Abstract
AIM
To clarify the association between aldo-keto reductase family 1 member B10 (AKR1B10) expression and hepatocarcinogenesis after hepatitis C virus eradication.
METHODS
In this study, we enrolled 303 chronic hepatitis C patients who had achieved sustained virological response (SVR) through interferon-based antiviral therapy. Pretreatment AKR1B10 expression in the liver was immunohistochemically assessed and quantified as a percentage of positive staining area by using image-analysis software. A multivariate Cox analysis was used to estimate the hazard ratios (HRs) of AKR1B10 expression for hepatocellular carcinoma (HCC) development after achieving SVR. The cumulative incidences of HCC development were evaluated using Kaplan-Meier analysis and the log-rank test.
RESULTS
Of the 303 chronic hepatitis C patients, 153 (50.5%) showed scarce hepatic AKR1B10 expression, quantified as 0%, which was similar to the expression in control normal liver tissues. However, the remaining 150 patients (49.5%) exhibited various degrees of AKR1B10 expression in the liver, with a maximal AKR1B10 expression of 73%. During the median follow-up time of 3.6 years (range 1.0-10.0 years), 8/303 patients developed HCC. Multivariate analysis revealed that only high AKR1B10 expression (≥ 8%) was an independent risk factor for HCC development (HR = 15.4, 95%CI: 1.8-132.5, P = 0.012). The 5-year cumulative incidences of HCC development were 13.7% and 0.5% in patients with high and low AKR1B10 expression, respectively (P < 0.001). During the follow-up period after viral eradication, patients expressing high levels of AKR1B10 expressed markedly higher levels of alanine aminotransferase and α-fetoprotein than did patients exhibiting low AKR1B10 expression.
CONCLUSION
Chronic hepatitis C patients expressing high levels of hepatic AKR1B10 had an increased risk of HCC development even after SVR.
Keywords: Human AKR1B10 protein, Hepatocellular carcinoma, Chronic hepatitis C, Immunohistochemistry, Risk factor, Sustained virological response
Core tip: Expression of a cancer-related oxidoreductase, aldo-keto reductase family 1 member B10 (AKR1B10) was upregulated in the liver in patients with chronic hepatitis C (CHC). High AKR1B10 expression was associated in a statistically significant manner with the risk of hepatocellular carcinoma (HCC) development even after sustained virological response (SVR) was achieved through interferon-based antiviral therapy. Pretreatment AKR1B10 expression of 8% was associated with a > 15-fold-increased risk of HCC development. Thus, AKR1B10 is not only a cancer biomarker but also a novel predictive marker for assessing the risk of HCC development in CHC patients who achieved SVR.
INTRODUCTION
Persistent hepatitis C virus (HCV) infection is one of the major causes of chronic liver disease leading to the development of hepatocellular carcinoma (HCC), which is the fifth most common cancer and the third most common cause of cancer-related death worldwide[1]. HCV is responsible for 27%-75% of the HCC cases in Europe and United States and > 80% of the cases in Japan[2,3]. Notably, HCV-positive patients present a 20-fold higher risk of developing HCC than do HCV-negative patients[4], which indicates a major carcinogenic role for persistent HCV infection. Given this association, chronic hepatitis C patients are frequently treated with interferon-based antiviral therapy, because the treatment not only eradicates HCV but also reduces the rate of HCC development. Interferon therapy most effectively lowers the risk of developing HCC in patients who achieve a sustained virological response (SVR)[5-7], and the recent emergence of direct-acting antiviral drugs (DAAs) against HCV has drastically increased the SVR rate of antiviral therapy[8,9]. However, the risk of HCC development persists after interferon therapy even in patients who achieve SVR[10]. Because assessment of the risk of developing HCC is clinically important in the management of patients with chronic hepatitis C, the requirement of predictors for HCC development in patients who achieve SVR is now increasing.
Aldo-keto reductase family 1 member B10 (AKR1B10), a cancer-related oxidoreductase, was originally identified as a gene whose expression was upregulated in human HCC but was low in normal liver tissues[11,12]. Recently, AKR1B10 upregulation was observed in several studies in certain chronic liver diseases such as chronic hepatitis B and C and steatohepatitis[13-16], which are widely recognized to represent a precancerous condition of HCC. AKR1B10 was upregulated in a stepwise manner from the surrounding liver tissues, which showed chronic hepatitis or cirrhosis, to HCC[17], and AKR1B10 upregulation was also demonstrated to be associated in a statistically significant manner with the risk of HCC development in chronic hepatitis B and C[13,15]. Furthermore, the results of in vitro and in vivo experiments demonstrated the involvement of AKR1B10 in cancer-cell proliferation[18,19].
The aforementioned findings collectively support the view that AKR1B10 upregulation is involved in the early stages of hepatocarcinogenesis. We further hypothesized that in patients in whom AKR1B10 is upregulated in the liver, the carcinogenic process has already progressed, and that these patients face a high risk of HCC even after successful viral eradication. If this is the case, then AKR1B10 expression could serve as a useful predictive marker for HCC development in chronic hepatitis C patients who achieve SVR. Thus, in this study, our aim was to clarify the association between pretreatment AKR1B10 expression and HCC development after SVR in patients with chronic hepatitis C.
MATERIALS AND METHODS
Patients
Between March 2004 and August 2014, a total of 605 patients with chronic HCV infection underwent interferon-based antiviral therapy at Juntendo University Shizuoka Hospital. Of the 605 patients, 401 achieved SVR, and these patients were considered for enrollment in this retrospective study. Chronic HCV infection was diagnosed based on continuous positivity for serum HCV RNA detected using reverse-transcription PCR. Exclusion criteria for this study were the following: (1) absence of liver biopsy within 6 mo before treatment; (2) positivity for hepatitis B surface antigen or HIV; (3) evidence of other chronic liver diseases (autoimmune hepatitis, primary biliary cirrhosis, hemochromatosis, and Wilson’s disease); (4) presence of HCC or any suspicious lesions detected through ultrasonography, dynamic computed tomography, or magnetic resonance imaging at enrollment; (5) history of previous treatment for HCC and liver transplantation; (6) a follow-up period of < 1.0 year after the end of treatment (EOT); and (7) development of HCC at < 1.0 year after the EOT, because HCC developed within 1.0 year might have existed before treatment. Based on these criteria, a total of 303 patients were finally enrolled in this study. Control normal liver tissues presenting no aberrant histological features were obtained from surgically resected specimens from 8 patients with liver metastasis from colorectal cancer.
This study was approved by the Ethics Committee of Juntendo University Shizuoka Hospital and performed in accordance with the Helsinki Declaration (as revised in Brazil, 2013). Written informed consent was obtained from all patients.
Laboratory investigations and liver histology
HCV was genotyped by performing PCR with the HCV Genotype Primer Kit (Institute of Immunology Co., Ltd., Tokyo, Japan) and classified into genotype 1, genotype 2, or other genotypes according to Simmonds’ classification system. Serum HCV viral load was determined with a Cobas Amplicor HCV monitor v2.0 by using the 10-fold-dilution method (Roche Diagnostics, Branchburg, NJ, United States). Patients who were negative for serum HCV RNA at 24 wk after the EOT were defined as having achieved SVR. The following laboratory data were collected immediately before treatment, at 24 wk after the EOT, and at every follow-up visit after SVR: complete blood count and levels of albumin, alanine aminotransferase (ALT), γ-glutamyl transpeptidase (GGT), α-fetoprotein (AFP), and des-γ-carboxy prothrombin. Histological staging and grading were evaluated by a pathologist, who was blinded to the patients’ clinical information, according to the Metavir classification system[20].
Patient follow-up
Each patient was examined for serum tumor markers and HCC by performing ultrasonography at least once every 6 mo after SVR. The absence of serum HCV RNA was annually reconfirmed. HCC was diagnosed predominantly through imaging studies, including dynamic computed tomography and magnetic resonance imaging. When the hepatic nodule did not show typical imaging features, diagnosis was confirmed by means of fine-needle aspiration biopsy followed by histological examination. Patient follow-up ended on March 31, 2016.
AKR1B10 immunohistochemistry
Immunohistochemical analysis of AKR1B10 was performed as described previously with certain modifications[14,21]. Briefly, deparaffinized and rehydrated sections were processed by performing heat-induced antigen retrieval in 0.1 mol/L citrate buffer at pH 6.0. After blocking endogenous peroxidase activity, sections were incubated with a mouse monoclonal antibody against AKR1B10 (1:100, Ab 57547; Abcam, Cambridge, United Kingdom) at room temperature, and then with a biotinylated secondary antibody (Ventana iVIEW DAB Universal Kit; Ventana Medical Systems Inc., Tucson, AZ, United States). Staining was visualized using 3,3’-diaminobenzidine tetrahydrochloride, and sections were counterstained with hematoxylin and eosin. AKR1B10 immunostaining was identified based on positive cytoplasmic staining and was quantified as the average percentage of AKR1B10-positive areas in 2 independent fields of hepatic parenchyma at 100 × magnification by using Lumina Vision 2.4 Bio-imaging software (Mitani Corporation, Tokyo, Japan). The average percentage of AKR1B10-positive areas was rounded to the integer by discarding fractions that were < 1%. Previous studies have established that the levels of AKR1B10 immunoreactivity and its mRNA levels are well correlated[14,15].
Statistical analyses
All statistical analyses were performed using PASW Statistics 18 (IBM SPSS, Chicago, IL, United States). The Mann-Whitney U test was used for continuous variables and the corrected χ2 method was used for categorical variables. Univariate and multivariate Cox proportional hazard models were used to evaluate factors that were significantly associated with HCC development. The Kaplan-Meier method was used to analyze the cumulative incidence of HCC development, and differences were tested using the log-rank test. The hazard ratio (HR) and 95%CI were calculated. P < 0.05 was considered statistically significant.
RESULTS
Patients’ characteristics and HCC development after SVR
The demographic, biochemical, and pathological characteristics of the 303 patients enrolled in this study are summarized in Table 1. Of the 303 patients, 260 received pegylated interferon plus ribavirin combination therapy for 24-72 wk, and 43 patients received protease inhibitor treatment (22 telaprevir, 10 simeprevir, and 11 faldaprevir) together with pegylated interferon plus ribavirin for 24 wk. During a median follow-up of 3.6 years (range, 1.0-10.0 years), 8 patients (2.6%) developed HCC. The estimated cumulative incidences of HCC were 1.2% and 3.7% at 3 and 5 years, respectively (Figure 1). As compared with patients who did not develop HCC, the patients who developed HCC more frequently presented the complication of diabetes mellitus (P = 0.019) and exhibited a higher degree of hepatic fibrosis (P < 0.001), lower albumin levels (P = 0.032), higher ALT levels (P = 0.017), lower platelet counts (P = 0.006), and higher AFP levels (P = 0.002) (Table 1).
Table 1.
Baseline characteristics of patients enrolled in the study
| Characteristics | All patients (n = 303) | With HCC development (n = 8) | Without HCC development (n = 295) | P value |
| Age, yr | 57 (20-85) | 62 (49-71) | 57 (20-85) | 0.1982 |
| Males | 182 (60.0) | 6 (75.0) | 176 (59.7) | 0.4843 |
| BMI (kg/m2) | 23.3 (15.3-39.5) | 23.8 (20.2-26.5) | 23.4 (15.3-39.5) | 0.8602 |
| Habitual drinker | 75 (24.8) | 2 (25.0) | 73 (24.7) | 1.0003 |
| Diabetes mellitus1 | 25 (8.3) | 3 (37.5) | 22 (7.5) | 0.0193 |
| HCV-RNA (logIU/mL)1 | 6.2 (1.2-7.6) | 5.7 (5.0-6.7) | 6.2 (1.2-7.6) | 0.2462 |
| HCV genotype 1 | 152 (50.2) | 6 (75.0) | 146 (49.5) | 0.2833 |
| Stage of fibrosis F3-F4 | 47 (15.5) | 6 (75.0) | 41 (13.9) | < 0.0013 |
| Grade of inflammation A2-A3 | 208 (68.6) | 8 (100.0) | 200 (67.8) | 0.0603 |
| Albumin (g/dL) | 4.2 (3.3-4.8) | 3.9 (3.3-4.4) | 4.2 (3.3-4.7) | 0.0322 |
| ALT (IU/L) | 52 (11-699) | 146 (31-209) | 52 (11-699) | 0.0172 |
| Platelet count (× 104/μL) | 17.5 (5.6-39.6) | 10.3 (7.9-19.3) | 17.6 (5.6-31.9) | 0.0062 |
| GGT (IU/L) | 37 (9-517) | 66 (28-161) | 36 (9-517) | 0.0532 |
| AFP (ng/mL)1 | 5 (1-1380) | 11 (5-870) | 5 (1-1380) | 0.0022 |
| PI use | 43 (14.2) | 1 (12.5) | 42 (14.2) | 1.0002 |
Data not available for all patients;
Mann Whitney-U test;
χ2 test. Data are expressed as medians (range) or numbers (%). P values are for comparisons between patients with and without HCC development. AFP: α-fetoprotein; ALT: Alanine aminotransferase; BMI: Body mass index; GGT: γ-glutamyl transpeptidase; HCV: Hepatitis C virus; PI: Protease inhibitor.
Figure 1.
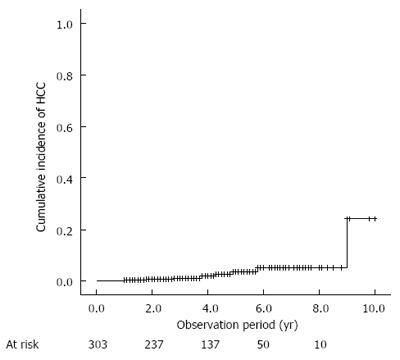
Cumulative incidence of hepatocellular carcinoma development after sustained virological response. HCC: Hepatocellular carcinoma.
Baseline AKR1B10 expression in the liver
Figure 2 shows representative immunohistochemical staining of AKR1B10 in liver tissues. In normal liver tissues, distinct positive staining for AKR1B10 was observed in bile-duct epithelia, but AKR1B10 immunoreactivity was either undetectable in the hepatic parenchyma, or faint immunoreactivity was observed in a few hepatocytes (Figure 2A and B). In some of the patients with chronic hepatitis C, AKR1B10 immunoreactivity in the liver was similar to that in control normal liver tissues: positive in bile-duct epithelia and negative in the hepatic parenchyma (Figure 2C and D). However, other patients showed prominent nucleocytoplasmic AKR1B10 immunoreactivity in scattered or clustered hepatocytes in the hepatic parenchyma (Figure 2E and F). Quantification of the AKR1B10-positive areas in the hepatic parenchyma revealed that none of the normal liver tissues showed AKR1B10 expression. Similarly, 153 patients (50.5%) presented scarce AKR1B10 expression, and their AKR1B10 positive staining area was quantified as 0%. By contrast, the remaining 150 patients (49.5%) presented various degrees of AKR1B10 expression in the liver parenchyma, with the maximal AKR1B10 expression area reaching 73% (Figure 3). The median AKR1B10-positive area in patients who developed HCC and did not develop HCC was 15% (range, 0%-60%) and 0% (range, 0%-73%), respectively, and this difference was statistically significant (P = 0.002).
Figure 2.

Representative AKR1B10 immunohistochemical staining of specimens. Normal liver tissue (A, B) and tissue from patients with chronic hepatitis C (C-F). Hematoxylin and eosin staining (A, C, E) and AKR1B10 immunostaining (B, D, F). Positive control, bile-duct epithelium; original magnification × 40.
Figure 3.
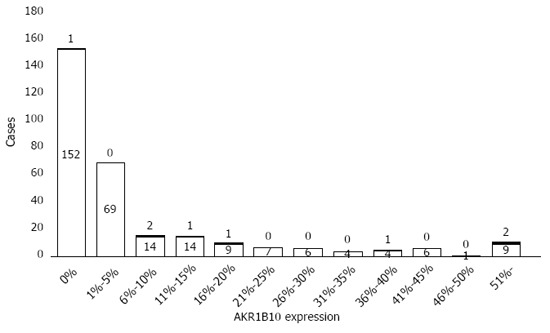
Distribution of AKR1B10 expression levels in the study cohort. Filled and blank patterns indicate patients with and without hepatocellular carcinoma development, respectively.
Baseline AKR1B10 expression and risk of HCC development after SVR
The results of univariate Cox logistic regression analysis identified 6 baseline variables that were significantly associated with HCC development after SVR: complication of diabetes mellitus, advanced fibrosis stage, low serum albumin levels and platelet counts, elevated serum AFP, and increased AKR1B10 expression. Multivariate Cox logistic regression analysis revealed that only high AKR1B10 expression was an independent risk factor for HCC development after SVR (Table 2). The area under the receiver operator characteristics curve analysis further revealed that an AKR1B10 expression level of 8% was the cutoff value for HCC development. The sensitivity and specificity for AKR1B10 expression of ≥ 8% were 0.750 and 0.766, respectively. The positive and negative predictive values were 0.080 and 0.991, respectively. The results of multivariate Cox logistic regression analysis indicated that the adjusted HR of high AKR1B10 expression (≥ 8%) for HCC development was 15.4 (95%CI: 1.8-132.5, P = 0.012). Kaplan-Meier plot analysis revealed that the 3- and 5-year cumulative incidence rates of HCC development in patients with high AKR1B10 expression were 3.4% and 13.7%, respectively, whereas those in patients with low AKR1B10 expression (< 8%) were 0.5% and 0.5%, respectively (P < 0.001; Figure 4). Only 2 patients expressing low levels of AKR1B10 developed HCC: one patient showed an AKR1B10 expression level of 7%, and developed HCC at 1.8 years, whereas the other did not show AKR1B10 expression (0%), and developed HCC at 9.0 years (Table 3).
Table 2.
Univariate and multivariate analyses for factors associated with hepatocellular carcinoma development
| Variables | HR (95%CI) | P value |
| Univariate analysis | ||
| Age (by each year) | 1.04 (0.31-1.11) | 0.347 |
| Male sex | 1.61 (0.32-8.04) | 0.564 |
| BMI (by each kg/m2) | 1.00 (0.80-1.25) | 0.982 |
| Habitual drinker | 1.26 (0.25-6.27) | 0.775 |
| Diabetes mellitus | 5.58 (1.19-26.21) | 0.030 |
| HCV-RNA (by each logIU/mL) | 0.87 (0.44-1.72) | 0.696 |
| HCV genotype 1 | 2.90 (0.58-14.35) | 0.195 |
| Stage of fibrosis | 3.59 (1.68-7.68) | 0.001 |
| Grade of inflammation | 4.32 (1.02-18.40) | 0.048 |
| Albumin (by each g/dL) | 0.03 (0.00-0.29) | 0.003 |
| ALT (by each IU/L) | 1.00 (1.00-1.01) | 0.196 |
| Platelet count (by each 104/mL) | 0.75 (0.63-0.90) | 0.002 |
| GGT (by each IU/L) | 1.00 (1.00-1.01) | 0.452 |
| AFP (by each ng/mL) | 1.00 (1.00-1.00) | 0.033 |
| PI use | 2.93 (0.31-27.70) | 0.347 |
| AKR1B10 (by each %) | 1.06 (1.03-1.10) | < 0.001 |
| Multivariate analysis | ||
| AKR1B10 (by each %) | 1.04 (1.03-1.10) | 0.001 |
AFP: α-fetoprotein; AKR1B10: Aldo-keto reductase family1 member B10; ALT: Alanine aminotransferase; BMI: Body mass index; GGT: γ-glutamyl transpeptidase; HCV: Hepatitis C virus; PI: Protease inhibitor.
Figure 4.
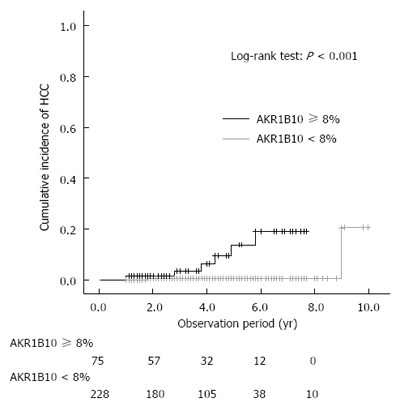
Cumulative incidence of hepatocellular carcinoma development after sustained virological response, shown according to AKR1B10 expression level.
Table 3.
Characteristics of the 8 patients with hepatocellular carcinoma development
| Age (yr) | Sex | Interval (yr) | F | A | Albumin (g/dL) | ALT (IU/L) | Platelet count (× 104/mL) | AKR1B10 | |
| 1 | 62 | F | 1.0 | 2 | 2 | 4.0 | 199 | 10.3 | 60% |
| 2 | 69 | F | 1.8 | 3 | 2 | 3.3 | 60 | 9.9 | 7% |
| 3 | 61 | M | 2.8 | 2 | 2 | 4.4 | 135 | 10.3 | 13% |
| 4 | 65 | M | 3.8 | 4 | 3 | 3.4 | 88 | 9.0 | 52% |
| 5 | 49 | M | 4.3 | 3 | 2 | 3.8 | 209 | 7.9 | 38% |
| 6 | 56 | M | 4.9 | 3 | 2 | 4.2 | 156 | 19.3 | 9% |
| 7 | 57 | M | 5.8 | 3 | 2 | 4.2 | 157 | 14.7 | 16% |
| 8 | 71 | M | 9.0 | 3 | 2 | 3.7 | 31 | 17.9 | 0% |
AKR1B10: Aldo-keto reductase family1 member B10; ALT: Alanine aminotransferase; HCC: Hepatocellular carcinoma.
Changes in biochemical-test results after SVR and baseline AKR1B10 expression
Serum aminotransferase levels are sensitive indicators of necroinflammatory activity in the liver, and an elevation of serum AFP levels without HCC is also related to liver-cell damage. To investigate sustained liver-cell damage after HCV eradication, we evaluated whether serum ALT and AFP levels were altered after SVR, and whether baseline AKR1B10 expression was associated with such changes. At 6 mo after the EOT, the ALT levels were markedly decreased (median: 52 IU/L at baseline, 16 IU/L at 6 mo, P = 0.001), and 264 (87.1%) patients achieved ALT normalization (defined as ALT ≤ 30 IU/mL). AFP levels also showed a notable decrease (median: 5 ng/mL at baseline, 3 ng/mL at 6 mo, P < 0.001), and 260 (85.8%) patients achieved an AFP level of ≤ 5 ng/mL. As compared with the low-AKR1B10 group (n = 228), the high-AKR1B10 group (n = 75) showed considerably higher levels of ALT (median: 15 IU/L vs 22 IU/L, P < 0.001) and AFP (median: 3 ng/mL vs 4 ng/mL, P < 0.001) at 24 wk after the EOT (Figure 5).
Figure 5.
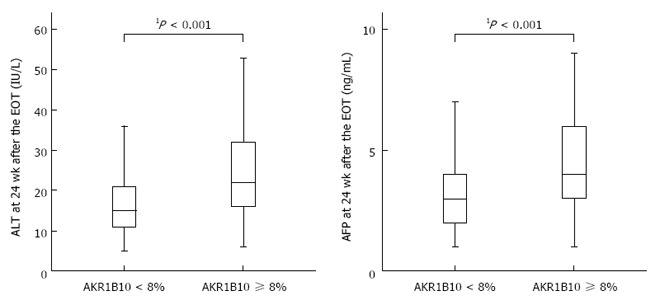
Relationships between baseline AKR1B10 expression levels and alanine aminotransferase and α-fetoprotein levels after sustained virological response. 1Mann-Whitney U test. EOT: End of treatment.
DISCUSSION
In this study, the incidence of HCC development was 3.7% at 5 years after SVR. This low incidence is comparable to that presented in recent reports (1.2%-5.8%)[22-25], and this is considered to confirm the relationship between SVR achievement and a reduced risk of subsequent HCC[26]. Approval of various DAAs, including protease inhibitors, has enabled most patients receiving the therapy to achieve SVR[8,9], and the safety of the all-oral combination therapy of DAAs could increase the number of patients receiving antiviral therapy[27,28]. Consequently, the number of SVR patients is now increasing drastically, and thus predictors of HCC development after SVR are becoming increasingly important.
To date, several factors have been reported to predict the risk of HCC development in patients with chronic hepatitis C, such as older age, male gender, alcohol intake, and hepatic fibrosis[2]. Before DAA became available, Genotype 1 infection was refractory to treatment, and may have been correlated with risk of HCC. The presence of advanced hepatic fibrosis prior to treatment is recognized as a significant risk factor for HCC development after achieving SVR, however, not all patients with advanced hepatic fibrosis develop HCC[29]. In the present study, our main finding is that high AKR1B10 expression in the liver is an independent predictor for HCC development even after achieving SVR. The adjusted HR demonstrated that a pretreatment AKR1B10 expression of ≥ 8% was associated with a > 15-fold-increased risk of HCC development after achieving SVR. Although the positive predictive value of AKR1B10 expression of ≥ 8% was only 0.080, the negative predictive value was extremely high: 0.991; this result suggests that the risk of HCC development was extremely low in patients showing an AKR1B10 expression of < 8%. Based on the assessment of baseline AKR1B10 expression, very-low-risk patients could be selected from the growing number of SVR patients, and thus an inefficient surveillance examination for HCC could be avoided.
The previous study also demonstrated that high AKR1B10 expression was a significant predictor of HCC development in patients with chronic hepatitis C. Interestingly, the ROC analysis-determined AKR1B10 cut-off value in the present study (8%) was higher than that in the previous report (6%)[14]. A fundamental difference between these studies was that the previous study included 42% non-SVR patients in the study cohort while the present study included only SVR patients. The difference in the AKR1B10 cut-off values might indicate the impact of SVR achievement on the subsequent changes in AKR1B10 expression. However, confirming this would be difficult since liver biopsy samples from patients who have completed treatment are rarely available
AKR1B10 emerged as a cancer biomarker because it is overexpressed in several cancers; however, the biological function of AKR1B10 and its potential involvement in carcinogenesis remain incompletely understood and are receiving increased attention. Because AKR1B10 is an efficient retinal reductase, the molecule is considered to inhibit retinoic acid signaling, which maintains epithelial cell differentiation[30,31]. Therefore, AKR1B10 upregulation has been hypothesized to play a pivotal role in promoting premature or neoplastic phenotypes in cancer cells[32,33]. In the case of human HCC, AKR1B10 upregulation was mainly observed in early-stage well-differentiated HCC and was considered to represent an early event in the hepatocarcinogenesis process[17,34]. Here, AKR1B10 upregulation occurred in patients with chronic hepatitis C, a preneoplastic condition of HCC, and reflected the risk of HCC development even after HCV eradication. Collectively, these data and those from previous studies suggest the involvement of AKR1B10 upregulation in the very early stages of hepatocarcinogenesis. In the patients in whom AKR1B10 was upregulated before treatment, the carcinogenic process might have already progressed, and in these patients, a high risk of HCC might remain even after successful viral eradication.
We also found that baseline AKR1B10 expression was related with ALT and AFP levels after SVR, both of which were identified as predictors of HCC development in several previous studies[35,36]. However, why ALT and AFP levels show sustained elevation even after HCV eradication is unknown. Intriguingly, baseline AKR1B10 expression was also associated with ALT levels in chronic hepatitis B patients who received successful antiviral therapy[13]. However, viral infection per se is unlikely to cause the sustained elevation of ALT and AFP after antiviral therapy: Recently, AKR1B10 expression was shown to be regulated by the transcription factor nuclear factor erythroid 2-related factor 2[37], which plays a pivotal role in the adaptive response to oxidative stress. Oxidative stress is a feature of steatohepatitis, and hepatic oxidative-stress markers have been correlated with the severity of hepatic necroinflammation[38-40]. Furthermore, AKR1B10 was identified as an upregulated gene in steatohepatits[16]. Given these findings, we suggest that oxidative stress might affect baseline AKR1B10 expression, sustain ALT and AFP elevation, and further HCC development after SVR.
The main limitations of this study were its monocentric aspect and retrospective nature. The number of cases of HCC development was very small because the incidence of HCC development after SVR was generally low. A future multicenter prospective analysis will be required to validate the association between AKR1B10 expression and the risk of HCC development in patients with chronic hepatitis C who achieve HCV eradication.
In conclusion, AKR1B10 upregulation is a major risk factor for HCC development in chronic hepatitis C patients who achieve SVR. Our findings not only identify AKR1B10 as a novel predictive marker of HCC, but also provide a new insight regarding AKR1B10 involvement in the molecular mechanism of hepatocarcinogenesis and suggest that AKR1B10 could serve as a novel therapeutic target for HCC prevention.
COMMENTS
Background
Persistent hepatitis C virus (HCV) infection is a major cause of chronic liver disease leading to hepatocellular carcinoma (HCC) development. Patients with chronic hepatitis C are frequently treated with interferon-based antiviral therapy, which most effectively lowers the risk of developing HCC in patients who achieve sustained virological response (SVR), and the use of direct-acting antiviral drugs against HCV has drastically increased the SVR rate of the therapy. However, the risk of HCC development persists after interferon therapy even in patients who achieve SVR. Therefore, the identification of predictors of HCC development after SVR is becoming increasingly important.
Research frontiers
The efficiency of surveillance examinations for HCC can be enhanced if we can identify reliable predictors of HCC development after HCV eradication.
Innovations and breakthroughs
In almost half of the chronic hepatitis C patients enrolled in this study, aldo-keto reductase family 1 member B10 (AKR1B10) was expressed in the liver, and high expression of AKR1B10 was found to be an independent risk factor for HCC development after SVR. Moreover, baseline AKR1B10 expression was correlated with the levels of two liver-damage markers after SVR.
Applications
The results of this study identify AKR1B10 as a novel predictive marker of HCC development. Furthermore, the findings suggest that AKR1B10 functions in the early stages of hepatocarcinogenesis, which raises the possibility that AKR1B10 could serve as a therapeutic target for HCC prevention.
Peer-review
Authors investigated the association between AKR1B10 expression and hepatocarcinogenesis after HCV eradication. They studied 303 chronic hepatitis c patients who had achieved SVR. This manuscript contains some interesting topics for prediction of HCC development after SVR.
Footnotes
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Japan
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
Institutional review board statement: This study was reviewed and approved by the Ethics Committee of Juntendo University Shizuoka Hospital.
Informed consent statement: Written informed consent was obtained from all the patients enrolled in the study.
Conflict-of-interest statement: The authors declare no conflict of interest related to this study.
Data sharing statement: No additional data are available.
Peer-review started: May 23, 2016
First decision: June 20, 2016
Article in press: August 1, 2016
P- Reviewer: Cerwenka HR, Kakizaki S, Mendez-Sanchez N S- Editor: Ma YJ L- Editor: A E- Editor: Ma S
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127:S35–S50. doi: 10.1053/j.gastro.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Kiyosawa K, Umemura T, Ichijo T, Matsumoto A, Yoshizawa K, Gad A, Tanaka E. Hepatocellular carcinoma: recent trends in Japan. Gastroenterology. 2004;127:S17–S26. doi: 10.1053/j.gastro.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Sun CA, Wu DM, Lin CC, Lu SN, You SL, Wang LY, Wu MH, Chen CJ. Incidence and cofactors of hepatitis C virus-related hepatocellular carcinoma: a prospective study of 12,008 men in Taiwan. Am J Epidemiol. 2003;157:674–682. doi: 10.1093/aje/kwg041. [DOI] [PubMed] [Google Scholar]
- 5.Cammà C, Giunta M, Andreone P, Craxì A. Interferon and prevention of hepatocellular carcinoma in viral cirrhosis: an evidence-based approach. J Hepatol. 2001;34:593–602. doi: 10.1016/s0168-8278(01)00005-8. [DOI] [PubMed] [Google Scholar]
- 6.Papatheodoridis GV, Papadimitropoulos VC, Hadziyannis SJ. Effect of interferon therapy on the development of hepatocellular carcinoma in patients with hepatitis C virus-related cirrhosis: a meta-analysis. Aliment Pharmacol Ther. 2001;15:689–698. doi: 10.1046/j.1365-2036.2001.00979.x. [DOI] [PubMed] [Google Scholar]
- 7.Shiratori Y, Ito Y, Yokosuka O, Imazeki F, Nakata R, Tanaka N, Arakawa Y, Hashimoto E, Hirota K, Yoshida H, et al. Antiviral therapy for cirrhotic hepatitis C: association with reduced hepatocellular carcinoma development and improved survival. Ann Intern Med. 2005;142:105–114. doi: 10.7326/0003-4819-142-2-200501180-00009. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi N, Okanoue T, Tsubouchi H, Toyota J, Chayama K, Kumada H. Efficacy and safety of telaprevir, a new protease inhibitor, for difficult-to-treat patients with genotype 1 chronic hepatitis C. J Viral Hepat. 2012;19:e134–e142. doi: 10.1111/j.1365-2893.2011.01528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayashi N, Izumi N, Kumada H, Okanoue T, Tsubouchi H, Yatsuhashi H, Kato M, Ki R, Komada Y, Seto C, et al. Simeprevir with peginterferon/ribavirin for treatment-naïve hepatitis C genotype 1 patients in Japan: CONCERTO-1, a phase III trial. J Hepatol. 2014;61:219–227. doi: 10.1016/j.jhep.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Morgan TR, Ghany MG, Kim HY, Snow KK, Shiffman ML, De Santo JL, Lee WM, Di Bisceglie AM, Bonkovsky HL, Dienstag JL, et al. Outcome of sustained virological responders with histologically advanced chronic hepatitis C. Hepatology. 2010;52:833–844. doi: 10.1002/hep.23744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scuric Z, Stain SC, Anderson WF, Hwang JJ. New member of aldose reductase family proteins overexpressed in human hepatocellular carcinoma. Hepatology. 1998;27:943–950. doi: 10.1002/hep.510270408. [DOI] [PubMed] [Google Scholar]
- 12.Cao D, Fan ST, Chung SS. Identification and characterization of a novel human aldose reductase-like gene. J Biol Chem. 1998;273:11429–11435. doi: 10.1074/jbc.273.19.11429. [DOI] [PubMed] [Google Scholar]
- 13.Mori M, Genda T, Ichida T, Murata A, Kamei M, Tsuzura H, Sato S, Narita Y, Kanemitsu Y, Ishikawa S, et al. Aldo-keto reductase family 1 member B10 is associated with hepatitis B virus-related hepatocellular carcinoma risk. Hepatol Res. 2016 doi: 10.1111/hepr.12725. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 14.Sato S, Genda T, Hirano K, Tsuzura H, Narita Y, Kanemitsu Y, Kikuchi T, Iijima K, Wada R, Ichida T. Up-regulated aldo-keto reductase family 1 member B10 in chronic hepatitis C: association with serum alpha-fetoprotein and hepatocellular carcinoma. Liver Int. 2012;32:1382–1390. doi: 10.1111/j.1478-3231.2012.02827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato S, Genda T, Ichida T, Murata A, Tsuzura H, Narita Y, Kanemitsu Y, Ishikawa S, Kikuchi T, Mori M, et al. Impact of aldo-keto reductase family 1 member B10 on the risk of hepatitis C virus-related hepatocellular carcinoma. J Gastroenterol Hepatol. 2016;31:1315–1322. doi: 10.1111/jgh.13295. [DOI] [PubMed] [Google Scholar]
- 16.Starmann J, Fälth M, Spindelböck W, Lanz KL, Lackner C, Zatloukal K, Trauner M, Sültmann H. Gene expression profiling unravels cancer-related hepatic molecular signatures in steatohepatitis but not in steatosis. PLoS One. 2012;7:e46584. doi: 10.1371/journal.pone.0046584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuzura H, Genda T, Sato S, Murata A, Kanemitsu Y, Narita Y, Ishikawa S, Kikuchi T, Mori M, Hirano K, et al. Expression of aldo-keto reductase family 1 member b10 in the early stages of human hepatocarcinogenesis. Int J Mol Sci. 2014;15:6556–6568. doi: 10.3390/ijms15046556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan R, Zu X, Ma J, Liu Z, Adeyanju M, Cao D. Aldo-keto reductase family 1 B10 gene silencing results in growth inhibition of colorectal cancer cells: Implication for cancer intervention. Int J Cancer. 2007;121:2301–2306. doi: 10.1002/ijc.22933. [DOI] [PubMed] [Google Scholar]
- 19.Satow R, Shitashige M, Kanai Y, Takeshita F, Ojima H, Jigami T, Honda K, Kosuge T, Ochiya T, Hirohashi S, et al. Combined functional genome survey of therapeutic targets for hepatocellular carcinoma. Clin Cancer Res. 2010;16:2518–2528. doi: 10.1158/1078-0432.CCR-09-2214. [DOI] [PubMed] [Google Scholar]
- 20.Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology. 1996;24:289–293. doi: 10.1002/hep.510240201. [DOI] [PubMed] [Google Scholar]
- 21.Heringlake S, Hofdmann M, Fiebeler A, Manns MP, Schmiegel W, Tannapfel A. Identification and expression analysis of the aldo-ketoreductase1-B10 gene in primary malignant liver tumours. J Hepatol. 2010;52:220–227. doi: 10.1016/j.jhep.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 22.Ogawa E, Furusyo N, Kajiwara E, Takahashi K, Nomura H, Maruyama T, Tanabe Y, Satoh T, Nakamuta M, Kotoh K, et al. Efficacy of pegylated interferon alpha-2b and ribavirin treatment on the risk of hepatocellular carcinoma in patients with chronic hepatitis C: a prospective, multicenter study. J Hepatol. 2013;58:495–501. doi: 10.1016/j.jhep.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 23.Huang CF, Yeh ML, Tsai PC, Hsieh MH, Yang HL, Hsieh MY, Yang JF, Lin ZY, Chen SC, Wang LY, et al. Baseline gamma-glutamyl transferase levels strongly correlate with hepatocellular carcinoma development in non-cirrhotic patients with successful hepatitis C virus eradication. J Hepatol. 2014;61:67–74. doi: 10.1016/j.jhep.2014.02.022. [DOI] [PubMed] [Google Scholar]
- 24.Yamashita N, Ohho A, Yamasaki A, Kurokawa M, Kotoh K, Kajiwara E. Hepatocarcinogenesis in chronic hepatitis C patients achieving a sustained virological response to interferon: significance of lifelong periodic cancer screening for improving outcomes. J Gastroenterol. 2014;49:1504–1513. doi: 10.1007/s00535-013-0921-z. [DOI] [PubMed] [Google Scholar]
- 25.Toyoda H, Kumada T, Tada T, Kiriyama S, Tanikawa M, Hisanaga Y, Kanamori A, Kitabatake S, Ito T. Risk factors of hepatocellular carcinoma development in non-cirrhotic patients with sustained virologic response for chronic hepatitis C virus infection. J Gastroenterol Hepatol. 2015;30:1183–1189. doi: 10.1111/jgh.12915. [DOI] [PubMed] [Google Scholar]
- 26.Morgan RL, Baack B, Smith BD, Yartel A, Pitasi M, Falck-Ytter Y. Eradication of hepatitis C virus infection and the development of hepatocellular carcinoma: a meta-analysis of observational studies. Ann Intern Med. 2013;158:329–337. doi: 10.7326/0003-4819-158-5-201303050-00005. [DOI] [PubMed] [Google Scholar]
- 27.Kumada H, Suzuki Y, Ikeda K, Toyota J, Karino Y, Chayama K, Kawakami Y, Ido A, Yamamoto K, Takaguchi K, et al. Daclatasvir plus asunaprevir for chronic HCV genotype 1b infection. Hepatology. 2014;59:2083–2091. doi: 10.1002/hep.27113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizokami M, Yokosuka O, Takehara T, Sakamoto N, Korenaga M, Mochizuki H, Nakane K, Enomoto H, Ikeda F, Yanase M, et al. Ledipasvir and sofosbuvir fixed-dose combination with and without ribavirin for 12 weeks in treatment-naive and previously treated Japanese patients with genotype 1 hepatitis C: an open-label, randomised, phase 3 trial. Lancet Infect Dis. 2015;15:645–653. doi: 10.1016/S1473-3099(15)70099-X. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida H, Shiratori Y, Moriyama M, Arakawa Y, Ide T, Sata M, Inoue O, Yano M, Tanaka M, Fujiyama S, et al. Interferon therapy reduces the risk for hepatocellular carcinoma: national surveillance program of cirrhotic and noncirrhotic patients with chronic hepatitis C in Japan. IHIT Study Group. Inhibition of Hepatocarcinogenesis by Interferon Therapy. Ann Intern Med. 1999;131:174–181. doi: 10.7326/0003-4819-131-3-199908030-00003. [DOI] [PubMed] [Google Scholar]
- 30.Crosas B, Hyndman DJ, Gallego O, Martras S, Parés X, Flynn TG, Farrés J. Human aldose reductase and human small intestine aldose reductase are efficient retinal reductases: consequences for retinoid metabolism. Biochem J. 2003;373:973–979. doi: 10.1042/BJ20021818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallego O, Ruiz FX, Ardèvol A, Domínguez M, Alvarez R, de Lera AR, Rovira C, Farrés J, Fita I, Parés X. Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10. Proc Natl Acad Sci USA. 2007;104:20764–20769. doi: 10.1073/pnas.0705659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Penning TM. AKR1B10: a new diagnostic marker of non-small cell lung carcinoma in smokers. Clin Cancer Res. 2005;11:1687–1690. doi: 10.1158/1078-0432.CCR-05-0071. [DOI] [PubMed] [Google Scholar]
- 33.Ruiz FX, Porté S, Parés X, Farrés J. Biological role of aldo-keto reductases in retinoic Acid biosynthesis and signaling. Front Pharmacol. 2012;3:58. doi: 10.3389/fphar.2012.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teramoto R, Minagawa H, Honda M, Miyazaki K, Tabuse Y, Kamijo K, Ueda T, Kaneko S. Protein expression profile characteristic to hepatocellular carcinoma revealed by 2D-DIGE with supervised learning. Biochim Biophys Acta. 2008;1784:764–772. doi: 10.1016/j.bbapap.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 35.Osaki Y, Ueda Y, Marusawa H, Nakajima J, Kimura T, Kita R, Nishikawa H, Saito S, Henmi S, Sakamoto A, et al. Decrease in alpha-fetoprotein levels predicts reduced incidence of hepatocellular carcinoma in patients with hepatitis C virus infection receiving interferon therapy: a single center study. J Gastroenterol. 2012;47:444–451. doi: 10.1007/s00535-011-0505-8. [DOI] [PubMed] [Google Scholar]
- 36.Asahina Y, Tsuchiya K, Nishimura T, Muraoka M, Suzuki Y, Tamaki N, Yasui Y, Hosokawa T, Ueda K, Nakanishi H, et al. α-fetoprotein levels after interferon therapy and risk of hepatocarcinogenesis in chronic hepatitis C. Hepatology. 2013;58:1253–1262. doi: 10.1002/hep.26442. [DOI] [PubMed] [Google Scholar]
- 37.Nishinaka T, Miura T, Okumura M, Nakao F, Nakamura H, Terada T. Regulation of aldo-keto reductase AKR1B10 gene expression: involvement of transcription factor Nrf2. Chem Biol Interact. 2011;191:185–191. doi: 10.1016/j.cbi.2011.01.026. [DOI] [PubMed] [Google Scholar]
- 38.Gambino R, Musso G, Cassader M. Redox balance in the pathogenesis of nonalcoholic fatty liver disease: mechanisms and therapeutic opportunities. Antioxid Redox Signal. 2011;15:1325–1365. doi: 10.1089/ars.2009.3058. [DOI] [PubMed] [Google Scholar]
- 39.Seki S, Kitada T, Yamada T, Sakaguchi H, Nakatani K, Wakasa K. In situ detection of lipid peroxidation and oxidative DNA damage in non-alcoholic fatty liver diseases. J Hepatol. 2002;37:56–62. doi: 10.1016/s0168-8278(02)00073-9. [DOI] [PubMed] [Google Scholar]
- 40.Chalasani N, Deeg MA, Crabb DW. Systemic levels of lipid peroxidation and its metabolic and dietary correlates in patients with nonalcoholic steatohepatitis. Am J Gastroenterol. 2004;99:1497–1502. doi: 10.1111/j.1572-0241.2004.30159.x. [DOI] [PubMed] [Google Scholar]


