Abstract
Histone proteins are subject to a host of posttranslational modifications (PTMs) that modulate chromatin structure and function. Such control is achieved by the direct alteration of the intrinsic physical properties of the chromatin fiber or by regulating the recruitment and activity of a host of trans-acting nuclear factors. The sheer number of histone PTMs presents a formidable barrier to understanding the molecular mechanisms at the heart of epigenetic regulation of eukaryotic genomes. One aspect of this multifarious problem, namely how to access homogeneously modified chromatin for biochemical studies, is well suited to the sensibilities of the organic chemist. Indeed, recent years have witnessed a critical role for synthetic protein chemistry methods in generating the raw materials needed for studying how histone PTMs regulate chromatin biochemistry. This review focuses on what is arguably the most powerful, and widely employed, of these chemical strategies, namely histone semisynthesis via the chemical ligation of peptide fragments.
Keywords: expressed protein ligation, posttranslational modification, epigenetics, histone
INTRODUCTION
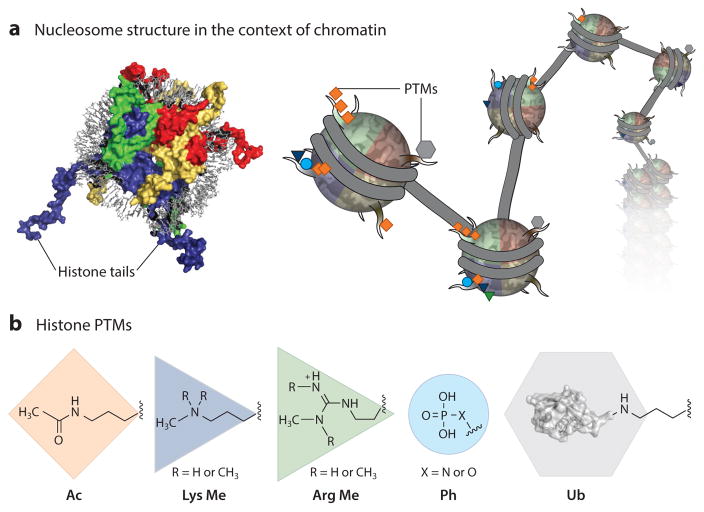
In eukaryotes, genomic DNA is segregated into the nucleus of a cell, where it is stored in the form of the nucleoprotein complex known as chromatin (Figure 1a) (1). The fundamental repeating unit of chromatin, the nucleosome, consists of an ~146-bp DNA duplex wrapped around a histone octamer composed of two copies each of histone H2A, histone H2B, histone H3, and histone H4 (Figure 1a) (2). Histones possess a canonical α-helical fold, which forms the structural scaffolding of the octamer core, and a basic N-terminal tail protruding from the nucleosome (3). The posttranslational modification (PTM) of histone proteins has emerged as a central component of epigenetic regulation, functioning within the nucleosome as a signal integration platform able to orchestrate chromatin-templated processes (4).
Figure 1.
Histone posttranslational modifications (PTMs) and chromatin regulation. (a) (Left) The nucleosome is the fundamental repeating unit of chromatin [Protein Data Bank (PDB) identifier 1KX5]. The N-terminal histone tails protrude from the nucleosome core and are a hot spot for PTMs (H2A, yellow; H2B, red; H3, blue; H4, green; DNA, gray). Nucleosomes disperse along the DNA into nucleosomal arrays (middle) and, with the help of PTMs, are able to recruit chromatin effectors and alter internucleosomal contacts. (b) Five of the many histone PTMs. Lysine acetylation (orange diamond ), lysine methylation (mono, di, or tri) (blue triangle), arginine methylation (mono, sym, or asym) ( green triangle), phosphorylation (serine, threonine, and histidine) (blue circle), and lysine ubiquitylation ( gray hexagon) differ in physiochemical complexity (ubiquitin, PDB 1UBQ3).
More than 15 chemotypes of histone PTMs, or marks, have been annotated across 100 distinct sites throughout the 4 core histones (1, 5, 6). These PTMs differ in physiochemical properties and complexity and include phosphorylation, acetylation, methylation, ubiquitylation, glycosylation, citrullination, SUMOylation, ADP ribosylation, proline isomerization, and higher forms of acylation (Figure 1b). Moreover, a subset of these modifications exist in multiple states (e.g., lysine can be monomethylated, dimethylated, or trimethylated), displaying functionally different physiological outcomes (Figure 1b) (7–9). Histone PTMs are installed, interpreted, and removed by chromatin effector proteins (which are colloquially referred to in the field as writers, readers, and erasers) that have different levels of genomic, nucleosomal, and amino acid residue specificity (7, 10).
Multiple PTMs can coexist on the same histone polypeptide; a recent mass spectrometry study employing HeLa cells identified 708 unique posttranslationally modified core histones (11). This observation, combined with the fact that each nucleosome contains eight histones, means that there may be a very large number of differentially modified nucleosomes in chromatin [the existence of asymmetric modification patterns within each histone pair increases the size of this potential inventory even further (12)]. The full extent of this combinatorial expansion (in either a “syntactic” or functional sense) remains unclear (13). However, there seems little doubt that chromatin-templated processes, including the installation and removal of histone marks themselves, must function in a shockingly complex biochemical landscape. Conceptual frameworks have been advanced that seek to make sense of this apparent complexity. The most famous hypothesis asserts that groups of histone PTMs may function synergistically, constituting a histone code, to bring about distinct downstream effects (14). In the broadest sense, this idea is supported by the finding that specific marks segregate and co-occupy with one another at genomic functional elements, and by the identification of trans-histone pathways, in which one histone mark leads to the establishment of an additional histone mark, as in the case of the ubiquitylation of histone H2B on Lys-120 (H2BK120Ub) being necessary for H3 methylations (15, 16). Whether or not phenomena of this type confirm the existence of a true code or are manifestations of a more traditional signaling hub remains an open question. However, there can be no debate that understanding the interplay between these modifications is paramount to our understanding of chromatin biology (14, 17–19).
Histone PTMs can alter the physiochemical properties of the chromatin fiber directly as well as recruit, expel, and modulate the activity of trans-acting chromatin effectors. For instance, the acetylation of H4K16 (H4K16ac) converts the basic amino group of the lysine side chain into a neutrally charged amide, disrupting internucleosomal histone–histone contacts and thereby enhancing nucleosomal accessibility (20, 21). Additionally, acetyl-lysine-binding modules within specific chromatin effectors engage this mark, resulting in the recruitment of these effectors to the acetylated chromatin substrate (22, 23). To a first approximation, it is thought that PTMs to the histone tails predominantly engage in chromatin signaling through chromatin effector recruitment (or expulsion), whereas modifications within the octamer core affect the physiochemical properties of chromatin directly (24, 25). However, as observed in the case of H4K16ac, specific histone PTMs can do both (20, 21).
Access to chemically homogeneous, modified chromatin is critical to elucidate the role that specific histone PTMs play in downstream biochemical processes. The purification of endogenous histones containing PTMs is possible (26, 27), but this process results in heterogeneous substrates (i.e., the isolated material will contain several distinct populations of differentially modified histones). This obfuscates biochemical analyses in studies that seek to unambiguously define causality in a process. In principle, modified histones can be obtained by enzymatic modification of unmodified recombinant histones. However, this approach has many drawbacks. The enzyme responsible for a given histone PTM may be unknown, or it may be difficult to isolate in an active form [many chromatin writers are multisubunit complexes that require premodified chromatins as substrates (28)]. Even in cases in which the enzyme is available, in vitro modification is often inefficient or causes heterogeneous modification of the chromatin substrate, as in the case of the chromatin effector p300 (29, 30). Due to the many shortcomings of the aforementioned approaches, (semi)synthetic methods for the generation of posttranslationally modified histones have gained much traction in the last decade and have been used extensively for the study of chromatin biochemistry (31). The main advantage of chemical synthesis is homogeneous substrate preparation, which enables precise biochemical analysis of both chromatin structure and function; that is, causality can be firmly established. In this review, we discuss recent advances in ligation-based technologies employed to access native PTM-containing histones. More expansive reviews that cover complementary genetic and bioconjugation methods to incorporate PTMs into histones can be found elsewhere (see the sidebars titled A Genetic Approach to Incorporate Post-translational Modifications into Histone Proteins and Cysteine-Directed Approaches to Access Histone Posttranslational Modifications) (31–33).
CONTEMPORARY SYNTHESIS OF POSTTRANSLATIONALLY MODIFIED HISTONES THROUGH HISTONE-DERIVED FRAGMENT LIGATION
Use of Synthetic Peptides to Probe the Role of Histone Posttranslational Modifications
Synthetic peptides containing PTMs have been used in the histone area for more than 40 years (48). In a landmark study, the specificity of deacetylase enzymes, which are responsible for removing histone lysine acetylation marks, was investigated with synthetic peptides containing acetyl-lysine synthesized via solid-phase peptide synthesis (SPPS) (48). SPPS has been extensively utilized to study histone PTMs in which the appropriated modified amino acid derivative is chemically accessible—as in, for instance, lysine acetylation; lysine methylation; and serine, threonine, and tyrosine phosphorylation (31, 49–52). This approach has been instrumental in the identification and structural characterization of histone PTM effectors, the generation of specific histone PTM antibodies, and the elucidation of the mechanistic and structural details of histone PTM incorporation and removal (e.g., 53–59). Additionally, libraries of modified histone peptides, generated using parallel or split-pool strategies, have proven invaluable in the rapid elucidation of chromatin effector and antibody-binding specificity (50, 51, 60–66). SPPS is generally limited to peptides that are shorter than 50 amino acid residues [or even shorter in the case of expedited combinatorial peptide strategies (67, 68)].
A GENETIC APPROACH TO INCORPORATE POSTTRANSLATIONAL MODIFICATIONS INTO HISTONE PROTEINS.
Genetic incorporation of PTMs into histones has been achieved through the amber codon suppression methodology (34, 35). This powerful method relies on the use of an orthogonal aminoacyl–tRNA (transfer RNA) synthetase/tRNA pair (generated by directed evolution methods) that accepts a posttranslationally modified amino acid. Subsequent ribosomal incorporation of this unnatural amino acid in response to an amber stop codon (UAG) within the gene of interest affords the corresponding modified recombinant protein. This technology has proven especially useful for the incorporation of acetyl-lysine into histones (34), which has allowed several biochemical and biophysical questions to be addressed (36). Moreover, amber suppression has been used to incorporate other PTMs into proteins, either directly [e.g., crotonylation (37, 38)] or as part of multistep processes [e.g., methylation, phosphorylation, and ubiquitylation (39–42)]. Modified histone production via amber suppression is operationally straightforward—at least once the necessary aminoacyl–tRNA synthetase/tRNA pair is generated. However, depending on the modified amino acid in question and the site of its incorporation within the histone, suppression efficiency may vary (35). Moreover, the generation of histones containing multiple modifications, whether homotypic or heterotypic, is currently impractical.
CYSTEINE-DIRECTED APPROACHES TO ACCESS HISTONE POSTTRANSLATIONAL MODIFICATIONS.
Cysteine-directed chemistry has been utilized to incorporate PTM mimics into recombinantly prepared histone proteins containing strategically incorporated cysteine mutations (43–45). This strategy is enabled by the sequences of the core histones; H2A, H2B, and H4 are devoid of cysteine, whereas H3 contains a single cysteine that can be removed without incident. Employing the cysteine sulfhydryl group as a unique reactive handle has led to the preparation of acetylated, methylated, and ubiquitylated histone mimics that can recapitulate the biological activity of the corresponding native posttranslationally modified histones (43–45). This methodology is both convenient and expedient because it typically involves the single-step conjugation of a thiol-reactive PTM-containing electrophile to a recombinant histone cysteine mutant. Furthermore, in the case of ubiquitylation, a disulfide approach allows for the chemical removal of this PTM, enabling study of the consequences of chemical deubiquitylation (44, 46). Although operationally simple, this approach does generate an analog of the native PTM, which could be a concern in some cases (47). Moreover, this approach does not allow multiple different (i.e., heterotypic) PTMs to be introduced site-specifically into the same histone.
Histone-derived peptides have obvious shortcomings when the process in question requires a more physiologically relevant substrate. Specifically, peptides are unable to address questions that involve a tertiary structural (or DNA-binding) element or multivalent interactions occurring outside the space of a linear peptide (e.g., between histones). Chromatin effectors often exist in multisubunit complexes in the size regime of the nucleosome (or greater), and are known to engage chromatin through multiple low-affinity epitopes (7, 28). Thus, characterization of these complexes using more physiological chromatin systems is often essential to obtain a more complete mechanistic understanding of these interactions (28). For example, the binding of the transcription factor BPTF (bromodomain PHD finger transcription factor) is modulated by H3K4 methylation and H4 acetylation through interactions with the PHD finger and bromodomain of the protein, respectively (23, 29). Clearly, such a multivalent interaction cannot be characterized using peptide substrates. The use of chromatin substrates also reveals aspects of enzyme regulation that are not evident with peptide substrates. For example, Set1C, a chromatin writer complex that methylates H3K4, is active on H3 peptides but requires the ubiquitylation of H2BK120 (H2BK120Ub) for activity on nucleosomal substrates (69, 70). Given the existence of these higher-order recognition processes, in recent years investigators have devoted considerable effort to generating full-length histones that contain PTMs and that can be reconstituted into physiologically relevant chromatin substrates.
Semisynthesis of Histone Proteins Employing Synthetic Peptides
Robust chemistries now exist for the incorporation of most commonly occurring PTMs into synthetic peptides. However, as noted above, a more physiological substrate is often required for the study of chromatin-related processes such as remodeling or PTM deposition and removal. Semisynthesis enables the production of posttranslationally modified full length histones that have been modified though SPPS. In this strategy, the PTM-containing part of the final protein product is of synthetic origin, whereas the remainder, the unmodified part, is produced by recombinant protein expression (71, 72). Implicit in this strategy is the idea that the recombinant building block, by being easy to produce, constitutes most of the final semisynthetic protein. Fortunately, fragment condensation strategies, featuring unprotected peptide building blocks and employing chemo- and regioselective chemical ligation steps, are available for this purpose (73). Histones are ideal candidates for the semisynthesis approach for several reasons.
Most histone PTMs are located near the N or C terminus of the protein, so relatively short synthetic peptides can be employed in the context of a two-building-block (i.e., one-ligation-step) assembly strategy.
Histones can be readily refolded, thereby allowing semisynthesis to be conducted in the presence of chaotropes, such as guanidinium chloride. This process allows the use of high concentrations of reactants, improving the kinetics and yields of the second-order chemical reaction.
Histones generally lack cysteine residues, a feature that is critical to the semisynthetic method. Not only does this property eliminate any problems associated with unwanted oxidation reactions, but also, because of the nature of the ligation chemistry used, it grants tremendous freedom in choosing a ligation site in the histone (elaborated on in the following two paragraphs).
Although several ligation chemistries have been developed that allow unprotected peptides to be assembled into proteins, in this review we focus on only one, namely the native chemical ligation reaction (NCL) (74). This approach and its semisynthetic extension, the expressed protein ligation reaction (EPL) (75), are by far the most commonly employed methods for the (semi)synthesis of proteins (76). Hundreds of proteins have been manufactured using these strategies, giving rise to a broad range of biochemical and biophysical probes for functional and structural studies (71, 72, 76). Moreover, unlike cysteine conjugation and amber suppression technologies (see the sidebars titled A Genetic Approach to Incorporate Posttranslational Modifications into Histone Proteins and Cysteine-Directed Approaches to Access Histone Posttranslational Modifications), NCL and EPL allow multiple different modifications to be incorporated into the same protein, a useful property for studies of the synergistic effects of histone modifications (11).
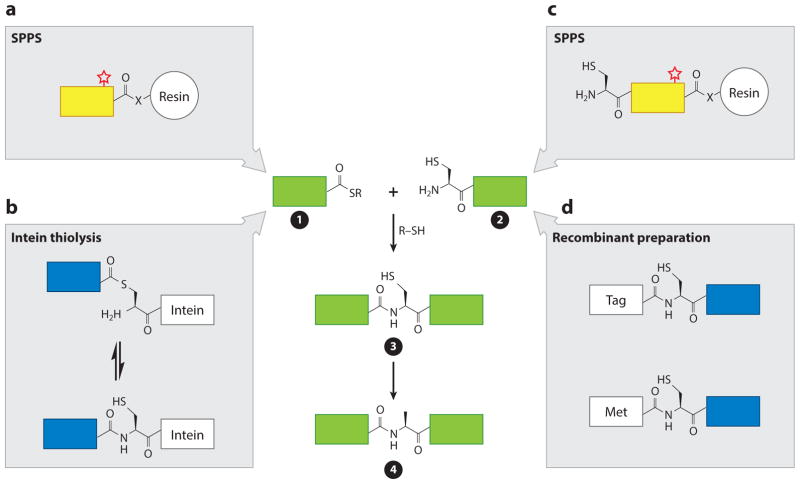
Native chemical ligation relies on the condensation in water at physiological pH of a peptide bearing a C-terminal thioester (α-thioester) with a second peptide containing an N-terminal 1,2-amino thiol moiety (e.g., cysteine) (Figure 2, constructs ➊ and ➋) (74). This process results in a native peptide bond between the two segments (Figure 2, construct ➌). If necessary, this process can be conducted in an iterative fashion until the complete protein is assembled (77). In the case of histones, this allows for the modular multimodification of a histone at distinct regions. The dearth of cysteine residues in histones might initially seem to pose a problem for the use of NCL and EPL. Indeed, early applications of the strategy to histones required the introduction of a strategically placed cysteine mutation to facilitate ligation (78, 79). This ligation remnant, or scar, remained in the final product, potentially complicating downstream experiments. However, with the advent of desulfurization strategies (80, 81), which extend the scope of NCL and EPL to noncysteine ligation junctions, histone sequences have become a much more pliable platform on which to perform NCL and EPL. For example, histone ligation sites are now often chosen to be adjacent to an alanine residue. This choice enables the substitution of this alanine with cysteine to support ligation. Following generation of the initial NCL or EPL product, this cysteine (the only such residue in the protein) is converted back to alanine by chemical desulfurization (Figure 2, construct ➍). This conversion renders the overall process traceless, thereby affording a native histone sequence containing a user-defined set of PTMs. Importantly, one can extend this strategy to many ligation junctions by employing thiol-containing analogs of other amino acids. Indeed, thiolated derivatives of several residues (leucine, phenylalanine, valine, threonine, aspartic acid, lysine, arginine) (82–88) have been employed in the traceless (semi)synthesis of many proteins, including several modified histones (89, 90).
Figure 2.
Histone (semi)synthesis by native chemical ligation reaction (NCL) and expressed protein ligation (EPL). Histone fragments containing α-thioesters (➊) are prepared by solid-phase peptide synthesis (SPPS) (a) or recombinantly through thiolysis from an intein fusion (b). Histone fragments containing 1,2-amino thiols (➋; cysteine is shown as a common example) are prepared synthetically via SPPS (c) or using recombinant DNA methods (d ); the latter involves either the proteolytic removal of an N-terminal tag in vitro or processing of the initiator methionine in vivo. Fragments are ligated together via NCL in the presence of exogenous thiols (R-SH) to yield full-length polypeptides containing a peptide bond at the ligation junction (➌). Because histones generally lack native cysteine residues, chemical ligation is typically followed by a desulfurization step to yield a cysteine-less histone (➍). Yellow indicates a building block of synthetic origin, blue indicates a fragment generated by recombinant DNA methods, a red star indicates that posttranslational modification incorporation is feasible, and green denotes that the fragment can be prepared by either chemical synthesis or recombinant DNA technology.
In NCL, the α-thioester-containing peptide fragment is prepared using chemical synthesis, regardless of whether the other fragment is of synthetic or recombinant origin. Thus, histones containing PTMs within the N-terminal region of the protein (i.e., the synthetic building block) are prepared via NCL. In EPL, however, the α-thioester-containing fragment is generated by recombinant protein expression (75), again regardless of whether the other fragment is of synthetic or recombinant origin. Accordingly, semisynthetic histones containing PTMs distal to the protein N terminus are typically generated using an EPL strategy. As discussed further below, advances in the production of the synthetic or recombinant building blocks, along with improvements in the ligation and postligation steps in the process, have allowed NCL and EPL to be used to generate multiple posttranslationally modified versions of all four canonical histones.
Preparation of Histone-Derived Fragments for Semisynthesis
Preparation of posttranslationally modified histones first involves the production of histone-derived fragments containing the appropriate ligation handles. A tremendous amount of effort has gone into the facile production of both synthetic and recombinant building blocks; multiple strategies exist to address the unique challenges of incorporating specific PTMs into an otherwise native histone template.
Synthetic building blocks
An absolute requirement of the NCL and EPL strategies is the presence of the α-thioester and 1,2-amino thiol (typically cysteine) moieties within the peptide building blocks. In terms of the synthetic peptide fragment, the latter presents little in the way of a challenge because the necessary residue can simply be incorporated during the last step of SPPS (Figure 2c). By contrast, the preparation of synthetic peptide α-thioesters, which are essential for generating histone proteins containing PTMs (or other noncoded elements) within the N-terminal tail regions, is more involved (Figure 2a). Of the two N-α-protection strategies commonly employed in SPPS, namely butyloxycarbonyl (Boc) and fluorenylmethyloxycarbonyl (Fmoc), the former naturally lends itself to the generation of peptide α-thioesters due to the stability of the thioester group to the reagents used during the coupling and deprotection steps in chain assembly. Thus, an α-thioester linkage between the growing peptide and the resin support can be carried through an entire Boc-SPPS. As a consequence, simple thiol-containing linkers, such as 3-mercaptopropionic acid (91), can be employed in the synthesis of peptide α-thioesters using Boc-SPPS (Table 1). Several modified histones have been generated via NCL strategies that involved the use of synthetic peptide α-thioester building blocks assembled by Boc-SPPS (see the section titled Semisynthesis Applied to Histones for specific examples).
Table 1.
Methodologies for histone (semi)synthesis
| (Semi)Synthetic step | Methodology | Notes |
|---|---|---|
| Peptide thioester preparation | Boc-based SPPS | Widely used; harsh acids required; not amenable to phosphorylated or glycosylated peptides NA |
| Mercaptopropionic acid linker | ||
| Fmoc-based SPPS | ||
| 2-Hydroxy-3-mercaptopropionic acid linker | Low yielding; one of the better routes to Fmoc-based thioester preparation | |
| Diaminobenzoic acid linker | NA | |
| Hydrazide-based SPPS | Prepared through Boc- and Fmoc-based SPPS; necessitates the in situ generation of a peptide thioester during chemical ligation | |
| N-terminal 1,2-amino thiol preparation | SPPS | Facile for the generation of short synthetic peptides, including unnatural thiolated amino acids |
| Recombinant preparation | More cost effective than SPPS | |
| Initiator methionine | Need to empirically test that the initiator methionine is processed correctly | |
| Protease-site/tag based | May aid in truncated histone stability; assists in purification | |
| Thioester preparation by recombinant DNA methods | Type of intein | NA |
| Mxe-GyrA | Standard intein used in a variety of contexts | |
| Ultrafast inteins | Results in quicker and more complete reactions and one-pot thiolysis/ligation | |
| Split intein column | Allows for the direct ligation and purification of histone constructs; works with denaturants | |
| Chemical ligation | Type of exogenous thiol | NA |
| MESNa | Thiol used in many chemical ligations | |
| MPAA | Significantly faster then MESNa; allows expedited ligation and hydrazide-based ligation | |
| TFET | Can be removed by degassing to enable one-pot desulfurization | |
| Desulfurization | Raney nickel | Low yielding due to aggregation |
| Homogeneous radical chain reaction | Facile method for the desulfurization of cysteine and thiolated amino acid derivatives |
Abbreviations: Boc, butyloxycarbonyl; Fmoc, fluorenylmethyloxycarbonyl; MESNa, sodium mercaptoethyl sulfonate; MPAA, mercaptophenylacetic acid; Mxe, Mycobacterium xenopi; NA, not applicable; SPPS, solid-phase peptide synthesis; TFET, 2,2,2-trifluoroethanethiol.
Boc-SPPS does, however, involve the use of a strong acid, typically anhydrous hydrogen fluoride (HF), during the cleavage/global deprotection step from the solid support. This requirement has two important consequences. The first is purely practical in that the specialized equipment needed to perform this step may not be available to every investigator or facility. The second limitation is chemical in that some functionalities are labile to HF, for example, phosphoserine and phosphothreonine, as well as O-glycosylated amino acids (92). By contrast, the Fmoc-SPPS strategy is compatible with a much broader range of PTMs and, because it does not require the use of HF, is more generally accessible to the average biochemist. These considerations have prompted tremendous effort toward the development of robust protocols for the generation of peptide α-thioesters via Fmoc-SPPS (92).
Unlike Boc-SPPS, the Fmoc-SPPS strategy is not directly compatible with a α-thioester due to the use of piperidine at every deprotection cycle of the chain assembly. Early efforts focused on modifying the deprotection conditions to minimize unwanted aminolysis/hydrolysis of the labile α-thioester (93). However, these strategies do not provide a general solution to the problem, particularly for longer sequences. Fortunately, more productive approaches to this problem have been developed, all of which involve the use of what is effectively a “latent” α-thioester. In these strategies, the growing peptide is linked to the resin via a nonthioester linkage that is completely stable to the reagents used in standard Fmoc-SPPS. Importantly, the chemistry of the linker is such that upon completion of the chain assembly and cleavage from the support, the modified C terminus of the peptide can be converted into a α-thioester using selective chemistry. Several latent thioester linkers are now available for Fmoc-SPPS of peptide α-thioesters (94–97). These include linkers based on 2-hydroxy-3-mercaptopropionic acid (94), diaminobenzoyl (95), and hydrazine (96) moieties, all of which have been applied to the (semi)synthesis of modified histones (Table 1) (90, 98, 99). Of these, the hydrazine linker, which yields a peptide hydrazide upon cleavage that is eventually converted into an active thioester via an oxidation/thiolysis process, is rapidly becoming the method of choice for the preparation of peptide α-thioesters by the Fmoc approach due to its ease of use and excellent efficiency (96).
Recombinant building blocks
Recombinant protein expression has been utilized to prepare unmodified histones fragments with either an N-terminal cysteine (in the case of a C-terminal fragment) or an α-thioester (in the case of an N-terminal fragment). Unlike the case for synthetic peptides, in which introduction of only one of the two reactive handles requires deviation from standard protocols, additional provisions are needed to prepare either class of reactive recombinant protein. The absolute requirement for an initiating methionine codon means that an N-terminal cysteine cannot be directly encoded in a protein, for example, a histone fragment. Several strategies to circumvent this problem have been developed (Table 1). Most trivially, the requisite cysteine can be placed adjacent to the initiating methionine with the expectation that it will become exposed as part of the normal in vivo processing of the protein. Although this strategy has been successfully applied to semisyntheses of several proteins, including histones (98, 100), it suffers from a few drawbacks. First, the processing is not always complete, and it can be hard to separate the desired protein from the unprocessed precursor. Second, the unmasked N-terminal cysteine can react with cellular metabolites such as pyruvate (101), and although protocols exist to reverse these adducts (102), the additional steps needed reduce overall yields. Third, this strategy precludes the introduction of an N-terminal affinity purification handle (e.g., polyhistidine or glutathione S-transferase tags), which is commonly used in recombinant protein production work flows.
Given these attendant issues, it is far more common to employ a fusion protein strategy as a route to N-terminal cysteine proteins. In this approach, the protein fragment of interest is directly linked via a specific protease recognition sequence to the desired affinity tag. Following purification, the fusion protein is treated with the corresponding protease to afford the mature fragment suitable for NCL and EPL. Key to this strategy is the use of a protease that tolerates cysteine at the S1′ site in the substrate. Fortunately, three commonly used proteases have this property, namely the Factor Xa protease (103), the SUMO protease (104), and the tobacco etch virus (TEV) protease (105). Although all of these enzymes have been used in NCL and EPL schemes, the last two are the most powerful because they can easily be overexpressed in Escherichia coli and, upon purification, have negligible nonspecific cleavage activity associated with them. An added benefit of the SUMO fusion strategy is that the protease recognition element, being a small and stable folded protein, often improves the expression levels and behavior of the protein fragment to which it is appended (104).
Introduction of an α-thioester into a recombinant protein also requires the use of a fusion protein strategy. Here, the protein fragment is linked through its C terminus to an engineered version of a protein domain called an intein (Figure 2b). Inteins are natural autoprocessing proteins that cut themselves out of a host polypeptide in a process known as protein splicing (76, 106). A detailed understanding of the mechanism of intein-mediated protein splicing has allowed the rational design of engineered inteins that can be used to install an α-thioester into essentially any recombinant protein. Operationally, an intein fusion protein is expressed in a suitable cell host and, following purification, is exposed to a small-molecule thiol to afford the desired protein α-thioester derivative, which can then be utilized for semisynthesis (75, 107). Parenthetically, engineered inteins have also been developed that allow the preparation of recombinant proteins bearing an N-terminal cysteine (108). However, this strategy is generally less efficient than the aforementioned protease approaches and thus has not been widely adopted (71).
Most EPL studies to date have employed protein α-thioesters generated from the corresponding fusion to an engineered Mycobacterium xenopi (Mxe)-GyrA intein. This intein is commercially available in the form of a bacterial expression plasmid (109). Despite widespread use, the Mxe-GyrA intein has a splicing half-life of several hours, which means that thiolysis reactions can be exceedingly slow [days are not uncommon (106, 110)] and fail to proceed to completion. The recent discovery of several ultrafast inteins with splicing half-lives in the tens of seconds (110, 111) promises to redress these issues. The utility of these inteins in the semisynthesis arena is further enhanced by the fact that they are naturally split; that is, the intein is fractured into two inactive pieces that must first noncovalently associate before protein trans splicing begins (106). Taking advantage of the low-nanomolar affinities between the split intein fragments, Vila-Perelló et al. (112) streamlined the entire EPL work flow and in the process improved the yield of recombinant protein α-thioester production (Table 1). The optimized protocol is illustrated through the semisynthesis of histone H2B containing an acetyl-lysine PTM at position 120. First, a fusion protein corresponding to H2B residues 1–116 directly linked though its C terminus to the N-terminal half of the split intein was expressed in E. coli. Note that because the split intein fragment has no splicing or thiolysis activity (without the complementary intein fragment around), premature cleavage of the fusion cannot occur, which is often an issue when working with intact inteins such as Mxe-GyrA (108). Second, purification was achieved following cell lysis by use of the affinity of the H2B(1–116) N-intein construct for the C-intein, the latter of which was immobilized on a solid support. Third, treatment of the immobilized fusion protein with a thiol-containing buffer solution afforded the H2B(1–116) α-thioester, which was then ligated in a one-pot operation to a synthetic peptide containing the desired PTM. Notably, this streamlined EPL strategy was compatible with strong denaturants that are often needed to solubilize histones from inclusion bodies (113). The improved speed and overall yields associated with this methodology should make it the method of choice for generating recombinant histone α-thioesters for use in EPL.
Improvements in Histone (Semi)Synthesis and Desulfurization
In this section, we describe highly optimized protocols for preparing semisynthetic histones. In tandem with advances in the production of the necessary building blocks for NCL and EPL, there have been important developments in the protocols employed in the actual ligation step, as well as the subsequent removal of the ligation scar by desulfurization. Beginning with the former, a key feature of NCL and EPL is the inclusion of a small-molecule thiol compound during the ligation step. This additive plays several roles, all of which exploit the fact that thioesters are soft electrophiles and hence more reactive to thiol-containing nucleophiles than to oxygen or nitrogen nucleophiles. Thus, a peptide can be produced and stored as a relatively unreactive alkyl α-thioester but then activated in situ by transthioesterification with the added thiol (Figure 2). By tuning the electronic properties of the incoming nucleophile, one can conveniently increase the reactivity of the thioester. Another beneficial consequence of facile transthioesterification is that it allows NCL and EPL to proceed when either (or both) of the peptide building blocks contains internal cysteine residues; although these residues can participate in transthioesterification to yield internal thioesters, the absence of a proximal amino group means that they are unable to proceed to amide bond formation and hence do not accumulate to a significant extent in the reaction mixture. Although a number of small-molecule thiols (e.g., thiophenol, benzylmercaptan, and 2-mercaptoethanesulfonate) have been used over the years in NCL and EPL, the field has now converged on the use of mercaptophenylacetic acid (MPAA). Initially described by the Kent group (114), MPAA significantly improves the kinetics of NCL and EPL reactions, a property that is attributed to an improved transthioesterification step between the two peptide fragments, leading to increased efficiency and better yields.
The last step in a contemporary NCL- or EPL-based histone semisynthesis is the reduction of the cysteine residue at the ligation site to a native alanine (or a thiolated amino acid derivative to the corresponding natural amino acid) (Figure 2) (80). This traceless ligation strategy was introduced by Blanco-Canosa & Dawson (95) and initially involved classical hydrogenolytic desulfurization using a Raney nickel catalyst. Indeed, the Raney nickel desulfurization protocol has been successfully applied to the semisynthesis of several PTM-containing histones (98, 100). As a response to concerns that Raney nickel might lead to protein losses through adsorption and aggregation processes, Wan & Danishefsky (81) subsequently introduced a more biopolymer-compatible desulfurization protocol based on a free radical–initiated process (referred to herein as free radical desulfurization). This reaction has proven to be incredibly powerful, typically affording high isolated yields of desulfurized products, and it is now the method of choice for removing NCL and EPL ligation scars through desulfurization. Notably, free radical desulfurization has been applied to the traceless semisynthesis of all four canonical histones (involving cysteine residues as well as various thiolated amino acid analogs) (29, 99, 115).
Finally, and most recently, Paine and colleagues (116) have reported a very useful advance that simultaneously affects both the ligation kinetics and the subsequent desulfurization steps. This research nicely addresses a shortcoming of the standard protocol of ligation followed by desulfurization, namely that commonly used thiol additives, including MPAA, are incompatible with free radical desulfurization due to quenching effects. Thus, a purification step needs to be inserted between the other steps, which reduces overall yields and obviously makes the process more cumbersome. The authors solved this problem by replacing MPAA in the ligation mixture with 2,2,2-trifluoroethanethiol (TFET). The authors showed that TFET is as effective as MPAA at improving the ligation kinetics but can be subsequently removed through a simple degassing step owing to its volatility. This process enables NCL and desulfurization to be conducted in a one-pot fashion without the purification of the initial ligation product. Although this streamlined strategy has yet to be directed toward histones, it is likely to be widely adopted and, as such, should increase yields and efficiency in future histone PTM syntheses.
SEMISYNTHESIS APPLIED TO HISTONES
Improvements in methods to generate histone-derived peptide/polypeptide fragments as well as innovations that improve ligation efficiency and ease of desulfurization have substantially reduced the time and effort needed to obtain a semisynthetic histone in biochemically useful quantities (summarized in Table 1). We emphasize that the incredible amount of work that has gone into the preparation of semisynthetic histones over the last several years will inform future syntheses as, in most cases, a suitable ligation junction/synthetic strategy has already been developed. This section explores the approaches taken to access histones with specific PTMs, focusing on more recent syntheses that have resulted in the facile generation of completely native PTM-containing histones.
Semisynthesis Applied to Histone H2A
Of the four core histones, H2A has received the least attention in terms of semisynthesis, perhaps reflecting the fact that H2A has comparatively fewer characterized PTMs relative to the other histones. Nonetheless, this histone nicely illustrates the sophistication of modern EPL strategies. Especially notable is the work of Fierz and colleagues (89, 117) on the preparation of H2A ubiquitylated at Lys-119 (H2AK119Ub). The semisynthesis was designed around three building blocks, two recombinant α-thioesters (produced by thiolysis of intein fusions) and a branched synthetic peptide. The overall route required two EPL reactions followed by desulfurization. The first step involved ligation of a ubiquitin α-thioester fragment (lacking the C-terminal glycine; i.e., residues 1–75) to a synthetic peptide corresponding to residues 114–126 to H2A. This peptide was equipped with two ligation handles, namely (a) a cysteine, which was attached to the ε-amino group of K119 and would eventually become the missing residue in ubiquitin, and (b) the non-proteinogenic amino acid, penicillamine, at the N terminus [this is effectively a thiolated version of valine (108)]. The penicillamine moiety was protected as a thiazolidine to ensure the desired regioselectivity in the first EPL step, namely ligation of the ubiquitin α-thioester exclusively to the cysteine-bearing lysine side chain within H2A. Subsequent unmasking of the penicillamine in this intermediate construct set up the second ligation reaction with the other histone building block, H2A(1–113) α-thioester. The final step involved free radical desulfurization, which converted the penicillamine to a valine, the native residue, and the cysteine to an alanine. The observant reader will have noted that this iterative EPL strategy was not completely traceless in that it caused the C-terminal glycine of ubiquitin to be changed to an alanine. This alteration was deemed acceptable for the biochemical and biophysical studies that were subsequently performed on the semisynthetic protein (29, 89, 117). However, there may be instances in which a completely native isopeptide linkage is desirable. Fortunately, strategies do exist for this purpose [see the section titled (Semi)Synthesis Applied to Histone H2B, below], although they are technically more complicated than the chemical ubiquitylation route outlined above for H2AK119Ub.
(Semi)Synthesis Applied to Histone H2B
Semisynthesis via NCL and EPL has been used to incorporate a variety of PTMs into histone H2B. Importantly, these efforts have afforded viable semisynthetic routes for the introduction of PTMs into the N and C termini of the protein, as well as internal regions.
Chiang et al. (98) were the first to report on the modification of the N terminus of H2B using semisynthesis. These authors synthesized a series of peptides corresponding to the first 16 amino acids of H2B and containing either four acetyl marks (K5ac, K11ac, K12ac, K15ac) or a phosphor-ylation mark at Ser-14 (S14ph), or these five PTMs together. The inclusion of a phosphorylated mark necessitated the use of Fmoc-SPPS for the preparation of the peptide α-thioesters—both latent thioester linkers, 2-hydroxy-3-mercaptopropionic acid (94) and diaminobenzoic acid (95), were employed successfully in the course of this study. The corresponding recombinant fragment, H2B residues 17–125 containing cysteine in place of Ala-17, was generated through in vivo removal of the initiating methionine. NCL followed by desulfurization afforded the desired set of modified H2B proteins. This study is notable in that a direct side-by-side comparison between the Raney nickel and free radical desulfurization protocols was performed, with the latter giving a twofold increase in isolated yield. Access to these constructs allowed the authors to show that H2BS14ph antibodies are sensitive to the co-occurring acetyl marks; in other words, S14ph is not recognized as an antigen in the presence of K5ac, K11ac, K12ac, and K15ac. This study highlights the potential of the epitope occlusion phenomenon to skew antibody-generated data sets, which are a mainstay of the epigenomics field (10).
H2B was one of the first proteins found to be ubiquitylated (27). Thus, it seems fitting that this protein would be the first to have the ubiquitin modification incorporated entirely through chemical means (15). The preparation of H2BK120Ub through semisynthesis demonstrated that extremely complex modifications could be site-specifically introduced into histones—ubiquitin is an ~8-kDa protein. The original semisynthetic route developed for H2BK120Ub is similar to that discussed above for H2AK119Ub; the latter was in fact inspired by the former. As in the H2A case, the site of ubiquitylation is near the C terminus of the protein, and the semisynthetic route employed involved sequential ligation steps utilizing three building blocks, two recombinant fragments, and a synthetic building block. However, there are a few significant differences between the syntheses, which ultimately resulted in the generation of ubiquitylated H2B without any ligation scars or mutations. The most important of these was the attachment of a photolytically removable NCL auxiliary to the ε-amino group of Lys-120 within the synthetic peptide.
Previous research on model peptides had demonstrated that this auxiliary can be used to link ubiquitin to the lysine side chain through a completely native isopeptide linkage (118). In effect, the auxiliary acts as a temporary cysteine residue (it contains a 1,2-amino thiol group) that, following ligation, is converted to a glycine through photolysis. Accordingly, the first step of the H2BK120Ub semisynthesis involved regioselective EPL between a recombinant Ub(1–75) α-thioester and a synthetic peptide corresponding to H2B residues 117–125 containing the aforementioned auxiliary at Lys-120 and an S-ortho-nitrobenzyl protected cysteine residue at the N terminus. UV irradiation of this initial ligation product simultaneously removed the auxiliary moiety and unmasked the cysteine, thereby allowing attachment of the remainder of the histone (residues 1–116) through a second EPL reaction. In the final step, Raney nickel–mediated desul-furization converted the cysteine residue at position 117 into the native alanine residue. Thus, the final product contained completely native sequences in both the histone and the ubiquitin portions.
Access to a homogeneous preparation of H2BK120Ub, enabled by semisynthesis, has proven to be extremely useful for dissecting a number of chromatin processes (15, 46, 70, 89, 119–121). For example, the presence of the H2BK120Ub in chromatin directly stimulates the methyltransferases Dot1L and Set1, leading to dramatically elevated methylation of histone H3 on Lys-4 and Lys-79, respectively (15, 70).
The disclosure of the H2BK120Ub synthesis opened the floodgates to a torrent of new methodologies centered on expedient routes to ubiquitylated proteins, including histones (122). The initial H2BK120Ub semisynthesis, while yielding the natively modified protein, was unwieldy (15), largely because of the need to synthesize the auxiliary through a multistep process (118). McGinty et al. (119) circumvented this hurdle by developing a second-generation semisynthesis, the key feature of which is the replacement of Gly-76 in ubiquitin with alanine. This route is entirely analogous to that outlined above for H2AK119Ub and, like that example, obviated the need for the ligation auxiliary (it is replaced by cysteine). The resulting semisynthesis is more expedient and allowed the authors to determine several aspects of the ubiquitylation–methylation cross talk between H2BK120Ub and H3K79me, including establishing that a specific surface of ubiquitin is required for methyltransferase stimulation (46, 119).
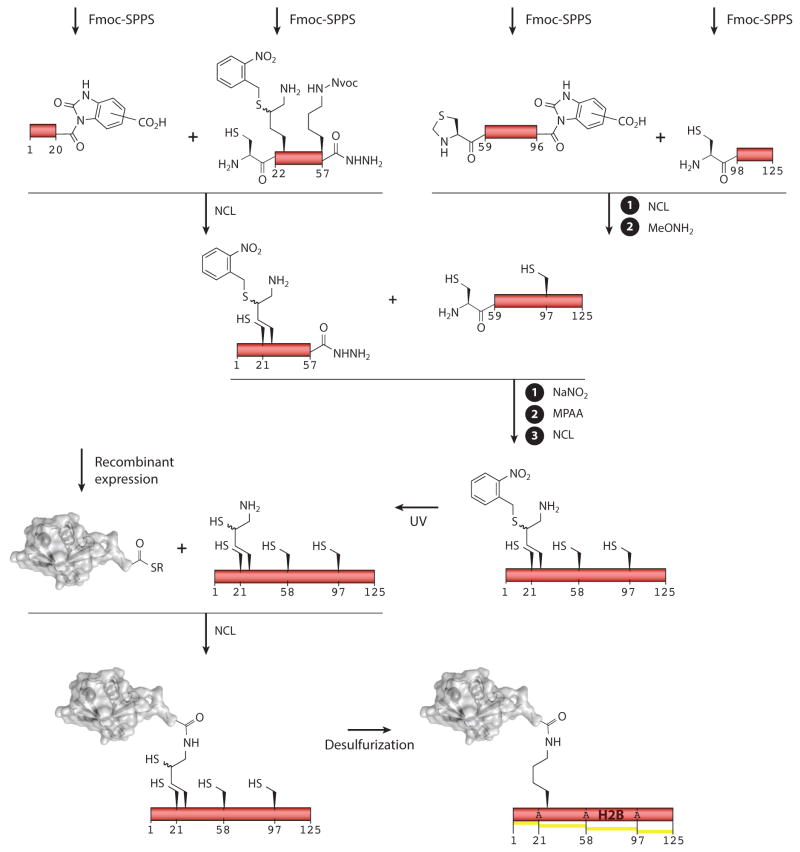
Arguably the most useful improvement in the chemical ubiquitylation area comes from the research of Brik and colleagues (123), who introduced the thiolated lysine derivative δ-mercaptolysine (Figure 3). This unnatural amino acid has a 1,2-amino thiol moiety embedded within the side chain, meaning that ubiquitin α-thioesters can be directly ligated to the ε-amino group via NCL and EPL. Subsequent desulfurization affords the ubiquitylated lysine. This powerful reagent has found use in many systems, including the generation of H2BK120Ub, in which an N-methylated isopeptide bond was deliberately installed to stabilize the PTM toward hydrolases present in the cell lysates to which it was exposed (120, 124). But perhaps the most noteworthy chemical ubiquitylation synthesis to date, at least as applied to histones, is the Brik group’s (90) preparation of H2B ubiquitylated at Lys-34 (Figure 3). This synthesis is a remarkable achievement for several reasons. First, the breadth of peptide chemistry employed is impressive, showcasing advances in the synthesis of peptide α-thioesters via Fmoc-SPPS (both the diaminobenzoic acid and hydrazine linkers are used) and the use of orthogonal protecting group strategies (both thiazolidine and an ortho-nitrobenzyl group are employed at various points). Second, the route developed was highly convergent. Thus, whereas the synthesis called for the assembly of five fragments, only three linear steps were involved (Figure 3). This simplification was, in part, made possible by the clever use of the α-hydrazide moiety (afforded by the hydrazine linker discussed in the section titled Synthetic Building Blocks, above), which was converted into an active α-thioester only at an advanced stage of the synthesis. Altogether, this research illustrates the tremendous range of tools now available to the synthetic protein chemist for use in the preparation of synthetically challenging modified histone targets.
Figure 3.
Synthesis of H2B site-specifically ubiquitylated at Lys-34. The target protein was assembled from five segments using a convergent iterative ligation strategy. Orthogonal protection of the δ-mercaptolysine residue at position 34 with a photolabile ortho-nitrobenzyl group enables regioselective conjugation of the ubiquitin protein. The final step involves simultaneous removal of all four ligation scars via radical-initiated desulfurization. Yellow indicates a building block of synthetic origin in the final construct. Abbreviations: Fmoc, fluorenylmethyloxycarbonyl; MPAA, mercaptophenylacetic acid; NCL, native chemical ligation reaction; Nvoc, 6-nitroveratryloxycarbonyl; SPPS, solid-phase peptide synthesis.
(Semi)Synthesis Applied to Histone H3
Extensive efforts have been devoted to the generation of histone H3 containing PTMs. Indeed, the first semisynthetic histone ever prepared was H3S10ph (78). In this case, the authors used Fmoc-SPPS to synthesize residues 1–31 of H3 containing the Ser-10 phosphorylation mark. The C terminus of this peptide was converted into a benzyl α-thioester post cleavage using standard activating agents. However, this method resulted in partial dephosphorylation of the peptide, so a metal affinity purification step was included to obtain the homogeneous H3S10ph peptide. A recombinant H3(32–135) fragment containing an N-terminal cysteine in place of the normal threonine residue was prepared through Factor Xa–mediated removal of a leader sequence. The ligation junction was chosen so that the nonnative cysteine in the ligated product was located outside the folded core of the protein, but far enough into the histone sequence to allow access to the major sites of PTM. The same ligation junction was utilized to prepare H3K4me3, although in this case the synthetic peptide was assembled by Boc-SPPS on a 3-mercaptopropionamide linker (91), and MPAA was used as the exogenous thiol to facilitate the ligation (23, 91). These semisynthetic H3 constructs were subsequently used in a series of biochemical studies that nicely highlight the importance of using full-length modified histones, as opposed to short histone-derived peptides; use of H3S10ph-containing nucleosomes as a substrate for the Spt–Ada–Gcn5–acetyltransferase (SAGA) complex revealed additional modes of regulation that were not evident with peptide substrates (78), and access to the H3K4me3 was instrumental in determining that the transcription factor BPTF engages chromatin via a multivalent interaction involving modified H3 and H4 (23).
McCafferty and colleagues (100) were the first to employ the cysteine-to-alanine desulfurization strategy to afford a traceless semisynthesis of a PTM-containing histone. Modified H3 variants containing a trimethyl mark at Lys-9 (H3K9me3) or five acetyl marks (K4Ac, K9Ac, K14Ac, K18Ac, and K23Ac) were prepared by NCL employing a ligation junction between residues 24 and 25. The synthetic α-thioester building blocks were prepared by Boc-SPPS using a mercaptopropionic acid linker, while the recombinant fragment containing an N-terminal cysteine was generated via in vivo processing of the initiating methionine. Following NCL, the authors used Raney nickel desulfurization to convert the cysteine back to the native alanine. This process made this pioneering synthesis traceless, albeit low yielding, likely due to the use of Raney nickel (10–30% after desulfurization). Circumventing the desulfurization step in exchange for a conservative H3S28C mutation, Ferreira et al. (79) generated a modified H3 containing acetyl marks at Lys-9, Lys-14, Lys-18, and Lys-23 and used this construct to probe the activities of histone remodelers. More recently, Kim et al. (115) described approaches to the study of N-terminal H3 modifications that extend further into the H3 tail and result in a native H3. These authors chose an H3A29 ligation site to enable the traceless semisynthesis of H3K27me3 via an NCL and desulfurization protocol.
Histone H3 contains a number of PTMs within the structured core of the protein. Access to these sites requires an iterative NCL and EPL strategy due to the size limitations on synthetic peptides: To a first approximation, residues that are more than ~40 residues from either terminus in the histone sequence cannot be modified using a standard two-building-block approach due to the size limitation of SPPS. Ottesen and colleagues (125) were the first to tackle this problem in the context of H3 by developing a synthesis of the protein containing an acetyl-lysine modification at position 56 (H3K56ac). The desired protein was assembled from three building blocks, all synthetic [H3(1–46), H3(47–90)K56ac, and H3(91–135)], via an iterative NCL process. These peptides were prepared by Boc-SPPS and appropriately armed with α-thioesters and N-terminal cysteines (replacing native alanines). The key feature of the synthesis was temporary protection of the cysteine in the middle fragment as a thiazolidine. This feature prevented the peptide from reacting with itself (leading to cyclization or polymerization), thereby allowing the controlled assembly of the fragments in the C-to-N direction. The final step in the synthesis was conversion of the two cysteines into alanines by desulfurization. Whereas the overall yield of this synthesis was relatively modest (7%), sufficient material was obtained to perform a series of biophysical experiments showing that the PTM promotes “breathing” of the DNA on the histone octamer.
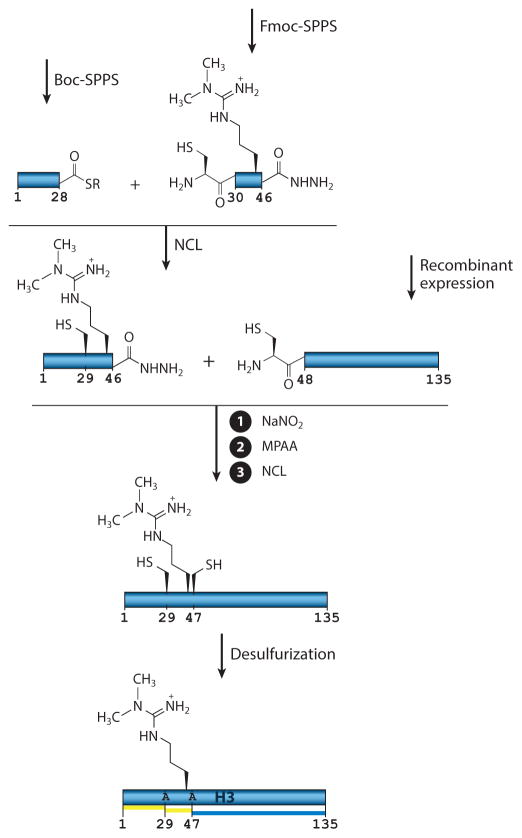
Recently, a more efficient semisynthesis was used to incorporate a dimethyl-arginine PTM at position 42 within the interior of H3 (H3R42me2) (Figure 4) (99). In this three-piece ligation strategy, Casadio et al. (99) used Boc- and Fmoc-SPPS to prepare peptides corresponding to H3(1–28) and H3(29–46), respectively. The latter peptide contained the PTM and was generated as a peptide hydrazide, allowing regioselective NCL between the two synthetic peptides to yield an intermediate product corresponding to residues 1–46. Subsequent conversion of the acyl-hydrazide to an α-thioester set up the second NCL step involving a recombinant H3 fragment corresponding to residues 47–135 and containing a cysteine at position 47. This recombinant building block was obtained by in vitro processing of a corresponding SUMO fusion. Finally, free radical desulfurization afforded the natively modified H3 protein, which was subsequently used in a series of biochemical studies that revealed a role for the PTM in transcription regulation (99). More generally, the convenient location of the ligation junctions in this synthesis, combined with the overall efficiency of this strategy, make this approach well suited to the generation of other internal modifications in H3 (e.g., H3K36 methylation).
Figure 4.
Multistep semisynthesis of histone H3 methylated at Arg-42. A three-piece sequential ligation scheme was used to assemble H3 containing a dimethyl arginine at position 42. Use of a peptide hydrazide in the central fragment allowed the synthesis to proceed in an N-to-C direction. Free radical desulfurization, as the final step, yielded the desired posttranslational modification–containing protein. Yellow indicates a building block of synthetic origin in the final construct, whereas blue indicates a recombinantly prepared fragment in the final construct. Abbreviations: Boc, butyloxycarbonyl; Fmoc, fluorenylmethyloxycarbonyl; MPAA, mercaptophenylacetic acid; NCL, native chemical ligation reaction; SPPS, solid-phase peptide synthesis.
Several semisynthetic routes allow modifications to be easily installed near the C terminus of the H3 sequence. H3 variants containing an acetyl-lysine modification at positions 115 and 122 (H3K115ac and H3K122ac) were prepared by EPL between a recombinant H3(1–109) α-thioester fragment, derived by thiolysis of a GyrA intein fusion, and suitably acetylated synthetic peptides corresponding to the remainder of the sequence and bearing the requisite N-terminal cysteine (126). These constructs were used to show that the interaction of the histone octamer with nucleosomal DNA is weakened by these modifications. H3 containing a phosphothreonine residue at position 118 (H3T118ph), generated using the same semisynthetic scheme, was subsequently found to have the same destabilizing effect (127). Notably, this effect was not observed using glutamine mutations to mimic the acetylation marks (126), further illustrating the need for native PTMs when studying the subtleties of chromatin regulation.
Semisynthesis Applied to Histone H4
To date, efforts to apply semisynthesis to H4 have focused on lysine acetylation. The first of these, again from the McCafferty group (100), involved traceless NCL between synthetic peptide α-thioesters corresponding to the first 14 residues of the protein, and containing various combinations of acetyl marks on residues Lys-5, Lys-8, and Lys-12, with the complementary recombinant fragment H4(15–103) (100). The peptide α-thioesters were generated by Fmoc-SPPS on a highly acid-labile 2-chlorotrityl support; this approach enabled the generation of a protected peptide intermediate, which was then thioesterified and globally deprotected in solution. Parenthetically, the rather complicated nature of this synthetic process illustrates the impact that modern latent thioester linkers have had on the preparation peptide α-thioesters by Fmoc-SPPS. The recombinant fragment was prepared via in vivo processing of an initiator methionine codon that was placed next to the cryptic N-terminal cysteine. The ligation products were desulfurized to yield native acetylated H4 variants, which could be reconstituted into chromatin. To access H4 tail modifications extending beyond position 15, Shogren-Knaak et al. (20) adapted the McCafferty group’s strategy to prepare a semisynthetic H4K16ac variant. Because the next alanine residue in H4 is at position 38 (which would require a more technically demanding peptide synthesis were it used as a ligation junction), these authors elected to fragment the protein between residues 22 and 23, which in turn meant mutating Arg-23 to cysteine. Despite this compromise, the desired protein was prepared and subsequently incorporated into nucleosomal arrays, where a seminal series of studies showed it to affect the folding behavior of chromatin (20). This semisynthetic strategy has been further extended to the study of H4K12ac, H4K16ac, and H4K20ac constructs in the multivalent recognition of the nucleosome by BPTF (23).
Semisynthetic strategies for introducing modifications into the N terminus of H4 continue to evolve. The most recent involves placing the ligation junction between residues 37 and 38 (46). NCL followed by cysteine-to-alanine conversion through radical-initiated desulfurization provides a traceless semisynthesis in which the entire N-terminal tail of the histone can be modified at will. This route strongly relies on highly optimized protocols for Boc- and Fmoc-SPPS, which make the synthesis of relatively long α-thioester peptides of the type required here relatively routine.
As we have attempted to emphasize in this review, most contemporary histone (semi)syntheses attempt to remove the ligation scar from the initial NCL or EPL product, typically by desul-furization. However, there is an alternative strategy that involves elaborating the cysteine thiol group post ligation, rather than erasing it. This strategy is nicely illustrated by Allahverdi et al. (128), who performed semisynthesis of a series of acetylated H4 constructs. These authors placed the ligation junction between residues 19 and 20, requiring mutation of Lys-20 to a cysteine. Following NCL between the appropriate synthetic and recombinant building blocks, the cysteine was reacted with 2-bromoethylamine, which converted it into an analog of lysine (43). Given the very large number of lysine residues in histones, this strategy could further expand the range of sites available to semisynthesis. Finally, C-terminal modifications have also been introduced into H4 by semisynthesis. Specifically, H4K77ac and H4K79ac constructs have been prepared via EPL between a synthetic H4(76–103) peptide and a recombinant H4(1–75) α-thioester generated using intein thiolysis (129). Subsequent desulfurization yielded the native constructs, which were used to reveal a role for these PTMs in modulating DNA breathing on the histone octamer.
CONCLUSIONS AND OUTLOOK
Protein (semi)synthesis has enabled the generation of dozens of posttranslationally modified his-tones over the last ~10 years. These reagents are instrumental in understanding the precise mechanisms by which chromatin-templated processes are regulated through the installation, readout, and removal of these marks. These efforts have improved our knowledge about how best to prepare these molecules—excellent strategies are available that afford synthetic access to most regions of histone primary sequences (Figure 5). Although there is always room for improvement in any multistep chemical process, we are now at a point where the manufacture of a modified histone is no longer a “PhD project” in and of itself. In fact, the preparation of many modified histones (e.g., acetylation and methylation) is now routine, and several modified histones are already commercially available, with many others sure to follow. Thus, the greatest challenges in the near future are likely to be related to how these reagents might be used to disentangle the formidable combinatorial complexity of chromatin (see the sidebar titled Accelerated Chromatin Biochemistry Using DNA-Barcoded Nucleosome Libraries), as well as how these building blocks can be incorporated into increasingly large reconstituted nucleosome arrays that are true facsimiles of native chromatin. Perhaps most exciting of all is the possibility that protein semisynthesis, with its unrivaled chemical precision and flexibility, might somehow be extended to manipulate the covalent structure of chromatin in a living cell. Thus, we imagine that the chromatin area will continue to be fertile ground for protein chemistry and that many new methodologies will emerge with applications even beyond the world of histones.
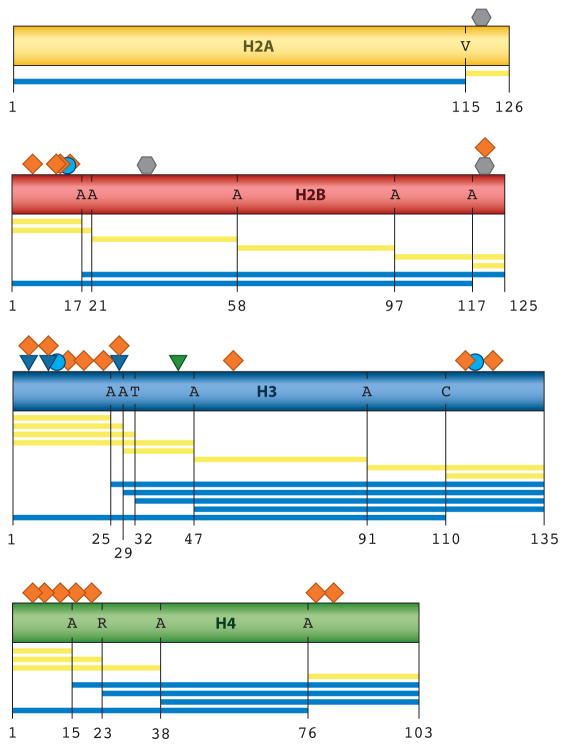
Figure 5.
Summary of established native chemical ligation (NCL) and expressed protein ligation (EPL) strategies to assemble the four canonical histones. Rectangles indicate the histone primary sequences, H2A ( yellow), H2B (red ), H3 (blue), and H4 ( green). Posttranslational modifications (PTMs) that have been installed by chemical ligation techniques are annotated above each histone: Ubiquitylation ( gray hexagon), acetylation (orange diamond ), lysine methylation (blue triangle), arginine methylation ( green triangle), and phosphorylation (blue circle) are displayed. Ligation sites are indicated by a black vertical line underneath each histone, and the one-letter amino acid code of the native amino acid on the C-terminal side of the junction is indicated. Synthetic and recombinant histone fragments employed in the assembly of the full-length protein (through either a single or iterative ligation strategy) are shown below each respective histone. Each yellow bar indicates a synthetic peptide, whereas each blue bar indicates the protein fragment was generated via recombinant DNA expression methods.
ACCELERATED CHROMATIN BIOCHEMISTRY USING DNA-BARCODED NUCLEOSOME LIBRARIES.
The reconstitution of individual PTM-containing histones into chromatin allows for the hypothesis-driven investigation of molecular mechanisms governing chromatin effectors (33). Although the approach is powerful, its throughput is slow (and sample intensive), which is especially problematic when one considers the sheer number of PTMs in histones and their combinatorial co-occurrence (10, 11). With this in mind, Nguyen et al. (29) realized that a potential solution to this problem might be to harness the extraordinary sensitivity, speed, and multiplexing nature of next-generation DNA sequencing. Accordingly, they put into place a synthetic work flow that allowed the rapid (within ~3 days) assembly of an entire nucleosome library, each member of which contains a unique combination of histone PTMs and is self-encoded by its associated DNA. The authors then employed this DNA-barcoded nucleosome library in a series of biochemical studies, which provided a wealth of information on the relative binding preferences and substrate preferences for a series of chromatin effectors. The extremely high throughput of this approach (in which many thousands of biochemical data points are obtained simultaneously), coupled with its remarkable sensitivity, suggests that this method will have broad application in the chromatin area as a hypothesis-generating tool.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Matthew Holt, Email: holtm3@gmail.com.
Tom Muir, Email: muir@princeton.edu.
LITERATURE CITED
- 1.Allis CD, Jenuwein T, Reinberg D. Epigenetics. Cold Spring Harbor, NY: Cold Spring Harb. Lab; 2007. [Google Scholar]
- 2.Kornberg RD. Structure of chromatin. Annu Rev Biochem. 1977;46:931–54. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 3.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 4.Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nat Rev Mol Cell Biol. 2013;14:211–24. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan M, Luo H, Lee S, Jin F, Yang JS, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–28. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Patel DJ, Wang Z. Readout of epigenetic modifications. Annu Rev Biochem. 2013;82:81–118. doi: 10.1146/annurev-biochem-072711-165700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–11. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 9.Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–89. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman MM, Ernst J, Wilder SP, Kundaje A, Harris RS, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41:827–41. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian Z, Tolic N, Zhao R, Moore RJ, Hengel SM, et al. Enhanced top-down characterization of histone post-translational modifications. Genome Biol. 2012;13:R86. doi: 10.1186/gb-2012-13-10-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voigt P, LeRoy G, Drury WJ, 3rd, Zee BM, Son J, et al. Asymmetrically modified nucleosomes. Cell. 2012;151:181–93. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rando OJ. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr Opin Genet Dev. 2012;22:148–55. doi: 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 15.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–16. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–8. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 17.Huff JT, Plocik AM, Guthrie C, Yamamoto KR. Reciprocal intronic and exonic histone modification regions in humans. Nat Struct Mol Biol. 2010;17:1495–99. doi: 10.1038/nsmb.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–85. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–47. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 21.Wilkins BJ, Rall NA, Ostwal Y, Kruitwagen T, Hiragami-Hamada K, et al. A cascade of histone modifications induces chromatin condensation in mitosis. Science. 2014;343:77–80. doi: 10.1126/science.1244508. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Y, Grummt I. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr Biol. 2005;15:1434–38. doi: 10.1016/j.cub.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 23.Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tropberger P, Schneider R. Scratching the (lateral) surface of chromatin regulation by histone modifications. Nat Struct Mol Biol. 2013;20:657–61. doi: 10.1038/nsmb.2581. [DOI] [PubMed] [Google Scholar]
- 25.Kebede AF, Schneider R, Daujat S. Novel types and sites of histone modifications emerge as players in the transcriptional regulation contest. FEBS J. 2014 doi: 10.1111/febs.13047. In press. [DOI] [PubMed] [Google Scholar]
- 26.Davies N, Lindsey GG. Histone H2B (and H2A) ubiquitination allows normal histone octamer and core particle reconstitution. Biochim Biophys Acta. 1994;1218:187–93. doi: 10.1016/0167-4781(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 27.West MH, Bonner WM. Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res. 1980;8:4671–80. doi: 10.1093/nar/8.20.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–6. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen UT, Bittova L, Müller MM, Fierz B, David Y, et al. Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries. Nat Methods. 2014;11:834–40. doi: 10.1038/nmeth.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang Z, Chen WY, Shimada M, Nguyen UT, Kim J, et al. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell. 2013;154:297–310. doi: 10.1016/j.cell.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fierz B, Muir TW. Chromatin as an expansive canvas for chemical biology. Nat Chem Biol. 2012;8:417–27. doi: 10.1038/nchembio.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voigt P, Reinberg D. Histone tails: ideal motifs for probing epigenetics through chemical biology approaches. ChemBioChem. 2011;12:236–52. doi: 10.1002/cbic.201000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allis CD, Muir TW. Spreading chromatin into chemical biology. ChemBioChem. 2011;12:264–79. doi: 10.1002/cbic.201000761. [DOI] [PubMed] [Google Scholar]
- 34.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding Nε-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–34. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 35.Davis L, Chin JW. Designer proteins: applications of genetic code expansion in cell biology. Nat Rev Mol Cell Biol. 2012;13:168–82. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 36.Neumann H, Hancock SM, Buning R, Routh A, Chapman L, et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–63. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gattner MJ, Vrabel M, Carell T. Synthesis of ε-N-propionyl-, ε-N-butyryl-, and ε-N-crotonyl-lysine containing histone H3 using the pyrrolysine system. Chem Commun. 2013;49:379–81. doi: 10.1039/c2cc37836a. [DOI] [PubMed] [Google Scholar]
- 38.Kim CH, Kang M, Kim HJ, Chatterjee A, Schultz PG. Site-specific incorporation of ε-N-crotonyllysine into histones. Angew Chem Int Ed. 2012;51:7246–49. doi: 10.1002/anie.201203349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen DP, Garcia Alai MM, Virdee S, Chin JW. Genetically directing ε-N, N-dimethyl-L-lysine in recombinant histones. Chem Biol. 2010;17:1072–76. doi: 10.1016/j.chembiol.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 40.Park HS, Hohn MJ, Umehara T, Guo LT, Osborne EM, et al. Expanding the genetic code of Escherichia coli with phosphoserine. Science. 2011;333:1151–54. doi: 10.1126/science.1207203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Fekner T, Ottesen JJ, Chan MK. A pyrrolysine analogue for site-specific protein ubiquitination. Angew Chem Int Ed. 2009;48:9184–87. doi: 10.1002/anie.200904472. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen DP, Elliott T, Holt M, Muir TW, Chin JW. Genetically encoded 1,2-aminothiols facilitate rapid and site-specific protein labeling via a bio-orthogonal cyanobenzothiazole condensation. J Am Chem Soc. 2011;133:11418–21. doi: 10.1021/ja203111c. [DOI] [PubMed] [Google Scholar]
- 43.Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–12. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatterjee C, McGinty RK, Fierz B, Muir TW. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat Chem Biol. 2010;6:267–69. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- 45.Li F, Allahverdi A, Yang R, Lua GB, Zhang X, et al. A direct method for site-specific protein acetylation. Angew Chem Int Ed. 2011;50:9611–14. doi: 10.1002/anie.201103754. [DOI] [PubMed] [Google Scholar]
- 46.Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011;7:113–19. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seeliger D, Soeroes S, Klingberg R, Schwarzer D, Grubmuller H, Fischle W. Quantitative assessment of protein interaction with methyl-lysine analogues by hybrid computational and experimental approaches. ACS Chem Biol. 2012;7:150–54. doi: 10.1021/cb200363r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krieger DE, Levine R, Merrifield RB, Vidali G, Allfrey VG. Chemical studies of histone acetylation. Substrate specificity of a histone deacetylase from calf thymus nuclei. J Biol Chem. 1974;249:332–34. [PubMed] [Google Scholar]
- 49.Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol. 2012;19:1155–60. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliver SS, Musselman CA, Srinivasan R, Svaren JP, Kutateladze TG, Denu JM. Multivalent recognition of histone tails by the PHD fingers of CHD5. Biochemistry. 2012;51:6534–44. doi: 10.1021/bi3006972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garske AL, Oliver SS, Wagner EK, Musselman CA, LeRoy G, et al. Combinatorial profiling of chromatin binding modules reveals multisite discrimination. Nat Chem Biol. 2010;6:283–90. doi: 10.1038/nchembio.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garske AL, Craciun G, Denu JM. A combinatorial H4 tail library for exploring the histone code. Biochemistry. 2008;47:8094–102. doi: 10.1021/bi800766k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 2001;8:473–79. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 54.Turner BM, Fellows G. Specific antibodies reveal ordered and cell-cycle-related use of histone H4 acetylation sites in mammalian cells. Eur J Biochem. 1989;179:131–39. doi: 10.1111/j.1432-1033.1989.tb14530.x. [DOI] [PubMed] [Google Scholar]
- 55.Smith BC, Denu JM. Mechanism-based inhibition of Sir2 deacetylases by thioacetyl-lysine peptide. Biochemistry. 2007;46:14478–86. doi: 10.1021/bi7013294. [DOI] [PubMed] [Google Scholar]
- 56.Smith BC, Denu JM. Acetyl-lysine analog peptides as mechanistic probes of protein deacetylases. J Biol Chem. 2007;282:37256–65. doi: 10.1074/jbc.M707878200. [DOI] [PubMed] [Google Scholar]
- 57.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9–methylated histone H3 tail. Science. 2002;295:2080–83. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 58.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 59.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–72. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 60.Bua DJ, Kuo AJ, Cheung P, Liu CL, Migliori V, et al. Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PLOS ONE. 2009;4:e6789. doi: 10.1371/journal.pone.0006789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem. 2007;282:2450–55. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu H, Galka M, Iberg A, Wang Z, Li L, et al. Systematic identification of methyllysine-driven interactions for histone and nonhistone targets. J Proteome Res. 2010;9:5827–36. doi: 10.1021/pr100597b. [DOI] [PubMed] [Google Scholar]
- 63.Rothbart SB, Krajewski K, Strahl BD, Fuchs SM. Peptide microarrays to interrogate the “histone code”. Methods Enzymol. 2012;512:107–35. doi: 10.1016/B978-0-12-391940-3.00006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, et al. Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev. 2013;27:1288–98. doi: 10.1101/gad.220467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Q, Chakravarty S, Ghersi D, Zeng L, Plotnikov AN, et al. Biochemical profiling of histone binding selectivity of the yeast bromodomain family. PLOS ONE. 2010;5:e8903. doi: 10.1371/journal.pone.0008903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rothbart SB, Lin S, Britton LM, Krajewski K, Keogh MC, et al. Poly-acetylated chromatin signatures are preferred epitopes for site-specific histone H4 acetyl antibodies. Sci Rep. 2012;2:489. doi: 10.1038/srep00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hilpert K, Winkler DF, Hancock RE. Peptide arrays on cellulose support: SPOT synthesis, a time and cost efficient method for synthesis of large numbers of peptides in a parallel and addressable fashion. Nat Protoc. 2007;2:1333–49. doi: 10.1038/nprot.2007.160. [DOI] [PubMed] [Google Scholar]
- 68.Kent SB. Chemical synthesis of peptides and proteins. Annu Rev Biochem. 1988;57:957–89. doi: 10.1146/annurev.bi.57.070188.004521. [DOI] [PubMed] [Google Scholar]
- 69.Kim J, Kim JA, McGinty RK, Nguyen UT, Muir TW, et al. The n-SET domain of Set1 regulates H2B ubiquitylation-dependent H3K4 methylation. Mol Cell. 2013;49:1121–33. doi: 10.1016/j.molcel.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, et al. RAD6-mediated transcription–coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–71. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muralidharan V, Muir TW. Protein ligation: an enabling technology for the biophysical analysis of proteins. Nat Methods. 2006;3:429–38. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- 72.Muir TW. Semisynthesis of proteins by expressed protein ligation. Annu Rev Biochem. 2003;72:249–89. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 73.Dawson PE, Kent SB. Synthesis of native proteins by chemical ligation. Annu Rev Biochem. 2000;69:923–60. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 74.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–79. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 75.Muir TW, Sondhi D, Cole PA. Expressed protein ligation: a general method for protein engineering. PNAS. 1998;95:6705–10. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vila-Perelló M, Muir TW. Biological applications of protein splicing. Cell. 2010;143:191–200. doi: 10.1016/j.cell.2010.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blaschke UK, Silberstein J, Muir TW. Protein engineering by expressed protein ligation. Methods Enzymol. 2000;328:478–96. doi: 10.1016/s0076-6879(00)28414-0. [DOI] [PubMed] [Google Scholar]
- 78.Shogren-Knaak MA, Fry CJ, Peterson CL. A native peptide ligation strategy for deciphering nucleosomal histone modifications. J Biol Chem. 2003;278:15744–48. doi: 10.1074/jbc.M301445200. [DOI] [PubMed] [Google Scholar]
- 79.Ferreira H, Flaus A, Owen-Hughes T. Histone modifications influence the action of Snf2 family remodelling enzymes by different mechanisms. J Mol Biol. 2007;374:563–79. doi: 10.1016/j.jmb.2007.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yan LZ, Dawson PE. Synthesis of peptides and proteins without cysteine residues by native chemical ligation combined with desulfurization. J Am Chem Soc. 2001;123:526–33. doi: 10.1021/ja003265m. [DOI] [PubMed] [Google Scholar]
- 81.Wan Q, Danishefsky SJ. Free-radical-based, specific desulfurization of cysteine: a powerful advance in the synthesis of polypeptides and glycopolypeptides. Angew Chem Int Ed. 2007;46:9248–52. doi: 10.1002/anie.200704195. [DOI] [PubMed] [Google Scholar]
- 82.Yang R, Pasunooti KK, Li F, Liu XW, Liu CF. Dual native chemical ligation at lysine. J Am Chem Soc. 2009;131:13592–93. doi: 10.1021/ja905491p. [DOI] [PubMed] [Google Scholar]
- 83.Malins LR, Cergol KM, Payne RJ. Peptide ligation–desulfurization chemistry at arginine. Chem-BioChem. 2013;14:559–63. doi: 10.1002/cbic.201300049. [DOI] [PubMed] [Google Scholar]
- 84.Thompson RE, Chan B, Radom L, Jolliffe KA, Payne RJ. Chemoselective peptide ligation–desulfurization at aspartate. Angew Chem Int Ed. 2013;52:9723–27. doi: 10.1002/anie.201304793. [DOI] [PubMed] [Google Scholar]
- 85.Harpaz Z, Siman P, Kumar KS, Brik A. Protein synthesis assisted by native chemical ligation at leucine. ChemBioChem. 2010;11:1232–35. doi: 10.1002/cbic.201000168. [DOI] [PubMed] [Google Scholar]
- 86.Chen J, Wang P, Zhu J, Wan Q, Danishefsky SJ. A program for ligation at threonine sites: application to the controlled total synthesis of glycopeptides. Tetrahedron. 2010;66:2277–83. doi: 10.1016/j.tet.2010.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haase C, Rohde H, Seitz O. Native chemical ligation at valine. Angew Chem Int Ed. 2008;47:6807–10. doi: 10.1002/anie.200801590. [DOI] [PubMed] [Google Scholar]
- 88.Crich D, Banerjee A. Native chemical ligation at phenylalanine. J Am Chem Soc. 2007;129:10064–65. doi: 10.1021/ja072804l. [DOI] [PubMed] [Google Scholar]
- 89.Fierz B, Kilic S, Hieb AR, Luger K, Muir TW. Stability of nucleosomes containing homogenously ubiquitylated H2A and H2B prepared using semisynthesis. J Am Chem Soc. 2012;134:19548–51. doi: 10.1021/ja308908p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Siman P, Karthikeyan SV, Nikolov M, Fischle W, Brik A. Convergent chemical synthesis of histone H2B protein for the site-specific ubiquitination at Lys34. Angew Chem Int Ed. 2013;52:8059–63. doi: 10.1002/anie.201303844. [DOI] [PubMed] [Google Scholar]
- 91.Camarero JA, Muir TW. Native chemical ligation of polypeptides. Curr Protoc Protein Sci. 2001;15:18.1–18.21. doi: 10.1002/0471140864.ps1804s15. [DOI] [PubMed] [Google Scholar]
- 92.Mende F, Seitz O. 9-Fluorenylmethoxycarbonyl-based solid-phase synthesis of peptide α-thioesters. Angew Chem Int Ed. 2011;50:1232–40. doi: 10.1002/anie.201005180. [DOI] [PubMed] [Google Scholar]
- 93.Clippingdale AB, Barrow CJ, Wade JD. Peptide thioester preparation by Fmoc solid phase peptide synthesis for use in native chemical ligation. J Pept Sci. 2000;6:225–34. doi: 10.1002/(SICI)1099-1387(200005)6:5<225::AID-PSC244>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 94.Botti P, Villain M, Manganiello S, Gaertner H. Native chemical ligation through in situ O to S acyl shift. Org Lett. 2004;6:4861–64. doi: 10.1021/ol0481028. [DOI] [PubMed] [Google Scholar]
- 95.Blanco-Canosa JB, Dawson PE. An efficient Fmoc-SPPS approach for the generation of thioester peptide precursors for use in native chemical ligation. Angew Chem Int Ed. 2008;47:6851–55. doi: 10.1002/anie.200705471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fang GM, Li YM, Shen F, Huang YC, Li JB, et al. Protein chemical synthesis by ligation of peptide hydrazides. Angew Chem Int Ed. 2011;50:7645–49. doi: 10.1002/anie.201100996. [DOI] [PubMed] [Google Scholar]
- 97.Kawakami T, Aimoto S. Peptide ligation via the in-situ transformation of an amide into a thioester at a cysteine residue. Adv Exp Med Biol. 2009;611:117–18. doi: 10.1007/978-0-387-73657-0_52. [DOI] [PubMed] [Google Scholar]
- 98.Chiang KP, Jensen MS, McGinty RK, Muir TW. A semisynthetic strategy to generate phosphorylated and acetylated histone H2B. ChemBioChem. 2009;10:2182–87. doi: 10.1002/cbic.200900238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Casadio F, Lu X, Pollock SB, LeRoy G, Garcia BA, et al. H3R42me2a is a histone modification with positive transcriptional effects. PNAS. 2013;110:14894–99. doi: 10.1073/pnas.1312925110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.He S, Bauman D, Davis JS, Loyola A, Nishioka K, et al. Facile synthesis of site-specifically acetylated and methylated histone proteins: reagents for evaluation of the histone code hypothesis. PNAS. 2003;100:12033–38. doi: 10.1073/pnas.2035256100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gentle IE, De Souza DP, Baca M. Direct production of proteins with N-terminal cysteine for site-specific conjugation. Bioconjug Chem. 2004;15:658–63. doi: 10.1021/bc049965o. [DOI] [PubMed] [Google Scholar]
- 102.Villain M, Vizzavona J, Rose K. Covalent capture: a new tool for the purification of synthetic and recombinant polypeptides. Chem Biol. 2001;8:673–79. doi: 10.1016/s1074-5521(01)00044-8. [DOI] [PubMed] [Google Scholar]
- 103.Erlanson DA, Chytil M, Verdine GL. The leucine zipper domain controls the orientation of AP-1 in the NFAT · AP-1 · DNA complex. Chem Biol. 1996;3:981–91. doi: 10.1016/s1074-5521(96)90165-9. [DOI] [PubMed] [Google Scholar]
- 104.Komarov AG, Linn KM, Devereaux JJ, Valiyaveetil FI. Modular strategy for the semisynthesis of a K+ channel: investigating interactions of the pore helix. ACS Chem Biol. 2009;4:1029–38. doi: 10.1021/cb900210r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tolbert TJ, Franke D, Wong CH. A new strategy for glycoprotein synthesis: ligation of synthetic glycopeptides with truncated proteins expressed in E. coli as TEV protease cleavable fusion protein. Bioorg Med Chem. 2005;13:909–15. doi: 10.1016/j.bmc.2004.06.047. [DOI] [PubMed] [Google Scholar]
- 106.Shah NH, Muir TW. Inteins: nature’s gift to protein chemists. Chem Sci. 2014;5:446–61. doi: 10.1039/C3SC52951G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Evans TC, Jr, Benner J, Xu MQ. Semisynthesis of cytotoxic proteins using a modified protein splicing element. Protein Sci. 1998;7:2256–64. doi: 10.1002/pro.5560071103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Southworth MW, Amaya K, Evans TC, Xu MQ, Perler FB. Purification of proteins fused to either the amino or carboxy terminus of the Mycobacterium xenopi gyrase A intein. Biotechniques. 1999;27:110–20. doi: 10.2144/99271st04. [DOI] [PubMed] [Google Scholar]
- 109.Chong S, Mersha FB, Comb DG, Scott ME, Landry D, et al. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene. 1997;192:271–81. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 110.Shah NH, Dann GP, Vila-Perelló M, Liu Z, Muir TW. Ultrafast protein splicing is common among cyanobacterial split inteins: implications for protein engineering. J Am Chem Soc. 2012;134:11338–41. doi: 10.1021/ja303226x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zettler J, Schütz V, Mootz HD. The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 2009;583:909–14. doi: 10.1016/j.febslet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 112.Vila-Perelló M, Liu Z, Shah NH, Willis JA, Idoyaga J, Muir TW. Streamlined expressed protein ligation using split inteins. J Am Chem Soc. 2013;135:286–92. doi: 10.1021/ja309126m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 114.Johnson EC, Kent SB. Insights into the mechanism and catalysis of the native chemical ligation reaction. J Am Chem Soc. 2006;128:6640–46. doi: 10.1021/ja058344i. [DOI] [PubMed] [Google Scholar]
- 115.Kim DH, Tang Z, Shimada M, Fierz B, Houck-Loomis B, et al. Histone H3K27 trimethylation inhibits H3 binding and function of SET1-like H3K4 methyltransferase complexes. Mol Cell Biol. 2013;33:4936–46. doi: 10.1128/MCB.00601-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thompson RE, Liu X, Alonso-Garciá N, Pereira PJ, Jolliffe KA, Payne RJ. Trifluoroethanethiol: an additive for efficient one-pot peptide ligation–desulfurization chemistry. J Am Chem Soc. 2014;136:8161–64. doi: 10.1021/ja502806r. [DOI] [PubMed] [Google Scholar]
- 117.Whitcomb SJ, Fierz B, McGinty RK, Holt M, Ito T, et al. Histone monoubiquitylation position determines specificity and direction of enzymatic cross-talk with histone methyltransferases Dot1L and PRC2. J Biol Chem. 2012;287:23718–25. doi: 10.1074/jbc.M112.361824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chatterjee C, McGinty RK, Pellois JP, Muir TW. Auxiliary-mediated site-specific peptide ubiquitylation. Angew Chem Int Ed. 2007;46:2814–18. doi: 10.1002/anie.200605155. [DOI] [PubMed] [Google Scholar]
- 119.McGinty RK, Kohn M, Chatterjee C, Chiang KP, Pratt MR, Muir TW. Structure–activity analysis of semisynthetic nucleosomes: mechanistic insights into the stimulation of Dot1L by ubiquitylated histone H2B. ACS Chem Biol. 2009;4:958–68. doi: 10.1021/cb9002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shema-Yaacoby E, Nikolov M, Haj-Yahya M, Siman P, Allemand E, et al. Systematic identification of proteins binding to chromatin-embedded ubiquitylated H2B reveals recruitment of SWI/SNF to regulate transcription. Cell Rep. 2013;4:601–8. doi: 10.1016/j.celrep.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 121.Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–47. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Abeywardana T, Pratt MR. Using chemistry to investigate the molecular consequences of protein ubiquitylation. ChemBioChem. 2014;15:1547–54. doi: 10.1002/cbic.201402117. [DOI] [PubMed] [Google Scholar]
- 123.Ajish Kumar KS, Haj-Yahya M, Olschewski D, Lashuel HA, Brik A. Highly efficient and chemo-selective peptide ubiquitylation. Angew Chem Int Ed. 2009;48:8090–94. doi: 10.1002/anie.200902936. [DOI] [PubMed] [Google Scholar]
- 124.Haj-Yahya M, Eltarteer N, Ohayon S, Shema E, Kotler E, et al. N-Methylation of isopeptide bond as a strategy to resist deubiquitinases. Angew Chem Int Ed. 2012;51:11535–39. doi: 10.1002/anie.201205771. [DOI] [PubMed] [Google Scholar]
- 125.Shimko JC, North JA, Bruns AN, Poirier MG, Ottesen JJ. Preparation of fully synthetic histone H3 reveals that acetyl-lysine 56 facilitates protein binding within nucleosomes. J Mol Biol. 2011;408:187–204. doi: 10.1016/j.jmb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Manohar M, Mooney AM, North JA, Nakkula RJ, Picking JW, et al. Acetylation of histone H3 at the nucleosome dyad alters DNA–histone binding. J Biol Chem. 2009;284:23312–21. doi: 10.1074/jbc.M109.003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.North JA, Javaid S, Ferdinand MB, Chatterjee N, Picking JW, et al. Phosphorylation of histone H3(T118) alters nucleosome dynamics and remodeling. Nucleic Acids Res. 2011;39:6465–74. doi: 10.1093/nar/gkr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Allahverdi A, Yang R, Korolev N, Fan Y, Davey CA, et al. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 2011;39:1680–91. doi: 10.1093/nar/gkq900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Simon M, North JA, Shimko JC, Forties RA, Ferdinand MB, et al. Histone fold modifications control nucleosome unwrapping and disassembly. PNAS. 2011;108:12711–16. doi: 10.1073/pnas.1106264108. [DOI] [PMC free article] [PubMed] [Google Scholar]