Abstract
We conducted a prospective study to identify genome-wide changes in peripheral gene expression before and after sports-related concussion (SRC). A total of 253 collegiate contact athletes underwent collection of peripheral blood mononuclear cells (PBMCs) before the sport season (baseline). Sixteen athletes who subsequently developed an SRC, along with 16 non-concussed teammate controls, underwent repeat collection of PBMCs within 6 h of injury (acutely). Concussed athletes underwent additional sample collection at 7 days post-injury (sub-acutely). Messenger RNA (mRNA) expression at baseline was compared with mRNA expression acutely and sub-acutely post-SRC. To estimate the contribution of physical exertion to gene changes, baseline samples from athletes who subsequently developed an SRC were compared with samples from uninjured teammate controls collected at the acute time-point. Clinical outcome was determined by changes in post-concussive symptoms, postural stability, and cognition from baseline to the sub-acute time-point. SRC athletes had significant changes in mRNA expression at both the acute and sub-acute time-points. There were no significant expression changes among controls. Acute transcriptional changes centered on interleukins 6 and 12, toll-like receptor 4, and NF-κB. Sub-acute gene expression changes centered on NF-κB, follicle stimulating hormone, chorionic gonadotropin, and protein kinase catalytic subunit. All SRC athletes were clinically back to baseline by Day 7. In conclusion, acute post-SRC transcriptional changes reflect regulation of the innate immune response and the transition to adaptive immunity. By 7 days, transcriptional activity is centered on regulating the hypothalamic-pituitary-adrenal axis. Future efforts to compare expressional changes in fully recovered athletes with those who do not recover from SRC could suggest putative targets for therapeutic intervention.
Key words: : gene network, IPA, longitudinal study, mild traumatic brain injury, mTBI, PBMC, sports-related concussion
Introduction
Despite 3.8 million sports-related concussions (SRC) in the United States annually, there is currently no approved treatment. This may be due in part to a limited understanding of concussion pathophysiology, as well as an inability to determine individuals at risk for long-term deficits.1 Genome-wide changes in peripheral gene expression using messenger RNA (mRNA) samples can provide valuable insights into the underlying pathophysiology and potential repair mechanisms of acute traumatic brain injury (TBI), including SRC. Gene expression changes in the brain have been previously characterized among severe TBI patients using post-mortem and post-operative brain tissue samples.2–4 However, little is known regarding gene expression after less severe forms of TBI, such as concussion. After severe TBI, differentially expressed genes were found to be related primarily to transcriptional regulation, energy metabolism, signal transduction, inflammation, and intercellular adhesion.2 Although differential gene expression after concussion has not been described, polymorphisms in several genes have been shown to influence outcome. These genes include the calcium channel subunit, brain-derived neurotrophic factor, dopamine D2 receptor, dopamine active transporter, and dopamine β-hydroxylase.5 While these polymorphisms could potentially be used to identify athletes at risk for poor outcome after SRC, it is unclear how they could be used to develop therapeutics for SRC.
A better understanding of the transcriptional and translational changes occurring in the brain after concussion is more likely to identify potential therapeutic targets. Progress on this front has been hampered by the inaccessibility of human brain tissue after concussion, as the mortality and need for neurosurgery from this injury is close to zero. Thus, in order to describe transcriptional changes after concussion, less invasive approaches are needed.
Several lines of evidence suggest that gene activities in peripheral immune cells are affected by central cellular activities in the setting of pathologic conditions such as TBI. TBI-associated breakdown of the blood brain barrier facilitates activation of brain microglia and astrocytes by peripheral leukocytes.6 This interaction is reflected in the observed correlation between mRNA expression in peripheral blood mononuclear cells (PBMCs) and in cerebrospinal fluid (CSF),7 as well as in brain tissue.8,9 Moreover, changes in peripheral gene expression have shed light on the central mechanisms underlying other neurologic disorders, such as autism, schizophrenia, and post-traumatic stress disorder.10–12 Taken together, these studies suggest that central neuronal damage is communicated to the periphery and can be detected through gene expression changes in peripheral blood cells. Our primary objective, therefore, was to determine changes in peripheral blood transcriptome using PBMCs before and after SRC.
Methods
Patients
Between 2010 and 2012, pre-season (baseline) PBMC samples were collected from 253 National Collegiate Athletic Association Division III contact sport athletes at two universities in Rochester, New York, and then banked. Contact sports included basketball, football, ice hockey, lacrosse and soccer. Both males and females ages 18 and older were invited to participate, and there was no restriction on race or ethnicity. Individuals unable to speak and read English or Spanish, due to availability of Immediate Post-Concussion Assessment and Cognitive Testing (ImPACT) in only these languages, were excluded. Additional exclusion criteria included use of drugs or alcohol on the day of consent and baselining, subjects who were pregnant, and history of TBI within 2 weeks prior to baseline. Regarding the latter, history of TBI >2 weeks prior to baseline was documented. These athletes were followed prospectively for the development of an SRC, which was defined as an injury witnessed by an on-field coach or certified athletic trainer and meeting the definition of concussion as defined by the Sport Concussion Assessment Tool 2.13 In brief, this tool provides a structured framework for evaluating 22 post-concussive symptoms, as well as orientation, memory, recall, balance, and gait. Deficiencies in any of these areas were used to confirm suspicion of concussion. In athletes with an SRC, a second PBMC sample was obtained within 6 h of injury (acute sample) and a third PBMC sample was obtained at 7 days post-injury (sub-acute sample).
Because physical exertion alone can potentially produce changes in gene expression,14 a comparison of gene changes before and after SRC potentially identifies genes not only related to SRC, but also to physical exertion. In other words, immediately after a concussion, gene expression changes reflect both brain injury and physical exertion. In order to isolate the effects of brain injury alone on gene expression changes, the effects of physical exertion must be measured and removed. In order to measure the effects of physical exertion on post-SRC gene changes, a non-injured control group was examined. Non-injured athlete controls were identified at the time of each SRC. Athletes who supplied a baseline pre-season PBMC sample were eligible to serve as controls when one of their teammates suffered an SRC. Controls were matched to the concussed athlete for gender, team, and sport. A further eligibility requirement for controls was that they must have provided the baseline pre-season PBMC sample in the same month and year as the concussed athlete, thus controlling for the time interval between baseline and injury.
Both the concussed athlete and the non-injured teammate control athlete underwent phlebotomy for PBMC sampling at the same time acutely (i.e. within 6 h) post injury. The concussed athlete, but not the uninjured teammate control, underwent repeat phlebotomy at the sub-acute time-point. Because prior studies have demonstrated the stability of gene transcription profiles among healthy adults sampled at intervals ranging from 1 week15 to several months,16 it was assumed that the mRNA expression among uninjured control athletes would not change significantly over the 6 days spanning the acute-to-sub-acute time period. Unlike the injured athletes, who ceased all physical exertion after concussion, there was no change in the exertional activities of controls between the acute and subacute time-points. The institutional review boards at University of Rochester and Rochester Institute of Technology approved this protocol; written informed consent was obtained from all study participants prior to subject participation.
Clinical outcome after SRC
Clinical outcome after SRC was determined by changes in cognitive performance, post-concussive symptoms, and postural stability, according to the recommendations of the 3rd International Conference on Concussion in Sport.17 All participating athletes underwent baseline, pre-season determination of cognition and postural stability with ImPACT and the Balance Error Scoring System (BESS), respectively. ImPACT and BESS testing were repeated in all subjects 7 days post-injury. ImPACT is a proprietary computer program that measures verbal memory, visual memory, reaction time, and visuomotor speed.18 ImPACT also includes a post-concussive symptom inventory. Normal day-to-day variation (termed “reliable change”) has been determined for each of these cognitive domains: verbal memory, 8.75 points; visual memory, 13.55 points; visuomotor speed, 4.98 points; post-concussion symptom score, 9.18 points; and reaction time, 0.06 sec.19 A significant change in a specific cognitive domain was thus defined as change exceeding the reliable change for that domain. BESS requires the athlete to stand in three different stances (double leg, single leg, and in tandem) for 20 sec with eyes closed. Each stance is performed on a firm surface and on a 10-cm thick foam pad. The BESS score is calculated by adding 1 error point for each performance error, with a maximum of 10 errors per stance.20 Unlike ImPACT, there are no accepted reliable change values for BESS.
PBMC and RNA isolation
PBMCs were isolated within 1 h of venous blood collection following the protocol described in detail elsewhere.21 Isolated PBMC pellets were suspended in complete RPMI-10 medium and moved to storage in a −80°C freezer, after which they were stored at −190°C until analysis. Total RNA was isolated from PBMCs using TRIzol® Plus RNA Purification Kits (Life Technologies, Grand Island, NY), and was treated with DNase I-Amplification Grade Kits (Life Technologies). The purity and concentration of RNA samples were verified using a NanoDrop DN-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). RNA integrity was determined on an Agilent bioanalyzer 2100 using the RNA 6000 Nano kit (Agilent, Sana Clara, CA). The quality of mRNA was evaluated in each sample. Samples with an RNA integrity number (RIN) less than 7.0 were excluded from analysis.
Microarray
Using the GeneChip 3′ IVT Expression kit, each RNA (100 ng) sample was reverse transcribed, converted to biotinylated complementary RNA, and hybridized to Affymetrix HG-U133 Plus 2.0 microarrays (Affymetrix Inc, Santa Clara, CA), which contain 54,675 probesets representing over 38,500 specific human genes. After staining with streptavidin-phycoerythrin and thorough washing, the raw data were obtained by laser scanning imaging.
Statistical analysis
Demographic variables were compared in concussed athletes and uninjured teammate controls using Student's t-test for age, and Fisher's exact test for gender, race, and sport. Clinically significant changes in cognitive function were determined by the percentage of athletes in each of the five cognitive domains displaying changes not exceeding reliable change. In addition, the mean score for each domain pre-injury was compared with the score post-injury using a paired t-test. Changes in postural stability were determined by comparing the mean score for each stance preinjury to the score post-injury using a paired t-test. Statistical significance was defined as a p value of less than or equal to 0.05.
Partek Genomics Suite software, version 6.6 (Partek Inc, St. Louis, MO), was utilized for all analyses performed on microarray data. Interrogating probes were imported, and corrections for background signal were applied using the robust multi-array average method, with additional corrections applied for the GC-content of probes. The probesets were standardized using quantile normalization, and expression levels of each probe underwent log-2 transformation to normalize data distributions. Parameters for identifying differentially expressed genes over time (i.e., within-subject comparison) were then identified using analyses of variance of each probe set's expression level as a function of time-point (baseline, acute, or sub-acute), while controlling for gender and race as covariates. Restricted maximum likelihood method was employed to fit the fixed and random effects of the design separately. In order to estimate the contribution of physical exertion to post-SRC gene changes, the same pre-post gene expression comparisons were planned among uninjured teammate controls athletes. Significant gene expression changes were defined as those increasing or decreasing by at least 1.5-fold (from baseline) and a p value threshold with a false discovery rate <0.05 corrected for multiple comparisons.
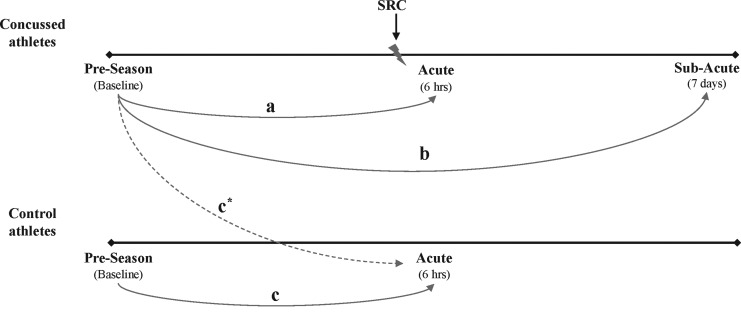
A Partek-generated heat map was used to display differential gene expression at baseline, as well as acutely and sub-acutely post-SRC. Significant changes in differentially expressed genes were identified by comparing gene expression at baseline with expression acutely after SRC, and by comparing gene expression at baseline with expression sub-acutely after SRC (Fig. 1).
FIG. 1.
Study design and analysis plan. Among athletes who suffered a sports-related concussion (SRC), gene expression was compared at baseline with acutely (within 6 h) post-SRC (a), and at baseline with sub-acutely (7 days) post-SRC (b). Among uninjured teammate athletes (controls), a piori analysis involved a comparison of gene expression at baseline with the same acute post-SRC time-point as the injured athlete to whom they were matched (c). Because none of the baseline samples from uninjured control athletes were suitable for messenger RNA (mRNA) analysis, baseline samples from athletes who subsequently suffered an SRC (i.e., before they were injured) were used as a surrogate for uninjured control baseline mRNA expression (c*). Based on the presumption that mRNA expression among uninjured teammate controls would not change significantly over the 6 days spanning the acute to sub-acute time period, mRNA expression at the acute time-point was applied to the sub-acute time-point.
The functional biologic networks associated with these significantly changed genes were identified using Ingenuity Pathway Analysis (IPA; Qiagen Ingenuity Systems Inc, Redwood City, CA). Central transcriptional nodes (“hubs”) were identified from the IPA-generated networks. Within each network, genes were ranked by the number of direct and indirect connections made with other genes. The top four genes with the most connections were considered transcriptional hubs.22
Results
Subjects and mRNA samples
Of the 253 athletes enrolled, 16 (6%) suffered a concussion during the study period. Sixteen uninjured teammate controls were enrolled during the same time period. History of TBI more than 2 weeks prior to baseline was similar in control and concussed athletes. Compared with control athletes, concussed athletes were older and more likely to be white (Table 1).
Table 1.
Demographics of Concussed (n = 16) and Control (n = 16) Athletes
| Athletes n (%) | Controls n (%) | p value | |
|---|---|---|---|
| Age (mean, SD) | 19.38 (1.47) | 18.53 (0.41) | 0.035 |
| Gender | 0.784 | ||
| Female | 8 (50%) | 9 (56%) | |
| Male | 8 (50%) | 7 (44%) | |
| Race | 0.018 | ||
| White | 16 (100%) | 10 (62.5%) | |
| Not reported | 0 (0%) | 6 (37.5%) | |
| Sport | 0.093 | ||
| Football | 6 (38%) | 7 (44%) | |
| Hockey | 4 (25%) | 0 (0%) | |
| Lacrosse | 1 (6%) | 0 (0%) | |
| Soccer | 5 (31%) | 9 (56%) | |
| Prior TBI | 7 (44%) | 2 (13%) | 0.113 |
SD, standard deviation; TBI, traumatic brain injury.
Of the samples obtained from the 16 concussed subjects, 15 had adequate mRNA (RIN >7) at baseline and the sub-acute time-point, while nine had adequate RNA at the acute time-point. Of the samples obtained from the 16 uninjured athlete controls, none had adequate mRNA at baseline but all 16 had adequate RNA integrity at the acute time period. Baseline samples from the 16 uninjured athlete controls were shipped separately from all other samples, leading us to speculate that adverse environmental conditions (e.g., heat) affected these samples. Because baseline samples from uninjured control athletes were unsuitable for analysis, baseline samples from athletes who subsequently suffered a concussion (i.e., before they were injured) were used as a surrogate for control baseline mRNA expression (Fig. 1).
Clinical outcomes
Among concussed athletes, there was no significant difference in mean cognitive performance on ImPACT pre-injury, compared with the sub-acute time-point (Table 2). No concussed athlete had changes exceeding reliable change in any of the five cognitive domains measured. Similarly, there was no significant difference in mean postural stability on BESS pre-injury, compared with sub-acutely post-injury (Table 3).
Table 2.
Mean (SD) ImPACT Performance Among Concussed Athletes (n = 16)
| Baseline | Day 7 | p value | |
|---|---|---|---|
| Verbal Memory Score | 88.13 (8.52) | 91.43 (6.88) | 0.211 |
| Visual Memory Score | 75.75 (15.81) | 75.79 (12.95) | 0.838 |
| Visual Motor Speed Score | 41.82 (5.90) | 43.91 (5.64) | 0.118 |
| Reaction Time (s) | 0.58 (0.07) | 0.54 (0.06) | 0.051 |
| Impulse Control Score | 5.13 (3.24) | 5.79 (2.94) | 0.459 |
| Total Symptom Score | 3.25 (7.49) | 2.57 (4.07) | 0.842 |
| Cognitive Efficiency Index | 0.39 (0.12) | 0.46 (0.08) | 0.059 |
SD, standard deviation; ImPACT, Immediate Post-Concussion Assessment and Cognitive Testing.
Table 3.
Mean (SD) Errors During BESS Assessment of Concussed Athletes (n = 16)
| Baseline | Day 7 | p value | |
|---|---|---|---|
| DL floor | 0 (0.00) | 0.06 (0.25) | 0.334 |
| SL floor | 3.31 (2.44) | 3.20 (1.86) | 0.506 |
| Tandom floor | 1.06 (1.12) | 1.20 (1.21) | 0.583 |
| DL foam | 0.13 (0.34) | 0.20 (0.41) | 0.671 |
| SL foam | 7.81 (2.37) | 7.00 (1.77) | 0.222 |
| Tandom foam | 4.56 (2.10) | 4.80 (2.88) | 0.628 |
| Total errors | 16.88 (5.38) | 16.47 (5.26) | 0.819 |
SD, standard deviation; BESS, Balance Error Scoring System; DL, double leg; SL, single leg.
Significant changes in gene expression before and after SRC
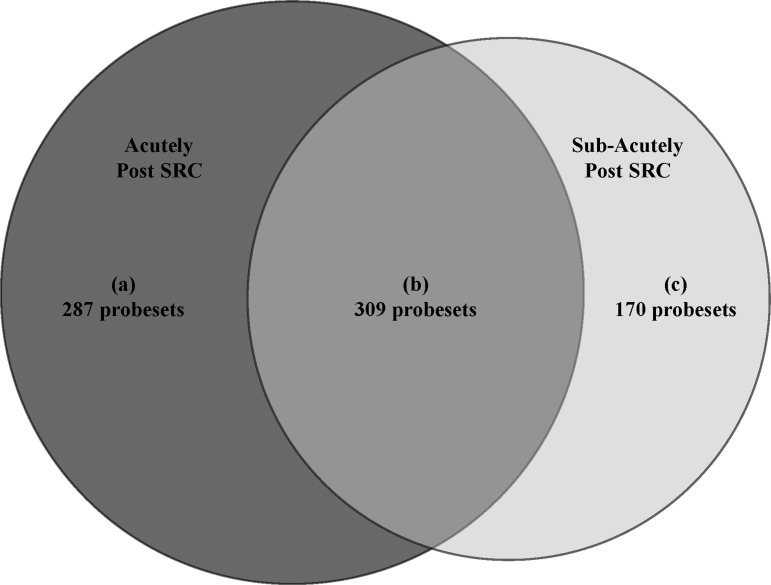
Of the 54,675 total probesets, 766 were found to have significant changes in expression level from baseline to post-SRC (either 1 day post-injury or 7 days post-injury) among concussed athletes. The expression of 596 probesets was significantly changed from baseline to the acute time-point, of which 287 were unique to this time-point. The expression of 479 probesets was significantly changed from the baseline to sub-acute time-point, of which 170 were unique to this time-point. The expression of 309 probesets was significantly changed from baseline to both the acute and sub-acute time-points (Fig. 2).
FIG. 2.
Probesets with significant changes in expression from baseline to post–sports-related concussion (SRC). Of the 54,675 total probesets, 766 were found to have significant changes in expression level from baseline to post-SRC. Specifically, 287 probesets were unique to (a) acutely after SRC, whereas 170 were unique to (c) sub-acutely after SRC. There were 309 probesets that were significantly differentially expressed from baseline to (b) both acutely and sub-acutely after SRC.
Twenty-five genes had > 2 fold change in mRNA expression between baseline and the acute post-SRC time-point (Supplementary Table 1; see online supplementary material at www.liebertpub.com). The genes with the largest decrease in expression were chemokine (C-C motif) ligand 4 (CCL4; 3.4-fold) and RAR-related orphan receptor A (RORA; 2.7-fold). The genes with the largest increase in expression were pyruvate dehydrogenase kinase, isozyme 4 (PDK4; 3.1-fold), and vacuole membrane protein 1 (VMP1; 2.6-fold).
Thirty-three genes had ≥2-fold change in mRNA expression between baseline and the subacute post-SRC time-point. (Supplementary Table 2; see online supplementary material at www.liebertpub.com). The genes with the largest decrease in expression were G0/G1switch 2 (G0S2; 7.7-fold), CCL3 (6.0-fold), and jun proto-oncogene (JUN; 4.7-fold). The genes with the largest increase in expression were EPM2A (laforin) interacting protein 1 (EPM2AIP1; 2.1-fold), and chemokine (C-X3-C motif) receptor 1 (CX3CR1; 1.9-fold).
There were no significant changes in gene expression between the concussed subjects at baseline (i.e., before they were injured) and the controls athletes at the acute time-point, even after controlling for race and gender.
Differential gene expression before and after SRC
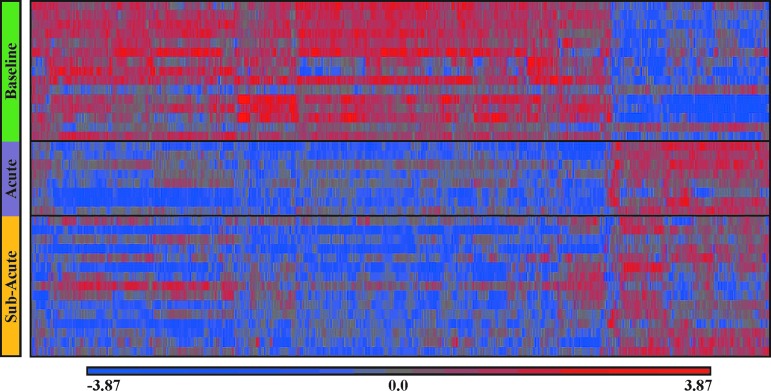
Among concussed athletes at baseline (i.e., before injury), the majority of differentially expressed gene transcripts displayed increased transcriptional activity (Fig. 3). However, acutely after SRC, of the 593 probesets that were differentially expressed, the majority (435; 73%) were down-regulated. This pattern persisted into the sub-acute time-point, where 408 of the 479 (85%) differentially expressed gene transcripts were down-regulated. Three hundred nine transcripts were differentially expressed at both the acute and sub-acute time-points.
FIG. 3.
Heat map of differential gene expression among athletes before (baseline) and after (acute and sub-acute) sports-related concussion (SRC). Color-coded expression levels of the 766 probesets that were significantly changed post-SRC (acute and sub-acute) relative to baseline (X-axis) among 16 SRC athletes, standardized to mean 0 and standard deviation of 1. Up-regulated genes are red, down-regulated genes are blue, genes with unchanged expression are colored gray. Individual SRC athletes are along the Y-axis, and grouped by the three indicated time-points. (There are fewer subjects at the acute time-point because of inadequate RNA in six peripheral blood mononuclear cell samples). A heat map of differential gene expression among uninjured athlete controls was not displayed because no significant pre-post changes in gene expression were detected. Color image is available online at www.liebertpub.com/neu
Canonical pathways and biologic networks associated with changes in gene expression after SRC
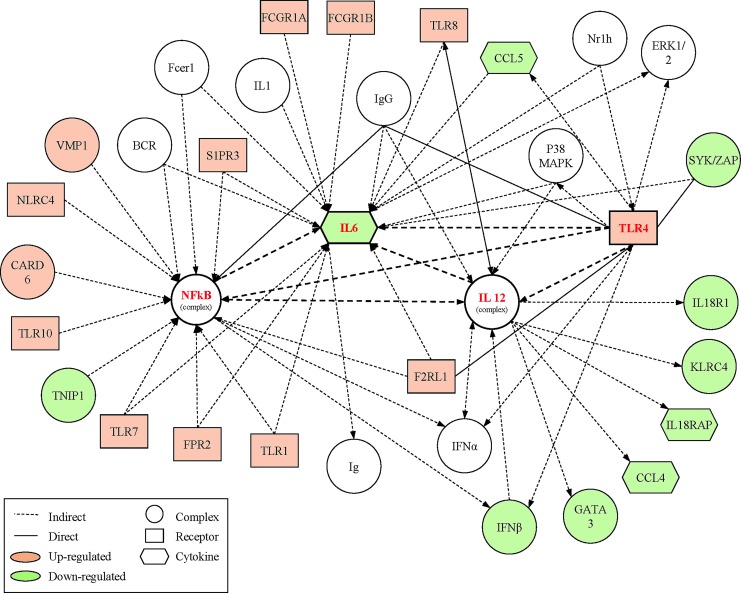
The top canonical pathways of genes that displayed significant changes from baseline to the acute post-SRC time-point involved “Immune Cell Functioning” and “Communication” that included the NF-κB and natural killer signaling pathways. The top network of gene changes were related to “Inflammatory Response, Infectious Disease, and Renal/Urological Disease” (Fig. 4). Genes identified as hubs in this network were interleukin 6 (IL-6; 19 connections), NF-κB (18 connections), IL-12 (13 connections), and toll-like receptor 4 (TRL4; 13 connections).
FIG. 4.
Top network of differentially expressed genes acutely (within 6 h) after sports-related concussion (SRC). Functional analysis of the top selected genes identified by microarray within 6 h of SRC centered on the “Inflammatory Response, Infectious Disease, Renal and Urological Disease” network. The network is graphically represented as nodes (genes) and lines (the biological relationship between genes). Red and green shaded nodes represent up- and down-regulated genes, respectively; empty nodes are those that are biologically linked to differentially expressed genes based on the evidence in the literature, but not differentially expressed in the analyzed samples. Solid lines represent a direct interaction between the two gene products while dotted lines indicates indirect interactions. Network hubs and their connections to each other are noted in bold. Only those genes with direct or indirect connections to one of the hubs were displayed in this figure for simplicity. Color image is available online at www.liebertpub.com/neu
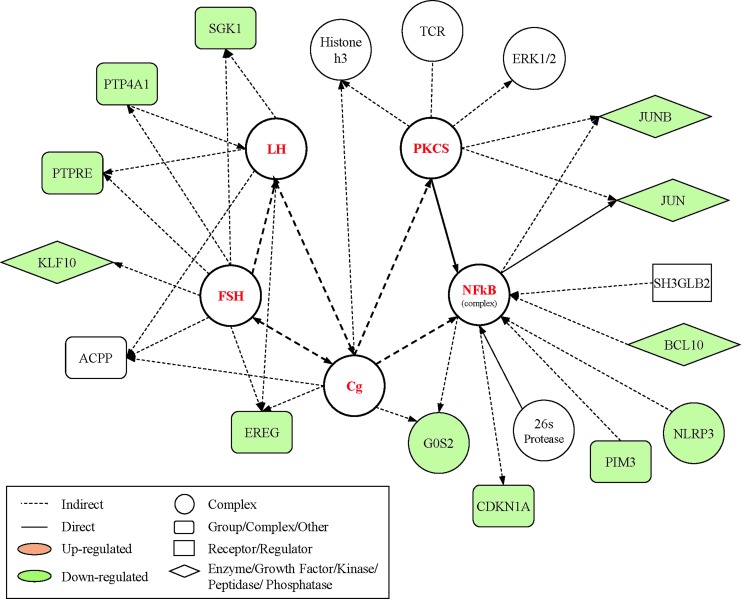
The top canonical pathways of genes that displayed significant changes from baseline to the sub-acute post-SRC time-point involved “Glucocorticoid Receptor Signaling,” and the top network of gene changes were related to “Neurological Disease, Cell Death and Survival” (Fig. 5). Genes identified as hubs in this network were NF-κB (11 connections), follicle-stimulating hormone (FSH; 8 connections), chorionic gonadotropin (Cg; 8 connections), luteinizing hormone (LH; 7 connections), and protein kinase catalytic subunit (PKCS; 7 connections).
FIG. 5.
Top network of differentially expressed genes sub-acutely (at 7 days) after sports-related concussion (SRC). Functional analysis of the top selected genes identified by microarray within 7 days after SRC centered on the “Neurological Disease, Cell Death and Survival, Cell Cycle” network. The network is graphically represented as nodes (genes) and lines (the biological relationship between genes). Red and green shaded nodes represent up- and down-regulated genes, respectively; empty nodes are those that are biologically linked to differentially expressed genes based on the evidence in the literature, but not differentially expressed in the analyzed samples. Solid lines represent a direct interaction between the two gene products while dotted lines indicate indirect interactions. Network hubs and their connections to each other are noted in bold. Only those genes with direct or indirect connections to one of the hubs were displayed in this figure for simplicity. Color image is available online at www.liebertpub.com/neu
Discussion
To our knowledge this is the first study to describe temporal changes in networks of altered genes after SRC, and indeed, after human TBI of any severity. Others have reported longitudinal changes in gene expression following experimental TBI in rodents. In these studies, early changes were related to transcriptional regulation, inflammation and cell signaling; later changes were related to complement system major histocompatibility complex class II pathway, and cell death/survival.23–26
Determining molecular causality and response to concussion is complex, but our findings suggest a distinct shift in gene expression activity between the acute and the sub-acute post-concussion periods. Because all SRC athletes were clinically back to baseline by Day 7, we interpret these gene changes to be adaptive. The cascade of molecular changes during the first 6 h following SRC is dominated by inflammatory activity centered around IL-6, IL-12, toll-like receptors (TLR), and NF-κB. What these four hub genes have in common is their effect on regulating the innate immune response, as well promoting the transition to an acquired, adaptive immune response.
Although the brain was once considered immunologically privileged, it is now known that it actively participates in inflammatory processes necessary for maintaining neural homeostasis. After experimental TBI, molecules such as heat shock proteins, high mobility group box-1, and hyaluronan released from damaged neurons have been shown to activate resident microglia via surface TLRs.27,28 TLRs are pattern recognition receptors that play an important role in the initiation of innate immunity.29 Using a similar mRNA pathway analysis, TLR signaling was found to be an important transcriptional hub 3 h after fluid percussion injury and 24 h after controlled cortical impact in rats.24
NF-κB is a major transcription factor that regulates genes responsible for both the innate and adaptive immune response. In support of our findings, two prior pathway analyses of mRNA expression in experimentally injured rodent brains also identified NF-κB signaling as an important transcriptional hub after TBI.24,30 NF-κB is known to be activated by stimulation of TLRs,31 which may explain why both were identified as key hubs after SRC. Moreover, downstream of NF-κB activation leads to expression of cytokines like IL-632 and IL-12.33 In the setting of more severe TBI, NF-κB expression has been found to be up-regulated in rodent models,34–36 as well as in humans.37 The precise role of NF-κB expression in modulating the innate and adaptive immune response after TBI has not been elucidated. However, genetically altered mice unable to up-regulate NF-κB have larger lesion volumes and blood-brain barrier breech after experimental TBI, suggesting that NF-κB activation may serve a neuroprotective function38 in this capacity.
IL-6 is an inflammatory cytokine that regulates the transition from innate to acquired immunity. The hallmark of this shift is a transition in the composition of inflammatory cells from neutrophils to mononuclear cells. IL-6 coordinates this transition by impacting cellular events that dampen innate immunity (e.g., suppressing chemokine release and promoting neutrophil apoptosis) while simultaneously promoting acquired immunity (e.g., promoting T-cell adhesion and blocking T-cell apoptosis).39 Several animal studies have demonstrated that TBI results in up-regulation of IL-6, and that a functioning IL-6 gene is necessary for recovery.40 In fact, a functional polymorphism (-174C/G) in the promoter region of IL-6 was found to be associated with increased mortality after severe TBI in humans.41 Using a mRNA pathway analysis, Redell and colleagues found the IL-6 signaling pathway to be an important hub within 3 h of both controlled cortical impact and fluid percussion injury in mice.24 As with animal studies, several human studies have shown that raised CSF levels of IL-6 correlate with improved post-TBI outcomes.42
Like IL-6, IL-12 is also a pro-inflammatory cytokine that participates in both innate (e.g., by inducing interferon-γ) and adaptive (e.g., by inducing a TH1 response in CD4+ cells) immunity.43 Two human studies have reported elevated IL-12 in the CSF and interstitial fluid after severe TBI.42,44 Although its role in TBI is less clear, IL-12 may function to shift microglial activation from the pro-inflammatory M1 phenotype (where IL-12 expression is typically high) to the anti-inflammatory M2 phenotype (where expression is reduced). In support of this idea, naturally-occurring45 and pharmacologically-induced46 reduction in IL-12 have been shown to be associated with reduced microglial activation after experimental TBI in mice.
Taken together, these findings suggest that in the acute phase, regulation of the innate immune response, and the transition to acquired immunity, is important for initiating recovery from SRC. Our results further suggest that 7 days post-injury, gene transcriptional activity shifts away from acute inflammation and toward the regulation of the hypothalamic-pituitary-adrenal (HPA) axis. We observed gene changes centering on glucocorticoid receptor signaling, with NF-κB, FSH, LH, Cg, and PKCS being the key transcriptional hubs. It is well known that more severe forms of TBI can disrupt the HPA axis, with reductions in growth hormone (GH) being the most commonly reported perturbation in humans.47 No prior reports have linked SRC to disturbances in the HPA axis, although retired boxers, kickboxers and professional football players have been shown to have various degrees of anterior pituitary dysfunction.48,49
Pre-clinical studies suggest that excessive glutamate receptor activation post-TBI impacts glucocorticoid mRNA transcription leading to its preferential down-regulation, especially in the hippocampus.50 Similar observations of the mesocorticolimbic system have been observed whereby excessive glucocorticoid receptor activation by cortisol increases the vulnerability of hippocampal neurons to damage from oxidative stress and excitotoxicity.51–53 Thus, down-regulation of glucocorticoid receptor signaling may serve to protect these vulnerable neurons during the sub-acute period, which may be necessary for eventual recovery. In the process of protecting these neurons, however, this down-regulation may reduce the production of important hormones such as GH, FSH, and LH, which can contribute to a variety of post-concussion symptoms. These gene activities are likely necessary to mitigate the excessive inflammation that develops during the acute period and present a shift in gene expression from neuronal proliferation to neuronal recovery, but may come at the expense of dysregulated hormonal control.
We observed that the majority of gene transcripts were down-regulated after SRC; 73% in the acute time period, and 85% at the sub-acute time period, relative to baseline. This finding is in contradiction to those in humans with severe TBI and many animal studies where up-regulation is more common. However, preclinical studies using rodent models of mTBI suggests that less severe brain injuries are associated with down- as opposed to up-regulation.25 Because all concussed athletes were clinically back to baseline at Day 7, suppression of inflammation and cell death cycles may be adaptive.
Limitations
An obvious limitation of this study is the relatively small sample size. In order to reduce the confounding effect of inter-individual variation in mRNA expression, we purposely sought to compare mRNA expression changes in individual athletes before and after concussion. This longitudinal study design necessitated obtaining baseline samples on hundreds of athletes at baseline/pre-season and then following them prospectively for the development of a concussion. The typical concussion rate is about 4–5% per year54 but in our study, 16 of 253 players (6%) suffered a concussion. Thus, in light of this study's unique design, the sample size may be relatively robust.
The use of isolated PBMCs rather than blood RNA tubes (e.g., PAXgene™, Tempus™) may have contributed to some samples having low RINs. Blood RNA tubes stabilize RNA at the point of venipuncture, thus reducing variability in manual processing associated with extracting RNA from PBMCs. Despite this advantage, the stability and reproducibly of gene transcript profiles using mRNA extracted from PBMCs has been well established. 15
Another limitation of the study was an inability to compare mRNA expression among uninjured athlete controls at baseline to expression during the contact sport season, which was obtained when a matched teammate suffered a concussion. This limitation resulted from the finding of inadequate RNA integrity among the baseline uninjured control samples and forced us to substitute baseline gene expression from the athletes who went on to be concussed (i.e., before they were injured). Despite this limitation, by using this methodology we found no significant changes in mRNA expression among uninjured teammate control athletes. However, a comparison among unrelated individuals likely has less power to detect significant changes in mRNA expression than a comparison among related individuals. Among controls, small group differences in mRNA expression due to differences in diet, sleep-wake cycles, and stress may have been missed.
In summary, we detected acute changes in peripheral gene expression following SRC, reflecting regulation of the innate immune response, as well as the transition to an acquired, adaptive immune response. By 7 days post-injury, transcriptional activity is centered on the regulation of the HPA axis. These findings illustrate a time-dependent shift in gene expression post-injury that may provide insight into the pathophysiology following SRC. An important next step is to verify translation of gene transcripts by measuring protein levels in an effort to provide a mechanistic insight into the pathophysiology of concussion. Future efforts to compare expressional changes among fully recovered athletes to those who do not recover from SRC could suggest putative targets for therapeutic intervention.
Supplementary Material
Acknowledgments
The authors wish to acknowledge head athletic trainers of University of Rochester and Rochester Institute of Technology, Eric Rozen and Ben Emke, for their assistance and support. The authors gratefully acknowledge the assistance of the athletic training staff, as well as Kirsten Ross, Stephanie Amalfe, and Nikita Bourque for their coordination efforts.
Author contributions: Design and conduct of the study—Bazarian and Merchant-Borna; collection, management, analysis, and interpretation of the data—Merchant-Borna, Lee, Wang, Gill, and Bazarian; manuscript preparation, review, and approval—Merchant-Borna, Lee, Gill, Bazarian, Bogner, and van Griensven.
Funding: This work was supported by funds from the NIH/NICHD (Award No. K24HD064754) and the NIH Intramural Research Program.
Author Disclosure Statement
Dr. Bazarian has a patent pending, “Method of Diagnosing Mild Traumatic Brain Injury.” Dr. Bazarian is a consultant for Banyan Biomarkers and Roche Diagnostics. For the other authors, no competing financial interests exist.
References
- 1.Langlois J.A., Rutland-Brown W., and Wald M.M. (2006). The epidemiology and impact of traumatic brain injury: a brief overview. J. Head Trauma Rehabil. 21, 375–378 [DOI] [PubMed] [Google Scholar]
- 2.Michael D.B., Byers D.M., and Irwin L.N. (2005). Gene expression following traumatic brain injury in humans: analysis by microarray. J. Clin. Neurosci. 12, 284–290 [DOI] [PubMed] [Google Scholar]
- 3.Liu H.D., Li W., Chen Z.R., Zhou M.L., Zhuang Z., Zhang D.D., Zhu L., and Hang C.H. (2013). Increased expression of ferritin in cerebral cortex after human traumatic brain injury. J. Neurol. Sci. 34, 1173–1180 [DOI] [PubMed] [Google Scholar]
- 4.Staffa K., Ondruschka B., Franke H., and Dresler J. (2012). Cerebellar gene expression following human traumatic brain injury. J. Neurotrauma 29, 2716–2721 [DOI] [PubMed] [Google Scholar]
- 5.McAllister T.W. (2010). Genetic factors modulating outcome after neurotrauma. PM R 2, S241–S252 [DOI] [PubMed] [Google Scholar]
- 6.Das M., Mohapatra S., and Mohapatra S.S. (2012). New perspectives on central and peripheral immune responses to acute traumatic brain injury. J. Neuroinflammation 9, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balakathiresan N., Bhomia M., Chandran R., Chavko M., McCarron R.M., and Maheshwari R.K. (2012). MicroRNA let-7i is a promising serum biomarker for blast-induced traumatic brain injury. J. Neurotrauma 29, 1379–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan P.F., Fan C., and Perou C.M. (2006). Evaluating the comparability of gene expression in blood and brain. Am. J. Med. Genet. B Neuropsychiatr. Genet. 141B, 261–268 [DOI] [PubMed] [Google Scholar]
- 9.Zhao W., Ho L., Varghese M., Yemul S., Dams-O'Connor K., Gordon W., Knable L., Freire D., Haroutunian V., and Pasinetti G.M. (2013). Decreased level of olfactory receptors in blood cells following traumatic brain injury and potential association with tauopathy. J. Alzheimers Dis. 34, 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segman R.H., Shefi N., Goltser-Dubner T., Friedman N., Kaminski N., and Shalev A.Y. (2005). Peripheral blood mononuclear cell gene expression profiles identify emergent post-traumatic stress disorder among trauma survivors. Mol Psychiatry 10, 500–513, 425. [DOI] [PubMed] [Google Scholar]
- 11.Yehuda R., Cai G., Golier J.A., Sarapas C., Galea S., Ising M., Rein T., Schmeidler J., Muller-Myhsok B., Holsboer F., and Buxbaum J.D. (2009). Gene expression patterns associated with posttraumatic stress disorder following exposure to the World Trade Center attacks. Biol. Psychiatry 66, 708–711 [DOI] [PubMed] [Google Scholar]
- 12.Glatt S.J., Tylee D.S., Chandler S.D., Pazol J., Nievergelt C.M., Woelk C.H., Baker D.G., Lohr J.B., Kremen W.S., Litz B.T., and Tsuang M.T.; Marine Resiliency Study Investigators. (2013). Blood-based gene-expression predictors of PTSD risk and resilience among deployed marines: a pilot study. Am. J. Med. Genet. B Neuropsychiatr. Genet. 162B, 313–326 [DOI] [PubMed] [Google Scholar]
- 13.McCrory P., Meeuwisse W., Johnston K., Dvorak J., Aubry M., Molloy M., and Cantu R. (2009). Consensus statement on concussion in sport: the 3rd International Conference on Concussion in Sport held in Zurich, November 2008. J. Athl. Train. 44, 434–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connolly P.H., Caiozzo V.J., Zaldivar F., Nemet D., Larson J., Hung S.P., Heck J.D., Hatfield G.W., and Cooper D.M. (2004). Effects of exercise on gene expression in human peripheral blood mononuclear cells. J. Appl. Physiol. 97, 1461–1469 [DOI] [PubMed] [Google Scholar]
- 15.Eady J.J., Wortley G.M., Wormstone Y.M., Hughes J.C., Astley S.B., Foxall R.J., Doleman J.F., and Elliott R.M. (2005). Variation in gene expression profiles of peripheral blood mononuclear cells from healthy volunteers. Physiol. Genomics 22, 402–411 [DOI] [PubMed] [Google Scholar]
- 16.De Boever P., Wens B., Forcheh A.C., Reynders H., Nelen V., Kleinjans J., Van Larebeke N., Verbeke G., Valkenborg D., and Schoeters G. (2014). Characterization of the peripheral blood transcriptome in a repeated measures design using a panel of healthy individuals. Genomics 103, 31–39 [DOI] [PubMed] [Google Scholar]
- 17.McCrory P., Meeuwisse W.H., Aubry M., Cantu R.C., Dvorak J., Echemendia R.J., Engebretsen L., Johnston K., Kutcher J.S., Raftery M., Sills A., Benson B.W., Davis G.A., Ellenbogen R., Guskiewicz K.M., Herring S.A., Iverson G.L., Jordan B.D., Kissick J., McCrea M., McIntosh A.S., Maddocks D., Makdissi M., Purcell L., Putukian M., Schneider K., Tator C.H., and Turner M. (2013). Consensus statement on concussion in sport: the 4th International Conference on Concussion in Sport, Zurich, November 2012. J. Athl. Train. 48, 554–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins M.W., Iverson G.L., Lovell M.R., McKeag D.B., Norwig J., and Maroon J. (2003). On-field predictors of neuropsychological and symptom deficit following sports-related concussion. Clin. J. Sport Med. 13, 222–229 [DOI] [PubMed] [Google Scholar]
- 19.Iverson G.L., Lovell M.R., and Collins M.W. (2003). Interpreting change on ImPACT following sport concussion. Clin. Neuropsychol. 17, 460–467 [DOI] [PubMed] [Google Scholar]
- 20.Guskiewicz K. (2001). Postural stability assessment following concussion: one piece of the puzzle. Clin. J. Sport Med. 11, 181–1189 [DOI] [PubMed] [Google Scholar]
- 21.Kanof M.E., Smith P.D., and Zola H. (2001). Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr. Protoc. Immunol. Chapter 7, Unit 7.1. [DOI] [PubMed] [Google Scholar]
- 22.Barabasi A.L., Gulbahce N., and Loscalzo J. (2011). Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 12, 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Almeida-Suhett C.P., Li Z., Marini A.M., Braga M.F., and Eiden L.E. (2014). Temporal course of changes in gene expression suggests a cytokine-related mechanism for long-term hippocampal alteration after controlled cortical impact. J. Neurotrauma 31, 683–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redell J.B., Moore A.N., Grill R.J., Johnson D., Zhao J., Liu Y., and Dash P.K. (2013). Analysis of functional pathways altered after mild traumatic brain injury. J. Neurotrauma 30, 752–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H.H., Lee S.M., Cai Y., Sutton R.L., and Hovda D.A. (2004). Differential gene expression in hippocampus following experimental brain trauma reveals distinct features of moderate and severe injuries. J. Neurotrauma 21, 1141–1153 [DOI] [PubMed] [Google Scholar]
- 26.Natale J.E., Ahmed F., Cernak I., Stoica B., and Faden A.I. (2003). Gene expression profile changes are commonly modulated across models and species after traumatic brain injury. J. Neurotrauma 20, 907–927 [DOI] [PubMed] [Google Scholar]
- 27.Park C., Cho I.H., Kim D., Jo E.K., Choi S.Y., Oh S.B., Park K., Kim J.S., and Lee S.J. (2008). Toll-like receptor 2 contributes to glial cell activation and heme oxygenase-1 expression in traumatic brain injury. Neurosci. Lett. 431, 123–128 [DOI] [PubMed] [Google Scholar]
- 28.Babcock A.A., Wirenfeldt M., Holm T., Nielsen H.H., Dissing-Olesen L., Toft-Hansen H., Millward J.M., Landmann R., Rivest S., Finsen B., and Owens T. (2006). Toll-like receptor 2 signaling in response to brain injury: an innate bridge to neuroinflammation. J. Neurosci. 26, 12826–12837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehnardt S. (2010). Innate immunity and neuroinflammation in the CNS: the role of microglia in Toll-like receptor-mediated neuronal injury. Glia 58, 253–263 [DOI] [PubMed] [Google Scholar]
- 30.White T.E., Ford G.D., Surles-Zeigler M.C., Gates A.S., Laplaca M.C., and Ford B.D. (2013). Gene expression patterns following unilateral traumatic brain injury reveals a local pro-inflammatory and remote anti-inflammatory response. BMC Genomics 14, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayden M.S., West A.P., and Ghosh S. (2006). NF-kappaB and the immune response. Oncogene 25, 6758–6780 [DOI] [PubMed] [Google Scholar]
- 32.Libermann T.A. and Baltimore D. (1990). Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol. Cell. Biol. 10, 2327–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy T.L., Cleveland M.G., Kulesza P., Magram J., and Murphy K.M. (1995). Regulation of interleukin 12 p40 expression through an NF-kappa B half-site. Mol. Cell. Biol. 15, 5258–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang K., Mu X.S., and Hayes R.L. (1995). Increased cortical nuclear factor-kappa B (NF-kappa B) DNA binding activity after traumatic brain injury in rats. Neurosci. Lett. 197, 101–104 [DOI] [PubMed] [Google Scholar]
- 35.Nonaka M., Chen X.H., Pierce J.E., Leoni M.J., McIntosh T.K., Wolf J.A., and Smith D.H. (1999). Prolonged activation of NF-kappaB following traumatic brain injury in rats. J. Neurotrauma 16, 1023–1034 [DOI] [PubMed] [Google Scholar]
- 36.Sanz O., Acarin L., Gonzalez B., and Castellano B. (2002). NF-kappaB and IkappaBalpha expression following traumatic brain injury to the immature rat brain. J. Neurosci. Res. 67, 772–780 [DOI] [PubMed] [Google Scholar]
- 37.Hang C.H., Chen G., Shi J.X., Zhang X., and Li J.S. (2006). Cortical expression of nuclear factor kappaB after human brain contusion. Brain Res. 1109, 14–21 [DOI] [PubMed] [Google Scholar]
- 38.Sullivan P.G., Bruce-Keller A.J., Rabchevsky A.G., Christakos S., Clair D.K., Mattson M.P., and Scheff S.W. (1999). Exacerbation of damage and altered NF-kappaB activation in mice lacking tumor necrosis factor receptors after traumatic brain injury. J. Neurosci. 19, 6248–6256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones S.A. (2005). Directing transition from innate to acquired immunity: defining a role for IL-6. J. Immunol. 175, 3463–3468 [DOI] [PubMed] [Google Scholar]
- 40.Erta M., Quintana A., and Hidalgo J. (2012). Interleukin-6, a major cytokine in the central nervous system. Int. J. Biol. Sci. 8, 1254–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dalla Libera A.L., Regner A., de Paoli J., Centenaro L., Martins T.T., and Simon D. (2011). IL-6 polymorphism associated with fatal outcome in patients with severe traumatic brain injury. Brain Inj. 25, 365–369 [DOI] [PubMed] [Google Scholar]
- 42.Helmy A., Carpenter K.L., Menon D.K., Pickard J.D., and Hutchinson P.J. (2011). The cytokine response to human traumatic brain injury: temporal profiles and evidence for cerebral parenchymal production. J. Cereb. Blood Flow Metab. 31, 658–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trinchieri G. (2003). Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3, 133–146 [DOI] [PubMed] [Google Scholar]
- 44.Stahel P.F., Kossmann T., Joller H., Trentz O., and Morganti-Kossmann M.C. (1998). Increased interleukin-12 levels in human cerebrospinal fluid following severe head trauma. Neurosci. Lett. 249, 123–126 [DOI] [PubMed] [Google Scholar]
- 45.Schwulst S.J., Trahanas D.M., Saber R., and Perlman H. (2013). Traumatic brain injury-induced alterations in peripheral immunity. J. Trauma Acute Care Surg. 75, 780–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gatson J.W., Liu M.M., Abdelfattah K., Wigginton J.G., Smith S., Wolf S., and Minei J.P. (2012). Resveratrol decreases inflammation in the brain of mice with mild traumatic brain injury. J. Trauma Acute Care Surg. 74, 470–474 [DOI] [PubMed] [Google Scholar]
- 47.Schneider H.J., Schneider M., Kreitschmann-Andermahr I., Tuschy U., Wallaschofski H., Fleck S., Faust M., Renner C.I., Kopczak A., Saller B., Buchfelder M., Jordan M., and Stalla G.K. (2011). Structured assessment of hypopituitarism after traumatic brain injury and aneurysmal subarachnoid hemorrhage in 1242 patients: the German interdisciplinary database. J. Neurotrauma 28, 1693–1698 [DOI] [PubMed] [Google Scholar]
- 48.Kelestimur F., Tanriverdi F., Atmaca H., Unluhizarci K., Selcuklu A., and Casanueva F.F. (2004). Boxing as a sport activity associated with isolated GH deficiency. J. Endocrinol. Invest. 27, RC28–RC32 [DOI] [PubMed] [Google Scholar]
- 49.Tanriverdi F., Unluhizarci K., Coksevim B., Selcuklu A., Casanueva F.F., and Kelestimur F. (2007). Kickboxing sport as a new cause of traumatic brain injury-mediated hypopituitarism. Clin. Endocrinol. (Oxf.) 66, 360–366 [DOI] [PubMed] [Google Scholar]
- 50.McCullers D.L., Sullivan P.G., Scheff S.W., and Herman J.P. (2002). Traumatic brain injury regulates adrenocorticosteroid receptor mRNA levels in rat hippocampus. Brain Res. 947, 41–49 [DOI] [PubMed] [Google Scholar]
- 51.McCullers D.L., Sullivan P.G., Scheff S.W., and Herman J.P. (2001). Mifepristone protects CA1 hippocampal neurons following traumatic brain injury in rat. Neuroscience 109, 219–230 [DOI] [PubMed] [Google Scholar]
- 52.McIntosh L.J. and Sapolsky R.M. (1996). Glucocorticoids may enhance oxygen radical-mediated neurotoxicity. Neurotoxicology 17, 873–882 [PubMed] [Google Scholar]
- 53.Goodman Y., Bruce A.J., Cheng B., and Mattson M.P. (1996). Estrogens attenuate and corticosterone exacerbates excitotoxicity, oxidative injury, and amyloid beta-peptide toxicity in hippocampal neurons. J. Neurochem. 66, 1836–1844 [DOI] [PubMed] [Google Scholar]
- 54.Meehan W.P., 3rd and Bachur R.G. (2009). Sport-related concussion. Pediatrics 123, 114–123 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.