Abstract
Background
RNA interference is a very effective approach for studies on gene function and may be an efficient method for controlling pests. Trehalase is a key gene in the chitin biosynthesis pathway in insects. Five trehalase genes have been cloned in Tribolium castaneum, though it is not known whether the detailed functions of these trehalases can be targeted for pest control.
Results
The functions of all five trehalase genes were studied using RNAi, and the most important results showed that the expression of all 12 genes decreased significantly from 12 to 72 h compared with the control groups, except GP1 at 72 h, when the expression of the TcTre2 gene was suppressed. The results also revealed different abnormal phenotypes, and the observed mortality rates ranged from 17 to 42 %. The qRT-PCR results showed that the expression of TPS, GS, two GP, CHS1a and CHS1b genes decreased significantly, while that of the CHS2 gene decreased or increased after RNAi after the five trehalases were silenced at 48 h. In addition, TPS gene expression decreased from 12 to 72 h after dsTcTre injection.
Conclusions
These results demonstrate that silencing of any individual trehalase gene, especially Tre1-4 and Tre2 gene can lead to moulting deformities and a high mortality rate through the regulation of gene expression in the chitin biosynthesis pathway and may be a potential approach for pest control in the future.
Electronic supplementary material
The online version of this article (doi:10.1186/s12896-016-0297-2) contains supplementary material, which is available to authorized users.
Keywords: Tribolium castaneum, Trehalase, RNA interference, Chitin biosynthesis pathway, Gene function
Background
RNA interference (RNAi), which is a robust and powerful experimental tool, has been widely used to study gene function through the suppression of gene expression [1]. Many reports on the use of double-stranded RNA (dsRNA) techniques to silence target genes that are resistant to insecticides have indicated that RNAi may be an efficient method for controlling insect pests, and novel pest management strategies involving the modification of plants are being developed [2–4]. In addition, RNAi technology has been widely used to investigate the functions of essential genes in insects [5]. RNAi has also been employed for the silencing of vital genes in Tribolium castaneum, Nilaparvata lugens and other insects [4, 6]. Furthermore, RNAi could be used in novel methods of pest management and may be more effective than other pest control measures once the problems of the efficiency and specificity of gene silencing are solved [7].
The chitin biosynthesis pathway is crucially important for insect life; the pathway regulates the main component of the cuticle of most insects and the peritrophic membranes of arthropods [8–13]. The first enzyme involved in the chitin biosynthesis pathway is trehalase (Tre, TRE or Treh), which hydrolyses trehalose into glucose as a source of the substrate for hexokinase (HK), while the last enzyme in the pathway is chitin synthase (CHS), which consists of two classes, CHS1 and CHS2, in the majority of insects [14, 15]. Glycogen phosphorylase (GP) in the decomposition pathway together with glycogen synthase (GS) in the synthetic pathway regulates glycogen levels. Glucose-6-phosphate generated through the hydrolysis of trehalose and degradation of glycogen is transformed into fructose-6-phosphate by glucose-6-phosphate isomerase (GPI), which plays different roles in the chitin biosynthesis pathway after the enzymatic action of glutamine: fructose-6-phosphate transaminase (GFAT) and in the glycolysis pathway after the enzymolysis of phosphofructokinase (PFK) (Additional file 1: Figure S1) [16–20].
In T. castaneum, there is one trehalose 6-phosphate synthase (TPS) that catalyses the synthesis of a trehalose from two glucose molecules, in addition to HKs (HK1 and HK2), one glycogen synthase (GS), two glycogen phosphorylases (GP1 and GP2), one glucose-6-phosphate isomerase (GPI), one fructose-6-phosphate transaminase (GFAT) and one phosphofructokinase (PFK). The two classes of chitin synthase (CHS1 and CHS2) are found in T. castaneum, and the CHS1 yields two alternatively spliced transcripts and the corresponding isoforms of the enzyme proteins, namely CHS1a and CHS1b [9]. CHS1 is responsible for moulting during the entire lifecycle of T. castaneum, and CHS2 is required for the formation of chitin in the peritrophic membrane (PM) and RNAi of this gene leads to reduced food uptake, growth arrest and failure to molt to the next instar insect. In addition, CHS1a is responsible for both the larval-pupal and pupal-adult moults, while CHS1b is only responsible for the latter [9].
Trehalase (a-glucoside-1-glucohydrolase, EC 3.2.1.28) is an anomer-inverting α-trehalose-1-D-glucosidase that hydrolyses a trehalose molecule into two glucose molecules. Two forms of trehalase, soluble trehalase (Tre1) and membrane-bound trehalase (Tre2), have been identified to date and cloned in several insect species, including Tenebrio molitor (T. molitor Coleoptera), Pimpla hypochondriaca (Hymenoptera), Bombyx mori (B. mori Lepidoptera), Apis mellifera (Hymenoptera), and Spodoptera exigua (S. exigua Lepidoptera) [21–27]. The first soluble and membrane-bound trehalases were found in T. molitor and B. mori, respectively [21, 23]. Previous research has shown that two types of trehalases regulate different CHS enzymes [28]. It has been reported that trehalase regulates the expression of chitin synthase in the insect cuticle and midgut, and inhibiting chitin biosynthesis kills pests when trehalase gene expression is suppressed or knocked down [28, 29]. However, the functions and relationships of many TcTres and whether and how they regulate different CHS enzymes are unknown, as indicated by Chen and Arakane [9, 28]. Trehalase has become an important target in pest management and control, and new and effective trehalase inhibitor products with practical significance as pesticides are being developed [30].
In the present study, five T. castaneum trehalase (TcTre) genes were identified and cloned, including four soluble trehalase genes (TcTre1-1, TcTre1-2, TcTre1-3 and TcTre1-4) and one membrane-bound trehalase gene (TcTre2); the results indicated that these trehalases may have different functions. The flour beetle, T. castaneum, is a member of the coleopteran order of insects. T. castaneum is a holometabolous insect in the larva, pupa and adult stages and represents a serious pest that damages grain in storage. It is thought to be an effective and reliable insect for conducting RNAi research [7, 31, 32]. It has also been reported that RNAi is highly effective in Tribolium, even in the postembryonic stages of Tribolium [9, 33]. Therefore, it will be useful to study the characteristics and functions of the five TcTre genes using RNAi. The transcription levels of the five TcTre genes were found to be substantially down-regulated after specific dsRNA injection targeting every TcTre. The present study revealed the specific functions of the five TcTres genes in chitin biosynthesis in insects as well as some functions that are common to all of them.
Methods
Insect cultures
The T. castaneum insects were kindly provided by Professor Chao-Ming Wei of Shanxi Normal University. The experimental beetle populations were raised in whole wheat flour with 5 % yeast powder at 30 ± 1 °C [7, 34]. The developmental stages were synchronised by providing fresh coarse wheat bran to replace the old coarse wheat bran every day, and the old coarse wheat bran mixture containing T. castaneum eggs was cultured for future experiments.
RNA extraction, cDNA synthesis and rapid amplification of full-length cDNA (PCR)
Total RNA was extracted from the whole body of T. castaneum larvae or adults using the TRIzol (Invitrogen, Carlsbad, California, USA) method. The RNA concentration was then determined by measuring the absorbance at 260 nm with a spectrophotometer [35]. The purified RNA was stored at -80 °C for future experiments. First-strand cDNA synthesis was performed using the PrimeScript® RT reagent Kit with gDNA Eraser (Takara). The obtained first-strand cDNA (1 μl) was used as the template for PCR.
Cloning of the fragments of five trehalase genes with the protein coding regions
Five pairs of specific primers based on the whole gene sequences published in GenBank were designed for TcTre1-1 cDNA (XM968798), TcTre1-2 cDNA (XM968883), TcTre1-3 cDNA (XM968859), TcTre1-4 cDNA (XM968826) and TcTre2 cDNA (EFA11183) (Table 1). The components of the PCR mixture included PCR buffer containing 0.1 mM dNTPs, 0.2 μM each primer, and 0.5 U of HiFi-Taq DNA polymerase (Transgene, China) in a total volume of 25 μl. The amplification reactions were performed with the specific primers according to the following conditions: 10 min at 94 °C, followed by 30 cycles of 30 s at 94 °C, 30 s at 55 °C and 120 s at 72 °C, and then 10 min at 72 °C [25].
Table 1.
Primers used in present study
| Gene | Application type | Primer set | Forwad (5’-3’) | Reverse (5’-3’) | Length of target fragment (bp) |
|---|---|---|---|---|---|
| TcTre1-1 | cDNA cloning | TcTre1-1 | GAACCACTTGTCCCTTGTC | TGCTAATCAAGTGGGCTT | 1626 |
| qRT-PCR | QTcTre1-1 | AACGACTCGCAATGGCTGG | CGGAGGCGTAGTGGAATAGAG | 127 | |
| dsRNA synthesis | dsTcTre1-1 | CGACCTGAAATTAGCCCAGAAG | GGCTCCCCCACTCTTTC | 357 | |
| TcTre1-2 | cDNA cloning | TcTre1-2 | CGTTTCATGTCGTTGTG | GAACACCACAGAGACGTAA | 1692 |
| qRT-PCR | QTcTre1-2 | GTGCCCAATGGGTTTATCG | CAACCACAACACTTCCTTCG | 261 | |
| dsRNA synthesis | dsTcTre1-2 | GGACAATGAGTTGGGGCAG | AACCGCGTAAAATCGCTCG | 493 | |
| TcTre1-3 | cDNA cloning | TcTre1-3 | ATGCCTTTCCTCCTACTTCTCACCC | GGTACTACGGCCGGGAAAAATAA | 1647 |
| qRT-PCR | QTcTre1-3 | CCTCTCATTCGTCACAAGCG | AAGCGTTTGATTTCTTTGCG | 205 | |
| dsRNA synthesis | dsTcTre1-3 | GGGACCGTGGGTTAAACCAAC | CTGTGGCCGCTGAACTGAA | 376 | |
| TcTre1-4 | cDNA cloning | TcTre1-4 | ATGAAGTCTCTCCTCGTTTCG | GATTTTACGCGGTTTCGG | 1662 |
| qRT-PCR | QTcTre1-4 | ACGGTGCCCGCATCTACTA | GTGTAGGTGGTCCCGTTCTTG | 187 | |
| dsRNA synthesis | dsTcTre1-4 | ACGATATCGAGCTCAGAGTCC | TACGTCTCTGTGGTGTTCGC | 462 | |
| TcTre2 | cDNA cloning | TcTre2 | TTAGTGTGTTGTGCGTTTGT | TGCGAAGAGCACGAGATAA | 1929 |
| qRT-PCR | QTcTre2 | CTCAGCCTGGCCCTTAGTTG | GGAGTCCTCGTAGATGCGTT | 120 | |
| dsRNA synthesis | dsTcTre2 | GGTGCGCTCCAACTACAAAG | GCTCCGCAGTTCCGTGTAG | 399 | |
| TcTPS | qRT-PCR | QTcTPS | CCTTGTTCCACTCAATGCCCGAC | GTCATGTATCCAGATCAAGGGAAC | 105 |
| TcHk1 | QTcHk1 | CGCACCGAATGCCAGAATC | GACCCACCCGACATCGATT | 141 | |
| TcHk2 | QTcHk2 | CGAATCGGCCTAATAGTTGGC | GACGGAGCCCTCGATTTCAT | 155 | |
| TcCHS1a | QTcCHS1a | CGTATAGCCGCCGACTTGAA | CCTATGACGAGAGCACCCAAGA | 173 | |
| TcCHS1b | QTcCHS1b | CCAGGATTGCAAAGGAGTTG | TTGAGGAAACGTCCGAGGTC | 179 | |
| TcCHS2 | QTcCHS2 | GAGTTGTGGCAATGTTCTCGC | GTGGTGTGGCCCTTGGTT | 116 | |
| TcGS | QTcGS | GGAGTTTCATGCACGGACTG | GTTGGCTTGTCGATGGGAA | 105 | |
| TcGP1 | QTcGP1 | GCGAGCGACTATGAACTGATG | TTGCGTGTCGGAAACTACAT | 104 | |
| TcGP2 | QTcGP2 | GGACGAATATTACACAAGTTACGAC | CAGACCCACGATTACGCTT | 119 | |
| TcGPI | QTcGPI | GTGATGCCGGAGGTGAAT | CACGTCGGTGATGGGCTT | 112 | |
| TcGFAT | QTcGFAT | GGAACTGGACATGGACCGTA | GAACGGTGAGGATGCGAGTT | 126 | |
| TcPFK | QTcPFK | GAGCAAGGACATGGAAGGGAA | CCAACCCAACCAGCCACTT | 158 | |
| Tcβ-actin | QTcactin | AGGGCGTCATGGTCGGTAT | TTCTACAACGAGCTCCGCG | 165 | |
| GFP | dsRNA synthesis | dsGFP | AAGGGCGAGGAGCTGTTCACCG | CAGCAGGACCATGTGATCGCGC | 206 |
The products were subjected to agarose gel electrophoresis. The true electrophoretic DNA bands corresponding to the expected size of approximately 1600–2000 bp were excised from the agarose gel and purified using a DNA gel extraction kit (OMEGA, USA). The purified DNA was ligated into the pMD18-T vector (TaKaRa, Japan) and sequenced using the dideoxynucleotide method.
Analysis of the TcTres tissue distribution using semi-quantitative PCR
cDNA synthesis and RT-PCR were performed to analyse the tissue distribution of TcTres using gene-specific primers [19]. The midgut, fat body, cuticle and Malpighian tubules from mixed larvae including the penultimate larval instar and last-instar larvae were dissected in a saline solution (0.75 % NaCl) and stored at -80 °C for future analysis. Total RNA was isolated from these tissues and reverse transcribed into cDNA. Five pairs of specific primers based on the previously cloned TcTre genomic DNAs and one pair of specific of Tcβ-actin primers were designed (Table 1), which were the same as the primers used in qRT-PCR. First, the content of cDNA in each tissue was adjusted to ensure that the brightness of every electrophoresis band was the same, based on the cloning of β-actin, and the corresponding amount of cDNA was then added to clone every TcTre under the same PCR conditions indicated above to produce specific expression in the four tissues.
RNAi targeting the five trehalase genes
The cDNA regions of the five TcTre genes showing the greatest sequence divergence were targeted for dsRNA production. One region was the target for the TcTre1-1, TcTre1-2, TcTre1-3 and TcTre1-4 cDNA regions, as shown in Additional file 1: Figure S2. The lengths of the homologous dsRNAs for TcTre1-1, TcTre1-2, TcTre1-3 and TcTre1-4 used in this study were 357, 493, 376 and 462 bp, respectively. The other region was the target for TcTre2, for which the dsRNA contained a unique sequence near the 3’end, and its length was 399 bp (Additional file 1: Figure S2). The highest and lowest nucleotide sequence identities between the sequences were 53.2 and 39.2 %, respectively. Five pairs of primers with the T7 RNA promoter sequence flanking the 5’-ends were designed according to these regions (Table 1), which were synthesized using the Promega T7 Expression kit. Templates for in vitro transcription were prepared via PCR using gene-specific primers with the T7 polymerase promoter sequence at both ends [7].
The NT-88-V3 series (Nikon) micromanipulator system was used. The dorsal side of the first abdominal segment of T. castaneum was selected as the injection point, and 0.5–0.7 μg of the dsRNAs (3 μg/μl) targeting the five TcTres sequences and GFP was injected into each insect [32].
Sample collection and phenotype observations
After the dsRNAs were injected into last-instar insects, we collected 12 larvae (showing malformation at best) and divided them into three tubes every 12 h for the first 48 h, which were then stored at -80 °C. Most of the larvae developed into the prepupa stage by 48 h after RNAi treatment. The abnormal pupae were collected according to the above method at 72 h after RNAi treatment, when most of the insects had formed pupae. We observed the phenotypes of the insects every 12 h. When the dsRNAs were injected into penultimate-instar insects, samples were collected at 48 and 72 h, and the penultimate-instar insects started to moult to become last-instar insects at 72 h.
qRT-PCR analyses of TcTres and the expression levels of related genes
Total RNA was isolated from T. castaneum on each day of the insect’s life cycle, including the larval, pupal and adult stages, and 1 μg of total RNA was extracted and used as the template to demonstrate the stability of Tcβ-actin. PCR was performed with the QTcactin-F and QTcactin-R primers under the following conditions: 95 °C for 5 min, followed by 28 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s, with a final extension at 72 °C for 10 min. The expression of TcTres was estimated through qRT-PCR using the Bio-Rad CFX96TM system (Bio-Rad, America) and SsoFastTM EvaGreen Supermix (Bio-Rad, America). All primers were designed to determine the expression of the corresponding homologous gene sequences, which included TcTre1-1, TcTre1-2, TcTre1-3, TcTre1-4, TcTre2, CHS1a, CHS1b, CHS2, GS, GP1, GP2, HK1, HK2, GPI, GFAT and PFK (Table 1); these primers were designed based on the unique regions of the sequences, which were identified in the alignment using Vector NTI Suite 7. Each reaction was performed in a final volume of 20 μl containing 1 μl of the cDNA sample (or standard), 1 μl (l0 μmol/μl) of each primer, 7 μl of RNAase-free and DNAase-free water and 10 μl of SsoFastTM EvaGreen Supermix. After 3 min of initial denaturation at 95 °C, the cycling protocol consisted of 40 cycles of denaturation at 95 °C for 5 s, annealing at 55 °C -62.5 °C for 20 s and, finally, the melting curve was performed at 65–95 °C (according to the instructions for SsoFastTM EvaGreen Supermix). Standard curves were obtained using ten-fold serial dilutions of pooled total RNA.
Statistical analyses
The mRNA expression levels in the non-injected group and the dsGFP-injected group were designated as the controls. All of the data obtained in this study are presented as the means ± standard errors (SEs) of 3–6 replicates and were analysed through one-way ANOVA. A P-value of less than 0.05 or 0.01 was considered significant or extremelysignificant in Duncan’s new multiple range test (DMRT) [36]. An asterisk indicates a significant difference in mRNA levels between the dsGFP group and each of the dsTcTres-injected groups measured at the same time (p, 0.05, T test), and a double asterisk indicates a highly significant difference (p, 0.01, T test).
Results
Analysis of the tissue distribution of the five TcTres
The tissue-specific expression of TcTres was determined through semi-quantitative reverse transcription PCR (RT-PCR). All five TcTres transcripts were detected in the midgut, fat body, cuticle and Malpighian tubules (Fig. 1). TcTre1-1 expression was higher in the cuticle, fat body and Malpighian tubule and lower in the midgut. The highest and lowest TcTre1-2 expression levels were observed in the fat body and Malpighian tubules, respectively. TcTre1-3 expression was higher in the midgut, fat body and Malpighian tubules than in the cuticle. The expression of TcTre1-4 was high in the midgut, fat body, cuticle and Malpighian tubules, whereas that of TcTre2 was low; the negative control exhibited no expression.
Fig. 1.
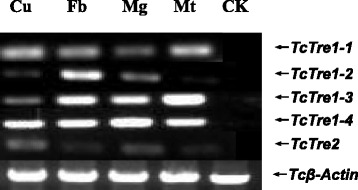
Analysis of the five TcTre transcripts in different tissues of last-instar larvae using semi-quantitative PCR (RT-PCR). Total RNA was extracted from various tissues: fat body (Fb), midgut (Mg), cuticle (Cu) and Malpighian tubules (Mt). The expression of Tcβ-actin in the four tissues was used to standardize the relative expression levels of the five TcTre genes. CK represents PCR products without a template used as the controls
Efficiency of TcTre RNAi in penultimate-instar larvae
RNAi targeting each TcTre was successful in each group, and the expression of the five trehalase genes had decreased significantly at 48 and 72 h after the corresponding dsRNA injections (Fig. 2). In addition, the transcript levels for any of these Tre genes are suppressed not only by the dsRNA for that gene, but also by dsRNA’s for other Tre genes as well (Fig. 2a, b, c and d). All of the soluble TcTre transcripts decreased significantly after dsTcTre2 injection, except for TcTre1-4 at 48 h. TcTre2 transcript levels increased after RNAi targeting all of the soluble dsTcTres (Fig. 2e). Following RNAi treatment, most of the penultimate-instar larvae exhibited sloughing and became last-instar larvae at 72 h.
Fig. 2.
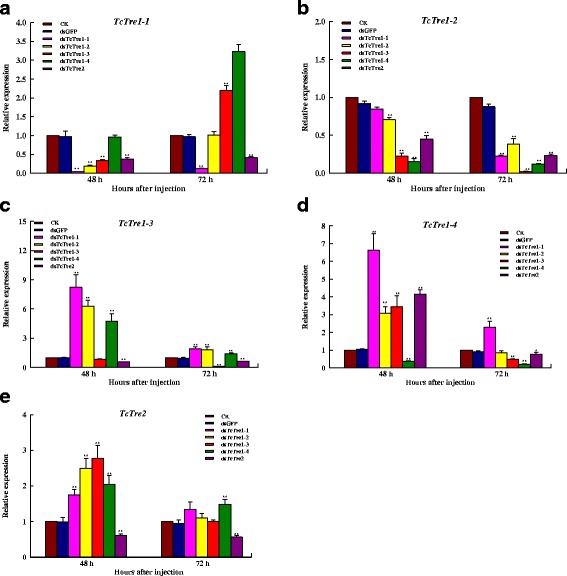
Changes in the mRNA levels of the five TcTres after specific RNAi in penultimate-instar larvae. The penultimate-instar larvae were selected as the targets for dsRNA injection. a, b, c, d and e represent the mRNA levels of TcTre1-1, TcTre1-2, TcTre1-3, TcTre1-4 and TcTre2 relative to Tcβ-actin mRNA level after RNAi targeting all five of the TcTres, analysed via qRT-PCR
Efficiency and specificity of RNAi targeting TcTres in last-instar larvae
To verify the specificity of RNAi targeting the TcTre genes, the alignment of the five dsTcTres was assessed, and no consecutive identical 19-bp sequences between pairs of fragments were found (Additional file 1: Figure S2). All of the TcTres decreased significantly from 12 to 72 h after dsRNA injection compared with the control group, and TcTre2 decreased most significantly after dsTcTre2 RNAi (Fig. 3). The corresponding RNAi efficiencies for TcTre1-1, TcTre1-2 and TcTre1-3 were 68, 56 and 93 % after 72 h, whereas those of TcTre1-4 and TcTre2 were 89 and 96 %, respectively, after just 48 h. The efficiencies obtained for TcTre1-3, TcTre1-4 and TcTre2 were higher than for the other TcTres.
Fig. 3.
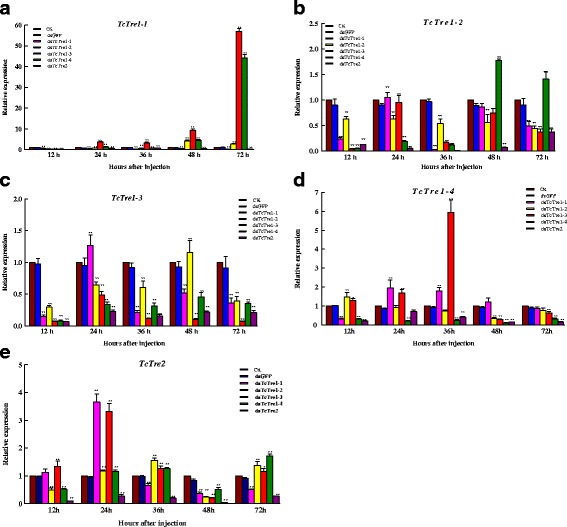
Changes in the mRNA transcripts of the five TcTre genes after specific RNAi in last-instar larvae. a, b, c, d and e represent the mRNA levels of TcTre1-1, TcTre1-2, TcTre1-3, TcTre1-4 and TcTre2 relative to the Tcβ-actin mRNA level after RNAi targeting all five of the TcTres, analysed via qRT-PCR. Last-instar larvae were chosen as the targets for dsRNA injection
The levels of the different TcTres showed both increases and decreases in every TcTre RNAi group (Fig. 3), whereas all of the TcTres showed decreases in the TcTre2 RNAi group. Also the transcript level of TcTre1-2 is suppressed not only by the conrresponding dsRNA, but also by other Tre dsRNA’s genes as well (Fig. 3b), the transcript level of TcTre2 increased after RNAi targeting the other soluble TcTres, except after 36 and 72 h in the TcTre1-1 RNAi group and 48 h in all of the soluble TcTre RNAi groups (Fig. 3e). The expression of TcTre1-1 increased and decreased after RNAi targeting the other soluble TcTres; however, it increased rapidly in the TcTre1-3 RNAi group at 24 h and in the TcTre1-4 RNAi group at 72 h (Fig. 3a). TcTre1-2 only showed an obvious increased at 48 and 72 h after TcTre1-4 RNAi (Fig. 3b). TcTre1-3 increased slightly at 24 h after TcTre1-1 RNAi and at 48 h after TcTre1-2 RNAi, although a significant reduction was detected in the other groups (Fig. 3c). There was variation in the expression of TcTre1-4 in the other soluble RNAi groups, and it peaked at 36 h in the TcTre1-3 RNAi group (Fig. 3d).
Analyses of phenotypes, aberrations and survival rates after RNAi
With the successful silencing of every TcTre, the insects subjected to RNAi exhibited a large number of deaths and presented various abnormal phenotypes (Fig. 4). The mortality rates recorded after RNAi targeting the five TcTres were as follows: TcTre1-1 (35 %), TcTre1-2 (17 %), TcTre1-3 (23 %), TcTre1-4 (42 %) and TcTre2 (40 %) (Table 2). These values corresponded to 35, 12, 18, 35 and 28 insects and were higher than the death rates of the naive insects (0 %) and the insects injected with dsGFP (7 %). The death rates differed in different insect stages. The following percentages were obtained for the number of deaths observed among every type of abnormal insect divided by the total number of abnormal insects in the same stage (larva, pupa or adult stages). From larva to pupa, the percentages were TcTre1-1 (20 %), TcTre1-2 (14 %), TcTre1-3 (13 %), TcTre1-4 (24 %) and TcTre2 (8 %), which corresponded to 20, 11, 10, 24 and 6 larvae that died before the pupal stage. During the pupa-adult stage, the percentages were TcTre1-1 (8 %), TcTre1-2 (0), TcTre1-3 (4 %), TcTre1-4 (13 %) and TcTre2 (24 %), which corresponded to 6, 0, 3, 7 and 16 pupae that died before becoming adults, and most of these numbers were higher than in the two control groups. There were some deaths recorded during the adult stage, when the following percentages were determined: TcTre1-1 (12 %), TcTre1-2 (2 %), TcTre1-3 (8 %), TcTre1-4 (6 %) and TcTre2 (12 %), which corresponded to just 9, 2, 5, 3 and 6 more adults than in the two control groups. Additional details are presented in Table 2, including the aberration rates, survival rates, pupation rates and eclosion rates (Table 2).
Fig. 4.
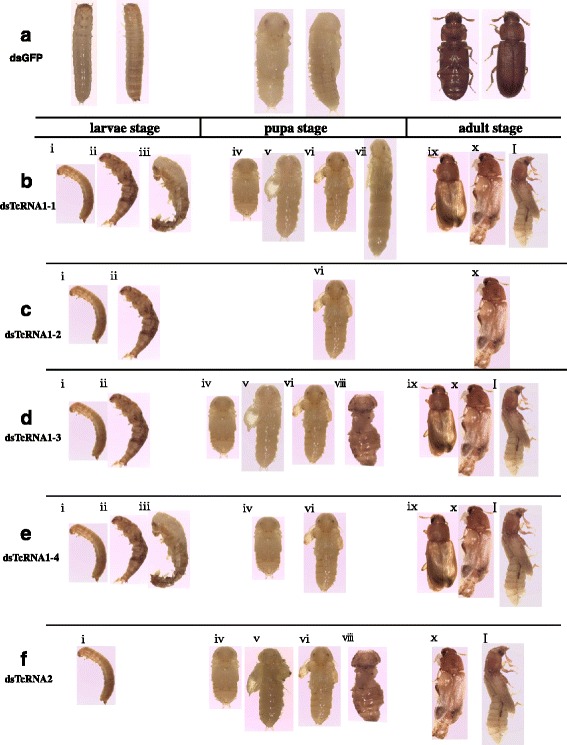
Abnormal phenotypes were caused by RNAi targeting the five dsTcTres. All abnormal insects in the three stages (larva-pupa stage, pupal stage, and adult stage) are displayed and are divided into eleven types, from i to xi, after RNAi. a dsGFP-injected group. b dsTcTre1-1-injected group. c dsTcTre1-2-injected group. d dsTcTre1-3-injected group. e dsTcTre1-4-injected group. f dsTcTre2-injected group. The percentage below each type of abnormal insect is the corresponding proportion among all of the abnormal insects in that stage, such as the larva-pupa stage
Table 2.
Tribolium castaneum phenotype rate after RNAi
| dsRNA | aberration rate (%) | survival rate (%) | pupation rate (%) | eclosion rate (%) |
|---|---|---|---|---|
| dsTcTre1-1 | 38 | 65 | 92 | 93 |
| dsTcTre1-2 | 17 | 83 | 100 | 100 |
| dsTcTre1-3 | 24 | 77 | 100 | 96 |
| dsTcTre1-4 | 45 | 58 | 95 | 88 |
| dsTcTre2 | 40 | 61 | 100 | 76 |
| dsGFP | 0 | 94 | 92 | 90 |
| no-injection | 0 | 100 | 100 | 100 |
Insects in the larval, pupal and adult stages were present after RNAi targeting dsGFP as well in every TcTre RNAi group (Fig. 5). Three abnormal phenotypes were observed in the larval stage, and the insects died with the abnormal phenotype: the body size of the larvae decreased (i), or the size of the larvae abdomen decreased (ii) (both of which were associated with a reduced food intake that mainly occurred from 24 to 36 h after TcTre RNAi), and some larvae could not exuviate the larval cuticle to become pupae (iii). During the pupal stage, five malformations were present, which included shrunken pupae that were much smaller than normal pupae (iv); pupae with one set or both sets of wings together with membranous bulging wings (v); pupae with one set or both sets of wings bulging, leaving normal membranous wings (vi); colossal pupae that were larger than normal pupae (vii); and disabled pupae that lacked some body parts (viii) (Fig. 4). All vii and viii pupae died with the abnormal phenotype. Three types of abnormal adults were observed: adults that survived with both sets of elytra slightly opened (ix); adults that partially shed their pupal cuticle with both sets of elytra open and membranous wings that were dislocated, leading to death in all of these individuals (x); and adults that showed combined characteristics of both pupae and adults, which were unable to extricate their appendages and died without completing adult eclosion (xi). All of the ix adults developed from “tiny abnormal” larvae that appeared abnormal in the larval stage and x adults that developed from most of the vi and v larvae; a small number of the iv pupae, vi pupae and v pupae became xi adults, and most of the iv larvae died with a burnt coloration on their body.
Fig. 5.
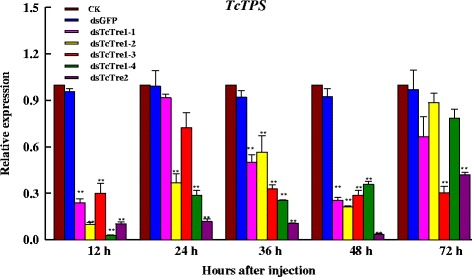
Effects of RNAi targeting the five TcTres on trehalose-6-phosphate synthase (TPS) in last-instar larvae. The levels of TPS mRNA relative to the Tcβ-actin mRNA level were measured via qRT-PCR
Effects of TcTre RNAi on the expression of the TPS, GS and GP genes
TPS, GS and GP are the key genes involved in energy metabolism during insect growth and development. qRT-PCR was used to detect mRNA levels after RNAi targeting the TPS, GS and two GP genes. The expression of TPS decreased from 12 to 72 h after TcTre RNAi, and its expression was decreased significantly at 12, 36 and 48 h (Fig. 5). In addition, TPS mRNA expression decreased significantly during the 24 h after TcTre1-2, TcTre1-4 and TcTre2 were knocked down, and its expression decreased significantly during the 72 h after TcTre1-3 and TcTre2 were silenced.
The results showed that the expression of the TPS, GS and the two GP genes was decreased significantly at 48 h after dsTcTre injection (Figs. 5 and 6a). The transcript levels of TPS, GS, GP1 and GP2 showed consecutive variation after RNAi targeting the five dsTcTres at 72 h. In addition, GP1 and GP2 exhibited high transcript levels at 72 h after dsTcTre1-2, dsTcTre1-3 and dsTcTre1-4 RNAi. The highest transcript levels of GP1 and GP2 were 79.6 and 6.6 times those in the respective control group injected with dsGFP (Fig. 6a). Moreover, the ascensional range of GP1 gene expression was much higher prior to GP2 gene expression.
Fig. 6.
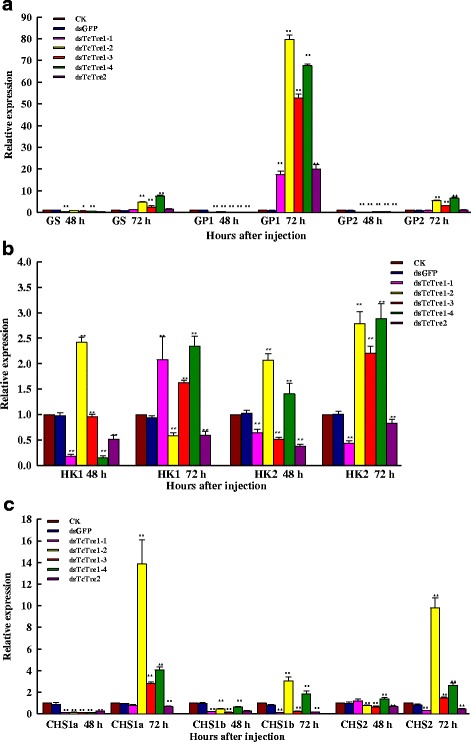
Effects on key genes in the chitin biosynthesis pathway after specific RNAi in last-instar larvae at 48 and 72 h. The levels of these genes relative to the Tcβ-actin mRNA level were measured via qRT-PCR. a mRNA levels of glycogen synthase (GS) and glycogen phosphorylase (GP). b mRNA levels of two hexokinases (HK1 and HK2). c mRNA levels of three chitin synthases (CHS1a, CHS1b and CHS2)
Knockdown of TcTres and the expression of genes in the chitin biosynthesis pathway
qRT-PCR was used to detect mRNA levels after RNAi targeting all of the TcTres, including eight genes in the chitin biosynthesis pathway (HK1, HK2, GPI, PFK, GFAT, CHS1a, CHS1b, CHS2). The expression of HK1 and HK2 decreased significantly in the 48 h after the injection of the TcTre1-1, TcTre1-3 and TcTre2 dsRNAs, whereas their expression increased after dsTcTre1-2 injection (Fig. 6b). The expression of HK1, HK2, CHS1a, CHS1b and CHS2 was decreased substantially after dsTcTre2 RNAi at both 48 and 72 h (Fig. 6b, c). The expression of CHS1a and CHS1b decreased slightly after RNAi targeting the four soluble dsTcTres at 48 h (Fig. 6c), and the expression of CHS2 either increased or decreased in the various soluble TcTre RNAi groups. The transcript levels of HK1, HK2, CHS1a, CHS1b and CHS2 exhibited consecutive variations after RNAi targeting the five dsTcTres at 72 h. The expression of HK2, CHS1a and CHS1b increased significantly at 72 h after dsTcTre1-2, dsTcTre1-3 and dsTcTre1-4 RNAi (Fig. 6b, c).
The results also showed that GPI, GFAT and PFK gene expression decreased significantly in the 24 and 36 h after TcTre2 gene expression was inhibited through RNAi (Fig. 7). The expression of GPI, GFAT and PFK was increased at 24 h after TcTre1-1, TcTre1-2 and TcTre1-3 RNAi but was decreased at 24 h after TcTre1-4, TcTre2 RNAi, with the exception of a decrease in PFK transcript levels after TcTre1-3 RNAi observed at 24 h (Fig. 7). In addition, three gene transcripts were decreased significantly at 36 h after RNAi targeting the five TcTres, except that there was an increase after TcTre1-2 RNAi.
Fig. 7.
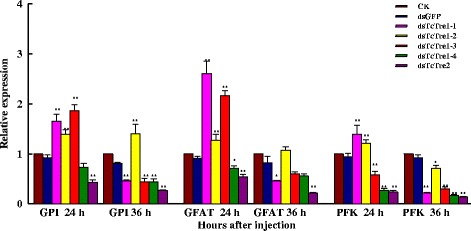
Effects on other genes in the chitin biosynthesis pathway after specific RNAi in last-instar larvae at 24 and 36 h. The mRNA levels of glucose phosphate isomerase (GPI), fructose phosphate transaminase (GFAT) and phosphofructokinase (PFK) relative to the Tcβ-actin mRNA level were measured via qRT-PCR
Discussion
RNAi has been extensively studied in insects in recent years and applied in pest control management [7, 37, 38]. In most cases, RNAi results in a low rate of deformed phenotypes in insects and shows varying efficiency among insect tissues [5, 39, 40]. A previous study revealed that dsRNA is more efficient in T. castaneum, maintaining the silencing effect for a longer period and inducing a higher rate of deformations [7]. In this study, the dsRNAs for the five TcTres were demonstrated to be highly selective and clearly discriminant, and they were verified to be accurate after cloning; the levels of the targeted mRNAs were greatly reduced, along with compensatory expression of the other TcTres transcripts after RNAi targeting any of the TcTres (Figs. 2 and 3). For example, TcTre1-1 transcripts are reduced not only by dsRNA for this gene but also by dsRNAs for Tre1-2, Tre1-3 and by Tre-2 genes at 48 h and by dsRNA for Tre-2 gene at 72 h (Fig. 2a). It is clear that there is compensating over-expression of Tre1-3 and Tre1-4 genes at 72 h (Fig. 2a). Similarly the Tre1-2 transcript suppression is not restricted to dsRNA for this gene but also by all other dsRNAs including those of the Tre-2 gene (Fig. 2b). The results indicate that one of Tre1-1, Tre1-2 and Tre1-3 was inhibited, and the other genes were also lower expression or compensating over-expression, while other Tre transcripts compensating over-expression after Tre1-4 or Tre2 knockdown and the aberration and mortality rates was more higher than other three Tre genes (Table 2). These results are consistent with other observations showing that RNAi is effective in Tribolium larvae, pupae and adults, and even in embryos [41, 42].
There are many reports describing the characteristics of the two forms of trehalases. The tissue distribution of the two forms of trehalases differs. Tre1 is mainly expressed in the midgut and Malpighian tubules in B. mori and is not expressed in the fat body, silk gland, ovary, trachea, or brain [23, 43–45]. Tre2 is expressed in ovary cells, the spermatophore and the midgut in locusts and in the visceral muscle surrounding the midgut in B. mori; Tre2 exhibits both inactive and latent forms that can be activated by being released after destruction of membrane integrity [23, 46, 47]. Zhang also reported that Tre1 is expressed in the midgut, Malpighian tubules and ovary cells, whereas Tre2 is expressed in the fat body, midgut and Malpighian tubules [48]. However, both Tre1 and Tre2 are expressed in the cuticle, midgut, Malpighian tubules, tracheae and fat body of S. exigua [28], which is very similar to T. castaneum, and all five Tres genes are expressed in the cuticle, fat body, midgut and Malpighian tubules (Fig. 1).
Trehalase participates in both homeostasis and development and is involved in blood sugar metabolism in insects as well as flight metabolism, chitin synthesis during moulting and cold tolerance [26, 27, 49–53]. In addition, Tre2 is expressed as a transmembrane enzyme with an active site on the outside of the cell membrane and mainly hydrolyses extracellular trehalose, whereas Tre1 is located within the cell and hydrolyses intracellular trehalose [44, 54]. Some studies based on injection of the hormone 20-hydroxyecdysone (20E) into B. mori have observed an increase in Tre1 transcript levels and no effect on the expression of Tre2, which indicates that insect moults are more closely associated with Tre1 [44]. Many studies have also shown that soluble Tre levels increase before insect moults and after injecting 20-hydroxyecdysone (20E) [19, 28, 44]. In the present study, GP1 and GP2 increased rapidly at 72 h (Fig. 5b), as did HK2 (Fig. 6a), most likely to compensate for the reduced level of the TcTre1-2 transcript, ensuring that the downstream CHS gene transcripts increased significantly (Fig. 6c). This result is consistent with the idea that conversion of glycogen into trehalose occurs in last-instar larvae [55]. At the same time, the CHS1b transcript decreased to a lower level, although its upstream gene transcripts GP1, GP2, HK1 and HK2 were increased substantially at 72 h after dsTcTre1-3 RNAi in the last-instar larvae (Figs. 5b and 6a, c). These results implied that the TcTre1-3 gene plays a role in energy metabolism by hydrolysing trehalose into glucose and regulating the expression of the CHS1b gene, which was demonstrated indirectly based on the 33 % yield of xi adults; the xi adults exhibited a phenotype similar to that resulting from loss of the CHS1b gene in Tribolium after CHS1b RNAi [22]. We found that after dsTcTre1-4 injection, the GFAT transcript level was decreased to 0.71-fold of that in the control group injected with dsGFP at 24 h and to 0.55-fold at 36 h, whereas the PFK transcript level was decreased more strongly, to 0.27-fold at 24 h and 0.17-fold at 36 h (Fig. 6b). However, 90 % (nine insects) of the iv pupae (a greater percentage than for the other TcTre RNAi groups) showed a new phenotype that did not appear in Arakane’s study due to knockdown of the CHS genes, which implied that this abnormal phenotype may be due to a lack of energy [9].
In addition, the 40 % abnormal insects and 39 % mortality recorded appeared to demonstrate that TcTre2 is important, and the uniform distribution of many abnormal phenotypes indicated that TcTre2 can adjust the expression of other TcTres (Fig. 4 and Table 2). All of the TcTres in the last-instar larvae were decreased sharply from 12 to 72 h after dsTcTre2 injection compared with the two control groups (Fig. 3); in contrast, TcTre2 expression increased after RNAi targeting the four soluble dsTcTres, except for TcTre1-2 and TcTre1-4 at 12 h, TcTre1-1 at 36 and 72 h and all of the soluble TcTres at 48 h (Fig. 3f). The same phenomenon was observed in the penultimate-instar larvae from 48 to 72 h after dsTcTre2 injection (Fig. 2), and a possible explanation for these findings is that the shortage of the TcTre2 transcript resulted in a substantial loss of intracellular glucose following the decline of intracellular trehalose. This explanation is consistent with the finding that the expression of TPS decreased from 12 to 72 h after dsTcTre2 RNAi (Fig. 5a), and all of the TcTres therefore decreased as a corresponding response. These results led to the hypothesis that TcTre2 is a trehalase that mainly hydrolyses extracellular trehalose into glucose flowing into the cell, which is not inconsistent with the idea that Tre2 is a transmembrane enzyme with an active site on the outside of the cell membrane in B. mori [44]. Tre2 protein expression was shown to increase substantially, and there was no Tre1 protein present due to the hydrolysis of trehalose into glucose for glycogen accumulation in B. mori injected with diapause hormone, which revealed that Tre2 plays a role in energy metabolism, similar to the finding that Tre2 targets the hydrolysis of trehalose into glucose that will flow into the oocytes of B. mori and Rhodnius prolixus [56–58]. In addition, Becker found that the relationship between GP and Tres in non-feeding insects shows a positive correlation, involving increased expression and activity of GP along with a decrease in Tres. Our qRT-PCR results revealed that some gene transcripts in both the chitin synthesis pathway and the energy synthesis pathway decreased, including HK, CHS, GPI, GFAT, and PFK, although GP1 increased substantially at 72 h and did not sufficiently compensate for the decline in glucose resulting from TcTre2 RNAi (Figs. 5b and 6). This finding is consistent with Becker’s observation that the insects lacked energy after dsTcTre2 injection [55, 59, 60].
Chitin biosynthesis is well established as a key target for pest control, and trehalase inhibitors are being developed for future pest management. It was reported that inhibition of the expression of the Laodelphax stritellus soluble and membrane-bound trehalase genes via the dsRNA feeding method resulted in mortality rates of 38.89 and 27.72 %, respectively [29]. In the pupal stage, the appearance of 60 % v and vi pupae among all of the abnormal pupae may also have resulted from the severe decline in the CHS1a transcript and CHS1b transcript, which caused a higher death rate (12 %) in the adult stage compared with the other TcTre RNAi groups (Fig. 4b). The abnormal pupal and adult phenotypes were consistent with Arakane’s findings that abnormal iii larvae and adults emerged after RNAi targeting CHS1a, and vi adults emerged after RNAi targeting CHS1b or other genes [9, 61, 62].
As it known that the same family genes which have relative high protein sequence identity, so the compensatory function or cross silencing have been found in gene function study by RNAi. In the study of Nilaparvata lugens TPS genes function, NLTPS2 gene’s expression followed decreased when NLTPS1 gene’s expression inhibited by RNAi [63]. In our study, the expression of TcTre1-1 also decreased when dsTcTre1-1, dsTcTre1-2, dsTcTre1-4 and dsTcTre2 were injected from 12 to 36 h, but it increased when dsTcTre1-3 injected from 24 to 72 h (Fig. 3a). As well as TcTre1-2 (Fig. 3b), TcTre1-4 (Fig. 3d) and TcTre2 (Fig. 3e) have compensatory function at some extent when other TcTre was knockdown. The seeming cross silences similar with TPS genes function study also been found [63], while it is also reported that N. lugens soluble trehalases (NlTre1-1 and NlTre1-2) and membrane-bound trehalase (NlTre2) also have the compensatory function [64]. The good inhibited effects and stronger phenotype were found when two Tre1 and one Tre2 gene in N. lugens was knockdown simultaneously [64]. However, double knockdown vti1a and vtilb did not produce a strong phenotype and it suggesting a compensatory mechanism exists within the secretory system though vti1a and vti1b by RNAi resulted in a significant decrease in the study of Vti family of SNARE proteins function [65]. Therefore, the crossing silencing and compensatory mechanism of the same gene family need more studied in the future.
Conclusions
These results demonstrate that the five trehalase genes exhibit different functions in chitin biosynthesis and energy metabolism by the way of RNAi. In particular, a mortality rate close to 40 % resulted from the silencing of TcTre1-4 or TcTre2 gene expression through RNAi, and it could be effective for pest control by a potential trehalase inhibitor [66]. Moreover, the development of anti-trehalase agents that target the chitin biosynthesis pathway or energy metabolism is an alternative method for exploring potential chemical insecticides.
Acknowledgements
We thank Dr. Chao-Ming Wei (College of Life Sciences, Shaanxi Normal University, Xi’an, China) for his kindly help.
Funding
This work was supported by National Natural Science Foundation of China (Grant Nos. 31371996 and 31672081), Visiting Scholar Foundation of State Key Laboratory of Biocontrol at Sun Yat-sen University (Grant No. SKLBC12F01), Hangzhou Science and Technology Development Program of China (Grant No. 20140432B01) and the Program for Excellent Young Teachers in Hangzhou Normal University (Grant No. JTAS 2011-01-031).
Availability of data and material
All the data supporting our findings is included within the manuscript and its Additional files.
Authors’ contributions
BT and SW designed the study, PW and LZ developed the RNAi methodology, PW, LZ and ZS performed experiments. QS, MY and GX performed the statistical analysis, PW, BT and SW wrote the manuscript. All authors have read and approved the final manuscript.
Competing interests
The authors declared that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- cDNA
Complementary DNA
- CHS
Chitin synthase
- CK
Control group
- Cu
Cuticle
- DMRT
Duncan’s new multiple range test
- dsGFP
Double-stranded GFP RNA
- dsRNA
Double-stranded RNA
- dsTcTre
Double-stranded trehalase RNA of Tribolium castaneum
- Fb
Fat body
- GFAT
Glutamine, fructose-6-phosphate transaminase
- GFP
Green fluorescent protein
- GP
Glycogen phosphorylase
- GPI
Glucose-6-phosphate isomerise
- GS
Glycogen synthase
- h
Hour
- HK
Hexokinase
- Mg
Midgut
- Mt
Malpighian tubules
- PCR
Polymerase chain reaction
- PFK
Phosphofructokinase
- PM
Peritrophic membrane
- qRT-PCR
Quantitative real-time polymerase chain reaction
- RNAi
RNA interference
- RT-PCR
Reverse transcription polymerase chain reaction
- SEs
Standard errors
- Tc
Tribolium castaneum
- TPS
Trehalose-6-phosphate synthase
- Tre
Trehalase
- μg
Microgramme
- μl
Microliter
Additional files
Synthesis and degradation of trehalose and glycogen in insects. Adapted from Rockstein [16], Elbein et al. [17], Montooth et al. [18], Kunieda et al. [19], and Tang et al. [20]. The red arrow and dotted arrow indicate that the enzyme in this pathway was analysed and that this pathway is unknown, respectively. Figure S2: Alignment of the nucleotide sequences of five TcTre genes in the region of the dsRNAs. (DOCX 21 kb)
Contributor Information
Bin Tang, Email: tbzm611@yahoo.com.
Ping Wei, Email: njnnwp@sina.cn.
Lina Zhao, Email: heping895784491@yeah.net.
Zuokun Shi, Email: szk1025@163.com.
Qida Shen, Email: shenqida@yeah.net.
Mengmeng Yang, Email: ymmwyz261752@163.com.
Guoqiang Xie, Email: gqxie520321@aliyun.com.
Shigui Wang, Email: sgwang@mail.hz.zj.cn.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Mao Y, Cai W, Wang J, Hong G, Tao X, Wang L, et al. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol. 2007;25:1307–13. doi: 10.1038/nbt1352. [DOI] [PubMed] [Google Scholar]
- 3.Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol. 2007;25:1322–26. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Li L, Hu Q, Zhang B, Wu W, Jin F, Jiang J. Expression of dsRNA in recombinant Isaria fumosorosea strain targets the TLR7 gene in Bemisia tabaci. BMC Biotechnol. 2015;15:64. doi: 10.1186/s12896-015-0170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belles X. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu Rev Entomol. 2010;55:111–28. doi: 10.1146/annurev-ento-112408-085301. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Fan HW, Huang HJ, Xue J, Wu WJ, Bao YY, et al. Chitin synthase 1 gene and its two alternative splicing variants from two sap-sucking insects, Nilaparvata lugens and Laodelphax striatellus (Hemiptera: Delphacidae) Insect Biochem Mol Biol. 2012;42:637–46. doi: 10.1016/j.ibmb.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Wang JD, Wu M, Wang BJ, Han ZJ. Comparison of the RNA interference effects triggered by dsRNA and siRNA in Tribolium castameum. Pest Manag Sci. 2013;69:781–786. doi: 10.1002/ps.3432. [DOI] [PubMed] [Google Scholar]
- 8.Arakane Y, Hogenkamp DG, Zhu YC, Kramer KJ, Specht CA, Beeman RW, et al. Characterization of two chitin synthase genes of the red flour beetle, Tribolium castaneum, and alternate exon usage in one of the genes during development. Insect Biochem Mol Biol. 2004;34:291–304. doi: 10.1016/j.ibmb.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Arakane Y, Muthukrishnan S, Kramer KJ, Specht CA, Tomoyasu Y, Lorenzen MD, et al. The Tribolium chitin synthase genes TcCHS1 and TcCHS2 are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix. Insect Mol Biol. 2005;14:453e463. doi: 10.1111/j.1365-2583.2005.00576.x. [DOI] [PubMed] [Google Scholar]
- 10.Arakane Y, Specht CA, Kramer KJ, Muthukrishnan S, Beeman RW. Chitin synthases are required for survival, fecundity and egg hatch in the red flour beetle, Tribolium castaneum. Insect Biochem Mol Biol. 2008;38:959e962. doi: 10.1016/j.ibmb.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Devine WP, Lubarsky B, Shaw K, Luschnig S, Messina L, Krasnow MA. Requirement for chitin biosynthesis in epithelial tube morphogenesis. Proc Natl Acad Sci U S A. 2005;102:17014e17019. doi: 10.1073/pnas.0506676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen XF, Tian HG, Zou LZ, Tang B, Hu J, Zhang WQ. Disruption of Spodoptera exigua larval development by silencing chitin synthase gene A with RNA interference. Bull Entomol Res. 2008;98:613e619. doi: 10.1017/S0007485308005932. [DOI] [PubMed] [Google Scholar]
- 13.Qu M, Yang Q. A novel alternative splicing site of class A chitin synthase from the insect Ostrinia furnacalise Gene organization, expression pattern and physiological significance. Insect Biochem Mol Biol. 2011;41:923e931. doi: 10.1016/j.ibmb.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Kramer KJ, Dziadik-Turner C, Koga D. Chitin metabolism in insects. Comp Insect Physiol Biochem Pharmacol. 1985;3:75–115. [Google Scholar]
- 15.Kramer KJ, Muthukrishnan S. Chitin metabolism in insects. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive molecular insect science. Oxford: Elsevier; 2005. pp. 111–144. [Google Scholar]
- 16.Rockstein M. Biochemistry of insect. San Francisco: Academic Press New York; 1978. [Google Scholar]
- 17.Elbein AD, Pan YT, Pastuszak I, Carroll D. New insights on trehalose: a multifunctional molecule. Glycobiology. 2003;13:17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- 18.Montooth KL, Marden JH, Clark AG. Mapping determinants of variation in energy metabolism, respiration and flight in Drosophila. Genetics. 2003;165:623–35. doi: 10.1093/genetics/165.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunieda T, Fujiyuki T, Kucharski R, Foret S, Ament SA, Toth AL, et al. Carbohydrate metabolism genes and pathways in insects: insights from the honey bee genome. Insect Mol Biol. 2006;15:563–76. doi: 10.1111/j.1365-2583.2006.00677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang B, Xu Q, Zou Q, Fang Q, Wang SG, Ye GY. Sequencing and characterization of glycogen synthase and glycogen phosphorylase genes from Spodoptera exigua and analysis of their function in starvation and excessive sugar intake. Arch Insect Biochem Physiol. 2012;80:42–62. doi: 10.1002/arch.21027. [DOI] [PubMed] [Google Scholar]
- 21.Takiguchi M, Niimi T, Su ZH, Yaginuma T. Trehalase from male accessory gland of an insect, Tenebrio molitor. cDNA sequencing and developmental profile of the gene expression. Biochem J. 1992;288:19–22. doi: 10.1042/bj2880019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkinson NM, Conyers CM, Keen JN, MacNicoll AD, Smith I, Weaver RJ. cDNAs encoding large venom proteins from the parasitoid wasp Pimpla hypochondriaca identified by random sequence analysis. Comp Biochem Physiol. 2003;134C:513–20. doi: 10.1016/s1532-0456(03)00041-3. [DOI] [PubMed] [Google Scholar]
- 23.Mitsumasu K, Azuma M, Niimi T, Yamashita O, Yamashita T. Membrane-penetrating trehalase from silkworm Bombyx mori. Molecular cloning and localization in larval midgut. Insect Mol Biol. 2005;14:501–508. doi: 10.1111/j.1365-2583.2005.00581.x. [DOI] [PubMed] [Google Scholar]
- 24.Lee JH, Saito S, Mori H, Nishimoto M, Okuyama M, Kim D, et al. Molecular cloning of cDNA for trehalase from the European honeybee, Apis mellifera L and its heterologous expression in Pichia pastoris. Biosci Biotechnol Biochem. 2007;71:2256–65. doi: 10.1271/bbb.70239. [DOI] [PubMed] [Google Scholar]
- 25.Tang B, Chen XF, Liu Y, Tian HG, Liu J, Hu J, et al. Characterization and expression patterns of a membrane-bound trehalase from Spodoptera exigua. BMC Mole Biol. 2008;9:51. doi: 10.1186/1471-2199-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tatun N, Singtripop T, Tungjitwitayakul J, Sakurai S. Regulation of soluble and membrane-bound trehalase activity and expression of the enzyme in the larval midgut of the bamboo borer Omphisa fuscidentalis. Insect Biochem Mol Biol. 2008;38:788–95. doi: 10.1016/j.ibmb.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Tatun N, Singtripop T, Sakurai S. Dual control of midgut trehalase activity by 20-hydroxyecdysone and an inhibitory factor in the bamboo borer Omhisa fuscidentalis Hampson. J Insect Physiol. 2008;54:351–57. doi: 10.1016/j.jinsphys.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Chen J, Tang B, Chen HX, Yao Q, Huang XF, Chen J, et al. Different functions of the insect soluble and membrane-bound trehalase genes in chitin biosynthesis revealed by RNA Interference. PLoS One. 2010;5:e10133. doi: 10.1371/journal.pone.0010133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q, Lu DH, Pu J, Wu M, Han ZJ. Cloning and RNA interference effects of trehalase genes in Laodelphax striatellus (Homoptera: Delphacidae) Acta Entomol Sin. 2012;55(8):911–20. [Google Scholar]
- 30.Tang B, Wei P, Chen J, Wang SG, Zhang WQ. Progress in gene features and functions of insect trehalases. Acta Entomol Sin. 2012;55(11):1315–21. [Google Scholar]
- 31.Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–55. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 32.Noh MY, Beeman RW, Arakane Y. RNAi-based functional genomics in Tribolium castaneum and possible application for controlling insect pests. Entomol Res. 2012;42:1–10. doi: 10.1111/j.1748-5967.2011.00437.x. [DOI] [Google Scholar]
- 33.Yoshinori T, Denell RE. Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev Genes Evol. 2004;214:575–78. doi: 10.1007/s00427-004-0434-0. [DOI] [PubMed] [Google Scholar]
- 34.Haliscak JP, Beeman RW. Status of malathion resistance in five genera of beetles infesting farm-stored corn, wheat, and oats in the United States. J Econ Entomol. 1983;76:717–22. doi: 10.1093/jee/76.4.717. [DOI] [Google Scholar]
- 35.Jaubert-Possamai S, Le Trionnaire G, Bonhomme J, Christophides GK, Rispe C, Tagu D. Gene knockdown by RNAi in the pea aphid Acyrthosiphon pisum. BMC Biotechnol. 2007;7:63. doi: 10.1186/1472-6750-7-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang T, Zhao CQ, Feng XY, Liu XY, Qiu LH. Knockdown of several components of cytochrome P450 enzyme systems by RNA interference enhances the susceptibility of Helicoverpa armigera to fenvalerate. Pest Manag Sci. 2012;68:1501–11. doi: 10.1002/ps.3336. [DOI] [PubMed] [Google Scholar]
- 37.Liu S, Ding Z, Zhang C, Yang B, Liu Z. Gene knockdown by intro-thoracic injection of double-stranded RNA in the brown planthopper, Nilaparvata lugens. Insect Biochem Mol Biol. 2010;40:666–71. doi: 10.1016/j.ibmb.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 38.Bellés X. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Ann Rev Entomol. 2010;55:111–28. doi: 10.1146/annurev-ento-112408-085301. [DOI] [PubMed] [Google Scholar]
- 39.Scott JG, Michel K, Bartholomay LC, Siegfried BD, Hunter WB, Smagghe G, et al. Towards the elements of successful insect RNAi. J Insect Physiol. 2013;59(12):1212–21. doi: 10.1016/j.jinsphys.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong Y, Friedrich M. Nymphal RNAi: systemic RNAi mediated gene knockdown in juvenile grasshopper. BMC Biotechnol. 2005;5:25. doi: 10.1186/1472-6750-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown SJ, Mahaffey JP, Lorenzen MD, Denell RE, Mahaffey JW. Using RNAi to investigate orthologous homeotic gene function during development of distantly related insects. Evol Dev. 1999;1:11–15. doi: 10.1046/j.1525-142x.1999.99013.x. [DOI] [PubMed] [Google Scholar]
- 42.Bucher G, Scholten J, Klingler M. Parental RNAi in Tribolium (Coleoptera) Curr Biol. 2002;5:R85–R86. doi: 10.1016/S0960-9822(02)00666-8. [DOI] [PubMed] [Google Scholar]
- 43.Yamashita O, Sumida M, Hasegawa K. Developmental changes in midgut trehalase activity and its localization in the silkworm, Bombyx mori. J Insect Physiol. 1974;20:1079–85. doi: 10.1016/0022-1910(74)90149-8. [DOI] [PubMed] [Google Scholar]
- 44.Mitsumasu K, Azuma M, Niim T, Yamashita O, Yaginuma T. Changes in the expression of soluble and integral-membrane trehalases in the midgut during metamorphosis in Bombyx mori. Zool Sci. 2008;25:693–98. doi: 10.2108/zsj.25.693. [DOI] [PubMed] [Google Scholar]
- 45.Sumida M, Yamashita O. Trehalase transformation in silkworm midgut during metamorphosis. J Comp Physiol. 1977;115:241–53. doi: 10.1007/BF00692534. [DOI] [Google Scholar]
- 46.Wegener G, Tschiedel V, Schlöder P, Ando O. The toxic and lethal effects of the trehalase inhibitor trehazolin in locusts are caused by hypoglycemia. J Exp Biol. 2003;206:1233–40. doi: 10.1242/jeb.00217. [DOI] [PubMed] [Google Scholar]
- 47.Kamei Y, Hasegawa Y, Niimi T, Yamashita O, Yaginuma T. Trehalase-2 protein contributes to trehalase activity enhanced by diapause hormone in developing ovaries of the silkworm, Bombyx mori. J Insect Physiol. 2011;57(5):608–13. doi: 10.1016/j.jinsphys.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 48.Zhang WQ, Chen XF, Tang B, Tian HG, Chen J, Yao Q. Insect chitin biosynthesis and its regulation. Chin J Appl Entomol. 2011;48(3):475–79. [Google Scholar]
- 49.Terra WR, Ferreira C. Insect digestive enzymes: properties, compartmentalization and function. Comp Biochem Physiol. 1994;109B:1–62. [Google Scholar]
- 50.Silva CPM, Terra RW, Ferreira C. The role of carboxyl, guanidine and imidazole groups in catalysis by a midgut trehalase purified from an insect larva. Insect Biochem Mol Biol. 2004;34:1089–99. doi: 10.1016/j.ibmb.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 51.Thompson SN. Trehalose—the insect ‘blood’ sugar. Adv Insect Physiol. 2003;31:203–85. [Google Scholar]
- 52.Friedman S. Trehalose regulation, one aspect of metabolic homeostasis. Annu Rev Entomol. 1978;23:389–407. doi: 10.1146/annurev.en.23.010178.002133. [DOI] [Google Scholar]
- 53.Clegg JS, Evans DR. Blood trehalose and flight metabolism in the blowfly. Science. 1961;134:54–5. doi: 10.1126/science.134.3471.54. [DOI] [PubMed] [Google Scholar]
- 54.Almeida FM, Bonini BM. Heterologous expression in Escherichia coli of Neurospora crassa neutral trehalase as an active enzyme. Protei Expre Purif. 2009;65:185–89. doi: 10.1016/j.pep.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 55.Lipiski Z, Ótowska K, Wawrowska J, Zaleska M. The concentration of carbohydrates in the body of development all stages of Apis mellifera carnica drone brood. J Apicult Sci. 2008;52:5–11. [Google Scholar]
- 56.Yuhki K, Yuh H, Teruyuki N, Okitsugu Y, Toshinobu Y. Trehalase-2 protein contributes to trehalase activity enhanced by diapause hormone in developing ovaries of the silkworm, Bombyx mori. J Insect Physiol. 2011;57:608–13. doi: 10.1016/j.jinsphys.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 57.Su ZH, Ikeda M, Sato Y, Saito H, Imai K, Isobe M, et al. Molecular characterization of ovary trehalase of the silkworm, Bombyx mori and its transcriptional activation by diapause hormone. Biochim Biophys Acta. 1994;1218:366–74. doi: 10.1016/0167-4781(94)90190-2. [DOI] [PubMed] [Google Scholar]
- 58.Santos R, Alves-Bezerra M, Rosas-Oliveira R, Majerowicz D, Meyer-Fernandes JR, Gondim KC. Gene identification and enzymatic properties of a membrane-bound trehalase from the ovary of Rhodnius prolixus. Arch Insect Biochem. 2012;81:199–213. doi: 10.1002/arch.21043. [DOI] [PubMed] [Google Scholar]
- 59.Becker A, Schlder P, Steele JE, Wegener G. The regulation of trehalose metabolism in insects. Experientia. 1996;52:433–39. doi: 10.1007/BF01919312. [DOI] [PubMed] [Google Scholar]
- 60.Żółtowska K, Lipiński Z, Łopieńska-Biernat E, Farjan M, Dmitryjuk M. The activity of carbohydrate-degrading enzymes in the development of brood and newly emerged workers and drones of the carniolan honeybee, Apis mellifera carnica. J Insect Sci. 2012;12:1–11. doi: 10.1673/031.012.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arakane Y, Dixit R, Begum K, Park Y, Specht CA, Merzendorfer H, et al. Analysis of functions of the chitin deacetylase gene family in Tribolium castaneum. Insect Biochem Mol Biol. 2009;39:355e365. doi: 10.1016/j.ibmb.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 62.Arakane Y, Baguinon MC, Jasrapuria S, Chaudhari S, Doyungan A, Kramer KJ, et al. Both UDP N-acetylglucosamine pyrophosphorylases of Tribolium castaneum are critical for molting, survival and fecundity. Insect Biochem Mol Biol. 2011;41:42e50. doi: 10.1016/j.ibmb.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 63.Yang MM, Zhao LN, Shen QD, Xie GQ, Wang SG, Tang B. Knockdown of two trehalose-6-phosphate synthases severely affects chitin metabolism gene expression in the rice brown planthopper Nilaparvata lugens. Pest Manag Sci. 2016; doi: 10.1002/ps.4287. [DOI] [PubMed]
- 64.Zhao LN, Yang MM, Shen QD, Liu XJ, Shi ZK, Wang SG, et al. Functional characterization of three trehalase genes regulating the chitin metabolism pathway in rice brown planthopper using RNA interference. Sci Repts. 2016;6:27841. doi: 10.1038/srep27841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Villarreal AM, Adamson SW, Browning RE, Budachetri K, Sajid MS, Karim S. Molecular characterization and functional significance of the Vti family of SNARE proteins in tick salivary glands. Insect Biochem Mol Biol. 2013;43(5):483–93. doi: 10.1016/j.ibmb.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ge LQ, Zhao KF, Huang LJ, Wu JC. The effects of triazophos on the trehalose content, trehalase activity and their gene expression in the brown planthopper Nilaparvata lugens (Stål) (Hemiptera: Delphacidae) Pestic Biochem Physiol. 2011;100:172–81. doi: 10.1016/j.pestbp.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]


