Summary
Motor behaviors such as walking or withdrawing the limb from a painful stimulus rely upon integrative multimodal sensory circuitry to generate appropriate muscle activation patterns. Both the cellular components and the molecular mechanisms that instruct the assembly of the spinal sensorimotor system are poorly understood. Here we characterize the connectivity pattern of a sub-population of lamina V inhibitory sensory relay neurons marked during development by the nuclear matrix and DNA binding factor Satb2 (ISRSatb2). ISRSatb2 neurons receive inputs from multiple streams of sensory information and relay their outputs to motor command layers of the spinal cord. Deletion of the Satb2 transcription factor from ISRSatb2 neurons perturbs their cellular position, molecular profile, and pre- and post-synaptic connectivity. These alterations are accompanied by abnormal limb hyperflexion responses to mechanical and thermal stimuli, and during walking. Thus, Satb2 is a genetic determinant that mediates proper circuit development in a core sensory-to-motor spinal network.
Introduction
The movements of animals are adapted to their environment by a highly integrative circuitry within the spinal cord that refines motor actions based on a variety of sensory cues (reviewed in Baldissera et al. 1981). Exteroceptive sensory information continuously streams into the central nervous system as an animal moves and includes detection of limb position, mechanical pressure and in the case of environmental threats, noxious pain. These sensory feedback pathways can rapidly and dynamically regulate motor actions via local spinal circuitry, even in the absence of supraspinal connections (Forssberg et al., 1975; Rossignol et al., 2006). Electrophysiological studies of sensorimotor circuits have found that both excitatory and inhibitory neurons within the dorsal spinal cord participate in the transformation of sensory cues into motor actions (Bannatyne et al., 2009; Cavallari et al., 1987; Schouenborg and Weng, 1994). Remarkable progress has been made using circuit mapping and genetics to identify neurons that gate and relay specific sensory modalities to higher brain centers (Bourane et al., 2015a; Bourane et al., 2015b; Braz et al., 2005; Duan et al., 2014; Fink et al., 2014), but less is known about the molecular and developmental features of spinal neurons that integrate different sensory streams for rapid modulation of the motor system.
It has long been recognized that multimodal sensory cues intersect within the medial deep dorsal horn of the spinal cord where direct-proprioceptive and indirect-nociceptive pathways converge (reviewed in Craig, 2003). Interestingly, circuit tracing has found that a significant fraction of spinal premotor neurons are likewise located in the medial deep dorsal horn (Levine et al., 2014; Stepien et al., 2010). Together these studies indicate that multiple types of sensory information including limb position and pain may target a somatotopically-organized sensorimotor circuit arrayed across lamina V that modulates motor activity (Schouenborg et al., 1995; Tripodi et al., 2011). Consistent with this view of sensorimotor integration, the ablation of broad groups of dorsal-spinal inhibitory neurons disrupts motor actions in response to sensory stimuli (Foster et al., 2015). Nevertheless, because the motor and sensory components of action are deeply intertwined (Rizzolatti and Sinigaglia, 2007), it has been difficult to study integrative sensorimotor circuitry without definitive cellular markers of the constituent components.
A subset of premotor cells in the deep dorsal horn can be identified by the expression of the transcription factor Satb2 (Levine et al., 2014). Satb2 is a DNA binding protein that interacts with nuclear matrix attachment regions and controls gene expression and chromatin remodeling. Functional studies of Satb2 have identified roles for this gene in musculoskeletal development, and shown this factor is necessary for establishing the proper identity and axon projections of callosal neurons (Alcamo et al., 2008; Britanova et al., 2008; Dobreva et al., 2006; Leone et al., 2014; Zhao et al., 2014). Interestingly, case studies of human mutations in the Satb2 gene have reported developmental delays in motor skill acquisition, coordination, and sensorimotor behavior (Balasubramanian et al., 2011; FitzPatrick et al., 2003; Van Buggenhout et al., 2005).
Here we describe the connectivity pattern of Satb2+ interneurons, and establish that the Satb2 gene is required for the proper development of spinal circuitry that mediates the transformation of sensory cues into motor behavior. Satb2+ neurons are predominantly inhibitory, receive monosynaptic input from proprioceptive neurons, and are activated by noxious stimuli via polysynaptic pathways. Satb2+ neurons form synaptic connections onto ventral locomotor-interneurons and motor neurons. Based on their neurotransmitter profile and connectivity we refer to the discrete population of Satb2+ cells in the dorsal horn as inhibitory sensory relay (ISRSatb2) neurons. When the Satb2 gene was mutated the cell position, molecular profile, synaptic inputs and synaptic outputs of ISRSatb2 neurons were markedly altered. Satb2 mutant mice display abnormal limb hyperflexion of the ankle during walking and hold the hindlimb in a prolonged flexed position following noxious thermal or mechanical stimulation of the paw. Our findings demonstrate that Satb2 is necessary for the connectivity of ISRSatb2 neurons, thereby revealing an intrinsic genetic program that contributes to the formation of a discrete subunit of intraspinal sensorimotor circuitry.
Results
Satb2 is expressed in a restricted population of deep dorsal horn inhibitory neurons
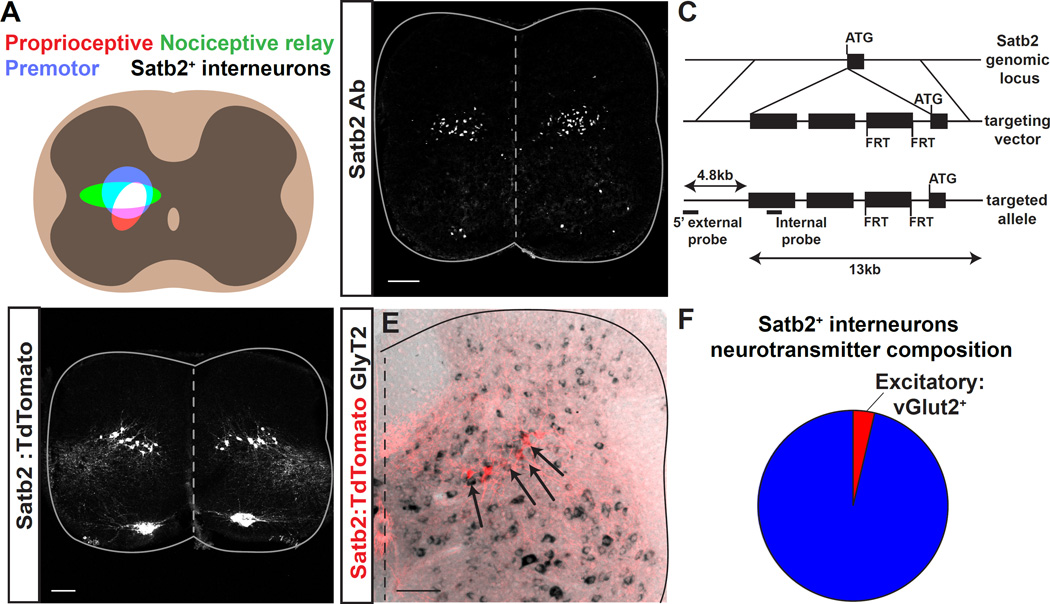
Both proprioceptive and nociceptive sensory neurons relay through the deep dorsal horn (lamina IV-VI) of the spinal cord, an area that was recently shown to contain a significant proportion of neurons that are directly upstream of motor neurons (Coulon et al., 2011; Levine et al., 2014; Stepien et al., 2010). We therefore sought to identify genes that are expressed in a specific group of neurons within this nexus of sensory-motor information. Satb1/2 was recently identified as a marker of a subset of premotor neurons located in spinal lamina V (Levine et al., 2014). We performed immunohistochemistry with a Satb2-specific antibody and found that, in contrast to many of the dorsal horn molecular markers (Lbx1, Lmx1b, Pax2, Tlx3, GlyT2) that label broad heterogeneous groups of neurons, Satb2 labels a discrete population of neurons in lamina V of the deep dorsal horn (Figure 1A, B).
Figure 1. Expression of Satb2 in inhibitory neurons in the spinal cord deep dorsal horn.
A. Schematic illustrating the alignment of Satb2+ interneurons with the termination domains of multiple motor control pathways. Left, Premotor neurons (blue), proprioceptive fiber termination (red), and nociceptive relay neurons (green) are located in the spinal cord deep dorsal horn. The overlap between these pathways is shown in white. Right, Location of Satb2+ interneurons aligns with the overlap in left panel. B. Immunohistochemistry using a Satb2-specific antibody (Satb2 Ab) reveals Satb2 expression in a subset of motor neurons in the medial motor column (MMC) and a band of spinal interneurons at embryonic day 15.5. C. Schematic for generation of Satb2-CreERT2 mouse line. Targeted insertion of CreERT2-WPRE-pA-FRT-neo-FRT cassette into the ATG of the Satb2 locus. D. Crossing Satb2-CreERT2 with the reporter line Rosa-CAG-LSL-TdTomato (Satb2:TdTomato) recapitulates the pattern of expression seen in B. Lumbar, e18.5. See also Figure S1. E. Dorsal spinal cord following in situ hybridization for the inhibitory neurotransmitter marker GlyT2 (black) in Satb2:Tomato animals (red). Double positive neurons are indicated with arrows throughout the Satb2+ interneuron population. F. Quantification of inhibitory (blue) and excitatory (red) neurotransmitter markers from Figure 1E and Figure S1. Percentages correspond to mean values. Spinal cords were analyzed at P13 for in situ hybridization. Scale bars in B, D and E, 100um.
Satb2 expression is first detectable at embryonic day e12 and continues through the first postnatal week (Figure 1B, S1). Satb2+ interneurons span the mediolateral extent of lamina V (Figure 1B), corresponding to a region where cells that are activated during the nociceptive withdrawal reflex (NWR) have been detected with electrophysiology (Schouenborg and Weng, 1994). We also observed transient expression of Satb2 in motor neurons of the medial and lateral motor columns as early as e10.5, which perdured in a subset of medial motor neurons into the first postnatal week (Figure S1). Satb2 expression is also detectable in the developing neocortex and specific regions of the musculoskeletal system (data not shown). These findings are consistent with reports of Satb2 expression in the nervous system and other tissues (Britanova et al., 2005; Dobreva et al., 2006; Szemes et al., 2006).
To extend previous studies and to determine whether Satb2 marks interneurons of known lineage, we performed colocalization experiments with antibodies for dorsal interneuron subtypes (Alaynick et al., 2011; Gross et al., 2002). Satb2 is expressed in the Lbx1+ lineage of dorsal interneurons (Britanova et al., 2005), a marker that is expressed broadly in the dorsal spinal cord including interneuron subtypes dI4–6 and dILA and dILB (Gross et al., 2002; Muller et al., 2002; Schubert et al., 2001). Satb2+ interneurons also co-express the LIM homeodomain markers Lim1/2 (Britanova et al., 2005), and a subset of Satb2+ interneurons coexpresses Ptf1a, a marker of dI4 and dIL lineages (Figure S2). However, when we performed colocalization experiments with markers that subdivide these broad lineage classes (Pax2, Tlx3, Brn3a, Lmx1b, Bhlhb5, or Isl1/2), we found that the Satb2+ interneuron population did not express additional markers of dI4, dI5, dIL, or dI6 lineages (Figure S2). These findings indicate that the classification of Satb2+ interneurons does not align with previously described populations of dorsal interneurons.
To gain genetic access to Satb2+ interneurons and define their circuit properties, we devised a temporally inducible Cre labeling strategy. To generate a Satb2:CreERT2 knock-in mouse line, CreERT2 was inserted into the ATG of the Satb2 locus (Figure 1C, and data not shown). Satb2:CreERT2 animals were crossed with the Cre-dependent reporter Rosa-CAG-LSL-TdTomato to generate Satb2:TdTomato animals. Although Satb2 is downregulated during the first postnatal week, tamoxifen administration at embryonic or early postnatal timepoints indelibly labels Satb2+ interneurons. Embryonic induction of Satb2:CreERT2 with tamoxifen at e12 labels spinal interneurons as well as a restricted population of motor neurons in the medial motor column (Figure 1D). Thus, TdTomato expression driven by our transgenic line accurately recapitulates Satb2 protein expression (compare Figure 1B to 1D, Figure S1).
Genetic labeling of Satb2+ interneurons facilitated further descriptive analyses of this population. To determine the neurotransmitter status of Satb2+ interneurons we performed double labeling experiments in which we examined the overlap of TdTomato expression with in situ hybridization probes for excitatory or inhibitory neurotransmitter markers. Only 3.8% of Satb2+ interneurons express the excitatory marker vGlut2, whereas the vast majority of cells express the inhibitory markers Gad65, Gad67, and/or GlyT2 (Figure 1E, F, S2). These experiments reveal that Satb2+ neurons represent a spatially restricted population of inhibitory dorsal interneurons arrayed along the mediolateral axis of the deep dorsal horn.
Satb2+ interneurons receive multimodal sensory input
Tracing experiments have noted that peripheral sensory commands converge on spinal neurons in the deep dorsal horn of the spinal cord, representing the first point of intersection between the nociceptive and proprioceptive systems (Levine et al., 2014). Primary fibers of proprioceptive sensory neurons and second order neurons that are recruited during the hindlimb nociceptive NWR reside in overlapping domains within the deep dorsal horn (Craig, 2003). Although the approximate site of convergence for these two pathways overlaps with the position of Satb2+ interneurons (Figure 1A), their connectivity has not been described.
To test whether Satb2+ interneurons receive proprioceptive input, we used a genetic labeling strategy (Parvalbumin-Cre x Rosa-CAG-LSL-Syp-TdTomato-deltaNeo; abbreviated PV:Syn-Tom) in which the presynaptic terminals of proprioceptive fibers are labeled with a synaptophysin-TdTomato fusion protein (Levine et al., 2014). We identified TdTomato+ contacts on the cell bodies of Satb2+ interneurons (Figure 2A, B), indicating that Satb2+ interneurons receive direct input from proprioceptive fibers. We next used a second strategy to visualize the full cell morphology of Satb2+ interneurons. Immunohistochemistry for parvalbumin and vGlut1 revealed proprioceptive contacts along the soma and processes of Satb2:TdTomato+ spinal interneurons. We found that 97 +/− 2.6% of Satb2:TdTomato+ spinal interneurons received direct contacts from proprioceptive fibers (Figure S3, n=29 cells in 3 spinal cords), however we noted quantitative differences within the Satb2+ interneuron population, which are characterized in more detail below.
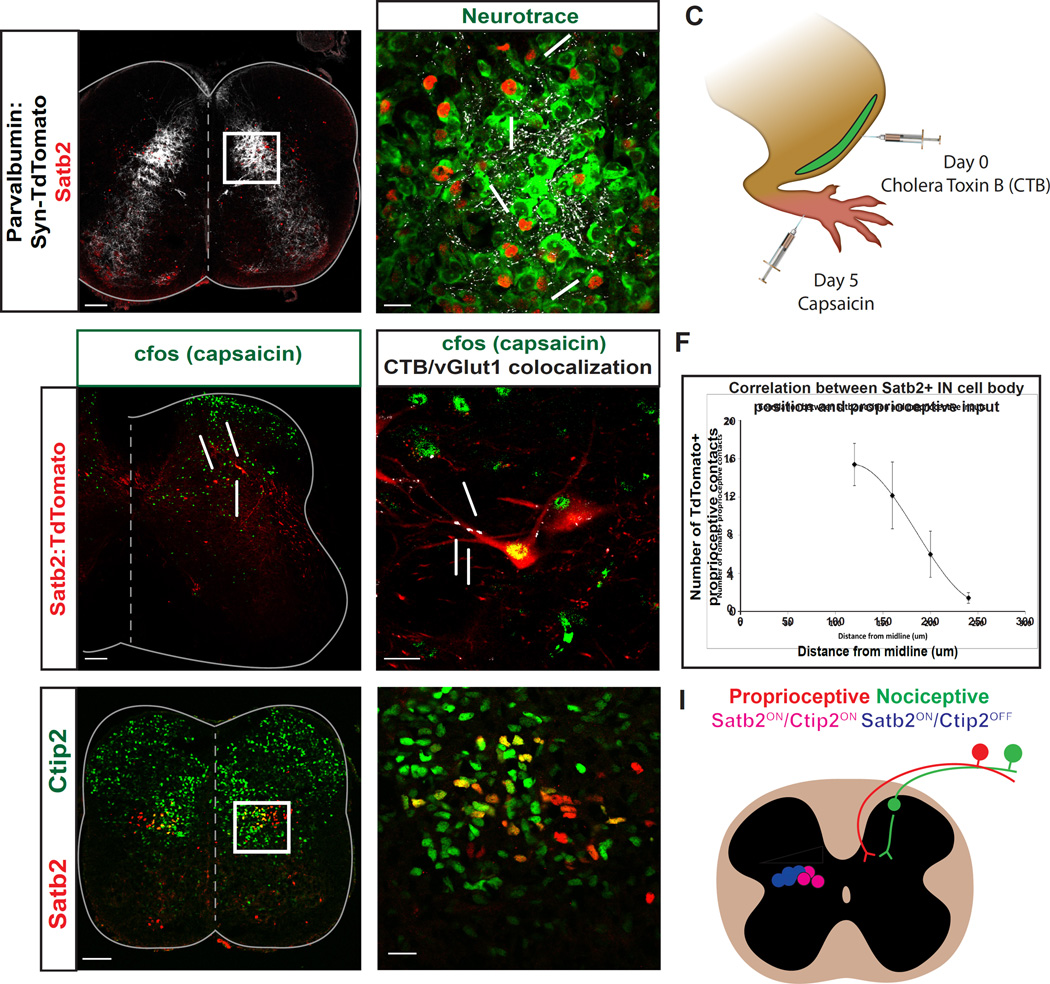
Figure 2. Satb2+ interneurons receive convergent sensory inputs and display heterogeneity along the mediolateral axis.
A, B. Proprioceptive fibers have a dense termination in the deep dorsal horn. Presynaptic terminals of proprioceptive fibers (white) were identified by crossing Parvalbumin-Cre to Rosa-LSL-Syp-TdTomato-deltaNeo (PV:Syn-TdTomato). Proprioceptive contacts (white) were identified on neurotrace+ cell bodies (green) of Satb2+ interneurons (red), highlighted by arrows. White box in A refers to high magnification image in B. P1 spinal cords in A, B. C. Schematic demonstrating experimental technique used to identify nociceptive relay neurons that also receive input from proprioceptive sensory fibers. Cholera Toxin-B was injected into the tibialis anterior (TA) muscle of Satb2:TdTomato animals to identify proprioceptive afferents. 5 days later, capsaicin was injected into the footpads and spinal cords were collected for labeling. D. c-Fos immunoreactivity (green) was used to identify cells in the spinal cord that are recruited following painful stimulation. Capsaicin was injected into the footpad of adult Satb2:TdTomato animals, as in C. Yellow cells that are co-positive for Satb2 (red) and c-Fos (green) are indicated by arrows. n=4 spinal cords. E. Satb2:TdTomato+ neurons (red) that receive multimodal sensory input were identified based on c-Fos immunoreactivity (green), and identification of proprioceptive synaptic terminals with vGlut1/CTB coexpression (white). F. Mediolateral position of Satb2+ interneurons corresponds to the number of contacts from proprioceptive fibers. Medial cells positioned closer to the midline had a greater number of contacts and lateral cells had fewer contacts. Quantified cells were binned by mediolateral position and the number of synaptic contacts is represented as mean +/− SEM. n=34 cells in 3 spinal cords. G, H. Ctip2 expression subdivides Satb2+ interneurons along the mediolateral axis. Medial Satb2+ interneurons (red) co-express Ctip2 (green), as indicated by yellow labeling in G, H. e15.5 spinal cords used for G, H. Scale bars in A, D, G, 100um. Scale bars in B, E, H, 20um. I. Summary diagram depicting features of Satb2+ interneurons. Left side: mediolateral organization of Satb2+ interneurons according to Ctip2 coexpression (blue/pink) and proprioceptive input (white wedge). Right side: multimodal sensory input from nociceptive (green) and proprioceptive pathways (red) onto Satb2+ interneurons (white).
Nociceptive sensory fibers predominantly target superficial spinal laminae, but relay information through the deep dorsal horn via spinal interneurons. To explore whether Satb2+ interneurons are a site of convergence for multimodal sensory inputs, we performed double labeling experiments of proprioceptive fibers and nociceptive relay neurons in Satb2:TdTomato animals. Intramuscular injection of cholera toxin (CTB) into the tibialis anterior (TA) muscle was performed to label proprioceptive afferents of a muscle that is recruited following the NWR (Schouenborg, 2004). Capsaicin was then injected into the footpads of these animals, and c-Fos immunoreactivity was used to identify cells that are recruited in response to a painful stimulus (Figure 2C). As expected, c-Fos+ neurons were located in superficial laminae ipsilateral to the injection site, as well as in the deep dorsal horn. At spinal levels of peak c-Fos labeling, 32.3% of Satb2:TdTomato+ neurons coexpress c-Fos in response to capsaicin injection into the footpad (Figure 2D; 42 of 130 TdTomato+ cells, n=5 spinal cords), indicating that a significant fraction of Satb2+ interneurons are activated even in response to focal stimulation of nociceptive sensory neurons that innervate the foot. In addition, we identified Satb2:TdTomato+ interneurons that express c-Fos following noxious stimulation and receive vGlut1+ contacts from CTB-labeled proprioceptive afferents (Figure 2E). Thus, individual Satb2+ interneurons represent a convergence point for multimodal sensory input. Based on Satb2 expression, neurotransmitter identity, and sensory input, we refer to them as inhibitory sensory relay (ISRSatb2) neurons.
ISRSatb2 neuron diversity along the mediolateral axis
Sensory-motor circuit components are known to display a topographic organization along the mediolateral axis of the spinal cord (Arber, 2012; Levine et al., 2012). The convergence of synaptic inputs onto ISRSatb2 neurons and their distribution along the mediolateral axis prompted us to ask whether ISRSatb2 neurons are composed of a homogeneous population of neurons or whether there is diversity in their identity and connectivity. To test whether interneuron cell body location is a determinant of the amount of proprioceptive input, we quantified the number of PV:Synaptophysin-TdTomato+ contacts on neurotrace+ cell bodies of ISRSatb2 neurons, and mapped the number of contacts per cell along the mediolateral axis. These experiments revealed an inverse relationship between cell body distance from the midline and the amount of proprioceptive input: cells positioned closer to the midline had a greater number of proprioceptive contacts, while lateral cells had fewer contacts (Figure 2F). These findings indicate that mediolateral position is an organizational feature that correlates with proprioceptive sensory connectivity onto ISRSatb2 neurons.
Although interneurons are topographically organized along the mediolateral axis of the deep dorsal horn, genetic markers that subdivide these groups have not been described. In the cortex, Satb2 participates in an antagonistic genetic interaction with Ctip2 to subdivide cortical progenitors into functionally distinct pools (Alcamo et al., 2008; Britanova et al., 2008). To test whether Satb2 and Ctip2 have mutually exclusive expression in the spinal cord, we performed immunohistochemistry for both markers. Surprisingly, in the spinal cord Satb2 and Ctip2 are not mutually exclusive and define two subpopulations of ISRSatb2 cells; medial neurons are Satb2+/Ctip2+, while lateral neurons are Satb2+/Ctip2- (Figure 2G, H). This molecular code is detectable as early as e13.5 and is maintained into postnatal stages (data not shown). These experiments reveal that sensory connectivity and molecular profile subdivide ISRSatb2 neurons along the mediolateral axis of the spinal cord (Figure 2I).
ISRSatb2 neurons target ventral circuitry associated with motor function
There is a significant representation of premotor neurons in the deep dorsal horn (Coulon et al., 2011; Hongo et al., 1989; Levine et al., 2014; Puskar and Antal, 1997; Stepien et al., 2010), and neurons that target supraspinal motor control centers are also known to reside in dorsal laminae (Matsushita and Hosoya, 1979; Surmeier et al., 1988). To test whether ISRSatb2 neurons may serve as a cellular substrate for transmitting sensory cues to either ascending or ventral motor control centers, we used a combination of circuit tracing approaches.
First, we used an unbiased genetic labeling approach to identify synaptic targets of ISRSatb2 neurons. Satb2:CreERT2 x Rosa-CAG-LSL-Syp-TdTomato-deltaNeo (abbreviated Satb2:Syn-Tom) provided, cell-type specific expression of the Synaptophysin-TdTomato presynaptic fusion protein. Visualization of the overall distribution of synaptic terminals of ISRSatb2 neurons revealed a specific pattern of labeling across the ventral spinal cord. Syn-Tom+ terminals were detected throughout the ventral spinal cord, but enriched in a diagonal region covering lamina V-VII and IX (Figure 3A, B).
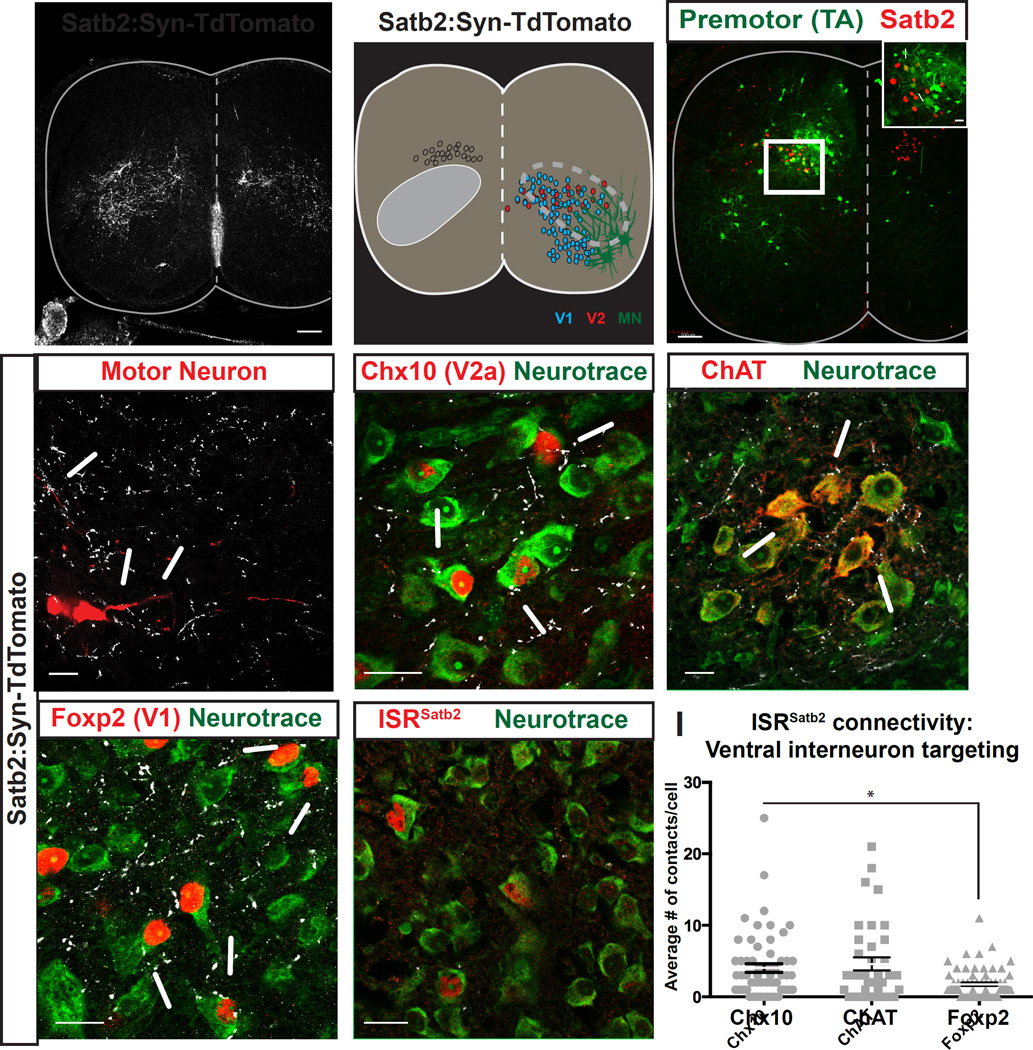
Figure 3. ISRSatb2 neurons synaptically target ventral motor circuitry.
A, B. Presynaptic terminals of ISRSatb2 neurons (A) broadly target the ventral spinal cord, overlapping with a region where components of ventral motor circuitry reside (B). Distribution of presynaptic terminals of ISRSatb2 was identified by genetic labeling (Satb2:Synaptophysin-TdTomato). Left side of spinal cord in B shows ISRSatb2 neuron cell bodies (white) and termination zone of presynaptic terminals (gray oval). Right side of spinal cord in B shows approximate cell body locations of V1 (blue), V2 (red) interneurons, motor neurons (MN, green). Dotted lines encompass the area of dense labeling of presynaptic terminals of ISRSatb2 neurons and the position of ventral neuron populations. C. Satb2 labels a subset of premotor neurons in the deep dorsal horn. Monosynaptically restricted Rabies labeling identifies premotor neurons (green) from tibialis anterior muscles that co-express Satb2 (red). White box in C corresponds to high magnification image shown in inset. Arrows in inset identify yellow cells that are positive for Rabies to identify premotor neurons (green) and Satb2 (red). D-G. ISRSatb2 neurons target cellular components of ventral motor circuitry. High magnification images from the ventral spinal cord showing contacts from presynaptic terminals of ISRSatb2 neurons (white) onto (D) virally labeled motor neurons (red), and onto neurotrace+ cell bodies (green) of (E) Chx10+ V2a interneurons (red), (F) ChAT+ motor neuron cell bodies (red), (G) Foxp2+ V1 interneurons (red). (H) In contrast, presynaptic terminals of ISRSatb2 neurons (white) on neurotrace+ cell bodies (green) were not identified onto ISRSatb2 neurons (red). (I) Quantification of the number of contacts onto ventral interneurons. There are significantly more contacts on ChAT+ motor neurons than on Foxp2+ V1 interneurons. Data points represent individual neurons; averages are represented as mean +/− SEM. See also Figure S3. Scale bars in A, C, 100um. Scale bars in C (inset), D-H, 20um.
We compared the location of Satb2:Syn-Tom labeling with the presynaptic targets of other dorsal interneuron classes. In contrast to the specific pattern of labeling in ventral lamina that we observed in Satb2:Syn-Tom spinal cords, both Ptf1a: and Lmx1b:Syn-Tom expressing interneurons broadly target dorsal spinal laminae in which their cell bodies reside (Figure S3). The location and distribution of synaptic terminals from ISRSatb2 neurons indicates their connectivity patterns are distinct from those of other dorsal interneuron classes implicated in sensory transduction.
To test whether ISRSatb2 neurons target ventral cell types indiscriminately or with absolute specificity for synaptic partners, we performed labeling experiments for neuronal populations in the ventral spinal cord of Satb2:Syn-Tom animals. Viral labeling of motor neurons for tibialis anterior (TA, flexor), gastrocnemius (GS, extensor), and hamstring (H, flexor) muscles provided a clear visualization of specific motor neuron soma and processes (see Methods) and revealed contacts from ISRSatb2 neurons onto hindlimb motor pools (Figure 3D, and data not shown). We next used an independent strategy to confirm synaptic contacts onto motor neurons in which monosynaptic connections between spinal interneurons and motor neurons were identified by Rabies transsynaptic labeling (Stepien et al., 2010; Wickersham et al., 2007). These experiments confirmed that ISRSatb2 neurons target motor neurons (Figure 3C). Next we characterized the ventral interneuron populations that receive inputs from ISRSatb2 neurons. We found that many ventral (lamina VII) interneuron classes implicated in motor control receive input from ISRSatb2 neurons (Figure 3E–H; 82.1 +/− 6.2% of Chx10+ V2a, 66.8 +/− 1.8% Foxp2+ V1, and 69 +/− 9.4% of ChAT+ motor neurons).
To test whether ISRSatb2 neurons display quantitative differences in their connectivity, we determined the average number of ISRSatb2 synaptic terminals on each cell type. Chx10+ V2a interneurons receive numerous contacts on their soma (Figure 3E; 4.35 +/− 0.84 TdTomato+ contacts/Chx10+ neuron). Synaptophysin:TdTomato+ contacts were also identified on ChAT+ motor neurons (Figure 3F, 4.28+/−0.97 TdTomato+ contacts/ChAT+ neuron) as well as Foxp2+ V1 interneurons (Figure 3G, 1.83+/−0.14 TdTomato+ contacts/Foxp2+ neuron). Although contacts were less abundant, we also identified TdTomato+ contacts onto Evx1+ V0 interneurons, Calbindin+ ventral interneurons of mixed identity, and Bhlhb5+ ventral interneurons of mixed identity (Figure S3). We next compared the average number of Syn-Tom+ contacts onto various ventral interneuron populations. Chx10+ V2a interneurons receive significantly more contacts than Foxp2+ V1 interneurons (Figure 3I, p<0.05, Mann Whitney), suggesting that synaptic input from Satb2+ interneurons is weighted among various ventral interneuron populations. Syn-Tom+ contacts were not identified onto ISRSatb2 neurons (Figure 3H).
Next we examined whether Satb2+ interneurons have ascending projections that target supraspinal motor control centers. scAAV containing a Cre-dependent fluorescent reporter protein (scAAV-Flex-GFP) was injected into the lumbar spinal cord of neonatal Satb2-CreERT2 animals. GFP labeling was evident in a pattern consistent with that of Satb2:TdTomato labeling. GFP+ fibers were not detected at the pyramidal decussation nor found within the ventral brainstem where ascending fiber tracts normally reside (Figure S3). These findings suggest that the predominant targets of ISRSatb2 neurons are motor neurons and ventral interneurons that regulate motor activity within the spinal cord.
Basic motor coordination is intact in the absence of Satb2
Given the established role of the Satb2 gene in developmental events that are critical for circuit formation, and the broad connectivity of ISRSatb2 neurons with ventral motor command neurons, we considered how loss of Satb2 gene function would impact motor activity. We first tested whether basic motor circuitry is intact in Satb2 null animals (Satb2lacZ/lacZ; Dobreva et al., 2006) using a fictive locomotor preparation that activates the central pattern generator (CPG) in the absence of sensory and descending brain input (Kiehn and Kjaerulff, 1996; Smith and Feldman, 1987). Patterned motor activity was normally coordinated in Satb2 null mutants between left-right and flexor-extensor pools (Figure 4A–C). Satb2 null animals also displayed normal cycle period length, cycle period variation, and burst duration (Figure S5 and data not shown), parameters that are altered when ventral interneuron function is perturbed (Alaynick et al., 2011; Goulding, 2009; Grillner and Jessell, 2009; Kiehn, 2016). These experiments indicate that the core components of ventral motor circuitry develop and function properly in the absence of Satb2.
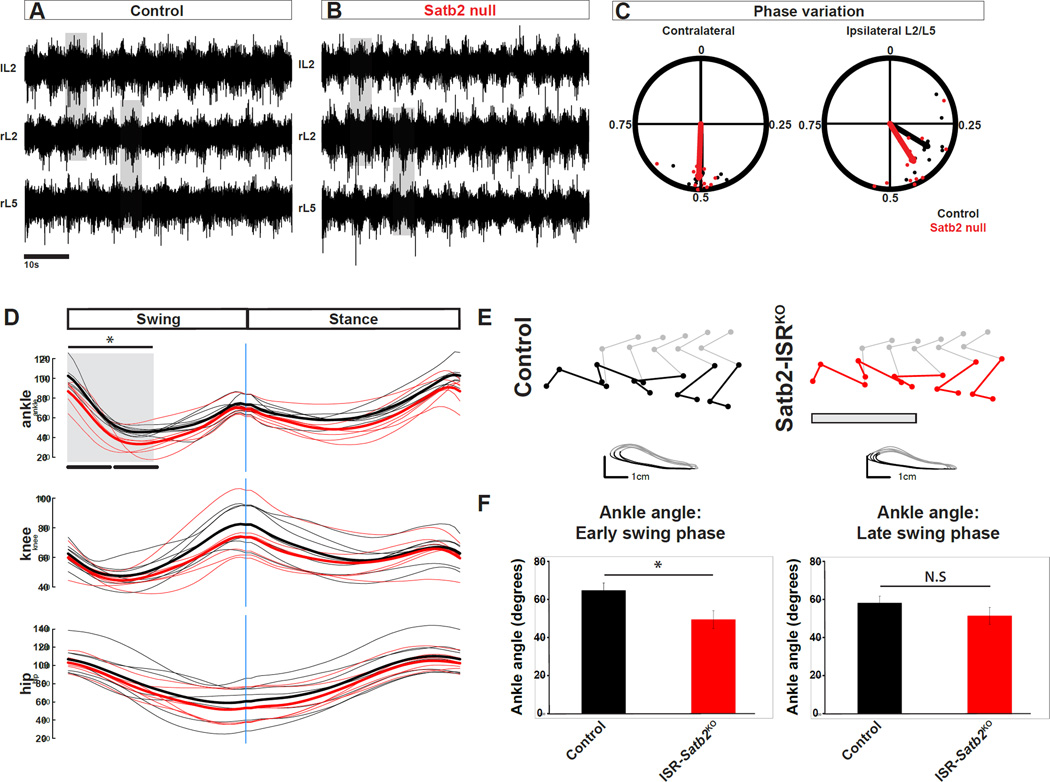
Figure 4. Motor output in response to loss of Satb2.
A, B. Example traces for left L2 (lL2), right L2 (rL2), and right L5 (rL5) ventral root recordings in control (A) or Satb2 null (B) spinal cords at e18.5. Neurochemically induced fictive locomotor activity in control spinal cords is characterized by alternating rhythmic bursts of motor neuron activity between contralateral L2 ventral roots and ipsilateral L2-L5 ventral roots. Neurochemical induction of fictive locomotor activity in Satb2 null spinal cords evokes rhythmic alternating bursts of motoneuron activity indistinguishable from wild type littermates. C. Quantification of phase analysis from contralateral and ipsilateral ventral root pairs reveals that fictive locomotor activity is normally coordinated in Satb2 null (red points) relative to wild type control littermates (black points). Points near 0.5 represent alternating activity. D. Kinematic analysis of the hindlimb during runway walking. Joint angle traces for ankle (top panel), knee (middle panel), and hip (bottom panel) are shown for swing and stance phases of a single gait cycle. Individual traces (average of two trials per individual) for control (black) and Satb2-ISRKO (red) are shown as dim lines, and average traces for each genotype are shown in bold. E. Representative stick diagrams and limb endpoint trajectories for control (left) and Satb2-ISRKO animals (right) Ankle angle is shown in color for control (black) and Satb2-ISRKO animals (red). Grey box in Satb2-ISRKO limb reconstructions shows an approximation for early swing phase (see also grey box in D, and quantification in left panel of F) where ankle hyperflexion is observed. In contrast, limb endpoint trajectories showing swing (grey) and stance (black) phases indicate normal movement of the foot marker. F. Ankle angle was significantly reduced in the first half of the swing cycle in Satb2-ISRKO animals. Ankle angle measurements are represented as mean +/− S.E.M. * indicates significant difference (p<0.05 for early swing phase); N.S indicates no significant difference. See also Figure S5.
To test whether targeted deletion of Satb2 from ISRSatb2 neurons impacts motor and sensory function in the behaving animal, we used a conditional Satb2flx allele. Lbx1:Cre was used as the deleter based on its coexpression with Satb2 in the spinal cord, and its exclusion from other Satb2+ cell types (Figure S2, S4, and data not shown). Use of Lbx1:Cre/Satb2flx/flx (Satb2-ISRKO) mutant animals allowed us to circumvent perinatal lethality in Satb2 null animals, allowing for behavioral analysis of sensory and motor function. We confirmed that Satb2-ISRKO animals lack Satb2 protein expression within spinal interneurons (Figure S4). Satb2-ISRKO mutant animals are born in normal numbers, have a normal lifespan, body weight and appearance at 8 and 20 weeks of age (data not shown). Behavioral tests that broadly assay motor function, including open field to test baseline motor activity and grip strength to test muscle strength, were normal in Satb2-ISRKO mutants (Figure S5). Satb2-ISRKO mutant animals also performed normally on the rotarod test measuring general motor coordination. Thus, Satb2 gene function appears to be dispensable in ISRSatb2 neurons for basic motor tasks.
Satb2-ISRKO animals display aberrant flexion behavior during walking and pain withdrawal
Recent studies have shown that proprioceptive defects alter limb position and the timing of muscle recruitment during walking (Akay et al 2014). The dense proprioceptive input onto ISRSatb2 neurons prompted us to examine limb kinematics of Satb2-ISRKO animals. Satb2-ISRKO mutants had a visible disruption in limb position during the swing phase of the step cycle. In particular, the ankle joint was held in a hyperflexed position beginning at swing onset, resulting in a reduced ankle angle throughout early swing phase (Figure 4D–F, Figure S5).
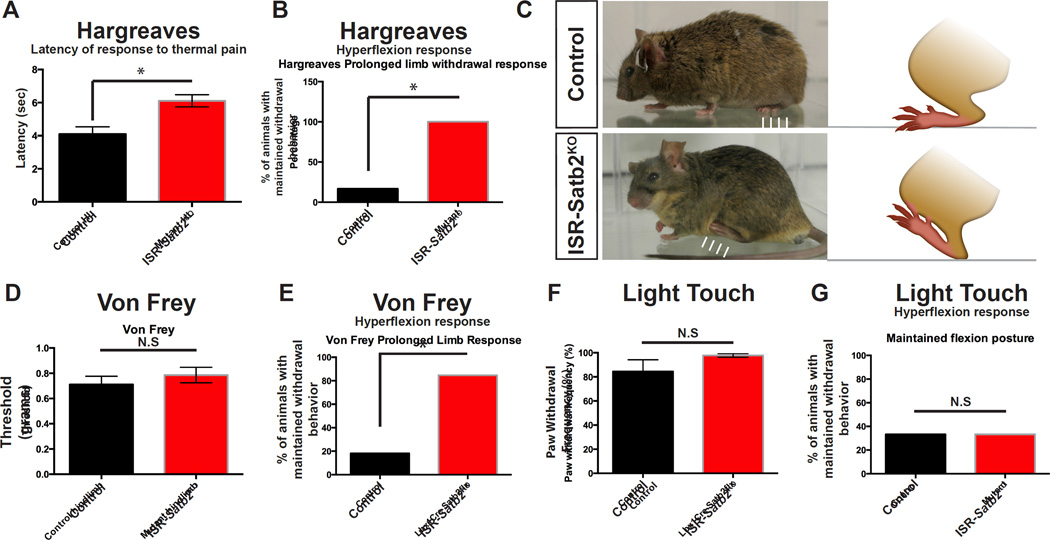
Next, to test whether Satb2 is required for sensory-evoked behaviors, we examined motor responses of Satb2-ISRKO animals to noxious and non-noxious stimuli. To perform the Hargreaves thermal pain test, a high intensity light source was shined on the footpad of test animals, eliciting an isolated, reproducible nociceptive withdrawal response of the foot (Allen and Yaksh, 2004; Hargreaves et al., 1988). We performed this test to measure two key components of the pain response: latency to the withdrawal response, and the dynamics of the withdrawal behavior, including the initial flexion response and replacement of the limb following withdrawal. We observed a small but significant increase in hindlimb response latency in Satb2-ISRKO mutant animals when compared with control littermates (Figure 5A). Interestingly, Satb2-ISRKO animals maintained a flexed posture following thermal pain stimulation (Figure 5B, C; Videos S1 and S2), revealing an aberrant flexion behavior following deletion of Satb2 from ISRSatb2 neurons.
Figure 5. Loss of Satb2 in spinal interneurons perturbs the motor response to noxious stimuli.
A. In the Hargreaves test for thermal pain, conditional loss of Satb2 in spinal interneurons in Satb2-ISRKO animals leads to an increase in the latency to withdrawal compared to control (Lbx1Cre-) animals. Data is represented as mean latency (seconds) +/− S.E.M. * indicates significant difference (p<0.05; Mann Whitney). B. Satb2-ISRKO animals display a prolonged withdrawal posture following Hargreaves stimulation (hyperflexion response), compared with control animals. Data is represented as mean (number of animals that maintain withdrawn posture) * indicates significant difference (p=0.01; Fischer’s exact test). C. Representative images of Control (top) and Satb2-ISRKO (bottom) animals following withdrawal response to Hargreaves thermal pain test. Following the withdrawal response, control animals replace the limb to a resting position (arrows in top panel), while Satb2-ISRKO animals maintain the withdrawn posture (arrows in bottom panel). D. Threshold to induce mechanical pain response in the Von Frey test was comparable between control and Satb2-ISRKO. Data is represented as mean threshold (grams of force) +/− S.E.M. N.S. indicates no significant difference (Mann Whitney). E. Satb2-ISRKO animals display a prolonged withdrawal posture following Von Frey mechanical pain stimulation. Data is represented as mean (number of animals maintaining withdrawn posture) * indicates significant difference (p=0.003; Fischer’s exact test). F, G. Satb2-ISRKO had a comparable withdrawal response (F) to light brush stimulation of the footpad, and light touch did not elicit the aberrant hyperflexion response in Satb2-ISRKO compared with controls (G). N.S. indicates no significant difference (F, Mann Whitney; G, Fischer’s exact test). See also Video S1, S2.
To test whether Satb2 expression in ISRSatb2 neurons is required to transduce other pain modalities, we performed the Von Frey test in Satb2-ISRKO animals to measure the response to mechanical stimulation. The threshold for producing the nociceptive withdrawal behavior following Von Frey fiber stimulation was normal in Satb2-ISRKO animals when compared with control littermates (Figure 5D). However, we observed a maintained flexion posture in Satb2-ISR animals following Von Frey stimulation (Figure 5E). In contrast, Satb2-ISRKO mutant animals have a normal withdrawal response to light touch (Figure 5F), and there are no differences in the withdrawn posture of the hindlimb following light brush stimulation (Figure 5G). Deletion of the Satb2 gene unmasks a genetic role for this transcription factor in ISRSatb2 neurons that leads to very selective defects in motor activity associated with hyperflexion while walking and retracting the limb from painful thermal and mechanical stimuli.
Loss of Satb2 perturbs ISRSatb2 neuron position and molecular profile
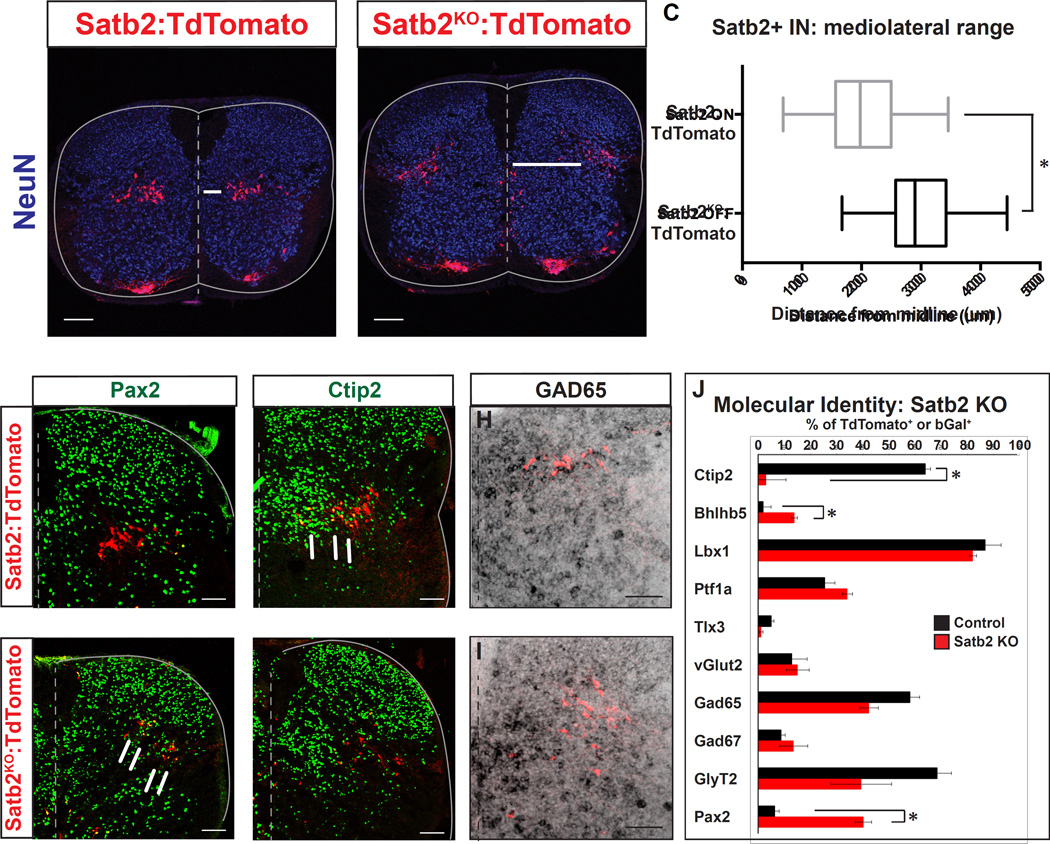
To understand how loss of the Satb2 gene contributes to the behavioral phenotype observed in Satb2-ISRKO animals, we monitored ISRSatb2 cell development using a lineage tracing strategy. Insertion of CreERT2 into the ATG of Satb2 generates a null allele (Figure S4), providing the opportunity to visually track the Satb2 null-interneurons in Satb2Cre/Cre:TdTomato animals (Satb2KO:Tom+). Satb2Cre/Cre (Satb2KO) animals phenocopy the null mouse line, in which there are defects in mandibular development and perinatal lethality (Dobreva et al., 2006; data not shown). We found that Satb2KO:Tom+ interneurons are generated in normal numbers (Figure 6A, B, and data not shown), however there were marked changes in cellular position along the mediolateral axis. ISRSatb2 (Satb2Cre/WT:TdTomato; abbreviated Satb2:Tom+) interneurons are normally organized in a tight band in the deep dorsal horn, spanning the mediolateral axis. In contrast, Satb2KO:Tom+ interneurons were shifted to a lateral position, and were more loosely organized with cells scattered dorsally (Figure 6A, B). The mean distance of Satb2:Tom+ neurons from the midline at lumbar levels was 201.6 +/− 4.7um, while Satb2:Tom+ neurons were located 301.5+/−4.4um from the midline (Figure 6C; p-value: <0.0001, Mann-Whitney), resulting in a final settling position at the lateral margin of the spinal cord. In addition to the main cluster of interneurons in the deep dorsal horn, we also observed a scattered population along the ventral midline in Satb2KO:TdTomato spinal cords. This population may represent a subset of neurons that were unable to exit the ventricular zone or that have an altered migration pattern (Figure 6B, arrowheads). These results indicate that Satb2 is necessary for establishing the medio-lateral position of ISRSatb2 neurons.
Figure 6. Loss of Satb2 leads to significant changes in cell body position and an altered molecular profile.
A-C. Cell body position is shifted to the lateral margin of the spinal cord in Satb2 null interneurons. A. Examination of Satb2:TdTomato control spinal cords reveals that ISRSatb2 neurons (red) are positioned throughout the mediolateral extent of lamina V in the deep dorsal horn. NeuN, blue. B. Examination of Satb2KO:TdTomato neurons (red) reveals a lateral shift in cell body position in response to loss of Satb2. Arrows in A,B represent the distance from the midline to the medial edge of the TdTomato+ population. Arrowheads in B emphasize the position of a subset of Satb2 null interneurons that are positioned near the central canal. C. Quantification of cell body position from A, B. Control (Satb2:TdTomato) and Satb2 null (Satb2KO:TdTomato) data are presented as box and whisker plots indicating the median and middle 50% of the range of cell body position. The middle 50% of cell body position encompassed a range of 156.6–249.8um for Satb2ON interneurons and 257.4–342.0um for Satb2KO interneurons. * indicates significant difference in the median (p<0.05; Mann-Whitney). D-I. Changes in molecular profile in response to loss of Satb2. Comparison of control Satb2+ (Satb2:TdTomato) and Satb2 null (Satb2KO:TdTomato) interneurons. D, E. Pax2 expression (green) is upregulated in Satb2 null interneurons (red in E) compared with control (red) interneurons. F, G. Loss of Ctip2 expression (green) in Satb2 null (red in G) neurons compared with control (red in F). Arrows in F highlight the presence of yellow double labeled cells in control Satb2:TdTomato interneurons. H, I. Colabeling of GAD65 (black) and TdTomato labeling is comparable in control (red in H) and Satb2KO:TdTomato (red in I). J. Quantification of colabeling of Tomato+ neurons in control Satb2:TdTomato and Satb2KO:TdTomato spinal cords. Data is represented as mean +/− S.E.M. * indicates significant difference (p<0.05; Mann Whitney). See also Supplemental Figure S6, Table S1. Scale bars in A, B, 100um. Scale bars in D-G, 50um. Scale bars in H, I, 200um.
To compare the molecular profile of Satb2:Tom+ and Satb2KO:Tom+ spinal neurons, we performed double labeling experiments with immunohistochemistry or in situ hybridization. A panel of transcription factor markers for dorsal and ventral spinal interneuron populations were selected based on their coexpression within ISRSatb2 cells and the surrounding neuron populations (Figure S1; Gross et al 2002). Immunohistochemistry for these markers revealed a number of gene expression changes (Figure 6D–J, Figure S6, Table S1, and data not shown). Ptf1a and Lbx1, expressed by many dorsal interneuron populations, including subsets of ISRSatb2 neurons, were unchanged in response to loss of Satb2. In contrast, there were significant changes in the expression of Pax2 (Figure 6D, E) and Bhlhb5 (Figure S6), genes that regulate neuronal circuit assembly and inhibitory neurotransmitter identity, respectively (Brohl et al., 2008; Huang et al., 2008). In addition, Ctip2 expression is almost completely lost in the absence of Satb2 (Figure 6F, G).
The changes we observed in the molecular profile of Satb2KO:Tom+ neurons prompted us to test whether there were corresponding alterations in neurotransmitter identity, as this would likely alter the function of these cells within a circuit. To test this we performed in situ hybridization for neurotransmitter markers in Satb2:Tom+ and Satb2KO:Tom+ animals. Importantly, the ratio of inhibitory/excitatory status of Tom+ neurons is unchanged in the absence of Satb2 gene function (Figure 6H–J and Figure S6, Table S1). Consistent with the role of Satb2 in cortical neuron migration (Alcamo et al., 2008; Britanova et al., 2008), these results demonstrate that Satb2 regulates gene expression and cell position in ISRSatb2 spinal neurons.
Loss of Satb2 disrupts ISRSatb2 neuron circuit integration
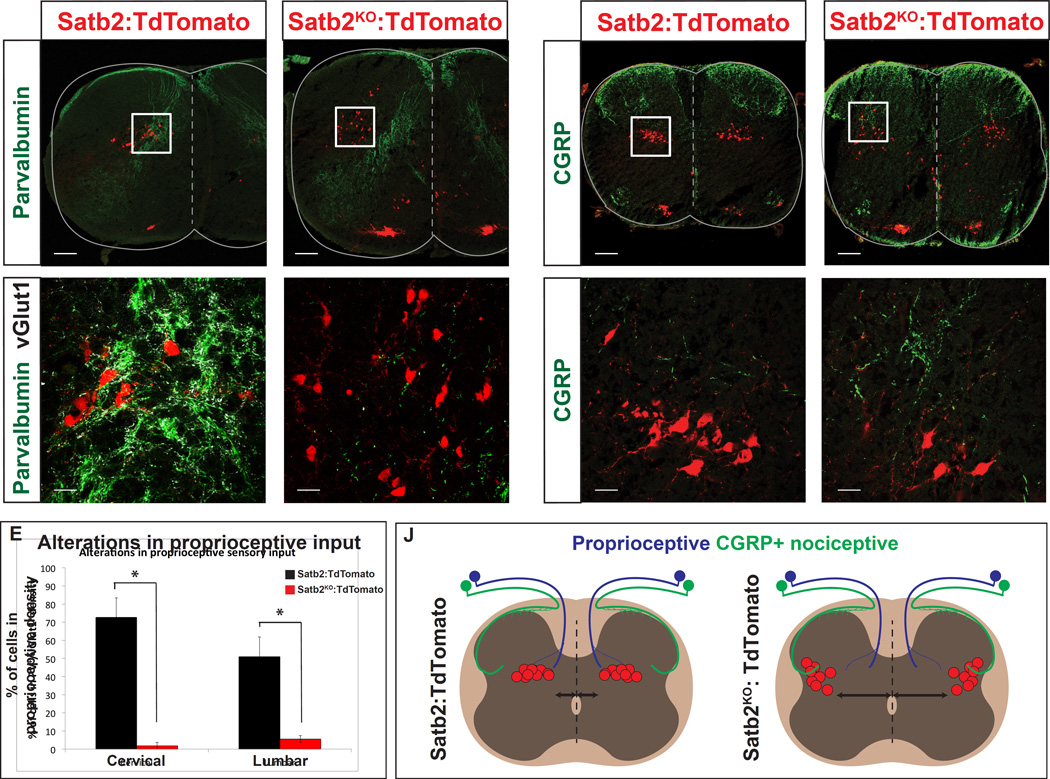
Cell body positioning within the spinal cord is an important regulator of synaptic connectivity (Arber, 2012). To determine whether the mislocalized Satb2KO:Tom+ cells had alterations in proprioceptive input we performed immunohistochemistry for parvalbumin and vGlut1. In Satb2:Tom+ animals, a majority of ISRSatb2 neurons are located within the pathway intersected by proprioceptive fibers. In contrast, the percentage of Satb2KO:Tom+ cells receiving proprioceptive input at cervical and lumbar levels was greatly reduced (Figure 7A–E). Interestingly, the stereotypical trajectory and density of proprioceptive fibers in the spinal cord was unchanged despite the mislocalization of one of their target cell types in Satb2 mutants (Figure 7A, C, and data not shown).
Figure 7. Alterations in sensory connectivity in Satb2 null spinal cords.
A-E. Loss of proprioceptive input in Satb2 null interneurons. A, B. Wild type ISRSatb2 neurons (Satb2:TdTomato) are located in the dense termination zone of proprioceptive afferents. White box in A corresponds to high magnification image shown in B. Proprioceptive contacts, identified by coexpression of parvalbumin (green) and vGlut1 (white), were identified on TdTomato+ neurons (red) in control Satb2:TdTomato (in A, B) or Satb2KO:TdTomato (in C, D) spinal cords. C, D. Satb2 null interneurons (Satb2KO:TdTomato) located at the lateral margin of the spinal cord are no longer positioned in the proprioceptive targeting domain. White box in C corresponds to high magnification image shown in D. E. Significant loss of Satb2KO:TdTomato interneurons (black) in the proprioceptive density, compared with control Satb2:TdTomato neurons (grey). Number of cells in the main density of proprioceptive fibers were quantified. At cervical levels, 72.6 +/− 2.5% of Satb2:TdTomato interneurons were located in the proprioceptive density (n=4 spinal cords). 1.8 +/− 1.8% of Satb2KO:TdTomato interneurons were located in the proprioceptive density (n=4 spinal cords). At lumbar levels, 51.0 +/− 3.4% of Satb2:TdTomato interneurons were located in the proprioceptive density (n=5 spinal cords). In contrast, 5.6 +/− 2.6% of Satb2KO:TdTomato interneurons were located in the proprioceptive density (n=6 spinal cords). Data is represented as mean +/− S.E.M. * indicates significant difference (p<0.05; Mann Whitney). F-I. Satb2 null interneurons occupy targeting domain of CGRP+ nociceptors. A, B. Consistent with previous reports, CGRP+ fibers (green) were identified in a dense termination in spinal lamina I and II, with few projections in lateral lamina V (Guo et al., 2011). Wild type ISRSatb2 neurons (red) in Satb2:TdTomato animals do not overlap with the CGRP targeting domain. White box in F corresponds to high magnification image shown in G. H, I. Satb2 null interneurons (red) in Satb2KO:TdTomato animals located in the lateral deep dorsal horn are positioned in an area overlapping with CGRP+ nociceptors (green). White box in H corresponds to high magnification image shown in I. J. Schematic showing alterations in sensory targeting in response to loss of Satb2. Left panel, ISRSatb2 neurons (red) in Satb2:TdTomato animals are located in the dense termination zone of proprioceptive fibers, but do not normally overlap with the fibers in lateral lamina V that originate from a subset of nociceptors expressing CGRP+. Right panel, Satb2 null interneurons (red) in Satb2KO:TdTomato animals are shifted to a lateral position, losing their position in the proprioceptive targeting domain. Instead, Satb2 null interneurons are overlapping with the termination zone of CGRP+ nociceptors. Scale bars in A, C, F, H, 100um. Scale bars in B, D, G, I, 20um
To determine whether laterally shifted Satb2KOTom+ neurons might receive new inputs, we examined primary afferents of peptidergic nociceptors expressing CGRP that are known to target spinal neurons in lateral lamina V (Guo et al., 2011). Under normal conditions Satb2:Tom+ neurons are located medial to the CGRP+ fiber density (Figure 7F, G). The projection of CGRP+ fibers was unchanged in Satb2 mutants, however the lateral Satb2KO:Tom+ neurons ectopically overlap with the termination domain of CGRP+ fibers (Figure 7H–J).
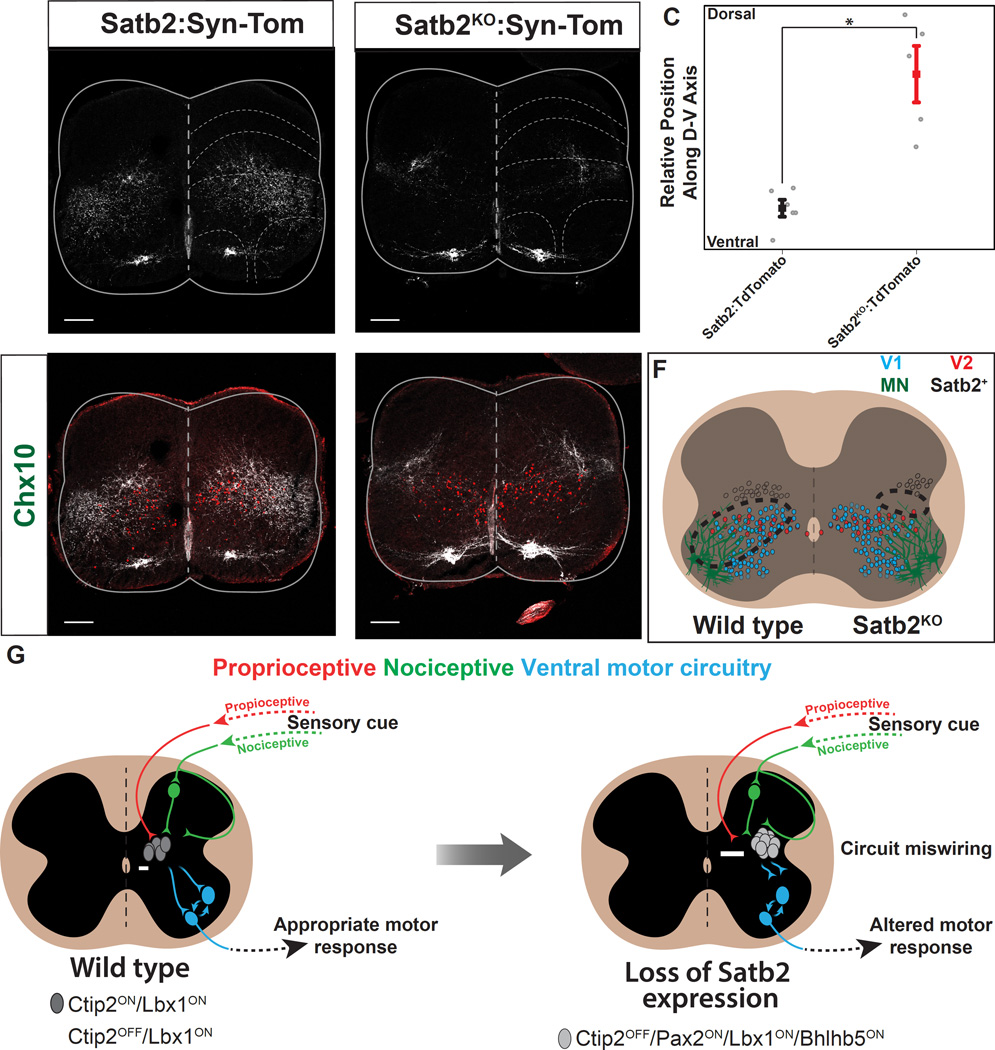
To test whether Satb2 is necessary to regulate the connectivity of ISRSatb2 cells with postsynaptic motor-circuit targets, we compared the distribution of terminals in Satb2:Synaptophysin-TdTomato (Satb2:Syn-Tom+) and Satb2KO:Synaptophysin-TdTomato (Satb2KO:Syn-Tom+) animals. In Satb2:Syn-Tom+ animals, synaptic termini were distributed throughout the ventral spinal cord including contacts with V2a, V1 and motor neurons (Figure 8A, D; see also Figure 3C–G). In contrast, the overall distribution of terminals were less abundant and shifted to dorsal targets above the V2a cell domain in Satb2KO:Syn-Tom+ animals (Figure 8A–E). These results demonstrate that Satb2 gene function is required to ensure the proper connectivity between ISRSatb2 neurons and their postsynaptic partners within the ventral spinal cord that regulate motor output (Figure 8F).
Figure 8. Loss of Satb2 reduces synaptic input to ventral motor circuitry.
A. Synaptic target domain of ISRSatb2 neurons (red), identifiable by genetic labeling with Satb2:Synaptophysin-TdTomato (Satb2:Syn-Tomato). Synaptophysin-TdTomato labeling (white) was observed throughout lamina V-VII and IX in Satb2:Syn-Tom animals. B. In Satb2 null spinal cords, Synaptophysin-TdTomato labeling (white) was dorsally restricted in Satb2KO:Syn-Tom animals C. Quantification of fluorescent pixel intensity for control (Satb2:Syn-Tom, black) and Satb2 null (Satb2KO:Syn-Tom, red) animals reveals a significant shift in the relative position of presynaptic terminals along the dorsal-ventral axis. Data is represented as mean +/− S.E.M. * indicates significant difference (p<0.05; Student’s t-test). D. In control animals (Satb2:Syn-Tom), the presynaptic terminals of ISRSatb2 neurons (red) are intermingled with Chx10+ V2a interneurons (green), one of the known target populations of ISRSatb2 neurons. See also Figure 3E. E. In Satb2 null animals (Satb2KO:Syn-Tom), the density of presynaptic terminals (red) is instead shifted dorsal to the Chx10+ V2a interneurons (green). F. Schematic showing the wild type distribution of presynaptic terminals of ISRSatb2 neurons (left panel, dotted line) overlapping with ventral interneurons (blue, red) and motor neurons (green). In contrast, the density of presynaptic terminals of Satb2 null interneurons (right panel, dotted line) is shifted dorsally to a position that no longer encompasses cellular components of ventral motor circuitry. G. Summary diagram of cell body position and circuitry changes that result from loss of Satb2. In wild type spinal cord, ISRSatb2 neurons (grey/white) are normally located at the intersection of proprioceptive (red) and indirect nociceptive (green) modalities, and form synaptic connections with motor circuitry in the ventral spinal cord (blue). In response to loss of Satb2, interneurons are shifted to a lateral position, and synaptic inputs and outputs are altered. These cellular and circuit-level changes are accompanied by an altered behavioral response during specific phases of walking and in response to noxious stimulation. Arrows indicate cell body distance from the midline. Scale bars in A, B, D, E, 100um.
Discussion
Commands from different types of sensory neurons that monitor limb position and pain converge onto spinal neurons within the deep dorsal horn, which are thought to serve as a local hub for integrating multiple sensory modalities (Craig, 2003). The output of this local circuit is capable of directly modulating the motor system to produce rapid state-dependent behaviors such as reflexive withdrawal movements. The cellular composition and developmental mechanisms that give rise to this sensorimotor system within the spinal cord are not well understood, but have focused primarily on plasticity based learning mechanisms (Schouenborg, 2004). We found that a discrete subpopulation of lamina V inhibitory sensory relay neurons (ISRSatb2) require the transcription factor Satb2 to establish their proper input- and output-pattern of connectivity. Deletion of the Satb2 gene from ISRSatb2 neurons does not disrupt most types of motor activity. Rather, we observed specific behavioral abnormalities in which hyperflexion defects were apparent during specific phases of walking and in response to sensory stimuli. Our studies demonstrate that Satb2 controls the development of a discrete component of the sensorimotor system.
Satb2 function unmasks a sensory-motor microcircuit
Within the deep dorsal horn sensorimotor neurons have diverse properties including wide dynamic range responses and distinct patterns of inputs from cutaneous and proprioceptive afferents (Harrison and Jankowska, 1985; Hongo et al., 1989). Furthermore, both inhibitory and excitatory sensorimotor interneurons are intermingled and some project locally while others project to supraspinal targets. Using Satb2 as a marker we found that ISRSatb2 neurons express inhibitory neurotransmitters and are arrayed along the mediolateral axis of lamina V. They receive input from proprioceptive and nociceptive pathways and target multiple cellular components of the ventral motor network, but appear to lack supraspinal projections. Thus, ISRSatb2 neurons represent a specific subset of cells within the sensory-to-motor pathway (Figure 8G).
Mutation of Satb2 does not perturb the function of the core circuitry associated with central pattern generation. Rather, Satb2 mutation disrupts both the location and connectivity of ISRSatb2 neurons: they reposition to the lateral region of lamina V, they have a greatly diminished number of synaptic inputs from proprioceptive fibers, and they lack outputs to the ventral spinal cord where motor actions are controlled (Figure 8G). Interestingly, the ablation of all proprioceptive sensory neurons leads to severe locomotor deficits including rotarod defects (Ilieva et al., 2008; Oliveira Fernandes and Tourtellotte, 2015), whereas Satb2 mutant mice had normal rotarod function. Taken together, these findings suggest that the remaining proprioceptive connections in Satb2 mutants are sufficient for many motor tasks.
We found that Satb2 expression in spinal interneurons is required for specific motor and sensory-evoked behaviors including proper limb position during runway walking, and limb flexion-withdrawal following thermal and mechanical stimulation. Studies in the cat suggest that muscle afferents help to set the timing of limb flexion and the transitions from swing to stance (Hiebert et al., 1996; Stecina et al., 2005). Perhaps the hyperflexion phenotypes observed in Satb2 mutants are caused by a defect in how proprioceptors control the timing of limb flexion in specific behavioral contexts such as when sensory cues activate a limb flexion response.
Satb2 mutants display an increased latency to withdrawal from thermal stimulation, revealing a breakdown in the transformation of sensory input into motor output (Figure 8G). The aberrant hindlimb hyperflexion that was observed following thermal and mechanical stimulation was not produced by light touch stimulation, indicating that Satb2 is required for the execution of a specific set of sensory-motor behaviors. Although it is unclear whether the behavioral changes observed in Satb2 mutants represent a loss- or aberrant gain-of-function of ISRSatb2 neurons, our findings demonstrate that Satb2 is necessary to ensure the proper development of a subcomponent of the sensorimotor circuitry whose function is only apparent during specific behavioral contexts. Importantly, the defects that arise from the targeted deletion of Satb2 from ISRSatb2 neurons functionally-reveals a cellular pathway that transforms multi-modal sensory stimuli into motor action.
Satb2 gene regulation and cellular position
We report an altered molecular profile within ISRSatb2 neurons in response to loss of the Satb2 gene. Transcription factors such as Lbx1 and Ptf1a that specify progenitor domains within dorsal interneurons were unchanged in Satb2 null interneurons. We instead observed changes in the expression of genes that regulate subsequent phases of neural development. For instance, Bhlhb5, a gene that is involved in neuronal circuit assembly (Ross et al., 2012), is upregulated in a subset of Satb2 null interneurons that are positioned near the central canal. Although Satb2 null interneurons retain their inhibitory neurotransmitter identity, Pax2 expression is significantly upregulated. Pax2 is a downstream target of Lbx1 and Ptf1a, and is known to regulate glycinergic cell fate as well as the expression of neuropeptides used for inhibitory neurotransmission (Brohl et al., 2008; Cheng et al., 2004; Gross et al., 2002; Huang et al., 2008). Although not extensively investigated in this study, the genetic changes we observed raise the possibility that Satb2 mutant ISRSatb2 neurons may have an abnormal composition of neurotransmitters and/or neuropeptides.
Satb2 is considered a direct repressor of Ctip2 in cortical neurons, however recent studies have identified a population of Satb2/Ctip2 coexpressing cortical neurons (Harb et al., 2016). Likewise, we found that medial ISRSatb2 neurons coexpress Ctip2, whereas lateral ISRSatb2 cells lack Ctip2. Unlike the cortex, when Satb2 is mutated Ctip2 expression is not upregulated in ISRSatb2 neurons, instead Ctip2 expression is lost. Our findings reveal two critical aspects of gene regulatory networks in the spinal cord. First, ISRSatb2 neurons lack the antagonistic gene interaction that segregates most Satb2+ and Ctip2+ cortical neurons (Alcamo et al., 2008; Britanova et al., 2008). Second, the Ctip2 status of ISRSatb2 neurons reveals a genetic heterogeneity among ISRSatb2 neurons along the mediolateral axis of lamina V. The loss of Satb2 appears to disrupt this cellular heterogeneity. Future studies will be important for understanding whether Ctip2 is a stringent indicator of synaptic input and output patterns within the ISRSatb2 neuron population.
Cell position is an important organizational feature within the diverse neuronal types that occupy the deep dorsal horn. For example, a subset of spinal interneurons in lamina V, termed “reflex encoders”, are topographically organized such that cellular position along the mediolateral axis correlates with patterns of synaptic input and output (Schouenborg et al., 1995). In addition, lamina V premotor neurons display a medial-extensor and lateral-flexor bias (Tripodi et al., 2011). We found that the medial ISRSatb2 neurons receive more proprioceptive inputs than the lateral cells. Thus, the subdivision of ISRSatb2 neurons based on their mediolateral Ctip2 status represents a molecular correlate that aligns with the orderly mapping relationships between pre- and/or post-synaptic targets. While past studies have focused on activity-based mechanisms during a critical window around birth (Schouenborg, 2004; Tripodi et al., 2011), our findings demonstrate that a complementary mechanism is also involved whereby Satb2 is an essential genetic regulator of the cellular connectome for spinal sensorimotor circuitry.
Experimental Procedures
Full methods can be found in the Supplemental Experimental Procedures
Supplementary Material
Acknowledgments
We gratefully acknowledge the generosity and advice of M. Goulding, L. Bachmann, C. Farrokhi, S. Bourane, and L. Garcia-Campmany; Q. Ma, R. Johnson, and C. Birchmeier for providing reagents, Y. Dayn for help with generation of the Satb2-CreERT2 transgenic mouse line. K. Lettieri, and C. Heller provided technical support and advice. K.L.H was supported as a National Science Foundation Graduate Research Fellow and the Chapman Foundation. A.J.L was supported by George E. Hewitt Foundation for Medical Research and Christopher and Dana Reeve Foundation. C.A.H. was supported by a US National research Service Award Fellowship from US National Institutes of Health NINDS. M.H was supported by the Timken-Sturgis Foundation and the Japanese Ministry of Education, Culture, Sports, Science, and Technology Long-Term Student Support Program. S.L.P is supported as a Howard Hughes Medical Institute Investigator and as a Benjamin H. Lewis chair in neuroscience. This research was supported by funding from the Howard Hughes Medical Institute, the Marshall Foundation and the Sol Goldman Charitable Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
K.L.H and S.L.P designed the study and wrote the manuscript. K.L.H and A.J.L designed and carried out the experiments with help from C.A.H and M.H. J.M.M and M.G assisted with behavioral testing design and execution. S.P.D conducted statistical analysis. R.G. provided the Satb2lacz/lacZ and Satb2flx/flx mouse lines. Y.K and T.K.S provided the Satb2 antibody.
References
- Alaynick WA, Jessell TM, Pfaff SL. SnapShot: spinal cord development. Cell. 2011;146:178–178. doi: 10.1016/j.cell.2011.06.038. e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Farinas I, Grosschedl R, McConnell SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Allen JW, Yaksh TL. Assessment of acute thermal nociception in laboratory animals. Methods in molecular medicine. 2004;99:11–23. doi: 10.1385/1-59259-770-x:139. [DOI] [PubMed] [Google Scholar]
- Arber S. Motor circuits in action: specification, connectivity, and function. Neuron. 2012;74:975–989. doi: 10.1016/j.neuron.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Balasubramanian M, Smith K, Basel-Vanagaite L, Feingold MF, Brock P, Gowans GC, Vasudevan PC, Cresswell L, Taylor EJ, Harris CJ, et al. Case series: 2q33.1 microdeletion syndrome--further delineation of the phenotype. Journal of medical genetics. 2011;48:290–298. doi: 10.1136/jmg.2010.084491. [DOI] [PubMed] [Google Scholar]
- Bannatyne BA, Liu TT, Hammar I, Stecina K, Jankowska E, Maxwell DJ. Excitatory and inhibitory intermediate zone interneurons in pathways from feline group I and II afferents: differences in axonal projections and input. The Journal of physiology. 2009;587:379–399. doi: 10.1113/jphysiol.2008.159129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourane S, Duan B, Koch SC, Dalet A, Britz O, Garcia-Campmany L, Kim E, Cheng L, Ghosh A, Ma Q, et al. Gate control of mechanical itch by a subpopulation of spinal cord interneurons. Science. 2015a;350:550–554. doi: 10.1126/science.aac8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourane S, Grossmann KS, Britz O, Dalet A, Del Barrio MG, Stam FJ, Garcia-Campmany L, Koch S, Goulding M. Identification of a spinal circuit for light touch and fine motor control. Cell. 2015b;160:503–515. doi: 10.1016/j.cell.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braz JM, Nassar MA, Wood JN, Basbaum AI. Parallel “pain” pathways arise from subpopulations of primary afferent nociceptor. Neuron. 2005;47:787–793. doi: 10.1016/j.neuron.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. The European journal of neuroscience. 2005;21:658–668. doi: 10.1111/j.1460-9568.2005.03897.x. [DOI] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Brohl D, Strehle M, Wende H, Hori K, Bormuth I, Nave KA, Muller T, Birchmeier C. A transcriptional network coordinately determines transmitter and peptidergic fate in the dorsal spinal cord. Developmental biology. 2008;322:381–393. doi: 10.1016/j.ydbio.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Cavallari P, Edgley SA, Jankowska E. Post-synaptic actions of midlumbar interneurones on motoneurones of hind-limb muscles in the cat. The Journal of physiology. 1987;389:675–689. doi: 10.1113/jphysiol.1987.sp016677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Arata A, Mizuguchi R, Qian Y, Karunaratne A, Gray PA, Arata S, Shirasawa S, Bouchard M, Luo P, et al. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nature neuroscience. 2004;7:510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- Coulon P, Bras H, Vinay L. Characterization of last-order premotor interneurons by transneuronal tracing with rabies virus in the neonatal mouse spinal cord. The Journal of comparative neurology. 2011;519:3470–3487. doi: 10.1002/cne.22717. [DOI] [PubMed] [Google Scholar]
- Craig AD. Pain mechanisms: labeled lines versus convergence in central processing. Annual review of neuroscience. 2003;26:1–30. doi: 10.1146/annurev.neuro.26.041002.131022. [DOI] [PubMed] [Google Scholar]
- Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Farinas I, Karsenty G, Grosschedl R. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:971–986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Duan B, Cheng L, Bourane S, Britz O, Padilla C, Garcia-Campmany L, Krashes M, Knowlton W, Velasquez T, Ren X, et al. Identification of spinal circuits transmitting and gating mechanical pain. Cell. 2014;159:1417–1432. doi: 10.1016/j.cell.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink AJ, Croce KR, Huang ZJ, Abbott LF, Jessell TM, Azim E. Presynaptic inhibition of spinal sensory feedback ensures smooth movement. Nature. 2014;509:43–48. doi: 10.1038/nature13276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzPatrick DR, Carr IM, McLaren L, Leek JP, Wightman P, Williamson K, Gautier P, McGill N, Hayward C, Firth H, et al. Identification of SATB2 as the cleft palate gene on 2q32-q33. Human molecular genetics. 2003;12:2491–2501. doi: 10.1093/hmg/ddg248. [DOI] [PubMed] [Google Scholar]
- Forssberg H, Grillner S, Rossignol S. Phase dependent reflex reversal during walking in chronic spinal cats. Brain research. 1975;85:103–107. doi: 10.1016/0006-8993(75)91013-6. [DOI] [PubMed] [Google Scholar]
- Foster E, Wildner H, Tudeau L, Haueter S, Ralvenius WT, Jegen M, Johannssen H, Hosli L, Haenraets K, Ghanem A, et al. Targeted ablation, silencing, and activation establish glycinergic dorsal horn neurons as key components of a spinal gate for pain and itch. Neuron. 2015;85:1289–1304. doi: 10.1016/j.neuron.2015.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding M. Circuits controlling vertebrate locomotion: moving in a new direction. Nature reviews Neuroscience. 2009;10:507–518. doi: 10.1038/nrn2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillner S, Jessell TM. Measured motion: searching for simplicity in spinal locomotor networks. Current opinion in neurobiology. 2009;19:572–586. doi: 10.1016/j.conb.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross MK, Dottori M, Goulding M. Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron. 2002;34:535–549. doi: 10.1016/s0896-6273(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Guo T, Mandai K, Condie BG, Wickramasinghe SR, Capecchi MR, Ginty DD. An evolving NGF-Hoxd1 signaling pathway mediates development of divergent neural circuits in vertebrates. Nature neuroscience. 2011;14:31–36. doi: 10.1038/nn.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harb K, Magrinelli E, Nicolas CS, Lukianets N, Frangeul L, Pietri M, Sun T, Sandoz G, Grammont F, Jabaudon D, et al. Area-specific development of distinct projection neuron subclasses is regulated by postnatal epigenetic modifications. eLife. 2016:5. doi: 10.7554/eLife.09531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves K, Dubner R, Brown F, Flores C, Joris J. A new and sensitive method for measuring thermal nociception in cutaneous hyperalgesia. Pain. 1988;32:77–88. doi: 10.1016/0304-3959(88)90026-7. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Jankowska E. Sources of input to interneurones mediating group I non-reciprocal inhibition of motoneurones in the cat. The Journal of physiology. 1985;361:379–401. doi: 10.1113/jphysiol.1985.sp015651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiebert GW, Whelan PJ, Prochazka A, Pearson KG. Contribution of hind limb flexor muscle afferents to the timing of phase transitions in the cat step cycle. Journal of neurophysiology. 1996;75:1126–1137. doi: 10.1152/jn.1996.75.3.1126. [DOI] [PubMed] [Google Scholar]
- Hongo T, Kitazawa S, Ohki Y, Xi MC. Functional identification of last-order interneurones of skin reflex pathways in the cat forelimb segments. Brain research. 1989;505:167–170. doi: 10.1016/0006-8993(89)90132-7. [DOI] [PubMed] [Google Scholar]
- Huang M, Huang T, Xiang Y, Xie Z, Chen Y, Yan R, Xu J, Cheng L. Ptf1a, Lbx1 and Pax2 coordinate glycinergic and peptidergic transmitter phenotypes in dorsal spinal inhibitory neurons. Developmental biology. 2008;322:394–405. doi: 10.1016/j.ydbio.2008.06.031. [DOI] [PubMed] [Google Scholar]
- Ilieva HS, Yamanaka K, Malkmus S, Kakinohana O, Yaksh T, Marsala M, Cleveland DW. Mutant dynein (Loa) triggers proprioceptive axon loss that extends survival only in the SOD1 ALS model with highest motor neuron death. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12599–12604. doi: 10.1073/pnas.0805422105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehn O. Decoding the organization of spinal circuits that control locomotion. Nature reviews Neuroscience. 2016;17:224–238. doi: 10.1038/nrn.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehn O, Kjaerulff O. Spatiotemporal characteristics of 5-HT and dopamine-induced rhythmic hindlimb activity in the in vitro neonatal rat. Journal of neurophysiology. 1996;75:1472–1482. doi: 10.1152/jn.1996.75.4.1472. [DOI] [PubMed] [Google Scholar]
- Leone DP, Heavner WE, Ferenczi EA, Dobreva G, Huguenard JR, Grosschedl R, McConnell SK. Satb2 Regulates the Differentiation of Both Callosal and Subcerebral Projection Neurons in the Developing Cerebral Cortex. Cerebral cortex. 2014 doi: 10.1093/cercor/bhu156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Hinckley CA, Hilde KL, Driscoll SP, Poon TH, Montgomery JM, Pfaff SL. Identification of a cellular node for motor control pathways. Nature neuroscience. 2014;17:586–593. doi: 10.1038/nn.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Lewallen KA, Pfaff SL. Spatial organization of cortical and spinal neurons controlling motor behavior. Current opinion in neurobiology. 2012;22:812–821. doi: 10.1016/j.conb.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita M, Hosoya Y. Cells of origin of the spinocerebellar tract in the rat, studied with the method of retrograde transport of horseradish peroxidase. Brain research. 1979;173:185–200. doi: 10.1016/0006-8993(79)90620-6. [DOI] [PubMed] [Google Scholar]
- Muller T, Brohmann H, Pierani A, Heppenstall PA, Lewin GR, Jessell TM, Birchmeier C. The homeodomain factor lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron. 2002;34:551–562. doi: 10.1016/s0896-6273(02)00689-x. [DOI] [PubMed] [Google Scholar]
- Oliveira Fernandes M, Tourtellotte WG. Egr3-dependent muscle spindle stretch receptor intrafusal muscle fiber differentiation and fusimotor innervation homeostasis. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2015;35:5566–5578. doi: 10.1523/JNEUROSCI.0241-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puskar Z, Antal M. Localization of last-order premotor interneurons in the lumbar spinal cord of rats. The Journal of comparative neurology. 1997;389:377–389. doi: 10.1002/(sici)1096-9861(19971222)389:3<377::aid-cne2>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Rizzolatti G, Sinigaglia C. Mirror neurons and motor intentionality. Functional neurology. 2007;22:205–210. [PubMed] [Google Scholar]
- Ross SE, McCord AE, Jung C, Atan D, Mok SI, Hemberg M, Kim TK, Salogiannis J, Hu L, Cohen S, et al. Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron. 2012;73:292–303. doi: 10.1016/j.neuron.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol S, Dubuc R, Gossard JP. Dynamic sensorimotor interactions in locomotion. Physiological reviews. 2006;86:89–154. doi: 10.1152/physrev.00028.2005. [DOI] [PubMed] [Google Scholar]
- Schouenborg J. Learning in sensorimotor circuits. Current opinion in neurobiology. 2004;14:693–697. doi: 10.1016/j.conb.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Schouenborg J, Weng HR. Sensorimotor transformation in a spinal motor system. Experimental brain research. 1994;100:170–174. doi: 10.1007/BF00227291. [DOI] [PubMed] [Google Scholar]
- Schouenborg J, Weng HR, Kalliomaki J, Holmberg H. A survey of spinal dorsal horn neurones encoding the spatial organization of withdrawal reflexes in the rat. Experimental brain research. 1995;106:19–27. doi: 10.1007/BF00241353. [DOI] [PubMed] [Google Scholar]
- Schubert FR, Dietrich S, Mootoosamy RC, Chapman SC, Lumsden A. Lbx1 marks a subset of interneurons in chick hindbrain and spinal cord. Mechanisms of development. 2001;101:181–185. doi: 10.1016/s0925-4773(00)00537-2. [DOI] [PubMed] [Google Scholar]
- Smith JC, Feldman JL. In vitro brainstem-spinal cord preparations for study of motor systems for mammalian respiration and locomotion. Journal of neuroscience methods. 1987;21:321–333. doi: 10.1016/0165-0270(87)90126-9. [DOI] [PubMed] [Google Scholar]
- Stecina K, Quevedo J, McCrea DA. Parallel reflex pathways from flexor muscle afferents evoking resetting and flexion enhancement during fictive locomotion and scratch in the cat. The Journal of physiology. 2005;569:275–290. doi: 10.1113/jphysiol.2005.095505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien AE, Tripodi M, Arber S. Monosynaptic rabies virus reveals premotor network organization and synaptic specificity of cholinergic partition cells. Neuron. 2010;68:456–472. doi: 10.1016/j.neuron.2010.10.019. [DOI] [PubMed] [Google Scholar]
- Surmeier DJ, Honda CN, Willis WD., Jr Natural groupings of primate spinothalamic neurons based on cutaneous stimulation. Physiological and anatomical features. Journal of neurophysiology. 1988;59:833–860. doi: 10.1152/jn.1988.59.3.833. [DOI] [PubMed] [Google Scholar]
- Szemes M, Gyorgy A, Paweletz C, Dobi A, Agoston DV. Isolation and characterization of SATB2, a novel AT-rich DNA binding protein expressed in development-and cell-specific manner in the rat brain. Neurochemical research. 2006;31:237–246. doi: 10.1007/s11064-005-9012-8. [DOI] [PubMed] [Google Scholar]
- Tripodi M, Stepien AE, Arber S. Motor antagonism exposed by spatial segregation and timing of neurogenesis. Nature. 2011;479:61–66. doi: 10.1038/nature10538. [DOI] [PubMed] [Google Scholar]
- Troullos ES, Goldstein DS, Hargreaves KM, Dionne RA. Plasma epinephrine levels and cardiovascular response to high administered doses of epinephrine contained in local anesthesia. Anesth Prog. 1987;34:10–13. [PMC free article] [PubMed] [Google Scholar]
- Van Buggenhout G, Van Ravenswaaij-Arts C, Mc Maas N, Thoelen R, Vogels A, Smeets D, Salden I, Matthijs G, Fryns JP, Vermeesch JR. The del(2)(q32.2q33) deletion syndrome defined by clinical and molecular characterization of four patients. European journal of medical genetics. 2005;48:276–289. doi: 10.1016/j.ejmg.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nature methods. 2007;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Qu Z, Tickner J, Xu J, Dai K, Zhang X. The role of SATB2 in skeletogenesis and human disease. Cytokine & growth factor reviews. 2014;25:35–44. doi: 10.1016/j.cytogfr.2013.12.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.