Summary
Post‐translationally modified peptides, such as those containing either phosphorylated or O‐glycosylated serine/threonine residues, may be presented to cytotoxic T lymphocytes (CTLs) by MHC class I molecules. Most of these modified peptides are captured in the MHC class I groove in a similar manner to that for unmodified peptides. N‐Myristoylated 5‐mer lipopeptides have recently been identified as a novel chemical class of MHC class I‐presented antigens. The rhesus classical MHC class I allele, Mamu‐B*098, was found to be capable of binding N‐myristoylated lipopeptides and presenting them to CTLs. A high‐resolution X‐ray crystallographic analysis of the Mamu‐B*098:lipopeptide complex revealed that the myristic group as well as conserved C‐terminal serine residue of the lipopeptide ligand functioned as anchors, whereas the short stretch of three amino acid residues located in the middle of the lipopeptides was only exposed externally with the potential to interact directly with specific T‐cell receptors. Therefore, the modes of lipopeptide–ligand interactions with MHC class I and with T‐cell receptors are novel and fundamentally distinct from that for MHC class I‐presented peptides. Another lipopeptide‐presenting MHC class I allele has now been identified, leading us to the prediction that MHC class I molecules may be separated on a functional basis into two groups: one presenting long peptides and the other presenting short lipopeptides. Since the N‐myristoylation of viral proteins is often linked to pathogenesis, CTLs capable of sensing N‐myristoylation may serve to control pathogenic viruses, raising the possibility for the development of a new type of lipopeptide vaccine.
Keywords: antigen presentation/processing, major histocompatibility complex, structural biology/crystallography
Introduction
Sensing cytosolic non‐self and altered self proteins is critical for host defences against microbial infections and cancer. This immune surveillance pathway leads to the specific elimination of abnormal cells, such as virus‐infected cells and transformed cells, for which MHC‐encoded class I molecules play a pivotal role by assisting CD8+ cytotoxic T lymphocytes (CTLs) to precisely detect abnormal cells. Almost all nucleated cells maintain moderate MHC class I expression levels on the cell surface. In these cells, a significant fraction of cytosolic self proteins is constantly degraded into peptides by proteolytic activity mediated by the proteasome complex, and transported to the lumen of the endoplasmic reticulum (ER) through the ER membrane‐associated transporter associated with antigen processing (TAP).1 The ER aminopeptidases trim the peptides into a form that is optimal for binding to β 2‐microglobulin‐associated MHC class I heavy chains, and the fully assembled MHC class I trimer complex then exits the ER and traffics to the plasma membrane. Cytosolic endogenous proteins comprise a major source of MHC class I ligands in normal cells, as indicated by the profoundly impaired expression of MHC class I molecules on the surface of cells that are deficient in TAP function.2 Similarly, peptides derived from non‐self and altered self proteins in abnormal cells are captured by MHC class I molecules in the ER, delivered to the cell surface, and exposed externally.3, 4 Therefore, abnormal cells are specifically recognized and eliminated by peptide‐specific CTLs, providing the rationale for peptide vaccines against human cancer.5
The widely accepted concept of the MHC class I‐mediated presentation of peptides to CTLs may now need some modifications to incorporate the novel MHC class I function of ‘lipopeptide’ antigen presentation.6 N‐Myristoylation occurs for a sizable fraction of self and viral proteins, and our recent findings have indicated that this post‐translational modification (PTM) of proteins is monitored by the MHC class I system, which is capable of presenting N‐myristoylated short (4‐ to 6‐mer) peptides (lipopeptides) to lipopeptide‐specific CTLs.7, 8 In this brief review, we start with an overview of known examples of MHC class I‐presented PTM peptides, and then focus on the discovery of N‐myristoylated viral lipopeptides as a novel chemical class of antigens recognized by MHC class I‐restricted T cells.
MHC class I‐presented peptides with PTMs
Phosphorylation, glycosylation and other covalent modifications to proteins that occur during or after their ribosomal synthesis are collectively referred to as PTMs. These PTMs play an essential role in protein structures and functions in normal cells.9 Furthermore, alterations in the quality and quantity of PTMs are often induced in abnormal cells;10, 11, 12 therefore, monitoring PTMs is of relevance to host protection. An array of peptides with PTMs is known to be presented by the MHC class I system (Table 1). In most cases, T cells that specifically recognize peptides with a particular PTM do not respond to the corresponding unmodified peptides,13, 14, 15 thereby predicting their ability to monitor PTMs.
Table 1.
Examples of post‐translational modifications (PTMs) monitored by MHC class I molecules
| PTMs | Sites of modifications | Crystallized MHC class I alleles | References |
|---|---|---|---|
| Phosphorylation | Ser, Thr | HLA‐A2 | 10, 16, 17 |
| O‐linked glycosylation | Ser, Thr | H‐2Kb | 11, 12, 18, 20 |
| Acetylation | N terminus | HLA‐B39 | 24, 56 |
| Formylation | N terminus | H2‐M3 | 45, 57 |
| N‐myristoylation | N‐terminal Gly | Mamu‐B*098 | 6, 7, 8 |
| Citrullination | Arg | HLA‐B27 | 22, 58 |
| Methylation | Arg | Not reported | 59 |
| Deamidation | Asn | Not reported | 60 |
| Cysteinylation | Cys | Not reported | 61 |
Phosphorylation
In mammalian cells, serine‐threonine kinases and tyrosine kinases play a major role in protein phosphorylation. Peptides containing phosphorylated serine/threonine residues, but not those containing phosphorylated tyrosine, have been identified as MHC class I‐presented ligands.16 Similar to unmodified peptides, phosphorylated peptides produced in the cytosol use TAP for their entry into the lumen of the ER and are captured by newly synthesized MHC class I molecules.15 X‐ray crystallographic analyses of the HLA‐A2:phosphorylated serine‐containing peptide complex indicate that the phosphate groups are directed upward and appear to be favourably positioned for interactions with T‐cell receptors (TCRs).17 The potential contribution of phosphate groups to MHC class I–ligand interactions has also been noted. Phosphorylated peptides containing a phosphorylated serine at position 4 are favourably accommodated in the groove of HLA‐A2 with the negatively charged phosphate group interacting with the positively charged Arg65 and Lys66 residues located at the portal of the A pocket.
Glycosylation
O‐linked glycosylation to serine/threonine residues is also a common PTM known to be detected by the MHC class I system, whereas N‐linked glycosylation has not been reported, possibly because of the size of N‐linked glycans being unfavourable for interactions with TCRs.12 Purcell et al. predicted on theoretical grounds that ~ 1–5% of peptide ligands may bear a glycan.18 O‐linked glycan‐containing peptides appear to be generated in the cytosol through cytosolic glycosyltransferases and transported through the TAP transporter into the lumen of the ER for interactions with MHC class I molecules.19 The crystal structure of H‐2Kb accommodating a vesicular stomatitis virus‐related synthetic 8‐mer peptide with an O‐glycan at position 6 indicates that the glycan moiety is fully exposed externally, thereby functioning as a major T‐cell epitope rather than contributing to MHC class I–ligand interactions.20
Citrullination
The conversion of arginine within proteins into citrulline is termed citrullination (or deimidation), and is catalysed by enzymes of the peptidylarginine deimidase family, which are capable of replacing the positively charged imine group of arginine with an uncharged ketone group.21 The crystal structures of citrullinated peptide‐bound HLA‐B27 molecules indicate that the loss of the positively charged group of arginine results in a marked conformational change in bound peptides, thereby allowing CTLs to differentially recognize citrullinated and non‐citrullinated peptides.22
Acetylation
Coupling the α‐amino group of the N‐terminal amino acid residue with an acetyl group is termed N‐terminal acetylation. N‐terminal acetylation is a major PTM of eukaryotic proteins, which is catalysed by N‐terminal acetyltransferases that use acetyl‐Coenzyme A as the donor substrate.23 The crystal structure of HLA‐B39 that binds an N‐terminally acetylated peptide derived from RNA helicase indicates that the attached acetyl moiety of the N‐terminal serine residue sticks out of the antigen‐binding groove for potential interactions with TCRs.24 On the other hand, N‐terminal acetylation compromises the stability of the MHC class I–peptide complex due to the lack of molecular interactions involving the N‐terminal amino group of bound peptides. Alternatively, N‐terminal acetylation induces the spatial rotation of the N‐terminal serine residue, allowing its side chain to establish interactions with tyrosine residues located in the F pocket.
N‐Myristoylated lipopeptides: a novel antigen repertoire
N‐Myristoylation is a PTM conserved in eukaryotes, and approximately 0·8% of mammalian proteins are estimated to be N‐myristoylated.25 N‐Myristoyltransferases (NMT‐1 and NMT‐2) catalyse the modification reaction, in which the saturated C14 fatty acid (myristic acid) of the donor substrate, myristoyl‐Coenzyme A, is transferred to the glycine residue that is exposed by the removal of the N‐terminal methionine residue.26, 27 Internal glycine residues may also receive the acyl chain when they are N‐terminally exposed during protein degradation processes.28 N‐Myristoylation serves to anchor the modified proteins to the cell membrane with the hydrophobic acyl chain embedded in lipid layers, thereby regulating a number of key cellular events, as indicated by the embryonic lethality of NMT‐1‐deficient mice.29 Approximately 3·7% of viral proteins are estimated to undergo N‐myristoylation by borrowing the host cellular machinery for N‐myristoylation (Table 2) (http://mendel.imp.ac.at/myristate/myrbase/), and N‐myristoylated viral proteins are often associated with pathogenesis.30 For example, the retroviral gag protein requires N‐myristoylation to function, and the G2A mutant of the gag protein that lacks the glycine residue for N‐myristoylation fails to stably interact with the cell membrane, resulting in the impaired assembly of viral particles.31
Table 2.
Examples of N‐myristoylated viral proteins
| Viruses | Proteins | N‐terminal sequencesa | References |
|---|---|---|---|
| Human immunodeficiency virus 1 | Nef | C14‐GGKWSK | 62 |
| Gag | C14‐GARASV | 31 | |
| Simian immunodeficiency virus | Nef | C14‐GGAISM | 62 |
| Gag | C14‐GVRNSV | 31 | |
| Herpes simplex virus 1 | UL11 | C14‐GLSFSG | 63 |
| Hepatitis B virus | Pre‐S1 | C14‐GQNLST | 64 |
| L protein | C14‐GGWSSK | 51 | |
| Simian virus 40 | VP2 | C14‐GAALTL | 65 |
| Poliovirus | VP4 | C14‐GAQVSS | 66 |
| Mouse mammary tumour virus | Gag | C14‐GVSGSK | 67 |
| Sabia virus | RING finger Z | C14‐GNSKSK | 52 |
| Lassa virus | Z protein | C14‐GNKQAK | 52 |
The serine or threonine residues at position 5 (underlined) are shared among most N‐myristoylated proteins.
The Nef protein of human and simian immunodeficiency viruses (HIV/SIV) also requires N‐myristoylation to exert its key functions, which include: (i) down‐regulation of the cell surface expression of MHC class I, MHC class II, CD1d, CD4, CD80 and CD86 molecules,32, 33, 34 (ii) enhancement of virion infectivity,35, 36 (iii) induction of apoptotic cell death,37 and (iv) inhibition of the production of class switched immunoglobulins.38, 39 Based on these pathogenesis‐related functions, we predicted that the N‐myristoylation reaction of the Nef protein may be a vital target that needs to be monitored by the immune system. We recently discovered that N‐myristoylated peptides (lipopeptides) constitute a new chemical class of the antigen repertoire recognized by CD8+ CTLs.7 Our initial assessment of SIV‐infected rhesus monkeys revealed that, following infection, T cells that produced interferon‐γ in response to N‐myristoylated 5‐mer and 6‐mer peptides (C14‐GGAIS and C14‐GGAISM; see Table 1) derived from the SIV Nef protein expanded in the circulation. Furthermore, the plasma viral load correlated reciprocally with the number of lipopeptide‐specific T cells, suggesting their role in the control of infection. These unexpected observations were further substantiated by the establishment of the two rhesus CD8+ TCR‐αβ + cytotoxic T‐cell lines, 2N5.1 and SN45. These T cells produced interferon‐γ and perforin in response to C14‐GGAIS (C14nef5), but failed to respond to the GGAIS peptide, myristic acid, or a mixture of the GGAIS peptide and myristic acid, which led us to the conclusion that the covalent conjugation of the 5‐mer peptide with myristic acid is an absolute requirement for antigenic activity. We initially considered molecules of the CD1 family (CD1a, ‐b, ‐c, and ‐d) to mediate the presentation of C14nef5 to T cells because of their known ability to bind a number of lipid antigens, including C20:1 fatty acid‐containing and C18:0 fatty acid‐containing lipopeptides presented by CD1a and CD1c, respectively.40, 41, 42, 43 However, this prediction was not substantiated because none of the anti‐CD1 antibodies blocked the presentation of C14nef5 to T cells mediated by peripheral blood mononuclear cells, and none of the cell transfectants expressing each CD1 isoform replaced peripheral blood mononuclear cells as lipopeptide antigen‐presenting cells.8
Molecular and structural bases underlying lipopeptide antigen presentation
The recognition of C14nef5 by 2N5.1 T cells was shown to be restricted by the MHC class I‐encoded molecule, Mamu‐B*098.6 The alignment of the amino acid sequences of Mamu‐B*098 and other MHC class I molecules indicated that Mamu‐B*098 was similar to previously reported peptide‐presenting MHC class I molecules, and Mamu‐B*098 was identified as a member of the classic, but not non‐classic, MHC class I family by a phylogenetic tree analysis (Fig. 1). The X‐ray crystal structure of the Mamu‐B*098:C14nef5 complex revealed that the overall structure exhibited a high degree of structural similarity with other peptide‐bound MHC class I molecules. Similar to previously identified MHC class I molecules,44 six pockets, termed A through F, were readily detected in the groove of Mamu‐B*098; however, these pocket structures were elaborately designed to bind N‐myristoylated short peptides rather than conventional long peptides (Figs 2 and 3).
Figure 1.
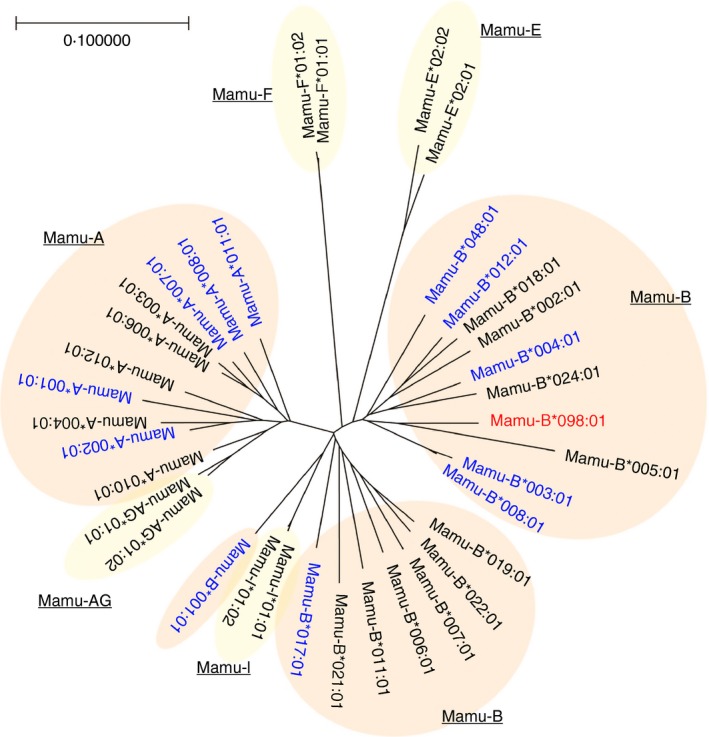
Phylogenetic tree of Mamu alleles. A phylogenetic tree was constructed by a neighbour‐joining method with bootstrap values of 5000 replications. The α1 and α2 domains of representative alleles belonging to the classical (Mamu‐A and ‐B) and non‐classical (Mamu‐AG, ‐I, ‐E, and ‐F) MHC class I families were analysed. Mamu‐B*098 is shown in red and Mamu alleles known to present peptide antigens are indicated in blue.55
Figure 2.
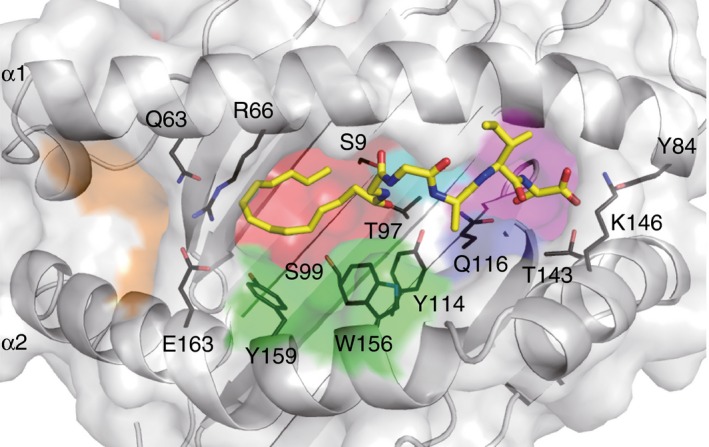
Crystal structure of the Mamu‐B*098:C14nef5 complex. The surface of the antigen‐binding groove of Mamu‐B*098 as well as the bound lipopeptide (yellow stick) are shown. The side chains of some amino acid residues critically interacting with the lipopeptide ligand are also indicated with black lines.
Figure 3.
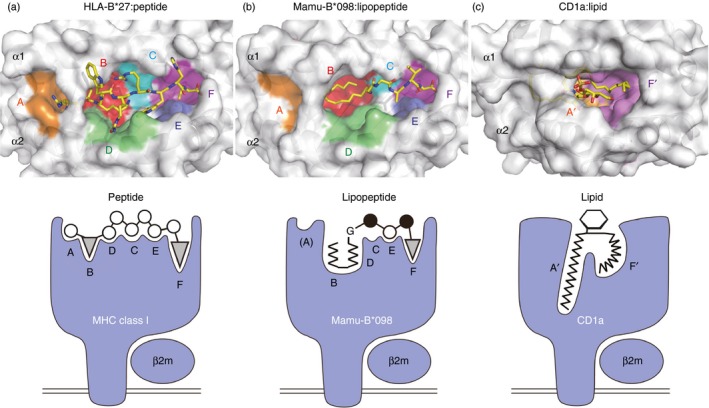
Groove structures for accommodating peptide, lipopeptide, and lipid antigens. Top views of HLA‐B27 (a, PDB code 3B6S), Mamu‐B*098 (b, PDB code 4ZFZ), and CD1a (c, PDB code 1ONQ) molecules are shown in the upper panels. Six pockets (A–F) of HLA‐B27 and Mamu‐B*098 as well as two pockets (A′ and F′) of CD1a molecules are indicated, and bound ligands are shown in yellow sticks. Side views of each complex are illustrated in the corresponding lower panels.
A pocket
The A pocket was spatially disconnected from the B and other pockets in the groove of Mamu‐B*098. This contrasts sharply with the A pockets of known peptide‐presenting MHC molecules that accommodate the N‐terminal amino acid residues of ligands.44 The salt bridge formed between Arg66 and Glu163 as well as the presence of the relatively bulky amino acid (Gln) at position 63 narrowed the channel connecting the A and B pockets to a size that hardly accommodated conventional peptides. The mouse H2‐M3 molecule capable of binding N‐formylated bacterial peptides was found to possess a salt bridge between Lys66 and Glu163, and the side chains of Leu167 and Phe171 protruded into the groove, thereby contributing to the size reduction of the A pocket.45 The X‐ray crystal structure of another lipopeptide‐presenting MHC class I allele revealed an apparently collapsed A pocket (D. Morita and M. Sugita, manuscript in preparation); therefore, ‘non‐functional’ A pockets may be a unique feature of MHC class I molecules that specifically capture peptides with N‐terminal PTMs.
B pocket
The myristoyl group of C14nef5 was accommodated in the B pocket of Mamu‐B*098. The B pocket was lined with an array of hydrophobic or non‐polar amino acid residues, serving to construct the hydrophobic environment. In addition, the small amino acid residues, Ser9, Thr97, and Ser99, located on the floor contributed to the spatial expansion of the pocket. These unique features are essential for accommodating the myristic group of the lipopeptide. We assume that these structural characteristics are not reminiscent of CD1 because the myristic acid‐binding B pocket of Mamu‐B*098 and the lipid‐binding pockets of CD1 differ significantly in their shape, size, and spatial localization. Furthermore, most of the amino acid residues of Mamu‐B*098 critically establishing Van der Waals interactions with the acyl chain are absent in CD1 molecules.6 Hence, the B pocket of Mamu‐B*098 may have evolved independently of CD1.
D pocket
The D pocket of Mamu‐B*098 also contributed to the accommodation of the myristoyl group of the ligand with Trp156 and Tyr159, particularly by establishing a number of Van der Waals contacts with the acyl chain. Nevertheless, the ligand occupancy of the D pocket of the Mamu‐B*098:C14nef5 complex was significantly lower than that of the B pocket, and its precise role remains unclear. However, the D pocket may provide extra space for accommodating longer acyl chains such as palmitic acid (C16).
F pocket
The F pocket of Mamu‐B*098 is one of the smallest F pockets identified to date in MHC class I molecules. The side chains of Tyr114 and Gln116 located on the floor of the F pocket were found to protrude upwards into the pocket, serving to reduce the volume of the pocket. Hence, it is conceivable that, unlike most MHC class I molecules with a large F pocket suitable for binding a bulky amino acid residue,46 only small amino acid residues may fit in well with the F pocket of Mamu‐B*098. The small side chain of the C‐terminal Ser residue (Ser5) of C14nef5 was found to be accommodated in this pocket. As is the case with peptide‐bound MHC class I molecules,44 the main chain of Ser5 established a hydrogen bond network with the conserved Tyr84, Thr143 and Lys146 residues of Mamu‐B*098. The side chain of Ser5 was wedged into the bottom of the F pocket by establishing a direct hydrogen bond with Gln116. Most N‐myristoylated proteins contain the prototypic N‐myristoylation motif, Gly‐X‐X‐X‐(Ser/Thr);47 therefore, the structural features of the B and F pockets predict that Mamu‐B*098 may sample lipopeptides derived from a wide variety of self‐ and foreign N‐myristoylated proteins (Table 2). It has also been predicted that Mamu‐B*098 is unable to bind conventional long peptides in a manner similar to that of other known MHC class I molecules.
Limited epitopic diversity of the lipopeptide antigen repertoire: implications for autoimmunity
The crystal structure of the Mamu‐B*098:C14nef5 complex has also provided valuable insights into the mechanisms by which T cells recognize lipopeptides. The three amino acid residues flanked by the conserved N‐terminal glycine and C‐terminal serine residues protruded out of the antigen‐binding groove and were exposed externally, suggesting that these residues primarily constituted the major T‐cell epitope. Peptide‐specific TCRs often interact with as many as six or seven amino acid residues of MHC class I‐presented peptide ligands,48, 49 allowing peptide‐specific T cells to easily discriminate foreign peptides from self‐peptides. Due to the apparently limited epitopic diversity achieved by the lipopeptide antigen repertoire, it may be challenging for lipopeptide‐specific T cells to precisely discriminate self‐ and non‐self‐peptides, suggesting that viral lipopeptide‐specific T cells cross‐react with self‐lipopeptides. Highly stringent negative selection in the thymus may be executed to eliminate self‐lipopeptide‐specific T cells; otherwise, autoimmune disorders may develop, as is often observed in patients with viral infections.50
Conclusion
Besides peptides and lipids, lipopeptides may constitute a distinct antigen repertoire recognized by αβ CTLs. Despite the potential risks of developing autoimmunity, CTL responses capable of specifically sensing the N‐myristoylation of viral proteins may be beneficial to host defences against viral infections because most N‐myristoylated viral proteins are associated directly with pathogenesis.51, 52 Furthermore, it is difficult for viruses to mutate the N‐terminal amino acid residues constituting the N‐myristoylation motif without affecting protein function;53, 54 therefore, it may be challenging for pathogenic viruses to evade lipopeptide‐specific CTL responses. ‘Classical’ MHC class I molecules have the capacity to mediate the ‘new’ function of lipopeptide antigen presentation. The findings of our recent studies have indicated that classical MHC class I molecules may be separated into at least two groups; binding peptide antigens and binding lipopeptide antigens. MHC biology has been a major focus of immunology research over the past three decades, and extensive efforts have been made to scrutinize it from every possible aspect; nevertheless, studies do not appear to be completed yet.
Disclosures
The authors have no competing interests to declare.
Acknowledgements
This work was supported by KAKENHI Grant Numbers 16K19151 (to D.M.), 15H01257, and 16K15517 (to M.S.). It was also supported by the Cooperation Research Program of the Primate Research Institute, Kyoto University.
References
- 1. Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol 2011; 11:823–36. [DOI] [PubMed] [Google Scholar]
- 2. Van Kaer L, Ashton‐Rickardt PG, Ploegh HL, Tonegawa S. TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4–8+ T cells. Cell 1992; 71:1205–14. [DOI] [PubMed] [Google Scholar]
- 3. Harty JT, Bevan MJ. Responses of CD8+ T cells to intracellular bacteria. Curr Opin Immunol 1999; 11:89–93. [DOI] [PubMed] [Google Scholar]
- 4. Lu X, Gibbs JS, Hickman HD, David A, Dolan BP, Jin Y et al Endogenous viral antigen processing generates peptide‐specific MHC class I cell‐surface clusters. Proc Natl Acad Sci U S A 2012; 109:15407–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Knutson KL, Schiffman K, Disis ML. Immunization with a HER‐2/neu helper peptide vaccine generates HER‐2/neu CD8 T‐cell immunity in cancer patients. J Clin Invest 2001; 107:477–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morita D, Yamamoto Y, Mizutani T, Ishikawa T, Suzuki J, Igarashi T et al Crystal structure of the N‐myristoylated lipopeptide‐bound MHC class I complex. Nat Commun 2016; 7:10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morita D, Igarashi T, Horiike M, Mori N, Sugita M. Cutting edge: T cells monitor N‐myristoylation of the Nef protein in simian immunodeficiency virus‐infected monkeys. J Immunol 2011; 187:608–12. [DOI] [PubMed] [Google Scholar]
- 8. Morita D, Yamamoto Y, Suzuki J, Mori N, Igarashi T, Sugita M. Molecular requirements for T cell recognition of N‐myristoylated peptides derived from the simian immunodeficiency virus Nef protein. J Virol 2013; 87:482–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mann M, Jensen ON. Proteomic analysis of post‐translational modifications. Nat Biotechnol 2003; 21:255–61. [DOI] [PubMed] [Google Scholar]
- 10. Zarling AL, Obeng RC, Desch AN, Pinczewski J, Cummings KL, Deacon DH et al MHC‐restricted phosphopeptides from insulin receptor substrate‐2 and CDC25b offer broad‐based immunotherapeutic agents for cancer. Cancer Res 2014; 74:6784–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stepensky D, Tzehoval E, Vadai E, Eisenbach L. O‐glycosylated versus non‐glycosylated MUC1‐derived peptides as potential targets for cytotoxic immunotherapy of carcinoma. Clin Exp Immunol 2006; 143:139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wolfert MA, Boons GJ. Adaptive immune activation: glycosylation does matter. Nat Chem Biol 2013; 9:776–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Engelhard VH, Altrich‐Vanlith M, Ostankovitch M, Zarling AL. Post‐translational modifications of naturally processed MHC‐binding epitopes. Curr Opin Immunol 2006; 18:92–7. [DOI] [PubMed] [Google Scholar]
- 14. Lakshminarayanan V, Thompson P, Wolfert MA, Buskas T, Bradley JM, Pathangey LB et al Immune recognition of tumor‐associated mucin MUC1 is achieved by a fully synthetic aberrantly glycosylated MUC1 tripartite vaccine. Proc Natl Acad Sci U S A 2012; 109:261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Andersen MH, Bonfill JE, Neisig A, Arsequell G, Sondergaard I, Valencia G et al Phosphorylated peptides can be transported by TAP molecules, presented by class I MHC molecules, and recognized by phosphopeptide‐specific CTL. J Immunol 1999; 163:3812–8. [PubMed] [Google Scholar]
- 16. Zarling AL, Ficarro SB, White FM, Shabanowitz J, Hunt DF, Engelhard VH. Phosphorylated peptides are naturally processed and presented by major histocompatibility complex class I molecules in vivo . J Exp Med 2000; 192:1755–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mohammed F, Cobbold M, Zarling AL, Salim M, Barrett‐Wilt GA, Shabanowitz J et al Phosphorylation‐dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self. Nat Immunol 2008; 9:1236–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Purcell AW, van Driel IR, Gleeson PA. Impact of glycans on T‐cell tolerance to glycosylated self‐antigens. Immunol Cell Biol 2008; 86:574–9. [DOI] [PubMed] [Google Scholar]
- 19. Haurum JS, Hoier IB, Arsequell G, Neisig A, Valencia G, Zeuthen J et al Presentation of cytosolic glycosylated peptides by human class I major histocompatibility complex molecules in vivo . J Exp Med 1999; 190:145–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Speir JA, Abdel‐Motal UM, Jondal M, Wilson IA. Crystal structure of an MHC class I presented glycopeptide that generates carbohydrate‐specific CTL. Immunity 1999; 10:51–61. [DOI] [PubMed] [Google Scholar]
- 21. Cuthbert GL, Daujat S, Snowden AW, Erdjument‐Bromage H, Hagiwara T, Yamada M et al Histone deimination antagonizes arginine methylation. Cell 2004; 118:545–53. [DOI] [PubMed] [Google Scholar]
- 22. Beltrami A, Rossmann M, Fiorillo MT, Paladini F, Sorrentino R, Saenger W et al Citrullination‐dependent differential presentation of a self‐peptide by HLA‐B27 subtypes. J Biol Chem 2008; 283:27189–99. [DOI] [PubMed] [Google Scholar]
- 23. Menzies KJ, Zhang H, Katsyuba E, Auwerx J. Protein acetylation in metabolism – metabolites and cofactors. Nat Rev Endocrinol 2016; 12:43–60. [DOI] [PubMed] [Google Scholar]
- 24. Sun M, Liu J, Qi J, Tefsen B, Shi Y, Yan J et al Nα‐terminal acetylation for T cell recognition: molecular basis of MHC class I‐restricted nα‐acetylpeptide presentation. J Immunol 2014; 192:5509–19. [DOI] [PubMed] [Google Scholar]
- 25. Maurer‐Stroh S, Gouda M, Novatchkova M, Schleiffer A, Schneider G, Sirota FL et al MYRbase: analysis of genome‐wide glycine myristoylation enlarges the functional spectrum of eukaryotic myristoylated proteins. Genome Biol 2004; 5:R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boutin JA. Myristoylation. Cell Signal 1997; 9:15–35. [DOI] [PubMed] [Google Scholar]
- 27. Wright MH, Heal WP, Mann DJ, Tate EW. Protein myristoylation in health and disease. J Chem Biol 2010; 3:19–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zha J, Weiler S, Oh KJ, Wei MC, Korsmeyer SJ. Posttranslational N‐myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science 2000; 290:1761–5. [DOI] [PubMed] [Google Scholar]
- 29. Yang SH, Shrivastav A, Kosinski C, Sharma RK, Chen MH, Berthiaume LG et al N‐myristoyltransferase 1 is essential in early mouse development. J Biol Chem 2005; 280:18990–5. [DOI] [PubMed] [Google Scholar]
- 30. Maurer‐Stroh S, Eisenhaber F. Myristoylation of viral and bacterial proteins. Trends Microbiol 2004; 12:178–85. [DOI] [PubMed] [Google Scholar]
- 31. Bryant M, Ratner L. Myristoylation‐dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci U S A 1990; 87:523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alexander M, Bor YC, Ravichandran KS, Hammarskjold ML, Rekosh D. Human immunodeficiency virus type 1 Nef associates with lipid rafts to downmodulate cell surface CD4 and class I major histocompatibility complex expression and to increase viral infectivity. J Virol 2004; 78:1685–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chaudhry A, Das SR, Hussain A, Mayor S, George A, Bal V et al The Nef protein of HIV‐1 induces loss of cell surface costimulatory molecules CD80 and CD86 in APCs. J Immunol 2005; 175:4566–74. [DOI] [PubMed] [Google Scholar]
- 34. Chen N, McCarthy C, Drakesmith H, Li D, Cerundolo V, McMichael AJ et al HIV‐1 down‐regulates the expression of CD1d via Nef. Eur J Immunol 2006; 36:278–86. [DOI] [PubMed] [Google Scholar]
- 35. Zheng YH, Plemenitas A, Fielding CJ, Peterlin BM. Nef increases the synthesis of and transports cholesterol to lipid rafts and HIV‐1 progeny virions. Proc Natl Acad Sci U S A 2003; 100:8460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL et al HIV‐1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 2015; 526:212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu XN, Laffert B, Screaton GR, Kraft M, Wolf D, Kolanus W et al Induction of Fas ligand expression by HIV involves the interaction of Nef with the T cell receptor ζ chain. J Exp Med 1999; 189:1489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qiao X, He B, Chiu A, Knowles DM, Chadburn A, Cerutti A. Human immunodeficiency virus 1 Nef suppresses CD40‐dependent immunoglobulin class switching in bystander B cells. Nat Immunol 2006; 7:302–10. [DOI] [PubMed] [Google Scholar]
- 39. Xu W, Santini PA, Sullivan JS, He B, Shan M, Ball SC et al HIV‐1 evades virus‐specific IgG2 and IgA responses by targeting systemic and intestinal B cells via long‐range intercellular conduits. Nat Immunol 2009; 10:1008–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Van Rhijn I, Young DC, De Jong A, Vazquez J, Cheng TY, Talekar R et al CD1c bypasses lysosomes to present a lipopeptide antigen with 12 amino acids. J Exp Med 2009; 206:1409–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sugita M, Cernadas M, Brenner MB. New insights into pathways for CD1‐mediated antigen presentation. Curr Opin Immunol 2004; 16:90–5. [DOI] [PubMed] [Google Scholar]
- 42. Morita D, Katoh K, Harada T, Nakagawa Y, Matsunaga I, Miura T et al Trans‐species activation of human T cells by rhesus macaque CD1b molecules. Biochem Biophys Res Commun 2008; 377:889–93. [DOI] [PubMed] [Google Scholar]
- 43. Morita D, Hattori Y, Nakamura T, Igarashi T, Harashima H, Sugita M. Major T cell response to a mycolyl glycolipid is mediated by CD1c molecules in rhesus macaques. Infect Immun 2013; 81:311–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Madden DR. The three‐dimensional structure of peptide‐MHC complexes. Annu Rev Immunol 1995; 13:587–622. [DOI] [PubMed] [Google Scholar]
- 45. Wang CR, Castano AR, Peterson PA, Slaughter C, Lindahl KF, Deisenhofer J. Nonclassical binding of formylated peptide in crystal‐structure of the MHC Class‐Ib molecule H2‐M3. Cell 1995; 82:655–64. [DOI] [PubMed] [Google Scholar]
- 46. Shastri N, Schwab S, Serwold T. Producing nature's gene‐chips: the generation of peptides for display by MHC class I molecules. Annu Rev Immunol 2002; 20:463–93. [DOI] [PubMed] [Google Scholar]
- 47. Utsumi T, Nakano K, Funakoshi T, Kayano Y, Nakao S, Sakurai N et al Vertical‐scanning mutagenesis of amino acids in a model N‐myristoylation motif reveals the major amino‐terminal sequence requirements for protein N‐myristoylation. Eur J Biochem 2004; 271:863–74. [DOI] [PubMed] [Google Scholar]
- 48. Borbulevych OY, Santhanagopolan SM, Hossain M, Baker BM. TCRs used in cancer gene therapy cross‐react with MART‐1/Melan‐A tumor antigens via distinct mechanisms. J Immunol 2011; 187:2453–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cole DK, Bulek AM, Dolton G, Schauenberg AJ, Szomolay B, Rittase W et al Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross‐reactivity. J Clin Invest 2016; 126:2191–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fujinami RS, von Herrath MG, Christen U, Whitton JL. Molecular mimicry, bystander activation, or viral persistence: infections and autoimmune disease. Clin Microbiol Rev 2006; 19:80–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gripon P, Le Seyec J, Rumin S, Guguen‐Guillouzo C. Myristylation of the hepatitis B virus large surface protein is essential for viral infectivity. Virology 1995; 213:292–9. [DOI] [PubMed] [Google Scholar]
- 52. Perez M, Greenwald DL, de la Torre JC. Myristoylation of the RING finger Z protein is essential for arenavirus budding. J Virol 2004; 78:11443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shugars DC, Smith MS, Glueck DH, Nantermet PV, Seillier‐Moiseiwitsch F, Swanstrom R. Analysis of human immunodeficiency virus type 1 nef gene sequences present in vivo . J Virol 1993; 67:4639–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brenner M, Munch J, Schindler M, Wildum S, Stolte N, Stahl‐Hennig C et al Importance of the N‐distal AP‐2 binding element in Nef for simian immunodeficiency virus replication and pathogenicity in rhesus macaques. J Virol 2006; 80:4469–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Budde ML, Lhost JJ, Burwitz BJ, Becker EA, Burns CM, O'Connor SL et al Transcriptionally abundant major histocompatibility complex class I alleles are fundamental to nonhuman primate simian immunodeficiency virus‐specific CD8+ T cell responses. J Virol 2011; 85:3250–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yague J, Alvarez I, Rognan D, Ramos M, Vazquez J, de Castro JA. An N‐acetylated natural ligand of human histocompatibility leukocyte antigen (HLA)‐B39. Classical major histocompatibility complex class I proteins bind peptides with a blocked NH2 terminus in vivo . J Exp Med 2000; 191:2083–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Colmone A, Wang CR. H2‐M3‐restricted T cell response to infection. Microbes Infect 2006; 8:2277–83. [DOI] [PubMed] [Google Scholar]
- 58. Anderton SM. Post‐translational modifications of self antigens: implications for autoimmunity. Curr Opin Immunol 2004; 16:753–8. [DOI] [PubMed] [Google Scholar]
- 59. Yague J, Vazquez J, Lopez de Castro JA. A post‐translational modification of nuclear proteins, N(G), N(G)‐dimethyl‐Arg, found in a natural HLA class I peptide ligand. Protein Sci 2000; 9:2210–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dalet A, Robbins PF, Stroobant V, Vigneron N, Li YF, El‐Gamil M et al An antigenic peptide produced by reverse splicing and double asparagine deamidation. Proc Natl Acad Sci U S A 2011; 108:E323–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pierce RA, Field ED, den Haan JM, Caldwell JA, White FM, Marto JA et al Cutting edge: the HLA‐A*0101‐restricted HY minor histocompatibility antigen originates from DFFRY and contains a cysteinylated cysteine residue as identified by a novel mass spectrometric technique. J Immunol 1999; 163:6360–4. [PubMed] [Google Scholar]
- 62. Rose JJ, Janvier K, Chandrasekhar S, Sekaly RP, Bonifacino JS, Venkatesan S. CD4 down‐regulation by HIV‐1 and simian immunodeficiency virus (SIV) Nef proteins involves both internalization and intracellular retention mechanisms. J Biol Chem 2005; 280:7413–26. [DOI] [PubMed] [Google Scholar]
- 63. Kim IJ, Chouljenko VN, Walker JD, Kousoulas KG. Herpes simplex virus 1 glycoprotein M and the membrane‐associated protein UL11 are required for virus‐induced cell fusion and efficient virus entry. J Virol 2013; 87:8029–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Le Duff Y, Blanchet M, Sureau C. The pre‐S1 and antigenic loop infectivity determinants of the hepatitis B virus envelope proteins are functionally independent. J Virol 2009; 83:12443–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Krauzewicz N, Streuli CH, Stuart‐Smith N, Jones MD, Wallace S, Griffin BE. Myristylated polyomavirus VP2: role in the life cycle of the virus. J Virol 1990; 64:4414–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Paul AV, Schultz A, Pincus SE, Oroszlan S, Wimmer E. Capsid protein VP4 of poliovirus is N‐myristoylated. Proc Natl Acad Sci U S A 1987; 84:7827–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hizi A, Henderson LE, Copeland TD, Sowder RC, Krutzsch HC, Oroszlan S. Analysis of gag proteins from mouse mammary tumor virus. J Virol 1989; 63:2543–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


