thymocytes using a CD4+CD25+ Regulatory T cell Isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's protocol. The cells obtained were about 80% pure, as determined via FACS analysis.
Membrane protein extraction
Isolated Treg cells were centrifuged at 850×g for 2 min, and the supernatant was then removed. Membrane proteins were extracted using a Mem-PERTM Eukaryotic Membrane Protein Extraction Reagent Kit (Thermo Scientific, Waltham, MA, USA). In brief, Reagent A was added to the pellet in order to lyse the cells. Two parts Reagent C along with one part Reagent B was then added, and after centrifugation and incubation, the hydrophilic top layer was discarded.
TMT labeling for proteomic analysis
After membrane protein extraction, 100 µg of protein was equally divided into two halves and labeled with two different TMT reagents (Thermo Scientific) following the manufacturer's standard protocol. The membrane protein samples from the spleens of six-week-old AD mice were labeled with TMT-126 and TMT-130, whereas those from the thymuses of normal mice were labeled with TMT-127 and TMT-131. The four samples labeled with different TMT reagents were mixed, dried, and then re-solubilized with water containing 0.5% formic acid for one-dimensional liquid chromatography/tandem mass spectrometry (1DLC/MS/MS) analysis.
Nano-LC-MS/MS analysis
The resultant peptides were analyzed using 1DLC-MS/MS. Peptides were identified using MS/MS with a nano-LC-MS system consisting of a Nano Acquity U7PLC system (Waters, Milford, MA, USA) and an LTQ Orbitrap elite mass spectrometer (Thermo Scientific) equipped with a nanoelectrospray source. An autosampler was used to load 5-µL aliquots of the peptide solutions onto a C18 trap-column (i.d. 300 µm, length 5 mm, and particle size 5 µm; Waters). The peptides were desalted and concentrated on the column at a flow rate of 5 µL/min. Then, the trapped peptides were back-flushed and separated on a 200-mm homemade microcapillary column, consisting of C18 matrix (Aqua; particle size 3 µm) packed into 100-µm silica tubing with an orifice i.d. of about 6 µm. The mobile phases, A and B, were composed of 0 and 100% acetonitrile, respectively, and each contained 0.1% formic acid. The LC gradient began with 5% B for 5 min and was increased to 15% B over 5 min, to 50% B over 100 min, and then to 95% B over 5 min, at which point it remained at 95% B for 5 min and then decreased to 5% B for another 5 min. The column was re-equilibrated to 5% B for 15 min before the next run. The voltage applied to produce the electrospray was 2.2 kV. During the chromatographic separation, the LTQ Orbitrap Elite was operated in a data-dependent mode under direct control of Xcalibur software (Thermo Scientific). The MS data were acquired using the following parameters: 10 data-dependent collision-induced dissociation (CID) MS/MS scans per every full scan in label-free mode; 10 data-dependent higher energy collision-induced dissociation (HCD) MS/MS scans per every full scan in TMT; CID scans acquired in LTQ with two-microscan averaging; full scans and HCD scans acquired in Orbitrap at the resolutions of 30000 and 15000, respectively, with two-microscan averaging; 35% normalized collision energy in CID and in HCD; ±1.5-Da isolation window; and dynamic exclusion enabled with a ±1.5-Da exclusion window. All 1DLC-MS/MS analyses for TMT-labeling quantification were performed in duplicate for each sample.
Data analysis
A probability-based (and error-tolerant) protein database search of MS/MS spectra against the latest IPI rat protein database (IPI rat v3.70) was performed using a local MASCOT server (2.3, Matrix Science, London, UK) to identify and quantify the analyzed proteins. The rate of decoy hits in the combined forward and reverse database was less than 1% of the forward hits at both the peptide and the protein levels in each of these experiments. The following search criteria were used: 20 ppm precursor ion mass tolerance; 0.5-Da product ion mass tolerance; two missed cleavages; trypsin as the enzyme; TMT modification at the N-terminus and lysine residues as well as carbamidomethylation at the cysteine residues as static modifications; oxidation at methionine; phosphorylation at serine, threonine, and tyrosine as variable modifications; an ion score threshold of 20; and TMT-6 plex for quantification. Quantification was based on the averaged signal-to-noise ratio of TMT reporter product ions of more than two unique peptides. In TMT experiments, reporter ions for peptide identification were extracted from small windows (±20 ppm) around their expected m/z in the HCD scan. As a single sample was individually labeled with two TMT reagents, peptides with similar ratios in the comparison of the intensity of reporter ions within 30% were selected for protein quantitation. The abundance ratio of a protein was estimated using the ratio between the total intensities of 12 proteins in different reporter ion channels. Given the distributions of protein log2 ratios, proteins showing ≤-0.4 or ≥0.4 were considered to be differentially expressed.
Human samples
Human blood samples were obtained from three non-AD healthy controls and from four AD patients who were diagnosed according to the criteria of Hanifin and Rajka.15 The Institutional Review Board approved this study (IRB no: 4-2013-0624), and all subjects provided written informed consent to participate in the study. PBMCs from the subjects were isolated by centrifugation on a Lymphoprep gradient (density 1.077 g/mL) and centrifuged at 800×g for 15 min at 4℃. Cells from the interphase were then washed three times with phosphate-buffered saline (PBS) containing 5 mM EDTA. Isolated PBMCs were used for flow cytometric analysis.
Flow cytometry
Cells were washed with PBS and stained with a fixable viability dye. After washing, cells were labeled at 4℃ for 30 min with anti-CD3, -CD4, -CD8, and -CD25 antibodies conjugated with fluorescent dye (eBioscience, San Diego, CA, USA), anti-CD47 (eBioscience), and PerCP-Cy5.5 anti-rat secondary antibody. For intracellular labeling, cells were fixed and permeabilized with cytofix/cytoperm buffer (eBioscience) and labeled with anti-human FOXP3 antibody (eBioscience) conjugated with FITC. Labeled cells were quantified using a BD FACSVerse flow cytometer, and the data were analyzed using FlowJo Software (BD Bioscience, San Jose, CA, USA).
Western blotting
After membrane protein extraction as described above, equal amounts of cellular proteins were mixed with 5x sample buffer and heated at 100℃ for 5 min. Proteins were then resolved on an 8% SDS-polyacrylamide gel. After electrophoresis, the proteins were transferred onto an ECL nitrocellulose membrane (GE Healthcare, Buckinghamshire, UK) using Tris buffer [0.025 M Tris-HCI (pH 6.8), 0.192 M glycine, and 20% MeOH]. The membrane was blocked for 1 h at room temperature with 5% skim milk in TBS-Tween 20, incubated overnight at 4℃ with anti-CD47 antibody (BD Bioscience) and anti-GAPDH antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and then incubated with horseradish peroxidase-conjugated antirat secondary antibody (Santa Cruz Biotechnology) for 1 h at room temperature. Finally, the membrane was developed using enhanced chemiluminescence Western blotting detection reagents (Santa Cruz Biotechnology) and quantified via densitometry.
Statistical analyses
A one-way ANOVA was used to assess the SCORAD results (Fig. 1B) and CD47 expression in NC/Nga mice (Fig. 3B and D). Bonferroni correction was used for post-hoc analysis. A two-way ANOVA was used to assess the results for serum total IgE (Fig. 1C). Pearson correlation was used for the analysis of CD47 expression in AD patients (Fig. 4C). Deviations were considered statistically significant when p<0.05.
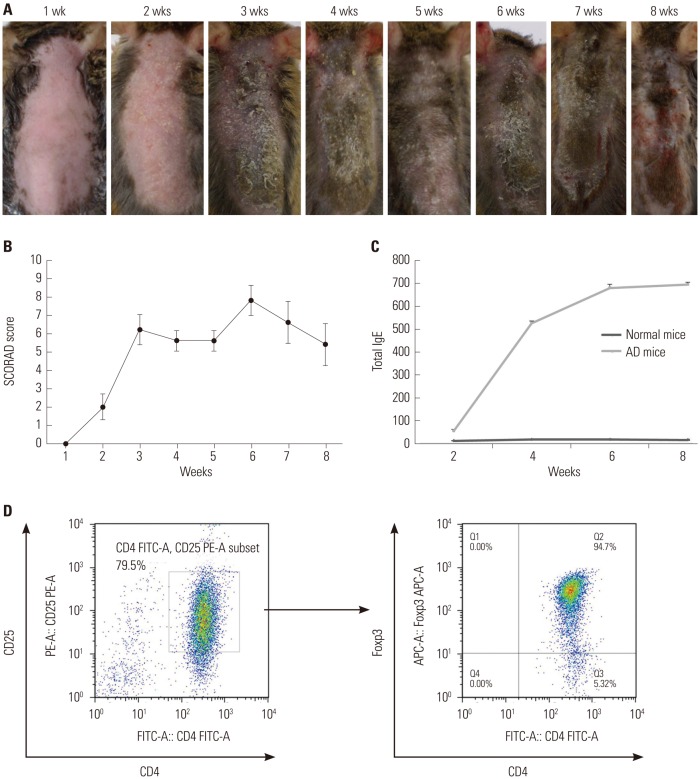
Fig. 1. Induction of AD-like skin lesions in NC/Nga mice and isolation of CD4+CD25+ Treg cells using MACS®. (A) Dorsal skin of mice treated with D. farinae extracts for 8 weeks. Significant erythema and crusts were observed after 3 weeks of D. farinae application. After 6 weeks of application, the most severe erythema, crusts, excoriation, and oozing were apparent. (B) SCORAD scores were plotted against the time of repeated topical application of D. farinae. (C) Serum IgE levels were measured via ELISA after repeated topical application of D. farinae ointment. All results are representative or mean±SD from groups that contained five mice. (D) CD4+CD25+ Treg cells were sorted using an AutoMACS cell sorter. The purity of isolated cells was 79.5%, and 94.7% of the isolated CD4+CD25+ Treg cells expressed Foxp3. Results are representative of three independent experiments. AD, atopic dermatitis; Treg, regulatory T; D. farinae, Dermatophagoides farinae; IgE, immunoglobulin E; ELISA, enzyme-linked immunosorbent assay.
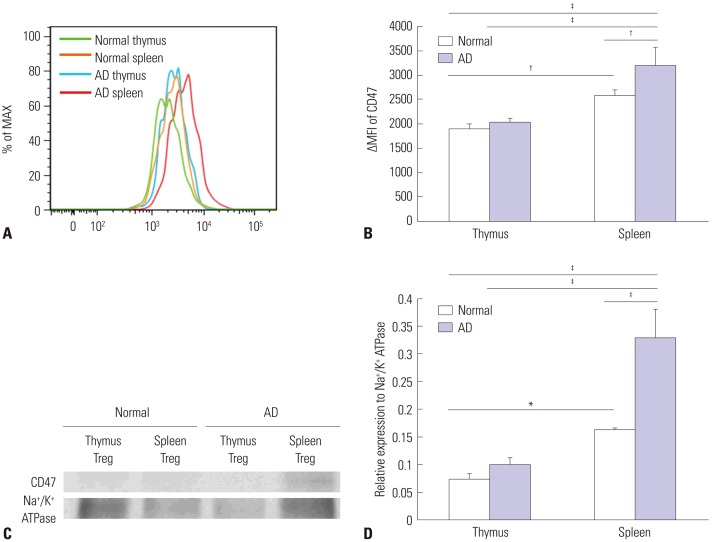
Fig. 3. Increased expression of CD47 in Treg cells from AD mice. (A) FACS analysis of expression of CD47 in Treg cells from thymuses and spleens from normal and AD mice. The ΔMFI of CD47 was determined in Treg cells. The green line indicates CD47 in a normal thymus, the orange line indicates CD47 in a normal spleen, the blue line indicates CD47 in an AD thymus, and the red line indicates CD47 in an AD spleen. (B) CD47 expression was more upregulated in Treg cells isolated from normal spleens than in those isolated from normal thymuses. Moreover, its expression was further upregulated in Treg cells from AD spleens. (C) Western blot analysis of the expression of CD47 on Treg cells from spleens and thymuses of AD mice and normal mice. The expression level of CD47 was similar to the results of the FACS analysis. (D) Western blot densitometry. Differences were determined via one-way ANOVA. For each group, mice n=4. *p<0.05, †p<0.01, ‡p<0.001. AD, atopic dermatitis; Treg, regulatory T.
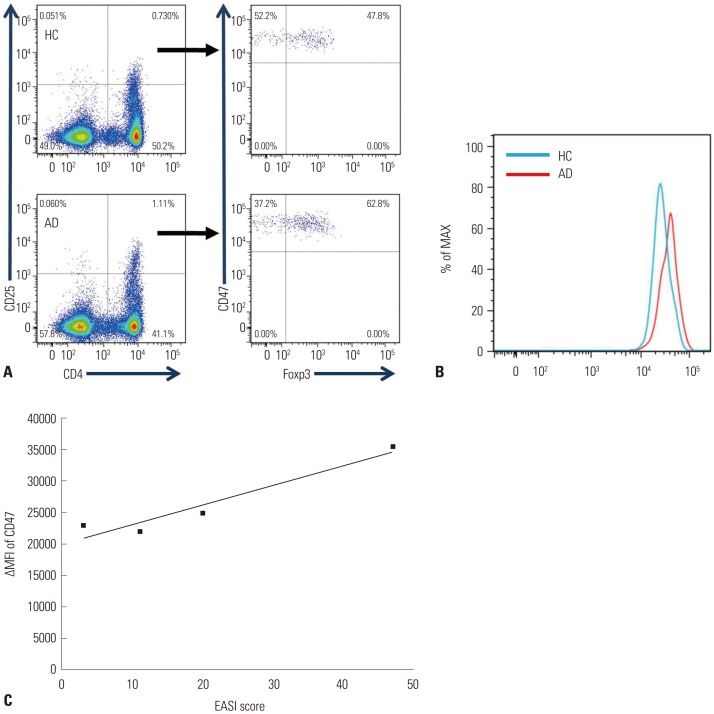
Fig. 4. Increased expression of CD47 in human Treg cells from AD patients. (A) All CD4+CD25+Foxp3+ Treg cells expressed CD47 in humans, a result similar to that for mice. (B) The expression level of CD47 was higher in Treg cells from AD patients than in those from healthy controls. (C) The severity of AD correlated with the expression of CD47 (Pearson correlation r=0.84, p=0.018). AD, atopic dermatitis; Treg, regulatory T.
RESULTS
AD-like skin lesions induced by application of D. farinae topical ointment in mice
Mild erythema was observed following a 2-week application of D. farinae ointment, and significant scarring and crusts were observed after 3 weeks of topical application of D. farinae extracts (Fig. 1A). The SCORAD scores exhibited a rapid and significant increase, with the highest score (7.8±0.73) noted after 6 weeks of D. farinae treatment. After the 6 weeks of D. farinae ointment treatment, a slight improvement in the skin was observed, and the SCORAD score decreased to 5.5±1.19 at 8 weeks of D. farinae topical application (p<0.001) (Fig. 1B).
In accordance with the clinical findings, repeated topical application of D. farinae caused a significant increase in serum IgE levels in NC/Nga mice compared to normal mice. The serum IgE level was 53±8.8 ng/mL after 2 weeks of application, and it increased at the 4- and 6-week time points before reaching a plateau (525.4±12.4 ng/mL after 4 weeks and 679.2±20.1 ng/mL after 6 weeks; p<0.001) (Fig. 1C).
Isolation of CD4+CD25+ Treg cells
To identify Treg cells specifically involved in AD pathogenesis, we isolated CD4+CD25+ T cells from AD spleens and normal thymuses with 80% purity. More than 90% of these cells expressed Foxp3, suggesting that these cells were Treg cells (Fig. 1D). Thus, we analyzed the cell surface proteins from these purified cells using TMT-labeling proteomic analysis.
Protein identification using TMT-label proteomic analysis
Quantification of 510 proteins was achieved via LC-MS/MS analysis and based on the peak area of precursor ions of identified peptides. Among the 510 quantified proteins, 63 were membrane proteins (Table 1), and 16 were plasma membrane proteins (Table 2). Considering the distributions of the protein log2 ratios, proteins that exhibited values of ≤-0.4 or ≥0.4 were considered to be differentially expressed. These criteria allowed for the identification of six upregulated proteins, including H-2 class II histocompatibility antigen, h-2 class I histocompatibility antigen K-W28 alpha chain-like isoform 1, D-P alpha chain-like isoform 4, protein tyrosine phosphatase receptor type C-associated protein, isoform 3 of receptor-type tyrosine-protein phosphatase C, and isoform 1 of the leukocyte surface antigen CD47 (Table 2) in Treg cells in AD. Among these, CD47 has been reported to be associated with immune regulation, particularly T cell costimulation16 and phagocytosis.17,18 Therefore, we further evaluated CD47 expression in Treg cells.
Table 1. Summary of the 63 Membrane Proteins Identified on TMT-Labeling Proteomic Analysis.
| No. | IPI No. | Protein | AD Treg log2 | Normal Treg log2 | Fit_ratio (AD Treg/normal Treg) |
|---|---|---|---|---|---|
| 1 | IPI00225390.5 | Tax=10090 GS=Cox6b1 Cytochrome c oxidase subunit 6B1 | 13.2891 | 13.6851 | −0.5458 |
| 2 | IPI00626994.3 | Tax=10090 GS=Ipo5 Isoform 1 of Importin-5 | 8.6202 | 8.9498 | −0.4795 |
| 3 | IPI00116877.5 | Tax=10090 GS=Tnpo2 transportin-2 | 10.0495 | 10.2952 | −0.3955 |
| 4 | IPI00798527.1 | Tax=10090 GS=Tnpo1 Isoform 2 of Transportin-1 | 8.9195 | 9.1111 | −0.3415 |
| 5 | IPI00109727.1 | Tax=10090 GS=Thy1 Thy-1 membrane glycoprotein | 12.6675 | 12.7869 | −0.2693 |
| 6 | IPI00165694.1 | Tax=10090 GS=Tomm34 Isoform 1 of Mitochondrial import receptor subunit TOM34 | 10.8784 | 10.9784 | −0.2498 |
| 7 | IPI00230540.1 | Tax=10090 GS=Vdac1 Isoform Mt-VDAC1 of Voltage-dependent anion-selective channel protein 1 | 11.784 | 11.8828 | −0.2487 |
| 8 | IPI00938467.1 | Tax=10090 GS=Lck proto-oncogene tyrosine-protein kinase LCK isoform a | 11.7831 | 11.8807 | −0.2475 |
| 9 | IPI00229935.3 | Tax=10090 GS=Ptprb Receptor-type tyrosine-protein phosphatase beta | 11.5368 | 11.5056 | −0.1187 |
| 10 | IPI00120466.1 | Tax=10090 GS=Cd5 T-cell surface glycoprotein CD5 | 8.0688 | 7.973 | −0.0541 |
| 11 | IPI00121309.2 | Tax=10090 GS=Ndufs3 NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial | 8.9542 | 8.8549 | −0.0505 |
| 12 | IPI00130344.3 | Tax=10090 GS=Clic1 Chloride intracellular channel protein 1 | 11.6859 | 11.5823 | −0.0463 |
| 13 | IPI00125929.2 | Tax=10090 GS=Ndufa4 NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 4 | 13.1399 | 13.0186 | −0.0286 |
| 14 | IPI00118986.1 | Tax=10090 GS=Atp5o;LOC100047429 ATP synthase subunit O, mitochondrial | 11.6007 | 11.4692 | −0.0184 |
| 15 | IPI00311682.5 | Tax=10090 GS=Atp1a1 Sodium/potassium-transporting ATPase subunit alpha-1 | 11.5598 | 11.4171 | −0.0072 |
| 16 | IPI00930882.1 | Tax=10090 GS=Slc3a2 4F2 cell-surface antigen heavy chain isoform a | 9.1199 | 8.973 | −0.0029 |
| 17 | IPI00119138.1 | Tax=10090 GS=Uqcrc2 Cytochrome b-c1 complex subunit 2, mitochondrial | 10.6442 | 10.4834 | 0.011 |
| 18 | IPI00323881.2 | Tax=10090 GS=Kpnb1 Importin subunit beta-1 | 11.0277 | 10.8536 | 0.0243 |
| 19 | IPI00845575.2 | Tax=10090 GS=Atl3 atlastin-3 isoform 1 | 9.676 | 9.4848 | 0.0413 |
| 20 | IPI00404182.2 | Tax=10090 GS=Rhob Rho-related GTP-binding protein RhoB | 11.4687 | 11.2678 | 0.0511 |
| 21 | IPI00133240.1 | Tax=10090 GS=Uqcrfs1 Cytochrome b-c1 complex subunit Rieske, mitochondrial | 9.7465 | 9.5288 | 0.0679 |
| 22 | IPI00129792.1 | Tax=10090 GS=Kpna4 Importin subunit alpha-4 | 11.9586 | 11.7313 | 0.0775 |
| 23 | IPI00111885.2 | Tax=10090 GS=Uqcrc1 Cytochrome b-c1 complex subunit 1, mitochondrial | 11.6341 | 11.4064 | 0.0778 |
| 24 | IPI00133706.1 | Tax=10090 GS=Rab1b Ras-related protein Rab-1B | 12.626 | 12.3771 | 0.0991 |
| 25 | IPI00121288.5 | Tax=10090 GS=Ndufb10 NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 | 13.8617 | 13.6027 | 0.1091 |
| 26 | IPI00467062.2 | Tax=10090 GS=Tapbp Isoform Long of Tapasin | 11.0743 | 10.8151 | 0.1093 |
| 27 | IPI00116770.1 | Tax=10090 GS=Rab18 Ras-related protein Rab-18 | 9.8218 | 9.5597 | 0.1122 |
| 28 | IPI00130280.1 | Tax=10090 GS=Atp5a1 ATP synthase subunit alpha, mitochondrial | 13.2884 | 13.015 | 0.1235 |
| 29 | IPI00122548.4 | Tax=10090 GS=Vdac3 Voltage-dependent anion-selective channel protein 3 | 10.805 | 10.5292 | 0.126 |
| 30 | IPI00622837.1 | Tax=10090 GS=Gm6265 cytochrome c oxidase subunit 6C-like | 13.0492 | 0012.7682 | 0.1311 |
| 31 | IPI00989100.1 | Tax=10090 GS=Rab1 22 kDa protein | 12.8755 | 12.5816 | 0.144 |
| 32 | IPI00411115.1 | Tax=10090 GS=Rab8b Ras-related protein Rab-8B | 10.2913 | 9.9681 | 0.1733 |
| 33 | IPI00990016.1 | Tax=10090 GS=Rab14 protein variant | 9.3363 | 8.9936 | 0.1928 |
| 34 | IPI00127983.1 | Tax=10090 GS=Tmed2;Gm10698 Transmembrane emp24 domain-containing protein 2 | 9.7858 | 9.4415 | 0.1944 |
| 35 | IPI00119618.1 | Tax=10090 GS=Canx Calnexin | 14.1173 | 13.7627 | 0.2048 |
| 36 | IPI00987138.1 | Tax=10090 GS=LOC100047577 cytochrome b5 type B-like | 14.909 | 14.5461 | 0.213 |
| 37 | IPI00224518.2 | Tax=10090 GS=Rab5c Ras-related protein Rab-5C | 10.0028 | 9.6327 | 0.2203 |
| 38 | IPI00407954.2 | Tax=10090 GS=Rap1b Ras-related protein Rap-1b | 13.2249 | 12.8544 | 0.2207 |
| 39 | IPI00759999.1 | Tax=10090 GS=Prdx5 Isoform Cytoplasmic+peroxisomal of Peroxiredoxin-5, mitochondrial | 10.2007 | 9.812 | 0.2389 |
| 40 | IPI00110849.1 | Tax=10090 GS=H2-Aa H-2 class II histocompatibility antigen, A-K alpha chain | 8.9564 | 8.5546 | 0.2519 |
| 41 | IPI00131176.1 | Tax=10090 GS=mt-Co2 Cytochrome c oxidase subunit 2 | 15.1602 | 14.7242 | 0.2861 |
| 42 | IPI00408892.2 | Tax=10090 GS=Rab7 Ras-related protein Rab-7a | 9.3238 | 8.8811 | 0.2928 |
| 43 | IPI00555000.2 | Tax=10090 GS=Uqcrb Cytochrome b-c1 complex subunit 7 | 13.7627 | 13.3125 | 0.3003 |
| 44 | IPI00341282.2 | Tax=10090 GS=Atp5f1 ATP synthase subunit b, mitochondrial | 14.4398 | 13.9238 | 0.3661 |
| 45 | IPI00403079.4 | Tax=10090 GS=Cd47 Isoform 1 of Leukocyte surface antigen CD47 | 14.8106 | 14.2506 | 0.4102 |
| 46 | IPI00128856.2 | Tax=10090 GS=Ptprc Isoform 3 of Receptor-type tyrosine-protein phosphatase C | 12.3368 | 11.7648 | 0.4221 |
| 47 | IPI00114377.1 | Tax=10090 GS=Cox7a2 Cytochrome c oxidase subunit 7A2, mitochondrial | 15.4368 | 14.8582 | 0.4287 |
| 48 | IPI00466570.4 | Tax=10090 GS=Tmed10 Isoform 1 of Transmembrane emp24 domain-containing protein 10 | 14.3041 | 13.6975 | 0.4567 |
| 49 | IPI00230241.6 | Tax=10090 GS=Atp5e ATP synthase subunit epsilon, mitochondrial | 11.4311 | 10.8022 | 0.4791 |
| 50 | IPI00117978.1 | Tax=10090 GS=Cox4i1 Cytochrome c oxidase subunit 4 isoform 1, mitochondrial | 13.5709 | 12.9405 | 0.4805 |
| 51 | IPI00985828.1 | Tax=10090 GS=LOC100045864 h-2 class I histocompatibility antigen, D-P alpha chain-like isoform 4 | 13.513 | 12.847 | 0.5161 |
| 52 | IPI00316976.3 | Tax=10090 GS=Ptprcap Protein tyrosine phosphatase receptor type C-associated protein | 11.7475 | 11.0731 | 0.5245 |
| 53 | IPI00850057.1 | Tax=10090 GS=LOC100044874 h-2 class I histocompatibility antigen, K-W28 alpha chain-like isoform 1 | 13.431 | 12.7493 | 0.5319 |
| 54 | IPI00108844.1 | Tax=10090 GS=M6pr Cation-dependent mannose-6-phosphate receptor | 12.1324 | 11.4145 | 0.568 |
| 55 | IPI00953773.1 | Tax=10090 GS=Ssr4 translocon-associated protein subunit delta isoform 1 precursor | 12.3014 | 11.5799 | 0.5716 |
| 56 | IPI00857192.1 | Tax=10090 GS=Fis1 mitochondrial fission 1 protein isoform 2 | 12.1282 | 11.4031 | 0.5752 |
| 57 | IPI00120719.4 | Tax=10090 GS=Cox5a Cytochrome c oxidase subunit 5A, mitochondrial | 14.0156 | 13.2366 | 0.6292 |
| 58 | IPI00674006.3 | Tax=10090 GS=Tmem173 Isoform 1 of Transmembrane protein 173 | 10.2513 | 9.444 | 0.6575 |
| 59 | IPI00323897.3 | Tax=10090 GS=Rab11a Ras-related protein Rab-11A | 10.9208 | 10.048 | 0.723 |
| 60 | IPI00759904.1 | Tax=10090 GS=Cyb5r3 Isoform 2 of NADH-cytochrome b5 reductase 3 | 12.5679 | 11.641 | 0.7771 |
| 61 | IPI00128450.1 | Tax=10090 GS=Esyt1 Isoform 1 of Extended synaptotagmin-1 | 10.771 | 9.8392 | 0.782 |
| 62 | IPI00120346.3 | Tax=10090 GS=Rab27b Ras-related protein Rab-27B | 10.8696 | 9.876 | 0.8437 |
| 63 | IPI00172243.1 | Tax=10090 GS=H2-Ab1 H-2 class II histocompatibility antigen, A-K beta chain | 11.5565 | 9.9744 | 1.4322 |
TMT, Tandem Mass Tags; Treg, regulatory T; AD, atopic dermatitis.
Table 2. Summary of the 16 Plasma Membrane Proteins Identified on TMT-Labeling Proteomic Analysis.
| No. | IPI No. | Protein | AD Treg log2 | Normal Treg log2 | Fit_ratio (AD Treg/normal Treg) |
|---|---|---|---|---|---|
| 1 | IPI00109727.1 | Tax=10090 GS=Thy1 Thy-1 membrane glycoprotein | 12.6675 | 12.7869 | −0.2693 |
| 2 | IPI00938467.1 | Tax=10090 GS=Lck proto-oncogene tyrosine-protein kinase LCK isoform a | 11.7831 | 11.8807 | −0.2475 |
| 3 | IPI00229935.3 | Tax=10090 GS=Ptprb Receptor-type tyrosine-protein phosphatase beta | 11.5368 | 11.5056 | −0.1187 |
| 4 | IPI00120466.1 | Tax=10090 GS=Cd5 T-cell surface glycoprotein CD5 | 8.0688 | 7.973 | −0.0541 |
| 5 | IPI00130344.3 | Tax=10090 GS=Clic1 Chloride intracellular channel protein 1 | 11.6859 | 11.5823 | −0.0463 |
| 6 | IPI00311682.5 | Tax=10090 GS=Atp1a1 Sodium/potassium-transporting ATPase subunit alpha-1 | 11.5598 | 11.4171 | −0.0072 |
| 7 | IPI00930882.1 | Tax=10090 GS=Slc3a2 4F2 cell-surface antigen heavy chain isoform a | 9.1199 | 8.973 | −0.0029 |
| 8 | IPI00404182.2 | Tax=10090 GS=Rhob Rho-related GTP-binding protein RhoB | 11.4687 | 11.2678 | 0.0511 |
| 9 | IPI00127983.1 | Tax=10090 GS=Tmed2;Gm10698 Transmembrane emp24 domain-containing protein 2 | 9.7858 | 9.4415 | 0.1944 |
| 10 | IPI00110849.1 | Tax=10090 GS=H2-Aa H-2 class II histocompatibility antigen, A-K alpha chain | 8.9564 | 8.5546 | 0.2519 |
| 11* | IPI00403079.4 | Tax=10090 GS=Cd47 Isoform 1 of Leukocyte surface antigen CD47 | 14.8106 | 14.2506 | 0.4102 |
| 12* | IPI00128856.2 | Tax=10090 GS=Ptprc Isoform 3 of Receptor-type tyrosine-protein phosphatase C | 12.3368 | 11.7648 | 0.4221 |
| 13* | IPI00985828.1 | Tax=10090 GS=LOC100045864 h-2 class I histocompatibility antigen, D-P alpha chain-like isoform 4 | 13.513 | 12.847 | 0.5161 |
| 14* | IPI00316976.3 | Tax=10090 GS=Ptprcap Protein tyrosine phosphatase receptor type C-associated protein | 11.7475 | 11.0731 | 0.5245 |
| 15* | IPI00850057.1 | Tax=10090 GS=LOC100044874 h-2 class I histocompatibility antigen, K-W28 alpha chain-like isoform 1 | 13.431 | 12.7493 | 0.5319 |
| 16* | IPI00172243.1 | Tax=10090 GS=H2-Ab1 H-2 class II histocompatibility antigen, A-K beta chain | 11.5565 | 9.9744 | 1.4322 |
TMT, Tandem Mass Tags; Treg, regulatory T; AD, atopic dermatitis.
*Six plasma membrane proteins upregulated specifically in AD Treg cells.
Validation of CD47 expression in Treg cells
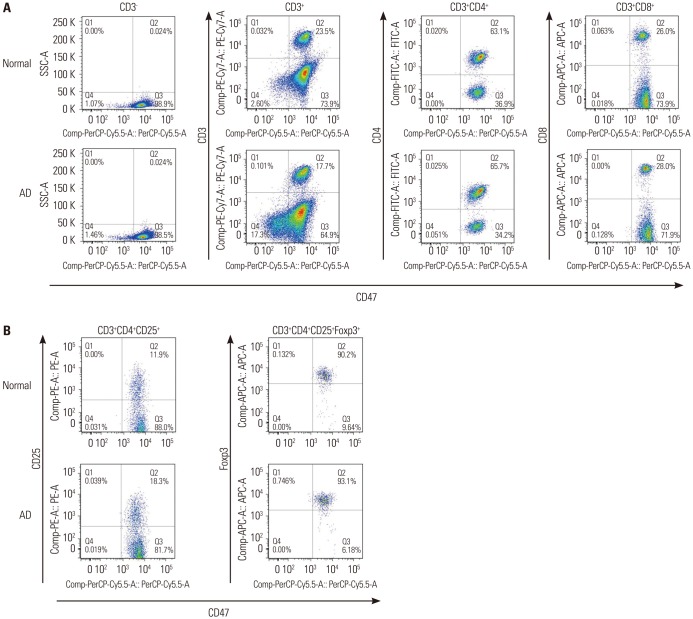
CD47 was expressed in all CD4+ T cells and CD8+ T cells (Fig. 2A). In addition, 99% of the CD4+CD25+Foxp3+ Treg cells also expressed CD47 (Fig. 2B), and there was no difference between AD mice and control mice. However, the expression level of CD47 was significantly higher in Treg cells from the AD mice than in those from the control mice (Fig. 3A). Moreover, its expression was much higher in Treg cells from the AD spleens than in those from the AD thymuses (Fig. 3B). Increased expression of CD47 in the Treg cells of AD mice relative to controls, especially in the spleen, were also confirmed on Western blot analysis (Fig. 3C and D).
Fig. 2. Expression of CD47 in various T cell subtypes. (A) FACS analysis of expression of CD47 in T cells, CD4+ T cells, and CD8+ T cells. All CD4+ T cells and CD8+ T cells expressed CD47. (B) FACS analysis of CD47 expression in CD3+CD4+CD25+ T cells and CD3+CD4+CD25+Foxp3+ T cells. Most CD4+CD25+Foxp3+ Treg cells expressed CD47. Treg, regulatory T; AD, atopic dermatitis.
Increased CD47 expression in Treg cells of AD patients
To provide support for the mice data results, the expression of CD47 was also analyzed in human PBMC. All CD4+CD25+Foxp3+ Treg cells expressed CD47 in both AD and healthy controls (Fig. 4A). However, the expression level of CD47 was higher in Treg cells in AD than in healthy controls (Fig. 4B). We also analyzed the correlation of the expression level of CD47 with AD severity. The expression of CD47 in Treg cells showed positive correlation with AD severity measured via EASI score (Pearson correlation r=0.84) (Fig. 4C). Higher levels of CD47 were shown to correlate significantly more severe AD.
DISCUSSION
Proteomics is the systematic analysis of protein profiles in a biological sample. Considering the discrepancies among profiles of genes, RNA transcripts, and proteins, proteomics is an ideal tool for the identification of new biomarkers, as the proteins are the main actors in the ongoing pathophysiology of a disease. The gel-free TMT approach uses isobaric labels to allow the genome-wide quantitation of the proteome. We employed this method to identify differentially expressed membrane proteins in induced Treg cells in AD. We initially induced AD-like skin lesions in NC/Nga mice using D. farinae ointment for 8 weeks and then utilized ELISA to measure the induced serum IgE to pinpoint the optimal time for proteomics analysis. After the application of D. farinae for 6 weeks, severe AD-like skin lesions appeared on the dorsal skin of the NC/Nga mice. Total IgE in the serum markedly increased and reached a plateau after 6 weeks of application of D. farinae. Based on these findings, mice treated for 6 weeks with D. farinae were used for proteomic analysis of membrane proteins in Treg cells. With the TMT-labeling method, we quantitated 510 proteins and ultimately identified six significantly upregulated plasma membrane proteins, including CD47, expressed in the Treg cells of AD mice.
CD47, an immunoglobulin-like protein, interacts functionally with integrins,19 thrombopondin-1,20 and signal regulatory protein α (SIRPα).18,19 This factor has been implicated in the regulation of neutrophil migration,21 axon extension,22 T cell costimulation,16 and phagocytosis.17,18 SIRPα/CD47 ligation inhibits phagocytosis by antigen-presenting cells, and a lack of CD47 expression results in the phagocytosis of red blood cells,18 T cells,23 and bone marrow cells.24 CD47 can also be transiently regulated by inflammatory stimuli in hematopoietic stem cells, and the presence of CD47 determines the probability of engulfment in vivo.23 Thus, CD47 is thought to function as an anti-phagocytosis signal. The thrombospondin-1/CD47 interaction inhibits interleukin (IL)-12 production by dendritic cells,25,26 IL-12 responsiveness,27 and Th1 differentiation.24,28 Furthermore, CD47 promotes the differentiation of Treg cells29 and regulates activated CD103+ Treg cell homeostasis;30 however, a deficiency of CD47 does not alter the inhibitory function of Treg cells.30 We showed in this study that CD47 expression was significantly higher in peripheral Treg cells in AD mice, particularly in spleen samples. Considering the function of CD47 as a signal of anti-phagocytosis and the increased population of Treg cells in AD patients, increased expression of CD47 in peripheral Treg cells in AD might expand the life span of these cells and result in the increased population of Treg cells in AD.
Treg cell populations primarily fall into two categories: naturally occurring Treg (nTreg) cells, which constitutively express Foxp3, and induced Treg (iTreg) cells, which are induced in the periphery by antigen stimulation or under tolerogenic conditions.10,31 These iTreg cells can be expanded and differentiated from nTreg cells or from CD4+CD25- effector T cells after stimulation by cytokines.10,32,33 Several experiments were conducted by a number of groups to determine the developmental and functional differences between nTreg and iTreg cells.34,35,36 Haribhai, et al.35 revealed a large number of transcripts that were differentially expressed between iTreg cells and nTreg cells. In other studies, Ikzf2 (Helios) and Nrp1 (neuropilin-1) expression were found to be more enhanced in nTreg cells than in iTreg cells, and these studies suggested that Helios may serve as a possible marker for nTreg cells.37,38 In our study, CD47 was more highly expressed in Treg cells from spleens than in those from thymuses in normal mice, and its expression was much higher in spleens from AD mice. However, there was no significant difference in CD47 expression in Treg cells from thymuses between normal and AD mice. Considering that most Treg cells in the thymus are nTreg cells and that those from the spleen are composed of both nTreg cells and iTreg cells, we suggest that CD47high Treg cells are likely iTreg cells and that the status of CD47 expression may be a marker that differentiates iTreg cells from nTreg cells.
In this study, we found that CD47 expression was upregulated in Treg cells in AD. Although CD47 is a ubiquitous membrane protein, increased expression of this factor in AD may play a role in the increased population of Treg cells and the consequent dominant Th2 immune response in AD. Further investigation will be necessary to determine the precise functional role of CD47 in Treg cells and to validate this factor as a differential marker between iTreg cells and nTreg cells; however, in this study, we clearly showed an increased expression of CD47 in peripheral Treg cells of both AD mice and AD patients, suggesting that CD47 is a valuable candidate molecule.
ACKNOWLEDGEMENTS
This study was supported by a grant from the Ministry of Health & Welfare, Republic of Korea (grant number: HI11C1671 and H13C0010). COP was supported by Soodang overseas training grant from the Yonsei University College of Medicine, Seoul, Korea.
KJY and YKN were supported by KBSI grant (T36413, P.I. KJY), Seoul, Korea.
Footnotes
The authors have no financial conflicts of interest.
References
- 1.Berke R, Singh A, Guralnick M. Atopic dermatitis: an overview. Am Fam Physician. 2012;86:35–42. [PubMed] [Google Scholar]
- 2.Furue M. Atopic dermatitis--immunological abnormality and its background. J Dermatol Sci. 1994;7:159–168. doi: 10.1016/0923-1811(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 3.Guttman-Yassky E, Nograles KE, Krueger JG. Contrasting pathogenesis of atopic dermatitis and psoriasis--part I: clinical and pathologic concepts. J Allergy Clin Immunol. 2011;127:1110–1118. doi: 10.1016/j.jaci.2011.01.053. [DOI] [PubMed] [Google Scholar]
- 4.Guttman-Yassky E, Nograles KE, Krueger JG. Contrasting pathogenesis of atopic dermatitis and psoriasis--part II: immune cell subsets and therapeutic concepts. J Allergy Clin Immunol. 2011;127:1420–1432. doi: 10.1016/j.jaci.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 5.Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity. 2013;38:414–423. doi: 10.1016/j.immuni.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Ochs HD, Ziegler SF, Torgerson TR. FOXP3 acts as a rheostat of the immune response. Immunol Rev. 2005;203:156–164. doi: 10.1111/j.0105-2896.2005.00231.x. [DOI] [PubMed] [Google Scholar]
- 7.Bacchetta R, Gambineri E, Roncarolo MG. Role of regulatory T cells and FOXP3 in human diseases. J Allergy Clin Immunol. 2007;120:227–235. doi: 10.1016/j.jaci.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 8.Ito Y, Adachi Y, Makino T, Higashiyama H, Fuchizawa T, Shimizu T, et al. Expansion of FOXP3-positive CD4+CD25+ T cells associated with disease activity in atopic dermatitis. Ann Allergy Asthma Immunol. 2009;103:160–165. doi: 10.1016/S1081-1206(10)60170-6. [DOI] [PubMed] [Google Scholar]
- 9.Gáspár K, Baráth S, Nagy G, Mócsai G, Gyimesi E, Szodoray P, et al. Regulatory T-cell subsets with acquired functional impairment: important indicators of disease severity in atopic dermatitis. Acta Derm Venereol. 2015;95:151–155. doi: 10.2340/00015555-1882. [DOI] [PubMed] [Google Scholar]
- 10.Samochocki Z, Alifier M, Bodera P, Jeziorkowska R, Rosiak E, Jurkiewicz B, et al. T-regulatory cells in severe atopic dermatitis: alterations related to cytokines and other lymphocyte subpopulations. Arch Dermatol Res. 2012;304:795–801. doi: 10.1007/s00403-012-1290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szegedi A, Baráth S, Nagy G, Szodoray P, Gál M, Sipka S, et al. Regulatory T cells in atopic dermatitis: epidermal dendritic cell clusters may contribute to their local expansion. Br J Dermatol. 2009;160:984–993. doi: 10.1111/j.1365-2133.2009.09035.x. [DOI] [PubMed] [Google Scholar]
- 12.Ou LS, Goleva E, Hall C, Leung DY. T regulatory cells in atopic dermatitis and subversion of their activity by superantigens. J Allergy Clin Immunol. 2004;113:756–763. doi: 10.1016/j.jaci.2004.01.772. [DOI] [PubMed] [Google Scholar]
- 13.Reefer AJ, Satinover SM, Solga MD, Lannigan JA, Nguyen JT, Wilson BB, et al. Analysis of CD25hiCD4+ "regulatory" T-cell subtypes in atopic dermatitis reveals a novel T(H)2-like population. J Allergy Clin Immunol. 2008;121:415–422. doi: 10.1016/j.jaci.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, et al. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal Chem. 2008;80:2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 15.Hanifin JM, Rajka G. Diagnostic features of atopic dermatitis. Acta Derm Venereol Suppl (Stockh) 1980;92:44–47. [Google Scholar]
- 16.Reinhold MI, Lindberg FP, Kersh GJ, Allen PM, Brown EJ. Costimulation of T cell activation by integrin-associated protein (CD47) is an adhesion-dependent, CD28-independent signaling pathway. J Exp Med. 1997;185:1–11. doi: 10.1084/jem.185.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han X, Sterling H, Chen Y, Saginario C, Brown EJ, Frazier WA, et al. CD47, a ligand for the macrophage fusion receptor, participates in macrophage multinucleation. J Biol Chem. 2000;275:37984–37992. doi: 10.1074/jbc.M002334200. [DOI] [PubMed] [Google Scholar]
- 18.Oldenborg PA, Zheleznyak A, Fang YF, Lagenaur CF, Gresham HD, Lindberg FP. Role of CD47 as a marker of self on red blood cells. Science. 2000;288:2051–2054. doi: 10.1126/science.288.5473.2051. [DOI] [PubMed] [Google Scholar]
- 19.Brown EJ, Frazier WA. Integrin-associated protein (CD47) and its ligands. Trends Cell Biol. 2001;11:130–135. doi: 10.1016/s0962-8924(00)01906-1. [DOI] [PubMed] [Google Scholar]
- 20.Gao AG, Lindberg FP, Finn MB, Blystone SD, Brown EJ, Frazier WA. Integrin-associated protein is a receptor for the C-terminal domain of thrombospondin. J Biol Chem. 1996;271:21–24. doi: 10.1074/jbc.271.1.21. [DOI] [PubMed] [Google Scholar]
- 21.Lindberg FP, Bullard DC, Caver TE, Gresham HD, Beaudet AL, Brown EJ. Decreased resistance to bacterial infection and granulocyte defects in IAP-deficient mice. Science. 1996;274:795–798. doi: 10.1126/science.274.5288.795. [DOI] [PubMed] [Google Scholar]
- 22.Miyashita M, Ohnishi H, Okazawa H, Tomonaga H, Hayashi A, Fujimoto TT, et al. Promotion of neurite and filopodium formation by CD47: roles of integrins, Rac, and Cdc42. Mol Biol Cell. 2004;15:3950–3963. doi: 10.1091/mbc.E04-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaiswal S, Jamieson CH, Pang WW, Park CY, Chao MP, Majeti R, et al. CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell. 2009;138:271–285. doi: 10.1016/j.cell.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouguermouh S, Van VQ, Martel J, Gautier P, Rubio M, Sarfati M. CD47 expression on T cell is a self-control negative regulator of type 1 immune response. J Immunol. 2008;180:8073–8082. doi: 10.4049/jimmunol.180.12.8073. [DOI] [PubMed] [Google Scholar]
- 25.Demeure CE, Tanaka H, Mateo V, Rubio M, Delespesse G, Sarfati M. CD47 engagement inhibits cytokine production and maturation of human dendritic cells. J Immunol. 2000;164:2193–2199. doi: 10.4049/jimmunol.164.4.2193. [DOI] [PubMed] [Google Scholar]
- 26.Doyen V, Rubio M, Braun D, Nakajima T, Abe J, Saito H, et al. Thrombospondin 1 is an autocrine negative regulator of human dendritic cell activation. J Exp Med. 2003;198:1277–1283. doi: 10.1084/jem.20030705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Latour S, Tanaka H, Demeure C, Mateo V, Rubio M, Brown EJ, et al. Bidirectional negative regulation of human T and dendritic cells by CD47 and its cognate receptor signal-regulator protein-alpha: down-regulation of IL-12 responsiveness and inhibition of dendritic cell activation. J Immunol. 2001;167:2547–2554. doi: 10.4049/jimmunol.167.5.2547. [DOI] [PubMed] [Google Scholar]
- 28.Avice MN, Rubio M, Sergerie M, Delespesse G, Sarfati M. CD47 ligation selectively inhibits the development of human naive T cells into Th1 effectors. J Immunol. 2000;165:4624–4631. doi: 10.4049/jimmunol.165.8.4624. [DOI] [PubMed] [Google Scholar]
- 29.Grimbert P, Bouguermouh S, Baba N, Nakajima T, Allakhverdi Z, Braun D, et al. Thrombospondin/CD47 interaction: a pathway to generate regulatory T cells from human CD4+ CD25- T cells in response to inflammation. J Immunol. 2006;177:3534–3541. doi: 10.4049/jimmunol.177.6.3534. [DOI] [PubMed] [Google Scholar]
- 30.Van VQ, Darwiche J, Raymond M, Lesage S, Bouguermouh S, Rubio M, et al. Cutting edge: CD47 controls the in vivo proliferation and homeostasis of peripheral CD4+ CD25+ Foxp3+ regulatory T cells that express CD103. J Immunol. 2008;181:5204–5208. doi: 10.4049/jimmunol.181.8.5204. [DOI] [PubMed] [Google Scholar]
- 31.Raimondi G, Turner MS, Thomson AW, Morel PA. Naturally occurring regulatory T cells: recent insights in health and disease. Crit Rev Immunol. 2007;27:61–95. doi: 10.1615/critrevimmunol.v27.i1.50. [DOI] [PubMed] [Google Scholar]
- 32.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huber S, Stahl FR, Schrader J, Lüth S, Presser K, Carambia A, et al. Activin a promotes the TGF-beta-induced conversion of CD4+ CD25- T cells into Foxp3+ induced regulatory T cells. J Immunol. 2009;182:4633–4640. doi: 10.4049/jimmunol.0803143. [DOI] [PubMed] [Google Scholar]
- 34.Bilate AM, Lafaille JJ. Induced CD4+Foxp3+ regulatory T cells in immune tolerance. Annu Rev Immunol. 2012;30:733–758. doi: 10.1146/annurev-immunol-020711-075043. [DOI] [PubMed] [Google Scholar]
- 35.Haribhai D, Lin W, Edwards B, Ziegelbauer J, Salzman NH, Carlson MR, et al. A central role for induced regulatory T cells in tolerance induction in experimental colitis. J Immunol. 2009;182:3461–3468. doi: 10.4049/jimmunol.0802535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh SH, Park CO, Wu WH, Kim JY, Jin S, Byamba D, et al. Corticotropin-releasing hormone downregulates IL-10 production by adaptive forkhead box protein 3-negative regulatory T cells in patients with atopic dermatitis. J Allergy Clin Immunol. 2012;129:151–159. 159.e1–159.e6. doi: 10.1016/j.jaci.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Sugimoto N, Oida T, Hirota K, Nakamura K, Nomura T, Uchiyama T, et al. Foxp3-dependent and -independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNA microarray analysis. Int Immunol. 2006;18:1197–1209. doi: 10.1093/intimm/dxl060. [DOI] [PubMed] [Google Scholar]
- 38.Hill JA, Feuerer M, Tash K, Haxhinasto S, Perez J, Melamed R, et al. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]