Abstract
A genome-wide SNP survey was used to identify chromosomal regions that showed linkage disequilibrium with respect to ascites susceptibility and ventricular hypertrophy in an F2 cross between previously described ascites-resistant and -susceptible lines. Variable number tandem repeats were used to obtain genotype data to further characterize these regions. A region on chromosome 9 (12 to 13 Mbp in 2011 assembly) shows association with ascites in the ascites lines and in several commercial broiler breeder lines with a significant sex effect. There are 2 candidate genes, AGTR1 (an angiotensin II type 1 receptor) and UTS2D (urotensin 2 domain containing), in this region that have been associated with hypertension and hypoxic response in mammals.
Keywords: ascites, hypertension, quantitative trait locus, commercial broiler
INTRODUCTION
Idiopathic pulmonary arterial hypertension is 1 of 5 types of pulmonary hypertension recognized by the American Lung Association. It is characterized in humans by right ventricular hypertrophy, elevated pulmonary arterial pressure, fatigue, chest pain, shortness of breath, and ultimately, right ventricular failure. A research group at the University of Arkansas has been investigating the genetic basis of ascites in the chicken, a disease analogous to idiopathic pulmonary arterial hypertension in humans. Ascites is a concern for the commercial poultry broiler industry. Selection for meat production through higher feed conversion and rapid production of body mass has increased demands on the cardiovascular system to support the growing tissues (Wideman et al., 2007; Lorenzoni et al., 2008). Many have speculated on how many genes are responsible for determining resistance or susceptibility to ascites. In fact, previous work has shown that ascites susceptibility is amenable to genetic selection (Anthony and Balog, 2003). Heritability estimates of 0.4 to 0.5 and rapid selection for pulmonary arterial hypertension resistance led Wideman and French (2000) to speculate that only a few major genes were involved. Analysis of blood oxygen saturation data from 15 generations led others to suggest an overdominant gene model for ascites (Navarro et al., 2006). Short-term divergent selection and F1 crosses led others to suggest there were 2 major dominant genes affecting ascites resistance (Druyan and Cahaner, 2007). Others have proposed a complex polygenic basis for the ascites syndrome (de Greef et al., 2001; Rabie et al., 2005; Hamal et al., 2010a,b). Accurate prediction of mode of inheritance of ascites has been facilitated through development of mechanisms for consistently inducing ascites through both invasive (Wideman and Kirby, 1995; Wideman et al., 1997, 2002, 2003) and noninvasive protocols (Wideman et al., 1998; Julian and Mirsalimi, 1992; Julian, 2000; Deeb et al., 2002; Balog et al., 2003; Pavlidis et al., 2007). Through this work, moderate to high heritabilities have been reported for ascites (Huchzermeyer et al., 1988; Peacock et al., 1989; Lubritz et al., 1995; Shlosberg et al., 1998; Wideman and French, 1999, 2000; Moghadam et al., 2001; Deeb et al., 2002; Pakdel et al., 2002; Balog et al., 2003; Ledur et al., 2006; Druyan et al., 2007a; Druyan and Cahaner, 2007; Pavlidis et al., 2007). These selection protocols have been applied to both research and commercial populations (Shlosberg et al., 1996; Wideman and French, 1999, 2000; Balog et al., 2003; Druyan et al., 2007a,b; Druyan and Cahaner, 2007; Pavlidis et al., 2007; Druyan et al., 2008, 2009). For a comprehensive review of the pathology, immunology, pharmacology, and genetics of ascites, see Wideman et al. (2013).
Divergently selected lines have been generated from a former pedigree elite broiler line (REL) using sib selection based on data generated through hypobaric challenge (Pavlidis et al., 2007). These lines were used to identify chromosomal regions linked to ascites susceptibility. One of these regions shows significant association with ascites susceptibility in the ascites research lines as well as in commercial broiler breeder lines.
MATERIALS AND METHODS
Genome Data
All genome positions are presented relative to the November 2011 assembly of the Gallus gallus genome GenBank accession ID: GCA_000002315.2.
Bird Stocks
All animal procedures were preapproved by the University of Arkansas Institutional Animal Care and Use Committee, approval number 09032. Birds used for this study represented 3 research lines and 3 commercial elite lines. The research lines consist of the ascites-resistant (RES) and -susceptible (SUS) lines divergently selected for ascites susceptibility using hypobaric chamber challenge (Pavlidis et al., 2007). Commercial lines W and Y are male elite lines selected primarily for growth, yield, and feed conversion, whereas line Z is a female elite line selected primarily for reproduction and growth traits.
F2 Population
An F1 reciprocal cross was performed using 3 RES sires individually inseminated onto 4 SUS dams and 2 SUS sires onto 4 RES dams. The F2 cross was also reciprocal using 7 RES×SUS sires on 18 SUS×RES dams and 6 SUS×RES sires on 16 RES×SUS dams to generate 1,756 chicks. A total of 820 of these chicks were challenged in the hypobaric chamber and scored for ascites susceptibility, and resistance as above. Samples submitted for genome-wide SNP analyses comprised all 34 F2 families ranging from 1 to 11 chicks per family with an average of 5.1 chicks per F2 family analyzed for whole genome SNP panel.
Hypobaric Chamber Trials
The hypobaric chamber at the University of Arkansas poultry research facility has been described previously (Pavlidis et al., 2007). The SUS, REL, RES, and F2 chicks were hatched at the University of Arkansas hatchery, whereas 1-d-old commercial line birds were obtained directly from a commercial hatchery. From the hatchery, chicks were wing banded and vaccinated for Marek's virus. Each trial was limited to a single line. Chicks were randomly assigned at 12 to 13 each to identical custom stainless steel battery cages (2 foot square and 1 foot high) within the hypobaric chamber. Each battery cage was fitted with trough feeders and nipple waterers. Birds were warm room brooded and temperature was decreased weekly (Pavlidis et al., 2007). The chamber was continuously monitored for simulated altitude, ventilation, and temperature. Ventilation was set to maintain a constant airflow at the rate of 17 m3/min and a simulated altitude of 9,500 ft above sea level (500 mm of Hg). The photoperiod was 24 h of light. Access to the chamber was through an airlock.
Mortalities were recorded every 24 h for the first 3 wk and every 12 h in the last 3 wk. Morbid birds were humanely euthanized. All mortalities were necropsied and examined for cause of death. Ascites susceptibility during the trials was based on death in the hypobaric chamber where there was excess abdominal fluid with specific heart characteristics: flaccidity, round shape, increased size, and right ventricle to total ventricle (RV/TV) ratio. Survivors of the 6-wk hypobaric trials were weighed, euthanized by cervical dislocation, and scored as above. Birds without the susceptible heart characteristics were designated resistant.
DNA
Blood samples (10 µL) were collected from all birds at 3 d of age via wing vein lancet puncture. The DNA was isolated from blood samples using a rapid method (Bailes et al., 2007).
Microsatellite Primers
The PCR primers were designed to amplify 2 VNTR (variable number tandem repeat) from the region on chromosome 9. The PHS009 is a GA repeat at 11,811,967 bp amplified with forward 5′-GGGGCTATCACATCTACTTT-3′ and reverse 5′-AGTTCAGAACCAGGAGAAGA-3′. The PHS010 is a AC repeat at 11,929,898 bp amplified with forward 5′-ACAGAATTAGGGTGGGTTTTT-3′ and reverse 5′-GCGGTGTGCCTGCTTTCT-3′. Primers were synthesized (Eurofins MWG Operon Inc., Huntsville, AL) with PHS009 forward primer labeled with Cy3 and PHS010 forward primer labeled with Fam.
PCR
Polymerase chain reaction was performed in 96-well plates using either an MJ Research PTC-100 (BioRad Laboratories, Hercules, CA) or an Eppendorf Mastercycler Gradient (Eppendorf North America, Hauppauge, NY). Reactions (20 µL) contained 1× Taq Buffer (50 mM Tris-Cl pH 8.3, 1 mM MgCl2, 30 µg/mL of BSA), 0.2 mM dNTP, 0.4 µM each forward and reverse primers, 2.5 units of Taq polymerase, and 2 μL of DNA (approximately 50–100 ng). The PCR conditions were an initial denaturation at 90°C for 30 s, followed by 44 cycles of 90°C for 15 s, 52°C for 25 s, and 72°C for 60 s, followed by a final 72°C for 3 min.
Gel Electrophoresis
The PCR products were resolved in 6% denaturing polyacrylamide gels (38:2 acrylamide:MBA, 50% urea) in 1×TEB (100 mM Tris, 10 mM boric acid, 2 mM EDTA, pH 9.2). The PCR product (3 µL) was mixed with 3 of µL loading buffer (95% formamide, 1× TEB, 0.02% bromophenol blue). Samples were heat denatured at 90°C for 5 min, quick chilled on ice, then 2 μL was loaded per lane. The CXR ladder (Promega Corp., Madison, WI) was used as a size marker.
Gel Imaging
Gels were scanned with a model 9600 Typhoon imager (GE Healthcare Life Sciences, Piscataway, NJ) and images analyzed using ImageQuant software. Alleles were identified by size relative to the markers and given letter designations (A through F) for each of the 2 loci, in the order in which they were detected in the analyses (based on chronological order rather than size order).
Statistical Methods
Allele and genotype distributions were tabulated and analyzed using Microsoft Excel (Microsoft Corp., Redmond, WA). Each locus in each line was analyzed for deviation from Hardy-Weinberg (HW) based on computing expected homozygote and heterozygote counts, which were then compared with the observed using the CHITEST function in Excel. Allele frequencies were computed for the entire line and used to compute expected allele and genotype frequencies for the line and for the resistant and susceptible subpopulations based on the hypobaric challenge. Expected genotype counts were computed using the HW equation. Expected haplotype counts for the subpopulations were computed based on the haplotype frequency in the entire population. Genotype data for those birds that died but where the necropsy was inconclusive for ascites were included in the overall allele and genotype frequencies but were not included in the resistant or susceptible subpopulations. For each allele, genotype, or haplotype, P-values were determined using the Excel CHITEST function comparing the observed counts for the resistant and susceptible subpopulations to the expected counts. The P-values were only calculated for alleles or genotypes where the frequency was greater than 10%. Calculated P-values were then multiplied by the number of computed CHITEST performed for either that allele or genotype analysis to generate a Bonnferoni adjusted P-value (e.g., if for a given locus there were 3 alleles present at greater than 10% then all computed P-values were multiplied by 3 to compute the adjusted P-value). Significant deviation from expected was assumed for adjusted P-value <0.05.
RESULTS
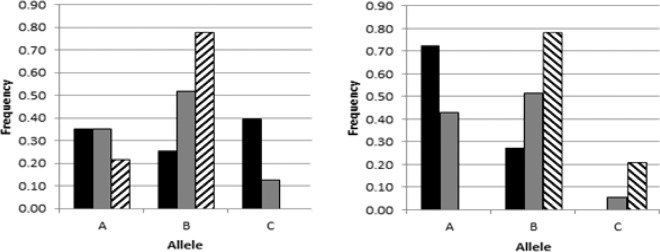
A genome-wide SNP panel was conducted in 2006 (Muir et al., 2008). As part of this project, DNA from the University of Arkansas were included representing 183 F2 birds from double reciprocal crosses of the RES and SUS lines (see Materials and Methods). The samples included 84 resistant and 89 highly susceptible F2 progeny. For the F2 samples, 970 SNP were uninformative, whereas 1,763 were polymorphic. The SNP frequencies were analyzed with respect to low vs. high RV/TV (hypertrophy) and resistance vs. susceptibility to ascites. The data were analyzed by chi-squared to detect deviation of observed from expected for both ascites and RV/TV. From this analysis several regions were identified as potentially linked to hypertrophy and ascites including several on chromosome 9. The region around 12 to 13 Mbp showed a strong association for both hypertrophy and ascites but almost exclusively for females (Figure 1). The PCR assays were developed for 2 VNTR (designated PHS009 and PHS010) from that region to further genotype additional DNA samples. The RES, SUS, and REL research lines were genotyped for both loci in generation 14 to determine how selection had affected allele and genotype frequencies. The PHS009 primers amplified 3 major alleles (designated A, B, and C) in the SUS, REL, and RES lines. Analysis of all 3 lines for this locus determined that SUS and REL showed expected heterozygosity and homozygosity counts (SUS, P = 0.72; REL, P = 0.46). For RES we detected deviation (P = 0.047) from HW deriving from fewer homozygotes (166 observed vs. 181 expected) and an overabundance of heterozygotes (110 observed vs. 95 expected). In the SUS line we detected an additional allele (designated F) that was a very minor allele at a frequency of 0.01. In the RES line we only observed the A and B alleles. The PHS010 primers also amplified 3 alleles (A, B, C) in the REL line. In the SUS line we only observed the A and B alleles, whereas in the RES line we only observed the B and C allele. The SUS, REL, and RES lines all conformed to HW for homozygosity and heterozgosity counts for PHS010 (SUS, P = 0.39; REL, P = 0.46; RES, P = 0.77). Comparison of the frequencies of the 3 major alleles for PHS009 and PHS010 in these 3 research lines revealed that 14 generations of divergent selection have resulted in markedly different allele frequencies (Figure 2), consistent with a QTL for ascites being present in this region. For PHS009, selection for resistance has resulted in a loss of the C allele with concomitant increase for the frequency of the B allele. Conversely, the B allele frequency has decreased in the SUS line with an increase in the C allele frequency. Likewise, for PHS010 the B allele frequency has increased in the RES and decreased in the SUS, whereas the A allele was not detected in the RES and increased in frequency in the SUS. Therefore, the PHS009 B allele appears to be associated with resistance and the C allele with susceptibility, whereas for PHS010 the B allele is apparently associated with resistance and the A allele with susceptibility. Because, in hypobaric challenges, the SUS line has an ascites incidence of approximately 95% whereas the RES line has an incidence of approximately 5% (Anthony and Balog, 2003), it is difficult to test for association within these lines. The REL line has an approximate ascites incidence of 50%, so the REL line is amenable for genetic association studies for ascites. Note that the original SNP mapping was in a reciprocal F2 cross between the RES and SUS. No significant allele or genotype associations were found for PHS009 (Table 1) in the REL line. Conversely, for PHS010 (Table 2) we detected a statistically significant deviation from expected for the BC genotype, where this genotype was 13% in the resistant phenotype and only 6% in the susceptible phenotype (adjusted P = 0.003). This deviation was most pronounced in females, where the BC genotype was 17% in resistant and 2% in susceptible (adjusted P = 0.001).
Figure 1.

Sex-specific SNP association analysis of chicken chromosome 9 for ascites and cardiac hypertrophy. Samples for an F1 and F2 cross of the ascites-resistant (RES) and ascites-susceptible (SUS) lines were genotyped for 59 informative SNP on chromosome 9. Chi-square P-values for observed vs. expected were plotted as 1 − logP for ascites phenotype (gray lines) or cardiac hypertrophy (black lines) for males (dashed lines) or females (solid lines).
Figure 2.
Shifts in allele frequencies resulting from selection, comparing the ascites-resistant (RES) and ascites-susceptible (SUS) lines to the former pedigree elite broiler (REL) line. Allele frequencies for PHS009 (left panel) or PHS010 (right panel) are plotted for the SUS (black), REL (gray), and RES (cross-hatch) lines at generation 14.
Table 1.
Allele and genotype data for PHS009 for the ascites experimental lines1
| SUS | RES | REL | REL male | REL female | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size (bp) | Item | All freq | All freq | All freq | R freq | S freq | Adj Pval | R freq | S freq | Adj Pval | R freq | S freq | Adj Pval |
| Allele | |||||||||||||
| A | 146 | 0.35 | 0.22 | 0.35 | 0.31 | 0.41 | 0.268 | 0.33 | 0.42 | 0.466 | 0.30 | 0.38 | 0.791 |
| B | 136 | 0.25 | 0.78 | 0.52 | 0.56 | 0.46 | 0.345 | 0.56 | 0.46 | 0.786 | 0.57 | 0.45 | 0.791 |
| C | 144 | 0.39 | 0.00 | 0.13 | 0.12 | 0.14 | 1.000 | 0.11 | 0.12 | 1.000 | 0.13 | 0.16 | 0.983 |
| F | 150 | 0.00 | 0.00 | 0.00 | 0.01 | 0.00 | 0.01 | 0.00 | 0.01 | 0.00 | |||
| Count | 506 | 552 | 542 | 280 | 212 | 162 | 126 | 118 | 86 | ||||
| Genotype | |||||||||||||
| AA | 0.10 | 0.02 | 0.10 | 0.09 | 0.14 | 0.855 | 0.12 | 0.16 | 1.000 | 0.03 | 0.12 | 0.254 | |
| BB | 0.06 | 0.58 | 0.26 | 0.31 | 0.18 | 0.233 | 0.31 | 0.19 | 0.825 | 0.31 | 0.16 | 0.733 | |
| CC | 0.17 | 0.00 | 0.03 | 0.04 | 0.02 | 0.02 | 0.02 | 0.05 | 0.02 | ||||
| AB | 0.22 | 0.40 | 0.41 | 0.39 | 0.42 | 1.000 | 0.36 | 0.43 | 1.000 | 0.44 | 0.42 | 1.000 | |
| AC | 0.28 | 0.00 | 0.08 | 0.05 | 0.10 | 0.536 | 0.04 | 0.10 | 0.597 | 0.07 | 0.12 | 1.000 | |
| AF | 0.00 | 0.00 | 0.01 | 0.01 | 0.00 | 0.01 | 0.00 | 0.02 | 0.00 | ||||
| BC | 0.18 | 0.00 | 0.12 | 0.11 | 0.13 | 1.000 | 0.14 | 0.11 | 1.000 | 0.08 | 0.16 | 1.000 | |
| Count | 253 | 276 | 271 | 140 | 106 | 81 | 63 | 59 | 43 | ||||
1The ascites susceptible (SUS), former pedigree elite broiler (REL), and ascites resistant (RES) lines were genotyped for PHS009. Allele and genotype frequencies (freq) were determined for the entire line (all) or for the ascites resistant (R) or susceptible (S) subpopulations based on a hypobaric challenge. The total number of alleles or genotypes is indicated below the frequencies (count). The REL samples were also analyzed according to sex. The P-values for a simple Bonnferoni correction (see Materials and Methods) of chi-squared test of observed vs. expected (Adj Pval) are presented for those alleles or genotypes where the frequency was greater than or equal to 0.10.
Table 2.
Allele and genotype data for PHS010 for the ascites experimental lines1
| SUS | RES | REL | REL male | REL female | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Item | Size (bp) | All freq | All freq | All freq | R freq | S freq | Adj Pval | R freq | S freq | Adj Pval | R freq | S freq | Adj Pval |
| Allele | |||||||||||||
| A | 161 | 0.72 | 0.00 | 0.43 | 0.40 | 0.46 | 1.000 | 0.46 | 0.46 | 1.000 | 0.37 | 0.45 | 0.783 |
| B | 151 | 0.28 | 0.78 | 0.51 | 0.52 | 0.50 | 1.000 | 0.50 | 0.48 | 1.000 | 0.55 | 0.52 | 1.000 |
| C | 171 | 0.00 | 0.21 | 0.06 | 0.07 | 0.04 | 0.549 | 0.04 | 0.06 | 1.000 | 0.08 | 0.02 | 0.206 |
| Count | 504 | 556 | 550 | 284 | 214 | 102 | 126 | 120 | 88 | ||||
| Genotype | |||||||||||||
| AA | 0.51 | 0.00 | 0.19 | 0.17 | 0.21 | 1.000 | 0.21 | 0.22 | 1.000 | 0.12 | 0.20 | 0.810 | |
| BB | 0.06 | 0.59 | 0.24 | 0.23 | 0.24 | 1.000 | 0.24 | 0.22 | 1.000 | 0.22 | 0.27 | 1.000 | |
| CC | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||
| AB | 0.43 | 0.00 | 0.46 | 0.46 | 0.46 | 1.000 | 0.43 | 0.44 | 1.000 | 0.50 | 0.48 | 1.000 | |
| AC | 0.00 | 0.00 | 0.02 | 0.01 | 0.03 | 0.02 | 0.03 | 0.00 | 0.02 | ||||
| BC | 0.00 | 0.40 | 0.09 | 0.13 | 0.06 | 0.003 | 0.10 | 0.08 | 0.389 | 0.17 | 0.02 | 0.001 | |
| Count | 252 | 278 | 275 | 142 | 107 | 82 | 63 | 60 | 44 | ||||
1The ascites susceptible (SUS), former pedigree elite broiler (REL), and ascites resistant (RES) lines were genotyped for PHS010. Allele and genotype frequencies (freq) were determined for the entire line (all) or for the ascites resistant (R) or susceptible (S) subpopulations based on a hypobaric challenge. The total number of alleles or genotypes is indicated below the frequencies (count). The REL samples were also analyzed according to sex. The P-values for a simple Bonnferoni correction (see Materials and Methods) of chi-squared test of observed vs. expected (Adj Pval) are presented for those alleles or genotypes where the frequency was greater than or equal to 0.10.
To determine linkage of specific PHS009 and PHS010 alleles, haplotypes were assigned where either or both of the loci were homozygous. For example, when a bird was homozygous BB for PHS009 and heterozygous BC for PHS010 then we could impute a BB haplotype and a BC haplotype. Totaling observed haplotypes could therefore determine which alleles for each locus were in linkage. For the REL line the BB haplotype (PHS009-PHS010 was the most frequent (48%) with the PHS009 B allele much less frequently associated with any other PHS010 allele (Table 3). Similarly, the A alleles for each locus seem to be in strong linkage. However, none of the major haplotypes showed significant association with ascites phenotype.
Table 3.
Haplotype (Hap) data for PHS009 and PHS010 for the former pedigree elite broiler (REL) and the commercial lines1
| REL | Line Y | Line Z | Line W | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hap | All freq | R freq | S freq | Adj Pval | Hap | All freq | R freq | S freq | Adj Pval | Hap | All freq | R freq | S freq | Adj Pval | Hap | All freq | R freq | S freq | Adj Pval |
| BB | 0.48 | 0.52 | 0.41 | 0.572 | AA | 0.39 | 0.35 | 0.44 | 1.000 | AA | 0.72 | 0.67 | 0.85 | 0.396 | AA | 0.57 | 0.61 | 0.55 | 1.000 |
| AA | 0.25 | 0.21 | 0.31 | 0.316 | AD | 0.20 | 0.15 | 0.27 | 0.343 | FB | 0.16 | 0.21 | 0.04 | 0.008 | AD | 0.20 | 0.16 | 0.24 | 0.938 |
| CA | 0.11 | 0.09 | 0.12 | 1.000 | BB | 0.18 | 0.26 | 0.09 | 0.014 | AB | 0.07 | 0.07 | 0.07 | 1.000 | CD | 0.09 | 0.08 | 0.10 | 1.000 |
| AB | 0.05 | 0.02 | 0.09 | BA | 0.12 | 0.11 | 0.13 | 1.000 | AE | 0.03 | 0.03 | 0.03 | CE | 0.08 | 0.11 | 0.06 | 1.000 | ||
| BA | 0.04 | 0.06 | 0.03 | CA | 0.10 | 0.12 | 0.09 | 1.000 | FA | 0.01 | 0.01 | 0.01 | CA | 0.03 | 0.03 | 0.04 | |||
| BC | 0.04 | 0.06 | 0.01 | AE | 0.01 | 0.01 | 0.02 | ||||||||||||
| CB | 0.01 | 0.02 | 0.01 | BA | 0.00 | 0.01 | 0.00 | ||||||||||||
| AC | 0.01 | 0.01 | 0.02 | ||||||||||||||||
| CC | 0.01 | 0.01 | 0.00 | ||||||||||||||||
| Cnt | 272 | 140 | 106 | 212 | 118 | 94 | 252 | 178 | 72 | 214 | 104 | 110 | |||||||
1Line designations are as described in the text. Haplotypes were imputed as described in the text, and frequencies determined for the entire line (All) or for the ascites resistant (R) or susceptible (S) subpopulations based on a hypobaric challenge. The total number of haplotypes is indicated below the frequencies (Cnt). The P-values for a simple Bonnferoni correction (see Materials and Methods) of chi-squared test of observed versus expected (Adj Pval) are presented for those haplotypes where the frequency was greater than or equal to 0.10.
The DNA for 192 birds from each of 3 modern commercial elite lines (designated W, Y, and Z) previously phenotyped in the hypobaric chamber were genotyped for PHS009 (Table 4) and PHS010 (Table 5). Line Y was the original source for development of the ascites experimental lines beginning in 1996. Line Y also maintained the same 4 alleles (A, B, C, and F) for PHS009 that were identified in the SUS, REL, and RES lines. In fact, the allele frequencies for PHS009 were similar to observed allele frequencies in the REL line, consistent with a common ancestry 15 yr ago. Comparison of heterozygote/homozygote counts to HW estimates showed no statistical deviation (P = 0.28). For PHS010, 2 major alleles (A and B) were identified in the SUS, REL, and RES lines and commercial lines; however, 2 additional alleles (D and F) were not observed in the SUS, REL, or RES experimental lines. Heterozygosity/homozygosity values for PHS010 were also consistent with HW estimates (P = 0.72).
Table 4.
Allele and genotype data for PHS009 for the commercial lines1
| Line Y | Line Y male | Line Y female | Line Z | Line Z male | Line Z female | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Item | Size (bp) | All freq | R freq | S freq | Adj Pval | R freq | S freq | Adj Pval | R freq | S freq | Adj Pval | All freq | R freq | S freq | Adj Pval | R freq | S freq | Adj Pval | R freq | S freq | Adj Pval |
| Allele | |||||||||||||||||||||
| A | 146 | 0.51 | 0.47 | 0.58 | 0.283 | 0.46 | 0.61 | 0.325 | 0.48 | 0.55 | 1.000 | 0.71 | 0.70 | 0.74 | 1.000 | 0.67 | 0.68 | 0.926 | 0.76 | 0.77 | 0.888 |
| B | 136 | 0.35 | 0.39 | 0.29 | 0.172 | 0.41 | 0.24 | 0.090 | 0.38 | 0.34 | 1.000 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||
| C | 144 | 0.14 | 0.14 | 0.13 | 1.000 | 0.13 | 0.16 | 1.000 | 0.14 | 0.11 | 1.000 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||
| F | 150 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.29 | 0.30 | 0.26 | 0.979 | 0.33 | 0.32 | 0.496 | 0.24 | 0.23 | 0.457 | |||
| Count | 376 | 226 | 150 | 122 | 76 | 104 | 74 | 382 | 252 | 130 | 156 | 56 | 90 | 70 | |||||||
| Genotype | |||||||||||||||||||||
| AA | 0.28 | 0.22 | 0.37 | 0.211 | 0.23 | 0.34 | 1.000 | 0.22 | 0.41 | 0.353 | 0.52 | 0.53 | 0.49 | 1.000 | 0.49 | 0.39 | 1.000 | 0.62 | 0.54 | 0.781 | |
| BB | 0.14 | 0.18 | 0.09 | 0.341 | 0.20 | 0.05 | 0.201 | 0.16 | 0.14 | 1.000 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||
| CC | 0.02 | 0.02 | 0.01 | 0.02 | 0.03 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||||||
| FF | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.09 | 0.13 | 0.02 | 0.017 | 0.15 | 0.04 | 0.058 | 0.11 | 0.00 | 0.204 | ||||
| AB | 0.32 | 0.34 | 0.28 | 1.000 | 0.33 | 0.32 | 1.000 | 0.35 | 0.24 | 1.000 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||
| AC | 0.14 | 0.15 | 0.13 | 1.000 | 0.13 | 0.21 | 1.000 | 0.18 | 0.05 | 0.593 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||
| AF | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.39 | 0.33 | 0.49 | 0.269 | 0.36 | 0.57 | 0.395 | 0.27 | 0.46 | 0.353 | ||||
| BC | 0.10 | 0.09 | 0.11 | 1.000 | 0.10 | 0.05 | 1.000 | 0.08 | 0.16 | 0.845 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||
| Count | 187 | 112 | 75 | 61 | 38 | 51 | 37 | 191 | 126 | 65 | 78 | 28 | 45 | 35 | |||||||
| Line W | Line W male | Line W female | |||||||||||||||||||
| All freq | R freq | S freq | Adj Pval | R freq | S freq | Adj Pval | R freq | S freq | Adj Pval | ||||||||||||
| Allele | |||||||||||||||||||||
| A | 0.66 | 0.65 | 0.67 | 1.000 | 0.65 | 0.73 | 0.898 | 0.65 | 0.63 | 1.000 | |||||||||||
| B | 0.02 | 0.02 | 0.02 | 0.02 | 0.01 | 0.02 | 0.03 | ||||||||||||||
| C | 0.32 | 0.33 | 0.31 | 1.000 | 0.33 | 0.26 | 0.656 | 0.33 | 0.34 | 1.000 | |||||||||||
| F | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||||||||||
| Count | 382 | 192 | 190 | 106 | 78 | 86 | 112 | ||||||||||||||
| Genotype | |||||||||||||||||||||
| AA | 0.40 | 0.39 | 0.42 | 1.000 | 0.40 | 0.49 | 1.000 | 0.37 | 0.38 | 0.988 | |||||||||||
| BB | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||||||||||
| CC | 0.07 | 0.08 | 0.06 | 0.624 | 0.09 | 0.03 | 0.417 | 0.07 | 0.09 | 1.000 | |||||||||||
| FF | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||||||||||
| AB | 0.04 | 0.04 | 0.03 | 0.04 | 0.03 | 0.05 | 0.04 | ||||||||||||||
| AC | 0.48 | 0.49 | 0.47 | 0.545 | 0.47 | 0.46 | 1.000 | 0.51 | 0.48 | 0.715 | |||||||||||
| AF | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||||||||||
| BC | 0.01 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.02 | ||||||||||||||
| Count | 191 | 96 | 95 | 53 | 39 | 43 | 56 | ||||||||||||||
1Line designations are as described in the text. Allele and genotype frequencies (freq) were determined for the entire line (all) or for the ascites resistant (R) or susceptible (S) subpopulations based on a hypobaric challenge. The total number of alleles or genotypes is indicated below the frequencies (count). The samples for each line were also analyzed according to sex. The P-values for a simple Bonnferoni correction (see Materials and Methods) of chi-squared test of observed vs. expected (Adj Pval) are presented for those alleles or genotypes where the frequency was greater than or equal to 0.10.
Table 5.
Allele and genotype data for PHS010 for the commercial lines1
| Line Y | Line Y male | Line Y female | Line Z | Line Z male | Line Z female | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Item | Size (bp) | All freq | R freq | S freq | Adj Pval | R freq | S freq | Adj Pval | R freq | S freq | Adj Pval | All freq | R freq | S freq | Adj Pval | R freq | S freq | Adj Pval | R freq | S freq | Adj Pval |
| Allele | |||||||||||||||||||||
| A | 161 | 0.51 | 0.49 | 0.55 | 1.000 | 0.48 | 0.57 | 1.000 | 0.51 | 0.53 | 1.000 | 0.65 | 0.63 | 0.69 | 0.963 | 0.62 | 0.59 | 0.915 | 0.68 | 0.76 | 0.514 |
| B | 151 | 0.27 | 0.32 | 0.18 | 0.025 | 0.32 | 0.14 | 0.062 | 0.33 | 0.22 | 0.430 | 0.32 | 0.34 | 0.29 | 0.852 | 0.36 | 0.39 | 0.520 | 0.29 | 0.23 | 0.252 |
| D | 159 | 0.21 | 0.18 | 0.27 | 0.195 | 0.20 | 0.28 | 0.646 | 0.15 | 0.26 | 0.371 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||
| E | 157 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.00 | 0.02 | 0.03 | 0.02 | 0.02 | 0.02 | 0.03 | 0.01 | ||||||
| Count | 376 | 226 | 150 | 120 | 76 | 104 | 74 | 382 | 252 | 130 | 134 | 56 | 90 | 70 | |||||||
| Genotype | |||||||||||||||||||||
| AA | 0.24 | 0.20 | 0.31 | 0.900 | 0.20 | 0.34 | 0.827 | 0.21 | 0.27 | 1.000 | 0.43 | 0.44 | 0.42 | 1.000 | 0.40 | 0.25 | 0.427 | 0.51 | 0.51 | 0.688 | |
| BB | 0.07 | 0.11 | 0.03 | 0.262 | 0.11 | 0.00 | 0.184 | 0.10 | 0.05 | 1.000 | 0.12 | 0.16 | 0.03 | 0.027 | 0.17 | 0.07 | 0.237 | 0.16 | 0.00 | 0.087 | |
| DD | 0.05 | 0.02 | 0.09 | 0.100 | 0.02 | 0.08 | 0.02 | 0.11 | 0.272 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||||
| EE | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||||||
| AB | 0.29 | 0.34 | 0.23 | 0.804 | 0.30 | 0.18 | 1.000 | 0.38 | 0.27 | 0.741 | 0.41 | 0.35 | 0.52 | 0.226 | 0.38 | 0.64 | 0.188 | 0.27 | 0.46 | 0.301 | |
| AD | 0.23 | 0.22 | 0.25 | 1.000 | 0.25 | 0.26 | 1.000 | 0.19 | 0.24 | 1.000 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||
| AE | 0.01 | 0.02 | 0.00 | 0.02 | 0.00 | 0.02 | 0.00 | 0.04 | 0.04 | 0.03 | 0.03 | 0.04 | 0.07 | 0.03 | |||||||
| BD | 0.09 | 0.10 | 0.08 | 1.000 | 0.11 | 0.11 | 1.000 | 0.08 | 0.05 | 1.000 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||
| BE | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.01 | 0.02 | 0.00 | 0.03 | 0.00 | 0.00 | 0.00 | |||||||
| DE | 0.01 | 0.00 | 0.01 | 0.00 | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||||||
| Count | 188 | 113 | 75 | 61 | 38 | 52 | 37 | 191 | 126 | 65 | 78 | 28 | 45 | 35 | |||||||
| Line W | Line W male | Line W female | |||||||||||||||||||
| All freq | R freq | S freq | Adj Pval | R freq | S freq | Adj Pval | R freq | S freq | Adj Pval | ||||||||||||
| Allele | |||||||||||||||||||||
| A | 0.52 | 0.52 | 0.52 | 1.000 | 0.60 | 0.59 | 0.366 | 0.52 | 0.46 | 1.000 | |||||||||||
| B | 0.02 | 0.02 | 0.02 | 0.01 | 0.01 | 0.02 | 0.03 | ||||||||||||||
| D | 0.30 | 0.26 | 0.34 | 0.373 | 0.24 | 0.30 | 0.504 | 0.24 | 0.38 | 0.220 | |||||||||||
| E | 0.16 | 0.21 | 0.12 | 0.102 | 0.15 | 0.09 | 0.415 | 0.22 | 0.13 | 0.446 | |||||||||||
| Count | 380 | 194 | 186 | 136 | 74 | 88 | 112 | ||||||||||||||
| Genotype | |||||||||||||||||||||
| AA | 0.26 | 0.27 | 0.25 | 1.000 | 0.28 | 0.32 | 1.000 | 0.25 | 0.20 | 1.000 | |||||||||||
| BB | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||||||||||
| DD | 0.06 | 0.04 | 0.09 | 0.541 | 0.06 | 0.08 | 1.000 | 0.02 | 0.09 | 0.821 | |||||||||||
| EE | 0.04 | 0.04 | 0.03 | 0.02 | 0.00 | 0.07 | 0.05 | 0.218 | |||||||||||||
| AB | 0.03 | 0.03 | 0.02 | 0.02 | 0.03 | 0.05 | 0.02 | ||||||||||||||
| AD | 0.35 | 0.29 | 0.42 | 0.278 | 0.26 | 0.38 | 1.000 | 0.32 | 0.45 | 0.414 | |||||||||||
| AE | 0.14 | 0.19 | 0.10 | 0.407 | 0.19 | 0.14 | 1.000 | 0.18 | 0.07 | 0.441 | |||||||||||
| BD | 0.01 | 0.00 | 0.02 | 0.00 | 0.00 | 0.00 | 0.04 | ||||||||||||||
| BE | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||||||||||
| DE | 0.11 | 0.14 | 0.08 | 0.527 | 0.17 | 0.05 | 0.302 | 0.11 | 0.09 | 1.000 | |||||||||||
| Count | 190 | 97 | 93 | 53 | 37 | 44 | 56 | ||||||||||||||
1Line designations are as described in the text. Allele and genotype frequencies (freq) were determined for the entire line (all) or for the ascites resistant (R) or susceptible (S) subpopulations based on a hypobaric challenge. The total number of alleles or genotypes is indicated below the frequencies (count). The samples for each line were also analyzed according to sex. The P-values for a simple Bonnferoni correction (see Materials and Methods) of chi-squared test of observed vs. expected (Adj Pval) are presented for those alleles or genotypes where the frequency was greater than or equal to 0.10.
Analysis of line Y with PHS009 showed no significant associations with ascites phenotype for any allele or genotype (Table 4). With PHS010 (Table 5) we detected a significant association of the B allele with resistance for the allele counts (32% in resistant vs. 18% in susceptible, adjusted P = 0.025). This appeared to be largely a male effect (32% resistant vs. 14% susceptible), but the counts were low enough that the adjusted P value (0.062) did not quite reach threshold. Haplotype analysis (Table 3) indicates that as in the REL line the A alleles and the B alleles for each locus are in linkage; the AA and BB haplotypes are among the most frequent. However, unlike in the REL the BB haplotype in line Y is apparently associated with resistance to ascites (26% in resistant vs. 9% in susceptible; adjusted P = 0.014).
For line Z, only 2 alleles were identified for PHS009, A and F, where the F allele is almost exclusively found in this line (Table 4), which may relate to the fact that line Z is a female line, whereas the other commercial lines are male lines. Interestingly, PHS009 showed probable deviation from HW estimates (P = 0.04) with fewer homozygotes observed, 99, than expected, 113, suggesting there could be a null allele in this line. There was no deviation for either allele from expected with respect to phenotype, but the FF homozygote was overrepresented in the resistant phenotype (13% resistant vs. 2% susceptible; adjusted P = 0.017). The deviation with respect to phenotype was most pronounced in males, but the counts were low enough that the significance threshold was not quite met (adjusted P = 0.058). Only 2 major alleles (A and B) were detected for PHS010 in line Z. This locus did conform to HW estimates (P = 0.7). The BB genotype in line Z (Table 5) showed significant association (16% resistant vs. 3% susceptible, adjusted P = 0.027) with the greatest association in females (16% in resistant vs. 0% in susceptible), but once again the counts were low enough that the adjusted P (0.087) did not quite reach threshold. For line Z, the A alleles for PHS009 and PHS010 appear to be primarily found in linkage as the AA haplotype constitutes 72% of all haplotypes that could be imputed (Table 3). The PHS009 B allele appears to be most frequently linked with the PHS010 F allele, and the FB haplotype shows significant association with ascites phenotype (21% in resistant vs. 4% in susceptible, adjusted P = 0.008).
For line W with PHS009, 3 alleles were observed. Analysis of this locus for HW showed a slight reduction in homozygotes observed, 91, versus the expected, 103, but the deviation was not great enough for statistical significance (P = 0.08). We detected no association of any allele or genotype for PHS009 with ascites phenotype in line W. Four alleles (A, B, D, and E) were observed for PHS010 in line W. Whereas the B allele was a major allele in lines Y, Z, and REL, the B allele is only a minor allele in line W. The E allele for PHS010 is a minor allele in the other lines but much more prevalent in line W. Thus, line W may be more distinct from the other 3 lines. The PHS010 locus data for line W were in accordance with HW (P = 0.43). As for PHS009, we detected no significant deviations for allele or genotype with respect to phenotype in line W. The major haplotype for line W was AA at 57% (Table 3). The A allele for PHS009 is also associated with the D allele of PHS010 because the AD haplotype was found at 20%. The other haplotypes were found at less than 10%, and no haplotype showed any association with ascites.
DISCUSSION
The region of 11.9 to 13.6 Mbp on Gga9 was originally identified based on significant association for 3 successive SNP with respect to ascites and cardiac hypertrophy in an F2 cross of the RES and SUS lines. The SNP-based linkage was significant only in females. A VNTR analysis using PHS009 and PHS010 in the SUS, REL, and RES lines showed that allele frequencies shifted dramatically, perhaps as a result of selection. For PHS009 the frequencies of the B and C alleles shifted in opposite directions with the B allele favoring resistance. For PHS010 there was a similar shift affecting the A and B alleles with the B allele increasing in the RES and decreasing in the SUS. Only for PHS010 did we detect a statistically significant deviation with respect to phenotype, where the BC genotype was associated with resistance but only for females. Association only with a genotype and not an allele suggests that the effect is mediated either in a multi-subunit complex (e.g., homodimer, heterotetramer, or larger complex), epistatic interactions on different chromatids, or it could be some threshold of activity mediated by the expression levels of 2 specific alleles. That this region showed significant association with ascites phenotype in an F2 cross of the RES and SUS but not in the REL indicates that the reciprocal F2 crosses do not simply recapitulate the REL but instead create unique QTL based on new epistatic interactions. A recently completed new genome-wide SNP analysis in samples from the REL showed no association of this specific region with ascites (N. B. Anthony and D. D. Rhoads, unpublished).
Examination of the commercial lines for this same region revealed that line Y, from which the SUS, REL, and RES lines were derived, has a similar allele distribution as the REL line for both VNTR but that PHS010 allele D appears to have either increased in frequency in line Y or decreased in REL since the SUS, REL, and RES lines were established. Therefore, the REL line and line Y have diverged since 1996, from either drift or, more likely, from continued selection for commercially important traits in line Y. Based on haplotype analysis, after at least 15 generations the AA and BB haplotypes are still overrepresented in the REL and Y lines, indicating that the 118 kbp separating the 2 VNTR has a low recombination frequency.
Overall the analyses of the REL and 3 commercial lines for the 2 VNTR loci, PHS009 and PHS010, detected significant associations in lines Y and Z, and not in REL or line W. These associations are in line Y with the B allele for PHS010 and in line Z with the FF genotype for PHS009 and BB genotype for PHS010 (Tables 4 and 5). This is also reflected in the haplotype analysis for PHS009 combined with PHS010 where in line Y there is an association with the BB haplotype and in line Z with the FB haplotype (Table 3). In line Y the B allele and the BB haplotypes favor resistance, and the association is stronger in males. In line Z there is a statistical association with the resistant phenotype for the FF genotype for PHS009 and the BB genotype for PHS010, so it is not surprising that the FB haplotype is also associated with resistance (21% resistant vs. 4% susceptible). Interestingly, the PHS009 FF genotype association is greater in males, whereas the PHS010 association is greater in females.
Examination of the chicken QTL that have been mapped to this region in the ChickenQTLdb at http://www.animalgenome.org identified breast muscle weight in an F2 Leghorn × Fayoumi cross (Zhou et al., 2006), BW at 200 d in a Leghorn × Red Jungle Fowl cross (Carlborg et al., 2003), BW at 63 d in a cross between a high growth and low growth line (Nadaf et al., 2009), and BW in a cross of high and low antibody response lines (Siwek et al., 2004). Therefore, this region might be expected to be under selection in commercial lines for increased BW and that selection might also affect ascites susceptibility in some lines. Indeed, recent unpublished data (S. Krishnamoorthy, D. D. Rhoads, and N. B. Anthony) suggest that genotypes for PHS009 and PHS010 loci show statistical deviation for breast yield in one commercial line.
Whereas the original genome-wide association study showed an association only in females of the F2 cross this region appears to have different sex effects in different lines. Examination of the differences in susceptibility to ascites in the 4 lines (REL, Y, Z, and W) shows that in all except REL the females are more susceptible to ascites in the hypobaric chamber (Table 6). Under other ascites-inducing conditions and in commercial broiler production, males are considered about twice as likely to develop ascites (Moghadam et al., 2001), so there are clear differences between induction of ascites in commercial production and in the hypobaric chamber. Females appear to be physiologically and anatomically predisposed to earlier development of ascites symptoms than males in the hypobaric challenge (N. B. Anthony, unpublished results). In an evaluation of 4 commercial strains at 1,350 m, others found no difference in ascites susceptibility with regard to sex (Huchzermeyer et al., 1988). Whether the sex specificity observed is mediated through interactions of one or more loci on chromosome Z is a current focus of current research. A recent genome-wide association study in the REL with respect to hypobaric challenge identified association with a single region on chromosome Z (N. B. Anthony and D. D. Rhoads, unpublished).
Table 6.
Ascites susceptibilities (%S) by sex for the former pedigree elite broiler (REL) and 3 commercial lines1
| All | Male | Female | |||
|---|---|---|---|---|---|
| Line | %S | Count | %S | Count | %S |
| REL | 39 | 288 | 44 | 204 | 42 |
| Y | 40 | 198 | 38 | 178 | 42 |
| Z | 34 | 212 | 26 | 160 | 44 |
| W | 50 | 184 | 42 | 198 | 57 |
1Count is the number of birds of that sex for which ascites phenotype was determined in the hypobaric chamber.
Examination of the genes in the region from the original SNP analysis identifies 2 possible candidate genes. Approximately 1.2 Mbp distal to PHS010 is the gene for UTS2D that encodes a urotensin 2 related peptide that is an alternative ligand for the urotensin II receptor. Urotensin II is a potent vasoconstrictor known to be involved in essential hypertension in humans (Watanabe et al., 2006). In hypertensive rats, UTS2D gene expression is increased in the kidney and increased expression is associated with congestive heart failure (Hirose et al., 2009; Nakayama et al., 2008). The gene for AGTR1, an angiotensin II type 1 receptor, is located approximately midway between PHS009 and PHS010. In humans, a SNP in the 3′-untranslated region of AGTR1 is associated with early onset of pulmonary arterial hypertension in humans (Elton et al., 2010; Chung et al., 2009). The 3′-untranslated region SNP is theorized to disrupt a miR155 target site which might disrupt regulation of AGTR1 expression. The vasoconstrictor angiotensin is regulated in humans by angiotensin-converting enzyme (ACE), which is required to cleave the precursor angiotensinogen into angiotensins I and II. Only angiotensin II is known to be biologically active (Chung et al., 2009). In humans, alleles of ACE have been associated with primary pulmonary hypertension (Abraham et al., 2003). The region containing the chicken ACE gene was also identified in the genome-wide SNP analysis (data not shown). Current research is focused on resequencing of the AGTR1, UTS2D, and ACE genes to identify SNP for further genetic analysis in the SUS, REL, and RES lines. Gene expression analysis of alternative alleles could also determine the basis of the effects of these loci on ascites susceptibility and resistance.
ACKNOWLEDGMENTS
Anthony and Rhoads contributed equally to the design and direction of this research. Support was provided by grants to Rhoads and Anthony from the Arkansas Biosciences Institute (Little Rock, AR) and Grant number: 1R15HL092517–0 from National Institutes of Health/National Heart Lung Blood Institute (Bethesda, MD) to Wideman. Al-Rubaye was supported by a fellowship from the Iraqi Ministry of Higher Education and Scientific Research (Baghdad, Iraq).
REFERENCES
- Abraham W. T., Raynolds M. V., Badesch D. B., Wynne K. M., Groves B. M., Roden R. L., Robertson A. D., Lowes B. D., Zisman L. S., Voelkel N. F., Bristow M. R., Perryman M. B. Angiotensin-converting enzyme DD genotype in patients with primary pulmonary hypertension: Increased frequency and association with preserved haemodynamics. J. Renin Angiotensin Aldosterone Syst. 2003;4:27–30. doi: 10.3317/jraas.2003.003. [DOI] [PubMed] [Google Scholar]
- Anthony N. B., Balog J. M. Divergent selection for ascites: Development of susceptible and resistant lines; Proc. 52nd Annu. Natl. Breed. Roundtable; Tucker, GA: US Poultry and Egg Association; 2003. pp. 39–58. [Google Scholar]
- Bailes S., Devers J., Kirby J. D., Rhoads D. An inexpensive, simple protocol for DNA isolation from blood for high-throughput genotyping by polymerase chain reaction or restriction endonuclease digestion. Poult. Sci. 2007;86:102–106. doi: 10.1093/ps/86.1.102. [DOI] [PubMed] [Google Scholar]
- Balog J. M., Kidd B. D., Huff W. E., Huff G. R., Rath N. C., Anthony N. B. Effect of cold stress on broilers selected for resistance or susceptibility to ascites syndrome. Poult. Sci. 2003;82:1383–1387. doi: 10.1093/ps/82.9.1383. [DOI] [PubMed] [Google Scholar]
- Carlborg Ö., Kerje S., Schütz K., Jacobsson L., Jensen P., Andersson L. A global search reveals epistatic interaction between QTL for early growth in the chicken. Genome Res. 2003;13:413–421. doi: 10.1101/gr.528003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W., Deng L., Carroll J., Mallory N., Diamond B., Rosenzweig E., Barst R., Morse J. Polymorphism in the angiotensin II type 1 receptor (AGTR1) is associated with age at diagnosis in pulmonary arterial hypertension. J. Heart Lung Transplant. 2009;28:373–379. doi: 10.1016/j.healun.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Greef K., Kwakernaak C., Ducro B., Pit R., Gerritsen C. Evaluation of between-line variation for within-line selection against ascites in broilers. Poult. Sci. 2001;80:13–21. doi: 10.1093/ps/80.1.13. [DOI] [PubMed] [Google Scholar]
- Deeb N., Shlosberg A., Cahaner A. Genotype-by-environment interaction with broiler genotypes differing in growth rate. 4. Association between responses to heat stress and to cold-induced ascites. Poult. Sci. 2002;81:1454–1462. doi: 10.1093/ps/81.10.1454. [DOI] [PubMed] [Google Scholar]
- Druyan S., Ben-David A., Cahaner A. Development of ascites-resistant and ascites-susceptible broiler lines. Poult. Sci. 2007a;86:811–822. doi: 10.1093/ps/86.5.811. [DOI] [PubMed] [Google Scholar]
- Druyan S., Cahaner A. Segregation among test-cross progeny suggests that two complementary dominant genes explain the difference between ascites-resistant and ascites-susceptible broiler lines. Poult. Sci. 2007;86:2295–2300. doi: 10.3382/ps.2007-00018. [DOI] [PubMed] [Google Scholar]
- Druyan S., Hadad Y., Cahaner A. Growth rate of ascites-resistant versus ascites-susceptible broilers in commercial and experimental lines. Poult. Sci. 2008;87:904–911. doi: 10.3382/ps.2008-00003. [DOI] [PubMed] [Google Scholar]
- Druyan S., Shinder D., Shlosberg A., Cahaner A., Yahav S. Physiological parameters in broiler lines divergently selected for the incidence of ascites. Poult. Sci. 2009;88:1984–1990. doi: 10.3382/ps.2009-00116. [DOI] [PubMed] [Google Scholar]
- Druyan S., Shlosberg A., Cahaner A. Evaluation of growth rate, body weight, heart rate, and blood parameters as potential indicators for selection against susceptibility to the ascites syndrome in young broilers. Poult. Sci. 2007b;86:621–629. doi: 10.1093/ps/86.4.621. [DOI] [PubMed] [Google Scholar]
- Elton T. S., Sansom S. E., Martin M. M. Cardiovascular disease, single nucleotide polymorphisms; and the renin angiotensin system: Is there a microRNA connection? Int. J. Hypertens. 2010 doi: 10.4061/2010/281692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamal K. R., Wideman R. F., Anthony N. B., Erf G. F. Differential expression of vasoactive mediators in microparticle-challenged lungs of chickens that differ in susceptibility to pulmonary arterial hypertension. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010a;298:R235–R242. doi: 10.1152/ajpregu.00451.2009. [DOI] [PubMed] [Google Scholar]
- Hamal K. R., Wideman R. F., Anthony N. B., Erf G. F. Differential gene expression of proinflammatory chemokines and cytokines in lungs of ascites-resistant and -susceptible broiler chickens following intravenous cellulose microparticle injection. Vet. Immunol. Immunopathol. 2010b;133:250–255. doi: 10.1016/j.vetimm.2009.07.011. [DOI] [PubMed] [Google Scholar]
- Hirose T., Takahashi K., Mori N., Nakayama T., Kikuya M., Ohkubo T., Kohzuki M., Totsune K., Imai Y. Increased expression of urotensin II, urotensin II-related peptide and urotensin II receptor mRNAs in the cardiovascular organs of hypertensive rats: Comparison with endothelin-1. Peptides. 2009;30:1124–1129. doi: 10.1016/j.peptides.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Huchzermeyer F. W., De Ruyck A. C., Van Ark H. Broiler pulmonary hypertension syndrome. III. Commercial broiler strains differ in their susceptibility. Onderstepoort J. Vet. Res. 1988;55:5–9. [PubMed] [Google Scholar]
- Julian R. J. Physiological, management and environmental triggers of the ascites syndrome: A review. Avian Pathol. 2000;29:519–527. doi: 10.1080/03079450020016751. [DOI] [PubMed] [Google Scholar]
- Julian R. J., Mirsalimi S. M. Blood oxygen concentration of fast-growing and slow-growing broiler chickens, and chickens with ascites from right ventricular failure. Avian Dis. 1992;36:730–732. [PubMed] [Google Scholar]
- Ledur M. C., Melo C. M. R., Nones K., Zanella E. L., Ninov K., Bonassi C. A., Jaenisch F. R. F., Moura A. S. A. M. T., Coutinho L. L., Schmidt G. S. Genetic and phenotypic parameters for organs, body and carcass weights, and haematocrit value, in a broiler × layer cross resource population; Proceedings of the 8th World Congress on Genetics Applied to Livestock Production; Minas Gerais, Brazil: Instituto Prociência; 2006. pp. 7–24. [Google Scholar]
- Lorenzoni A. G., Anthony N. B., Wideman R. F., Jr. Transpulmonary pressure gradient verifies pulmonary hypertension is initiated by increased arterial resistance in broilers. Poult. Sci. 2008;87:125–132. doi: 10.3382/ps.2007-00178. [DOI] [PubMed] [Google Scholar]
- Lubritz D. L., Smith J. L., McPherson B. N. Heritability of ascites and the ratio of right to total ventricle weight in broiler breeder male lines. Poult. Sci. 1995;74:1237–1241. doi: 10.3382/ps.0741237. [DOI] [PubMed] [Google Scholar]
- Moghadam H., McMillan I., Chambers J. R., Julian R. J. Estimation of genetic parameters for ascites syndrome in broiler chickens. Poult. Sci. 2001;80:844–848. doi: 10.1093/ps/80.7.844. [DOI] [PubMed] [Google Scholar]
- Muir W. M., Wong G. K.-S., Zhang Y., Wang J., Groenen M. A. M., Crooijmans R. P. M. A., Megens H.-J., Zhang H., Okimoto R., Vereijken A., Jungerius A., Albers G. A. A., Lawley C. T., Delany M. E., MacEachern S., Cheng H. H. Genome-wide assessment of worldwide chicken SNP genetic diversity indicates significant absence of rare alleles in commercial breeds. Proc. Natl. Acad. Sci. USA. 2008;105:17312–17317. doi: 10.1073/pnas.0806569105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadaf J., Pitel F., Gilbert H., Duclos M. J., Vignoles F., Beaumont C., Vignal A., Porter T. E., Cogburn L. A., Aggrey S. E., Simon J., Le Bihan-Duval E. QTL for several metabolic traits map to loci controlling growth and body composition in an F2 intercross between high- and low-growth chicken lines. Physiol. Genomics. 2009;38:241–249. doi: 10.1152/physiolgenomics.90384.2008. [DOI] [PubMed] [Google Scholar]
- Nakayama T., Hirose T., Totsune K., Mori N., Maruyama Y., Maejima T., Minagawa K., Morimoto R., Asayama K., Kikuya M., Ohkubo T., Hashimoto J., Kohzuki M., Takahashi K., Imai Y. Increased gene expression of urotensin II-related peptide in the hearts of rats with congestive heart failure. Peptides. 2008;29:801–808. doi: 10.1016/j.peptides.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Navarro P., Visscher P. M., Chatziplis D., Koerhuis A. N. M., Haley C. S. Segregation analysis of blood oxygen saturation in broilers suggests a major gene influence on ascites. Br. Poult. Sci. 2006;47:671–684. doi: 10.1080/00071660601077931. [DOI] [PubMed] [Google Scholar]
- Pakdel A., Van Arendonk J., Vereijken A., Bovenhuis H. Direct and maternal genetic effects for ascites-related traits in broilers. Poult. Sci. 2002;81:1273–1279. doi: 10.1093/ps/81.9.1273. [DOI] [PubMed] [Google Scholar]
- Pavlidis H. O., Balog J. M., Stamps L. K., Hughes J. D., Jr., Huff W. E., Anthony N. B. Divergent selection for ascites incidence in chickens. Poult. Sci. 2007;86:2517–2529. doi: 10.3382/ps.2007-00134. [DOI] [PubMed] [Google Scholar]
- Peacock A. J., Pickett C., Morris K., Reeves J. T. The relationship between rapid growth and pulmonary hemodynamics in fast-growing broiler chicken. Am. Rev. Respir. Dis. 1989;139:1524–1530. doi: 10.1164/ajrccm/139.6.1524. [DOI] [PubMed] [Google Scholar]
- Rabie T. S. K. M., Crooijmans R. P. M. A., Bovenhuis H., Vereijken A. L. J., Veenendaal T., van der Poel J. J., Van Arendonk J. A. M., Pakdel A., Groenen M. A. M. Genetic mapping of quantitative trait loci affecting susceptibility in chicken to develop pulmonary hypertension syndrome. Anim. Genet. 2005;36:468–476. doi: 10.1111/j.1365-2052.2005.01346.x. [DOI] [PubMed] [Google Scholar]
- Shlosberg A., Bellaiche M., Berman E., Perk S., Deeb N., Neumark E., Cahaner A. Relationship between broiler chicken haematocrit-selected parents and their progeny, with regard to haematocrit, mortality from ascites and bodyweight. Res. Vet. Sci. 1998;64:105–109. doi: 10.1016/s0034-5288(98)90004-2. [DOI] [PubMed] [Google Scholar]
- Shlosberg A., Bellaiche M., Zeitlin G., Ya’acobi M., Cahaner A. Hematocrit values and mortality from ascites in cold-stressed broilers from parents selected by hematocrit. Poult. Sci. 1996;75:1–5. doi: 10.3382/ps.0750001. [DOI] [PubMed] [Google Scholar]
- Siwek M., Cornelissen S., Buitenhuis A., Nieuwland M., Bovenhuis H., Crooijmans R., Groenen M., Parmentier H., van der Poel J. Quantitative trait loci for body weight in layers differ from quantitative trait loci specific for antibody responses to sheep red blood cells. Poult. Sci. 2004;83:853–859. doi: 10.1093/ps/83.6.853. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Kanome T., Miyazaki A., Katagiri T. Human urotensin II as a link between hypertension and coronary artery disease. Hypertens. Res. 2006;29:375–387. doi: 10.1291/hypres.29.375. [DOI] [PubMed] [Google Scholar]
- Wideman R. F., Chapman M., Owens C., Devabhaktuni M., Cavitt L., Wang W., Erf G. Broiler survivors of intravenous micro-particle injections: Evaluation of growth, livability, meat quality, and arterial blood gas values during a cyclic heat challenge. Poult. Sci. 2003;82:484–495. doi: 10.1093/ps/82.3.484. [DOI] [PubMed] [Google Scholar]
- Wideman R. F., Erf G., Chapman M., Wang W., Anthony N., Xiaofang L. Intravenous micro-particle injections and pulmonary hypertension in broiler chickens: Acute post-injection mortality and ascites susceptibility. Poult. Sci. 2002;81:1203–1217. doi: 10.1093/ps/81.8.1203. [DOI] [PubMed] [Google Scholar]
- Wideman R. F., French H. Broiler breeder survivors of chronic unilateral pulmonary artery occlusion produce progeny resistant to pulmonary hypertension syndrome (ascites) induced by cool temperatures. Poult. Sci. 1999;78:404–411. doi: 10.1093/ps/78.3.404. [DOI] [PubMed] [Google Scholar]
- Wideman R. F., French H. Ascites resistance of progeny from broiler breeders selected for two generations using chronic unilateral pulmonary artery occlusion. Poult. Sci. 2000;79:396–401. doi: 10.1093/ps/79.3.396. [DOI] [PubMed] [Google Scholar]
- Wideman R. F., Kirby Y., Owen R., French H. Chronic unilateral occlusion of an extrapulmonary primary bronchus induces pulmonary hypertension syndrome (ascites) in male and female broilers. Poult. Sci. 1997;76:400–404. doi: 10.1093/ps/76.2.400. [DOI] [PubMed] [Google Scholar]
- Wideman R. F., Wing T., Kirby Y., Forman M., Marson N., Tackett C., Ruiz-Feria C. Evaluation of minimally invasive indices for predicting ascites susceptibility in three successive hatches of broilers exposed to cool temperatures. Poult. Sci. 1998;77:1565–1573. doi: 10.1093/ps/77.10.1565. [DOI] [PubMed] [Google Scholar]
- Wideman R. F., Chapman M. E., Hamal K. R., Bowen O. T., Lorenzoni A. G., Erf G. F., Anthony N. B. An inadequate pulmonary vascular capacity and susceptibility to pulmonary arterial hypertension in broilers. Poult. Sci. 2007;86:984–998. doi: 10.1093/ps/86.5.984. [DOI] [PubMed] [Google Scholar]
- Wideman R. F., Kirby Y. K. Evidence of a ventilation-perfusion mismatch during acute unilateral pulmonary artery occlusion in broilers. Poult. Sci. 1995;74:1209–1217. doi: 10.3382/ps.0741209. [DOI] [PubMed] [Google Scholar]
- Wideman R. F., Rhoads D. D., Erf G. F., Anthony N. B. Pulmonary arterial hypertension (ascites syndrome) in broilers: A review. Poult. Sci. 2013;92:64–83. doi: 10.3382/ps.2012-02745. [DOI] [PubMed] [Google Scholar]
- Zhou H., Deeb N., Evock-Clover C. M., Ashwell C. M., Lamont S. J. Genome-wide linkage analysis to identify chromosomal regions affecting phenotypic traits in the chicken. II. Body composition. Poult. Sci. 2006;85:1712–1721. doi: 10.1093/ps/85.10.1712. [DOI] [PubMed] [Google Scholar]