Abstract
Bacillus amyloliquefaciens H57 is a bacterium isolated from lucerne for its ability to prevent feed spoilage. Further interest developed when ruminants fed with H57-inoculated hay showed increased weight gain and nitrogen retention relative to controls, suggesting a probiotic effect. The near complete genome of H57 is ~3.96 Mb comprising 16 contigs. Within the genome there are 3,836 protein coding genes, an estimated sixteen rRNA genes and 69 tRNA genes. H57 has the potential to synthesise four different lipopeptides and four polyketide compounds, which are known antimicrobials. This antimicrobial capacity may facilitate the observed probiotic effect.
Keywords: Bacillus amyloliquefaciens, Probiotic, Antimicrobials, Illumina
Introduction
Bacillus amyloliquefaciens species have been taxonomically classified as part of the Bacillus subtilis group. Members of this group share substantial morphological similarities and near identical (98.1 %–99.8 %) 16S rRNA gene sequences [1]. Other members of the Bacillus subtilis group include B. subtilis, B. atrophaeus, B. licheniformis, B. sonorensis, B. tequilensis, B. vallismortis, and the B. mojavensis subgroup. The production of bioactive metabolites, the ability to form spores and a lack of pathogenicity make members of the Bacillus subtilis group ideal candidates for use as probiotics. Strains of B. amyloliquefaciens synthesise non-ribosomal bioactive lipopeptides such as surfactin, fengycin, bacillomycin D and members of the iturin family [2–4]. These lipopeptides have demonstrated activity as antimicrobials and inhibit a wide range of bacterial and fungal pathogens [3, 5].
The strain B. amyloliquefaciens H57 (H57 hereafter) was first isolated in the search for a biological control agent to prevent fungal spoilage of hay [6]. Due to its spore forming ability and production of antimicrobial compounds, H57 was revealed as the best candidate of a panel of isolates for commercialisation as a spoilage control agent under the product name HayRite™. Importantly, sheep and cattle fed on HayRite™ treated feed showed an increase in digestibility and nitrogen retention leading to increased live weight gain [6]. This new development into the potential of H57 to act as a probiotic has led to further investigation of this strain.
Here, we present a summary description of the classification and features of H57, along with a sequencing description and annotation summary. The availability of a genome sequence for H57 will facilitate research into the probiotic effects observed in animals treated with this bacterium.
Organism information
Classification and features
A near-complete 16S rRNA gene was identified in the H57 genome, which by BLAST [7] is most closely related (99 % identical) to other B. amyloliquefaciens strains including FZB42 (B. amyloliquefaciens subsp. plantarum; acc. NR075005.1), HPCAQB14 (acc. KF861603.1) and SB 3200 (acc. GU191911.1). Comparison of the average read coverage of the genome and 16S rRNA gene, suggests that H57 has 13 copies of the rRNA operon. A concatenated alignment of 99 single copy marker genes obtained from publicly available Bacillus genomes using HMMER [8] confirmed the classification of strain H57 as a member of the species B. amyloliquefaciens (Fig. 1).
Fig. 1.
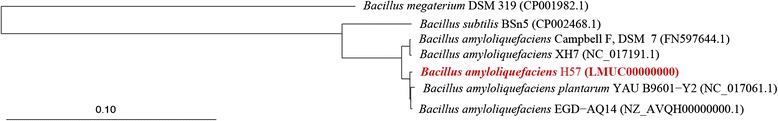
Maximum likelihood tree showing the alignment of H57 with other Bacillus genomes. Alignment was performed using HMMER [8] whilst maximum likelihood was inferred using FastTree version 2.7.7 [32]. The inferred tree was visualised using ARB version 6.0.2 [33]. Bar: 0.1 substitutions per nucleotide position
H57 is a Gram-positive rod shaped bacterium averaging 2.5 μm in length and 1 μm in width (Fig. 2d). It is an aerobic spore forming bacterium that is motile with peritrichous flagella. H57 spores are centrally located and average 1.25 μm in length (Fig. 2b). Optimum growth occurs at a temperature of 29 °C and pH 7.0 (Table 1). The colony morphology of strain H57 is circular convex with undulate margins. When grown on a nutrient agar plate, colonies are an off-white colour as shown in Fig. 2c.
Fig. 2.
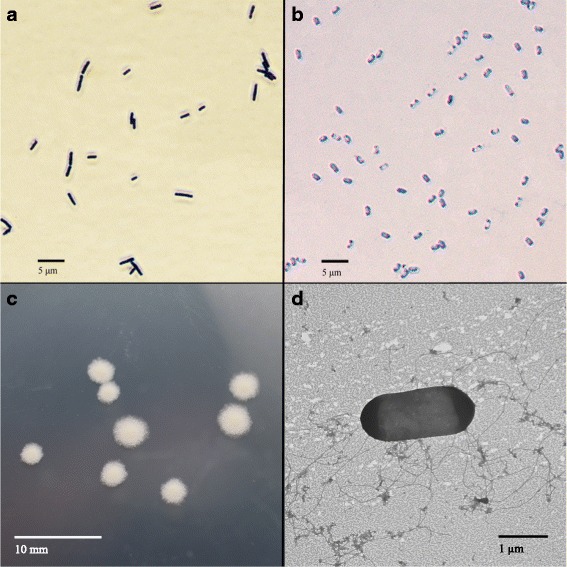
Cellular and colony morphology of B. amyloliquefaciens H57. a Vegetative H57 cells at 1000x magnification captured with a Nikon DS-Ri1 camera attached to a Nikon Eclipse 80i microscope under phase contrast. b H57 spores at 1000x magnification captured with a Leica DFC 500 camera attached to a Leica DM5500B compound microscope with Nomarski differential interference contrast. c Pure culture of H57 grown on nutrient agar plate. d Electron microscope image of a vegetative H57 cell showing numerous peritrichous flagella, negatively stained with phosphotungstic acid
Table 1.
Classification and general features of Bacillus amyloliquefaciens strain H57 [10]
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [34] | |
| Phylum Firmicutes | TAS [35–37] | ||
| Class Bacilli | TAS [38, 39] | ||
| Order Bacillales | TAS [40, 41] | ||
| Family Bacillaceae | TAS [40, 42] | ||
| Genus Bacillus | TAS [40, 43, 44] | ||
| Species Bacillus amyloliquefaciens | TAS [45–47] | ||
| Strain: H57 | |||
| Gram stain | Positive | IDA | |
| Cell shape | Rod/chains | IDA | |
| Motility | Motile | IDA | |
| Sporulation | Sporulating | IDA | |
| Temperature range | Mesophilic | IDA | |
| Optimum temperature | 29 °C | IDA | |
| pH range; Optimum | 5-9; 7 | IDA | |
| Carbon source | Glucose, fructose, mannitol, sucrose, trehalose | IDA | |
| MIGS-6 | Habitat | Leaves of Medicago sativa | TAS [6] |
| MIGS-6.3 | Salinity | Up to 6 % (w/v) | IDA |
| MIGS-22 | Oxygen requirement | Aerobe | IDA |
| MIGS-15 | Biotic relationship | Symbiotic (beneficial) | TAS [6] |
| MIGS-14 | Pathogenicity | Non-pathogen | NAS |
| MIGS-4 | Geographic location | Gatton, QLD, Australia | IDA |
| MIGS-5 | Sample collection | 2001 | IDA |
| MIGS-4.1 | Latitude | 27° 32' 24'' S | IDA |
| MIGS-4.2 | Longitude | 152° 20' 24'' E | IDA |
| MIGS-4.4 | Altitude | 89 m | IDA |
a Evidence codes - IDA Inferred from Direct Assay, TAS Traceable Author Statement (i.e., a direct report exists in the literature), NAS Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [48]
Genome sequencing information
Genome project history
Strain H57 was selected for sequencing due to its ability to act as a probiotic in agricultural animals. The draft genome was deposited in GenBank under the accession number LMUC00000000. Genome sequencing and assembly was performed at the Australian Centre for Ecogenomics, The University of Queensland. Gene annotation was performed using the AnnotateM script [9]. A summary of the project is shown in Table 2 using MIGS version 2.0 [10] criteria.
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS 31 | Finishing quality | Draft |
| MIGS 28 | Libraries used | Illumina paired end library (256 bp insert size) |
| MIGS 29 | Sequencing platforms | Illumina MiSeq |
| MIGS 31.2 | Fold coverage | 49× |
| MIGS 30 | Assemblers | Spades 3.0.0. |
| MIGS 32 | Gene calling method | PROKKA |
| Locus tag | Ga0082361 | |
| Genbank ID | LMUC00000000 | |
| GenBank Date of Release | 04/04/2016 | |
| GOLD ID | Ga0082361 | |
| BIOPROJECT | PRJNA300579 | |
| MIGS 13 | Source material identifier | Bacillus amyloliquefaciens H57 |
| Project relevance | Probiotic, Agriculture |
Growth conditions and genomic DNA preparation
Genomic DNA of H57 was isolated from a freeze-dried product of H57 spores combined with sodium bentonite (1:1). DNA was extracted from the H57 spores using the ‘Repeated Bead-beating and Column Extraction’ method described by Yu and Forster (2005) [11]. In brief, 0.1 g of sporulated product was added to 1 mL of lysis buffer (2.9 % NaCl, 0.6 % Tris, 0.05 M EDTA pH 8.0 and 4 % SDS) in a cryotube containing 0.5 g zirconia beads (BioSpec Products Inc., Bartlesville, USA). The sample was then homogenised in a mini bead beater 16 (BioSpec Products Inc., Bartlesville, USA) for 2 cycles of 3 min. Between cycles the samples were incubated for 15 min at 70 °C, centrifuged (13,200 rpm for 5 min at 4 °C) and supernatant transferred to a fresh tube. Following bead beating further extraction was performed on the supernatant using the QIAGEN QIAmp DNA Mini Kit as per kit instructions (QIAGEN, Doncaster, VIC).
Genome sequencing and assembly
The genome of H57 was sequenced on an Illumina MiSeq sequencing platform (Illumina, Inc. San Diego, CA). DNA libraries were prepared using the Nextera® XT DNA Library Preparation Kit (Illumina, San Diego, CA) according to the manufacturer’s instructions. An input of 1 ng was used to prepare DNA libraries, which was then cleaned using Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA). The purified PCR product was then size selected for amplicons with a size between 300 bp and 800 bp. Illumina paired-end sequencing was performed, producing a total of 1,351,526 reads. Primer and adaptor sequences were removed using Trimmomatic v0.32 [12] resulting in an average read length of 256 bp. Reads were assembled using SPAdes 3.0.0. [13]. The H57 genome was obtained in 16 contigs ranging in size from 701,147 bp to 10,158 bp with a combined length of 3,958,833 bp. Genome completeness and contamination was estimated using CheckM version 1.0.0, indicating that the genome was near complete (99.51 %) with no detectable contamination (0 %) [14].
Genome annotation
Gene annotation was achieved using a combination of protein databases via AnnotateM Version 6.0 [9]. Open reading frames were initially generated using PROKKA [15]. The resulting protein sequence was then searched against the IMG, Uniref, COG, PFAM and TIGRfam databases [16–20] to identify homologous genes. The software ProteinOrtho [21] was used to identify orthologous genes to other known B. amyloliquefaciens strains for further comparison. Genes unique to H57 were compared against the KEGG gene database [22] to identify metabolic functions.
Genome properties
The draft genome assembly of H57 consists of sixteen contigs totalling 3,958,833 bp and a G + C content of 46.42 %, which is likely a slight underestimate of its genome size due to unresolved collapsed repeats, primarily rRNA operons (Table 3). With a coding region of 3,549,557 bp, this assembly represents a total of 3,945 ORFs. Of those genes, 3,836 encode proteins and the remainder encode sixteen rRNAs (7 × 5S, 7 × 16S and 2 × 23S), 69 tRNAs and 24 other RNA genes (Table 3). Of the annotated genes, the majority were assigned a putative function (80.66 %) with 69.81 % assigned into Clusters of Orthologous Groups, presented in Table 4. Of the 3,945 ORFs in the H57 genome, 3,751 were inferred to be orthologous to other B. amyloliquefaciens strains, including strains CC178, DSM7, XH7, TF28, Y2, IT-45, LFB112 and B. amyloliquefaciens subsp plantarum strains UCMB5113, FZB42, NAU-B3, YAU B9601-Y2, and TrigoCor1448. Of the 194 genes unique to H57, several appear to be involved in the degradation of aromatic compounds, more specifically the breakdown of 4-hydroxyphenylacetic acid.
Table 3.
Genome statistics
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 3,958,833 | 100.00 |
| DNA coding (bp) | 3,549,557 | 89.66 |
| DNA G + C (bp) | 1,837,549 | 46.42 |
| DNA scaffolds | 16 | 100.00 |
| Total genes | 3,945 | 100.00 |
| Protein coding genes | 3,836 | 97.24 |
| RNA genes | 109 | 2.76 |
| Pseudo genes | 0 | 0.00 |
| Genes with internal clusters | 387 | 9.81 |
| Genes with function prediction | 3,182 | 80.66 |
| Genes assigned to COGs | 2,754 | 69.81 |
| Genes with Pfam domains | 3,364 | 85.27 |
| Genes with signal peptides | 191 | 4.84 |
| Genes with transmembrane helices | 1,046 | 26.51 |
| CRISPR repeats | 0 | 0.00 |
Table 4.
Number of genes associated with general COG functional categories
| Code | Value | %agea | Description |
|---|---|---|---|
| J | 136 | 3.48 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.00 | RNA processing and modification |
| K | 89 | 2.23 | Transcription |
| L | 95 | 2.43 | Replication, recombination and repair |
| B | 1 | 0.03 | Chromatin structure and dynamics |
| D | 22 | 0.56 | Cell cycle control, Cell division, chromosome partitioning |
| V | 17 | 0.44 | Defence mechanisms |
| T | 58 | 1.48 | Signal transduction mechanisms |
| M | 97 | 2.48 | Cell wall/membrane biogenesis |
| N | 40 | 1.02 | Cell motility |
| U | 37 | 0.95 | Intracellular trafficking and secretion |
| O | 64 | 1.64 | Posttranslational modification, protein turnover, chaperones |
| C | 92 | 2.35 | Energy production and conversion |
| G | 109 | 2.79 | Carbohydrate transport and metabolism |
| E | 160 | 4.09 | Amino acid transport and metabolism |
| F | 62 | 1.59 | Nucleotide transport and metabolism |
| H | 93 | 2.38 | Coenzyme transport and metabolism |
| I | 53 | 1.36 | Lipid transport and metabolism |
| P | 93 | 2.38 | Inorganic ion transport and metabolism |
| Q | 30 | 0.77 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 203 | 5.19 | General function prediction only |
| S | 238 | 6.09 | Function unknown |
| - | 2169 | 55.49 | Not in COGs |
aThe total is based on the total number of protein coding genes in the genome
Insights from the genome sequence
Comparative analysis of the H57 genome indicates that its central metabolism is consistent with other strains of B. amyloliquefaciens. The presence of a complete TCA cycle and electron transport chain indicates the potential for aerobic respiration. H57 has a narGHJI operon and the transcriptional regulator fnr, suggesting that it is also capable of growing anaerobically using nitrate as an electron acceptor [23]. This capability would be required for H57 to grow in anoxic environments.
The genome of H57 also encodes a number of enzymes involved in carbohydrate metabolism. A search against the carbohydrate-active enzyme database [24] reveals that H57 is dominant in glycoside hydrolase families 1, 43 and 13 (Table 5). The GH 1 and GH 43 families comprise enzymes that degrade the various sugar monomers of hemicellulose. This suggests that H57 may contribute to breaking down the less fibrous components of the plant cell wall. The abundance of GH 13 enzymes, which are a family of α-amylases, suggests that H57 also contributes to the breakdown of starch. The presence of these carbohydrate-activated enzymes alludes to the notion that H57 may assist in the digestion of animal feeds by breaking down certain polysaccharides of the plant cell wall.
Table 5.
Carbohydrate activated enzyme profile of glycoside hydrolases in H57
| Family | Known activity | %a |
|---|---|---|
| GH16 | Xyloglucan, keratan-sulfate, endo-1,4-β-galactosidase, endo-1,3- β-glucanase, and others | 2.5 |
| GH4 | Maltose-6-phosphate glucosidase, α-glucosidase, α-galactosidase, and others | 7.5 |
| GH5 | Chitosanase, β-mannosidase, cellulase, glucan 1,3-β-glucosidase, and others | 2.5 |
| GH13 | α-amylase, pullulanase, cyclomaltodextrin glucanotransferase and others | 10 |
| GH11 | Xylanase | 2.5 |
| GH23 | Lysozyme type G and peptidoglycan lyase | 2.5 |
| GH3 | β-glucosidase, xylan 1,4-β-xylosidase, β-N-acetylhexosaminidase, and others | 2.5 |
| GH126 | Other | 2.5 |
| GH18 | Chitinase, endo-β-N-acetylglucosaminidase, and others | 7.5 |
| GH26 | β-mannanase and β-1,3-xylanase | 2.5 |
| GH53 | Endo-β-1,4-galactanase | 2.5 |
| GH51 | α-L-arabinofuranosidase and endoglucanase | 5 |
| GH1 | β-glucosidase, β-galactosidase, β-mannosidase, and others | 12.5 |
| GH73 | Peptidoglycan hydrolase with endo-β-N-acetylglucosaminidase specificity | 5 |
| GH30 | Glucosylceramidase, β-1,6-glucanase, β-xylosidase | 5 |
| GH32 | Endo-inulinase, endo-levanase, exo-inulinase, and others | 7.5 |
| GH46 | Chitosanase | 2.5 |
| GH109 | α-N-acetylgalactosaminidase | 5 |
| GH43 | Arabinases and xylosidases | 10 |
| GH68 | Levansucrase, β-fructofuranosidase and inulosucrase | 2.5 |
| Total GH hits: | 40 | |
| Total ORFs: | 3,828 | |
| % GH ORFs: | 1.04 |
aPercentage of total GH hits
Consistent with observed anti-fungal activity, the H57 genome encodes a broad range of antimicrobial compounds. These include genes for non-ribosomal synthesis of antimicrobial lipopeptides such as surfactin (srfABCD), iturin (ituABCD), bacillomycin D (bmyABC) and fengycin (fenABCDE). Surfactin is capable of inhibiting a wide range of microorganisms due to its ability to insert itself into the cell wall creating ion pores [25]. Bacillomycin D, iturin and fengycin all have demonstrated antifungal properties primarily based on their ability to disrupt the fungal cell wall [26–28]. The genes for the expression of antibiotic polyketides are also present on the H57 genome. These include the operons mlnABCDEFGHI, dfnABCDEFGHIJ and baeEDLMNJRS, which encode macrolactin, difficidin and bacillaene respectively. These compounds inhibit a wide range of microorganisms acting chiefly on preventing protein synthesis [29–31].
Conclusions
The ~3.96 Mbp genome of B. amyloliquefaciens H57 reveals the basis of its antimicrobial nature and potential to survive and reproduce in anoxic animal gastrointestinal tracts. In common with other B. amyloliquefaciens strains, H57 encodes a wide range of antimicrobial compounds that explain its effectiveness as a biocontrol agent for fungi and other feed spoilage organisms. The production of these compounds may also contribute to the observed probiotic effect by inhibiting potentially pathogenic organisms creating a healthier microbial ecosystem.
Acknowledgements
The authors acknowledge the facilities, and the scientific and technical assistance, of the Australian Microscopy & Microanalysis Research Facility at the Centre for Microscopy and Microanalysis, The University of Queensland. We thank Roger Shivas of the Department of Agriculture and Fisheries, QLD for assistance in microscopy. The authors also thank Serene Low and Nicola Angel of the Australian Centre for Ecogenomics for library preparation and sequencing. This work was supported by funding from Ridley AgriProducts and the Australian Research Council (LP120200837).
Authors’ contributions
BJS cultivated the bacterium, contributed to bioinformatic analysis, submitted the genome and drafted the manuscript. AS completed genome assembly, gene annotation and revised the manuscript. NL performed DNA library preparation, participated in the design of the study and revised the manuscript. DO and AVK participated in the design and supervision of the study, assisted with the interpretation of results and helped to draft the manuscript. PD isolated the bacterium, conceived of the study and participated in its design. PH participated in the studies conception, design and coordination. PH provided support with interpretation of the data and helped draft the manuscript. All authors revised and approved of the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- GH
Glycoside hydrolase
References
- 1.Wang L-T, Lee F-L, Tai C-J, Kasai H. Comparison of gyrB gene sequences, 16S rRNA gene sequences and DNA–DNA hybridization in the Bacillus subtilis group. Int J Syst Evol Microbiol. 2007;57(8):1846–1850. doi: 10.1099/ijs.0.64685-0. [DOI] [PubMed] [Google Scholar]
- 2.Chen XH, Koumoutsi A, Scholz R, Schneider K, Vater J, Süssmuth R, Piel J, Borriss R. Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J Biotechnol. 2009;140(1–2):27–37. doi: 10.1016/j.jbiotec.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Koumoutsi A, Chen X-H, Henne A, Liesegang H, Hitzeroth G, Franke P, Vater J, Borriss R. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J Bacteriol. 2004;186(4):1084–1096. doi: 10.1128/JB.186.4.1084-1096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu GY, Sinclair JB, Hartman GL, Bertagnolli BL. Production of iturin A by Bacillus amyloliquefaciens suppressing Rhizoctonia solani. Soil Biol Biochem. 2002;34(7):955–963. doi: 10.1016/S0038-0717(02)00027-5. [DOI] [Google Scholar]
- 5.Huang X, Suo J, Cui Y. Optimization of antimicrobial activity of surfactin and polylysine against Salmonella enteritidis in milk evaluated by a response surface methodology. Foodborne Pathog Dis. 2011;8(3):439–443. doi: 10.1089/fpd.2010.0738. [DOI] [PubMed] [Google Scholar]
- 6.Dart P, Brown S. Testing hay treated with mould-inhibiting, biocontrol inoculum: microbial inoculant for hay: a report for the Rural Industries Research and Development Corporation. no. 05/103: Rural Industries Research and Development Corporation; 2005.
- 7.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 8.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue):W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haroon MF. AnnotateM. https://github.com/fauziharoon/annotateM. Accessed 23rd April 2014.
- 10.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotech. 2008;26(5):541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu Z, Forster RJ. Nucleic acid extraction, oligonucleotide probes and PCR methods. In: Makkar HPS, McSweeney CS, editors. Methods in Gut Microbial Ecology for Ruminants. Netherlands: Springer; 2005. pp. 81–104. [Google Scholar]
- 12.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comp Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Peer J PrePrints. 2014;2:e554v1. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 16.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(D1):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31(1):371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, et al. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012;40(Database issue):D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23(10):1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 20.Tatusov R, Fedorova N, Jackson J, Jacobs A, Kiryutin B, Koonin E, Krylov D, Mazumder R, Mekhedov S, Nikolskaya A, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4(1):41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lechner M, Findeiss S, Steiner L, Marz M, Stadler P, Prohaska S. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12(1):124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez DS. KEGG. Brief Bioinform. 2002;3(3):316. doi: 10.1093/bib/3.3.314. [DOI] [Google Scholar]
- 23.Nakano MM, Zuber P. Anaerobic growth of a "strict aerobe" (Bacillus subtilis) Annu Rev Microbiol. 1998;52:165–190. doi: 10.1146/annurev.micro.52.1.165. [DOI] [PubMed] [Google Scholar]
- 24.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(Database issue):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peypoux F, Bonmatin JM, Wallach J. Recent trends in the biochemistry of surfactin. Appl Microbiol Biotechnol. 1999;51(5):553–563. doi: 10.1007/s002530051432. [DOI] [PubMed] [Google Scholar]
- 26.Deleu M, Paquot M, Nylander T. Effect of fengycin, a lipopeptide produced by Bacillus subtilis, on model biomembranes. Biophys J. 2008;94(7):2667–2679. doi: 10.1529/biophysj.107.114090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maget-Dana R, Peypoux F. Iturins, a special class of pore-forming lipopeptides: biological and physicochemical properties. Toxicology. 1994;87(1–3):151–174. doi: 10.1016/0300-483X(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 28.Nasir MN, Besson F. Conformational analyses of bacillomycin D, a natural antimicrobial lipopeptide, alone or in interaction with lipid monolayers at the air-water interface. J Colloid Interface Sci. 2012;387(1):187–193. doi: 10.1016/j.jcis.2012.07.091. [DOI] [PubMed] [Google Scholar]
- 29.Patel PS, Huang S, Fisher S, Pirnik D, Aklonis C, Dean L, Meyers E, Fernandes P, Mayerl F. Bacillaene, a novel inhibitor of procaryotic protein synthesis produced by Bacillus subtilis: production, taxonomy, isolation, physico-chemical characterization and biological activity. J Antibiot. 1995;48(9):997–1003. doi: 10.7164/antibiotics.48.997. [DOI] [PubMed] [Google Scholar]
- 30.Romero-Tabarez M, Jansen R, Sylla M, Lünsdorf H, Häußler S, Santosa DA, Timmis KN, Molinari G. 7-O-malonyl macrolactin A, a new macrolactin antibiotic from Bacillus subtilis active against methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococci, and a small-colony variant of Burkholderia cepacia. Antimicrob Agents Chemother. 2006;50(5):1701–1709. doi: 10.1128/AAC.50.5.1701-1709.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zweerink MM, Edison A. Difficidin and oxydifficidin: novel broad spectrum antibacterial antibiotics produced by Bacillus subtilis. III. Mode of action of difficidin. J Antibiot. 1987;40(12):1692–1697. doi: 10.7164/antibiotics.40.1692. [DOI] [PubMed] [Google Scholar]
- 32.Price MN, Dehal PS, Arkin AP. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26(7):1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfgang Ludwig OS, Ralf W, Lothar R, Harald Meier Y, Arno B, Tina L, Susanne S, Gangolf J, Wolfram F, Igor B, Stefan G, Ginhart AW, Oliver G, Silke G, Stefan H, Ralf J, Andreas K, Thomas L, Ralph L, Michael M, Björn N, Boris R, Robert S, Alexandros S, Norbert S, Alexander V, Michael L, Thomas L, Arndt B, Karl-Heinz S. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32(4):1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garrity G, Holt J. The road map to the manual. In: Garrity G, Boone D, Castenholz R, editors. Bergey’s manual of systematic bacteriology. 2. New York: Springer; 2001. pp. 119–169. [Google Scholar]
- 36.Gibbons NE, Murray RGE. Proposals concerning the higher taxa of bacteria. Int J Syst Evol Microbiol. 1978;28(1):1–6. [Google Scholar]
- 37.Murray R. The higher taxa, or, a place for everything…? In: Holt J, editor. Bergey's Manual of Systematic Bacteriology. 1. Baltimore: The Williams and Wilkins Co; 1984. pp. 31–34. [Google Scholar]
- 38.Oren A, Garrity GM. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol. 2016;66(3):1603–6. [DOI] [PubMed]
- 39.Ludwig W, Schleifer K, Whitman W. ClassI. Bacilli class nov. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey F, Schleifer K, Whitman W, editors. Bergey's Manual of Systematic Bacteriology. 2. New York: Springer; 2009. pp. 19–20. [Google Scholar]
- 40.Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Bacteriol. 1980;30(1):225–420. doi: 10.1099/00207713-30-1-225. [DOI] [PubMed] [Google Scholar]
- 41.Prévot AR. Bacillales. In: Hauderoy P, Ehringer G, Guillot G, Magrou J, Prévot AR, Rosset D, Urbain A, editors. Dictionnaire des Bactéries Pathogènes. 2nd ed. Paris: Masson et Cie; Paris 1953. p. 1-692.
- 42.Fischer A. Untersuchungen über bakterien. Jahrbücher für Wissenschaftliche Botanik. 1985;27:1–163. [Google Scholar]
- 43.Gibson T, Gordon R, Genus I. Bacillus Cohn 1872, 174; Nom. gen. cons. Nomencl. Comm. Intern. Soc. Microbiol. 1937, 28; Opin. A. Jud. Comm. 1955, 39. In: Buchanan R, Gibbons N, editors. Bergey’s Manual of Determinative Bacteriology. 8th. Baltimore: The Williams and Wilkins Co; 1974. pp. 529–550. [Google Scholar]
- 44.Cohn F. Untersuchungen über Bakterien. Beitr Biol Pflanz. 1872;1:127–224. [Google Scholar]
- 45.Fukomoto J. Studies on the production of bacterial amylase. I. Isolation of bacteria secreting potent amylase and their distribution. Nippon Nogeikagaku Kaishi. 1943;19:487–503. doi: 10.1271/nogeikagaku1924.19.7_487. [DOI] [Google Scholar]
- 46.Priest FG, Goodfellow M, Shute LA, Berkeley RCW. Bacillus amyloliquefaciens sp. nov., nom. rev. Int J Syst Bacteriol. 1987;37(1):69–71. doi: 10.1099/00207713-37-1-69. [DOI] [Google Scholar]
- 47.Wang L-T, Lee F-L, Tai C-J, Kuo H-P. Bacillus velezensis is a later heterotypic synonym of Bacillus amyloliquefaciens. Int J Syst Evol Microbiol. 2008;58(3):671–675. doi: 10.1099/ijs.0.65191-0. [DOI] [PubMed] [Google Scholar]
- 48.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]


