Abstract
Beyond the canonical role in efficient ATP production through oxidative metabolism, mitochondria are increasingly recognized as critical in defining stem cell function and fate. Implicating a fundamental interplay within the epigenetics of eukaryotic cell systems, the integrity of mitochondria is found vital across the developmental/differentiation spectrum from securing pluripotency maintenance to informing organotypic decisions. This overview will discuss recent progress on examining the plasticity of mitochondria in enabling the execution of programming and reprogramming regimens, as well as the application of nuclear reprogramming and somatic cell nuclear transfer as rescue techniques to generate genetically and functionally corrected pluripotent stem cells from patients with mitochondria DNA-based disease.
Introduction
Pluripotent stem cells are defined by their capacity for indefinite self-renewal and the ability to give rise to all cell types within the three embryonic germ layers. This unique class of cells is epitomized by embryonic stem cells (ESCs), which are derived from the inner cell mass of the blastocyst stage of the embryo. Beyond the natural potential in deriving diverse lineages, the ability to revert the fate of a specialized differentiated cell back to a pluripotent ground state has the potential to revolutionize the fields of stem cell biology and regenerative medicine, and hence garnered Sir John Gurdon and Shinya Yamanaka the Nobel Prize in Physiology and Medicine. From Gurdon’s initial work cloning frogs to optimization of the technique in sheep and other mammals has laid the groundwork for recent developments in utilizing somatic cell nuclear transfer (SCNT) into human oocytes for the derivation of stem cells [1]. This approach has been complemented by the discovery that nuclear reprogramming induced through expression of four embryonic transcription factors, Oct4, Sox2, KLF4 and cMyc, is sufficient to reset differentiated cells back into induced pluripotent stem cells (iPSCs), which mimic the features of their embryonic counterparts [2]. These discoveries have enabled the generation of patient-specific pluripotent stem cells, which offer unique platforms to examine mechanisms of disease pathogenesis in a dish, to screen small molecules to identify novel therapeutics for difficult to treat diseases, as well as for toxicity screening in lineages specified from pluripotent stem cells. Ultimately, these cells offer an unlimited and autologous cell source for regenerative applications across degenerative diseases for which curative therapies are currently lacking (Table 1).
Table 1.
Comparison of different pluripotent stem cell types
| IVF-ESCs | NT-ESCs | iPSCs | |
|---|---|---|---|
| Stem cell-based therapy | Heterologous | Autologous | Autologous |
| mtDNA origin | Oocyte | Oocyte | Somatic cells |
| mtDNA mutation frequency | Germline, Low | Germline Low | Somatic, accumulate with age |
| Reprogramming | Natural oocyte factors | Natural oocyte factors | Induced by transcription factors |
| de novo copy number variations (per cell line) | 0.5 | 0.8 | 1.8 |
| CG differentially methylated regions (DMRs) | Baseline control | 212 | 619 |
| Recurrent (hotspot) CG DMRs | Baseline control | 50 | 110 |
| Non-CG mega DMRs | Baseline control | 7 | 70 |
| Differentially expressed genes | Baseline control | 48 | 629 |
Over the past decade the field has made great strides in understanding the genetic and epigenetic mechanism by which nuclear reprogramming can reset the fate of a differentiated cell. Complementing these studies, is the emerging appreciation that mitochondrial function and energy metabolism are tightly linked to the fate and function of a stem cell. In this review we will discuss recent findings underscoring the enabling role of mitochondria and their dynamics in the acquisition of the pluripotent state and how nuclear reprogramming and SCNT can be leveraged to derive pluripotent stem cells from patients with mitochondrial DNA (mtDNA)-based disease.
Mitochondria as stemness regulators
Fundamental to nuclear reprogramming is the reduction in mtDNA copy numbers and regression in mitochondrial density, distribution and ultrastructure [3–5], events that collectively recapitulate the mitochondrial features of ESCs [6]. Indeed recent evidence indicates that mitochondrial clearance through Atg5-independent autophagy is essential for pluripotency induction and generation of iPSCs [7]. Moreover, proteomic profiling has identified a reduction in subunit expression of complex I and IV and an increase in II, III, V of the mitochondrial electron transport chain as an early reprogramming event preceding remodeling of other metabolic pathways and expression of pluripotency genes, indicating that mitochondrial remodeling is not simply a consequence of transition between cell identities, but may represent an initiating event [3,8]. Functionally, this transition manifests as a suppression of cellular respiration in favor of glycolysis in iPSCs, with somatic sources having a greater glycolytic and lower oxidative capacity displaying a higher reprogramming efficiency [3,5]. Although on the surface mitochondria-associated plasticity may be interpreted to indicate that pluripotent stem cells may minimize their requirement for mitochondria, it has been demonstrated that mitochondrial homeostasis is necessary for maintenance of the pluripotent state as excessive mitochondrial fission or knockdown of the mtDNA specific polymerase gamma leads to loss of pluripotency [9,10]. Stem cells actually appear to actively maintain their mitochondria, potentially even hydrolyzing ATP through ATP synthase to support high mitochondrial membrane potential [3,11–14]. Consistent with these observations, stem-like cells asymmetrically segregate their mitochondria during cell division, with a greater proportion of young mitochondria observed in daughter cells displaying stem cell traits, while impaired segregation leading to loss of stem cell properties in the cell progeny [15]. Therefore stem cells may repurpose mitochondria from their canonical role of energy generators to alternative functions in support of stem cell function and maintenance of pluripotency.
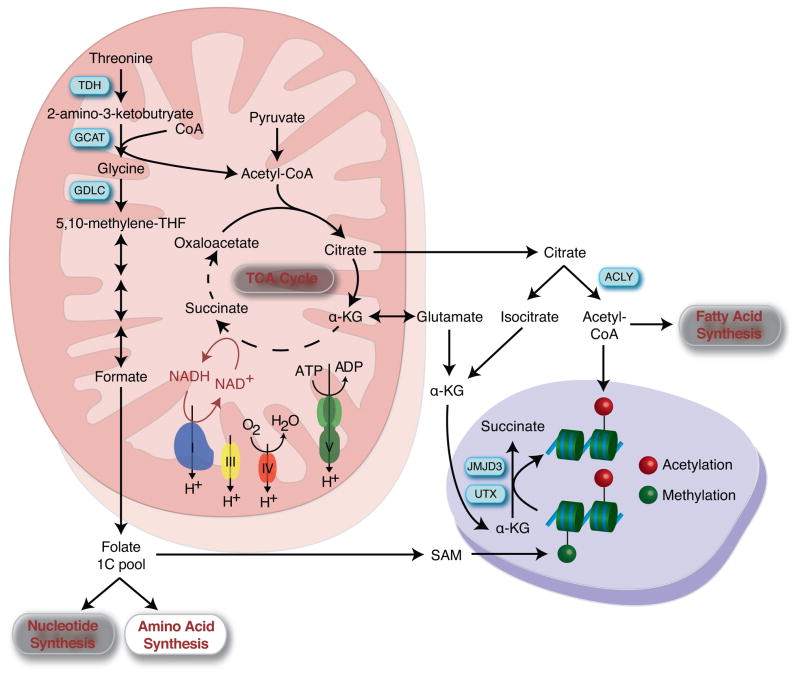
In pluripotent stem cells, like other populations of rapidly proliferating cells, the demand for anabolic precursors for biosynthesis of cellular component may outpace their requirement for energy in the form of ATP [16]. Therefore reducing their dependence on mitochondria-dependent ATP generation may enable stem cells to commission alternative mitochondrial pathways to support stemness, such as elevated cataplerosis, the extraction of incompletely oxidized substrates from the mitochondria, to serve as substrates for anabolic pathways and post-translational protein modifications [17]. ESCs do not appear to completely oxidize glycolysis-derived pyruvate to CO2 and H20 in the tricarboxylic acid cycle (TCA), but utilize pyruvate to generate TCA intermediates that can be exported out of the mitochondria and utilized for alternative purposes [18]. For example, mitochondrial-derived citrate can be exported from the mitochondria and subsequently metabolized by ATP-citrate lyase to generate cytosolic acetyl-CoA, which serves as a substrate for protein and histone acetylation, as well as for fatty acid and cholesterol biosynthesis [19]. Early differentiation of ESCs is associated with a reduction in acetyl-CoA production and loss of histone H3 lysine 9 and lysine 27 acetylation, indicating that TCA-derived cytosolic acetyl-CoA may be critical for maintaining histone acetylation and an open chromatin state during pluripotency [18]. Functionally, inhibition of ATP citrate lyase reduces acetyl-CoA content and histone acetylation results in induction of myogenic differentiation [20], while acetate supplementation impairs both early differentiation capacity and histone deacetylation [18]. However this axis may not be exclusive to the pluripotent state as ATP-citrate lyase derived acetyl-CoA and increased histone acetylation is also associated with adipogenesis [19]. ESCs also metabolize both glucose and glutamine to generate alpha-ketoglutarate, another key metabolite that connects metabolism with the epigenetic regulation of the pluripotent state [21]. A high alpha-ketoglutarate/succinate ratio is critical in regulating a number of enzymes that couple the conversion of alpha-ketoglutarate to succinate with their specific functions, such as HIFα prolyl hydroxylase and a class of alpha-ketoglutarate dependent dioxygenases, which include Jumonji C-domain-containing histones demethylases and the ten-eleven translocation (TET) family of DNA demethylases. Therefore high levels of alpha-ketoglutarate in ESCs favors demethylation of repressive chromatin marks including histone 3 lysine 9 and lysine 27 timethylation and histone 4 lysine 20 trimethylation and maintenance of the pluripotent state [21]. Although these alternate roles of TCA cycle intermediates are established in maintaining pluripotency, it remains unexamined how they contribute to the acquisition of the pluripotent state during nuclear reprogramming, where global changes in DNA methylation and histone marks are occurring.
Nuclear reprogramming does significantly modify other metabolic pathways that regulate the epigenetic state, including increases in the expression of mitochondrial enzymes required for threonine catabolism, such as threonine dehydrogenase (TDH), glycine C-acetyltransferase (GCAT) and glycine decarboxylase (GLDC) [22]. Activation of this pathway results in elevated conversion of threonine into acetyl-CoA and glycine, the latter of which is metabolized to 5,10-methylenetetrahydrofolate to supply single carbon equivalents to the folate pool. This pool then provides these one-carbon units to support the biosynthesis of nucleotides and amino acids, as well as generation of S-adenosylmethionine (SAM), which serves as the substrate for histone H3 lysine 4 trimethylation, a critical component for self-renewal of pluripotent stem cells [22]. Compromising this pathway either through threonine restriction or modulation of TDH activity leads to loss of ESC colony growth and impairs nuclear reprogramming [22–25]. Although humans do not express functional TDH, a parallel pathway relying on methionine appears to fuel the SAM pathway [26]. Taken together, by minimizing their reliance on mitochondrial oxidative ATP generation, pluripotent stem cells can exploit alternative mitochondrial functions to maintain stemness.
Correcting mitochondrial DNA defects in pluripotent stem cells
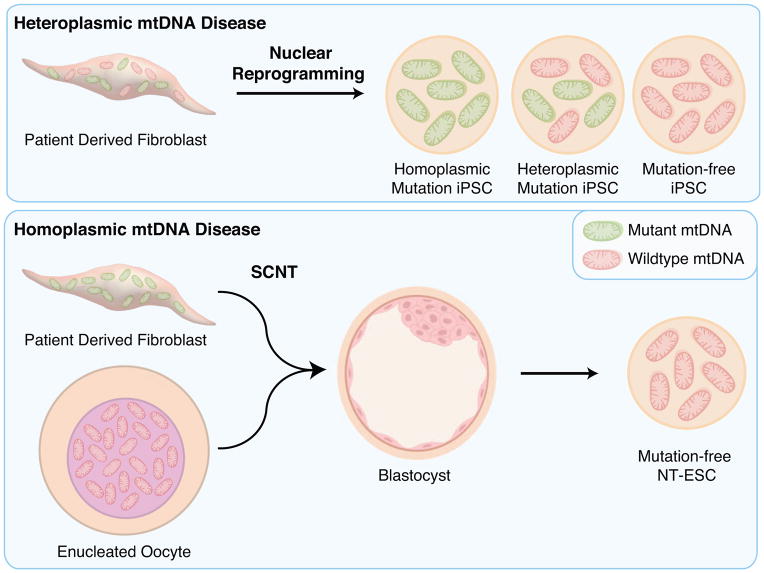
Mutations in mtDNA contribute to a wide range of life-threatening conditions. Among well-described mtDNA diseases are maternally inherited germline mutations associated with defects in oxidative energy metabolism [27]. Pathogenic mtDNA mutations may reside in tRNA, rRNA or protein coding genes and often persist in each cell as a mixture of mutated and healthy wild type molecules, a state known as heteroplasmy. Often, these two mtDNA types are unevenly segregated during mitosis, resulting in daughter cells with significantly different levels of heteroplasmy, which offers an opportunity to select against cells with high mutations loads. Mitochondrial dynamics underlying pluripotent induction, including a significant mtDNA copy number reduction followed by clonal cell expansion, in essence recapitulates a genetic bottleneck during nuclear reprogramming. As a consequence of this bottleneck, mtDNA type can drift toward homoplasmic mutant or wild type in individual iPSC clones. This phenomenon provides the opportunity to recover iPSC clones with normal mtDNA from patients with heteroplasmic mtDNA mutations [28–31]. However, this option is not sufficient when mtDNA mutation is homoplasmic or high heteroplasmic, and requires complementary approaches.
One strategy would be to correct the mtDNA mutation, and although initial proof of concept studies has demonstrated that mitochondrial-targeted endonucleases or TALENs may be utilized to shift heteroplasmy of mtDNA haplotypes/mutations in cells [32,33] and mouse embryos [34], whether these techniques can be utilized to generate mutation-free pluripotent stem cells have not been examined. Alternatively, a complete mitochondrial replacement strategy through SCNT may be required. Transplantation of a somatic cell nucleus into enucleated cytoplasm from unfertilized metaphase 2 (MII) arrested oocyte induces rapid epigenetic and cell cycle changes resulting in the erasure of somatic cell identity and formation of oocyte-like meiotic chromosomes. Reconstructed SCNT oocytes are functional as they can be triggered to develop into a diploid zygote and ultimately have been demonstrated to produce viable offspring in numerous mammalian species [35–38]. In addition, human SCNT oocytes can also be used to derive patient matched embryonic stem cells (NT-ESCs) [39]. A hallmark of NT-ESCs is that they are cytoplasmic hybrids (cybrids), which contain a somatic cell nuclear genome and the oocyte’s mtDNA. This fundamental feature of SCNT allows replacement of the entire mtDNA complement in patient somatic cells carrying mtDNA mutations while producing high quality pluripotent cells [39]. NT-ESCs carry almost exclusively oocyte mtDNA ensuring that stem cells regain normal OXPHOS function, irrespective of mtDNA mutation type or heteroplasmy level in starting somatic cells. Technically, a whole somatic cell is fused to a cytoplast resulting in a small somatic cell/oocyte mtDNA heteroplasmy. However, due to significant mtDNA copy number differences between somatic cells and MII oocytes (ratio <1:100) and a bottleneck effect during embryo and ESC development, the mutated mtDNA is diluted and often becomes practically undetectable in NT-ESCs [31]. A potential limitation of this approach is that the donor mtDNA haplotype in NT-ESCs may be different than the original in parental somatic cells due to natural sequence variation between the oocyte donors and patients affected by mtDNA mutations. Hypothetically, this may result in metabolic insufficiency in NT-ESCs due to inadequate interactions between the donor mitochondrial and patient nuclear genomes [40]. However, there is no direct evidence of mtDNA incompatibility between human populations including in children born in families from distant haplogroups. This suggests that normal nuclear-to-mitochondrial interactions are highly conserved within species and any human haplotype may serve as a functional donor mtDNA for mitochondrial replacement therapy [40]. Indeed, NT-ESCs carrying “unmatched” donor mtDNA displayed differentiation potential and transcriptome profiles similar to control ESCs produced from in vitro fertilized embryos (IVF-ESCs)[31]. In addition their metabolic functions were also normal suggesting that donor mtDNA interacts normally with patient nuclear DNA [31]. Although functionally similar, recent evidence indicates that murine NT-ESCs induced an immune response that leads to rejection when administered to recipients that have matched nuclear genomes, but mismatched mtDNA, indicating that non-cell-autonomous effects must also be considered [41] (Table 2).
Table 2.
Methods for eliminating mtDNA mutations
| Nuclear transfer (MRT) | Mito TALEN | |
|---|---|---|
| Efficiency of eliminating mutations | 80–99% | 65–85% |
| Applicable for germ line therapy | Yes | Yes |
| Applicable for heteroplasmic mutations | Yes | Yes |
| Applicable for homoplasmic mutations | Yes | No |
| Applicable for pluripotent stem cells | Yes | Not tested |
| Potential side effects | mtDNA haplotype mismatch | Off-target effects |
Both nuclear reprogramming and SCNT-based approaches to derive mutation free pluripotent stem cells from patients with mitochondrial disease offer potential benefits and limitations. While nuclear reprogramming is relatively simple to perform and does not require the same technical expertise as required for SCNT, NT-ESCs may more closely resemble IVF-ESCs, the ‘gold standard’ of pluripotency. Indeed, initial studies of transcription-factor-based nuclear reprogramming demonstrated that incomplete DNA methylation resulted in significant differences in the transcriptional signature of iPSCs versus ESCs [42–44]. In addition, iPSCs show significant differences in DNA methylation patterns when compared to genetically matched IVF-ESCs and NT-ESCs, including a three- and ten-fold exaggerated aberrancy in CG and non-CG methylation than NT-ESCs, respectively [45]. These studies suggest that oocyte-based reprogramming is more robust and capable of resetting the DNA methylation and corresponding gene expression program of somatic cells. While a separate study recapitulated the distinct clustering of these cell lines, they did not detect differences in coding mutations and loss of imprinting between iPSCs and NT-ESCs derived from other cells lines [46]. These different observations may be due to the iPSC derivation strategy and ages of the somatic cell source, but indicates that additional studies are required on a greater number of cell lines to examine the functional differences between iPSCs and NT-ESCs.
While the incidence of inherited mtDNA disease is relatively rare, acquired, somatic mtDNA mutations are more common and implicated in normal aging itself and in the etiology of diverse age-onset diseases including neurodegenerative conditions such as Parkinson’s and Alzheimer’s disease [47–51]. With age, mtDNA heteroplasmy can drift toward mutant types, particularly in post-mitotic neurons. When the proportion of mutant mtDNA reaches a critical threshold, the cell’s ability to supply minimum energy for vital functions is affected, leading to disease symptoms. Due to the random nature of somatic mtDNA mutations, the mutation type may differ among individual cells within tissues, making difficult to detect mtDNA mutations in pooled cells or tissues and may pose a risk of uncovering rare mitochondrial mutations due to the clonal nature of iPSC generation. In contrast, female germ cells have relatively quiescent mitochondria that mainly rely on the glycolytic energy supply rather than on OXPHOS. Therefore, a female germline may evade accumulation of mutations and provide purifying selection to generate oocytes and subsequent SCNT-ESCs with less mtDNA mutation burden.
Summary
The present overview highlights advances in mitochondrial biology in the context of an emerging understanding of mitochondria-dependent control of stemness in health and disease. Decoding intimate events that repurpose mitochondria from traditional energy generators to enablers of bi-directional transitions between somatic versus pluripotent cell identities has now open new avenues in developmental biology with potential applications in regenerative medicine. Case in point, resetting or replacing disease-corrupted mitochondria, through nuclear reprogramming and somatic cell nuclear transfer respectively, to produce healthy patient derived stem cells underscore the most recent breakthrough in the field paving the way towards targeting the cause and potentially establishing future cures for patients with inherited or age-related mtDNA disorders.
Figure 1.
Mitochondria contribute to stemness maintenance. By reducing their reliance on oxidative metabolism for ATP generation and potentially utilizing ATP to maintain mitochondria membrane potential, stem cells can repurpose their mitochondria to cataplerosis. These mitochondria-derived metabolites are utilized as precursors for the anabolic generation of cell building blocks and serve as substrates for reactions that add or remove epigenetic marks that are critical for directing cell fate.
Figure 2.
Creating disease-free pluripotent stem cells from patients with mitochondrial DNAN (mtDNA)-based disease. Nuclear reprogramming segregates wildtype and mutant mt-DNA enabling the clonal expansion of mutation-free induced pluripotent stem cells (iPSC) from patients with heteroplasmic mt-DNA disease. Homoplasmic mtDNA disease requires a complete mitochondria replacement strategy using somatic cell nuclear transfer (SCNT) to generate mutation free nuclear transfer embryonic stem cells (NT-ESC).
Acknowledgments
This work was supported by grants from the National Institutes of Health (K99-HL121079, R01-HD063276, R01-HD057121, R01-HD059946, P51-OD011092), Leducq Foundation, Marriott Foundation, Mayo Clinic Center for Regenerative Medicine and OHSU institutional funds.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gurdon J. Harveian Oration 2014: Stem cells and cell replacement prospects. Clin Med (Lond) 2015;15:160–167. doi: 10.7861/clinmedicine.15-2-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3**.Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. Nuclear reprogramming induces mitochondrial and metabolic remodeling resulting in a metabolic shift from oxidative metabolism to glycolysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28:721–733. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- 5.Panopoulos AD, Yanes O, Ruiz S, Kida YS, Diep D, Tautenhahn R, Herrerias A, Batchelder EM, Plongthongkum N, Lutz M, et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Research. 2012;22:168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Folmes CD, Dzeja PP, Nelson TJ, Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell. 2012;11:596–606. doi: 10.1016/j.stem.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma T, Li J, Xu Y, Yu C, Xu T, Wang H, Liu K, Cao N, Nie BM, Zhu SY, et al. Atg5-independent autophagy regulates mitochondrial clearance and is essential for iPSC reprogramming. Nat Cell Biol. 2015;17:1379–1387. doi: 10.1038/ncb3256. [DOI] [PubMed] [Google Scholar]
- 8.Hansson J, Rafiee MR, Reiland S, Polo JM, Gehring J, Okawa S, Huber W, Hochedlinger K, Krijgsveld J. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep. 2012;2:1579–1592. doi: 10.1016/j.celrep.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Facucho-Oliveira JM, Alderson J, Spikings EC, Egginton S, St John JC. Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J Cell Sci. 2007;120:4025–4034. doi: 10.1242/jcs.016972. [DOI] [PubMed] [Google Scholar]
- 10.Todd LR, Damin MN, Gomathinayagam R, Horn SR, Means AR, Sankar U. Growth factor erv1-like modulates Drp1 to preserve mitochondrial dynamics and function in mouse embryonic stem cells. Mol Biol Cell. 2010;21:1225–1236. doi: 10.1091/mbc.E09-11-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung S, Dzeja PP, Faustino RS, Terzic A. Developmental restructuring of the creatine kinase system integrates mitochondrial energetics with stem cell cardiogenesis. Ann N Y Acad Sci. 2008;1147:254–263. doi: 10.1196/annals.1427.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Khvorostov I, Hong JS, Oktay Y, Vergnes L, Nuebel E, Wahjudi PN, Setoguchi K, Wang G, Do A, et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO Journal. 2011;30:4860–4873. doi: 10.1038/emboj.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armstrong L, Tilgner K, Saretzki G, Atkinson SP, Stojkovic M, Moreno R, Przyborski S, Lako M. Human induced pluripotent stem cell lines show similar stress defence mechanisms and mitochondrial regulation to human embryonic stem cells. Stem Cells. 2010;28:661–673. doi: 10.1002/stem.307. [DOI] [PubMed] [Google Scholar]
- 14.Prigione A, Hossini AM, Lichtner B, Serin A, Fauler B, Megges M, Lurz R, Lehrach H, Makrantonaki E, Zouboulis CC, et al. Mitochondrial-associated cell death mechanisms are reset to an embryonic-like state in aged donor-derived iPS cells harboring chromosomal aberrations. PLoS One. 2011;6:e27352. doi: 10.1371/journal.pone.0027352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15*.Katajisto P, Dohla J, Chaffer CL, Pentinmikko N, Marjanovic N, Iqbal S, Zoncu R, Chen W, Weinberg RA, Sabatini DM. Stem cells. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science. 2015;348:340–343. doi: 10.1126/science.1260384. Stemlike cells asymmetrically segregate mitochondria during cell division, with daughter cells receiving a greater porportion of young mitochondria maintaining stem cell traits. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 17.Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nature Reviews Molecular Cell Biology. 2012;13:270–276. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- 18*.Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, Nemirovski A, Shen-Orr S, Laevsky I, Amit M, et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015;21:392–402. doi: 10.1016/j.cmet.2015.02.002. Reduced glycolysis and acetyl-CoA production leads to loss of histone acetylation during early differentiation of embryonic stem cells. [DOI] [PubMed] [Google Scholar]
- 19.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bracha AL, Ramanathan A, Huang S, Ingber DE, Schreiber SL. Carbon metabolism-mediated myogenic differentiation. Nature Chemical Biology. 2010;6:202–204. doi: 10.1038/nchembio.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. Embryonic stem cells metabolize both glucose and glutamine to maintain a high alpha-ketoglutarate to succinate ratio, which favors demethylation of repressive chromatin and histone marks through Jumonji C-domain-containing histones demethylases and the ten-eleven translocation (TET) family of DNA demethylases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22**.Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. Mouse pluripotent stem cells depend on high levels of threonine catabolism to provide one carbon equivalents for the folate pool, enabling the generation of S-adenosylmethionine and histone methylation in support of pluripotency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325:435–439. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexander PB, Wang J, McKnight SL. Targeted killing of a mammalian cell based upon its specialized metabolic state. Proc Natl Acad Sci U S A. 2011;108:15828–15833. doi: 10.1073/pnas.1111312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han C, Gu H, Wang J, Lu W, Mei Y, Wu M. Regulation of L-threonine dehydrogenase in somatic cell reprogramming. Stem Cells. 2013;31:953–965. doi: 10.1002/stem.1335. [DOI] [PubMed] [Google Scholar]
- 26.Shiraki N, Shiraki Y, Tsuyama T, Obata F, Miura M, Nagae G, Aburatani H, Kume K, Endo F, Kume S. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014;19:780–794. doi: 10.1016/j.cmet.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 27.Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat Rev Genet. 2012;13:878–890. doi: 10.1038/nrg3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujikura J, Nakao K, Sone M, Noguchi M, Mori E, Naito M, Taura D, Harada-Shiba M, Kishimoto I, Watanabe A, et al. Induced pluripotent stem cells generated from diabetic patients with mitochondrial DNA A3243G mutation. Diabetologia. 2012;55:1689–1698. doi: 10.1007/s00125-012-2508-2. [DOI] [PubMed] [Google Scholar]
- 29.Folmes CD, Martinez-Fernandez A, Perales-Clemente E, Li X, McDonald A, Oglesbee D, Hrstka SC, Perez-Terzic C, Terzic A, Nelson TJ. Disease-causing mitochondrial heteroplasmy segregated within induced pluripotent stem cell clones derived from a patient with MELAS. Stem Cells. 2013;31:1298–1308. doi: 10.1002/stem.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamalainen RH, Manninen T, Koivumaki H, Kislin M, Otonkoski T, Suomalainen A. Tissue- and cell-type-specific manifestations of heteroplasmic mtDNA 3243A>G mutation in human induced pluripotent stem cell-derived disease model. Proc Natl Acad Sci U S A. 2013;110:E3622–3630. doi: 10.1073/pnas.1311660110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31**.Ma H, Folmes CD, Wu J, Morey R, Mora-Castilla S, Ocampo A, Ma L, Poulton J, Wang X, Ahmed R, et al. Metabolic rescue in pluripotent cells from patients with mtDNA disease. Nature. 2015;524:234–238. doi: 10.1038/nature14546. Demonstrated that nuclear reprogramming and SCNT can generate genetically and functionally corrected pluripotent stem cells from patients with heteroplasmic and homoplasmic mtDNA-based disease. [DOI] [PubMed] [Google Scholar]
- 32.Alexeyev MF, Venediktova N, Pastukh V, Shokolenko I, Bonilla G, Wilson GL. Selective elimination of mutant mitochondrial genomes as therapeutic strategy for the treatment of NARP and MILS syndromes. Gene Ther. 2008;15:516–523. doi: 10.1038/gt.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bacman SR, Williams SL, Pinto M, Peralta S, Moraes CT. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat Med. 2013;19:1111–1113. doi: 10.1038/nm.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34*.Reddy P, Ocampo A, Suzuki K, Luo J, Bacman SR, Williams SL, Sugawara A, Okamura D, Tsunekawa Y, Wu J, et al. Selective elimination of mitochondrial mutations in the germline by genome editing. Cell. 2015;161:459–469. doi: 10.1016/j.cell.2015.03.051. Demonstrated that gene editing using mitochondrial-targeted restriction endonucleases and TALENs can shift levels of mtDNA heteroplasmy and prevent germline transmission of specific mtDNA haplotypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campbell KH, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- 36.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 37.Solter D. Mammalian cloning: advances and limitations. Nat Rev Genet. 2000;1:199–207. doi: 10.1038/35042066. [DOI] [PubMed] [Google Scholar]
- 38.Wilmut I, Beaujean N, de Sousa PA, Dinnyes A, King TJ, Paterson LA, Wells DN, Young LE. Somatic cell nuclear transfer. Nature. 2002;419:583–586. doi: 10.1038/nature01079. [DOI] [PubMed] [Google Scholar]
- 39**.Tachibana M, Amato P, Sparman M, Gutierrez NM, Tippner-Hedges R, Ma H, Kang E, Fulati A, Lee HS, Sritanaudomchai H, et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell. 2013;153:1228–1238. doi: 10.1016/j.cell.2013.05.006. First generation of diploid human ESCs using somatic cell nuclear transfer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chinnery PF, Craven L, Mitalipov S, Stewart JB, Herbert M, Turnbull DM. The challenges of mitochondrial replacement. PLoS Genet. 2014;10:e1004315. doi: 10.1371/journal.pgen.1004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deuse T, Wang D, Stubbendorff M, Itagaki R, Grabosch A, Greaves LC, Alawi M, Grunewald A, Hu X, Hua X, et al. SCNT-derived ESCs with mismatched mitochondria trigger an immune response in allogeneic hosts. Cell Stem Cell. 2015;16:33–38. doi: 10.1016/j.stem.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 42.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, Qi Z, Downey SL, Manos PD, Rossi DJ, et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13:541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma H, Morey R, O’Neil RC, He Y, Daughtry B, Schultz MD, Hariharan M, Nery JR, Castanon R, Sabatini K, et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature. 2014;511:177–183. doi: 10.1038/nature13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johannesson B, Sagi I, Gore A, Paull D, Yamada M, Golan-Lev T, Li Z, LeDuc C, Shen Y, Stern S, et al. Comparable frequencies of coding mutations and loss of imprinting in human pluripotent cells derived by nuclear transfer and defined factors. Cell Stem Cell. 2014;15:634–642. doi: 10.1016/j.stem.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 47.Khusnutdinova E, Gilyazova I, Ruiz-Pesini E, Derbeneva O, Khusainova R, Khidiyatova I, Magzhanov R, Wallace DC. A mitochondrial etiology of neurodegenerative diseases: evidence from Parkinson’s disease. Ann N Y Acad Sci. 2008;1147:1–20. doi: 10.1196/annals.1427.001. [DOI] [PubMed] [Google Scholar]
- 48.Bratic A, Larsson NG. The role of mitochondria in aging. J Clin Invest. 2013;123:951–957. doi: 10.1172/JCI64125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaturvedi RK, Flint Beal M. Mitochondrial diseases of the brain. Free Radic Biol Med. 2013;63:1–29. doi: 10.1016/j.freeradbiomed.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 50.Lin MT, Simon DK, Ahn CH, Kim LM, Beal MF. High aggregate burden of somatic mtDNA point mutations in aging and Alzheimer’s disease brain. Hum Mol Genet. 2002;11:133–145. doi: 10.1093/hmg/11.2.133. [DOI] [PubMed] [Google Scholar]
- 51.Wallace DC. Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science. 1992;256:628–632. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]