Abstract
The aggregation of intrinsically disordered peptides and proteins is associated with a wide range of highly debilitating neurological and systemic disorders. In this work we explored the potential of a structure-based drug discovery procedure to target one such system, the soluble monomeric form of the Aβ42 peptide. We utilised for this purpose a set of structures of the Aβ42 peptide selected from clusters of conformations within an ensemble generated by molecular dynamics simulations. Using these structures we carried out fragment mapping calculations to identify binding ‘hot spots’ on the monomeric form of the Aβ42 peptide. This procedure provided a set of hot spots with ligand efficiencies comparable to those observed for structured proteins, and that are clustered into binding pockets. We verified that such pockets exhibit a propensity to bind small molecules known to interact with the Aβ42 peptide. Taken together these results provide an initial indication that fragment-based drug discovery may represent a potential therapeutic strategy for diseases associated with the aggregation of intrinsically disordered proteins.
Introduction
The aggregation of intrinsically disordered peptides and proteins is associated with a wide range of human disorders, including Alzheimer's and Parkinson's diseases1, 2. These diseases, for which at present there are no effective treatments, are increasingly common in our ageing society3, prompting a variety of therapeutic strategies to be proposed and pursued4, 5. Among such strategies, increasing attention has been devoted to finding drug-like small molecules capable of interfering with the aggregation process of intrinsically disordered proteins, and of promoting their normal behaviour6–10. In this context, stabilizing the soluble monomeric form of these proteins is appealing because it can influence downstream aggregation events7, 11, including the formation of small oligomeric species that are increasingly recognised as the origin of neuronal damage2, 12, 13. It has indeed been suggested that intrinsically disordered proteins may be targeted by identifying specific sequence regions that exhibit specific “molecular recognition features” (MoRFs)9, 14, 15.
In this work we investigate an alternative approach to this problem, which is based on the structure-based search of potential binding pockets in intrinsically disordered proteins. Although structure-based drug discovery is a strategy that has been effective in identifying small-molecule ligands that bind to the native states of globular proteins16, 17, there are two major challenges in the extension of this approach to intrinsically disordered proteins. The first is the existence of very substantial technical difficulties in acquiring accurate information about the structure and dynamics of disordered proteins by experimental methods18–20 and the second is that the binding pockets in these molecules are likely to be present only transiently. Despite these problems, recent evidence indicates that disordered binding interfaces can be effectively targeted by small molecules8–10.
In order to explore the potential of this structure-based approach for disordered monomeric polypeptide chains, we have considered the 42-residue Aβ peptide (Aβ42), whose aggregation process is associated with the pathogenesis of Alzheimer's disease1, 2. This peptide is highly disordered in solution, populating a heterogeneous ensemble of conformations21, 22, a situation in sharp contrast to that of the fibrillar state of the peptide, which is ordered and has been characterised in general terms, notably by X-ray fibre diffraction23, electron microscopy and solid-state NMR24 studies.
Here, we have adopted a strategy for identifying small-molecule binding sites in the Aβ42 peptide in which molecular dynamics simulations are combined with fragment-based drug design25–28. Given the challenges in obtaining atomistic descriptions of the conformational ensembles populated by intrinsically disordered regions18–20, molecular dynamics simulations represent a vital tool in elucidating the structures and dynamics of these systems29–40. In this view, the approach that we describe extends to intrinsically disordered proteins a type of strategy that, from the relaxed complex method41 to subsequent methods42–48, has been aimed at introducing flexibility in docking. In fragment-based drug design a library of small-molecule fragments is screened to find those that have a propensity to bind to specific “hot spot” regions in a given conformation of a protein25–28. By using this approach we present evidence that in the case of the Aβ42 peptide it is possible to identify clusters of binding hot spots that could serve as binding sites for drug-like small molecules assembled from fragments.
Methods
Replica-exchange molecular dynamics simulations
There are several different procedures that can be used to generate structural ensembles representing the conformational space of intrinsically disordered peptides and proteins, including molecular modelling49, 50, molecular dynamics simulations29–38, and molecular simulations with NMR restraints18–20, 51, 52. In the present case the ensemble was obtained by performing replica-exchange molecular dynamics (REMD) simulations53 using the GROMACS molecular dynamics simulations package54, following a protocol similar to that used previously55. The REMD method enhances the sampling of the conformational space of polypeptide chains by overcoming energy barriers that could otherwise trap the simulations in local minima53. The duration of the simulation was 100ns, using an integration step of 2fs, at 48 temperatures ranging from 276.1-376.9K with the AMBER99SB force field56 and the TIP3P water model57; this force field has been shown to reproduce with rather good accuracy NMR parameters in molecular dynamics simulations of other peptides and proteins58, 59. The monomeric form of the Aβ42 peptide with charged termini was placed in a box of 7x7x7 nm3 in periodic boundary conditions, with about 10,000 water molecules and three Na+ counterions to neutralize the net charge of the peptide. The protonation state of titratable groups is know to affect molecular interactions60–62; here in all the simulations Glu and Asp residues were assumed to be protonated, while His residues were assumed to be not protonated, as detected experimentally at neutral pH63. The system was equilibrated for 500ps before the production run. Replica exchange attempts were made every 250ps resulting in a success rate of about 13%. We verified the structures obtained at 278K with NMR results reported in the literature at similar temperatures21, 36, 64; chemical shifts were back-calculated from the structures using the CamShift method65.
Cluster analysis
20,000 structures were taken from a 60-100 ns portion of the trajectory at 309.4 K at 2 ps intervals, and clustered by means of the GROMACS g_cluster tool using the single linkage algorithm, by which a structure is added to a cluster when its RMSD on all Cα atoms to any member of the cluster is less than cut-off of 2 Å. We thus identified 45 clusters with populations ranging from 0.05% to 2%, which were included in the docking analysis. Side-chain contacts were calculated as the pairwise average distance between all the atoms (other than Cα) of a side-chain with that of another.
Fragment-based mapping of binding hot spots
A wide range of approaches are available to perform fragment-based computational mapping of potentially druggable binding hot spots66–70, including the GRID method71, the multiple copy simultaneous search (MCSS) method72, the ROSETTALIGAND method73 and the mixed-solvent molecular dynamics (MixMD) method74. In this work we identified binding hotspots of small molecular fragments by combining the FTMap66 and FRED75 methods. The FTMap method, which is based on a Fast Fourier Transform (FFT) correlation approach, was used as an initial screen of the 45 structures representative of the corresponding clusters that we identified, and the top ten structures were selected for a further fragment-based docking analysis. In order to perform the docking, among possible alternatives42, 43, including GLIDE44, DOCK45, MolDock46, and GOLD76, we used here FRED75, which is a protein structure-based docking program that performs an exhaustive search that systematically samples multiple possible poses to a given resolution, and is thus more computationally expensive than FTMap.
We used a library of ten small organic molecular fragments (benzene, cyclohexane, cyclopropyl, dymethyl ketone, furan, imidazole, methanol, methylamide, oxazole and pyrazole), which commonly appears in fragment libraries26, 77 and have the hydrophobic character expected to favour the binding to the hydrophobic regions of the Aβ42 peptide. For each fragment, exhaustive docking was performed on the surface of the Aβ42 peptide with a rotation step of 1.25 Å and translation step of 1 Å, and the 10,000 top ranked poses were retained and optimized based on a shape-based Gaussian scoring function78. The top 300 poses were then selected based on a consensus score of four scoring functions: Shapegauss, PLP (Piecewise Linear Potential), OEChemscore and Screenscore, which are implemented in FRED75. In order to further optimise the structures, among a wide range of possible alternative methods79–84, we used SZYBKI (OpenEye Scientific, www.eyesopen.com) and the Merck Molecular Force Field MMFF94s85, 86, where the partial charges of ligands were first calculated by Molcharge using the AM1BCC charges (OpenEye Scientific, www.eyesopen.com).
A binding hot spot is defined in this study as a small surface area capable of binding multiple ligand fragments. In order to estimate the quality of a given hot spot we considered the potential ligand efficiency of the fragments that bind to it. The ligand efficiency is defined as87 Fp/Np, where Np is the number of heavy atoms in a ligand probe p, and Fp (in kcal/mol) is the binding free energy of the probe. In our calculations we considered the potential energy Ep in the MMFF94s force field85, rather than the binding free energy Fp of the probe. This approach represents an approximation, as the binding free energy could not generally be expected to be very accurately approximated by the potential energy of binding88. This type strategy is primarily adopted because of its computational efficiency with the aim of generating a small number of candidate fragments and small molecules to facilitate subsequent experimental studies of binding. The validity of this approximation has been discussed in the case of folded proteins, where enthalpic contributions were found to be larger than entropic ones in the binding of small fragments of the type considered here89. We should emphasise, however, that the role of entropic contributions in the case of disordered proteins may be greater and will require further studies to be fully clarified.
The normalisation of the binding free energy by the number of heavy atoms has been suggested to be a useful means of evaluating the quality of hot spots because larger fragments tend to have better binding energy just because of their larger sizes87. We then defined the average potential ligand efficiency of a given hot spot as Le= Σp Lep/N, where Lep = Ep/Np is the potential ligand efficiency and N is the total number of ligand probes that bind to the hot spot. More negative potential ligand efficiency values are thus indicative of better binding hot spots. For comparison, we considered the cases of structured proteins, the ATP binding site of p38 MAP kinase and the active site of β-secretase, finding similar potential ligand efficiency values (see Tables S1 and S2 in the Supplementary Information90). Structural representations of structures of the Aβ42 peptide and its ligands were created using PyMOL91.
Identification of binding pockets
As is common in fragment-based drug design procedures25–28, we identified potential small-molecule binding pockets as clusters of neighbouring binding hot spots.
Docking and molecular dynamics studies of possible binding modes between Aβ42 and curcumin, and Aβ42 and Congo red
Conformers of curcumin and Congo red were first generated by Omega v2.4.3, a multi-conformer structure database generation program by OpenEye Scientific92, using a 0.5 Å RMSD cut-off between conformers. Docking at the binding sites of Aβ42 of the two compounds were performed by FRED, using the scoring functions and method mentioned above. For each compound the most highly ranked binding mode was then used as the starting structure in molecular dynamics simulations performed using GROMACS with settings similar to those of the REMD method described above, but in this case at constant temperature (298K) for 80 ns. We used force field parameters and topologies based on General Amber Force Field (GAFF) and AMBER99SB as prepared by ACPYPE/Antechamber for GROMACS (http://www.ccpn.ac.uk/software/ACPYPE-folder). The system was first energy minimized in vacuo and then in TIP3P water, equilibrated by slowly heating from 272K to 298 over a 500 ps period, before a production run of 80 ns was carried out. In total, we ran 20 trajectories of 80 ns, one for each of the 10 binding pockets and the 2 small molecules.
Results
Generation and validation of an ensemble of conformations of the Aβ42 peptide
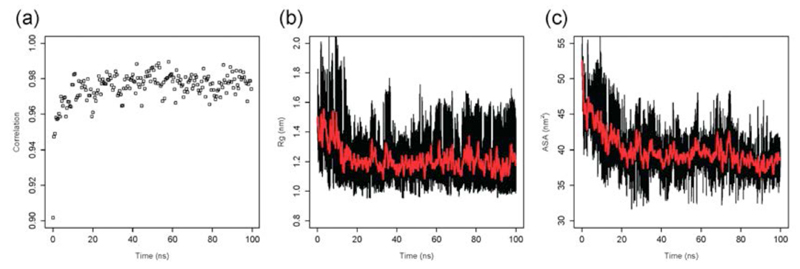
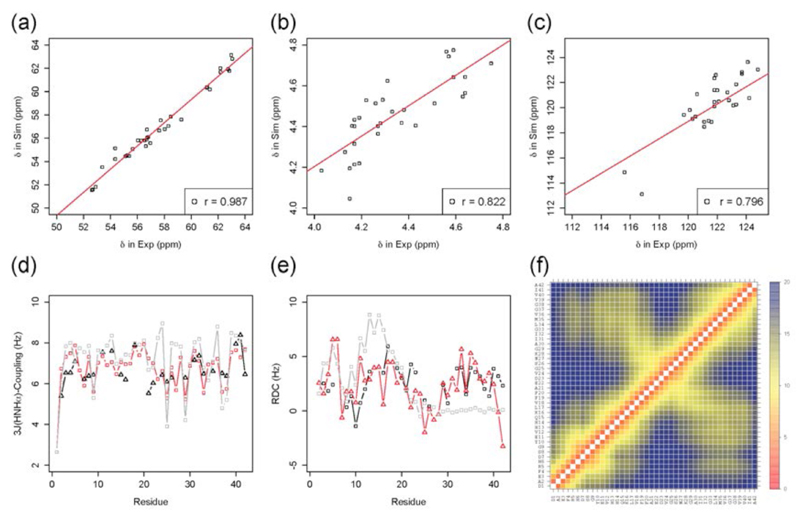
The first step of the procedure that we discuss in this work is the generation of an ensemble of conformations that represents the soluble monomeric form of the Aβ42 peptide (Figure 1a), which was carried out by replica-exchange molecular dynamics (REMD) simulations53 in explicit solvent (see Methods). In order to establish whether the Aβ42 structural ensemble generated by the procedure described above provides a good representation of the conformations that this peptide populates in solution, we investigated the agreement between various experimental and back-calculated structural parameters, where the latter were obtained from the ensemble of structures. A good correlation was found between experimental and calculated chemical shifts of the Cα atoms21 (Figure 2a), as well as the evolution of radius of gyration (Figure 2b) and solvent accessible surface area (Figure 2c). This analysis was performed for the conformations determined at 278K, the temperature at which the experimental chemical shifts that we considered were measured21 (Figure 3a-c). We found good correlations between experimental and back-calculated chemical shifts (the coefficients of correlation were 0.987 for Cα, 0.822 for Hα and 0.796 for N). We also compared experimental and back-calculated 3J-couplings36, as well as residual dipolar couplings (RDCs)64, finding a good agreement also in these cases (Figure 3d-e). In particular, the level of such an agreement was found to be higher than that provided by the statistical coil model (SCM, Figure 3d-e), which has been found to describe accurately the dimension and structures populated by highly disordered states of proteins49; for the SCM ensemble and the present ensemble the RMSD values were, respectively, 1.22Hz and 0.82Hz for the 3J-couplings, and 2.91Hz and 2.07Hz for the RDCs.
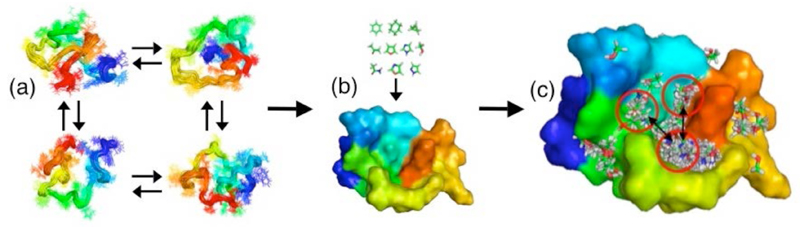
Figure 1.
Scheme illustrating the strategy discussed in this work in which molecular dynamics simulations are combined with computer-based fragment-based hot spot mapping to identify potential binding sites on the soluble monomeric form of the Aβ42 peptide. (a) Representative examples are selected by a clustering procedure within an ensemble of conformations representing the natively unfolded state of the Aβ42 peptide; (b) Hot spot regions are mapped on these structures using a set of small molecule fragments; (c) Neighbouring hot spot regions are identified as potential small-molecule binding sites.
Figure 2.
Analysis of the convergence of the molecular dynamics simulations used in this work to generate an ensemble of structures representing the soluble monomeric form of the Aβ42 peptide: (a) Time series of the correlation between experimental and calculated Cα chemical shifts, which indicate that after about 40 ns (out of a total of 100 ns) a good correlation is reached between experimental and calculated chemical shifts. (b) Time series of the radius of gyration. (c) Time series of the solvent accessible surface area (SASA).
Figure 3.
Validation of the ensemble of conformations representing the soluble monomeric form of the Aβ42 peptide used in this work. (a-c) Correlation between experimental21 and back-calculated chemical shifts: Cα (a), Hα (b) and N (c). (d) Comparison between experimental36(black) and back-calculated (red) 3J couplings (Hz). (e) Comparison between experimental64 (black) and back-calculated (red) residual dipolar couplings (RDCs, Hz). For reference, 3J couplings and RDCs are also shown as predicted by the statistical coil model49 (grey). (f) Inter-residue distance map (Å).
Structural analysis of the Aβ42 ensemble
The simulations indicate that, under the conditions that we have investigated, Aβ42 populates a restricted but highly dynamical ensemble of conformations that is significantly more compact than that expected for a random coil49, in agreement with previous conclusions on this system21, 93. In addition, the average value of the radius of gyration of the Aβ42 peptide in our simulations is of about 13 Å, compared to the SCM value of about 20 Å. The presence of transient structural motifs in Aβ42 is also indicated by the differences between the present results and those obtained by SCM (Figure 3d-e).
Analysis of the inter-residue distances in the ensemble of conformations that we have generated here (Figure 3f) suggests that the overall structure and dynamics of the Aβ42 peptide are particularly strongly affected by the behaviour of a few specific regions of the amino acid sequence that have a high tendency to form turns, in particular Asp7-Tyr10, Asp23-Ser26 and Gly37-Val40, as well as by the interactions between the two main hydrophobic regions (residues Leu17-Ala21 and Ile31-Val36) of the polypeptide chain. In particular, the combination of this latter tertiary contact, which is observed in several clusters, with the transient formation of a turn in the Asp23-Ser26 region appears to be the main driving factors for the quasi-hairpin-like structures often reported for the Aβ42 peptide, where the turn formation could be assisted by electrostatic interactions between Glu22, Asp23 and Lys28. These findings are in good agreement with structural insights drawn previous from nuclear Overhauser enhancement (NOE) data21 as well as from molecular dynamics simulations35, 37.
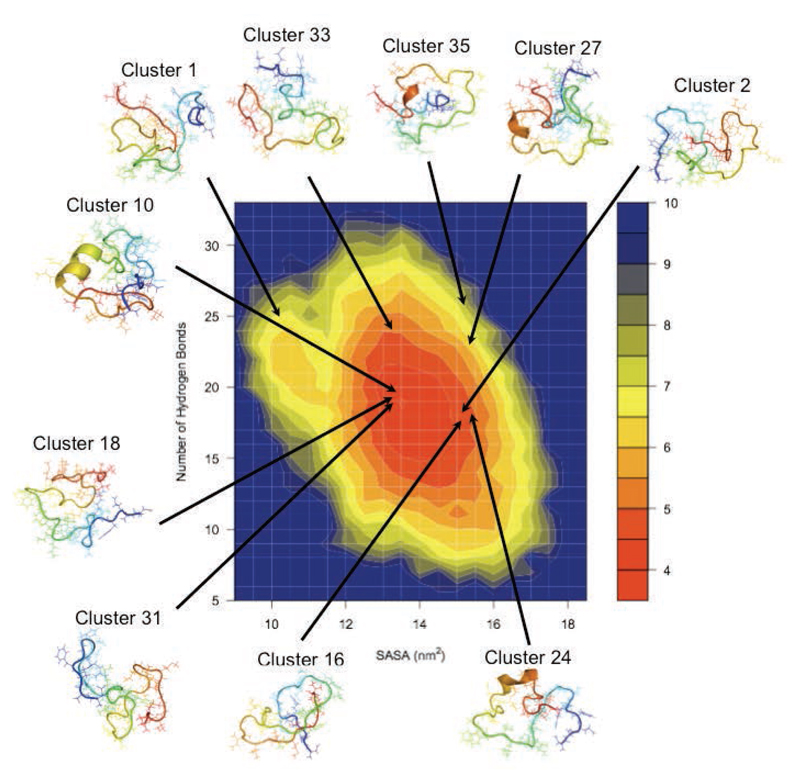
We next used a cluster analysis to find families of similar structures in the Aβ42 ensemble (see Methods). This procedure resulted in 396 clusters, the 45 most populated of which were selected for further analysis. Taken together, these 45 clusters include about 67% of the members of the structural ensemble, illustrating its heterogeneity; the most populated clusters are shown in Figure 4 on a free energy landscape plotted as a function of the number of hydrogen bonds (backbone-backbone, backbone-sidechain and sidechain-sidechain) and of the solvent-exposed surface area of hydrophobic residues. The secondary structure elements, side-chain distance maps and long-range contacts most frequently observed in eight of the most populated clusters are shown as examples in Figure 5.
Figure 4.
Free energy landscape of the Aβ42 peptide as a function of the number of hydrogen bonds (backbone-backbone, backbone-sidechain and sidechain-sidechain) and of the solvent-exposed surface area of hydrophobic residues. Hydrogen bonds were defined using the GROMACS g_hbond function, when hydrogen donors and acceptors are within 3.5 Å and the hydrogen-donor-acceptor angles are within 30 degrees. The most populated clusters are found in different regions of the free energy landscape.
Figure 5.
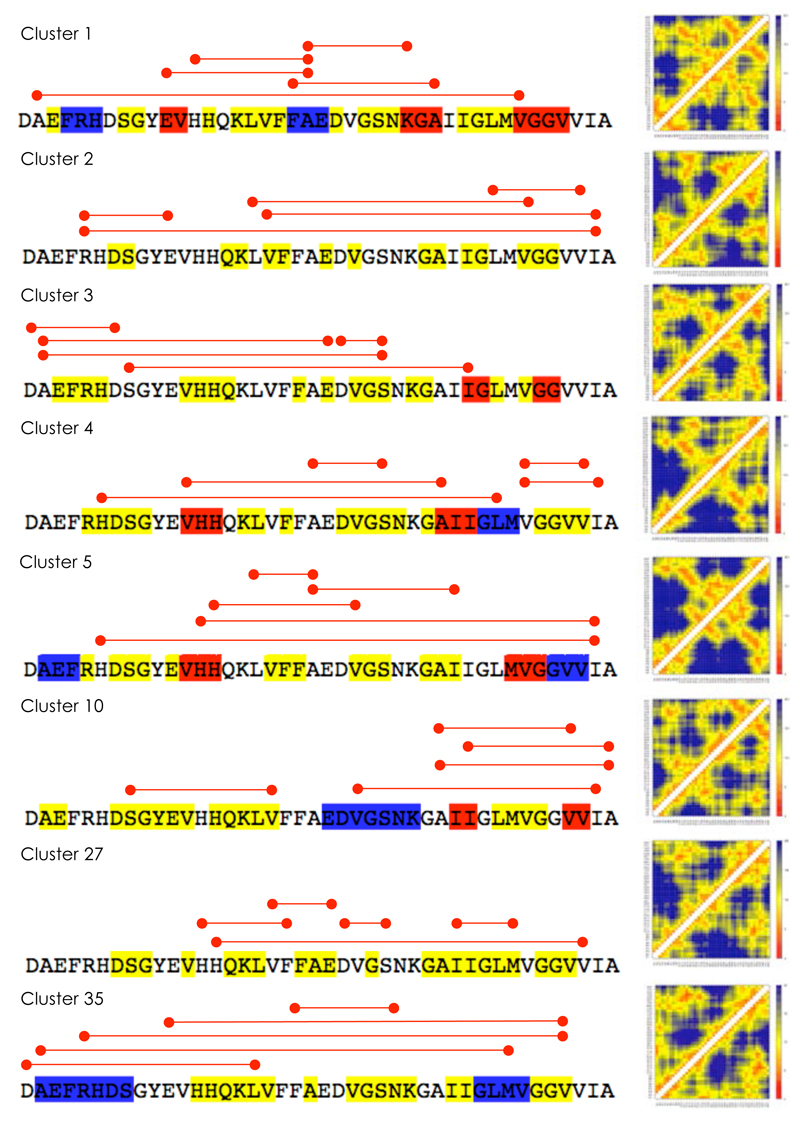
Characterisation of eight representative clusters of structures within the ensemble of conformations of the soluble monomeric form of the Aβ42 peptide used in this work. We present here the five most populated clusters (cluster 1-5) and five examples of clusters found to contain binding pockets (clusters 1, 2, 10, 27 and 35). Highly populated clusters may (as clusters 1 and 2) or may not (as clusters 3, 4 and 5) exhibit binding pockets. For each cluster we report the secondary structure elements determined by DSSP106 (yellow: turns; blue: α-helices; red: β-sheets) and the side-chain distance maps. The five shortest long-range side-chain contacts (i.e. more than three residues apart along the amino acid sequence) are indicated by red lines.
The features in these individual distance maps of clusters resemble in part the overall features exhibited in the average distance map (Figure 3f). For example, in cluster 1 the turns around Ser8-Gly9 and Gly25-Asn27 are very close to the corresponding regions of the first two turns identified in the general distance map, whereas cluster 2 exhibits the very prominent turn around residues Asp7-Try10 in the N-terminal region. Further, the characteristic contacts between the two hydrophobic regions (residues Leu17-Ala21 and Ile31-Val36) are also noticeable in all the distance maps shown in Figure 5. The identification of geometrically similar families of conformations of the Aβ42 peptide suggests that distinct sets of structurally related conformations exist in the ensemble.
Fragment-based hot spot mapping
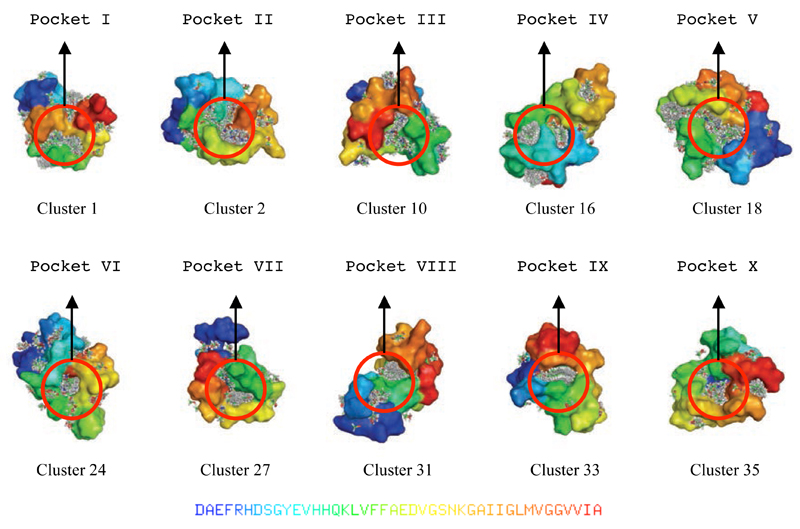
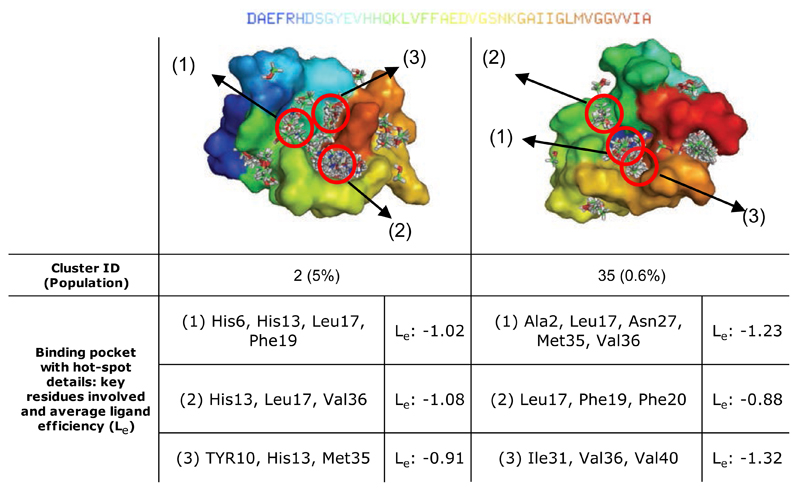
Individual representative structures were selected from the top 45 clusters and used to identify binding hot spots using FTMap calculations66 (see Methods). The ten structures found to contain multiple binding hot spots in close proximity to each other were selected for further analysis (Table 1 and Figure 6) and used in an exhaustive rigid-body docking procedure using FRED75 (see Methods). Two representative structures are shown in Figure 7, where key residues forming the hot spots and the corresponding potential ligand efficiency values87 (Le, see Methods) are listed.
Table 1.
List of the ten binding pockets (in Roman numerals, column 2) and corresponding binding hot spots (in Arabic numerals, column 3) identified within ten clusters of conformations (column 1) in the Aβ42 structural ensemble described in this work. Each of these ten clusters exhibits one binding pocket comprising between two and five binding hot spots; for example the binding pocket VI is found in cluster 24 and comprises four hot spots. The remaining 35 clusters did not exhibit binding pockets. The specific residues in the hot spots are also reported (column 4). The structures of the ten binding pockets are shown in Figure 6.
| Cluster | Pocket ID | Hot Spot ID | Key Residues Involved |
|---|---|---|---|
| Cluster 1 | I | 1 | Leu17, Val18, Phe20 |
| Cluster 1 | I | 2 | Ala30, Gly33, Lys16, Gly33 |
| Cluster 2 | II | 1 | His6, His13, Leu17, Phe19 |
| Cluster 2 | II | 2 | His13, Leu17, Val36 |
| Cluster 2 | II | 3 | Tyr10, His13, Met35 |
| Cluster 10 | II | 1 | Leu17, Phe19, Phe20, Ile41, Val40 |
| Cluster 10 | II | 2 | His6, Phe20, Val39, Val40, Val18 |
| Cluster 16 | IV | 1 | Glu11, His13, Lys16, Leu17 |
| Cluster 16 | IV | 2 | Arg5, Asp7, Glu11, Lys16, Val24, Val36 |
| Cluster 16 | IV | 3 | Ala21, Asp23, Val24 |
| Cluster 18 | V | 1 | Leu17, His6, Ile3, Tyr10, Gly25, Asn27 |
| Cluster 18 | V | 2 | Lys16, Leu17, Phe19, Val24, Gly25 |
| Cluster 18 | V | 3 | Phe19, Glu22, Asp23, Val24 |
| Cluster 18 | V | 4 | His6, Try10, Ile31,Arg5 |
| Cluster 18 | V | 5 | Val24, Ile31, Gly33 |
| Cluster 24 | VI | 1 | His13, His14, Val18, Val24, Val39 |
| Cluster 24 | VI | 2 | His14, Met35, Val39, Val40 |
| Cluster 24 | VI | 3 | Val18, Phe19, Asp23 |
| Cluster 24 | VI | 4 | Val18, Asp23, Met35 |
| Cluster 27 | VI | 1 | Lys28, Val39, Ile31, Met35 |
| Cluster 27 | VI | 2 | Val13, His13, Val18, Asp23, Ser26, Lys28, Val39 |
| Cluster 27 | VI | 3 | His14, Leu17, Val39, Val40 |
| Cluster 27 | VI | 4 | Tyr10, Val12, Phe20 |
| Cluster 31 | VII | 1 | Lys16, Val18, Ile32, Leu34, Gln15 |
| Cluster 31 | VII | 2 | His14, Gln15, Lys16 |
| Cluster 31 | VII | 3 | His14, Ile41, Gln15 |
| Cluster 33 | IX | 1 | Gln15, Leu17, Gly33 |
| Cluster 33 | IX | 2 | Gln15, Leu17, Ser8, Gly9 |
| Cluster 33 | IX | 3 | Asp7, Ser8, Val18, Leu34 |
| Cluster 35 | X | 1 | Ala2, Leu17, Asn27, Met3, Val36 |
| Cluster 35 | X | 2 | Leu17, Phe19, Phe20 |
| Cluster 35 | X | 3 | Ile31, Val36, Val40 |
Figure 6.
Illustration of the ten binding pockets identified by fragment-probe mapping in the ten most populated clusters (see Table 1) within the Aβ42 structural ensemble used in this work.
Figure 7.
Examples of adjacent binding pockets in the soluble monomeric form of the Aβ42 peptide identified through the approach described in this work. Results for clusters 2 and 35 (see Fig. 4) are shown together with a characterisation of the corresponding hot spots, the potential ligand efficiency (Le) (in kcal/mol, see Methods).
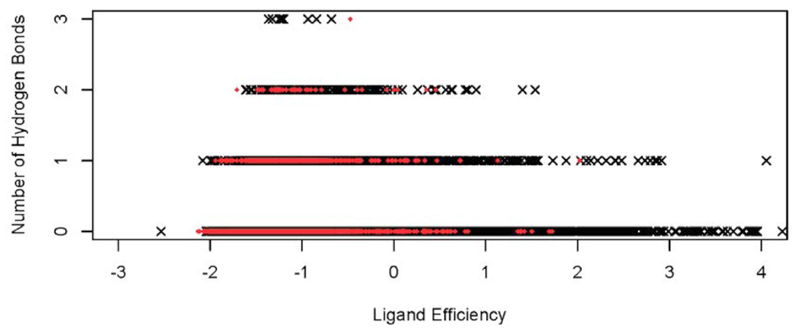
We then examined those hot spots that were found to bind three or more different fragments. These hot spots exhibited potential ligand efficiency values (Le) ranging from -0.8 to -1.5 kcal/mol, which are comparable to the ones that we observed for the model globular proteins that we studied (-1.3 to -1.5 kcal/mol, see Methods and Tables S1 and S2 in the Supplementary Information90), and to those reported in the literature94. Consistent with previous observations89, we also found the number of hydrogen bonds formed by the fragments to be comparable to the number typically found in folded proteins, thus providing insight into the origin of the enthalpic contributions to binding (Figure 8). On average, we found a value of slightly less than 1 hydrogen bond per fragment per hot spot for the Aβ42 peptide, which is similar to that observed for fragments of the same size extracted from high resolution structures in the PDB89.
Figure 8.
Comparison between the potential ligand efficiency (Le) (x-axis, in kcal/mol, see Methods) and the number of hydrogen bonds (y axis) for all the poses of the fragments in the binding hot spots identified in this work within the Aβ42 structural ensemble and those in model globular proteins (see Methods and Tables S1 and S2 in the Supplementary Information90); the red circles and numbers correspond to the hot-spot IDs listed in Table 1.
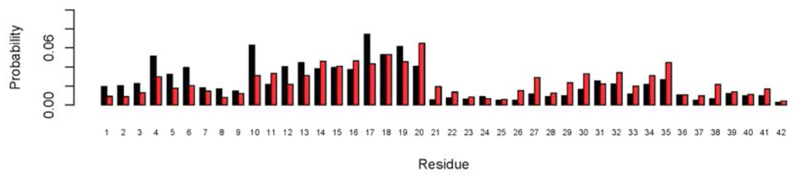
Further examination of these results revealed that the Aβ42 peptide exhibits regions of the amino acid sequence with different propensity to bind small molecular fragments (Figure 9). In particular, the central hydrophobic cluster (CHC) region (residues Leu17-Ala21) has a high propensity to form binding hot spots and to bind small molecular fragments. We found that residues Phe4, Tyr10, Leu17, Phe19, Ile31 and Met35 are involved in many of the hot spots identified in the mapping. A list of the binding sites, corresponding hot spots and key residues involved is provided in Table 1. It is interesting to note that the type of residues (in particular Phe, Tyr, Leu, Ile and Met) involved in forming binding sites identified here for an intrinsically disordered peptide correspond quite closely with those that have been described for structured proteins95, 96.
Figure 9.
Residue-specific probability of binding small molecular fragments in hot spots of the Aβ42 peptide, calculated by FTMap (see Methods). Non-bonded (black bars) and hydrogen bond (red bars) interactions are shown separately. The central hydrophobic region (CHC, horizontal red bar, residues Leu17-Ala21) is particular involved in hot spot formation.
Small-molecule interactions with potential binding pockets
We identified potential binding pockets by clustering neighbouring binding hot spots. To examine the significance of these potential binding pockets, we performed docking studies of two compounds, curcumin and Congo red, that have been shown to inhibit the aggregation of the Aβ peptide97, 98. Although the mechanism of action of these compounds on the behaviour of the Aβ peptide is still unclear, there is evidence suggesting that the peptide may bind to them either individually or in small oligomeric assemblies formed through detergent-like interactions97–99.
We performed docking calculations for the two compounds for possible binding modes in a systematic manner within the binding pockets that we identified in this work (Table 1 and Figure 6). The resulting top binding modes exhibit peptide-ligand interaction energies comparable with those seen in studies of globular proteins (see Methods). Two examples of such top binding modes are illustrated in Figure 10. We found that the aromatic rings of these compounds play an important role in binding to the hot spots, in particular through ring stacking interactions with the side-chains of residues in the hot spots themselves (e.g. Phe4, His6, Tyr10, Phe19 and Phe20). These interactions have been suggested as being an important feature in other small molecules capable of binding the Aβ peptide, such as polyphenols100, catechins101 and ketones102.
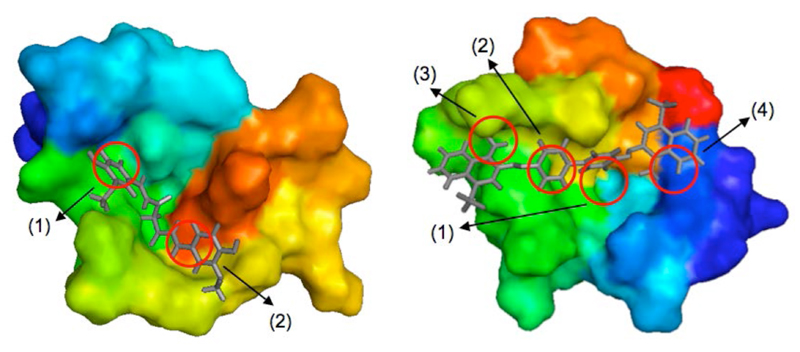
Figure 10.
Top binding modes of curcumin (left) and Congo red (right) with the Aβ42 peptide, which were identified through the analysis of the fragment-based mapping of the binding hot spots; hot spot labels refer to Table 1 and Figure 6 (pocket II in cluster 2 for curcumin and pocket V in cluster 18 for Congo red). Molecular dynamics simulations of the complexes show that the ligands remain bound over a 80 ns period.
We then carried out molecular dynamics simulations in order to probe the tendency of the small molecules to remain bound to the identified binding pockets (see Methods). These simulations revealed that at least in some of these complexes the ligands remain bound to the Aβ42 peptide over a period of 80 ns at room temperature. The results of these simulations indicate that the interactions of the ligands with many of the residues identified in the hot spots are key to strong binding.
Discussion and Conclusions
The drug discovery approach that we have described in this work is based on the idea of extending to intrinsically disordered proteins the well-established observation that a significant proportion of the free energy of binding in conventional protein-ligand complexes derives from relatively small regions of the protein surface, known as hot spots103. Ligands that bind simultaneously to multiple hot spots could result in higher binding affinities and better specificity. In the case of intrinsically disordered peptides and proteins, such as the Aβ42 peptide considered here, the additional complication is that these systems do not populate a small number of specific conformations, but rather experience conformational fluctuations of large amplitude. In these cases, the existence of hot spots requires careful examination, and so the aim of this work has been to explore this idea and identify compounds capable of binding to specific pockets in particular conformations. We have therefore carried out screens of small molecules by identifying a series of representative highly-populated clusters of conformations within the Aβ42 structural ensemble.
The concept that intrinsically disordered proteins are potentially druggable is relatively recent6–10. Our results indicate that the conformational space populated by the Aβ42 peptide may contain specific structures with significant statistical weights, and that such conformations may contain binding pockets that can be targeted by small molecules. The overall dimension of the Aβ42 peptide in its monomeric form in solution is rather more compact than a random coil, and transient hydrophobic pockets exist most likely as a result of the high propensity of certain regions to form turns and of interactions between hydrophobic regions in the amino acid sequence of the peptide. The compactness and long-range contacts are important for forming potential binding pockets because they provide the environment for favourable hydrogen bond, electrostatic, hydrophobic or van der Waals interactions as well as shape complementarity with a ligand. One could also expect the type of binding pockets that we have identified here for the monomeric form of the Aβ42 peptide to appear in more structured assemblies formed by this peptide, including oligomeric, membrane-bound and fibrillar conformations. It is possible that small molecules designed to bind the monomeric form would also, and less transiently, bind such assemblies, as their more ordered nature could reduce the entropic penalty of binding.
We found that hot spot formation is assisted particularly by the N-terminal and CHC regions, as illustrated in Figure 9. Phe4 and Tyr10 are closely involved in hot spot formation, together with Leu17, Phe19, Ile31 and Met35, which interact with each other to form favourable pockets that could be suitable for binding small molecules. Two of the residues that we identified as involved in hot spots formation, Phe19 and Met35, are known to be particularly important in the aggregation process of the Aβ42 peptide, as mutation of either of the two residues has been shown to inhibit oligomer and fibril formation104, 105. It is intriguing to speculate that finding brain-penetrable small molecules that could bind to a pocket formed by these residues may have a significant effect in inhibiting the aggregation of the Aβ42 peptide.
From a methodological point of view, the identification of clusters of conformations populated by the Aβ42 peptide in its monomeric form in solution allows the number of structures to be searched to find potential binding sites to be significantly reduced. Such reduction in search space is crucial since the screening procedure is computationally costly. The application of this strategy to identify of binding pockets in the case of the Aβ42 peptide (Table 1 and Figure 6) has enabled us to provide a list of candidate binding pockets, which will in turn make it possible to perform virtual screening of large numbers of drug-like molecules that are likely to bind better at these sites.
The work presented here represents an initial step toward targeting the Aβ42 peptide in its monomeric form, by demonstrating that it exhibits potential small molecule binding sites. One development of this approach will be to improve the accuracy of the Aβ42 structural ensemble by incorporating experimental data in the molecular dynamics simulations in a way similar to that used for example for α-synuclein18, 19, 51. Using the potential binding sites identified on different representative structures, the next step of this drug design strategy will be to conduct a structure based high-throughput docking screening of small-molecules to these, and verification of in silico hits by in vitro and in vivo studies.
Supplementary Material
Acknowledgements
This work was supported by grants from Alzheimer’s Research UK (YC) and BBSRC (CMD and MV). We are grateful to Tobin Sosnick and his group for providing the SCM structures of the Aβ42 peptide.
References
- [1].Chiti F, Dobson CM. Annu Rev Bioch. 2006;75:333. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- [2].Haass C, Selkoe DJ. Nat Rev Mol Cell Biol. 2007;8:101. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- [3].Prince M, Bryce R, Ferri C. Alzheimer's Disease lnternational. 2011 [Google Scholar]
- [4].Balch WE, Morimoto RI, Dillin A, Kelly JW. Science. 2008;319:916. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- [5].Dobson CM. Science. 2004;304:1259. doi: 10.1126/science.1093078. [DOI] [PubMed] [Google Scholar]
- [6].Cheng Y, LeGall T, Oldfield CJ, Mueller JP, Van YYJ, Romero P, Cortese MS, Uversky VN, Dunker AK. Trends Biotech. 2006;24:435. doi: 10.1016/j.tibtech.2006.07.005. [DOI] [PubMed] [Google Scholar]
- [7].Citron M. Nat Rev Drug Disc. 2010;9:387. doi: 10.1038/nrd2896. [DOI] [PubMed] [Google Scholar]
- [8].Dunker AK, Uversky VN. Curr Op Pharmac. 2010;10:782. doi: 10.1016/j.coph.2010.09.005. [DOI] [PubMed] [Google Scholar]
- [9].Metallo SJ. Curr Op Chem Biol. 2010;14:481. doi: 10.1016/j.cbpa.2010.06.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tompa P. Curr Op Struct Biol. 2011;21:419. doi: 10.1016/j.sbi.2011.03.012. [DOI] [PubMed] [Google Scholar]
- [11].Yamin G, Ono K, Inayathullah M, Teplow DB. Curr Pharm Des. 2008;14:3231. doi: 10.2174/138161208786404137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo JS, Taddei N, Ramponi G, Dobson CM, Stefani M. Nature. 2002;416:507. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- [13].Cremades N, Cohen SIA, Deas E, Abramov AY, Chen AY, Orte A, Sandal M, Clarke RW, Dunne P, Aprile FA, Bertoncini CW, et al. Cell. 2012;149:1048. doi: 10.1016/j.cell.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. J Mol Biol. 2006;362:1043. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- [15].Dosztanyi Z, Meszaros B, Simon I. Bioinformatics. 2009;25:2745. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Arkin MR, Wells JA. Nat Rev Drug Disc. 2004;3:301. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- [17].Whittle PJ, Blundell TL. Annu Rev Bioph Biomol Struct. 1994;23:349. doi: 10.1146/annurev.bb.23.060194.002025. [DOI] [PubMed] [Google Scholar]
- [18].Bertoncini CW, Jung YS, Fernandez CO, Hoyer W, Griesinger C, Jovin TM, Zweckstetter M. Proc Natl Acad Sci USA. 2005;102:1430. doi: 10.1073/pnas.0407146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dedmon MM, Lindorff-Larsen K, Christodoulou J, Vendruscolo M, Dobson CM. J Am Chem Soc. 2005;127:476. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- [20].Mukrasch MD, Markwick P, Biernat J, von Bergen M, Bernado P, Griesinger C, Mandelkow E, Zweckstetter M, Blackledge M. J Am Chem Soc. 2007;129:5235. doi: 10.1021/ja0690159. [DOI] [PubMed] [Google Scholar]
- [21].Hou LM, Shao HY, Zhang YB, Li H, Menon NK, Neuhaus EB, Brewer JM, Byeon IJL, Ray DG, Vitek MP, Iwashita T, et al. J Am Chem Soc. 2004;126:1992. doi: 10.1021/ja036813f. [DOI] [PubMed] [Google Scholar]
- [22].Zhang S, Iwata K, Lachenmann MJ, Peng JW, Li S, Stimson ER, Lu Y, Felix AM, Maggio JE, Lee JP. J Struct Biol. 2000;130:130. doi: 10.1006/jsbi.2000.4288. [DOI] [PubMed] [Google Scholar]
- [23].Makin OS, Serpell LC. FEBS J. 2005;272:5950. doi: 10.1111/j.1742-4658.2005.05025.x. [DOI] [PubMed] [Google Scholar]
- [24].Tycko R. Annu Rev Phys Chem. 2011;62:279. doi: 10.1146/annurev-physchem-032210-103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hajduk PJ, Greer J. Nat Rev Drug Disc. 2007;6:211. doi: 10.1038/nrd2220. [DOI] [PubMed] [Google Scholar]
- [26].Congreve M, Chessari G, Tisi D, Woodhead AJ. J Med Chem. 2008;51:3661. doi: 10.1021/jm8000373. [DOI] [PubMed] [Google Scholar]
- [27].Murray CW, Rees DC. Nat Chem. 2009;1:187. doi: 10.1038/nchem.217. [DOI] [PubMed] [Google Scholar]
- [28].Murray CW, Blundell TL. Curr Op Struct Biol. 2010;20:497. doi: 10.1016/j.sbi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- [29].Ball KA, Phillips AH, Nerenberg PS, Fawzi NL, Wemmer DE, Head-Gordon T. Biochemistry. 2011;50:7612. doi: 10.1021/bi200732x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chebaro Y, Mousseau N, Derreumaux P. J Phys Chem B. 2009;113:7668. doi: 10.1021/jp900425e. [DOI] [PubMed] [Google Scholar]
- [31].Convertino M, Vitalis A, Caflisch A. J Biol Chem. 2011;286:41578. doi: 10.1074/jbc.M111.285957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Krone MG, Baumketner A, Bernstein SL, Wyttenbach T, Lazo ND, Teplow DB, Bowers MT, Shea JE. J Mol Biol. 2008;381:221. doi: 10.1016/j.jmb.2008.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lam AR, Teplow DB, Stanley HE, Urbanc B. J Am Chem Soc. 2008;130:17413. doi: 10.1021/ja804984h. [DOI] [PubMed] [Google Scholar]
- [34].Mitternacht S, Staneva I, Hard T, Irback A. Proteins. 2010;78:2600. doi: 10.1002/prot.22775. [DOI] [PubMed] [Google Scholar]
- [35].Sgourakis NG, Merced-Serrano M, Boutsidis C, Drineas P, Du ZM, Wang CY, Garcia AE. J Mol Biol. 2011;405:570. doi: 10.1016/j.jmb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sgourakis NG, Yan YL, McCallum SA, Wang CY, Garcia AE. J Mol Biol. 2007;368:1448. doi: 10.1016/j.jmb.2007.02.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Urbanc B, Cruz L, Yun S, Buldyrev SV, Bitan G, Teplow DB, Stanley HE. Proc Natl Acad Sci USA. 2004;101:17345. doi: 10.1073/pnas.0408153101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Vitalis A, Caflisch A. J Mol Biol. 2010;403:148. doi: 10.1016/j.jmb.2010.08.003. [DOI] [PubMed] [Google Scholar]
- [39].Herrera FE, Chesi A, Paleologou KE, Schmid A, Munoz A, Vendruscolo M, Gustincich S, Lashuel HA, Carloni P. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cuchillo R, Michel J. Biochem Soc Trans. 2012;40:1004. doi: 10.1042/BST20120086. [DOI] [PubMed] [Google Scholar]
- [41].Lin JH, Perryman AL, Schames JR, McCammon JA. J Am Chem Soc. 2002;124:5632. doi: 10.1021/ja0260162. [DOI] [PubMed] [Google Scholar]
- [42].Kitchen DB, Decornez H, Furr JR, Bajorath J. Nat Rev Drug Disc. 2004;3:935. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- [43].Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, et al. J Med Chem. 2006;49:5912. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- [44].Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, et al. J Med Chem. 2004;47:1739. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- [45].Ewing TJA, Makino S, Skillman AG, Kuntz ID. J Comp Aid Mol Des. 2001;15:411. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- [46].Thomsen R, Christensen MH. J Med Chem. 2006;49:3315. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- [47].Sinko W, Lindert S, McCammon JA. Chem Biol Drug Des. 2013;81:41. doi: 10.1111/cbdd.12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kaufmann KW, Meiler J. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jha AK, Colubri A, Freed KF, Sosnick TR. Proc Natl Acad Sci USA. 2005;102:13099. doi: 10.1073/pnas.0506078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ozenne V, Bauer F, Salmon L, Huang J-R, Jensen MR, Segard S, Bernado P, Charavay C, Blackledge M. Bioinformatics. 2012;28:1463. doi: 10.1093/bioinformatics/bts172. [DOI] [PubMed] [Google Scholar]
- [51].Allison JR, Varnai P, Dobson CM, Vendruscolo M. J Am Chem Soc. 2009;131:18314. doi: 10.1021/ja904716h. [DOI] [PubMed] [Google Scholar]
- [52].Cavalli A, Camilloni C, Vendruscolo M. J Chem Phys. 2013;138 doi: 10.1063/1.4793625. [DOI] [PubMed] [Google Scholar]
- [53].Hansmann UHE, Okamoto Y. Curr Op Struct Biol. 1999;9:177. doi: 10.1016/S0959-440X(99)80025-6. [DOI] [PubMed] [Google Scholar]
- [54].Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. J Comp Chem. 2005;26:1701. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- [55].De Simone A, Derreumaux P. J Chem Phys. 2010;132 doi: 10.1063/1.3385470. [DOI] [PubMed] [Google Scholar]
- [56].Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Proteins. 2006;65:712. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926. [Google Scholar]
- [58].Best RB, Hummer G. J Phys Chem B. 2009;113:9004. doi: 10.1021/jp901540t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Showalter SA, Bruschweiler R. J Am Chem Soc. 2007;129:4158. doi: 10.1021/ja070658d. [DOI] [PubMed] [Google Scholar]
- [60].Elcock AH, McCammon JA. Bioph J. 2001;80:613. doi: 10.1016/S0006-3495(01)76042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Park H, Lee S. J Am Chem Soc. 2003;125:16416. doi: 10.1021/ja0304493. [DOI] [PubMed] [Google Scholar]
- [62].ten Brink T, Exner TE. J Chem Inf Mod. 2009;49:1535. doi: 10.1021/ci800420z. [DOI] [PubMed] [Google Scholar]
- [63].Newby F. University of Cambridge: 2012. PhD Thesis. [Google Scholar]
- [64].Lim KH, Henderson GL, Jha A, Louhivuori M. ChemBioChem. 2007;8:1251. doi: 10.1002/cbic.200700194. [DOI] [PubMed] [Google Scholar]
- [65].Kohlhoff KJ, Robustelli P, Cavalli A, Salvatella X, Vendruscolo M. J Am Chem Soc. 2009;131:13894. doi: 10.1021/ja903772t. [DOI] [PubMed] [Google Scholar]
- [66].Brenke R, Kozakov D, Chuang GY, Beglov D, Hall D, Landon MR, Mattos C, Vajda S. Bioinformatics. 2009;25:621. doi: 10.1093/bioinformatics/btp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ivetac A, McCammon JA. Chem Biol Drug Des. 2010;76:201. doi: 10.1111/j.1747-0285.2010.01012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hoffer L, Renaud JP, Horvath D. Comb Chem High Throughput Screening. 2011;14:500. doi: 10.2174/138620711795767884. [DOI] [PubMed] [Google Scholar]
- [69].Lexa KW, Carlson HA. Q Rev Bioph. 2012;45:301. doi: 10.1017/S0033583512000066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kumar A, Voet A, Zhang KYJ. Curr Med Chem. 2012;19:5128. doi: 10.2174/092986712803530467. [DOI] [PubMed] [Google Scholar]
- [71].Goodford PJ. J Med Chem. 1985;28:849. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- [72].Miranker A, Karplus M. Proteins. 1991;11:29. doi: 10.1002/prot.340110104. [DOI] [PubMed] [Google Scholar]
- [73].Meiler J, Baker D. Proteins. 2006;65:538. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- [74].Lexa KW, Carlson HA. J Am Chem Soc. 2011;133:200. doi: 10.1021/ja1079332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].McGann M. J Chem Inf Mod. 2011;51:578. doi: 10.1021/ci100436p. [DOI] [PubMed] [Google Scholar]
- [76].Jones G, Willett P, Glen RC, Leach AR, Taylor R. J Mol Biol. 1997;267:727. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- [77].Boyd SM, Turnbull AP, Walse B. WIREs Comput Mol Sci. 2012;2:868. [Google Scholar]
- [78].McGann MR, Almond HR, Nicholls A, Grant JA, Brown FK. Biopolymers. 2003;68:76. doi: 10.1002/bip.10207. [DOI] [PubMed] [Google Scholar]
- [79].Deng Y, Roux B. J Phys Chem B. 2009;113:2234. doi: 10.1021/jp807701h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Gallicchio E, Lapelosa M, Levy RM. J Chem Theor Comp. 2010;6:2961. doi: 10.1021/ct1002913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Mobley DL, Dill KA. Structure. 2009;17:489. doi: 10.1016/j.str.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Muddana HS, Varnado CD, Bielawski CW, Urbach AR, Isaacs L, Geballe MT, Gilson MK. J Comp Aid Mol Des. 2012;26:475. doi: 10.1007/s10822-012-9554-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Wereszczynski J, McCammon JA. Q Rev Bioph. 2012;45:1. doi: 10.1017/S0033583511000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Purisima EO, Hogues H. J Phys Chem B. 2012;116:6872. doi: 10.1021/jp212646s. [DOI] [PubMed] [Google Scholar]
- [85].Halgren TA. J Comp Chem. 1999;20:720. doi: 10.1002/(SICI)1096-987X(199905)20:7<720::AID-JCC7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- [86].Wlodek S, Skillman AG, Nicholls A. J Chem Theor Comp. 2010;6:2140. doi: 10.1021/ct100095p. [DOI] [PubMed] [Google Scholar]
- [87].Hopkins AL, Groom CR, Alex A. Drug Disc Today. 2004;9:430. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- [88].Reynolds CH, Holloway MK. ACS Med Chem Lett. 2011;2:433. doi: 10.1021/ml200010k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ferenczy GG, Keseru GM. J Chem Inf Mod. 2012;52:1039. doi: 10.1021/ci200608b. [DOI] [PubMed] [Google Scholar]
- [90].See Supplementary Material Document No.______ for the results on the potential ligand efficiency values.
- [91].DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
- [92].Kirchmair J, Wolber G, Laggner C, Langer T. J Chem Inf Mod. 2006;46:1848. doi: 10.1021/ci060084g. [DOI] [PubMed] [Google Scholar]
- [93].Danielsson J, Jarvet J, Damberg P, Graslund A. Mag Res Chem. 2002;40:S89. [Google Scholar]
- [94].Kuntz ID, Chen K, Sharp KA, Kollman PA. Proc Natl Acad Sci USA. 1999;96:9997. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Ma BY, Elkayam T, Wolfson H, Nussinov R. Proc Natl Acad Sci USA. 2003;100:5772. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Keskin O, Ma BY, Nussinov R. J Mol Biol. 2005;345:1281. doi: 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- [97].Lendel C, Bolognesi B, Wahlstrom A, Dobson CM, Graslund A. Biochemistry. 2010;49:1358. doi: 10.1021/bi902005t. [DOI] [PubMed] [Google Scholar]
- [98].Yang FS, Lim GP, Begum AN, Ubeda OJ, Simmons MR, Ambegaokar SS, Chen PP, Kayed R, Glabe CG, Frautschy SA, Cole GM. J Biol Chem. 2005;280:5892. doi: 10.1074/jbc.M404751200. [DOI] [PubMed] [Google Scholar]
- [99].Pedersen MO, Mikkelsen K, Behrens MA, Pedersen JS, Enghild JJ, Skrydstrup T, Malmendal A, Nielsen NC. J Phys Chem B. 2010;114:16003. doi: 10.1021/jp108035y. [DOI] [PubMed] [Google Scholar]
- [100].Porat Y, Abramowitz A, Gazit E. Chem Biol Drug Des. 2006;67:27. doi: 10.1111/j.1747-0285.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- [101].Ehrnhoefer DE, Bieschke J, Boeddrich A, Herbst M, Masino L, Lurz R, Engemann S, Pastore A, Wanker EE. Nat Struct Mol Biol. 2008;15:558. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- [102].Zheng XY, Gessel MM, Wisniewski ML, Viswanathan K, Wright DL, Bahr BA, Bowers MT. J Biol Chem. 2012;287:6084. doi: 10.1074/jbc.C111.328575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Clackson T, Wells JA. Science. 1995;267:383. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- [104].Bernstein SL, Wyttenbach T, Baumketner A, Shea JE, Bitan G, Teplow DB, Bowers MT. J Am Chem Soc. 2005;127:2075. doi: 10.1021/ja044531p. [DOI] [PubMed] [Google Scholar]
- [105].Bitan G, Tarus B, Vollers SS, Lashuel HA, Condron MM, Straub JE, Teplow DB. J Am Chem Soc. 2003;125:15359. doi: 10.1021/ja0349296. [DOI] [PubMed] [Google Scholar]
- [106].Kabsch W, Sander C. Biopolymers. 1983;22:2577. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.