Abstract
Celiac disease (CD) remains one of the most significant autoimmune diseases worldwide. The pathogenesis of CD is not clearly understood and is probably attributed to genomic variations and host genetic make-up. Case-control and cohort studies of the association between the TNF-α -308 G > A (rs1800629) polymorphism and CD susceptibility have yielded inconsistent results. In this study, PubMed, EMBASE, and Google Scholar web-databases were searched for pertinent reports showing association of TNF-α -308 G > A gene with CD risk. A total of eleven reports involving 1774 controls and 1147 CD cases were included. Significant associations in four genetic models, viz. variant allele (A vs. G: p = 0.001; OR = 2.051, 95% CI = 1.452–2.895), variant homozygous (AA vs. GG: p = 0.001; OR = 6.626, 95% CI = 3.569–12.300), recessive (AA vs. GG + AG: p = 0.001; OR = 4.766, 95% CI = 3.177–7.152) and dominant (AA + AG vs. GG: p = 0.008; OR = 1.910, 95% CI = 1.181–3.088) were found in comparison with wild type homozygous GG genotype. However, heterozygous genetic model did not show any association. Sensitivity analysis revealed stable and statistically robust results. Our results suggest that TNF-α -308 G > A gene polymorphism significantly contributes to CD susceptibility.
Celiac disease (CD; also known as celiac sprue, gluten-sensitive enteropathy, and nontropical sprue) is a chronic inflammatory autoimmune disorder characterized by abnormalities in the small intestine caused by permanent intolerance to dietary gluten or related proteins from wheat and rye1. Although few CD patients may suffer primarily from gastrointestinal symptoms, CD may be related with extra-intestinal disorders2. Genetic, immunological and environmental factors have been attributed for the disease. Till date, the etiology of CD has not yet been fully elucidated. However, CD has shown a strong genetic relation with HLA (human leucocyte antigen) class II gene, contributing no more than 40% to the disease risk, which shows involvement of genetic components in the development of CD3,4. Recently published genome-wide association study (GWAS) has identified several non-HLA genes as new susceptibility factors for CD. So far, 39 loci with 57 independent association signals have been defined and many of these loci harbour genes associated with immunity5. This suggests that genetic susceptibility to CD is also conferred by some other non HLA genes. Earlier, studies have revealed that genes encoding for the pro-inflammatory cytokines could predispose to immunological mediated lesion of CD6. Cytokines are important mediators of immunity and their responses due to imbalance or deficiency in the cytokine network may largely determine autoimmune disease susceptibility and severity.
Tumor necrosis factor-alpha (TNF-α) is a potentpro-inflammatory and immunoregulatory cytokine mapped on chromosome 6 (6p21.31) spanning about 3 kb and contains 4 exons. TNF-α stimulates many other cytokines and mediates the cytokine cascade that causes inflammation7. TNF-α gene has close linkage between HLA class I and class II8, and tightly regulated at the level of transcription9. Previous studies reported that sequence variation in the regulatory region of TNF-α gene has been correlated with various autoimmune diseases10. Several biallelic single nucleotide polymorphisms (SNPs) have been noted in the TNF-α gene11. Among them one G (guanine) >A (adenine) polymorphism is located upstream of the gene at -308 and is known to influence TNF-α levels. In comparison with the TNF-α -308G allele, A allele has higher transcriptional activity and often connected to autoimmune diseases12,13. The location of the gene within the major histocompatibility complex and the putative role of -308 G > A polymorphism on the promoter activity of TNF-α gene has raised the possibility that this polymorphism may influence immunologic homeostasis and contribute to the pathogenesis of CD.
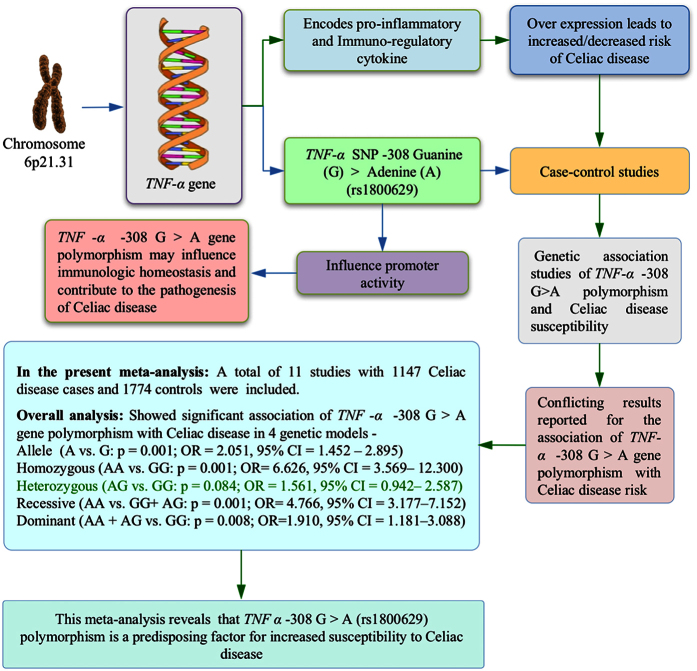
Keeping aforesaid information in view, to date a number of case control and cohort studies have been performed to investigate the association of TNF-α -308 G > A gene polymorphism and CD susceptibility14,15,16,17,18,19,20,21,22,23,24. However, the results are inconsistent and controversial because of small sample size of individual study and possible selective bias. In order to overcome the limitations of single studies, we conducted this meta-analysis by pooling previous single studies to increase the statistical power and to derive more precise and comprehensive relationship between TNF-α -308 G > A gene polymorphism and CD susceptibility. The schematic representation of the entire pooled study is presented as Graphical Abstract (Fig. 1).
Figure 1. Graphical abstract of the meta-analysis performed to evaluate the association of TNF-α -308 G > A (rs1800629) polymorphism and CD susceptibility.
Results
Characteristics of the selected published studies
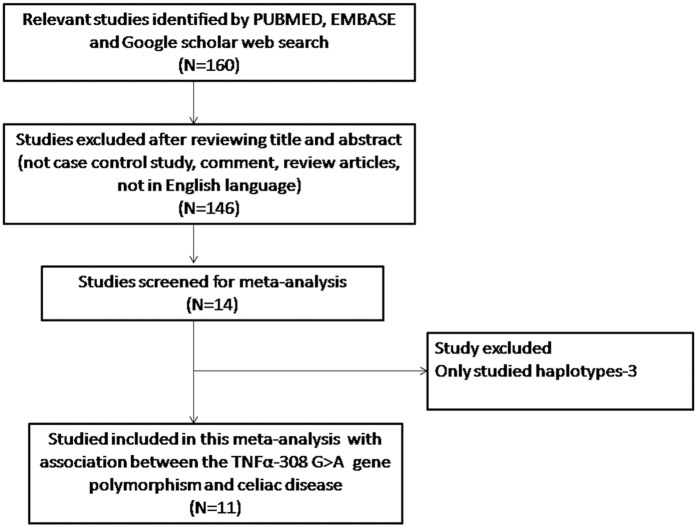
The number of hits obtained by doing a literature search via PubMed (Medline), EMBASE and Google Scholar search database were one hundred and sixty. All the retrieved hits (articles) were checked by reading their titles and abstracts, and the full texts for the possibly relevant publications. In addition, the articles were further scrutinized for their appropriateness for this meta-analysis (Fig. 2: PRISMA Flow-diagram). Likewise, the references listed in the retrieved publications were also screened for other possible apposite articles. In order to derive a precise conclusion from this pooled analysis, very stringent criteria were followed for searching and selecting the pertinent publications, for e.g., only case-control or cohort design studies with frequencies of all the three genotypes were included. After thorough analysis and following the stern criteria of selection (inclusion or exclusion), eleven originally published studies representing the above mentioned possible association were found eligible and included in this study (Table 1). A PRISMA flow-diagram showing the selection process (inclusion/exclusion) of the studies for this meta-analysis is given in Fig. 2. Information regarding distribution of genotypes, HWE p-values in the controls, and susceptibility to CD is provided in Table 2. All the eleven studies were assessed for the quality according to the Newcastle-Ottawa Scale and most studies (80%) scored 5 stars or more, suggesting a moderate to good quality (Table 3).
Figure 2. PRISMA flow-diagram showing the selection process (inclusion/exclusion) of the pertinent studies of TNF-α -308 G > A (rs1800629) polymorphism and CD risk.
Table 1. Summary of major characteristics of all the studies included in the present meta-analysis.
| First author and Year of Publication | Country | Ethnicity | Controls | Cases | Study | Technique used | Association Yes/No |
|---|---|---|---|---|---|---|---|
| Rossi et al.14 | Brazil | Caucasian | 267 | 244 | HB | RT-PCR | Yes |
| de Albuquerque et al.15 | Italy | Caucasian | 96 | 192 | HB | PCR-RFLP | Yes |
| Kekik et al.16 | Turkey | Caucasian | 93 | 33 | HB | PCR-SSP | No |
| Capilla et al.17 | Spain | Caucasian | 256 | 144 | HB | GPC | Yes |
| Hermann et al.18 | Hungary | Caucasian | 277 | 19 | HB | PCR-RFLP | No |
| Barisani et al.19 | Italy | Caucasian | 202 | 155 | HB | PCR-RFLP | No |
| Garrote et al.20 | Spain | Caucasian | 99 | 50 | HB | PCR-SSP | No |
| Lio et al.21 | Italy | Caucasian | 220 | 110 | HB | ARMS-PCR | Yes |
| Cataldo et al.22 | Italy | Caucasian | 96 | 66 | HB | PCR Hybridization | Yes |
| Hahn Zoric et al.23 | Sweden | Caucasian | 103 | 89 | HB | PCR-RFLP | No |
| Garrote et al.24 | Spain | Caucasian | 65 | 45 | HB | PCR-RFLP | Yes |
HB = Hospital based.
Table 2. Genotypic distribution of TNF-α -308G > A gene polymorphism included in the present meta-analysis.
| Authors and year | Controls | Cases | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | Minor allele | Genotype | Minor allele | ||||||
| GG | GA | AA | MAF | GG | GA | AA | MAF | p-value | |
| Rossi et al.14 | 206 | 58 | 3 | 0.11 | 107 | 117 | 20 | 0.32 | 0.62 |
| deAlbuquerque et al.15 | 42 | 46 | 8 | 0.32 | 100 | 63 | 29 | 0.31 | 0.34 |
| Kekik et al.16 | 15 | 78 | 0 | 0.41 | 16 | 14 | 3 | 0.30 | 0.01 |
| Capilla et al.17 | 191 | 60 | 5 | 0.13 | 90 | 44 | 10 | 0.22 | 0.90 |
| Hermann et al.18 | 148 | 118 | 11 | 0.25 | 8 | 9 | 2 | 0.34 | 0.03 |
| Barisani et al.19 | 157 | 44 | 1 | 0.11 | 85 | 55 | 15 | 0.27 | 0.25 |
| Garrote et al.20 | 72 | 26 | 1 | 0.14 | 31 | 18 | 1 | 0.2 | 0.41 |
| Lio et al.21 | 163 | 53 | 4 | 0.13 | 61 | 34 | 15 | 0.29 | 0.89 |
| Cataldo et al.22 | 73 | 22 | 1 | 0.12 | 44 | 12 | 12 | 0.26 | 0.64 |
| Hahn zoric et al.23 | 70 | 27 | 6 | 0.18 | 23 | 44 | 22 | 0.49 | 0.13 |
| Garrote et al.24 | 51 | 14 | 0 | 0.10 | 20 | 23 | 2 | 0.30 | 0.33 |
MAF: Minor Allele Frequency; HWE: Hardy Weinberg Equilibrium.
Table 3. Quality assessment conducted according to the Newcastle-Ottawa criteria for all the included studies in this meta-analysis.
| First author | Year | Quality indicators | ||
|---|---|---|---|---|
| Selection | Comparability | Outcome | ||
| Rossi et al. | 2015 | *** | ** | ** |
| de Albuquerque et al. | 2015 | **** | *** | ** |
| Kekik et al. | 2011 | ** | * | ** |
| Capilla et al. | 2007 | *** | ** | *** |
| Hermann et al. | 2007 | *** | * | *** |
| Barisani et al. | 2006 | *** | ** | *** |
| Garrote et al. | 2005 | ** | ** | *** |
| Lio et al. | 2005 | *** | * | *** |
| Cataldo et al. | 2003 | *** | * | *** |
| Hahn zoric et al. | 2003 | ** | * | *** |
| Garrote et al. | 2002 | **** | ** | *** |
Evaluation of publication bias
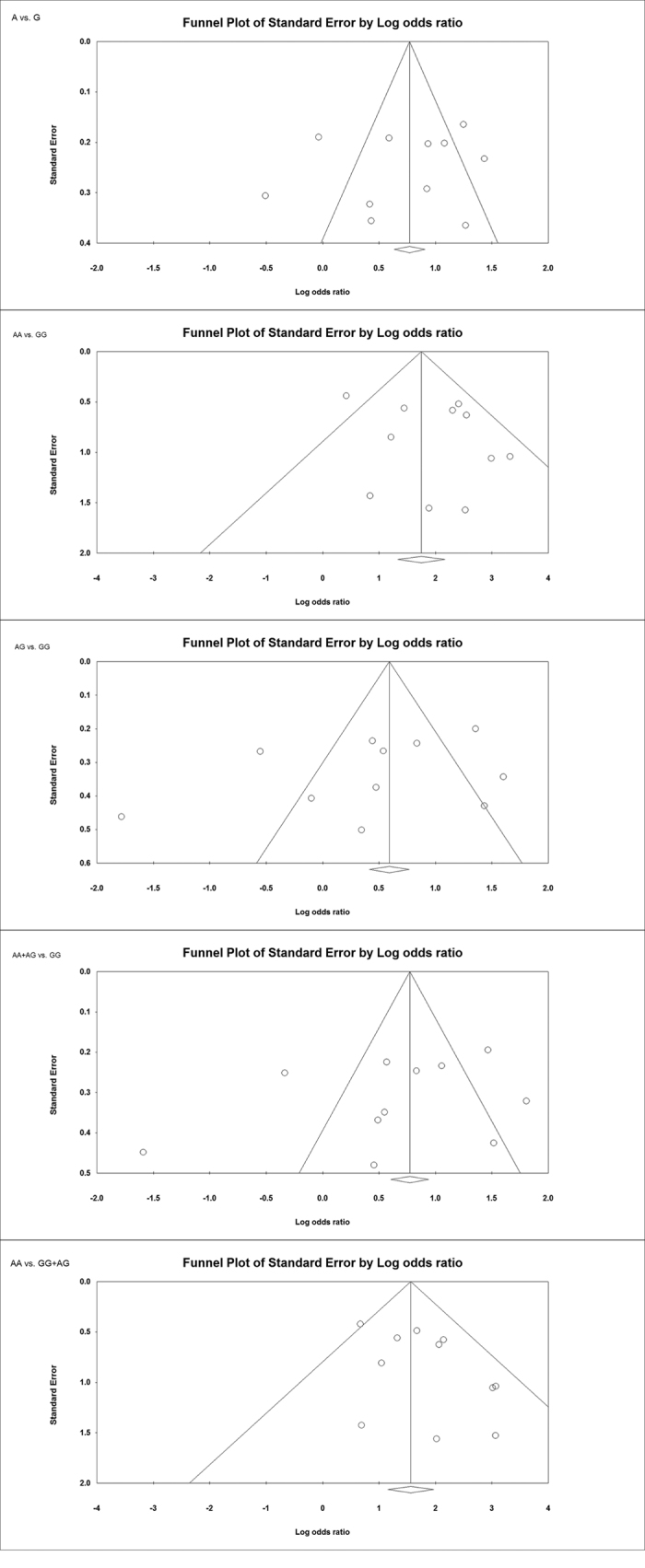
Funnel plot and Egger’s test were employed to test publication bias among the selected studies (Table 4; Fig. 3). The Funnel plots remained symmetric and Egger’s linear regression test also indicated no evidence of publication bias among the studies testing -308 G > A polymorphism of TNF-α gene and CD susceptibility (Fig. 3).
Table 4. Overall statistics to test publication bias and heterogeneity in the present meta-analysis.
| Comparisons | Egger’s regression analysis | Heterogeneity analysis | Model used for the present meta-analysis | ||||
|---|---|---|---|---|---|---|---|
| Intercept | 95% Confidence Interval | p-value | Q-value | Pheterogeneity | I2 (%) | ||
| A vs. G | −1.96 | −8.68 to 4.76 | 0.52 | 60.14 | 0.001 | 83.37 | Random |
| AA vs. GG | 1.16 | −1.18 to 3.51 | 0.29 | 18.29 | 0.050 | 45.35 | Fixed |
| AG vs. GG | −3.37 | −9.91 to 3.17 | 0.27 | 76.58 | 0.001 | 86.94 | Random |
| AA + AG vs. GG | −3.08 | −9.82 to 3.66 | 0.32 | 76.96 | 0.001 | 87.00 | Random |
| AA vs. GG + AG | 1.34 | −0.37 to 3.07 | 0.11 | 12.21 | 0.271 | 18.10 | Fixed |
Figure 3. Assessment of publication bias shown for all the genetic models (allele: A vs. G, homozygous: AA vs. GG, heterozygous: AG vs. GG, dominant: AA + AG vs. GG, recessive: AA vs. GG + AG) with Funnel plots in studies assaying odds of CD associated with TNF-α -308 G > A polymorphism.
Test of heterogeneity
Q-test and I2 statistics were used to evaluate the inter- and intra-study variations, and based upon the significance value different models were selected for the present meta-analysis (Table 4). Heterogeneity was observed in three genetic models, i.e., variant allele (A vs. G) heterozygous (AG vs. GG) and recessive (AA + AG vs. GG), hence random effects model was applied.
Meta-analysis of TNF-α -308 G > A polymorphism and CD susceptibility
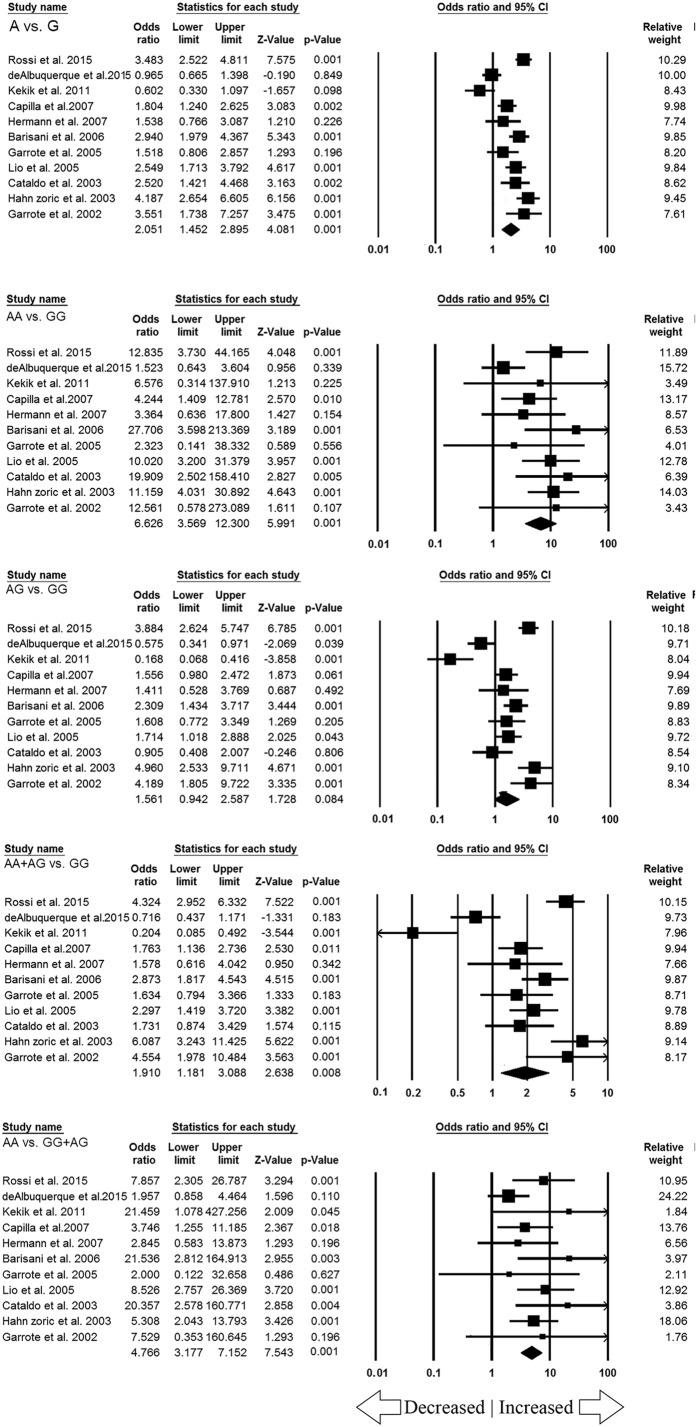
After cautious appraisal, all the data from the selected eleven studies were pooled together that lead to 1774 controls and 1147 CD cases were included to evaluate the overall association between the TNF-α -308 G > A gene polymorphism and the risk of CD. Overall significant increased risk of CD susceptibility was found in variant allele (A vs. G: p = 0.001; OR = 2.051, 95% CI = 1.452–2.895), homozygous variant (AA vs. GG: p = 0.001; OR = 6.626, 95% CI = 3.569–12.300), recessive (AA vs. GG + AG: p = 0.001; OR = 4.766, 95% CI = 3.177–7.152) and dominant (AA + AG vs. GG: p = 0.008; OR = 1.910, 95% CI = 1.181–3.088) genetic models in comparison with wild type GG genotype. However heterozygous (AG vs. GG: p = 0.084; OR = 1.561, 95% CI = 0.942–2.587) genetic model did not show any association. The Forest plots for all the genetic models are shown in Fig. 4.
Figure 4. Forest plot for overall analysis (allele: A vs. G, homozygous: AA vs. GG, heterozygous: AG vs. GG, dominant: AA + AG vs. GG, recessive: AA vs. GG + AG) showing OR with 95% CI to evaluate the association of TNF-α -308 G > A (rs1800629) polymorphism and CD risk.
Note: Black square represents the value of OR and the size of the square indicates the inverse proportion relative to its variance. Horizontal line is the 95% CI of OR.
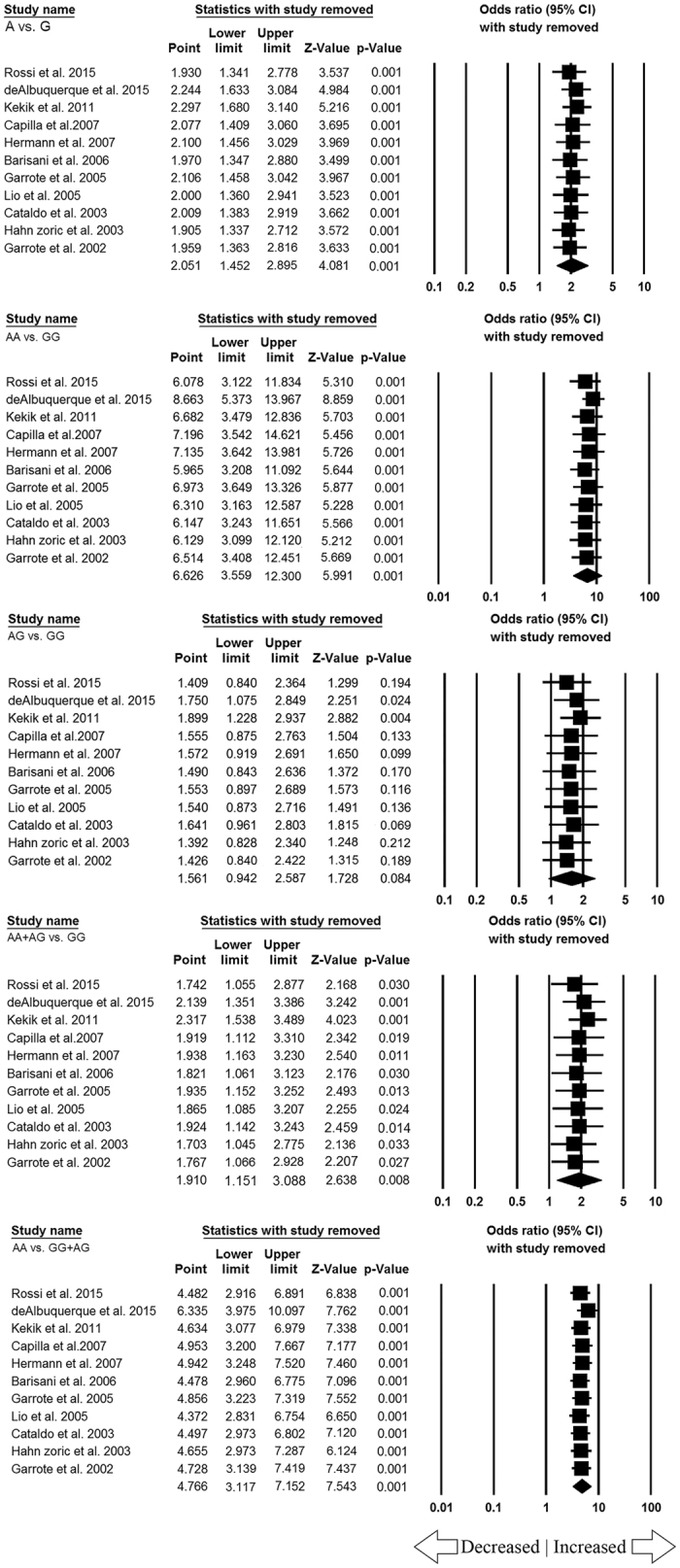
Sensitivity analysis
One way sensitivity analysis was performed to appraise the effect of each individual study on the pooled ORs by serially deleting one single study each time. When omitted each study in the current meta-analysis, the pooled ORs were always remains the same and no other single study influenced the pooled ORs. This suggests that the results of TNF-α -308 G > A gene polymorphism and CD susceptibility were stable and statistically robust (Fig. 5).
Figure 5. Sensitivity analysis showing all the genetic models (allele: A vs. G, homozygous: AA vs. GG, heterozygous: AG vs. GG, dominant: AA + AG vs. GG, recessive: AA vs. GG + AG).
Discussion
Celiac disease (CD) is one of the best understood autoimmune disorders and genetic factors are considered to be strong determinants of this disease25, which encourage researchers to search for the responsible genes for CD susceptibility. TNF-α is pleiotropic cytokine mediates broad range of proinflammatory and inflammatory responses and alter levels of TNF-α among different individuals can be a major risk for differences in the susceptibility and severity of CD. Most of the evidences suggest that the endogenous production of TNF-α may be influenced by TNF-α promoter polymorphisms, thereby affecting messenger RNA (mRNA) and protein expression levels26. In view of facts about the significant role of TNF-α gene in modulation of acute inflammation and host innate immunity, various studies have been conducted to analyze the relationship between TNF-α -308 G > A gene polymorphism and the development of CD, but the results from different published studies were contradictory and inconclusive14,15,16,17,18,19,20,21,22,23,24. In order to improve the statistical power, we performed the present meta-analysis to assess the said association of TNF-α -308 G > A polymorphism and CD risk from eleven case-control studies, as combining of the data from different studies has an advantage of reducing the random error27. In the current meta-analysis, we separately extracted different genotype frequencies of the TNF-α -308 G > A polymorphism in CD and estimated the pooled ORs and their corresponding 95% CI in all the genetic models. Our meta-analysis showed that ORs of the four genetic models were on the right side of the vertical line and the corresponding 95% CI of the pooled ORs were also on the right side of the vertical line with a significant p-value. This suggest that TNF-α -308 G > A gene polymorphism might contribute to functional role in the pathogenesis of CD. Earlier studies have suggested the role of TNF-α gene in CD development, and it has been observed that TNF-α -308 A allele directly affects the transcriptional activity that leads to higher production of TNF-α in in-vitro experiment28 and higher levels of TNF-α transcription has been reported to facilitate the inflammatory response to gluten29. Genetic variants in the TNF-α gene might increase the production of TNF-α for prolonged duration and cause uncontrolled or severe inflammation and influences the increased risk of CD. Also, up-regulated TNF-α expression has been observed in epithelial cells and intraepithelial lymphocytes in the mucosa of CD patients30. TNF-α also triggers a proteolytic cascade mediated by matrix metalloproteinases (MMPs) secretion from intestinal myofibroblasts and results in intestinal architectural alteration31. Therefore, blockade of this cytokine may prevent the activation of proteolytic MMPs and ultimately resumes the intestinal haemostasis. Earlier studies have reported the use of monoclonal antibodies against TNF-α (infliximab), beneficial for patients with severe refractory CD and uncontrolled sprue32,33. However, larger scale clinical trial studies are necessary to demonstrate the clinical utility of TNF-α -308 G > A polymorphism for the diagnosis and the treatment of CD.
The genetics involved in the development of CD is very complex, with evidence for the participation of multiple intrinsic (genetic) and extrinsic (environmental) factors34, hence, a single genetic variant is generally inadequate and failed to interpret the risk of this disease. Notwithstanding, the significant findings achieved from this study, we still have to acknowledge some of the limitations of this meta-analysis. Firstly, we found heterogeneity in this pooled study, which might be attributed to one or more of the following reasons, the ethnic origin of the patients, recruiting control samples and quality of studies. Second, language bias may exist because reports published in the English language were only considered for this meta-analysis. Third, we only included published articles available in PubMed, EMBASE, and Google Scholar web databases, pertinent articles published in other databases, print only journals, and unpublished (i.e., studies reported other than research articles in journals, for e.g., doctoral/academic thesis etc.) studies may have been missed. Fourth, meta-analysis remains a retrospective research, which is subject to the methodological deficiencies or selection bias of the included studies and may possibly influence or deviate the reliability of our study results35.
Nevertheless, our current meta-analysis has some advantages, which could be summarizes as follows. First, we tried our best to find as many published studies by means of various searching approaches. Second, publication bias was not detected by funnel plot and Egger’slinear regression test, thus the all results were statistically robust. Third, sensitivity analysis also demonstrated that the results were not influenced by any single study. Fourth, we used strict data extraction and analysis to make satisfactory and reliable conclusion.
In conclusion, we can say that meta-analysis is an extremely valuable and powerful tool for numerical data-analysis that takes in to account both statistically significant and non-significant data from the individual studies and results into a cumulative precise conclusion. To the best of our knowledge, this is the very first meta-analysis showing association of TNF-α -308 G > A polymorphism with increased CD susceptibility. Hence, determining TNF-α -308A genotype is a probable determinant for the development of CD. Further, well designed large-scale studies with the considerations of gene-gene and gene-environment interactions are warranted to investigate this association. Future studies are encouraged to validate our current findings and to prove the clinical relevance of TNF-α -308 G > A polymorphism in the development of CD. Here, we analyzed the -308 G > A variant of TNF-α gene for the risk of CD without considering the interaction between several other SNPs. In future, we intend to explore other pertinent interactions to facilitate the discovery of CD development.
Methods
Identification of eligible studies
Pertinent studies were cautiously identified by multiple comprehensive search of PubMed (Medline), EMBASE and Google Scholar online web databases covering all research articles published with a combination of the following key words: ‘Tumor necrosis factor-alpha gene OR TNF-α gene OR TNF gene AND polymorphism OR mutation OR variant AND -308 rs1800629 AND Celiac OR Coeliac disease’ (last updated on May 2016). All the references present in the retrieved studies were also checked by hand search to identify the studies that possibly have not been included in these databases. All the published studies matching with the above mentioned eligibility criteria were selected and included in the present meta-analysis.
Inclusion and exclusion criteria for the selection of studies
In order to minimize heterogeneity and facilitate the accuracy of the findings, studies included in the present meta-analysis had to follow the preset criteria: (i) must evaluated TNF-α -308 G > A polymorphism and CD risk, (ii) reported original data from the case-control and cohort study, (iii) enrolled pathologically confirmed CD cases and CD-free controls, (iv) presented useful genotype frequency in all the cases as well as in the controls, and (v) must be published in the English language. The leading reasons for the exclusion of the studies were, overlapping of the data, review articles, abstract only, and case-only studies. The studies of TNF polymorphism to predict survival and expression level considering it as an indicator for the response to therapy were also excluded.
Data extraction
For all the selected publications, the methodological quality assessment and the data extraction were independently abstracted in duplicate by two independent investigators using a standard protocol. The data accuracy was determined by using a data-collection form according to the pre-set inclusion criteria as stated in the above section. The quality assessment of the case-control studies for this meta-analysis was done by two independent investigators (AJ & SAD) based on the pre-set eligibility criteria of the present study, and sequential exclusion of the inappropriate studies. In case of disagreements between the two investigators on any item regarding the data collected from the retrieved studies, the issue was openly discussed in detail and an agreement was reached following a discussion with the adjudicator (RKM). The abstracted characteristics from the selected studies included the name of first author, the year of publication, the country of origin, sources of study, the number of cases/controls, genotyping methods and frequencies and association with CD.
Quality assessment of the selected studies
Methodological quality assessment of the included studies was done separately by two independent researchers using the Newcastle-Ottawa Scale (NOS) criteria36. The NOS criteria included three aspects: (1) subject selection: 0–4 points; (2) comparability of subject: 0–2 points; (3) clinical outcome: 0–3 points. Studies that were awarded 5 stars or more can be considered as of medium to high quality37.
Statistical analysis
The pooled odds ratios (ORs) and their respective 95% confidence intervals (CIs) were calculated to evaluate the relation between TNF-α -308 G > A gene polymorphism and CD risk. Heterogeneity belief associated with the selected studies was investigated by the chi-square based Q-test and I2 test38. The heterogeneity was considered significant if the p-value was less than 0.1. While calculating the pooled ORs, fixed-effects models were used if the I2 value was less than 70%; otherwise, random-effects models were adopted39,40. Hardy-Weinberg equilibrium (HWE) is an application of the binomial theorem to population genetics and stating that the genetic variation in a population will remain constant from one generation to the next in the absence of disturbing factors. The assessment of departure from HWE is performed by chi-square test in the controls because a deviation from HWE in the cases might indicate a genetic association, and the difference of HWE between the cases and the controls can be used to test for association41. Minor allele frequency (MAF) is the frequency of the less (or least) frequent allele in a given locus and a first reported population. The funnel plot asymmetry was calculated by the Egger’s linear regression test, which is a linear regression methodology to measure the funnel plot asymmetry on the natural logarithm scale of the OR. Student’s t-test (p-value < 0.05 was maintained as a sign of statistically significant publication bias) was used to determine the significance of the intercept42. The entire statistical analysis for the present meta-analysis (pooled analysis) was performed with the help of Comprehensive Meta-analysis (CMA) Version 2 software program (Biostat, USA).
Additional Information
How to cite this article: Khan, S. et al. TNF-α -308 G > A (rs1800629) Polymorphism is Associated with Celiac Disease: A Meta-analysis of 11 Case-Control Studies. Sci. Rep. 6, 32677; doi: 10.1038/srep32677 (2016).
Acknowledgments
The authors are grateful to the Deanship of Scientific Research, Jazan University, and Deanship of Scientific Research, University of Ha’il, Saudi Arabia, for providing the necessary dry-lab facility for this study.
Footnotes
The authors declare no competing financial interests.
Author Contributions Conceived and designed the study and experiments: S.K., R.K.M., A.J., S.A.D., M.W., A.K.P., M.Y.A., M.E.A.K. and S.H. Performed the experiments: S.K., R.K.M., A.K.P., M.Y.A., M.E.A.K. and S.H. Analyzed the data: S.K., R.K.M., A.J., S.A.D., M.W., A.K.P. and M.E.A.K. Contributed reagents/materials/analysis tools: M.W., M.Y.A. and S.H. Wrote the paper: R.K.M., A.J., S.A.D., M.W., M.Y.A. and S.H. All authors reviewed the manuscript.
References
- Green P. H. & Cellier C. Celiac disease. N. Engl. J. Med. 357, 1731–1743 (2007). [DOI] [PubMed] [Google Scholar]
- Holmes G. K. Coeliac disease and type 1 diabetes mellitus the case for screening. Diabet. Med. 18, 169–177 (2001). [DOI] [PubMed] [Google Scholar]
- Gjertsen H. A., Lundin K. E. A., Sollid L. M., Eriksen J. A. & Thorsby E. T cells recognise a peptide derived from α-gliadin presented by the celiac disease-associated HLA-DQ (α1*0501, β1*0201) heterodimer. Hum. Immunol. 39, 243–252 (1994). [DOI] [PubMed] [Google Scholar]
- Greco L., Corazza G., Babron M.-C., Clot F., Fulchignoni-Lataud M.-C. et al. Genome search in celiac disease.Am. J. Hum. Gen. 62, 669–675 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollid L. M. & Jabri B. Triggers and drivers of autoimmunity: lessons from coeliac disease. Nat. Rev. Immunol. 13, 294–302 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahat N., Shapiro S., Karban A., Gerstein R., Kinarty A. et al. Cytokine profile in coeliac disease. Scand. J. Immunol. 49, 441–446 (1999). [DOI] [PubMed] [Google Scholar]
- Nemec P., Pavkova-Goldbergova M., Stouracova M., Vasku A., Soucek M. et al. Polymorphism in the tumor necrosis factor-alpha gene promoter is associated with severity of rheumatoid arthritis in the Czech population. Clin. Rheumatol. 27, 59–65 (2008). [DOI] [PubMed] [Google Scholar]
- Nedwin G. E., Naylor S. L., Sakaguchi A. Y., Smith D., Jarrett-Nedwin J. et al. Human lymphotoxin and tumor necrosis factor genes: structure, homology and chromosomal localization. Nucleic Acids Res. 13, 6361–6373 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsytsykova A. V. & Goldfeld A. E. Inducer-specific enhanceosome formation controls tumor necrosis factor alpha gene expression in T lymphocytes. Mol. Cell Biol. 22, 2620–2631 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel D. A., Udalova I. A., De Silva A. P., Mc Govern D. P., Kinouchi Y. et al. Inflammatory bowel disease is associated with a TNF polymorphism that affects an interaction between the OCT1 and NF-kB transcription factors. Hum. Mol. Genet. 11, 1281–1289 (2002). [DOI] [PubMed] [Google Scholar]
- Richardson A., Sisay-Joof F., Ackerman H., Usen S., Katundu P. et al. Nucleotide diversity of the TNF gene region in an African village. Genes Immun. 2, 343–348 (2001). [DOI] [PubMed] [Google Scholar]
- Wilson A. G., Symons J. A., McDowell T. L., Mc Devitt H. O. & Duff G. W. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc. Natl. Acad. Sci. USA 94, 3195–3199 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W., Maoqing W., Wangyang C., Fulan H., Dandan L. et al. Relationship between the polymorphism of tumor necrosis factor-a -308G > A and susceptibility to inflammatory bowel diseases and colorectal cancer: a meta-analysis. Eur. J. Hum. Genet. 19(4), 432–437 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi E., Basso D., Zambon C. F., Navaglia F., Greco E. et al. TNFA Haplotype genetic testing improves HLA in estimating the risk of celiac disease in children. PLoS One 10(4), e0123244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Albuquerque M. R. M., Martins E. F. A., Crovella S., Segat L. & Eleutério S. P. R. Tumor necrosis factor-α and interleukin-6 gene polymorphism association with susceptibility to celiac disease in Italian patients. Genet. Mol. Res. 14, 16343–18352 (2015). [DOI] [PubMed] [Google Scholar]
- Kekik C., Oguz F. S., Karahn G., Seyhun Y., Aslan E. et al. IL-10 and TNF-alpha gene polymorphisms in patients with Celiac disease. Turkish Journal of Immunology 16, 11–16 (2011). [Google Scholar]
- Capilla A., Donat E., Planelles D., Espinós C., Ribes-Koninckx C. et al. Genetic analyses of celiac disease in a Spanish population confirm association with CELIAC3 but not with CELIAC4. Tissue Antigens 70(4), 324–329 (2007). [DOI] [PubMed] [Google Scholar]
- Hermann C., Krikovszky D., Vásárhelyi B., Dezsofi A. & Madácsy L. Polymorphisms of the TNF-alpha gene and risk of celiac disease in T1DM children. Pediatr. Diabetes 8(3), 138–141 (2007). [DOI] [PubMed] [Google Scholar]
- Barisani D., Ceroni S., Meneveri R., Cesana B. M. & Bardella M. T. IL-10 polymorphisms are associated with early-onset celiac disease and severe mucosal damage in patients of Caucasian origin. Genet. Med. 8(3), 169–174 (2006). [DOI] [PubMed] [Google Scholar]
- Garrote J. A., Arranz E., Gómez-González E., León A. J., Farré C. et al. IL6, IL10 and TGFB1 gene polymorphisms in coeliac disease: differences between DQ2 positive and negative patients. Allergol. Immunopathol. (Madr) 33(5), 245–249 (2005). [DOI] [PubMed] [Google Scholar]
- Lio D., Scola L., Forte G. I., Accomando S., Giacalone A. et al. TNF alpha, IFN gamma and IL-10 gene polymorphisms in a sample of Sicilian patients with coeliac disease. Dig. Liver Dis. 37(10), 756–760 (2005). [DOI] [PubMed] [Google Scholar]
- Cataldo F., Lio D., Marino V., Scola L., Crivello A. et al. Cytokine genotyping (TNF and IL-10) in patients with celiac disease and selective IgA deficiency. Am. J. Gastroenterol. 98(4), 850–856 (2003). [DOI] [PubMed] [Google Scholar]
- Hahn-Zoric M., Hytönen A. M., Hanson L. A., Nilsson L. A. & Padyukov L. Association of -1087 IL10 and -308 TNFA gene polymorphisms with serological markers of coeliac disease. J. Clin. Immunol. 23(4), 291–296 (2003). [DOI] [PubMed] [Google Scholar]
- Garrote J. A., Arranz E., Tellería J. J., Castro J., Calvo C. et al. TNF alpha and LT alpha gene polymorphisms as additional markers of celiac disease susceptibility in a DQ2-positive population. Immunogenetics 54(8), 551–555 (2002). [DOI] [PubMed] [Google Scholar]
- Kumar V., Wijmenga C. & Withoff S. From genome-wide association studies to disease mechanisms: celiac disease as a model for autoimmune diseases. Semin. Immunopathol. 34, 567–580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A. G., Symons J. A., McDowell T. L., McDevitt H. O. & Duff G. W. Effects of a polymorphism in the human tumour necrosis factor alpha promoter on transcriptional activation. Proc. Natl. Acad. Sci. USA 94, 3195–3199 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis J. P., Boffetta P., Little J., O’Brien T. R., Uitterlinden A. G. et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int. J. Epidemiol. 37, 120–132 (2008). [DOI] [PubMed] [Google Scholar]
- Louka A. S., Lie B. A., Talseth B., Ascher H., Ek J. et al. Coeliac disease patients carry conserved HLA-DR3-DQ2 haplotypes revealed by association of TNF alleles. Immunogentics. 55, 339–343 (2003). [DOI] [PubMed] [Google Scholar]
- Kroeger K. M., Steer J. H., Joyce D. A. & Abraham L. J. Effects of stimulus and cell type on the expression of the -308 tumour necrosis factor promoter polymorphism. Cytokine 12, 110–119 (2000). [DOI] [PubMed] [Google Scholar]
- O’Keeffe J., Lynch S., Whelan A., Jackson J., Kennedy N. P. et al. Flow cytometric measurement of intracellular migration inhibition factor and tumour necrosis factor alpha in the mucosa of patients with coeliac disease. Clin. Exp. Immunol. 125, 376–382 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz I., Evans K. E., Papageorgiou V. & Sanders D. S. Are patients with coeliac disease seeking alternative therapies to a gluten-free diet? J. Gastrointestin. Liver. Dis. 20, 27–31 (2011). [PubMed] [Google Scholar]
- Gillett H. R., Arnott I. D., McIntyre M., Campbell S., Dahele A. et al. Successful infliximab treatment for steroid-refractory celiac disease: a case report. Gastroenterology 122, 800–805 (2002). [DOI] [PubMed] [Google Scholar]
- Costantino G., della T. A., Lo P. M. A., Caruso R., Mazzon E. et al. Treatment of lifethreatening type I refractory coeliac disease with long-term infliximab. Dig. Liver. Dis. 40, 74–77 (2008). [DOI] [PubMed] [Google Scholar]
- King A. L. & Ciclitira P. J. Celiac disease: strongly heritable, oligogenic, but genetically complex. Mol. Genet. Metab. 71, 70–75 (2000). [DOI] [PubMed] [Google Scholar]
- Stroup D. F., Berlin J. A., Morton S. C., Olkin I., Williamson G. D. et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 283, 2008–2212 (2000). [DOI] [PubMed] [Google Scholar]
- Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 25, 603–605 (2010). [DOI] [PubMed] [Google Scholar]
- Hu P., Huang M. Y., Hu X. Y., Xie X. J., Xiang M. X. et al. Meta-analysis of C242T polymorphism in CYBA genes: risk of acute coronary syndrome is lower in Asians but not in Caucasians. J. Zhejiang. Univ. Sci. B. 16, 370–379 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. & Li B. A multiplicative-epistatic model for analyzing interspecific differences in outcrossing species. Biometric. 55, 355–365 (1999). [DOI] [PubMed] [Google Scholar]
- Mantel N. & Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Ins. 22, 719–748 (1959). [PubMed] [Google Scholar]
- DerSimonian R. & Laird N. Meta-analysis in clinical trials. Control. Clin. Trials. 7, 177–188 (1986). [DOI] [PubMed] [Google Scholar]
- Song K. & Elston R. C. A powerful method of combining measures of association and Hardy–Weinberg disequilibrium for fine-mapping in case-control studies. Statistics. In. Medicine. 25, 105–126 (2006). [DOI] [PubMed] [Google Scholar]
- Egger M., Davey Smith G., Schneider M. & Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 315, 629–634 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]